Abstract
Background
Neuropeptide precursors are traditionally viewed as proteins giving rise to small neuropeptide molecules. Prodynorphin (PDYN) is the precursor protein to dynorphins, endogenous ligands for the κ-opioid receptor. Alternative mRNA splicing of neuropeptide genes may regulate cell- and tissue-specific neuropeptide expression and produce novel protein isoforms. We here searched for novel PDYN mRNA and their protein product in the human brain.
Methods
New PDYN transcripts were identified using nested PCR amplified from oligo(dT) selected full-length capped mRNA. Gene expression was analyzed by qRT-PCR, PDYN protein by western blotting and confocal imaging, dynorphin peptides by radioimmunoassay. Neuronal nuclei were isolated using fluorescence-activated nuclei sorting from postmortem human striatal tissue. Immunofluorescence staining and confocal microscopy was performed for human caudate nucleus.
Results
Two novel human PDYN mRNA splicing variants were identified. Expression of one of them was confined to the striatum where its levels constituted up to 30% of total PDYN mRNA. This transcript may be translated into ΔSP-PDYN protein lacking 13 N-terminal amino acids, a fragment of signal peptide (SP). ΔSP-PDYN was not processed to mature dynorphins and surprisingly, was targeted to the cell nuclei in a model cellular system. The endogenous PDYN protein was identified in the cell nuclei in human striatum by western blotting of isolated neuronal nuclei, and by confocal imaging.
Conclusions and general significance
High levels of alternatively spliced ΔSP-PDYN mRNA and nuclear localization of PDYN protein suggest an essential nuclear function for this isoform of the opioid peptide precursor in human striatum.
Keywords: Neuropeptide precursor protein, prodynorphin, alternative splicing, nuclear localization, human brain
1. INTRODUCTION
A function of neuropeptide precursor proteins is to give rise to neuropeptides, small peptide molecules that serve as neurotransmitters, neuromodulators or neurohormones acting through the G-protein coupled receptors. The precursor molecules translocate into the endoplasmic reticulum where they fold and undergo posttranslational modifications, then transported to the Golgi apparatus and targeted to the regulated secretory pathway [1–4]. In endocrine cells, precursor proteins are cleaved to mature neuropeptides in the trans-Golgi network and secretory vesicles [1–6]. In neurons, the neuropeptide precursors are processed during transport from the perikaryon to axon terminals, or at these terminals and dendrites where they are stored in unprocessed form [7].
Splicing of precursor mRNA is essential for eukaryotic gene expression especially in the brain in which the most complex pattern of alternative splicing contributes to cell differentiation, neuronal migration and synaptogenesis [8]. Little is known about alternative mRNA splicing of neuropeptide genes that may regulate cell- and tissue-specific neuropeptide expression, subcellular localization of precursor proteins and neuronal plasticity, or produce novel protein isoforms.
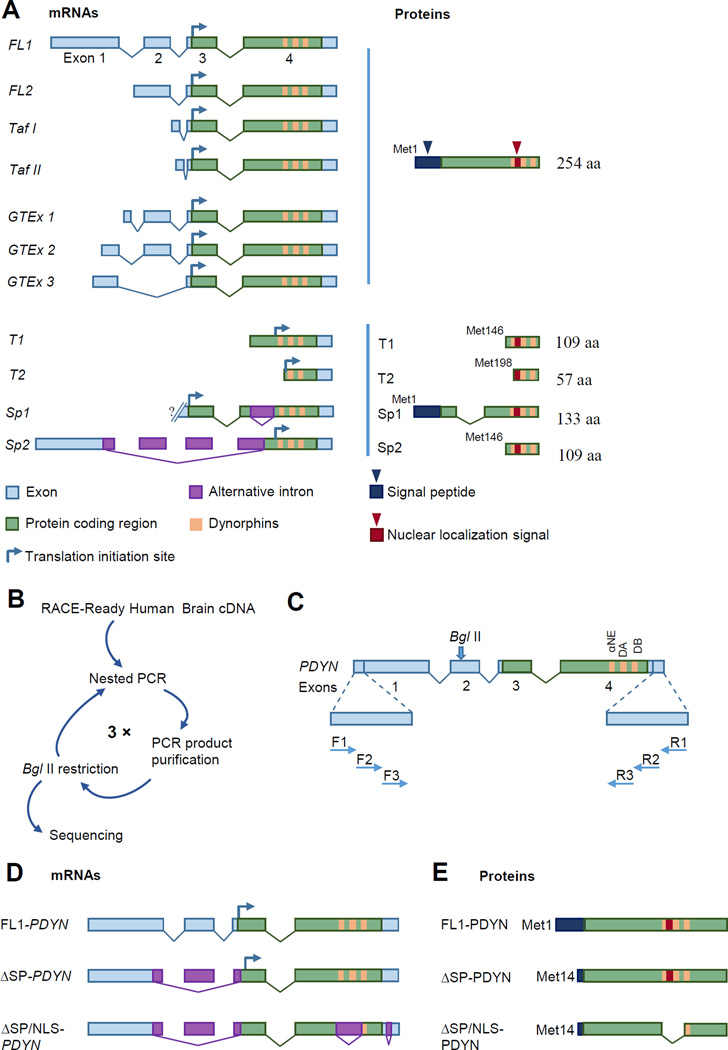
The prodynorphin gene (PDYN) gives rise to dynorphins, the endogenous opioid peptides that are ligands for the κ-opioid receptor [9]. Dynorphins are expressed in the basal ganglia, hippocampus, cerebral cortex and spinal cord [10] and are essential for regulation of sensory processes [1, 5, 6], modulation of reward [2, 3], motor control [4] and stress-induced behavioral responses [11]. The canonical form of PDYN pre-mRNA consists of three introns, and four exons of which the 3rd and 4th encode for the full-length protein (FL-PDYN mRNA) [12, 13] (Fig. 1A). Altogether, eight human PDYN transcripts FL1, FL2, splicing variants Sp1 and Sp2, and 5´-truncated Taf1, Taf2, T1, and T2 mRNAs were described in previous studies [13, 14]. Three variants (GTEx 1–3) of dominant FL-PDYN transcripts that differ in the length of exon 1, the presence of exon 2 and distribution in human brain were identified by RNA-seq analysis (GTEx Portal, http://www.gtexportal.org/home/) (Fig. 1A). Searching for novel PDYN mRNA, we here identified two PDYN splice variants, and characterized the expression levels and distribution in the human brain of one of them, i.e. ΔSP-PDYN. This transcript gives rise to a protein that lacks the 13 N-terminal aa, a fragment of the signal peptide. Surprisingly, ΔSP-PDYN ectopically expressed in a model cell line and the endogenous protein in human striatum, is located in the cell nuclei suggesting a nuclear function for this neuropeptide precursor protein.
FIGURE 1. Structure of known and identification of novel PDYN transcripts and proteins.
A. Previously described PDYN transcripts coding for the full-length (FL) and truncated proteins. The dominant FL1-PDYN and transcripts identified using RNA-Seq that differ in the length of the first exon and the presence of the second exon (GTEx 1–3), as well as three transcripts with alternative TSS, FL2 and testis-specific Taf I and Taf II transcripts, encode FL-PDYN protein. Other PDYN isoforms such as alternatively spliced Sp1 and Sp2 and transcripts initiated within the coding part of exon 4 (T1 and T2) may produce truncated PDYN proteins. Curved arrows show position of the translation initiation (TI).
B. Scheme of novel transcript identification. Human brain RACE-Ready cDNA was amplified in three rounds of nested PCR followed by digestion with BglII to eliminate the dominant FL-PDYN transcripts. The procedure allowed amplification of minor PDYN mRNA isoforms which then were analyzed by sequencing.
C. Structure of PDYN mRNAs consisting of four exons (boxes) and three introns (lines), and the localization of the primers used in the nested PCR are shown, not to scale. F and R depict forward and reverse primers (not to scale). αNE, DA and DB are the α-neoendorphin, dynorphin A and dynorphin B encoding sequences.
D, E. FL-PDYN and alternatively spliced ΔSP- and ΔSP/NLS-PDYN mRNA isoforms (D), and corresponding protein products (E). 3´-fragment of exon 1, exon 2 and 5´-fragment of exon 3 are spliced out in ΔSP- and ΔSP/NLS-transcripts. Two new introns are present in ΔSP/NLS mRNA isoform in exon 4 region; one of them overlaps with the dynorphin encoding domain, while another is located in the 3´-UTR (D). Signal peptide is truncated in both ΔSP- and ΔSP/NLS-PDYN. Putative NLS sequence is located in the dynorphin domain in FL- and ΔSP-PDYN but is absent in ΔSP/NLS-PDYN (E).
