Abstract
Phytoplankton, with an estimated 30 000 to 1 000 000 species clustered in 12 phyla, presents a high taxonomic and ecophysiological diversity, reflected by the complex distribution of pigments among the different algal classes. High performance liquid chromatography is the gold standard method for qualitative and quantitative analysis of phytoplankton pigments in seawater and culture samples, but only a few pigments can be used as robust chemotaxonomic markers. A major challenge is thus to identify new ones, characteristic of a strain, species, class or taxon that cannot be currently identified on the basis of its pigment signature. Using an optimized extraction process coupled to a HPLC de-replication strategy, we examined the pigment composition of 37 microalgae strains, representative of the broad taxonomic diversity of marine and freshwater species (excluding cyanobacteria). For each species, the major pigments already described were unambiguously identified. We also observed the presence of several minor unidentified pigments in each chromatogram. The global analysis of pigment compositions revealed a total of 124 pigments, including 98 pigments or derivatives unidentified using the standards. Absorption spectra indicated that 35 corresponded to chlorophyll/porphyrin derivatives, 57 to carotenoids and six to derivatives having both spectral signatures. Sixty-one of these unidentified or new carotenoids and porphyrin derivatives were characteristic of particular strains or species, indicating their possible use as highly specific chemotaxonomic markers capable of identifying one strain out of the 37 selected. We developed a graphical analysis using Gephi software to give a clear representation of pigment communities among the various phytoplankton strains, and to reveal strain-characteristic and shared pigments. This made it possible to reconstruct the taxonomic evolution of microalgae classes, on the basis of the conservation, loss, and/or appearance of pigments.
Introduction
Photosynthetic microorganisms have evolved a wide range of photoprotective and photosynthetic pigments capable of collectively harvesting most of the wavelengths of visible light available in underwater marine habitats [1]. This chemodiversity reflects the diverse molecular adaptations to the multiple photic conditions met over the evolution of microalgae taxa in marine ecosystems. In spite of their lability, complex distribution among phytoplankton classes, and variable expression, pigments are of great interest as chemotaxonomic markers to identify species or taxa, and assess their abundance, productivity and biodiversity in seawater samples containing different phytoplankton communities [2–4] The identification and dosage of pigments and derivatives in sediments also provides a useful way to assess the ocean productivity, model the spatial and seasonal sedimentation and hydrodynamic processes, and demonstrate local or global marine ecosystem changes [5–7]. In recent decades, HPLC has emerged as the gold standard analytical tool for qualitative and quantitative analysis of phytoplankton pigments in seawater and culture samples because of its ease of use, rapidity, sensitivity, resolution, and potential for development on research vessels [2,8–12]. HPLC is the technique of choice for the standardized quantification of chlorophyll a and identification and quantification of minor pigments. It is also used as the reference method for the validation of other chlorophyll a measurement techniques, including remote sensing dosage. Since 1980, methodological recommendations and optimized protocols have been proposed by the SCOR/UNESCO research group and NASA for reliable phytoplankton pigment analysis and inter-calibration studies. Among others, the Van Heukelem & Thomas (2001) [13] method is currently one of the most efficient and recommended to analyse microalgal organic pigments. Methodological optimization of HPLC performance demonstrated that, in addition to the major pigments easily identified by their absorbance spectrum, band ratio and polarity, several minor unidentified chlorophyll and carotenoids derivatives are usually present in extracts from environmental samples or cultivated species. Most of the time, because of their very low abundance and fastidious purification, these minor pigments are not identified, despite their possible interest as chemotaxonomic markers or for biotechnological or biomedical applications. They can correspond to molecules effectively present in living algal cells, to biosynthetic precursors and intermediates, or to artefacts or natural derivatives/molecules produced by the alteration of chlorophylls or carotenoids in environmental conditions or during extraction and/or purification. One of the major challenges faced by scientists involved in phytoplankton research is to identify among these molecules those that unambiguously signal the presence of species, genera or classes, and can be used as robust chemotaxonomic markers for the molecular fingerprinting of phytoplankton. Another challenge is to develop standardized protocols allowing the rapid identification and dosage of these biomarkers for their convenient use in routine analysis of phytoplankton samples.
We recently developed and optimized an efficient phytoplankton pigment extraction method that limits pigment degradation and enhances extraction yields and access to pigments strongly linked to the thylakoid membranes [14]. By coupling this optimized extraction protocol to standardized Van Heukelem & Thomas HPLC analysis [13], we examined the pigment composition of 37 microalgae strains (excluding prokaryotes), representative of the broadest possible taxonomic diversity of marine and freshwater species. The objectives were (i) to provide a comprehensive analysis of the pigment chemodiversity of eukaryote photosynthetic microorganisms; (ii) to propose new potential chemotaxonomic markers to improve algal assemblage determination in open waters; (iii) to pinpoint ancillary carotenoid or porphyrin derivatives with original spectral properties for chemotaxonomy.
Methods
Phytoplankton strains
Thirty-seven phytoplankton strains belonging to 34 different species and 26 different genera were selected to provide as representative selection as possible of the taxonomic diversity of eukaryotic marine and freshwater species (Table 1). Strains were selected to include 15 major classes of microalgae, among the species the most studied in labs worldwide, and their taxonomic status was determined according to the international Algae Base taxonomic classification http://www.algaebase.org (accessed on 2016-03-27). All strains used in this study are referenced in international algae culture collections, including SAG, CCAP, CCMP, UTEX and IFREMER collections.
Table 1. Microalgae strain origin and culture condition.
| Phylum | Class | Family | Strain code | Short name | Origin of the strain | Culture condition |
|---|---|---|---|---|---|---|
| Eukaroyte Plantae | ||||||
| Rhodophyta | Porphyridiophyceae | Porphyridiaceae | Porphyridium purpureum SAG 1380-1d | Pp | SAG | SW—Walne’s medium |
| Rhodellophyceae | Rhodellaceae | Rhodella violacea SAG 115.79 | Rv | SAG | SW—Walne’s medium | |
| Cyanidiophyceae | Galdieriaceae | Galdieria sulphuraria | Gs | Ifremer | FW—Walne’s medium—SA | |
| Glaucophyta | Glaucophyceae | Glaucocystaceae | Cyanophora paradoxa SAG 29.80 | Cp | SAG | FW—Walne’s medium |
| Charophyta | Conjugatophyceae | Closteriaceae | Closterium baillyanum SAG 50.89 | Cb | SAG | FW—Walne’s medium—CaCO3 |
| Chlorophyta | Chlorophyceae | Scenedesmaceae | Scenedesmus acutus f. alternans UTEX 72 | Saa | UTEX | FW—Walne’s medium |
| Scenedesmus obliquus UTEX 1450 | So | UTEX | FW—Walne’s medium | |||
| Haematococcaceae | Haematococcus pluvialis CCAP 34/7 | Hp | CCAP | FW—Walne’s medium | ||
| Dunaliellaceae | Dunaliella sp. CCAP 19/19 | Dsp | CCAP | SW—Walne’s medium | ||
| Dunaliella salina SAG 19–3 | Ds | SAG | SW—Walne’s medium | |||
| Trebouxiophyceae | Chlorellaceae | Chlorella autotrophica CCMP 243 | Ca | CCMP | SW—Walne’s medium | |
| Chorella vulgaris SAG 2.80 | Cv | SAG | FW—Walne’s medium | |||
| Mamiellophyceae | Bathycoccaceae | Ostreococcus tauri H95 | Ot | Ifremer | SW—Walne’s medium | |
| Chlorodendrophyceae | Chlorodendraceae | Tetraselmis suecica CCMP 904 | Ts | CCMP | SW—Walne’s medium | |
| Eukaryote Chromista | ||||||
| Cercozoa | Chlorarachniophyceae | Chlorarachniaceae | Chlorarachnion reptans SAG 26.97 | Cr | SAG | SW—Walne’s medium—Si—SE |
| Bigelowiella natans CCMP 621 | Bn | CCMP | SW—F/2—Si | |||
| Haptophyta | Coccolithophyceae (Prymnesiophyceae) | Isochrysidaceae | Isochrysis galbana SAG 13.92 | Ig | CCAP | SW—Walne’s medium |
| Tisochrysis lutea CCAP 927/14 | Tl | CCAP | SW—Walne’s medium | |||
| Noelaerhabdaceae | Emiliania huxleyi SAG 33.90 | Eh | SAG | SW—Walne’s medium—CaCO3 | ||
| Bacillariophyta | Mediophyceae | Thalassiosiraceae | Thalassiosira pseudonana CCMP 1335 | Tp | CCMP | SW—Walne’s medium- Si |
| Skeletonemataceae | Skeletonema grethae CCAP 1077/4 | Sg | CCAP | SW—Walne’s medium- Si | ||
| Eupodiscacea | Odontella aurita 123 | Oa1 | Ifremer | SW—Walne’s medium- Si | ||
| Odontella aurita 122 | Oa2 | Ifremer | SW—Walne’s medium- Si | |||
| Chaetocerotaceae | Chaetoceros mulleri | Cmu | Ifremer | SW—Walne’s medium- Si | ||
| Chaetoceros minus | Cmi | Ifremer | SW—Walne’s medium- Si | |||
| Chaetoceros sp. Tenuissimus like | Ctl | Ifremer | SW—Walne’s medium- Si | |||
| Chaetoceros calcitrans f. pumillum CCAP 1010/11 | Ccp | CCAP | SW—Walne’s medium- Si | |||
| Chaetoceros calcitrans CCMP 1315 | Cc | CCMP | SW—Walne’s medium- Si | |||
| Chaetoceros gracilis UTEX LB 2658 | Cg | UTEX | SW—Walne’s medium- Si | |||
| Bacillariophyceae | Bacillariaceae | Nitzschia sp. CCMP 2526 | Nsp | CCMP | SW—Walne’s medium- Si | |
| Bacillariophyceae incertae sedis | Phaeodactylaceae | Phaeodactylum tricornutum CCMP 632 | Pt | CCMP | SW—Walne’s medium—Si | |
| Cryptophyta | Cryptophyceae | Pyrenomonadaceae | Rhodomonas salina CCAP 978/27 | Rs | CCAP | SW—Walne’s medium |
| Eukaryote protozoa | ||||||
| Miozoa | Dinophyceae | Goniodomataceae | Alexandrium tamarense MOG 835 (Toxic) | At1 | Ifremer | SW—ESP |
| Alexandrium tamarense PLY 497A (non toxic) | At2 | Ifremer | SW—ESP | |||
| Alexandrium minutum CM1002 (Non toxic) | Am1 | Marine Institute | SW—F/2 | |||
| Alexandrium minutum AM89BM (Toxic) | Am2 | Ifremer | SW—L1 | |||
| Euglenophyta | Euglenophyceae | Euglenaceae | Euglena proxima SAG 1224-11a | Ep | SAG | FW—Walne’s medium |
Classification was established according to Guiry, M.D. & Guiry, G.M. 2016. AlgaeBase. Worldwide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 27th March 2016. SW: Seawater; FW: Freshwater; Si: Silicates (0,4g.L-1); CaCO3: Calcium carbonate (0,25 mg.L-1); SE: Soil extract (0,5 mL.L-1); SA: Sulfuric acid (final medium pH = 2); ESP: Bold’s basal medium + proteose peptone + soil extract + vitamins.; F/2: Guillard medium; L1: Modified F/2 medium. Incertae sedis classification is displayed where phenotypic characteristics are not supported by genomic data. Colours distinguish the taxonomic lineages and outsider phyla in terms of pigmentation.
Culture and collection
Microalgae were grown in sterile 100 mL flasks under continuous light (120 μmol.m-2.s-1) provided by fluorescent lamps (Philips TLD 58W 865), at 20°C in normalized growth medium for each strain (Table 1). Standardization of the culture parameters allowed the quantitative interspecific comparison of pigment production yields (Chromatograms in Supporting Information S2 Fig). Cells were harvested by centrifugation in early stationary growth phase since in the open environment, phytoplankton populations are a mix of new cells and newly senescent cells.
Pigment extraction
Pigments were extracted according to a new original [13] extraction protocol, which allowed an efficient, environment-friendly, non-damaging extraction of pigments, even those strongly linked to the thylakoid membranes [14]. Briefly, a pellet of 50.106 algae cells was dispersed in 500 μL of ethanol (EtOH) and mixed with 0.728 g of 500 μm glass beads for 30 min at 30 Hz frequency in a mixer-mill (Retsch MM-400). The beads were then removed, the pigment extract centrifuged for 5 min at 4500 g at 4°C and the supernatant filtered on a 0.2 μM PTFE filter before immediate HPLC analysis.
HPLC analysis
Pigment extracts were analysed using the SCOR/UNESCO reference method described by Van Heukelem et al. (2001) [13], with a slight modification. Pigments were separated on an Eclipse XDB-C8 (4.6 × 150 mm, 3.5 μm particle size, Agilent) column, using an Agilent HPLC-UV-DAD series 1200. The elution gradient consisted in a 30 min gradient from 95% to 5% solvent A (70:30 methanol, 28 mM aqueous TBAA pH 6.5). At the end of the run, an isocratic flow of 5% solvent A was maintained for 10 min. The flow rate remained at 1 mL/min during the run. Pigment elution was acquired at 436, 450, 405 and 665 nm. Wavelength 436 nm is used to look for all pigments, 450 nm is specific to carotenoids, 405 nm allow the visualization of chlorophyll degradation products and 665 nm is specific to chlorophyll a and its derivatives. All peak spectra were acquired from 210 to 800 nm (Agilent Diode Array Detector G1315D) and the ChemStation software (Agilent Technologies) was used to create a spectral library.
Pigment identification
A chromatographic and spectral library was created from standard pigments provided by DHI lab products (Denmark). The pigments identified with a pure standard were: chlorophyll b (Chl b), chlorophyll a (Chl a), pheophorbide a methyl ester (Pheide a methyl ester), chlorophyllide a (Chlide a), chlorophyll c2 (Chl c2), chlorophyll c3 (Chl c3), pheophytin a (Phe a), astaxanthin (Asta), antheraxanthin (Anthe), lutein (Lut), 19’-hexanoylfucoxanthin (Hex-fuco), fucoxanthin (Fuco), 19’-but-fucoxanthin (19’-but-Fuco), 19’-hex-fucoxanthin (19’-hex-Fuco), violaxanthin (Viola), 9’-cis-neoxanthin (c-Neo), zeaxanthin (Zea), diadinoxanthin (Diadino), diatoxanthin (Diato), β,ε-carotene (β,ε-Car) and β,β-carotene (ββ-Car). Canthaxanthin (Cantha), dinoxanthin (Dino), chlorophyll a and b allomers (Chl a or b allo), peridinin (Peri), alloxanthin (Allo), peridinin isomer (Peri-iso), crocoxanthin (Croco) and prasinoxanthin (Pras) were identified according to DHI mix-108 and DHI mix-111 (DHI, Denmark). Standards were evaporated under nitrogen and solubilized in ethanol allowing comparison of spectral signatures, as samples were analysed in the same solvent. Pigments in microalgae extracts were identified by comparison of chromatographic and spectral data recorded with standard pigments, using ChemStation software (Agilent). Unidentified pigments were also stored in the spectral library. Identification of pigments was realised according to software retention time and match factor result. The software compared retention times and aligned absorption spectra (from 210 to 800 nm), to calculate a match factor comprised between 0 and 1000, representing the degree of similarity between spectra. The comparison of two spectra gives the match factor, which is defined as:
The x and y values are measured absorbance from the first and second spectrum respectively, at the same wavelength; n is the number of data points and ∑ the sum of the data. At the extremes, a match factor of 0 indicates no match whilst 1000 indicates identical spectra. In our case, values greater than 999 indicate that the spectra are similar. If the score value was between 995 and 999, a stringent alignment of spectra in the visible area was then checked (from 350 to 800 nm) and, if the match factor was higher than 999, the pigment was considered as similar and marked with an asterisk (*). All values below 995 indicate that the spectra are different. For very low abundance pigments, the match factor could be influenced by spectral noise level. In case of strong abundance of a well-known pigment, even if the score was slightly below 999, the pigment was identified according to the occurrence of the well-known pigment described in the species and marked with a cross (†). For each unidentified pigment which did not match a score >995, the spectrum was registered in the spectral library and annotated according to their specific retention time and spectral properties (i.e. carotenoid or porphyrin spectral signature).
