Abstract
Interstrand crosslinks (ICLs) are covalent lesions formed by cisplatin. The mechanism for the processing and removal of ICLs by DNA repair proteins involves nucleotide excision repair (NER), homologous recombination (HR) and fanconi anemia (FA) pathways. In this report, we monitored the processing of a flanking uracil adjacent to a cisplatin ICL by the proteins involved in the base excision repair (BER) pathway. Using a combination of extracts, purified proteins, inhibitors, functional assays and cell culture studies, we determined the specific BER proteins required for processing a DNA substrate with a uracil adjacent to a cisplatin ICL. Uracil DNA glycosylase (UNG) is the primary glycosylase responsible for the removal of uracils adjacent to cisplatin ICLs, whereas other uracil glycosylases can process uracils in the context of undamaged DNA. Repair of the uracil adjacent to cisplatin ICLs proceeds through the classical BER pathway, highlighting the importance of specific proteins in this redundant pathway. Removal of uracil is followed by the generation of an abasic site and subsequent cleavage by AP endonuclease 1 (APE1). Inhibition of either the repair or redox domain of APE1 gives rise to cisplatin resistance. Inhibition of the lyase domain of Polymerase β (Polβ) does not influence cisplatin cytotoxicity. In addition, lack of XRCC1 leads to increased DNA damage and results in increased cisplatin cytotoxicity. Our results indicate that BER activation at cisplatin ICLs influences crosslink repair and modulates cisplatin cytotoxicity via specific UNG, APE1 and Polβ polymerase functions.
Keywords: BER, cisplatin, ICL, resistance
Introduction
Cisplatin is a commonly used chemotherapeutic for the treatment of a wide variety of malignancies [1, 2]. Many cancers, however, develop resistance to treatment which limits clinical success [3]. Cisplatin forms several types of lesions on the DNA, and the dynamics of the formation of these lesions, and their subsequent removal, is crucial for understanding the mechanisms underlying drug resistance. The majority of the lesions generated by cisplatin are intrastrand adducts, formed between adjoining purines on the same strand of DNA. Nucleotide excision repair (NER) is the primary pathway responsible for the removal of the intrastrand adducts formed by the drug. In addition, cisplatin also forms covalent interstrand crosslinks (ICLs) between guanines on opposite strands of DNA. However, it has been postulated that cells utilize multiple DNA repair pathways for the removal of cisplatin ICLs. The formation of ICLs blocks fundamental cellular processes like DNA replication and RNA transcription, influencing cell survival. It is important to note that repair of ICLs formed by cisplatin is likely different from the repair of the ICLs formed by other crosslinking agents like nitrogen mustards, psoralen and mitomycin C (MMC), and that the same mechanisms cannot be generalized for the repair of all ICLs. Unique structural distortions created by ICLs formed by various drugs influence the choice of specific repair pathways as well as the physiological rates of their removal. Crystallographic data and NMR solution structures of a cisplatin ICL have provided key insights on the degree of distortion created by a cisplatin ICL DNA lesion [4]. A cisplatin ICL produces significant distortion of the double helical DNA compared to other crosslinking agents. Due to the crosslink the DNA is unwound by 70–80° and bent towards the minor groove. This DNA distortion forces the cytosines adjacent to the crosslinked guanines from the double helix, exposing them to the nuclear environment.
We have recently demonstrated that these flipped out cytosines have a propensity to undergo preferential oxidative deamination, creating a uracil adjacent to the platinum crosslink [5]. By using purified proteins, we further showed that these newly formed uracils adjacent to the ICL are processed by proteins involved in the BER pathway. Purified uracil DNA glycosylase (UDG) was able remove the uracil base, the abasic site was cleaved by apurinic/apyrimidinic endonuclease 1 (APE1), and subsequently DNA polymerase β (Polβ) filled in the gap. However, there are four other DNA glycosylases present in human cells that have the ability to excise uracil from DNA [6]. Our studies have shown a deficiency in UNG, the primary uracil DNA glycosylase in humans, gives rise to cisplatin resistance [5]. In addition to UNG, single-strand selective mono-functional uracil DNA glycosylase (SMUG1) also has the ability to excise uracil from DNA. In some scenarios, other glycosylases such as methyl-CpG domain protein 4 (MBD4) and thymine DNA glycosylase (TDG), which are involved in the removal of thymine bases generated by deamination of 5-methylcytosine, may also participate in the removal of uracil.