2. MATERIALS AND METHODS
2.1. Postmortem human brain tissue
Brain tissues were collected from ten deceased subjects with no evidence of neuropathology and no known history of neuropsychiatric disease by KI Donatum at Forensic Medicine, Department of Oncology and Pathology, Karolinska Institute, Stockholm, Sweden, by qualified pathologists under the full ethical clearance from the Stockholm Ethical Review Board. Donation was performed after an informed consent by the next of kin or by a registration as a donor by the deceased. Tissue samples of cerebellum, striatum (caudate nucleus, putamen and nucleus accumbens), dorsolateral prefrontal cortex and orbitofrontal cortex were obtained from seven subjects and used for gene expression validation by qRT-PCR (Table 1, subjects 1–7). Samples from cerebellum and striatum were used for fluorescence-activated nuclei sorting followed by western blot and radioimmunoassay (Table 1, subjects 8–10).
TABLE 1.
Demographic data and tissue characteristics of human subjects
| Subject | Age (years) |
sex | PMI (hours) |
Brain pH | RQI | Brain structure | Assay | Smoking history |
Cause of death | Toxicological findings at time of death |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (#71) | 23 | M | 41.7 | 6.96 | 8.1 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | Yes | muliple injuries, suicide |
None |
| 2 (#79) | 23 | M | 39.0 | 6.83 | 8.0 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | No | heart disease | None |
| 3 (#81) | 53 | F | 38.7 | 6.75 | 7.7 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | NA | multiple injuries, accidental death |
Ethanol, caffeine, zopiclone, oxazepam |
| 4 (#84) | 69 | M | 26.5 | 6.69 | 7.8 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | No | hanging, suicide | Oxazepam |
| 5 (#89) | 54 | M | 61.0 | 6.28 | 7.1 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | No | intoxalc, accidental death |
Ethanol, mirtazapine, promethazine, lactate, glucose |
| 6 (#80) | 59 | F | 47.3 | 6.27 | 6.8 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | Yes | perf duodenal ulcer | Ethanol, citalopram, desmethylcitalopram |
| 7 (#102) | 31 | M | 61.7 | 6.40 | 7.9 | NAc, Cd, Put, OFC, PFC, Cbl |
qRT-PCR | No | Rupture of the inferior vena, fracture liver damage |
Ketamine |
| 8 (#11) | 57 | M | 46.0 | NA | - | Str, Cbl | FANS, WB, RIA | NA | hypertensive cardiomyopathy |
NA |
| 9 (#184) | 21 | M | 44.0 | NA | - | Str, Cbl | FANS, WB, RIA | NA | hanging, suicide | Methylphenidate |
| 10 (#191) | 92 | M | 27.5 | 6.68 | - | Str, Cbl | FANS, WB, RIA | NA | hanging, suicide | NA |
| 11 (SW1) | 23 | F | 11.0 | 6.85 | - | Cd | IF | Yes | motor vehicle accident | None |
| 12 (SW2) | 46 | F | 24.0 | 6.32 | - | Cd | IF | No | homicide | None |
| 13 (SW3) | 42 | M | 20.0 | 6.82 | - | Cd | IF | NA | cardiac related | None |
| 14 (SW4) | 57 | M | 10.0 | 6.65 | - | Cd | IF | NA | cardiac related | None |
NAc, nucleus accumbens; Cd, caudate nucleus; Put, putamen; Str, striatum; OFC, orbitofrontal cortex; PFC, dorsolateral prefrontal cortex; Cbl, cerebellum.
qRT-PCR, quantitative real-time PCR; IF, immunofluorescence; FANS, fluorescent-activated nuclei sorting; WB, western blotting; RIA, radioimmunoassay; RQI, RNA quality indicator.
Toxicological parameters measured: ethanol, methanol, acetone, cannabis, amphetamines and opiates, cocaine metabolite, MDMA analogues, buprenorphine concentrations; drug screening.
Sections of caudate nucleus used for analysis of ΔSP-PDYN intracellular localization by confocal microscopy were obtained from four subjects from the Postmortem Brain Core, Center for Psychiatric Neuroscience, University of Mississippi Medical Center, Jackson, MS, USA. The protocol for recruitment, tissue collection, and interviews was approved by the Institutional Review Boards of University Hospitals Case Medical Center, Cleveland, OH, and University of Mississippi Medical Center. Written informed consent was obtained from legally-defined next of kin for tissue collection and informant-based retrospective diagnostic interviews (Table 1, subjects 11–14). No subjects had evidence of head trauma, neurologic, neuropathological, or psychiatric disease.
2.2. RNA isolation and cDNA synthesis
Total RNA was purified using RNeasy Lipid Tissue Mini Kit (QIAGEN, Maryland, USA), treated with RNase–free DNase I on-column for 30 min at room temperature and stored at −80°C for further use. Total RNA amount was quantified by Nanodrop® (Nanodrop Technologies, Inc., USA). RNA Quality Indicator was analyzed by Eukaryote Total RNA StdSens assay using a Bio-Rad Experion instrument (Bio-Rad Laboratories, Hercules, CA, USA). For cDNA synthesis, total mRNA was reverse transcribed with PDYN-specific primer (Table 2, Fig. S1) using iScript Select kit (Bio-Rad Laboratories, Hercules, CA, USA). PDYN specific 3´-primer is complementary to the exon 4 sequence that was spliced out as an intron in ΔSP/NLS-PDYN mRNA, thus allowing detection of only ΔSP- but not ΔSP/NLS-PDYN mRNA.
TABLE 2.
Primers sequences used for cDNA synthesis, identification of novel isoforms, qRT-PCR and plasmid construction.
| Primer name | Primer sequence (5′ – 3′) | Amplicon size (bp) |
Assay |
|---|---|---|---|
| ΔSP-PDYN | F: 5′ -GAGGCCCCTGTCCTGGCTGCC-3′ R: 5′ -GGGAGCAAATCAGGGGATTG -3′ |
130 | qRT-PCR |
| PDYN Ex3-4 | F: 5′ -CACCACAGCGGACTGCCTGT -3′ R: 5′ -AGCAGGGCAGCCTGGCATTG -3′ |
111 | qRT-PCR |
| 706R | 5′ - GGTCCTCCTCAGCGAGATAGAGT-3′ | - | Reverse transcription |
| DREmet43 PDYN1086 |
F: 5′-GGCTTTGGTGGTGTTCACAGCT-3′ R: 5′-GTACACAATGCTGAGCTGAGCAT-3′ |
1043 | Nested PCR, 1st round |
| PDYN53 PDYN1053 |
F: 5′-CCTCTTTGGCACCTCCTCCCAA-3′ R: 5′-GGCACATATAAGAGGATGAATGAATG-3′ |
1000 | Nested PCR, 2st round |
| PDYN68 PDYN1038 |
F: 5′-CCCAAGCCGGAGTCAAGGAG-3′ R: 5′-TGAATGAATGCACTCCAACCTGAA-3′ |
970 | Nested PCR, 3st round |
| ΔSP-pCMV4 | F: 5′ -CCGCCAGATCTACCATGTTCCCCTCCACCACAGC-3′ R: 5′-GGCGGAAGCTTTTATGCATCAAAAAGCTCTCCAG-3′ |
751 | Plasmid construction |
F, forward primer; R, reverse primer. PCR conditions are described in Methods
2.3. Nested PCR
cDNA from human brain (FirstChoice® RACE-Ready cDNA Kit, Ambion) which is synthesized from high quality total RNA enriched for mRNA content by three rounds of oligo(dT) selection and amplified only from full-length, capped mRNA was used as a source. First round amplification was performed with DREmet43F and PDYN1086R primers (Table 2; Fig. 1B, C) on a Mastercycler (Eppendorf, Berlin, Germany) followed by digestion with BglII to eliminate the dominant FL1- and FL2-PDYN transcripts. In the second round amplification was performed with PDYN53F and PDYN1053R primers (Table 2), the resulting PCR products were purified using a Illustra GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare, UK Limited, Little Chalfont, UK), digested with BglII and subjected to a third round of amplification with PDYN68F and PDYN1038R primers (Table 2). Two PCR products of 850 bp size (ΔSP-PDYN) and 560 bp size(ΔSP/NLS-PDYN), were excised from the gel, purified using an Illustra GFX™ PCR DNA and Gel Band Purification Kit and sequenced in both directions. The following conditions were applied for the endpoint PCR; 95°C for 3 min followed by 40 cycles at 95.0°C for 15 s, 60.0°C for 30 s and 72.0°C for 30 s.
2.4. Quantitative reverse transcription PCR (qRT-PCR)
qRT-PCR was performed on a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Primers were designed using Vector NTI advance 11 software (Table 2, Fig. S1). The following conditions were applied for the two steps RT-qPCR reaction; 95.0°C for 3 min followed by 40 cycles at 95.0°C for 10 s, annealing temperature for 61.5°C for 30 s. A relative quantification analysis was performed, based on an external calibration curve [15], constructed from purified ΔSP- and FL-PDYN PCR products. Melting curve analysis was performed to confirm the specificity of amplification and lack of primer dimers. To ensure correct amplification, PCR products were separated on agarose gel and sequenced in both directions.