All unidentified pigments were named according to their spectral signature and their elution order: Car(x) for carotenoid-like pigments, P(x) for porphyrin-like pigments and PCar(x) for pigments with both spectral properties and with a Red-Soret ratio higher than 5%.
Carotenoids and porphyrin band ratio were calculated according to [8]. Details for band ratio calculation are presented in supporting information S1 file for carotenoids (S1A Fig) and for porphyrins (S1B Fig).
Determination of pigment communities among phytoplankton strains
The inter-strain comparison of pigment composition was studied and graphically represented using the open source Gephi communities’ network software (http://gephi.github.io/) as recommended in [15]. All strains and pigments (whether identified or not) were entered in the software and pigments were associated with their respective strains only by a qualitative link (presence of the pigment in the strain). The software grouped the strains together in communities sharing common pigments. Circles with a high diameter represent strains, while circles with a small diameter represent pigments. The validation of the communities was illustrated by the central circles of beta-carotene and chlorophyll a (including Chl a epimer), which were common to all strains. To make the results easier to read, microalgae belonging to the red, green and brown lineages are presented using their respective colours.
Results
Pigment composition of the 37 phytoplankton strains
The pigment chromatograms recorded at 436 nm are presented in Supporting Information S2 Fig. Chlorophyll a (and epimer) and β,β-carotene were common to all strains. In addition, each strain contained approximately 10 major and minor pigments, identified using the standard pigment database. All pigments detected among the biodiversity of the 37 strains were classified according to their HPLC retention times (Table 2). This analysis revealed that, in addition to the 26 standard pigments, 98 molecules corresponding to unidentified pigments or standards derivatives were present in the chromatograms. According to their spectral properties (presence or absence of absorption bands I, II and III), 57 pigments were classified as carotenoid derivatives (Car), 35 as porphyrin derivatives (P) and six pigments with spectral properties of both were named PCar. These unidentified pigments were named according to their elution order in the HPLC system. Table 2 summarizes the retention time, maximal absorption wavelengths, band ratio and occurrence in the strains for each pigment detected in the chromatograms. Pigment standards are written in italic characters and pigments characteristic of one strain or one species are written in bold characters. Pigments identified on the basis of their polarity and spectral absorption are indicated in brackets. The number of unidentified pigments may be in fact slightly inferior to 98, as the real number of chlorophyll allomers is probably inferior to those discriminated by our analysis, according to the slight variability of retention times and spectrum comparison. A high diversity of pigment spectra was observed and rules of identification and differentiation were created, allowing the creation of Table 2. Some pigments have very close retention times and band ratios but were not a sufficiently good match to be merged. We determined band ratio for chlorophyll and carotenoid. These results could be used to make comparisons among our results and with descriptions in the literature.
Table 2. Elution order of all pigments identified in microalgae strains.
Maximum absorbance spectra in the eluent were detailed for porphyrins (P) and carotenoids (Car).
| Peak N° | Short name | Retention time [min] | Porphyrin | Carotenoid | Occurrence in strains | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Soret [blue] I [nm] | Red II [nm] | Soret:Red | I [nm] | II [nm] | III [nm] | %III/II | ||||
| 1 | P1 | 6.6 | 432 | 622 | 9.67 | Nsp | ||||
| 2 | Chl c3 | 6.7 | 456 | 589 | 8.79 | Eh* | ||||
| 3 | Car1 | 6.8 | 467 | At1; At2*; Am1; Am2 | ||||||
| 4 | P2 | 7.3 | 446 | 631 | 8.33 | Cmu; Ccp; Ctl; Oa1*; Ig*; Cg*; Pt; At1; At2 | ||||
| 5 | P3 | 7.7 | 441 | 630 | 6.73 | Nsp | ||||
| 6 | P4 | 8.1 | 434 | 628 | 5.83 | Gs | ||||
| 7 | P5 | 8.1 | 458 | 636 | 9.00 | At1 | ||||
| 8 | P6 | 8.2 | 436 | 651 | 4.40 | Cr | ||||
| 9 | P7 | 8.2 | 434 | 629 | 6.13 | Sg | ||||
| 10 | Chlide a | 8.4 | 432 | 666 | 0.98 | Cb | ||||
| 11 | P8 | 8.6 | 436 | 656 | 5.20 | Cr | ||||
| 12 | P9 | 8.8 | 440 | 631 | 6.56 | Ot | ||||
| 13 | Chl c2 | 8.9 | 447 | 633 | 7.90 | Rs; Ctl; Cmu; Oa2; Eh; Tl; At1†; At2; Am2; | ||||
| 14 | P10 (Chl c2 like) | 8.9 | 447 | 634 | 7.53 | Tp; Cg; Ig; Oa1; Cmi; Pt* | ||||
| 15 | P11 | 8.9 | 445 | 666 | 6.01 | Ccp | ||||
| 16 | P12 | 8.9 | 440 | 666 | 2.06 | Pt | ||||
| 17 | P13 | 8.9 | 435 | 666 | 1.31 | So | ||||
| 18 | P14 | 8.9 | 436 | 666 | 1.30 | Sg | ||||
| 19 | P15 | 8.9 | 445 | 666 | 3.06 | Cc | ||||
| 20 | P16 (Chl c1 like) | 9.2 | 444 | 633 | 6.48 | Tp; Pt†; Cg; Ccp; Ctl; Cmu; Oa1; Oa2; Tl; Ig | ||||
| 21 | P17 | 9.2 | 443 | 634 | 5.53 | Sg | ||||
| 22 | P18 | 9.2 | 445 | 633 | 9.2 | Cc | ||||
| 23 | P19 | 11.0 | 432 | 667 | 1.00 | Pt; Sg | ||||
| 24 | Car2 | 11.9 | 453 | Ig; Tl | ||||||
| 25 | Pheide a | 12.1 | 409 | 666 | 2.24 | Gs | ||||
| 26 | Peri | 12.3 | 476 | At1; At2; Am1; Am2 | ||||||
| 27 | P20 | 12.8 | 410 | 665 | 1.97 | Cr | ||||
| 28 | Peri-iso | 12.8 | 477 | At1; At2; Am1; Am2 | ||||||
| 29 | P21 | 13.8 | 431 | 665 | 1.00 | Cb; Pt; Sg; Gs; So | ||||
| 30 | Car3 | 14.1 | 470 | Ctl; Nsp | ||||||
| 31 | Car4 | 14.1 | 468 | At1 | ||||||
| 32 | PCar1 | 14.2 | 422 | 666 | 9.12 | 470 | Am2 | |||
| 33 | Car5 | 14.6 | 449 | Oa1 | ||||||
| 34 | Car6 | 15.0 | 454 | 474 | 218 | Ot | ||||
| 35 | Car7 | 15.5 | 462 | At1; At2; Am1; Am2 | ||||||
| 36 | Car 8 | 15.5 | 440 | 467 | 48 | Bn; Ts | ||||
| 37 | Car9 | 15.5 | 442 | 468 | 81 | Cr | ||||
| 38 | Car 10 | 15.9 | 468 | At1; At2; Am1; Am2 | ||||||
| 39 | Car 11 | 16.3 | 418 | 441 | 470 | 103 | Cr | |||
| 40 | Car12 (t-neo-like) | 16.3 | 417 | 441 | 470 | 89 | Bn; Ep; Ts; Ot; Cv; Ca; Dsp; Ds; Hp; So; Saa | |||
| 41 | Car13 | 16.7 | 484 | Am1 | ||||||
| 42 | Fuco | 16.9 | 453 | Tp; Pt; Cc; Cg; Ccp; Ctl; Cmi; Cmu; Nsp; Oa1; Oa2; Sg; Eh; Tl; Ig | ||||||
| 43 | Car14 | 17.5 | 479 | At2; Am1; Am2 | ||||||
| 44 | Car15 (Loro like) | 17.5 | 445 | 472 | 41 | Bn; Cr; Ts; So; Saa | ||||
| 45 | 9-cis Neo | 17.6 | 413 | 436 | 465 | 86 | Bn; Ep; Cr; Ts; Ot; Cv; Ca; Dsp; Ds; Hp; Cb; So; Saa | |||
| 46 | Car16 | 17.7 | 475 | At1*; At2; Am1; Am2 | ||||||
| 47 | Car17 | 17.9 | 425 | 447 | 476 | 62 | At1; At2; Am1; Am2 | |||
| 48 | 19-hex k Fuco | 17.9 | 448 | 469 | 85 | Eh | ||||
| 49 | Pras | 18.4 | 455 | Ot | ||||||
| 50 | Viola | 18.8 | 416 | 439 | 469 | 89 | Bn; Cr; Ig; Ts; Ot; Cv; Ca; Dsp; Ds; Hp; Cb; So; Saa | |||
| 51 | Asta | 19.4 | 479 | Cr; Saa | ||||||
| 52 | Car18 | 19.8 | 459 | Ot | ||||||
| 53 | 19’-Hex-Fuco | 19.2 | 447 | 462 | 468 | 9 | Eh | |||
| 54 | Car19 (cis-Fuco like) | 20.2 | 443 | Tp; Pt; Cc; Cg; Ccp; Ctl; Cmi; Cmu; Nsp; Sg; Tl; Ig; Oa1; Oa2 | ||||||
| 55 | Car20 | 20.4 | 403 | 428 | 455 | 64 | Am1; Am2; At1; At2 | |||
| 56 | Diadino | 20.6 | 422 | 446 | 475 | 54 | Ep; Tp†; Pt; Cc; Cg; Ccp; Ctl; Cmi†; Cmu; Nsp; Oa1*; Oa2; Sg; Eh; Tl; Ig; Am1; Am2; At1; At2 | |||
| 57 | Car21 | 20.8 | 411 | 434 | 462 | 68 | Bn; Ts; Dsp* | |||
| 58 | Car22 | 20.9 | 458 | Ep | ||||||
| 59 | Car23 | 21.5 | 440 | 466 | 55 | Cr | ||||
| 60 | Car24 | 21.5 | 442 | 467 | 42 | Ts | ||||
| 61 | Car25 | 21.5 | 443 | 469 | 22 | Ot; Ca; Ds | ||||
| 62 | Car26 | 21.5 | 415 | 437 | 464 | 82 | Cv | |||
| 63 | Car27 | 21.5 | 417 | 438 | 465 | 70 | Saa; Hp | |||
| 64 | Anthe | 21.7 | 445 | 473 | 60 | Dsp; Bn; Cb | ||||
| 65 | Car28 | 21.9 | 419 | 439 | 468 | 37 | Ep | |||
| 66 | Car29 | 22.1 | 471 | Cr | ||||||
| 67 | Allo | 22.1 | 452 | 479 | 32 | Rs; Am1 | ||||
| 68 | Car30 | 22.4 | 421 | 441 | 470 | 67 | Ep | |||
| 69 | Car31 | 22.4 | 452 | At2 | ||||||
| 70 | Car32 | 22.6 | 420 | 441 | 470 | 58 | Ep | |||
| 71 | Car33 | 23.1 | 445 | 462 | 475 | 58 | Rs | |||
| 72 | Diato | 23.2 | 469 | 478 | 32 | Tp; Cc; Cg; Ccp; Ctl; Cmi; Cmu*; Nsp; Oa1; Oa2; Sg; Eh; Tl; Ig; Ep*; Am1; Am2*; At1 | ||||
| 73 | P22 | 23.6 | 429 | 659 | 1.00 | Gs | ||||
| 74 | Zea | 23.9 | 451 | 477 | 25 | Bn; Cr; Cb; Cp; Ts*; Ot; Cv; Ca; Dsp; Ds; Hp; So; Saa; Rv; Pp; Gs; Cg* | ||||
| 75 | Lut | 24.2 | 445 | 472 | 60 | Bn; Cr; Ts; Cv; Ca; Ds; Dsp; Hp; Cb; So; Saa | ||||
| 76 | Car34 | 24.6 | 427 | 453 | 59 | Ot | ||||
| 77 | P23 | 24.8 | 438 | 688 | 5.01 | Ts | ||||
| 78 | Car35 | 24.8 | 420 | 438 | 465 | 41 | Dsp; Ds; Hp; Cb; So; Saa; Cv | |||
| 79 | PCar 2 | 24.8 | 438 | 688 | 19.3 | 416 | 438 | 465 | 9 | Ca |
| 80 | Car36 | 25.0 | 444 | 469 | 10 | Cp; Gs; Rv; Pp* | ||||
| 81 | Cantha | 25.5 | 476 | No presence in analyzed strains | ||||||
| 82 | Car37 | 25.8 | 419 | 439 | 467 | 61 | Ts*; Hp*; So; Saa | |||
| 83 | Car38 | 25.9 | 419 | 440 | 467 | 59 | Cv; Ca; Ds; Dsp; | |||
| 84 | Car39 | 26.0 | 445 | 471 | 31 | Gs; Rv; Pp | ||||
| 85 | Car40 | 26.5 | 461 | Ep | ||||||
| 86 | Car41 | 27.6 | 446 | 473 | 37 | Ts; Bn | ||||
| 87 | P24 | 27.6 | 420 | 655 | 1.84 | Cg*; Cmu; Nsp | ||||
| 88 | P25 | 27.6 | 420 | 655 | 1.88 | Ccp; Ctl | ||||
| 89 | P26 | 27.7 | 456 | 635 | 1.84 | Hp; Saa | ||||
| 90 | P27 | 28.6 | 436 | 661 | 1.23 | Bn | ||||
| 91 | Car42 | 29.0 | 446 | 473 | 38 | Bn; Cr; Ts | ||||
| 92 | Chl b | 29.7 | 468 | 651 | 2.80 | Bn; Ep; Cr; Ts; Ot; Cv; Ca; Dsp; Ds; Hp; So; Saa; Cb | ||||
| 93 | Car43 | 29.9 | 468 | Tl | ||||||
| 94 | Car44 (Croco like) | 30.2 | 446 | 472 | 50 | Rs | ||||
| 95 | Chl b-epi | 30.3 | 468 | 651 | 2.80 | Bn; Ep; Ts; Ot; Cv; Ca; Dsp; Ds; So; Hp | ||||
| 96 | Car45 | 31.0 | 445 | 471 | 25 | Rv | ||||
| 97 | P28 (chl a allomer) | 31.1 | 420 | 655 | 1.78 | Tp; Pt; Cc; Cg; Ccp; Ctl; Cmi; Cmu; Nsp; Oa1; Hp; Saa; At1; At2 | ||||
| 98 | Car46 | 31.2 | 446 | 472 | 50 | Bn | ||||
| 99 | Car47 (Cryp) | 31.2 | 451 | 477 | 24 | Gs; Rv; Pp | ||||
| 100 | P29 (chl a allomer) | 31.3 | 419 | 662 | 1.63 | Pt*; Cc*; Ccp; Ot | ||||
| 101 | P30 (chl a allomer) | 31.5 | 430 | 664 | 1.11 | Ts | ||||
| 102 | P31 (chl a allomer) | 31.5 | 432 | 665 | 1.10 | Cv | ||||
| 103 | P32 (chl a allomer) | 31.5 | 422 | 664 | 1.20 | Pt; Cc*; Ccp; Ctl; Ot | ||||
| 104 | P33 (chl a allomer) | 31.5 | 429 | 665 | 1.11 | Nsp*; Oa1*; Cmi; Cmu*; Ca; Hp*; Saa; At1; At2 | ||||
| 105 | Car48 | 31.8 | 474 | Tl | ||||||
| 106 | Chl a | 32.3 | 431 | 665 | 1.00 | All strains | ||||
| 107 | P34 | 32.6 | 422 | 666 | 3.53 | Nsp; Ccp | ||||
| 108 | Chl a -epi | 32.8 | 431 | 665 | 1.00 | All strains with At1*; At2*; [excepted Eh] | ||||
| 109 | Car49 | 33.1 | 472 | Ep | ||||||
| 110 | Car50 | 33.4 | 425 | 451 | 44 | Gs | ||||
| 111 | P35 | 33.7 | 438 | 655 | 4.95 | Cr | ||||
| 112 | PCar3 | 34.5 | 445 | 666 | 6.67 | 416 | 445 | 474 | 56 | At1 |
| 113 | PCar4 | 34.5 | 415 | 666 | 5.16 | 415 | 444 | 474 | 37 | Am1 |
| 114 | PCar5 | 34.5 | 413 | 666 | 3.95 | 412 | 445 | 473 | 20 | Am2 |
| 115 | Car51 | 34.6 | 442 | 469 | 55 | Ot | ||||
| 116 | Car52 | 35.0 | 432 | 453 | 482 | 31 | Dsp | |||
| 117 | PCar6 | 35.2 | 468 | 653 | 8.66 | 441 | 468 | 498 | 9 | Ts |
| 118 | Car53 | 35.6 | 416 | 439 | 465 | 33 | Cv | |||
| 119 | Car54 | 35.7 | 438 | 459 | 488 | 47 | Ts; Ds; Dsp | |||
| 120 | Car55 | 35.8 | 444 | 467 | 9 | Bn; Pt*; Cmu; Cb*; Cp; Gs; Ot; Cv; Ca*; Rv; Pp; Hp; Sg*; Ccp*; Ctl*; Nsp*; Oa1*; Oa2* | ||||
| 121 | βε-Car | 36.4 | 444 | 472 | 56 | Rs; Ts; Cv; So; Cb; Cr | ||||
| 122 | ββ-Car | 36.5 | 450 | 475 | 15 | All strains | ||||
| 123 | Car56 | 37.1 | 440 | 466 | 28 | Cp; Dsp; So; Cb; Ts; Bn | ||||
| 124 | Car57 | 37.1 | 442 | 466 | 11 | Gs | ||||
Pigment standards are written in italic characters. Pigment characteristics of one strain or several strains within a particular species are written in bold characters. Strains marked with an asterisk cross (†) indicates that the pigment is abundant and fully characterized in literature. Strains marked with an asterisk (*) have a match factor of 999 in visible spectra.