After the removal of uracil by a glycosylase, an abasic site is formed and is recognized by APE1. APE1 is a multifunctional protein that has a wide variety of cellular functions in response to oxidative stress. APE1 facilitates DNA repair by cleaving the phosphodiester bond near the 5′ end of the apurinic/apyrimidinic (AP) site, which can be further processed by Polβ. APE1 contains both a redox domain responsible for transcriptional regulation of gene expression as well as the DNA repair domain required for AP site cleavage in the BER pathway. Studies show both repair and redox functions of APE1 are required for cell growth and survival [7]. Increased expression of APE1 in the A549 lung adenocarcinoma cell line has been associated with cisplatin resistance [8]. However, in the case of ovarian cancer, no significant difference was found between APE1 expression in platinum sensitive and platinum refractory cancers [9]. Targeting APE1 has been shown to mediate sensitivity to DNA damaging agents such as BCNU and MMC [10, 11]. Accumulation of abasic sites and subsequent generation of DNA double strand breaks (DSBs) are likely responsible for this cellular sensitivity. On the other hand, previous studies in our lab have shown that pretreatment with methoxyamine (MX), which binds to abasic sites and blocks the access of APE1, enhanced repair of cisplatin ICLs and induced cisplatin resistance in human cancer cells [5]. Structural differences in chemotherapeutic agents could be the basis for the activation of different repair pathways. Cisplatin and MMC both form ICLs on DNA, however, the resulting structural changes are unique to each DNA damaging agent.
Polβ is a 39 kDa protein with a well characterized function in the BER pathway and contains two catalytically active units. The polymerase activity is located in the 31-kDa C-terminal polymerase domain, while the 8-kDa lyase domain (on the N-terminus) is responsible for the dRP lyase removal activity. Our lab has demonstrated that the polymerase domain of Polβ is required for cisplatin sensitivity [12]. DNA synthesis by Polβ near cisplatin ICLs is prone to errors, leading to the subsequent recruitment of mismatch repair (MMR) proteins. We have shown that this non-productive repair by BER and MMR proteins inhibits repair of cisplatin ICLs leading to increased cellular sensitivity [12]. Moreover, Polβ D256A variant deficient in polymerase activity was sensitive to cisplatin treatment, suggesting the requirement of Polβ mediated DNA synthesis for mediating cisplatin sensitivity. Misincorporation by Polβ gives rise to DNA mismatches near ICL sites, and we have previously shown that MMR protein binding is dependent on DNA synthesis by Polβ [12]. However, the role of dRP lyase activity of Polβ in this mechanism has not been studied, and it is unclear whether the proposed cisplatin ICL processing mechanism occurs via short patch (SP-BER) or long patch BER (LP-BER) pathways. Inhibition of the dRP lyase activity of Polβ has been known to favor the long patch BER pathway [13].
X-ray repair cross-complementing gene 1 (XRCC1) is a molecular scaffold protein which plays a critical role in BER and single strand break repair pathways [14]. XRCC1 is recruited to single strand breaks by Poly-ADP ribose polymerase 1 (PARP1) where it provides a platform for the interaction of numerous BER proteins. Lack of XRCC1 can lead to accumulation of single strand breaks, resulting in replication stress and potential formation of DNA DSBs [15]. Studies show that down-regulation of XRCC1 leads to increased sensitivity to cisplatin in the hepatocellular carcinoma cell line, HepG2 [16]. In these cells, reduced DNA repair capacity and increased reactive oxygen species (ROS) production have been associated with enhanced cellular sensitivity to cisplatin. Moreover, increased XRCC1 expression has been associated with cisplatin resistance in gastric cancer cells [17, 18]. The thioredoxin like protein-1 (TXNL-1) mediated downregulation of XRCC1 has been shown to be a novel mechanism for cisplatin sensitivity in gastric cancer cells [17]. XRCC1 deficiency has also been shown to sensitize ovarian cancer cells to cisplatin and this sensitivity is further potentiated by ATR inhibition [19].