2.5. Plasmid construction
cDNAs encoding FL- and ΔSP-PDYN were amplified by PCR using FL-pCMV4 [14] and ΔSP-pCMV4 primers (see Table 2), generating a BglII site at the 5´ end and a HindIII site at the 3´ end. This facilitated cloning into the BglII/HindIII sites of pCMV4 vector (Clontech, San Diego, CA, USA) to construct pCMV-FL-PDYN and pCMV-ΔSP-PDYN. pCMV-FL-PDYN contains coding part of exons 3 and 4 and encodes FL-PDYN (254 aa), while pCMV-ΔSP-PDYN with a 5´-truncated exon 3 and intact exon 4 gives rise to ΔSP-PDYN (241 aa). Sequencing was performed to check the correctness of both plasmids.
2.6. Cell culture and transfection
Rat insulinoma RINm-5F cells were cultured in RPMI-1640 medium with GlutaMAX (Gibco, Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum, penicillin and streptomycin in 5% CO2 and at 37°C. 2.0 × 105 cells per well were plated on four-well chamber slides (Lab-Tek, Nunc Inc., IL,USA) for immunocytochemistry analyses. 2.9 × 105 cells were plated per 60-mm dish for the transfection procedure. Cells transfected with plasmid DNA using Lipofectamine 2000™ (Invitrogen, Carlsbad, CA, USA), were harvested for protein analysis and immunostaining. For radioimmunoassay transfected cells were cultured for 18 h prior harvesting in the absence of cell medium.
2.7. Fluorescence-activated nuclei sorting (FANS)
Isolation of neuronal nuclei was performed as previously described [16, 17]. In brief, 1 g of thawed tissue from each human striatum and cerebellum was disrupted in Dounce homogenizer in lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM Mg (CH3COO)2, 0.1 mM EDTA, 10 mM Tris HCl (pH 8.0), 0.1% Triton X-100, 1 mM DTT), the suspension was loaded onto a 15% sucrose cushion and centrifuged at 30,000 × g for 2.5 hours at 4°C. Nuclei pellet was resuspended in PBS, monoclonal anti-NeuN antibody (Millipore, Billerica, MA) conjugated with Zenon anti-mouse Alexa 647-IgG (Molecular Probes) was added to the suspension and incubated at 4°C for 45 min. Neuronal, NeuN positive (NeuN(+)) nuclei were separated from non-neuronal, NeuN negative (NeuN(−)) nuclei using the FACSDiVa high-speed cell sorter (Becton Dickinson, Heidelberg, Germany). Nuclear suspension was centrifuged at 30,000 × g for 3 min and nuclear pellets were resuspended in PBS and stored at −80°C.
Two quality controls for FANS procedure were performed. First control includes the reanalyzing of the sorted populations by flow cytometry. 20 µl of the sorted nuclei was resuspended in 80 µl of PBS with a following flow cytometry analysis using the same gate setting as for FANS. The purities of NeuN(+) and NeuN(−) nuclei fractions were 97.3 ± 0.8%, and 99.95 ± 0.1%, respectively. Second quality control performed by analysis of expression of neuronal markers, RBFOX3 coding for NeuN and ENO2 coding for enolase 2 as well as GFAP, an astroglial marker in neuronal and non-neuronal FAN-sorted fractions by droplet digital PCR [18]. This analysis demonstrated clear separation of neuronal and non-neuronal nuclei by FANS, with more than 90% of RBFOX3 and ENO2 expression confined to neuronal nuclei, and more than 97% of GFAP expression confined to non-neuronal nuclei (manuscript in preparation).
2.8. Anti-human PDYN and anti-dynorphin antibodies
Rabbit polyclonal antisera were generated against human PDYN C-terminal fragment (CTF; aa 241–254) and PDYN-derived opioid peptides (Dyn A1–17, Dyn B1–13, and Leu-enkephalin-Arg) all conjugated with keyhole limpet hemocyanin via Cys added to their N terminus [7, 14, 19]. IgG fractions were purified with Protein A-Sepharose and extensively characterized. Anti-PDYN CTF antibody demonstrating high specificity was used for western blot (WB) and immunohistochemistry (IHC) [7]. Anti-Dyn A antibody did not react with PDYN in WB and IHC experiments when HeLa or PC12 cells transfected with pCMV-PDYN plasmid were analyzed; these cells do not process PDYN to dynorphins. Negative controls included pre-immune IgG fractions, and primary antibody blocked with 10 nM of antigenic human PDYN CTF aa 241–254 or related rat Pdyn C-terminal fragment (aa 235–248), which has 7 mismatches in the 15 amino acid sequence from the human CTF. In WB and IHC experiments, the PDYN signal was blocked by preincubation of the antiserum with 10 nM of the human PDYN CTF, but not with rat Pdyn CTF. The specificity of these antibodies has been reported earlier [14, 20].
Anti-Dyn A antibody demonstrated 100% molar crossreactivity with Dyn A(9–17) and 0.1% molar crossreactivity with Dyn B, Dyn A(1–8), a-neoendorphin, Leu-enkephalin, and big dynorphin. Anti-Dyn B antiserum showed 100% molar crossreactivity with big dynorphin, 0.8% crossreactivity with Leu-morphine (29 aa C-terminally extended Dyn B), and 0.1% crossreactivity with Dyn A(1–17), Dyn A(1–8), a-neoendorphin, and Leu-enkephalin. Dyn A, Dyn B, and Leu-enkephalin-Arg RIA readily detected these peptides in brain tissues of wild-type (WT) mice [21] and rats [20], whereas did not detect any peptide immunoreactivity in the striatum, hippocampus, and frontal cerebral cortex of Pdyn knockout mice used as negative controls [22], demonstrating high specificity of this method and the absence of interference with other peptides present in tissue extracts.
2.9. Immunofluorescent cell staining
RINm-5F cells transfected with plasmid coding for ΔSP-PDYN or FL-PDYN cDNA, or mock transfected with CMV-vector plasmid were fixed and incubated with primary anti-human PDYN CTF, (1:600), and anti-dynorphin A (1:500) 18 hours at +4°C followed by incubation with secondary goat anti-rabbit antibody conjugated with Alexa Fluor 488 (1:1000) (Molecular Probes, Ltd, Paisley, UK).
2.10. PDYN immunofluorescent staining in human caudate nucleus
Snap frozen human post-mortem tissue samples of caudate nucleus obtained from four subjects (see Table 1, for demographic data and tissue characteristics) were cryosectioned (8 sections 20 µm each) and mounted on superfrost+ microscope slides. Slides were stored at −80°C until analysis. For the immunofluorescence (IF) staining sections were placed in a freshly prepared 0.1 M phosphate buffer (pH 7.4) solution containing 4% paraformaldehyde. After washing in phosphate buffered saline containing 0.03% H2O2 to block endogenous peroxide, sections were incubated with rabbit polyclonal antibody raised against human PDYN CTF (1:4,000) in 0.1 M phosphate buffer contained 0.3% TX-100 and 0.1% NaN3 for 48 hours at 4°C. Sections were repeatedly washed in Tris-buffered saline (pH 7.4) containing 0.5% Tween20 (TNT) and were blocked with Tris-buffered saline containing 2.5% blocking reagent (TNB, Perkin Elmer, Waltham, USA) for 30 minutes at room temperature. After that sections were incubated in TNB buffer containing horseradish peroxidase conjugated secondary anti-rabbit IgG antibody raised in swine (1:200; DAKO, Glostrup, Denmark). After washing in TNT buffer the sections were incubated in amplification diluent containing fluorescein or Cy5 conjugated tyramine (1:100; Perkin Elmer, Waltham, USA). Nuclei were visualized using Hoechst 33342 (1:10,000, Biotium, CA, USA) in combination with SYTOX green (1:300; Life Technologies, Stockholm, Sweden).
Lipofuscin-autofluorescence was blocked by incubating the sections in 70% EtOH containing 1% Sudan black. Slides were mounted using ProLong Gold antifade reagent (Invitrogen, Waltham, Massachusetts, USA) and precision cover glasses (Marienfeld, Lauda-Köningshofen, Germany).
The specificity of anti-PDYN antibody was determined by an adsorption test and immunostaining with preimmune rabbit IgG. Briefly, rabbit antibody against PDYN (1:4,000) were preincubated with PDYN antigen (human PDYN CTF 241–254 aa peptide, 100 nM) overnight at 4°C. The mixture was applied to a section of human caudate putamen and immunoreactivity was determines as described above. For comparison, adjacent sections were incubated with rabbit anti PDYN (1:4,000; positive control) and pre-immune rabbit IgG (1:4,000; negative control). Results of adsorption test were visualized with epifluorescence microscopy. Adsorption test demonstrated elimination of the specific immunoreactivity on human caudate nucleus tissue. Pre-immune IgG did not produce positive staining with minor reactivity in in capillaries.