Among the uncharacterized pigments, six could be identified on the basis of their polarity (retention time), spectral properties and by descriptions in the literature of ancillary pigments in strains. They corresponded to Chl c1 like (P16) [16,17], t-neoxanthin (Car12) [18] loroxanthin like (Car15) [19,20], crocoxanthin like (Car44) [21], β-cryptoxanthin like (Car47) [22,23] and cis-fucoxanthin like (Car19) [17,24]. P28 to P33 were identified as Chl a allomers, as they corresponded to minor pigments with polarity and maximal absorption wavelengths very close to Chl a, but eluting a few seconds before Chl a [25]. This is also the case for P26, which corresponds to a Chl b allomer. All pigments identified were checked against literature descriptions with the same analytical method [13,26]. Without validation by mass spectrometry analysis, unidentified Car or P designating the pigment were instead used for putative identification.
Detection of photoprotective pigments
As expected, the carotenoids involved in the xanthophyll cycles, ensuring thermic dissipation of high irradiance energy, were different between the red, green and brown lineages. Accumulation of zeaxanthin and absence of antheraxanthin and violaxanthin were observed in the Rhodophyceae (Pp, Rv and Gs), Glaucophyceae (Cp) and Charophyceae (Cb). Diadinoxanthin and diatoxanthin were detected in the Bacillariophyceae (Sg, Cc, Cg, Ccp, Ctl, Cmi, Cmu, Nsp, Oa1, Oa2, Pt and Tp) and Haptophyta (Tl, Ig and Eh). In some cases, diadinoxanthin was detected in the absence of diatoxanthin, indicating the total conversion of diatoxanthin into diadinoxanthin in our relatively low light culture conditions. Conversely, it is important to remember that diatoxanthin increases as cells become less healthy. Zeaxanthin/antheraxanthin/violaxanthin were detected in the chlorophytes Cb and Dsp and in the Chlorarachniophyceae Bn. Zeaxanthin/violaxanthin in absence of antheraxanthin were also detected in the other chlorophytes analysed, and in the Chlorarachniophyceae Cr. The presence of zeaxanthin and violaxanthin indicated the possibility for these strains to synthesize antheraxanthin, and its absence indicated a total conversion into zeaxanthin or violaxanthin in our culture conditions. Isochrysidaceae (Ig) and Skeletonemale (Cg) combined a diatoxanthin / diadinoxanthin photoprotective system with the ability to synthesize zeaxanthin, antheraxanthin and/or violaxanthin because of the conservation of an ancestral gene [27].
Presence of pigments currently used as chemotaxonomic markers
Among the 124 HPLC peaks analysed in all phytoplankton strains, 63 were strain-characteristic and six were species-characteristic (shared by several strains in one considered species, in our case dinoflagellates). We thus detected 63 pigments (Table 2. bold characters) that could be used as stringent specific chemotaxonomic markers to discern a strain or species over the 37 studied. Six of these 63 pigments corresponded to standard pigments. Chl c3, 19’-hexanoyloxy fucoxanthin and 19-hexanoyl-k-fucoxanthin pigments indicated the presence of Emiliania huxleyi SAG33.90, prasinoxanthin indicated the presence of the Prasinophyte Ostreococcus tauri H95 and pheophorbide a occurred in Galdieria sulphuraria. Pheide a usually occurs in senescent algae, while in our case all the cultures were in early stationary phase of growth but cultivated in acidic condition, at pH 2, which could explain the loss of Mg and the phytyl-chain of Chl a. Moreover, the very low detection of pheophorbide a (Pheide a) among the 37 strains studied confirmed the quality of our fresh pigment extraction process, that limited Chl a damage. Chlorophyllide (Chlide a) is present in Cb but cannot be used as a chemotaxonomic marker because this molecule participates in the turnover of chlorophyll a. Chlide a is potentially distributed universally in microalgae.
S1 Table lists the pigments currently used as chemotaxonomic markers for phytoplankton strains, species or classes, according to the classification proposed by Wright and co-workers [28]. The detection of a chemotaxonomic marker unambiguously signals the presence of a strain in a sample, but its absence should not be interpreted as an absolute demonstration of the absence of a strain, as light and nutrient conditions can silence pigment expression.
Canthaxanthin is described as a significant chemotaxonomic marker but not always present for the Cyanophyceae CYANO-1 class and may be present in Eustigmatophyceae EUSTIG-1 and Dinoflagellate Chloroplast type-1 classes. As we did not detect canthaxanthin, we concluded that no strain was able to synthesize canthaxanthin. The light conditions in our experiments were not favourable to its biosynthesis, as canthaxanthin is a secondary carotenoid, involved in algal photoprotection. Astaxanthin is described as a chemotaxonomic marker for the Chlorophyceae (CHLORO-1) and Trebouxiophyceae classes. Accordingly, we identified it in Scenendesmus acutus f. alternans UTEX72, and in the Chlorarachniophyceae Chlorarachnion reptans SAG26.87. No astaxanthin was detected in other Chlorophyta strains, suggesting that the light irradiance was too low to trigger astaxanthin biosynthesis in these strains [29]. Antheraxanthin is used as a chemotaxonomic marker for the Chrysophyceae CHRYSO-1 and Eustigmatophyceae EUSTIG-1 classes and belongs to the zeaxanthin/antheraxanthin/violaxanthin photoprotection system. None of the strains studied here belong to these classes but some belong to the Chlorophyta species with the xanthophyll cycle photoprotection system. Thus, we identified this pigment (as minority or trace pigment) in Dunaliella sp. CCAP19/19. We also detected Antheraxanthin in Closterium baillyanum SAG50.89 (Closteriaceae) and Bigelowiella natans CCMP243 (Chlorarachniophyceae). Pigment patterns in Dinophyta were described by Zapata et al. [30]. Here, we analysed four strains with a chloroplast known as dinoflagellate chloroplast type-1. These four strains contained common carotenoids Car1, Car7 and Car10 with lambda max respectively at 467, 462 and 468 nm in our system. Peridininol and dinoxanthin are described as markers of dinoflagellate chloroplast type-1. Car1 could be identified as Peridininol but not enough clues are available to confirm it. Eight haptophyte pigment types were defined by Zapata [31]. Emiliania huxleyi SAG33.90 belongs to haptophyte pigment type-6, its characteristic pigments are Chl c3, 19-hex-k fucoxanthin and 19’hex-fucoxanthin. Other haptophytes analysed, Ig and Tl, did not exhibit such stringent markers.
Analysis of phytoplankton pigment communities and detection of characteristic pigments
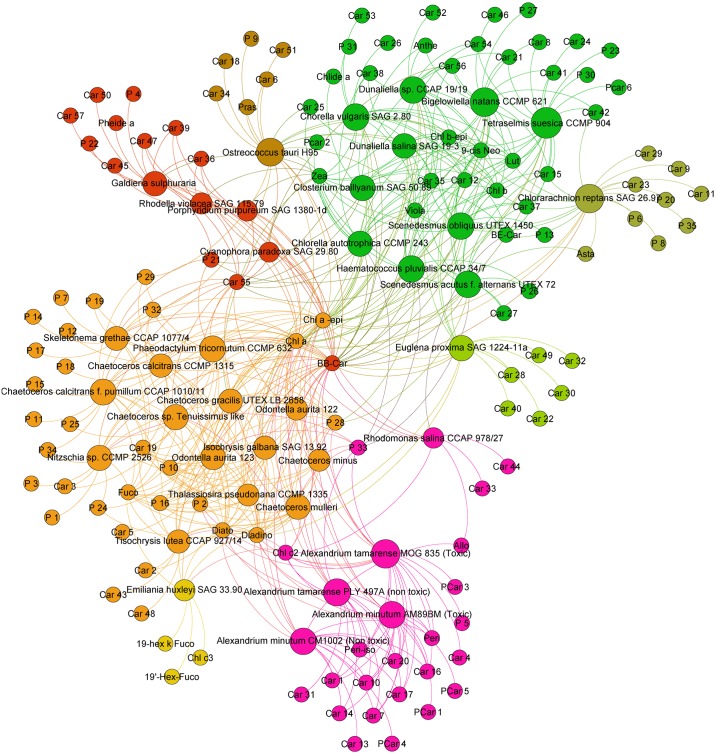
We performed a community analysis to highlight the taxonomic clusters defined by the commonly shared pigments in the diverse phytoplankton strains. Fig 1 presents the pigment communities obtained for all strains. Strains clustered together into four major communities that correspond to the green, red, brown lineages and Dinophyceae focus. The cluster only shows a link between algae and pigment without taking into account the quantity of pigment. This result confirmed that the common sharing of photoprotective pigments, respectively zeaxanthin in the red lineage, violaxanthin/antheraxanthin/zeaxanthin in the green lineages and diadinoxanthin/diatoxanthin in brown strains, is a major relevant taxonomic key to classify the strains on the basis of their pigments. The graphical analysis also confirmed that in addition to the common photosynthetic and photoprotective pigments, most strains contain characteristic pigments. The graphical representation allows a fast identification of these pigments, as they cluster in the most external sides of the Gephi figure, and are connected to their productive strain by a unique link. Using the Gephi representation, the number of pigment characteristics of one strain was also very easy to determine visually. For example, Ostreococcus tauri H95 contains five characteristic carotenoids (Car6, Car18, Car34, Car51 and Pras) and one characteristic porphyrin (P9), while Emiliania huxleyi SAG33.90 contains two characteristic carotenoids (19-hex-k-fuco, 19’-hex-fuco) and one characteristic chlorophyll (Chl c3).
Fig 1. Pigment communities among the 37 microalgae strains studied.
The Gephi analysis reveals that strains cluster together into four major communities corresponding to the green, red, brown lineages and a Dinophyceae clustering. Strain-characteristic pigments cluster at the edges of Fig 1, allowing a fast count and determination.
In the middle of the figure, key photosynthetic and photoprotective pigments are represented i.e., Chl a, its epimer and ββ-Car. In the yellow-orange group, Diato, Diadino and Fuco groups belong to the brown-yellow algae lineage. The green lineage is close to Chl b and its epimer, Viola, 9-cis-Neo and Lut. Zeaxanthin, because of its accumulation in red algae, is at the frontier between red lineage and green lineage. Dinoflagellates form a group with the presence of peridinin and its epimer.
As a second step, we separated the clusters, according to xanthophyll cycle, in order to perform a limited Gephi analysis. The aim is to obtain a better resolution for strain-characteristic pigments and a fast pigment determination shared by a short number of strains within one or several species.
Red lineage
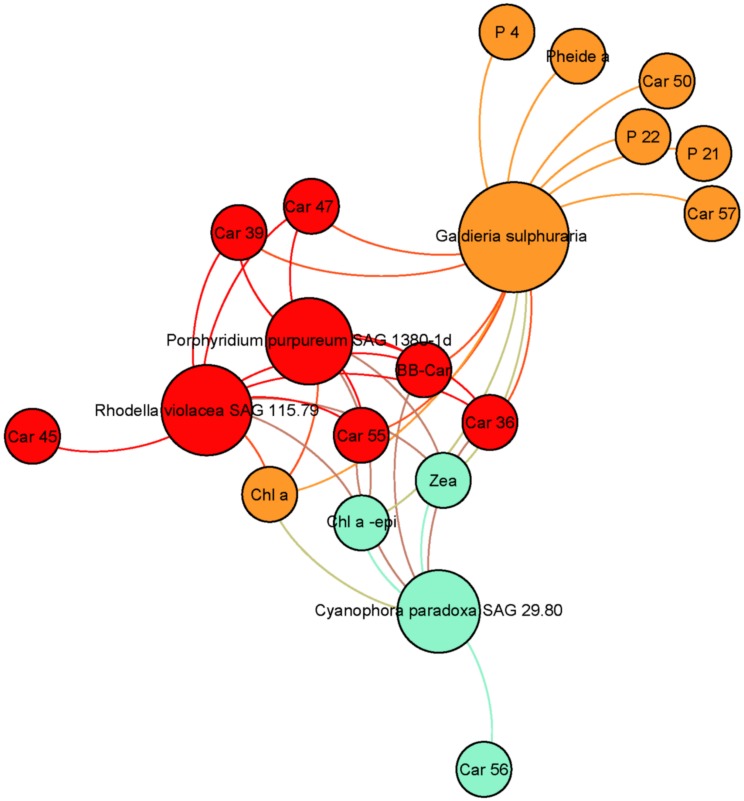
Fig 2 presents the pigment communities and contents among the red lineage. Rhodophytes (Pp, Rv, Gs) and glaucophytes (Cp) are characterized by the common synthesis of Zea, Chl a epimer, Car36 and Car39. It should also be noted that rhodophytes and glaucophytes strains synthesize phycobiliproteins, which are not presented in this study. Car47 was identified as β-cryptoxanthin and the presence of this pigment in three strains accumulating zeaxanthin was coherent because β-cryptoxanthin is the biosynthetic intermediate between β-carotene and zeaxanthin. Its presence in Porphyridium purpureum SAG1380-1d was also independently confirmed by a high-resolution mass spectrometric analysis [32]. In addition, Galdieria sulphuraria contained 1 characteristic carotenoid (Car50), and the three characteristic porphyrins P4, P21 and P22. Pheide a is only present in Gs in the red lineage but it cannot be considered as a marker because this pigment is a degradation pigment. This fact cannot be explained by the extraction process (gentle method) but by the acid culture conditions of this strain, which catabolises this kind of chlorophyll degradation product. Car45 was characteristic of Rv.