In our previous studies, we have shown that the extrahelical cytosines adjacent to cisplatin ICL sites are more prone to undergo oxidative deamination to uracil. The resulting uracils activate the BER pathway. We observed that the polymerase activity of Polβ is essential for maintaining cisplatin sensitivity, and error prone DNA synthesis by Polβ leads to misincorporation of nucleotides adjacent to cisplatin ICL sites [5]. Our results showed that Polβ misincorporates dATP at ~1.6 fold higher frequency as compared to the correct nucleotide base dCTP opposite guanine near the ICL site [5]. The generation of these mismatches leads to increased recruitment of MMR proteins and a subsequent role of MMR in blocking productive cisplatin specific ICL repair [12, 20]. The binding of BER and MMR proteins results in a futile cycle at the ICLs by blocking the productive ICL DNA repair pathways, resulting in the persistence of DNA ICLs which give rise to enhanced cisplatin cytotoxicity. Unlike the changes observed in cisplatin ICL DNA repair following BER or MMR loss, there are no differences in cisplatin intrastrand adduct DNA repair [5, 12]. Despite cisplatin intrastrand adducts playing a role in the efficacy of the drug, when it pertains to the BER and MMR pathways, the mediation of cisplatin efficacy is via ICL non-productive processing.
In this manuscript, the contribution of BER proteins in mediating cisplatin sensitivity and ICL processing was further assessed. UNG was found to be required to mediate cisplatin sensitivity while other uracil DNA glycosylases have no effect on cisplatin efficacy. In addition, we studied the individual roles of the DNA repair and redox domains of APE1 in mediating cisplatin sensitivity in human breast cancer cells. The data indicates that both activities of APE1 are important for mediating cisplatin sensitivity. It was also established that the 5′ dRP lyase activity of Polβ plays no role in cisplatin efficacy, while XRCC1 activity can mediate cisplatin sensitivity. Overall, these results highlight a role for UNG initiated canonical BER processing of uracils adjacent to cisplatin ICLs to mediate drug efficacy.
Materials and Methods
Chemicals, antibodies and enzymes
Cisplatin, oxaliplatin, methoxyamine, methyl methanesulfonate (MMS) and myricetin were purchased from Sigma Aldrich. APE1 redox inhibitor E3330 was purchased from Novus biologicals. All other chemicals were from standard suppliers. APE1 antibody was from Novus biologicals, antibody specific for phosphorylated H2AX was from Cell Signaling Technology, MSH6 antibody was from BD pharmigen and α-tubulin antibody was from Sigma Aldrich. Oligonucleotides used in these studies were purchased from IDT DNA Technologies. Uracil DNA glycosylase from E. coli (UDG) 2,000 U/ml, human apurinic/apyridimic endonuclease (APE1) 10,000 U/ml, and Uracil Glycosylase inhibitor (UGI) 2,000 U/ml were from New England Biolabs.
DNA constructs
The oligonucleotides (5′-CTCTTCCCCCTCTCCTTCTTGUCCTCTTCCTTCCCCTTCCCT-Cy3-3′ and 5′-AGGGAAGGGGAAGGAAGAGGGCAAGAAGGAGAGGGGGAAGAG-3′) were either left unprocessed to represent a uracil containing undamaged DNA substrate or were treated with cisplatin to create a single site-specific cisplatin interstrand crosslink (ICL), with uracil adjacent to the crosslink. Oligonucleotides containing cisplatin adducts were designed using a protocol optimized to generate single site specific defined lesions [21, 22]. One of the DNA oligonucleotides used for the preparation of the ICL substrate was designed to contain a single G to ensure the formation of a mono adduct. This single G strand also contains a 3′ Cy3 label. Platination reactions were carried out in the dark in 1 mM Sodium phosphate (pH 7.5) and 3 mM NaCl at a drug to single strand oligonucleotide ratio of 15:1 for 12–14 hours to ensure proper mono-adduct formation. Unreacted platinum was removed by passing the reaction mixture through a Tip 20 column (Qiagen). The platinated substrate eluted from the column was ethanol precipitated and annealed to the complementary oligonucleotide to generate duplex substrates containing site-specific lesions. To form the ICL, the annealed duplex was dialyzed overnight at 37ºC in 100 mM sodium perchlorate and 10mM Tris-HCl (pH 7.5) and then at RT for 8 hours to increase the efficiency of the ICL. The substrate was recovered from dialysis, ethanol precipitated and loaded onto a DNA sequencing gel. The presence of the crosslink retards the separation of the duplex oligonucleotide under denaturing conditions and runs as a smeared product on the gel. This smeared band is excised from the gel, DNA is eluted and ethanol precipitated to obtain the cisplatin ICL DNA substrate.
Cell lines
The human breast adenocarcinoma MDA-MB-231 (authenticated by the Karmanos Cancer Institute Genomics Core) cells were grown in RPMI 1640 containing 10% FBS and 1% pen-strep. The MDA-MB-231 Polβ K72A lyase dead cell line was grown in the presence of 700 μg/mL geneticin. The human cervical cancer HeLa cell line and mouse embryonic fibroblasts (MEFs) were maintained in DMEM- high glucose media with 10% FBS and 1% pen-strep. The MEFs were grown in an incubator with 10% CO2.