2.11. Epifluorescence microscopy
Entire slides were scanned using a slide-scanning microscope (Vslide, Metasystems, Altlussheim, Germany) equipped with 10 × and 20 × objectives and the appropriate excitation and emission filter sets for detection SYTOX green (504–523 nm) and cy5 (625–670 nm). After scanning individual field of view images were stitched and used for identification the regions of interest.
2.12. Confocal microscopy and image processing
Fluorescent images of the RINm-5F cells were acquired with a Nipkow spinning disk confocal system (Yokogawa CSU-10; Yokogawa Electric Corporation, Tokyo, Japan) attached to a Diaphot 200 microscope (Nikon, Kanagawa, Japan) and equipped with a 60×1.40/NA oil immersion objective (Nikon). Fluorescence was excited at 488 nm with argon ion laser (Melles-Griot, Didam, The Netherlands) and detected at 520/35 nm using cooled EM-CCD camera (iXon+ DU888; Andor, Belfast, Northern Ireland, UK) under MetaFluor software control (Molecular Devices Corp., Downingtown, PA).
Images of the human caudate nucleus were acquired by an Olympus FV1000 confocal laser scanning microscope (Olympus) equipped with a 20×/0.95W XLUMPlanFI objective. Emission spectra for each dye were limited as follows: Hoechst (361–497 nm) and Cy5 (630–647 nm). For co-localization analysis of PDYN and Hoechst z-stacks of 24 images were acquired with a depth interval of 0.5 µm. The open source platform Image J (NIH, MD) was used for 3D projections and orthogonal co-localization analysis.
2.13. Dynorphin radioimmunoassay (RIA)
The procedure was described elsewhere [7, 14, 23]. Briefly, cell extracts in 1 M acetic acid and cell culture medium mixed with acetic acid were boiled at 95°C for 5 min. After centrifugation, supernatant was loaded on a SP-Sephadex ion exchange C-25 column, peptides were eluted and analyzed by RIA. Protein concentrations were estimated by DC protein assay (Bio-Rad, Laboratories, Hercules, CA, USA).
Total content of PDYN protein and PDYN derived opioid peptides in the NeuN(+) and NeuN(−) nuclei isolated from human striatal and cerebellar tissues was determined using procedure based on liberation of Leu-enkephalin-Arg (LER), a PDYN marker from PDYN / PDYN fragments by trypsin digestion followed by LER RIA as described earlier [7]. Aliquots containing 1.0×106 of striatal or cerebellar NeuN(+) and NeuN(−) nuclei were heated up to 95°C (5 min). After centrifugation, supernatant was collected and incubated with immobilized TPCK-trypsin (Biochemical Corporation, Lakewood, NJ, USA) in 0.1 M NH4HCO3 buffer, pH 8.0 for 16 h at 37°C to liberate LER from PDYN protein and its fragments. Reaction was stopped by addition of acetic acid (1 M final concentration) and heating at 95°C for 5 min. After centrifugation, the mixture was subjected to SP-Sephadex C-25 chromatography and LER was analyzed by RIA. The content of PDYN was calculated in fmol of protein assuming that a PDYN molecule contains three LER copies; data was obtained in three independent experiments. Aliquots of nuclear extracts taken prior to the digestion, or trypsin alone did not produce any signal in LER RIA.
2.14. Western blotting
Cell nuclei isolated from striatal and cerebellar tissues and RINm-5F transfected cells for analysis of PDYN were extracted with SDS buffer (0.45 M Tris-HCl, pH 8.5, 2.5% glycerol and 4% SDS) supplemented with DTT and Compete Inhibitor Cocktail (Roche, Basel, Switzerland), as described previously [23]. Protein concentrations were determined with DC protein assay (Bio-Rad, Laboratories, Hercules, CA, USA) and Pierce assay (Thermo Scientific Life Science Research Products, Rockford, IL, USA) for sorted nuclei and transfected cells, respectively. Proteins were resolved by SDS-PAGE on 10% Tricine gels, transferred onto nitrocellulose membranes (Schleicher and Schuell, Keene, USA), and stained with MemCode reversible Protein Stain Kit (Pierce, Rockford, IL), as described previously [7, 14]. Membranes blocked with 5% nonfat dry milk were probed with anti-PDYN CTF antibody (1:1000) and were incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad Laboratories, Hercules, CA, USA) (1:12500).
2.15. Sequence analyses
FL-PDYN and ΔSP-PDYN sequences were analyzed for the presence of signal peptide (SP), putative nuclear localization signal (NLS), and leucine-rich nuclear export signal (NES) sequences (SignalP V4.0, cNLS Mapper, PSORT II, and NetNES 1.1).
3. RESULTS
The PDYN gene produces several RNA variants encoding for the full-length (FL) and truncated PDYN proteins (Fig. 1A). The dominant FL1-PDYN and three transcripts with alternative TSS (transcription start site), such as FL2 and testis-specific Taf I, and Taf II transcripts, encode full-length PDYN protein. RNA-seq analysis of the human transcriptome demonstrated that PDYN transcripts dominant in the brain differ in the length of the first exon and the presence of the second exon (GTEx Portal, http://www.gtexportal.org/home/). PDYN transcript Sp1 may produce PDYN protein isoforms that lack a segment between the signal peptide and the opioid sequences. Another PDYN transcript (Sp2) with an alternative TSS, alternative 5´ splice site (SS) selection in exon 1, exons 2 and 3 skipping and alternative 3´ SS selection in exon 4 could possibly be translated from third methionine codon (Met146). Other PDYN transcripts initiated within the coding part of exon 4 may result in PDYN protein isoforms that are N-terminally truncated T1 and T2 translated either from the third or fourth methionine codon (Met146 or Met198), respectively.
3.1. Novel human PDYN splice variants
Having identified several PDYN transcripts lacking exon 2 [13, 14], we searched for other such transcripts in the human brain as a part of strategy aimed to identify all types of PDYN mRNAs. Exon 2-containing transcripts including dominant FL1 and FL2 mRNAs were eliminated by treatment of cDNA with BglII endonuclease, which cleavage site is located in exon 2 (Fig. 1B, C). After three consecutive rounds of nested PCR each followed by BglII treatment, two novel alternatively spliced PDYN transcripts ΔSP-PDYN and ΔSP/NLS-PDYN were isolated (Fig. 1, B–E). ΔSP-PDYN is 850 nt long, consists of three exons and two introns with alternative 5´SS selection in exon 1 (lacking 71 nt), exon 2 skipping (60 nt) and alternative 3´ SS selection in exon 3 (lacking 36 nt). ΔSP-PDYN also retained non-canonical exon 3–4 junction. The consensus sequence presented in the exon 3–4 junction likely indicates the U12-dependent intron excision [24]. The exon 2–3 junction was absent in the ΔSP-PDYN mRNA, and a novel exon 1–3 junction is non-canonical (see Fig. S1). Alternative splice sites are usually weaker than constitutive sites, and alternative exons or introns often have an atypical composition [25, 26]. The ΔSP-PDYN mRNA lacking the first methionine codon (Met1) may be translated from the second methionine codon (M14) giving rise to a 241 aa protein in which the N-terminal 13 amino acids of the signal peptide sequence are absent (Fig. 1E).
The second novel PDYN splice variant in which sequences coding for SP and NLS (nuclear localization signal) are absent (ΔSP/NLS-PDYN) is 560 nt long. It resembles ΔSP-PDYN but is shorter due to the presence of two alternative introns in exon 4: 307 nt including the α-neoendorphin, dynorphin A (Dyn A) and 5´-part of dynorphin B (Dyn B) encoding sequences and 31 nt in the 3´-untranslated region (Fig. 1D). In ΔSP/NLS-PDYN the alternative intron junction in exon 4 is non-canonical (see Fig. S1). This mRNA may be translated into a 127 amino acid protein deficient in α-neoendorphin and dynorphin A sequences (Fig. 1E).