Fig 2. Pigment communities among red strains accumulating zeaxanthin for photoprotection (Rhodophyta, Glaucophyta).
The core of Fig 2 groups together photosynthetic pigments, with Chl a and its epimer, ββ-Car and Zea. Two other pigments included are Car55, which has an elution very close to ββ-Car, and Car36. Car36 is a key ancillary pigment binding Rhodophyta and Glaucophyta. Presence of these pigments in oceanographic analyses can indicate the presence of the corresponding phyla.
Synthesis of astaxanthin and lutein was previously reported in Gs in autotrophic conditions [33] but no traces of either of these pigments were detected in our study. Absence of these two pigments indicates that the relatively low light conditions in our experiments were not favourable to their biosynthesis in this strain. Nevertheless, Gs has a porphyrin (P21) in common with green lineage algae, this is also the case of a carotenoid Car57 in Cp.
Green lineage
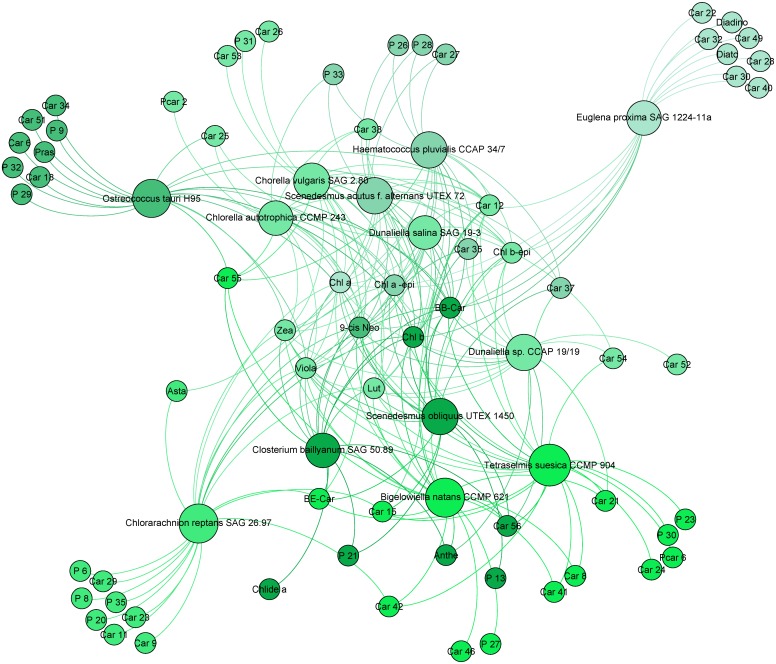
Fig 3 presents the pigment composition of strains containing a Viola/Anthera/Zea cycle. Ep was studied with the green lineage as its endosymbiont contains Chl b. All green strains contained Chl b, Chl b epimer and Neo. Lutein was also present in all strains except Ot and Ep. βε-Car is a lutein precursor, its occurrence in some strains is explained by the pathway suggested by Takaichi [18].
Fig 3. Pigment communities among green strains, using the zeaxanthin/antheraxanthin/violaxanthin cycle for photoprotection (Charophyta, Chlorophyta and Cercozoa).
Car12, identified as t-neoxanthin-like, was common to nearly all chlorophytes and the chlorarachniophyte Bn, but excluded Cr. This pigment is a close metabolic derivative from neoxanthin and 9-cis-violaxanthin. Car12, which is present in our chlorophyte strains, is quite a good marker for this phylum. Moreover, most strains contain one to eight characteristic pigments: Car52 in Dsp; Car46 and P27 in Bn; Car24, PCar6, P23 and P30 in Ts; Car26, Car53 and P31 in Cv; P13 in So; Car9, Car11, Car23, Car29, P6, P8, P20 and P35 in Cr; and PCar 2 in Ca. In our case, PCar 2 seems to be a co-elution of a porphyrin and a carotenoid. The carotenoid has the same lambda max as Car35 present in Cv and other green algae analysed.
Strain Cr contained characteristic porphyrin derivatives (P6, P8, P20 and P35) and four characteristic carotenoids (Car9, Car11, Car23 and Car29). They may belong to the species Nitzschia curvilineata SAG48.91, whose addition to the culture medium was required to feed Cr. Its presence in the Cr culture was minor but necessary for the normal growth of this strain. However, no fucoxanthin, diatoxanthin or diadinoxanthin (major markers from diatoms such as Nitzschia curvilineata) were detected in the analysis of the culture of Cr. As major markers from diatoms could not be detected, we assume that no minor pigments from Nitzschia curvilineata could be detected in the sample Cr. Then, a taxonomic curiosity with Cr analysis occurred. This phylum (Cercozoa) belongs to the green lineage (photoprotection cycle Zea/Anthe/Viola). Pigments common with Chlorophyta strains were detected but none from the photoprotection cycle (Diato/Diadino) were detected in the strains Cr and Bn. However, this phylum is still classified in the Chromista kingdom. By taking into account only pigment criteria, we think that Chlorarachniophyceae should belong to the Plantae kingdom. Closterium baillyanum is the only representative of the Charophyta phylum in this study. Chlide a, P21, 9-cis-Neo, Viola/Zea, Anthe, Lut, βε-Car and Chl b were detected in this strain. These pigments confirm that this phylum is in the correct position in the chemotaxonomic tree. Astaxanthin is not present in most chlorophytes (except Saa and Cr). Asta is considered as a marker but it cannot be used in all conditions. Our culture conditions were not favourable to biosynthesis of this marker. This photoprotective pigment is only highly expressed in stress conditions, as is the case for astaxanthin esters in Hp.
If we refer to the Zea/Anthe/Viola xanthophyll cycle in green algae, the low light culture conditions allow the presence of these three pigments. Nevertheless, only Dsp, Bn and Cb contain Anthe with a retention time of 21.5 min. Other algae from the green lineage contain pigments having a 21.5 min retention time but their spectral properties did not properly match with Anthe identification. This is the case for Car23 in Cr; Car24 in Ts; Car25 in Ot, Ca and Ds; Car26 in Cv; and Car27 in Saa and Hp. The low abundance of these peaks may explain the pigment diversity described, which could belong to Anthe derivatives. Anthe was not detected in Ep which contained six characteristic carotenoids Car22, Car28, Car30, Car32, Car40 and Car49 with the presence of Diato-Diadino which highlights the specificity of the Euglena lineage. The latter are also represented in the following figures. According to their retention time and spectra, Car30 and Car32 could be identified as vaucheriaxanthin esters.
Brown lineage and Dinophyceae clade
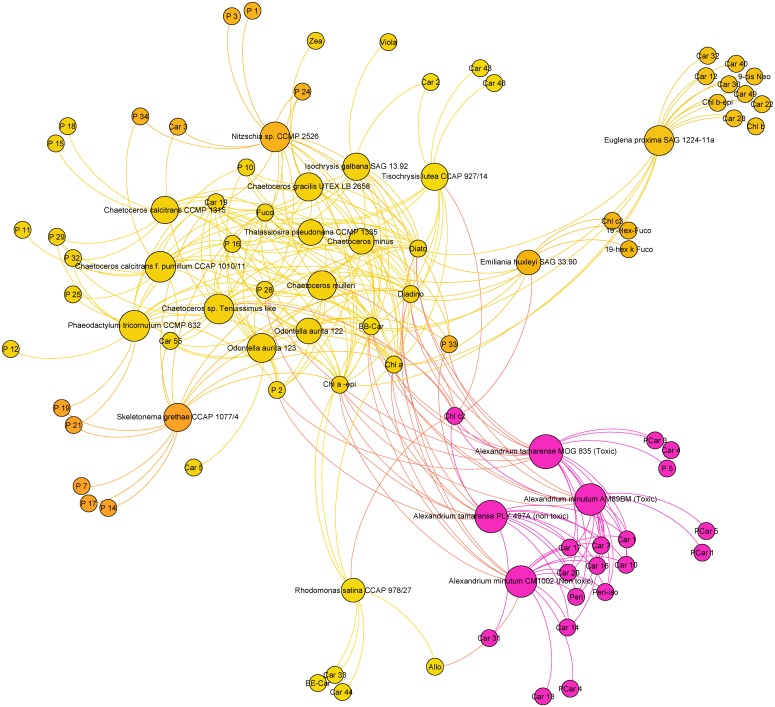
Figs 4 and 5 present the pigment communities and content among the strains containing a Diato/Diadino cycle.
Fig 4. Pigment communities among strains containing a Diato/Diadino cycle (Haptophyta, Ochrophyta, Dinophyta and Euglenozophyta).
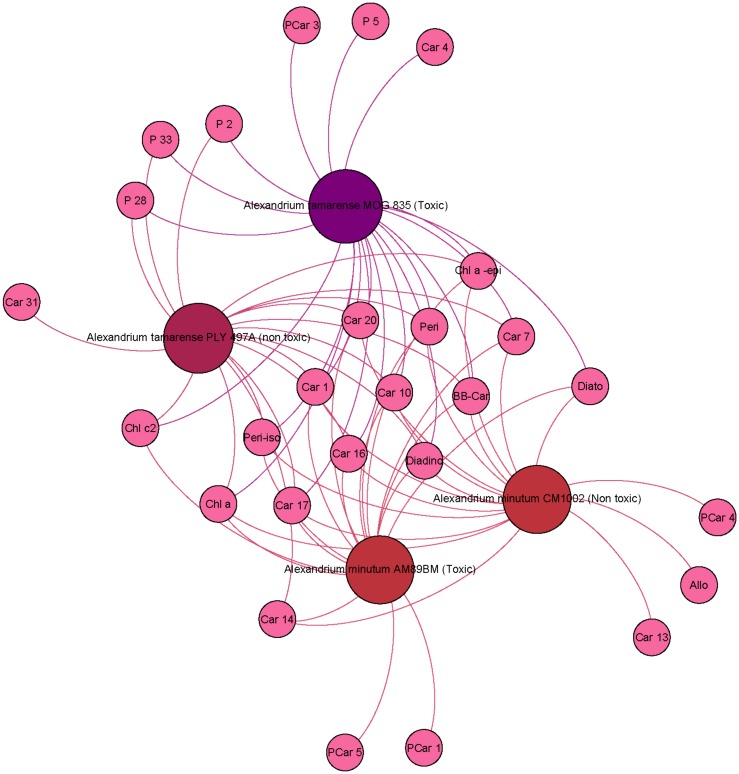
Fig 5. Pigment communities among Alexandrium strains.
All strains are grouped near Chl a and its epimer, ββ-Car, Diato and Diadino. Diatoms and Haptophyta are grouped near Fuco and Car19 (9-cis-fucoxanthin like). These microalgae have a strong diversity of chlorophyll pigment types (small circles at the periphery of the figure). Specific porphyrin markers are present in Sg (P7, P14 and P17), Nsp (P1 and P3), Ccp (P11), Cc (P15 and P18) and Pt (P12). P28 is also near the centre of the figure suggesting its closeness with Chl a biosynthesis. High chlorophyllide and pheophorbide contents, resulting from the hydrolysis of chlorophylls and pheophytins by active chlorophyllases, are usually described in bacillariophytes [34,35]. Accordingly, some porphyrin derivatives were exclusively found in certain Bacillariophyceae strains among which they were shared. P2 was detected in Cmu, Ccp, Ctl, Oa1, Pt and Cg (6 Bacillariophyceae) in At1 and At2 (dinoflagellates) and also in Ig (Coccolithophyceae). P24 and P25 have very close properties but it was not possible to merge these pigments using the matching factor. They are present in Cg, Cmu and Nsp for P24, and P25 was characteristic of two Chaetoceros strains (Ccp and Ctl).
A Chl c2 like pigment (P10), exhibiting a retention time very close to that of Chl c2 and also a close absorption spectrum (score >995) was also detected in several Chromista strains (Fig 4). This spectral variation may be linked with the co-elution with Chl c1 (P16) at different levels of expression. Other porphyrin derivatives were shared between strains from various classes. P29 and P32 were present in the chlorophyte Ot. P33 was also present in Ca, Hp and Saa.
Chl c2 is a key marker in the Chromista, as well as in dinoflagellates and Cryptophyta. Presence of Allo in Rs is a known taxonomic marker, but it is was not described in Dino-1 plastid type. Fig 4 reveals that Allo is present in Am1, a result not previously observed in literature because it is a marker of Dino plastid type-5 [30]: most photosynthetic dinoflagellates contain a chloroplast with peridinin as the major carotenoid. Chloroplasts from other algal lineages have been reported, suggesting multiple plastid losses and replacements through endosymbiotic events. The pigment composition of 64 dinoflagellate species (122 strains) was analysed by high-performance liquid chromatography. In addition to chlorophyll (chl) a, both Chl c2 and divinyl protochlorophyllide occurred in chl c-containing species. Chl c1 co-occurred with Chl c2 in some peridinin-containing (e.g., Gambierdiscus spp.) and fucoxanthin-containing dinoflagellates (e.g., Kryptoperidinium foliaceum). Chl c3 occurred in dinoflagellates whose plastids contained 19’-acyloxyfucoxanthins (e.g., Karenia mikimotoi). Chl b was present in green dinoflagellates (Lepidodinium chlorophorum). Based on unique combinations of chlorophylls and carotenoids, six pigment-based chloroplast types were defined: Type 1: peridinin/dinoxanthin/chl c2 (Alexandrium minutum); Type 2: fucoxanthin/19’-acyloxy fucoxanthins/4-keto-19’-acyloxy-fucoxanthins/gyroxanthin diesters/chl c2, c3, monogalactosyl-diacylglycerol-chl c2 (Karenia mikimotoi); Type 3: fucoxanthin/19’-acyloxyfucoxanthins/gyroxanthin diesters/chl c2 and c3 (Karlodinium veneficum); Type 4: fucoxanthin/Chl c1 and c2 (K. foliaceum); Type 5: alloxanthin/Chl c2/phycobiliproteins (Dinophysis tripos); Type 6: neoxanthin/violaxanthin/a major unknown carotenoid/Chl b (Lepidodinium chlorophorum). While plastids with peridinin, and probably those with Chl b, originated from secondary endosymbiosis, the other chloroplast types were obtained through tertiary endosymbiosis. Chloroplast types corresponded with evolutionary lineages within dinoflagellates. Caution must be observed when only peridinin is used for tracking photosynthetic dinoflagellates in field samples. The additional marker pigments offer oceanographers greater power for detecting dinophytes in mixed populations [32].
All Cercozoa, Haptophyta and Ochrophyta strains contained Fuco. Fuco ester derivatives were likely to be present in these strains [36], as confirmed by the detection of 19’-but-fuco and 19’-hex-fuco in Eh. It is most likely that other Fuco derivatives, including the 4-keto forms of both Fuco esters, corresponded to unidentified carotenoids detected in the chromatograms. Tl contained two characteristic carotenoids, Car43 and Car48. Car2 was common to Ig and Tl, the two Isochrysidaceae.