Preparation of cell extracts
HeLa whole cell extracts were prepared by using a modified protocol that resulted in 10–15 mg/ml protein [23]. Briefly, a HeLa cell pellet was incubated in hypotonic buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 5 mM DTT, 0.5 mM phenyl methyl sulfonyl fluoride, and 1 μg/ml each of leupeptin and pepstatin) for 30 minutes on ice. The cells were homogenized with a Dounce homogenizer followed by addition of a high salt buffer (50 mM Tris-HCl (pH 8.0), 10 mM MgCl2 2 mM DTT, 25% sucrose; 50% glycerol) to the lysed cell pellet mixture. 4.1 M saturated ammonium sulfate solution was added and the solution and stirred for 30 minutes. The solution was transferred to polycarbonate tubes (Beckman) and was centrifuged at 42,000 rpm for 3 hours. The supernatant was collected and 0.33 g/ml ammonium sulfate was added. The pellet was re-suspended in 25 mM HEPES-NaOH (pH 7.9), 0.1 M KCl, 12 mM MgCl2, 1 mM EDTA, 2 mM DTT and 10% glycerol and dialyzed overnight in the same buffer. Buffer was exchanged three times. After dialysis, insoluble material was removed by centrifugation at 5,000 rpm for 10 minutes. The concentration of the extract was measured by Bradford assay and the solution was aliquoted and frozen at −80ºC. Cell extracts from other cell lines used in the study were prepared using a secondary method. Briefly, cells growing in 10 cm plates were allowed to reach confluence and a cell pellet was obtained by scraping. The cells was resuspended in 3-times the volume of the pellet in TNN buffer (10 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA) in the presence of the protease inhibitors and kept on ice for 30 minutes, vortexing every 10 minutes. The pellet was centrifuged at 13,000 rpm for 15 minutes. The supernatant was removed and transferred to a new tube. Protein concentration was determined by Bradford assay.
In vitro Glycosylase assay
Cy3 labeled DNA substrates (10 nM) containing either uracil or uracil adjacent to a cisplatin ICL were incubated with HeLa whole cell extracts (0.4 mg/ml) in the glycosylase reaction buffer for 1 hour. The reactions were carried out in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml bovine serum albumin and 10% Glycerol (v/v). The reactions were quenched by adding sequencing dye, containing formamide, and were heated for 5 minutes at 95ºC and resolved on a 12% sequencing gel. The gels were visualized using a Typhoon FLA 7000 scanner (GE) by scanning for Cy3. For the UDG inhibition, UGI was preincubated with HeLa extract for 30 minutes before the addition of DNA substrate, and the reactions were carried out as described above. Whole cell extracts from MEFs were used for additional glycosylase experiments using the same method.
shRNA transfection
Mission shRNA lentiviral bacterial glycerol stocks were purchased from Sigma Aldrich. The plasmid DNA was transfected into HEK293FT cells along with lentiviral packaging plasmids to make viral supernatants as previously described [20]. At the time of experiment, total 2 mL of viral stocks were used to infect MDA-MB-231 cells for 24 hours in the presence of 8 μg/mL polybrene (Sigma Aldrich). Cells were harvested at 96 hour and 120 hour timepoints post infection for analyzing protein and transcript expression.
Western blot analysis
The cells were harvested at indicated time points and washed with 1X cold PBS and the pellets were stored at −80ºC until needed. The proteins were extracted using RIPA lysis buffer (25 mM Tris pH 7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1% SDS) containing protease inhibitors 0.5 M phenyl methyl sulphonyl fluoride PMSF, 1mg/ml leupeptin and 1 mg/ml pepstatin A and 10μL/mL phosphatase inhibitor cocktail 2 (Sigma Aldrich). The proteins were separated on polyacryalamide gels appropriate for the size of the protein of interest. The proteins were transferred onto Immobilon-P transfer membranes (Millipore). The membranes were then blocked using 5% milk in TBST. The primary antibody incubations were carried out overnight. The dilutions used were as follows: Anti-APE1 1:1000, Anti-MSH6 1:2,000, Anti γH2AX 1:1,000 and Anti-α-tubulin at 1:100,000. The secondary antibody dilutions were carried out at RT in blocking buffer at 1:2,000 dilutions. The signal was then detected using enhanced chemiluminescence detection system.