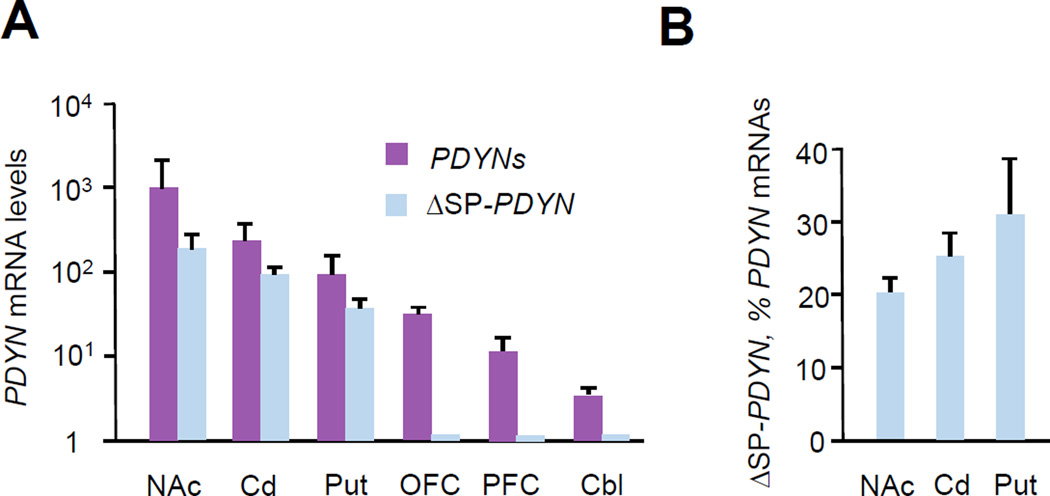
3.2. ΔSP-PDYN transcript expression is brain region-specific
For analysis of total PDYN transcripts, PCR primers targeted to the exon 3–4 junction present in the most dominant PDYN isoforms, were used (Fig. S1). To selectively detect ΔSP-PDYN transcript, forward PCR primer was designed to span the novel exon 1–3 junction (Fig. S1). The cDNA for this experiment was synthesised using a 3´-primer complementary to the exon 4 sequence that was spliced out as an intron in ΔSP/NLS-PDYN mRNA, thus allowing detection of ΔSP- but not of ΔSP/NLS-PDYN mRNA (Fig S1). The levels of ΔSP-PDYN mRNA were high in the human nucleus accumbens, caudate nucleus and putamen, while negligible in frontal lobes (orbitofrontal cortex and dorsa-lateral prefrontal cortex) and the hindbrain (cerebellum) despite robust PDYN mRNA expression (Fig. 2A). In the striatal subregions, ΔSP-PDYN mRNA constituted a substantial fraction, 20– 30% of the total PDYN mRNAs containing an exon 3–4 junction, including FL-PDYN mRNAs (Fig. 2B), and strongly correlated (R = 0.98; R, Pearson correlation coefficient) with the latter in all tree striatal subregions.
FIGURE 2. The ΔSP-PDYN expression profile in the human brain.
A. The expression levels of mRNAs for ΔSP-PDYN versus dominant PDYN transcripts. Levels were normalized to µg of total RNA. Data are shown as mean ± SEM for nucleus accumbens (NAc); caudate nucleus (Cd); putamen (Put); orbitofrontal cortex (OFC); dorsolateral prefrontal cortex (PFC); cerebellum (Cbl). For Put n = 5 human subject; for other brain areas n = 4 subjects.
B. ΔSP-PDYN mRNA as percentage of dominant PDYN mRNA levels, which have a canonical junction of exons 3–4 and include FL1-, FL2- and ΔSP-PDYN mRNAs. Data is shown as mean ± SEM.
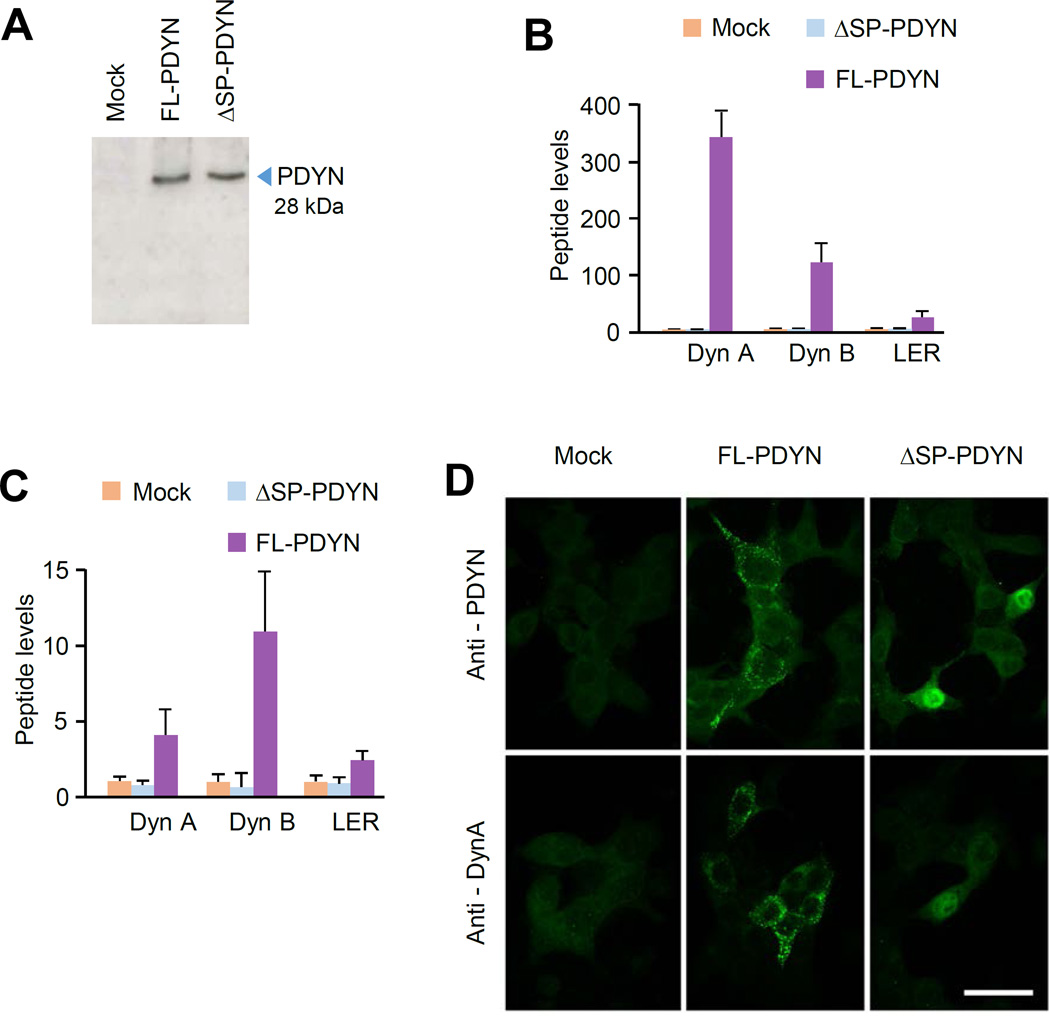
3.3. Unprocessed ΔSP-PDYN is targeted to the nuclei in model cell line
Initiation of ΔSP-PDYN translation from Met14, processing of this protein to opioid peptides and its subcellular localization was studied using the rat insulinoma cell line RINm5F as a model system. These cells do not express endogenous Pdyn but may process ectopically expressed neuropeptide precursor proteins to mature neuropeptides (36). RINm5F cells were transfected with pCMV-ΔSP-PDYN or pCMV-FL-PDYN plasmids, or pCMV vector resulting in synthesis of proteins with a very similar molecular weight about 28 kDa that corresponded to the calculated size of the PDYN molecule (Fig. 3A). FL-PDYN protein was processed to Dyn A, Dyn B and LER (leucine-enkephalin-arginine) that were identified in cell extracts and culture media by RIA (Fig. 3 B, C). Whereas, no opioid peptides were generated in the cells expressing the ΔSP-PDYN protein (Fig. 3B).
FIGURE 3. Processing and subcellular localization of ectopically expressed ΔSP- and FL-PDYN in RINm-5F cells.
A. ΔSP- and FL-PDYN proteins analyzed by western blotting with anti-PDYN CTF antibody in RINm-5F transfected cells.
B, C. The levels of prodynorphin derived dynorphin A (Dyn A), dynorphin B (Dyn B) and Leu-enkephalin-Arg (LER) in the RINm-5F transfected cells (B) and in the culture media (C) analyzed by RIA. The cells were transfected with pCMV vector (mock), pCMV-ΔSP-PDYN or pCMV-FL-PDYN. Data were normalized to the levels in mock transfected cells and are shown as mean ± SEM; n = 3 experiments.
D. Subcellular localization of ΔSP-PDYN protein in RINm-5F cells observed under confocal fluorescence microscopy. Transfected cultures were immunolabeled with antibodies against PDYN CTF or dynorphin A. Images were taken with × 100 objective and × 5 electron zoom factor. Scale bar, 30 µm.