Dinophyceae
For Fig 5, we selected two toxic strains (At1 and Am2) and two non-toxic strains (At2 and Am1). Detection of Peri and Peri-iso in the four strains confirmed that they belonged to the Dino-1 group defined by [28], but these pigments could not explain the toxicity. The conclusion is the same for Car1, Car7, Car10, Car16, Car17 and Car20. These pigments were present in all Alexandrium strains (At1, At2, Am1 and Am2) and in no others. These pigments are good markers of the genus Alexandrium. Phycotoxin analyses (P. Lassus, personal communication) showed the toxins of At1 are GTX4, C2, NeoSTX and GTX1 (by order of decreasing abundance), while the toxins of Am2 are GTX3, C2 and GTX2. Pigments can be toxic sometimes [37] but in the case of At1 and Am2, toxicity seems to be held by molecules other than carotenoids. Car4 was present only in toxic A. tamarense (At1), while PCar1 (similar retention time and maximum wavelength) was detected only in toxic A. minutum (Am2). Three strains contained characteristic porphyrin-carotenoid pigments, namely PCar4 in Am1, PCar1 and PCar3 in At1, PCar4 in Am1, PCar5 in Am2. It is important to note that PCar1, PCar3, PCar4 and PCar5 are uncommon molecules with spectral properties from both porphyrins and carotenoids.
Table 3 summarizes the pigment combinations that, according to the results of our study, can be used as stringent chemotaxonomic markers to discern one strain or one species over the 37 algae studied. Pigments in bold indicate the strain-characteristic pigments whose detection gives high robustness to the identification of the strain in the sample.
Table 3. Key pigments to identify the 37 phytoplankton strains.
| Phylum | Class | Family | Strain code | Short name | Characteristic pigment combination | Taxonomic group |
|---|---|---|---|---|---|---|
| Eukaroyte Plantae | ||||||
| Rhodophyta | Porphyridiophyceae | Porphyridiaceae | Porphyridium purpureum SAG 1380-1d | Pp | No stringent combination | RHODO-1 |
| Rhodellophyceae | Rhodellaceae | Rhodella violacea SAG 115.79 | Rv | Car45 | RHODO-1 | |
| Cyanidiophyceae | Galdieriaceae | Galdieria sulphuraria | Gs | P4—Pheide a—P22—Car50—Car57 | RHODO-1 | |
| Glaucophyta | Glaucophyceae | Glaucocystaceae | Cyanophora paradoxa SAG 29.80 | Cp | No stringent combination | GLAUCO-1 |
| Charophyta | Conjugatophyceae | Closteriaceae | Closterium baillyanum SAG 50.89 | Cb | No stringent combination | CHLORO-1 |
| Chlorophyta | Chlorophyceae | Scenedesmaceae | Scenedesmus acutus f. alternans UTEX 72 | Saa | No stringent combination | CHLORO-1 |
| Scenedesmus obliquus UTEX 1450 | So | P13 | CHLORO-1 | |||
| Haematococcaceae | Haematococcus pluvialis CCAP 34/7 | Hp | No stringent combination | CHLORO-1 | ||
| Dunaliellaceae | Dunaliella sp. CCAP 19/19 | Dsp | Car52 | CHLORO-1 | ||
| Dunaliella salina SAG 19–3 | Ds | No stringent combination | CHLORO-1 | |||
| Trebouxiophyceae | Chlorellaceae | Chlorella autotrophica CCMP 243 | Ca | PCar2 | TREBUXIO-1 | |
| Chorella vulgaris SAG 2.80 | Cv | Car26—Car53—P31 | TREBUXIO-1 | |||
| Mamiellophyceae | Bathycoccaceae | Ostreococcus tauri H95 | Ot | P9—Car6—Pras—Car18—Car34—Car51 | PRASINO-3A | |
| Chlorodendrophyceae | Chlorodendraceae | Tetraselmis suecica CCMP 904 | Ts | Car24—P23—P30—PCar6 | PRASINO-2A | |
| Eukaryote Chromista | ||||||
| Cercozoa | Chlorarachniophyceae | Chlorarachniaceae | Chlorarachnion reptans SAG 26.97 | Cr | P6—P8—P20—Car9—Car11—Car23—Car29—P35 | CHLORARAC-1 |
| Bigelowiella natans CCMP 621 | Bn | P27—Car46 | CHLORARAC-1 | |||
| Haptophyta | Coccolithophyceae (Prymnesiophyceae) | Isochrysidaceae | Isochrysis galbana SAG 13.92 | Ig | No stringent combination | HAPTO-3 |
| Tisochrysis lutea CCAP 927/14 | Tl | Car43—Car48 | HAPTO-3 | |||
| Noelaerhabdaceae | Emiliania huxleyi SAG 33.90 | Eh | Chl c3—19-hex-k-Fuco—19’-hex-Fuco | HAPTO-6 | ||
| Bacillariophyta | Mediophyceae | Thalassiosiraceae | Thalassiosira pseudonana CCMP 1335 | Tp | No stringent combination | DIATOM-1 |
| Skeletonemataceae | Skeletonema grethae CCAP 1077/4 | Sg | P7—P14—P17 | DIATOM-1 | ||
| Eupodiscacea | Odontella aurita 123 | Oa1 | Car5 | DIATOM-1 | ||
| Odontella aurita 122 | Oa2 | No stringent combination | DIATOM-1 | |||
| Chaetocerotaceae | Chaetoceros mulleri | Cmu | No stringent combination | DIATOM-1 | ||
| Chaetoceros minus | Cmi | No stringent combination | DIATOM-1 | |||
| Chaetoceros sp. Tenuissimus like | Ctl | No stringent combination | DIATOM-1 | |||
| Chaetoceros calcitrans f. pumillum CCAP 1010/11 | Ccp | P11 | DIATOM-1 | |||
| Chaetoceros calcitrans CCMP 1315 | Cc | P18 | DIATOM-1 | |||
| Chaetoceros gracilis UTEX LB 2658 | Cg | No stringent combination | DIATOM-1 | |||
| Bacillariophyceae | Bacillariaceae | Nitzschia sp. CCMP 2526 | Nsp | P1—P3 | DIATOM-3 | |
| Bacillariophyceae incertae sedis | Phaeodactylaceae | Phaeodactylum tricornutum CCMP 632 | Pt | P12 | DIATOM-1 | |
| Cryptophyta | Cryptophyceae | Pyrenomonadaceae | Rhodomonas salina CCAP 978/27 | Rs | Car33—Car44 | CRYPTO-1 |
| Eukaryote protozoa | ||||||
| Miozoa | Dinophyceae | Goniodomataceae | Alexandrium tamarense MOG 835 (Toxic) | At1 | P5—PCar1—PCar3 | DINO-1 |
| Alexandrium tamarense PLY 497A (non toxic) | At2 | No stringent combination | DINO-1 | |||
| Alexandrium minutum CM1002 (Non toxic) | Am1 | Car13—PCar4 | DINO-1 | |||
| Alexandrium minutum AM89BM (Toxic) | Am2 | PCar5 | DINO-1 | |||
| Euglenophyta | Euglenophyceae | Euglenaceae | Euglena proxima SAG 1224-11a | Ep | Car22—Car28—Car30—Car32—Car40—Car49 | EUGLENO-1 |
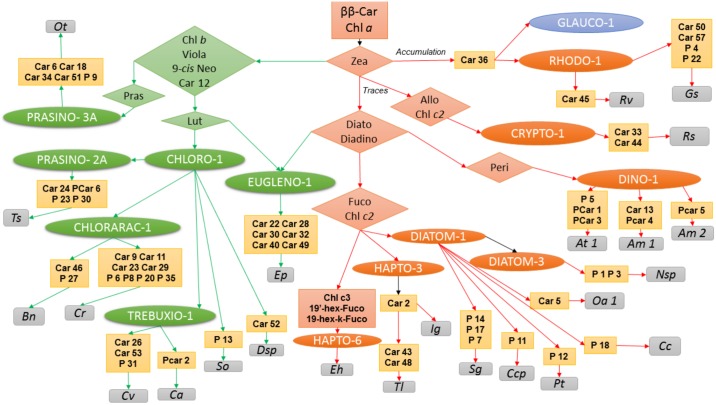
The resolution of the ancillary pigment in order to identify strain could be solved as a dichotomy key as represented in Fig 6. Colours indicate the different taxonomic lineages and outsider phyla in terms of pigmentation.
All strains contain Chl a and ββ-Car. Zea is also present in all strains but its quantity depends on the different lineages. Accumulation of Zea, in absence of other photoprotective pigments, is observed in Rhodophyta or Glaucophyta. Traces of Zea could be observed in Chromista. Its presence in the xanthophyll cycle is linked with the green lineage. The amount of Zea is a major decisional element for stringent identification of plastid types. According to Table 3 and Fig 6, 24 algae strains could be identified according to ancillary pigment.
Fig 6. Dichotomous key for microalgae strain identification by pigment composition.
This dichotomous key reveals that stringent identification of strains is possible according to pigment composition results presented in this paper.
In the red lineage, Car36 is a key pigment allowing the identification of Rhodophyta and Glaucophyta. For a stringent identification Rv could be identified via Car45 and Gs via Car50, Car58, P4 and P22. The Rs (Cryptophyta) pigment composition reveals specific pigments (Car33, Car44) and the brown lineage/Dinophyceae clustering (Chl c2). Allo is a carotenoid previously described to be a marker of Cryptophyta. Moreover, we have to notice Cryptophyceae class produces phycobiliproteins as well as Porphyridiophyceae, Rhodellophyceae, Cyanidiophyceae, Glaucophyceae and cyanobacteria. This phylum is derived from the red lineage, but actually classified in the Chromista kingdom. All other Chromista lost their ability to synthesize Phycobiliproteins in favour of the light harvesting complex with a Diato/Diadino xanthophyll cycle. Bacilliariophyta and Haptophyta have Fuco and Chl c2. Among three analysed haptophytes, Eh was the only strain showing Chl c3. Ig and Tl have Car2 in common. Tl could be discriminated with specific Car43 and Car48. In Bacilliariophyta (diatoms), porphyrin biodiversity generates a strong difference between strains. Indeed, Sg, Nsp, Ccp, Pt and Cc had unique porphyrins. Moreover, Car19 is a common pigment in the Mediophyceae class and strain Pt belonging to Bacillariophyceae incertae sedis. This carotenoid could be used a group marker as well as Car2, which is a common pigment for both Isochrysidaceae analysed (Ig and Tl). The dinoflagellates analysed have specific pigments mixing characteristics of porphyrins and carotenoids (PCar1,3,4, and 5), allowing for easy identification. Peri and Peri-iso are well known to be present in dinoflagellates. In the four Alexandrium strains analysed, Car1, Car7, Car16, Car17, and Car20 are common pigments for this Genus.
Pigment composition of the Euglenophyceae representative (Ep) showed pigments in common with the green lineage (Car12, 9-cis Neo, Chl b and Chl b-epi) whilst some other pigments are characteristic of the brown lineage (Diadino and Diato). This typical signature should be compared to the endosymbiosis processes during its evolution. Car22, Car28, Car30, Car32, Car40 and Car49 are all markers of the Euglenophyceae strain (Ep).
The green lineage is characterized by Chl b, 9-cis Neo Car12 and Lut. Among the six strains belonging to CHLORO-1 two could be identified easily: So with P13 and Dsp with Car52. In TREBUXIO-1 three specific pigments characterize Cv and one Ca. Chlorarachniophyceae presented two and seven specific markers for Bn and Cr, respectively. Not all Prasinophyceae chloroplast types contains Pras. This is the case for Ts which, moreover, has four specific pigments (Car24, PCar6, P23, P30). Ot belongs to the PRASINO-3A plastid type, which contains Pras and five specific pigments (Car6, Car18, Car34, Car51 and P9).
This overview of pigment-based taxonomic clusters allows a fast determination of strain-specific and shared pigments. These findings support the hypothesis that some minor photoprotective and photosynthetic pigments can constitute tools to cluster strains.
Discussion
The pigment composition of 37 phytoplankton strains from banks or environmental collections was examined using an optimized extraction process coupled to an automated HPLC dereplication method. This study revealed the presence of unidentified chlorophyll derivatives and carotenoids in most species studied, in addition to classical pigments already described as taxonomic markers of particular phyla. Interspecific comparisons revealed that some of these unidentified pigments are characteristic of phyla, classes or orders, and may have high interest as stringent taxonomic markers. The Carotenoids Handbook [38] reveals the occurrence of more than 800 carotenoids from natural sources. The same kind of comprehensive and updated analysis bas not really been performed with chlorophyll derivatives but it seems chlorophylls comprise a group of at least 50 tetrapyrrolic pigments [39]. With respect to the acceptance criteria, it was difficult to systematically match our unknown pigments to the literature, but our results can still be considered as a contribution to microalgae chemotaxonomy. With the high biodiversity of phytoplankton species, their diverse molecular adaptations to the photic conditions throughout the evolution of their lineages, and considering that only a few species have been studied, it is obvious that many pigments remain to be identified. This work gives an overview of the biodiversity of carotenoids and chlorophylls in an array of microalgae strains, shows how they can be used as chemotaxonomic markers, which may lead to some of them finding commercial applications.
Pigment analysis aspects
Some gold HPLC references for phytoplankton pigment analysis can be found in [2,9,40] and [13]. The described methods based on visible spectral analysis and applications are mainly thought to use pigment identifications and concentrations as chemotaxonomic markers, but none of them really exploit the UV part of their spectrum. These tools are powerful for well characterized pigment identifications. Nevertheless, visible spectral analysis is not sufficient when structural identification of pigments is needed. By coupling spectral properties to mass spectrometry analysis it becomes possible to confirm isomers or ester bonds. Studies with mass spectrometry are very powerful for single pigment identification and characterization. However, results depend on ionization rate. This identification technique generally fails when complex pigment mixes are addressed. It is still difficult to get a better overview than spectral analysis. NMR analyses are reliable for pigment characterization especially with the development in recent years of high magnetic field instruments and cryoprobes. Some authors developed a methodology based on HR MAS 1H NMR spectroscopy coupled with multivariate analysis aiming to classify microalgae [41]. However, this method is not currently widespread because of the high cost of such equipment. Thrane and co-workers [42] described a Gauss-peak spectra method to simplify algal pigment mixture spectra but this does not solve the community affiliation. It is therefore still useful to develop new tools for a better, faster and more reliable understanding of algal communities in open waters.
Reliable identification and quantification are necessary to unravel phytoplankton contribution to biological and biogeochemical processes affected by climate change. During the process that leads to algal community determination (i.e., taxonomic distribution), care should be taken of the production of potential artefact pigments during sample storage/extraction/analysis. Some methods can produce unstable, “non-existent natural products”. Methanol, for example, if it comes into contact with a hot point such as the tip of a sonication probe, is likely to produce methyl esters in presence of natural products. However, xanthophylls can also be linked naturally to the membranes as ester forms [38]. Another historical extraction is based on 90% acetone solvent, which is good to inhibit degradation enzymes, but its intrinsic characteristics favour apolar pigment extraction. Although some solvents provide a broad extraction spectrum (e.g., N,N’-dimethylformamide), it is probably better to choose another extraction strategy to limit the subsequent detection of artefacts [14]. With the growing interest in improving the extraction processes using GRAS solvents (Generally Recognized As Safe) [43] and taking into account the relatively good pigment extraction yield without significant studies reporting formation of degradation products, ethanol 100% based extraction seems to be a good start to deal with natural compounds in phytoplankton chemotaxonomy [13].