Transcript analysis using RT-PCR
The cells were harvested at indicated time points, washed with 1X cold PBS and pellets were stored at −80ºC. Total RNA was extracted using TriZol reagent (Life Technologies). The cDNA was synthesized using MMLV reverse transcriptase enzyme (Life Technologies) as per the manufacturer’s protocol. The primers were ordered from Integrated DNA Technologies (listed in Supplemental Information). The transcript levels were quantified using SYBR select master mix from Life Technologies in Step-One real time PCR system using GAPDH as an endogenous control. The percent transcript knockdown was determined from 2−ΔΔCT values.
Colony survival assay
~500 cells were plated in complete medium in 60mm dishes. The cells were allowed to attach overnight and next day were treated with increasing concentrations of cisplatin in serum free media. After 2 hours of treatment, media was changed to fresh complete media. The cells were allowed to grow colonies for 7–8 days. The colonies were fixed using 95% methanol and stained with 0.02% crystal violet. Colonies with >50 cells were counted and the results were represented as the number of colonies in untreated control normalized to 100%.
MTS assay
Cells (~7,000) were plated in 96 well plates and were treated with cisplatin for 2 hours in serum free media or for 24 hours in complete media. For experiments using APE1 inhibitors, 2 hours pretreatment with myricetin or E3330 was carried out before cisplatin treatment. The media was changed to fresh complete media and cells were incubated for 72 hours after the treatment. MTS assay was performed as per the manufacturer’s protocol (Promega). Briefly, 20 μL MTS dye was added to each well, followed by incubation for 2–3 hours at 37°C. The absorbance was measured at 490 nm and the survival was expressed by normalizing untreated cells to 100%. The experiment was done in triplicate for each drug concentration.
APE1 inhibition assay with Methoxyamine
The APE1 inhibition assay was conducted using same reaction conditions as the glycosylase assay. HeLa whole cell extracts (0.4 mg/ml) were pre-incubated with methoxyamine at 200 μM followed by a 60 minute incubation with DNA substrates (10 nM) containing a uracil or a uracil adjacent to cisplatin ICL at 37ºC. The reaction was stopped by adding formamide and heated at 95ºC for 5 minutes, loaded onto a 12% sequencing gel and visualized on the Typhoon gel scanner.
Modified alkaline comet assay using H2O2
Modified alkaline comet assay was used to analyze the repair of cisplatin interstrand cross-links. The cells were treated with cisplatin for 2 hours in serum free media and the assay was carried out at the indicated time points [5, 12, 24]. Before harvesting, cells were exposed to 50 μM H2O2 for 15 minutes. Cell suspensions (~10,000 cells) were embedded on a microscopic slide, lysed and incubated in ice-cold alkaline solution for 20 min to allow the DNA to unwind. Electrophoresis was carried out for 30 min at 28 V, 300 mA. Slides were neutralized (0.4M Tris pH 7.5) and stained with SYBR gold stain (Invitrogen). The comets were scored using a Nikon epifluorescence microscope. ~50 cells were analyzed per slide using Komet Assay Software 5.5F (Kinetic Imaging, Liverpool, UK). The data was expressed as the percentage of crosslinks that remained at that particular time point normalized to 100% at 0 hours which corresponds to 2 hours post cisplatin treatment.