The subcellular localization of ΔSP- and FL-PDYN proteins was visualized using confocal microscopy of transfected RINm-5F cells after cell labeling with antibody against the PDYN C-terminal fragment (CTF) that is present in both proteins, and with anti-Dyn A antibody directed against a mature opioid peptide. In transfected cells, FL-PDYN immunoreactivity identified with both antibody showed similar labeling patterns; immunoreactive PDYN formed punctate clusters characteristic for localization in the endoplasmic reticulum and Golgi lamellae (Fig. 3D). In contrast, the ΔSP-PDYN isoform was densely packed in the cell nucleus. Weak or no ΔSP-PDYN produced signal was seen with anti-Dyn A antibody. No labeled cells were observed in mock-transfected cells, with a prebleed IgG fraction used for immunostaining, or when antiserum was preincubated with 10 nM of the human CTF. Preincubation of the antiserum with rat CTF did not affect the labeling. Thus, ΔSP-PDYN ectopically expressed in RINm-5F cells is targeted to the cell nucleus where it is not processed to opioid peptides.
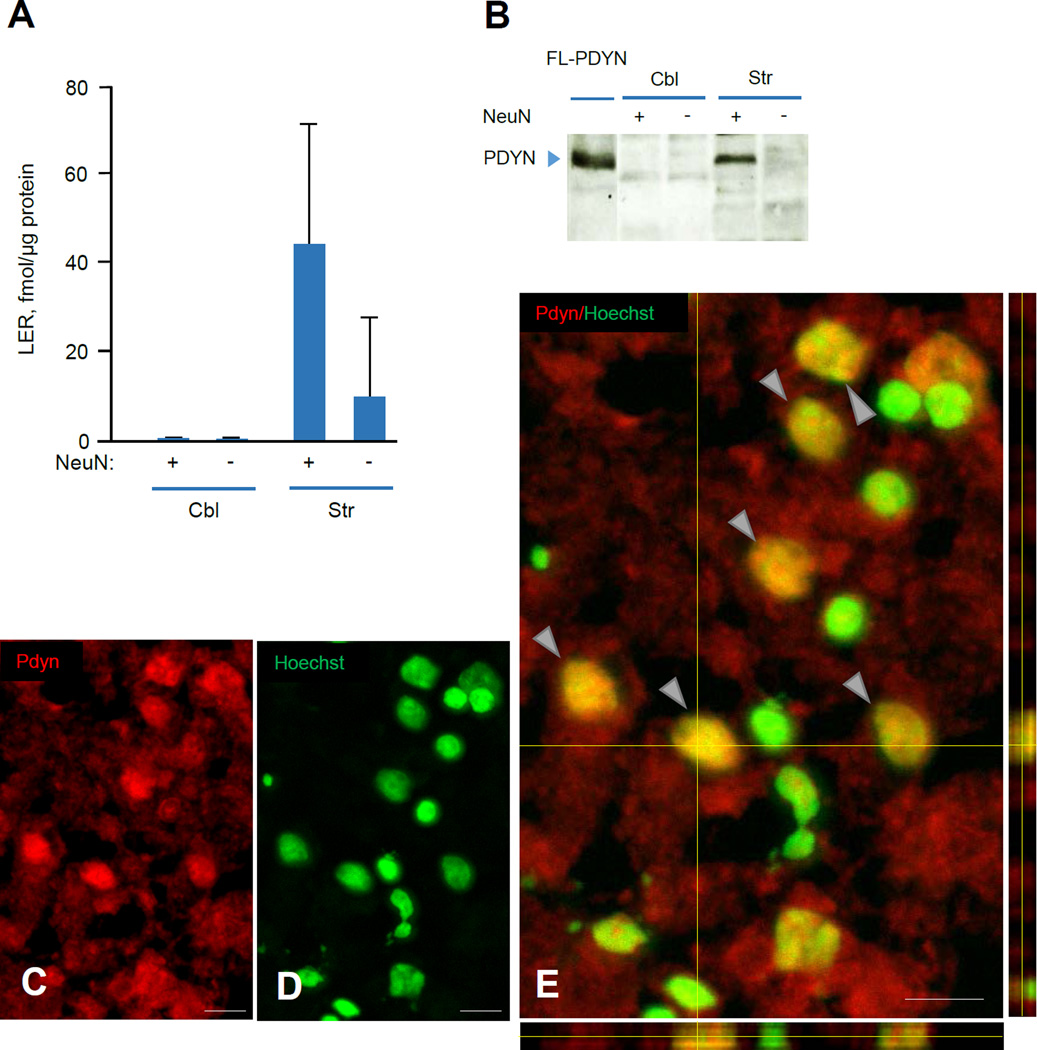
3.4. PDYN is localized in neuronal nuclei in human striatum
The NLS score for human PDYN predicts a high probability of nuclear localization for ΔSP-PDYN (Table S1). We therefore next examined whether PDYN is localized in the cell nuclei in human striatum (caudate nucleus), an area with high ΔSP-PDYN expression levels. Neuronal nuclei were isolated from frozen human tissues using fluorescence-activated nuclei sorting. Neuronal nuclei were labeled with antibody against NeuN, a neuronal marker. This antibody recognizes nuclei of mature neurons, but not those of neural progenitors and glial cells [27]. Human cerebellar tissue was used as negative control; PDYN expression is low in this tissue and no ΔSP-PDYN mRNA was detected (Fig. 2A).
PDYN protein in isolated neuronal and non-neuronal nuclei, was analyzed by RIA of LER, a PDYN marker [28] after treatment of nuclear extracts with trypsin, and by western blotting (Fig. 4A, B). Digestion with trypsin liberates LER from the precursor protein [7]. The levels of liberated LER in the fraction of neuronal nuclei were 4-fold higher than that in non-neuronal nuclei isolated from human striatum (Fig. 4A). The amount of the liberated LER in both neuronal and non-neuronal nuclei prepared from cerebellum was below the detection limit (Fig. 4A). Western blot analysis of striatal neuronal nuclei identified immunoreactive PDYN protein as a single band with molecular size of 28 kDa that corresponded to unprocessed FL- and ΔSP-PDYN proteins. No PDYN immunoreactivity was evident in neuronal and non-neuronal nuclei from cerebellum and non-neuronal nuclei from striatum (Fig. 4B).
FIGURE 4. ΔSP-PDYN is targeted to the neuronal nuclei in human striatum.
A. The levels of PDYN derived peptide LER in neuronal and non-neuronal nuclei isolated by FANS from human striatal (Str) and cerebellar (Cbl) tissue samples. LER, a PDYN marker was analyzed by RIA. The amount of the liberated LER in both neuronal and non-neuronal nuclei prepared from cerebellum was below the detection limit. Equal amount of neuronal and non-neuronal nuclei (106 nuclei) isolated from each tissue sample were analyzed. Data is shown as mean ± SEM, n = 2 subjects. Data was normalized to 1 µg of protein.
B. PDYN proteins analyzed by western blotting in neuronal and non-neuronal nuclei isolated by FANS from human striatal (Str) and cerebellar (Cbl) tissue samples. Equal amount of neuronal and non-neuronal nuclei (106 nuclei) isolated from each tissue sample were analyzed. Ectopically expressed in RINm-5F cells FL-PDYN was used as a positive control, producing a single band with molecular size of 28 kDa that corresponded to unprocessed PDYN protein.
C–E. PDYN immunoreactivity was detected in the neuronal cell nuclei and cytoplasm in the human caudate nucleus. The brain sections were stained for ΔSP-PDYN protein with anti-PDYN CTF antibody (red) (C) and for nuclei with Hoechst (green) (D). Representative 3D confocal reconstruction projections demonstrates double labeling (yellow) of neuronal nuclei (arrows) (E) indicating nuclear localization of PDYN isoform. Confocal images of Z-stacks of 24 images acquired with a depth interval of 0.5 µm of the human caudate nucleus. Analysis shows that PDYN is present in the nucleus. The center image is the X-Y view, the images below and right are orthogonal projections in the X-Z and Y-Z planes (cross section at the yellow line). n = 4 subjects. Images were taken with 20 × /0.95W XLUMPlanFI water objective. Scale bar, 20 µm.
Immunofluorescent labeling of human caudate nucleus sections of with anti-h-PDYN CTF antibody followed by confocal 3D reconstruction identified immunoreactive PDYN located in the neuronal cell nuclei and cytoplasm (Fig. 4C–E). The confocal microscope orthogonal projections on the acquired images demonstrated that PDYN immunoreactivity (red) was co-localized with the Hoechst (green) nuclear staining (Fig. 4E). No PDYN was seen when anti-PDYN CTF antibody was omitted (data not shown), replaced with rabbit IgG or was inactivated by antigen peptide adsorption (Fig. S2). Taken together molecular and immuno-labeling experiments strongly support the hypothesis on localization of PDYN protein in neuronal nuclei in human striatum.
4. DISCUSSION
The present study identified two novel alternatively spliced ΔSP- and ΔSP/NLS-PDYN mRNAs that both lack exon 2 and a fragment of exon 3. In the striatum, the expression levels of ΔSP-PDYN mRNA were substantial constituting up to 30% of total levels of PDYN mRNA molecules, while in the dl-PFC, orbitofrontal cortex and cerebellum were negligible. This marked difference suggests a specific function of ΔSP-PDYN in the striatum.