Pigment retention time constitutes another important parameter. Times are generally quite reliable within the same batch analysis (±0.05 min). However, variations can occur at the beginning of the elution when the analytic column has become too old and/or the TBAA salts become too oxidized. In this case, it is important to keep a watchful eye on wavelength maxima and the injection of a standard mix between a couple of samples and/or add a non-natural internal standard to each natural sample. Generally, we can consider that retention time requires a loose tolerance in the first 10 min of the elution with the Van Heukelem and co-authors method [13] and identification should rely more on spectra similarities. Finally, a data exploration can be based on different strategies that are more or less sophisticated and computationally managed [44,45]. For example, results can be managed in clusters [46]. Non-taxonomic interpretations of pigment datasets can be realized by pigment indices and by ecological similarity indices [47]. To compare UV-visible spectra, we worked with the algorithm provided by ChemStation (Agilent Technologies) to provide identification scores based on the shape of the spectrum and on relative intensity of the lambda max. However, X-hitting algorithm developed by [48] might be also adapted to discriminate pigments. In our study, we found that many chlorophylls and carotenoids are very similar. The tough task was to define the right criteria in term of spectrum variation making it possible to merge or to discriminate two unknown pigments. The goal was to be flexible enough to not discriminate two short variations related to the detection but selective enough to discriminate two slight structural variations. To help make the decision, the UV range of each spectrum was included in the intercomparison using the ChemStation algorithm. In our experiment, a score ≥999/1000 was considered high enough to merge two pigments. Between 995 and 999, and if the visible results were perfectly aligned, merging of the two pigments was accepted. If the alignment was not very accurate, merging of two pigments was rejected. A score <995 discriminated two pigments straight away. However, sometimes when spectrum intensity was very low (trace compounds), the UV range was not very helpful in the decision-making process due to noise. Only the analyst’s experience could then give an interpretation of the data. This basis allowed us to build consistent Gephi communities, which display the direct viewable links between pigments and related strains.
Then, Canonical Variates Analysis are efficient to discriminate microalgae subgroups [49] (i.e., the same goal as our study), but do not make it possible to pinpoint chemotaxonomic markers in communities analysis. This strategy is efficient only if molecule selectivity is high (i.e., sufficiently separated by the chromatographic system). With the latest methods, selectivity of the main pigments is quite good. However, chemodiversity is often hidden in the baseline [50], and some of these molecules are closely structurally related, which means they are not always well separated by a general analytical method. These minor analogues can slightly pollute a Gaussian (i.e., individual pigment spectra and background components as weighted sums of Gaussian functions), and have to be taken into account in data interpretation. This is sometimes still slightly trickier for minor peaks and traces. Several times, when dealing with a signal with both porphyrin and carotenoid spectral properties, we had to name some minor pigments PCar. These pigments seemed to be pure enough to use the apex as a reference (Retention time and λmax) in Table 2. These kinds of spectra are not very common but provide a consistent signal [51]. Carotenoids are involved in chlorophyll photoprotection. To carry out this function and the subsequent regulation of energy flow within the photosynthetic apparatus, carotenoids are bound in discrete pigment-protein complexes in the proximity of chlorophylls [52]. Light-harvesting complexes (LHC) display a wide range of architectures in terms of pigment composition, stoichiometry and location in the apparatus. In these complexes, there is a bridging molecule between the chlorophyll and the carotenoid moieties, which is the fifth ligand for Mg in the chlorophyll macrocycle. In dinoflagellates, the LHC is named PCP for Peridinin-chlorophyll a-Protein. This complex is water-soluble and the Chl/carotenoid ratio is in favour of carotenoids [53]. Because our method of extraction covers a wide range of polarity and a high extraction rate, it seems likely that we were able to extract this kind of complex in small amounts, even if it is probably not strictly peridinin that is detected in our PCar. Optimized extraction technique and extraction device are referred to [14]. Finally, we can conclude this part of discussion by saying that, in the mid and long term, technological breakthroughs will probably eventually make it possible to identify each trace compound in a complex pigment mix. Currently, the right question to ask is probably whether a peak/spectrum (even incompletely characterized or slightly polluted in the peak tail) useful to distinguish communities or not?
Chemotaxonomy and markers
Chemical taxonomy is based on investigations of the distribution of compounds or groups of biosynthetically related metabolites in series of related clades, taxa or species. A major challenge is to identify stringent chemotaxonomic markers that unambiguously signal the presence of taxonomic entities in natural samples. Despite the complexity of metabolism, even for photosynthetic unicellular organisms, significant advances have been made in recent decades, notably in genomics. Metabolism is the product of anabolism and catabolism reactions driven by genes and environmental factors. Some biosynthesis genes have been lost or acquired during lineage radiation and this helps us to distinguish clades and phenotypes. However, multiple factors can influence pigment expression. In terms of biogenetic classification, primary metabolites can be distinguished from secondary metabolites. The former are directly involved in physiology (growth, development, and reproduction), and the latter, as far as their functions are understood, are involved in allelopathy mechanisms, aiming to protect algae against environmental stresses, defend them from predators, and/or communicate intra- and interspecifically. The attribution of microalgal pigments to these classes remains unclear and is not very well documented but, according to the definitions, it is obvious that they can play both roles [54]. The analysis of our results raised the question of which kind of metabolites best indicate a plastid type clade, a phylum, a class, or finer taxonomic levels, which is quite difficult to answer based on present knowledge. However, our study has contributed some elements to this discussion.
First, we assume that ephemeral biosynthetic intermediates have little chemotaxonomic value since they cannot be reliable over time. This is probably the case for some pigments such as protochlorophyllide, chlorophyllide derivatives, β-cryptoxanthin and β-cryptoxanthin epoxide.
Our analysis also showed that major photosynthetic pigments (Chlorophyll a, b, and c) are often not suitable for accurately defining taxonomic groups (i.e., plastid type clades) as shown on Fig 6. Indeed, chlorophyll a is widely distributed because of its major role in both photosystems (II and I). Derivative forms of Chl a arise either from its anabolism or its catabolism. The various forms of enzymes responsible can be detected and are quite common in all taxa. However, some classes like Bacillariophyceae are known to synthesize large quantities of chlorophyllases, which are more or less specific [35]. This fact explains why we found a large chlorophyll chemodiversity in our bacillariophyte analyses. Moreover, epimerases are highly substrate-selective as well as promiscuous. Some traces of minor pigments likely result from cellular enzymes and oxidations, and the difficulty is to discern which of them are sufficiently specific to become reliable markers. Moreover, microalgae that are adapted for growth in extreme biotopes or with a particular metabolism might express specific enzymes modifying common signalling pathways and/or modifying common pigments. In this case, the pigments produced could be good candidates to serve as chemotaxonomic markers and possibly also as molecules of interest for biotechnologies. For example, this could explain why we found a stringent pigment combination for Galdieria sulphuraria (Gs) strain (Car50, Car57, P4, P22).
On the other hand, some degradation pigments (e.g., red pheophorbides, chlorophyll catabolites, primary fluorescent chlorophyll catabolites and non-fluorescent chlorophyll catabolites) cannot be used as markers. It is also the case with pheophytin a for which the sample storage and extraction process would likely increase the amount of this pigment. In our case—with collection at the end of exponential growth, no storage before analysis and an optimized soft extraction—conditions were not met to produce significant amounts of degradation products. Since pheophytin a is probably the most widely distributed degradation product in natural communities, it cannot be considered as a specific marker for microalgae chemotaxonomy but does however have an interest for monitoring phytoplankton populations senescence. In this case, when studying degradation pigments it is more appropriate to work at 405 nm rather than 436 nm.
Fluctuations in pigment quantities are usually a rapid response to environmental factors because most of pigments are involved in basic processes such as photosynthesis and photoprotection. Cells need to be reactive to survive in a very competitive and rough environment. The fast pigment conversion levels results without any changes in gene expression, although long-term acclimation causes changes in the transcript level of the genes, which encode proteins in biosynthetic pathways [55]. In Table 4, we propose a list of hypothetical markers resulting from pigment metabolism that are presumably genetically driven. We hypothesize that strong markers can be found in these metabolites [18,56]. Some of these molecules may not be detected by visible spectroscopy. Hence, acquisition of UV spectra has potential advantages, even though it has not been exploited in phytoplankton chemotaxonomy. This methodology has been applied to plant chemotaxonomy [57] but most of the time, UV detection is coupled with other detectors like mass spectrometry or ELSD [58] to cluster different taxa. The field needs innovative breakthroughs to identify next generation of markers. Hyphenated analytical tools like HPLC-UV DAD-MSn, which have a dereplication strategy, could become a future means of expanding microalgal chemotaxonomy beyond strict pigment detection and taking into account more secondary metabolites.
Table 4. List of genetically-driven molecules involved in pigment biosynthesis pathways.
| CHLOROPHYLL AND PORPHYRIN PATHWAYS |
| Biosynthetic intermediates |
| Porphobilinogen |
| Uroporphyrinogen |
| Protoporphyrinogen IX |
| Mg-Protoporphyrin IX 13-monomethyl ester |
| Billiverdin |
| Bilirubin |
| Mesobilirubinogen |
| Protochlorophyllide |
| Divinyl-protochlorophyllide |
| Divinyl-chlorophyllide a |
| Ending products |
| Uroporphyrin I |
| Coproporphyrin I |
| D-Urobilin |
| I-Urobilin |
| L-Stercobilin |
| Bacteriochlorophyll a |
| Bacteriochlorophyll b |
| Bacteriochlorophyll c |
| Bacteriochlorophyll d |
| Zn-Bacteriochlorophyll a |
| CAROTENOID PATHWAYS |
| Biosynthetic intermediates |
| 4,4'-Diapo-neurosporene |
| Hydroxy-spirilloxanthin |
| 3,4-Dihydro-anhydrorhodovibrin |
| 7,9,9'-tricis-Neurosporene |
| Phytofluene |
| Neurosporene |
| Zeinoxanthin |
| α- and β-Cryptoxanthin |
| ζ- and γ-Carotene |
| β-Zeacarotene |
| Spheroidene |
| [3R,2'S]-Myxol 2'-α-L-fucoside |
| 5-Deoxy-strigol |
| 3-Hydroxy-echineone |
| Caloxanthin |
| Xanthoxin |
| 3'-Hydroxy-abscisate |
| Ending products |
| Staphyloxanthin |
| 4,4'-Diapo-lycopenedial |
| Rhodopinal glucoside |
| 2,2'-Diketo-spirilloxanthin |
| Tetrahydro-spirilloxanthin |
| [2S,2'S]-Oscillol 2,2'-di[α-L-fucoside] |
| R.g.-Keto III |
| Okenone |
| Lutein |
| 3,4-Dihydro-spheroidene |
| 7,8-Dihydro-β-Carotene |
| Hydroxy-chlorobactene glucoside ester |
| [3R,2'S]-Myxol 2'-[2,4-di-O-methyl-α-L-fucoside |
| [3S,2'S]-4-Ketomyxol 2'-α-L-fucoside |
| Strigol |
| Astaxanthin diester |
| Isoreneriatene |
| Thermo-biszeaxanthin |
| Zeaxanthin diglucoside |
| Nostoxanthin |
| Capsanthin |
| Capsorubin |
| Dihydroxy-phaseic acid |
This non-exhaustive list was built by mining KEGG pathway maps, map 00906 for carotenoids and map 00860 for porphyrins and chlorophylls [accessible at www.genome.jp/kegg updated version on 18th April 2016].
Biosynthetic intermediates could be detected in microalgal analyses but, according to the discussion above, probably constitute weak markers for chemotaxonomy. Ending products could be also detected. These molecules are more likely able to be accumulated and remain stable in cells. They could constitute strong markers for photosynthetic microorganism chemotaxonomy. This hypothesis is strengthened by the fact that lutein is accumulated in some of our samples belonging to CHLORO-1 and EUGLENO-1 types. Similarly, our experience with biosynthetic intermediates showed that violaxanthin was not always detected in our Cb analyses. Moreover, β-cryptoxanthin was not present in our Cp analyses but detected in the same strain by Baudelet et al. [23].
Molecules listed for carotenoid biosynthesis belongs to terpenoid backbone biosynthesis, unusual spirilloxanthin pathway, normal spirilloxanthin pathway, okenone pathway, lutein biosynthesis, spheroiden pathway, myxol biosynthesis, and astaxanthin biosynthesis. Some pathways belong to cyanobacteria, alpha- and gamma- proteobacteria displaying photosynthetic pigments. Detection of markers belonging to these pathways is valuable since biologists are faced with a wide variety of microorganisms in natural samples, not only to eukaryotic microalgae. Because genes from metabolism were acquired or lost during evolution, microalgae clades do not express the same gene patterns and nor the same suites of pigments. Genomic data mining for microalga species is currently quite limited so it is not that simple to conclude whether a gene has been lost or if it is insufficiently expressed to be detected. For strains with which we have the most experience, intermittent presence of some pigments means that the related genes are conserved but where a pigment is absent, and background information is sparse, conclusions should be drawn with caution.
Among all the factors influencing pigment-related gene expression metabolism, UV has been particularly studied [59–62]. Irradiance and spectral distribution of light are also of course directly involved in photosynthesis [63–71]. The related aspects of photoperiod [72] and circadian rhythm [73] have an impact on microalgal pigment composition. Low temperature culture or an anoxic environment can lead to partial conversion of methyl-pyropheophorbide in meso-a pyropheophorbide [74]. Nutrients also affect the relative concentrations of pigments [75–78]. Mixing regimes of water masses, at a culture or meso scale level, are also influential factors [79,80]. According to species, pigmentation can change depending on the growth phase [81]. For example, xanthophyll to chlorophyll ratio increases throughout the microalgal growth phase and declines once in stationary phase in the genera Amphidinium and Euglena [82]. Thus, it is widely accepted that the environmental conditions of microorganisms can affect their metabolism. A methodology has thus been recommended that aims to vary systematically different culture parameters (composition of media, aeration, temperature, pH, addition of enzyme inhibitors etc.) to increase the number of available secondary metabolites from a single strain [83]. This methodology, called OSMAC (One Strain—Many Compounds), optimizes the investigation of chemodiversity produced by a single strain of a microorganism. Changes in culture conditions and inducing stresses would modulate different biosynthetic pathways, thereby increasing the metabolic diversity of a strain and the possibility of obtaining original compounds for biotechnology applications and chemotaxonomy purposes [84]. Fluctuations in pigment diversity (including minor ones) likely arise from both types of metabolism, but minor compounds probably belong more to secondary metabolites or participate in pigment turnover. To improve our general understanding of this phenomenon, and although the experimental logistics would be heavy, holistic studies could be performed mimicking ecosystem complexity in terms of biological communities (biocenosis) and biotopes. The present study has not explored these aspects so far but provides a way in which such studies could be pursued.
Micro-organism co-cultivation makes it possible to study different kinds of ecological behaviour. Symbiosis and predation can both affect phenotypes [85,86]. Among the analysed strains, Chlorarachnion reptans (Cr) illustrates that some species can predate other photosynthetic species. It is well known than some dinoflagellates can practice kleptoplastidy (i.e., retain plastids from different strains) [87]. Regarding the Cr strain, we were not able to determine whether this was really the case as we did not identify the major markers of its prey, the diatom Nitzschia curvilineata. It seems that the kleptochloroplast turnover is variable, but it might be relatively fast after feeding on a prey [88], which could be the case for Cr. This kind of behaviour should be taken into account for data interpretation as should some molecules that remain silent as long as there is no co-cultivation (or co-presence) with other microorganisms (i.e., allelopathy effects). Our results are mostly based on monospecific cultures, which may constitute a bias compared to multispecies samples collected in open waters. A validation of our ending markers (yellow boxes in Fig 6) by oceanographers would be welcome as this would modulate their scope, as Tamm and co-workers did with lake samples [89].
Regarding chemotaxonomy and lineage evolution, this study raises two main issues. The first question is about whether the Chlorarachniophyceae should belong to the Chromista as is presently accepted. Our findings support the hypothesis that the Chlorarachniophyceae could belong to CHLORO-1 plastid type and so, to the green lineage, rather than the brown lineage (Chromista) in terms of pigments. Indeed, Bn and Cr both contain all the markers belonging to the green lineage (i.e., Chl b, Viola, 9-cis Neo, Car12, and Lut). However, Chlorarachniophyceae plastids are surrounded by four membranes like the Chromista, which is a sign of a secondary endosymbiosis, while green alga plastids are bound by only two membranes [90]. A probable explanation that is now quite well accepted is that the original secondary host phagosome was not photosynthetic and thus different from the original Chromista secondary host [91]. Then, Euglenozoans belonging to Protozoa display Diato and Diadino pigments which are part of the Chromista photoprotection apparatus. They also harbour the violaxanthin photoprotection system (Viola-Anthera-Zea), which belongs to the green lineage. So, both known photoprotection systems seem to be viable. This could indicate a recent tertiary endosymbiosis in terms of evolution, a hypothesis that is strengthened by the fact that Euglenids display a three-membrane topology [92] like dinoflagellates, which are well known for their tertiary endosymbiosis (one membrane has been lost during the evolution). The taxonomy of this algae group is still contentious. Further corroborating studies concerning ultrastructural characters (membrane plastids, microtubules, starch, pyrenoid, etc.), biochemistry (isoprenoid biosynthetic enzymes, photorespiratory enzymes), and molecular phylogeny (single or combined genes, intron presence, genome arrangement, etc.) still need to be performed to elucidate the evolution of these groups.
As mentioned by Wright and Jeffrey [93], one of the issues in phytoplankton chemotaxonomy is that many taxa share the same pigment patterns and often cannot be identified based on pigment combination alone. Ten years after their article, this situation is more or less the same despite the exploration of minor pigment diversity. Indeed, among the 32 algal groups defined by [93], our study examined 14 and did not distinguish a stringent pigment combination identifying strains Pp, Cp, Cb, Saa, Hp, Ds, Ig, Oa2, Cmu, Cmi, Ctl, Cg, Tp or At2. However, we found a couple of markers for the other strains that we studied. Another relevant challenge is to identify stringent markers applicable to a particular plastid type clade, which this is much more complex. Two strategies can be considered. The first one, which is the weaker of the two, is to look for known stringent strain markers in natural multi-species samples to gain a qualitative overview of the clades present. However, without a stringent pigment combination for each species likely to develop in monitored environments it is risky to extend this conclusion further. For this strategy, Fig 6 provides 55 new (or unidentified) strain-specific pigments. The second strategy is to look for pigments that are common to a single clade. Our study highlighted Car36, which is common to the Glauco-1 and Rhodo-1 plastid types, as well as Car1, Car7, Car16, Car17, and Car20 for all Alexandrium strains analysed (Dino-1 plastid type). We did not identify other pigments specific to a taxon, even within the genera where we analysed several representatives (Chaetoceros, Odontella, Dunaliella and Scenedesmus). This latter strategy is interesting, but analyses of pure strains in monitored environments are still essential to strengthen the conclusions. Quantitative measurements are obviously important for understanding the dynamics of phytoplankton communities. However, traces or minor pigments are difficult to quantify accurately, especially when these pigments remain unidentified (i.e., no available standard). Moreover, the ratios with Chl -a or -b would have little chemotaxonomic value since chlorophyll cellular quota can vary widely among strains, the environmental conditions and the extraction technique [47]. This is why we did not include any quantification is the present study. It seems there is still a scientific gap to be overcome before we can fully use minor pigments in phytoplankton community monitoring. At the present time, the best way to move ahead could be to work from relative quantifications (peaks area) with a standard operating procedure for all the steps leading to pigment analysis.
To conclude this part, all these data illustrate part of the cellular plasticity of microalgae and could be used as references for qualitative data. To give a broad idea of pigment quantities and characteristics (traces, minor and major pigments), we have provided chromatograms and spectra in the Supplemental data (S2 and S3 Figs). These should be interpreted with care. Only a part of the microalgal biodiversity is known and described. It is important not to over-interpret natural sample analyses since many unknown algal types still remain to be identified from the global ocean [1]. Absence of some markers therefore cannot be used to conclude about the absence particular taxa from natural samples. Absence of evidence is not the same as evidence of absence. The untapped chemodiversity of microalgae remains a good opportunity to find chemotaxonomic markers. However, microalgal cellular plasticity, linked notably to the activation of silent or weakly expressed microbial gene clusters, still represents a challenge to the identification of strong markers. Hence this is a truly complex field.
Perspectives for future research
Major pigments of microalgae are well known and easily identified by UV-DAD detector in standard chromatographic conditions. However, when biologists or oceanographers conduct field studies on phytoplankton samples, they are confronted with a large chemodiversity including minor or variable pigments, pigments present in inconsistent quantities, and trace pigments. Using all this data in a rational way to understand the dynamics of phytoplankton populations remains a challenge. In this study, we proposed a guide to several specific chemotaxonomic markers that may be used to explore microalgae pigment diversity/complexity, and pinpointed important factors that can modulate data interpretations. Ninety-eight pigments were new or undescribed carotenoid or porphyrin derivatives with original spectral properties, but whose planar structure, relative structure and absolute configuration remain to be established. Nevertheless, our outcomes highlight that even non-fully characterized pigments could be used in oceanography and limnology. This microalgae community analysis can also be useful as a prioritization tool for aquaculture, drug discovery and other applications involving pigment chemodiversity. In our study, it was decided not to vary culture light irradiances in order to avoid increasing complexity in our analysis. Our results should be considered as pigment fingerprints (i.e., metabolic snapshot in defined conditions) that we could call the endopigmentome, since the exometabolome was not studied in this project. Using the same methodology, future work could focus on the same strains, varying environmental factors and experimental design. A meta-analysis of our study, combined with the following related studies and then an comparison with field data could be extremely relevant for discerning which markers are the most reliable.
Supporting information
(A) Calculation of carotenoid band ratio. UV-visible spectra showing wavelength I; II and III of a carotenoid. Band ratio corresponds to the % ratio Δ III/ Δ II. (B) Calculation of porphyrin band ratio. UV-visible spectra showing the location of I the Soret (blue maximum) and II the red bands of Chlorophyll -a.
(PDF)
(PDF)
(PDF)
(PDF)
(ZIP)
Acknowledgments
The authors thank Prof. Claire El-Fakir (University of Orléans, France) and Joséphine Ras (Oceanographic laboratory of Villefranche-sur-Mer, France) for their plentiful advice about pigment analysis. We thank also Dr. Patrick Lassus and Dr. Andrea Contreras (Laboratory of phycotoxins—IFREMER Nantes) for kindly providing the Alexandrium strains. We wish to thank our colleagues Marie-Lise Picquet, Catherine Rouxel and Nathalie Schreiber from the laboratory of Physiology and Biotechnology of Algae (IFREMER Nantes) for their assistance in the cultivation of several microalgae strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a doctoral fellowship grant from Région Pays de la Loire for BS’s PhD (N° 2008-09351), and by French National Agency for Research (ANR-08EBIO-0016) for PHOTOMER project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jeffrey SW, Wright SW, Zapata M. Microalgal class and their signature pigments In: Phytoplankton pigments: Characterization, chemotaxonomy and applications in oceanography. Cambridge University Press; 2011. p. 3–77. (Environmental chemistry). [Google Scholar]
- 2.Zapata M, Rodríguez F, Garrido J. Separation of chlorophylls and carotenoids from marine phytoplankton:a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Marine Ecology Progress Series. 2000;195:29–45. [Google Scholar]
- 3.Everitt DA, Wright SW, Volkman JK, Thomas DP, Lindstrom EJ. Phytoplankton community compositions in the western equatorial Pacific determined from chlorophyll and carotenoid pigment distributions. Deep Sea Research Part A Oceanographic Research Papers. 1990. June 1;37(6):975–97. [Google Scholar]
- 4.Wright SW, van den Enden RL, Pearce I, Davidson AT, Scott FJ, Westwood KJ. Phytoplankton community structure and stocks in the Southern Ocean (30–80°E) determined by CHEMTAX analysis of HPLC pigment signatures. Deep Sea Research Part II: Topical Studies in Oceanography. 2010. Mai;57(9–10):758–78. [Google Scholar]
- 5.Szymczak-Żyła M, Kowalewska G, Louda JW. Chlorophyll-a and derivatives in recent sediments as indicators of productivity and depositional conditions. Marine Chemistry. 2011. Juillet;125(1–4):39–48. [Google Scholar]
- 6.Rontani J-F, Volkman JK. Phytol degradation products as biogeochemical tracers in aquatic environments. Organic Geochemistry. 2003. January;34(1):1–35. [Google Scholar]
- 7.Reuss N, Conley DJ, Bianchi TS. Preservation conditions and the use of sediment pigments as a tool for recent ecological reconstruction in four Northern European estuaries. Marine Chemistry. 2005. Juin;95(3–4):283–302. [Google Scholar]
- 8.Jeffrey SW, Mantoura RFC, Wright SW. Phytoplankton pigments in oceanography: guidelines to modern methods. Mayenne: UNESCO; 1996. 661 p. [Google Scholar]
- 9.Wright S, Jeffrey S, Mantoura R, Llewellyn C, Bjornland T, Repeta D, et al. Improved Hplc Method for the Analysis of Chlorophylls and Carotenoids from Marine-Phytoplankton. Mar Ecol-Prog Ser. 1991. November;77(2–3):183–96. [Google Scholar]
- 10.Van Heukelem L, Lewitus AJ, Kana TM, Craft NE. Improved separations of phytoplankton pigments using temperature-controlled high performance liquid chromatography. Oceanographic Literature Review. 1995;5(42):376. [Google Scholar]
- 11.Van Heukelem L, Lewitus AJ, Kana TM, Craft NE. High-Performance Liquid Chromatography of Phytoplankton Pigments Using a Polymeric Reversed-Phase C18 Column1. Journal of Phycology. 1992;28(6):867–872. [Google Scholar]
- 12.Claustre H, Hooker SB, Van Heukelem L, Berthon J-F, Barlow R, Ras J, et al. An intercomparison of HPLC phytoplankton pigment methods using in situ samples: application to remote sensing and database activities. Marine Chemistry. 2004. Février;85(1–2):41–61. [Google Scholar]
- 13.Van Heukelem L, Thomas C. Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. J Chromatogr A. 2001. February 23;910(1):31–49. [DOI] [PubMed] [Google Scholar]
- 14.Serive B, Kaas R, Bérard J-B, Pasquet V, Picot L, Cadoret J-P. Selection and optimisation of a method for efficient metabolites extraction from microalgae. Bioresource Technology. 2012. November;124:311–20. 10.1016/j.biortech.2012.07.105 [DOI] [PubMed] [Google Scholar]
- 15.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media [Internet]. 2009; http://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154
- 16.Stauber JL, Jeffrey SW. Photosynthetic Pigments in Fifty-One Species of Marine Diatoms1. Journal of Phycology. 1988;24(2):158–172. [Google Scholar]
- 17.Mulders KJM, Weesepoel Y, Lamers PP, Vincken J-P, Martens DE, Wijffels RH. Growth and pigment accumulation in nutrient-depleted Isochrysis aff. galbana T-ISO. J Appl Phycol. 2013. October 1;25(5):1421–30. [Google Scholar]
- 18.Takaichi S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Marine Drugs. 2011. June 15;9(6):1101–18. 10.3390/md9061101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aitzetmüller K, Strain HH, Svec WA, Grandolfo M, Katz JJ. Loroxanthin, a unique xanthophyll from Scenedesmus obliquus and Chlorella vulgaris. Phytochemistry. 1969. September;8(9):1761–70. [Google Scholar]
- 20.Tukaj Z, Matusiak-Mikulin K, Lewandowska J, Szurkowski J. Changes in the pigment patterns and the photosynthetic activity during a light-induced cell cycle of the green alga Scenedesmus armatus. Plant Physiology and Biochemistry. 2003. Avril;41(4):337–44. [Google Scholar]
- 21.Rial P, Garrido JL, Jaén D, Rodríguez F. Pigment composition in three Dinophysis species (Dinophyceae) and the associated cultures of Mesodinium rubrum and Teleaulax amphioxeia. J Plankton Res. 2013. March 1;35(2):433–7. [Google Scholar]
- 22.Lohr M, Wilhelm C. Xanthophyll synthesis in diatoms: quantification of putative intermediates and comparison of pigment conversion kinetics with rate constants derived from a model. Planta. 2001. February 1;212(3):382–91. 10.1007/s004250000403 [DOI] [PubMed] [Google Scholar]
- 23.Baudelet P-H, Gagez A-L, Bérard J-B, Juin C, Bridiau N, Kaas R, et al. Antiproliferative Activity of Cyanophora paradoxa Pigments in Melanoma, Breast and Lung Cancer Cells. Marine Drugs. 2013. November 1;11(11):4390–406. 10.3390/md11114390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.William Louda J, Loitz JW, Rudnick DT, Baker EW. Early diagenetic alteration of chlorophyll-a and bacteriochlorophyll-a in a contemporaneous marl ecosystem; Florida Bay. Organic Geochemistry. 2000. Décembre;31(12):1561–80. [Google Scholar]
- 25.Murray AP, Gibbs CF, Longmore AR, Flett DJ. Determination of chlorophyll in marine waters: intercomparison of a rapid HPLC method with full HPLC, spectrophotometric and fluorometric methods. Marine Chemistry. 1986. Juillet;19(3):211–27. [Google Scholar]
- 26.Roy S, Llewellyn CA, Egeland Skarstad, Johnsen G. Phytoplankton pigments Characterization, chemotaxonomy and applications in oceanography. Cambridge: Cambridge University Press; 2011. 845 p. [Google Scholar]
- 27.Lohr M, Wilhelm C. Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc Natl Acad Sci U S A. 1999. July 20;96(15):8784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright SW, Jeffrey SW. Pigment Markers for Phytoplankton Production In: Volkman JK, editor. Marine Organic Matter: Biomarkers, Isotopes and DNA [Internet]. Springer; Berlin Heidelberg; 2006. [cited 2014 Nov 6]. p. 71–104. (The Handbook of Environmental Chemistry). http://link.springer.com/chapter/10.1007/698_2_003 [Google Scholar]