Novel PDYN mRNAs may give rise to PDYN proteins with the N-terminally truncated signal peptide. Analysis of the amino acid sequence the cNLS Mapper predicted the existence of a bipartite NLS in the opioid peptide domain of the ΔSP-PDYN. The PDYN NLS (179RKYPKRSSEVAGEGDGDSMGHEDLYKRYGG209) shows a high score and consists of a fragment joining α-neoendorphin and Dyn A sequences, four C-terminal and three N-terminal aa of these peptides, respectively, and two adjacent to them pairs of basic amino acids representing processing sites. The probability of ΔSP-PDYN to be located in the cell nuclei predicted by the PSORT II program is 39%, while that of FL-PDYN is 11%. The NLS encoding sequence is spliced out in the ΔSP/NLS-PDYN mRNA. Due to the signal peptide truncation, the cryptic bipartite NLS may become functional and target the ΔSP-PDYN protein into the cell nuclei. Indeed, ΔSP-PDYN was located in the cell nuclei when ectopically expressed in a model cell line, and in neuronal nuclei in human striatum. A functionality of the predicted NLS was validated in our previous study (see Fig. 7 in [14]). Specifically, Sp2-PDYN mRNA generated by alternative splicing and T1- and T2-PDYN mRNAs transcriptionally initiated within the coding part of exon 4 were identified in the human brain [14]. Sp2 and T1 mRNAs gave rise to the 12 kDa T1-PDYN protein translated from Met146 (Fig. 1A). T1-protein lacks a signal peptide but retains the predicted bipartite NLS and was located in the cell nucleus when ectopically expressed in COS1 cells. In contrast, the T2-PDYN protein translated from the Met198 codon and lacking the predicted NLS (Fig. 1A) resides in the cytoplasm [14]. Targeting of T1- but not T2-protein to the cell nuclei demonstrated functional efficacy of the predicted bipartite NLS.
Consistently with model cellular data, a nuclear PDYN isoform was identified by western blot and radioimmunoassay in neuronal nuclei isolated from human striatum using fluorescence-activated nuclei sorting. Immunofluorescence staining and confocal microscopy of the human caudate nucleus sections confirmed nuclear localization of the PDYN isoform. The molecular size of PDYN molecules in the neuronal nuclei was similar to that of FL-PDYN, suggesting that the nuclear PDYN immunoreactivity visualized in the human brain originates from ΔSP-PDYN, but not the shorter T1-protein.
Several previous studies support the hypothesis on a nuclear localization and nuclear functions of neuropeptide precursor proteins. An early bioinformatics analysis suggested that the opioid precursors PDYN and PENK may function as DNA-binding proteins on the basis of the presence of zinc-finger and helix-loop-helix domains, together with the similarity with hunchback, lil-1, tal and twist transcription factors [29]. Similar segments include enkephalin sequences which are parts of heptapeptide repeats characteristic of an alpha-helical coiled-coil structure distinctive of an amphipathic helix-loop-helix DNA-binding motif. The opioid precursors also have cystein-rich regions characteristic of zinc-finger domains in transcription factors. The cysteines forming zinc-finger domains are perfectly conserved in opioid peptide precursors.
Consistently with our human findings, our prior electron microscopic study identified immunoreactive Pdyn protein located on the nuclear membrane, and dynorphin A on the nuclear pore and inside the neuronal nuclei in rat nucleus accumbens [19]. In somata of the same neurons, Pdyn was located on the smooth endoplasmic reticulum and Golgi lamellae. Nuclear localization of another neuropeptide precursor protein, proenkephalin (PENK) in embryonic fibroblasts and in myoblasts in vitro was demonstrated by Spruce and co-workers [30]. Multiple antigenic domains of the endogenous nuclear PENK were unmasked in cells undergoing differentiation or entering growth arrest. PENK ectopically expressed in cell lines was localized exclusively in the cytoplasm, however, targeted to the nucleus when mutated at the first ATG codon, or when devoid of its signal peptide sequence. The authors speculated that alternate initiation of transcription, alternate splicing or initiation of translation from upstream in-frame CUG codons could lead to a PENK translation product devoid of a functional signal peptide, which is capable of nuclear translocation. The endogenous nuclear PENK may be physically associated with two transcription factors, p53 and the RelA(p65) subunit of NF-κB, and with the co-repressor histone deacetylase suggesting participation of this protein in transcriptional repression complexes involved in apoptosis and cell survival [31].
In conclusion, the concept of nuclear localization and functions of neuropeptide precursors complementing to the general view on these proteins as molecules destined to be cleaved into short neuropeptides, gains experimental support. A functional role of ΔSP-PDYN should be elucidated, and its molecular partners and targets identified. This is especially important in the light of the recent identification of dominant pathogenic PDYN missense mutations that cause profound neurodegeneration in the human brain [23, 32–34]. These mutations are clustered in the opioid domain overlapping with NLS sequence, and may affect the biogenesis and trafficking of this protein that in turn may contribute to neuropathological changes. In general terms, dysregulation of complex PDYN transcriptional and translational mechanisms affecting a generation of canonical and nuclear PDYN proteins may define human pathological conditions including chronic pain and neurodegenerative diseases, in which a pathogenic role of PDYN is firmly established but which mechanisms that are not mediated through opioid receptors have not yet been elucidated [35, 36].
Supplementary Material
HIGHLIGHTS.
Two novel splicing variants of human PDYN mRNA have been identified.
A novel ΔSP-PDYN splicing variant is mostly expressed in human striatum.
ΔSP-PDYN protein lacks a fragment of signal peptide (SP).
ΔSP-PDYN is not processed to mature dynorphins but targeted to the cell nuclei in a model cellular system.
Unprocessed, likely ΔSP-PDYN is localized in neuronal nuclei in human striatum.
Acknowledgments
This work was supported by grants from the Swedish Science Research Council and Swedish Council for Working Life and Social Research to GB. We like to thank Ms. Agnieszka Limiszewska for her technical assistance with the immunofluorescence processing of human samples. For the brain tissues from the University of Mississippi Medical Center, the authors acknowledge the invaluable contributions made by the families consenting to donate brain tissue and be interviewed. We also acknowledge the support of the Cuyahoga County Medical Examiner’s Office, Cleveland, Ohio, and the expert assistance of Drs. James C. Overholser and George Jurjus, and of Lesa Dieter and Gouri Mahajan in acquiring written consent, interviewing the family members, and in collecting the tissues. This work was funded in part by the Postmortem Brain Core of the Center for Psychiatric Neuroscience, funded through an IDeA COBRE award to CAS from The National Institute of General Medical Sciences (P30 GM103328).
Abbreviations
- Dyn A
dynorphin A
- Dyn B
dynorphin B
- CTF
C-terminal fragment
- FANS
fluorescence-activated nuclei sorting
- LER
leucine-enkephalin-arginine
- NLS
nuclear localization signal
- PDYN
prodynorphin
- PENK
proenkephalin
- RIA
radioimmunoassay
- SP
signal peptide
- SS
splice site
- TSS
transcription start site
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article
Author contributions: OK, IB, TY and GB designed research; OK, IB, HW, TY, GG, OD, DSV, KA, HD, MA, JM, AFS, GR, and CAS performed research; OK, IB, TY, GG, OD, DSV, KA, HD, MA, JM, AFS, OKr and GB, analysed and discussed data; and OK, IB, TY and GB wrote the paper.