- 29.Steinbrenner J, Linden H. Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control. Plant Mol Biol. 2003. May;52(2):343–56. [DOI] [PubMed] [Google Scholar]
- 30.Zapata M, Fraga S, Rodrguez F, Garrido JL. Pigment-based chloroplast types in dinoflagellates. Mar Ecol Prog Ser. 2012. September 28;465:33–52. [Google Scholar]
- 31.Zapata M, Jeffrey S, Wright S, Rodríguez F, Garrido J, Clementson L. Photosynthetic pigments in 37 species (65 strains) of Haptophyta: implications for oceanography and chemotaxonomy. Marine Ecology Progress Series. 2004;270:83–102. [Google Scholar]
- 32.Juin C, Bonnet A, Nicolau E, Bérard J-B, Devillers R, Thiéry V, et al. UPLC-MSE Profiling of Phytoplankton Metabolites: Application to the Identification of Pigments and Structural Analysis of Metabolites in Porphyridium purpureum. Marine Drugs. 2015. April 22;13(4):2541–58. 10.3390/md13042541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graziani G, Schiavo S, Nicolai MA, Buono S, Fogliano V, Pinto G, et al. Microalgae as human food: chemical and nutritional characteristics of the thermo-acidophilic microalga Galdieria sulphuraria. Food & Function. 2013;4(1):144. [DOI] [PubMed] [Google Scholar]
- 34.Holden M. The Purification and Properties of Chlorophyllase. Photochemistry and Photobiology. 1963;2(2):175–180. [Google Scholar]
- 35.Jeffrey S, Hallegraeff G. Chlorophyllase distribution in ten classes of phytoplankton: a problem for chlorophyll analysis. Mar Ecol Prog Ser. 1987;35(3):293–304. [Google Scholar]
- 36.Egeland ES, Garrido JL, Zapata M, Maestro MA, Liaaen-Jensen S. Algal carotenoids. Part 64. Structure and chemistry of 4-keto-19′-hexanoyloxyfucoxanthin with a novel carotenoid end group. J Chem Soc, Perkin Trans 1. 2000. January 1;(8):1223–30. [Google Scholar]
- 37.Mangoni O, Imperatore C, Tomas CR, Costantino V, Saggiomo V, Mangoni A. The New Carotenoid Pigment Moraxanthin Is Associated with Toxic Microalgae. Marine Drugs. 2011. February 10;9(2):242–55. 10.3390/md9020242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britton G, Liaaen-Jensen S, Pfander H. Carotenoids Handbook. Springer; 2004. 660 p. [Google Scholar]
- 39.Scheer H. Light-Harvesting Antennas in Photosynthesis. Green BR, Parson WW, editors. Dordrecht: Springer Netherlands; 2003. (Advances in Photosynthesis and Respiration; vol. 13). [Google Scholar]
- 40.Mantoura RFC, Llewellyn CA. The rapid determination of algal chlorophyll and carotenoid pigments and their breakdown products in natural waters by reverse-phase high-performance liquid chromatography. Analytica Chimica Acta. 1983;151:297–314. [Google Scholar]
- 41.Chauton MS, Optun OI, Bathen TF, Volent Z, Gribbestad IS, Johnsen G. HR MAS 1H NMR spectroscopy analysis of marine microalgal whole cells. Marine Ecology Progress Series. 2003;256:57–62. [Google Scholar]
- 42.Thrane J-E, Kyle M, Striebel M, Haande S, Grung M, Rohrlack T, et al. Spectrophotometric Analysis of Pigments: A Critical Assessment of a High-Throughput Method for Analysis of Algal Pigment Mixtures by Spectral Deconvolution. Schmitt FG, editor. PLOS ONE. 2015. September 11;10(9):e0137645 10.1371/journal.pone.0137645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez-Rojo S, Visentin A, Maestri D, Cocero MJ. Assisted extraction of rosemary antioxidants with green solvents. Journal of Food Engineering. 2012. March;109(1):98–103. [Google Scholar]
- 44.Wolf D, Siems K. Burning the Hay to Find the Needle Data Mining Strategies in Natural Product Dereplication. CHIMIA International Journal for Chemistry. 2007;61(6):339–45. [Google Scholar]
- 45.Schmidt B, Jaroszewski JW, Bro R, Witt M. Combining PARAFAC Analysis of HPLC-PDA Profiles and Structural Characterization Using HPLC-PDA-SPE-NMR-MS Experiments: Commercial Preparations of St. John’s Wort. Anal Chem. 2008. March 1;80(6):1978–87. 10.1021/ac702064p [DOI] [PubMed] [Google Scholar]
- 46.Böröczky K, Laatsch H, Wagner-Döbler I, Stritzke K, Schulz S. Cluster Analysis as Selection and Dereplication Tool for the Identification of New Natural Compounds from Large Sample Sets. Chemistry & Biodiversity. 2006;3(6):622–634. [DOI] [PubMed] [Google Scholar]
- 47.Higgins HW, Wright SW, Schlüter L. Quantitative interpretation of chemotaxonomic pigment data In: Phytoplankton pigments—Characterization, chemotaxonomy and applications in oceanography. Cambridge: Cambridge University Press; 2011. p. 257–313. [Google Scholar]
- 48.Hansen ME, Smedsgaard J, Larsen TO. X-Hitting: an algorithm for novelty detection and dereplication by UV spectra of complex mixtures of natural products. Anal Chem. 2005. November 1;77(21):6805–17. 10.1021/ac040191e [DOI] [PubMed] [Google Scholar]
- 49.Lang I, Hodac L, Friedl T, Feussner I. Fatty acid profiles and their distribution patterns in microalgae: a comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC plant biology. 2011;11(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camp D, Davis RA, Campitelli M, Ebdon J, Quinn RJ. Drug-like Properties: Guiding Principles for the Design of Natural Product Libraries. J Nat Prod. 2011;75(1):72–81. 10.1021/np200687v [DOI] [PubMed] [Google Scholar]
- 51.Carbonera D, Di Valentin M, Spezia R, Mezzetti A. The unique photophysical properties of the Peridinin-Chlorophyll-a-Protein. Current protein & peptide science. 2014;15(4):332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polivka T. Effects of self-assembled aggregation on excited states In: Carotenoids Physical, chemical, and biological functions and properties. Boca Raton: CRC Press; 2010. p. 137–57. [Google Scholar]
- 53.Salvadori E, Di Valentin M, Kay CWM, Pedone A, Barone V, Carbonera D. The electronic structure of the lutein triplet state in plant light-harvesting complex II. Phys Chem Chem Phys. 2012. September 21;14(35):12238–51. 10.1039/c2cp40877e [DOI] [PubMed] [Google Scholar]
- 54.Legrand C, Rengefors K, Fistarol GO, Granéli E. Allelopathy in phytoplankton—biochemical, ecological and evolutionary aspects. Phycologia. 2003. July;42(4):406–19. [Google Scholar]
- 55.Kuczynska P, Jemiola-Rzeminska M, Strzalka K. Photosynthetic Pigments in Diatoms. Marine Drugs. 2015. September 16;13(9):5847–81. 10.3390/md13095847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stonik V, Stonik I. Low-Molecular-Weight Metabolites from Diatoms: Structures, Biological Roles and Biosynthesis. Marine Drugs. 2015. June 9;13(6):3672–709. 10.3390/md13063672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernal FA, Delgado WA, Cuca LE. Fingerprint analysis of unfractionated Piper plant extracts by HPLC-UV-DAD coupled with chemometric methods. Journal of the Chilean Chemical Society. 2012;57(3):1256–1261. [Google Scholar]
- 58.Ivanišević J, Thomas OP, Lejeusne C, Chevaldonné P, Pérez T. Metabolic fingerprinting as an indicator of biodiversity: towards understanding inter-specific relationships among Homoscleromorpha sponges. Metabolomics. 2011. June;7(2):289–304. [Google Scholar]
- 59.Donkor VA, Häder D-P. Effects of ultraviolet irradiation on photosynthetic pigments in some filamentous cyanobacteria. Aquat Microb Ecol. 1996;11:143–9. [Google Scholar]
- 60.Donkor VA, Häder D-P. Ultraviolet radiation effects on pigmentation in the cyanobacterium Phormidium uncinatum. Acta Protozoologica. 1997;36:49–55. [Google Scholar]
- 61.Estevez MS, Malanga G, Puntarulo S. UV-B effects on Antarctic Chlorella sp cells. Journal of Photochemistry and Photobiology B: Biology. 2001. September 1;62(1–2):19–25. [DOI] [PubMed] [Google Scholar]
- 62.Figueroa FL, Nygård C, Ekelund N, Gómez I. Photobiological characteristics and photosynthetic UV responses in two Ulva species (Chlorophyta) from southern Spain. Journal of Photochemistry and Photobiology B: Biology. 2003. Décembre;72(1–3):35–44. [DOI] [PubMed] [Google Scholar]
- 63.Brody M, Emerson R. The quantum yield of photosynthesis in Porphyridium cruentum, and the role of chlorophyll a in the photosynthesis of red algae. The Journal of General Physiology. 1959;43:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gantt E. Pigmentation and photoacclimatation In: Biology of the red algae. Paris: Lavoisier; 1990. p. 203–19. [Google Scholar]
- 65.MacIntyre HL, Kana TM, Anning T, Geider RJ. Photoacclimatation of photosynthesis irradiance response curves and photosynthetic pigments in microalgae and cyanobacteria. Journal of Phycology. 2002;38(1):17–38. [Google Scholar]
- 66.Mouget J-L, Rosa P, Tremblin G. Acclimation of Haslea ostrearia to light of different spectral qualities—confirmation of “chromatic adaptation” in diatoms. Journal of Photochemistry and Photobiology B: Biology. 2004. Juillet;75(1–2):1–11. [DOI] [PubMed] [Google Scholar]
- 67.Mouget J-L, Rosa P, Vachoux C, Tremblin G. Enhancement of marennine production by blue light in the diatom Haslea ostrearia. Journal of Applied Phycology. 2005. October 18;17(5):437–45. [Google Scholar]
- 68.Dubinsky Z, Stambler N. Photoacclimation processes in phytoplankton: mechanisms, consequences, and applications. Aquat Microb Ecol. 2009. September;56(2–3):163–76. [Google Scholar]
- 69.Gao Y, Cui Yunluan, Xiong Wei, Li Xiaobo, Wu Qingyu. Effect of UV-C on Algal Evolution and Differences in Growth Rate, Pigmentation and Photosynthesis Between Prokaryotic and Eukaryotic Algae. Photochemistry and Photobiology. 2009;85(3):774–82. 10.1111/j.1751-1097.2008.00493.x [DOI] [PubMed] [Google Scholar]
- 70.Huang JJ, Cheung PC-K. +UVA treatment increases the degree of unsaturation in microalgal fatty acids and total carotenoid content in Nitzschia closterium (Bacillariophyceae) and Isochrysis zhangjiangensis (Chrysophyceae). Food Chemistry. 2011. Décembre;129(3):783–91. 10.1016/j.foodchem.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 71.Kuwahara SS, Cuello JL, Myhre G, Pau S. Growth of the green algae Chlamydomonas reinhardtii under red and blue lasers. Optics and Lasers in Engineering. 2011. March;49(3):434–8. [Google Scholar]
- 72.Sakshaug E, Andresen K. Effect of light regime upon growth rate and chemical composition of a clone of Skeletonema costatum from the Trondheimsfjord, Norway. J Plankton Res. 1986. January 1;8(4):619–37. [Google Scholar]
- 73.Tukaj Z, Matusiak-Mikulin K, Lewandowska J, Szurkowski J. Changes in the pigment patterns and the photosynthetic activity during a light-induced cell cycle of the green alga Scenedesmus armatus. Plant Physiology and Biochemistry. 2003. April;41(4):337–44. [Google Scholar]
- 74.Pickering MD, Keely BJ. Low temperature abiotic formation of mesopyrophaeophorbide a from pyrophaeophorbide a under conditions simulating anoxic natural environments. Geochimica et Cosmochimica Acta. 2011;75(2):533–40. [Google Scholar]
- 75.Lewitus AJ, Caron DA. Relative effects of nitrogen or phosphorus depletion and light intensity on the pigmentation, chemical composition, and volume of Pyrenomonas salina (Cryptophyceae). Mar Ecol Prog Series. 1990;61:171–81. [Google Scholar]
- 76.Latasa M, Berdalet E. Effect of nitrogen or phosphorus starvation on pigment composition of cultured Heterocapsa sp. Journal of Plankton Research. 1994. January 1;16(1):83–94. [Google Scholar]
- 77.Henriksen P, Riemann B, Kaas H, Sørensen HM, Sørensen HL. Effects of nutrient-limitation and irradiance on marine phytoplankton pigments. Journal of Plankton Research. 2002;24(9):835–58. [Google Scholar]
- 78.Figueroa FL, Israel A, Neori A, Martinez B, Malta EJ, Put A, et al. Effect of nutrient supply on photosynthesis and pigmentation to short-term stress (UV radiation) in Gracilaria conferta (Rhodophyta). Marine Pollution Bulletin. 2010. October;60(10):1768–78. 10.1016/j.marpolbul.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 79.MacIntyre HL, Kana TM, Geider RJ. The effect of water motion on short-term rates of photosynthesis by marine phytoplankton. Trends Plant Sci. 2000. January;5(1):12–7. [DOI] [PubMed] [Google Scholar]
- 80.Thompson PA, Pesant S, Waite AM. Contrasting the vertical differences in the phytoplankton biology of a dipole pair of eddies in the south-eastern Indian Ocean. Deep Sea Research Part II: Topical Studies in Oceanography. 2007. April;54(8–10):1003–28. [Google Scholar]
- 81.Klein B. Variations of pigment content in two benthic diatoms during growth in batch cultures. Journal of Experimental Marine Biology and Ecology. 1988. March 31;115(3):237–48. [Google Scholar]
- 82.Wilhelm C, Manns L. Changes in pigmentation of phytoplankton species during growth and stationary phase—consequences for reliability of pigment-based methods of biomass determination. Journal of Applied Phycology. 1991. December;3(4):305–10. [Google Scholar]
- 83.Bode HB, Bethe B, Höfs R, Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem. 2002;3(7):619–27. [DOI] [PubMed] [Google Scholar]
- 84.Knight V, Sanglier J-J, DiTullio D, Braccili S, Bonner P, Waters J, et al. Diversifying microbial natural products for drug discovery. Applied Microbiology and Biotechnology. 2003;62(5):446–58. [DOI] [PubMed] [Google Scholar]
- 85.Pietra F. Secondary metabolites from marine microorganisms: bacteria, protozoa, algae and fungi. Achievements and prospects. Nat Prod Rep. 1997. October;14(5):453–64. [DOI] [PubMed] [Google Scholar]
- 86.Abby SS, Touchon M, De Jode A, Grimsley N, Piganeau G. Bacteria in Ostreococcus tauri cultures—friends, foes or hitchhikers? Frontiers in Microbiology. 2014. November 7;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nishitani G, Nagai S, Hayakawa S, Kosaka Y, Sakurada K, Kamiyama T, et al. Multiple Plastids Collected by the Dinoflagellate Dinophysis mitra through Kleptoplastidy. Appl Environ Microbiol. 2012. February 1;78(3):813–21. 10.1128/AEM.06544-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skovgaard A. Role of chloroplast retention in a marine dinoflagellate. Aquatic Microbial Ecology. 1998;15:293–301. [Google Scholar]
- 89.Tamm M, Freiberg R, Tõnno I, Nõges P, Nõges T. Pigment-Based Chemotaxonomy—A Quick Alternative to Determine Algal Assemblages in Large Shallow Eutrophic Lake? Anil AC, editor. PLOS ONE. 2015. March 24;10(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Archibald JM, Keeling PJ. Recycled plastids: a “green movement” in eukaryotic evolution. Trends Genet. 2002. November;18(11):577–84. [DOI] [PubMed] [Google Scholar]
- 91.Delwiche CF. Tracing the Thread of Plastid Diversity through the Tapestry of Life. The American Naturalist. 1999. October;154(s4):S164–77. 10.1086/303291 [DOI] [PubMed] [Google Scholar]
- 92.Durnford DG, Gray MW. Analysis of Euglena gracilis Plastid-Targeted Proteins Reveals Different Classes of Transit Sequences. Eukaryotic Cell. 2006. December 1;5(12):2079–91. 10.1128/EC.00222-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wright SW, Jeffrey SW. Pigment Markers for Phytoplankton Production In: Volkman JK, editor. Marine Organic Matter: Biomarkers, Isotopes and DNA. Berlin/Heidelberg: Springer-Verlag; 2006. p. 71–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Calculation of carotenoid band ratio. UV-visible spectra showing wavelength I; II and III of a carotenoid. Band ratio corresponds to the % ratio Δ III/ Δ II. (B) Calculation of porphyrin band ratio. UV-visible spectra showing the location of I the Soret (blue maximum) and II the red bands of Chlorophyll -a.
(PDF)
(PDF)
(PDF)
(PDF)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.