REFERENCES
- 1.Ji RR, Befort K, Brenner GJ, Woolf CJ. ERK MAP kinase activation in superficial spinal cord neurons induces prodynorphin and NK-1 upregulation and contributes to persistent inflammatory pain hypersensitivity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:478–485. doi: 10.1523/JNEUROSCI.22-02-00478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vries TJ, Shippenberg TS. Neural systems underlying opiate addiction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:3321–3325. doi: 10.1523/JNEUROSCI.22-09-03321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreek MJ, LaForge KS, Butelman E. Pharmacotherapy of addictions. Nature reviews. Drug discovery. 2002;1:710–726. doi: 10.1038/nrd897. [DOI] [PubMed] [Google Scholar]
- 4.Cenci MA, Lee CS, Bjorklund A. L-DOPA-induced dyskinesia in the rat is associated with striatal overexpression of prodynorphin- and glutamic acid decarboxylase mRNA. The European journal of neuroscience. 1998;10:2694–2706. [PubMed] [Google Scholar]
- 5.Naranjo JR, Mellstrom B, Achaval M, Sassone-Corsi P. Molecular pathways of pain: Fos/Jun-mediated activation of a noncanonical AP-1 site in the prodynorphin gene. Neuron. 1991;6:607–617. doi: 10.1016/0896-6273(91)90063-6. [DOI] [PubMed] [Google Scholar]
- 6.Tan-No K, Takahashi H, Nakagawasai O, Niijima F, Sato T, Satoh S, Sakurada S, Marinova Z, Yakovleva T, Bakalkin G, Terenius L, Tadano T. Pronociceptive role of dynorphins in uninjured animals: N-ethylmaleimide-induced nociceptive behavior mediated through inhibition of dynorphin degradation. Pain. 2005;113:301–309. doi: 10.1016/j.pain.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Yakovleva T, Bazov I, Cebers G, Marinova Z, Hara Y, Ahmed A, Vlaskovska M, Johansson B, Hochgeschwender U, Singh IN, Bruce-Keller AJ, Hurd YL, Kaneko T, Terenius L, Ekstrom TJ, Hauser KF, Pickel VM, Bakalkin G. Prodynorphin storage and processing in axon terminals and dendrites. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:2124–2126. doi: 10.1096/fj.06-6174fje. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nature reviews. Neuroscience. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 9.Chavkin C, James IF, Goldstein A. Dynorphin Is a Specific Endogenous Ligand of the Kappa-Opioid Receptor. Science. 1982;215:413–415. doi: 10.1126/science.6120570. [DOI] [PubMed] [Google Scholar]
- 10.Mansour A, Fox CA, Meng F, Akil H, Watson SJ. Kappa 1 receptor mRNA distribution in the rat CNS: comparison to kappa receptor binding and prodynorphin mRNA. Molecular and cellular neurosciences. 1994;5:124–144. doi: 10.1006/mcne.1994.1015. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin JP, Marton-Popovici M, Chavkin C. Kappa opioid receptor antagonism and prodynorphin gene disruption block stress-induced behavioral responses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:5674–5683. doi: 10.1523/JNEUROSCI.23-13-05674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horikawa S, Takai T, Toyosato M, Takahashi H, Noda M, Kakidani H, Kubo T, Hirose T, Inayama S, Hayashida H, et al. Isolation and structural organization of the human preproenkephalin B gene. Nature. 1983;306:611–614. doi: 10.1038/306611a0. [DOI] [PubMed] [Google Scholar]
- 13.Telkov M, Geijer T, Terenius L. Human prodynorphin gene generates several tissue-specific transcripts. Brain research. 1998;804:284–295. doi: 10.1016/s0006-8993(98)00706-9. [DOI] [PubMed] [Google Scholar]
- 14.Nikoshkov A, Hurd YL, Yakovleva T, Bazov I, Marinova Z, Cebers G, Pasikova N, Gharibyan A, Terenius L, Bakalkin G. Prodynorphin transcripts and proteins differentially expressed and regulated in the adult human brain. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:1543–1545. doi: 10.1096/fj.05-3743fje. [DOI] [PubMed] [Google Scholar]
- 15.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Dammer EB, Duong DM, Diner I, Gearing M, Feng Y, Lah JJ, Levey AI, Seyfried NT. Neuron enriched nuclear proteome isolated from human brain. Journal of proteome research. 2013;12:3193–3206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wongjitrat C, Horthongkham N, Sutthent R, Srisurapanon S. Comparison of Droplet Digital PCR and Real Time PCR Method for HBV DNA Quantification. Journal of the Medical Association of Thailand = Chotmaihet thangphaet. 2015;98(Suppl 9):S140–S145. [PubMed] [Google Scholar]
- 19.Hara Y, Yakovleva T, Bakalkin G, Pickel VM. Dopamine D1 receptors have subcellular distributions conducive to interactions with prodynorphin in the rat nucleus accumbens shell. Synapse. 2006;60:1–19. doi: 10.1002/syn.20273. [DOI] [PubMed] [Google Scholar]
- 20.Christensson-Nylander I, Nyberg F, Ragnarsson U, Terenius L. A general procedure for analysis of proenkephalin B derived opioid peptides. Regulatory peptides. 1985;11:65–76. doi: 10.1016/0167-0115(85)90032-1. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen XV, Masse J, Kumar A, Vijitruth R, Kulik C, Liu M, Choi DY, Foster TC, Usynin I, Bakalkin G, Bing G. Prodynorphin knockout mice demonstrate diminished age-associated impairment in spatial water maze performance. Behavioural brain research. 2005;161:254–262. doi: 10.1016/j.bbr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Merg F, Filliol D, Usynin I, Bazov I, Bark N, Hurd YL, Yakovleva T, Kieffer BL, Bakalkin G. Big dynorphin as a putative endogenous ligand for the kappa-opioid receptor. Journal of neurochemistry. 2006;97:292–301. doi: 10.1111/j.1471-4159.2006.03732.x. [DOI] [PubMed] [Google Scholar]
- 23.Bakalkin G, Watanabe H, Jezierska J, Depoorter C, Verschuuren-Bemelmans C, Bazov I, Artemenko KA, Yakovleva T, Dooijes D, Van de Warrenburg BP, Zubarev RA, Kremer B, Knapp PE, Hauser KF, Wijmenga C, Nyberg F, Sinke RJ, Verbeek DS. Prodynorphin mutations cause the neurodegenerative disorder spinocerebellar ataxia type 23. American journal of human genetics. 2010;87:593–603. doi: 10.1016/j.ajhg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padgett RA. New connections between splicing and human disease. Trends in genetics : TIG. 2012;28:147–154. doi: 10.1016/j.tig.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E, Goren A, Ast G. Alternative splicing: current perspectives. BioEssays : news and reviews in molecular, cellular and developmental biology. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- 26.Thanaraj TA, Stamm S. Prediction and statistical analysis of alternatively spliced exons. Progress in molecular and subcellular biology. 2003;31:1–31. doi: 10.1007/978-3-662-09728-1_1. [DOI] [PubMed] [Google Scholar]
- 27.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 28.Nyberg F, Nylander I, Terenius L. Enkephalin-containing polypeptides in human cerebrospinal fluid. Brain research. 1986;371:278–286. doi: 10.1016/0006-8993(86)90363-x. [DOI] [PubMed] [Google Scholar]
- 29.Bakalkin G, Ponomariev D, Sarkisyan RA, Terenius L. Sequence similarity between opioid peptide precursors and DNA-binding proteins. FEBS letters. 1991;282:175–177. doi: 10.1016/0014-5793(91)80471-e. [DOI] [PubMed] [Google Scholar]
- 30.Bottger A, Spruce BA. Proenkephalin is a nuclear protein responsive to growth arrest and differentiation signals. The Journal of cell biology. 1995;130:1251–1262. doi: 10.1083/jcb.130.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McTavish N, Copeland LA, Saville MK, Perkins ND, Spruce BA. Proenkephalin assists stress-activated apoptosis through transcriptional repression of NF-kappaB- and p53-regulated gene targets. Cell death and differentiation. 2007;14:1700–1710. doi: 10.1038/sj.cdd.4402172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smeets CJ, Jezierska J, Watanabe H, Duarri A, Fokkens MR, Meijer M, Zhou Q, Yakovleva T, Boddeke E, den Dunnen W, van Deursen J, Bakalkin G, Kampinga HH, van de Sluis B, Verbeek DS. Elevated mutant dynorphin A causes Purkinje cell loss and motor dysfunction in spinocerebellar ataxia type 23. Brain : a journal of neurology. 2015;138:2537–2552. doi: 10.1093/brain/awv195. [DOI] [PubMed] [Google Scholar]
- 33.Jezierska J, Stevanin G, Watanabe H, Fokkens MR, Zagnoli F, Kok J, Goas JY, Bertrand P, Robin C, Brice A, Bakalkin G, Durr A, Verbeek DS. Identification and characterization of novel PDYN mutations in dominant cerebellar ataxia cases. Journal of neurology. 2013;260:1807–1812. doi: 10.1007/s00415-013-6882-6. [DOI] [PubMed] [Google Scholar]
- 34.Saigoh K, Mitsui J, Hirano M, Shioyama M, Samukawa M, Ichikawa Y, Goto J, Tsuji S, Kusunoki S. The first Japanese familial case of spinocerebellar ataxia 23 with a novel mutation in the PDYN gene. Parkinsonism & related disorders. 2015;21:332–334. doi: 10.1016/j.parkreldis.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Lai J, Ossipov MH, Vanderah TW, Malan TP, Jr, Porreca F. Neuropathic pain: the paradox of dynorphin. Molecular interventions. 2001;1:160–167. [PubMed] [Google Scholar]
- 36.Yakovleva T, Marinova Z, Kuzmin A, Seidah NG, Haroutunian V, Terenius L, Bakalkin G. Dysregulation of dynorphins in Alzheimer disease. Neurobiology of aging. 2007;28:1700–1708. doi: 10.1016/j.neurobiolaging.2006.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.