Results
Uracil DNA glycosylase is the primary enzyme for removing uracils adjacent to a cisplatin ICL
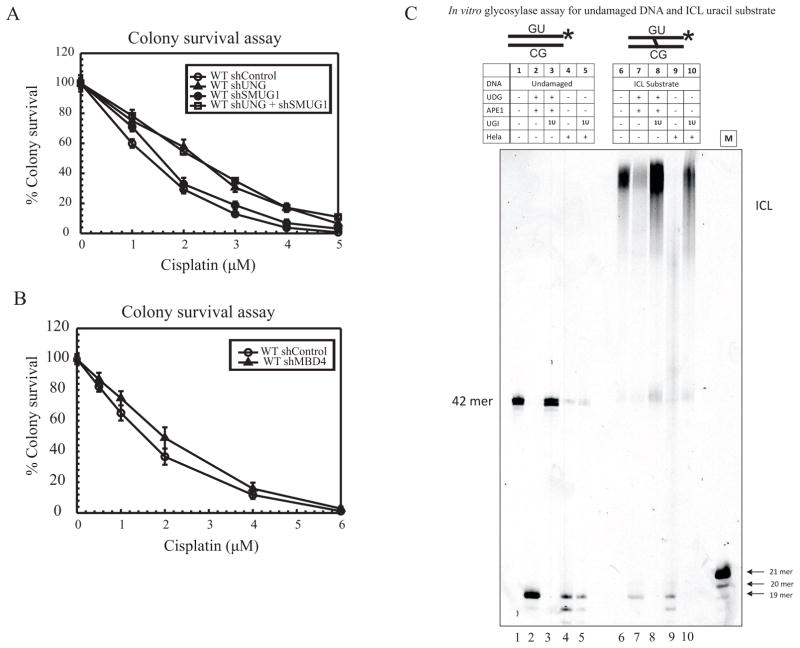
Removal of abnormal uracil in DNA is mediated by various DNA glycosylases (e.g., UNG, SMUG1, MBD4 and TDG). UNG is the enzyme responsible for the removal of uracils in most spontaneous deamination events occurring in a cell. We have previously shown that extra-helical cytosines adjacent to cisplatin ICLs undergo preferential oxidative deamination to uracil [5]. Our mechanistic model and previous data highlight a role for BER activation to enhance cisplatin sensitivity and thus, we hypothesized that a uracil glycosylase would initiate the BER activation adjacent to cisplatin ICLs [5]. To assess which uracil glycosylase is responsible for activation of the BER pathway in response to cisplatin or whether there was redundancy in uracil glycosylase function, UNG and SMUG1 were knocked down separately and together in MDA-MB-231 cells using shRNA containing lentiviral particles. The knockdown (KD) efficiency was determined by analyzing transcript levels using qRT-PCR. The transcript levels for both UNG and SMUG1 were shown to be reduced by ~85 to 90% compared to control (Supplementary Figures S1AB). Next, colony survival assays were performed using increasing concentrations of cisplatin. UNG KD cells showed resistance to cisplatin as compared to control cells, while SMUG1 KD did not affect cisplatin sensitivity. However, KD of both UNG and SMUG1 resulted in a resistant phenotype when compared with the control cells (Figure 1A). Previously, we have shown that UNG −/− MEFs display enhanced ICL DNA repair and increased resistance to cisplatin when compared with wild type (WT) cells [5]. Taken together, these results suggest that loss of UNG alone gives rise to decreased cisplatin sensitivity and the absence of SMUG1 has no effect and no redundant activity in regard to a cisplatin response. No significant effect on cisplatin cytotoxicity was seen with MBD4 KD, further supporting the evidence that UNG is the primary uracil glycosylase responsible for influencing the repair of cisplatin ICLs and mediating cisplatin cytotoxicity (Figure 1B, MBD4 KD shown in Supplementary Figure S1C).
Figure 1.
Cisplatin cytotoxicity and effect on glycosylase activity (A) Colony survival assay in MDA-MB-231 cells following UNG and SMUG1 knockdown: shControl (open circles), shUNG (closed triangles), shSMUG1 (closed circles) and shUNG + shSMUG1 (open squares). Results are represented as mean ± SE from 3 independent experiments. Cells were transfected with shRNA directed against UNG and SMUG1. (B) Colony survival assay in MDA-MB-231 cells following MBD4 knockdown with shControl (open circles) and shMBD4 (closed triangles). shRNA transfected cells were treated with increasing doses of cisplatin and cytotoxicity. Results are represented as mean ± SE from 3 independent experiments. (C) In vitro glycosylase assay, DNA (5nM) was incubated with either pure enzyme or HeLa extract. Lane 1, undamaged DNA alone.; lane 2, undamaged DNA treated with UDG and APE1 to generate a 19 mer product; lane 3, undamaged DNA substrate treated with UDG, APE1 and 1 unit of UGI; lane 4, undamaged DNA incubated with HeLa extract; lane 5 reactions in which HeLa extract was preincubated with 1 unit of UGI before adding the undamaged DNA substrate. Lanes 6–10 follow the same set up as lanes 1–5, but with ICL DNA substrate. Both undamaged and ICL substrates contain a central uracil and a 3′ Cy3 label. M is a 21-nt marker.
In addition to cell culture studies, we used synthetic ICL DNA substrates to assess the role of UNG in the removal of uracils that arise adjacent to a cisplatin ICL. Experiments were carried out with both undamaged and cisplatin ICL DNA substrates. The top strand of the 42 base pair duplex contained a single uracil at position 19, adjacent to the ICL, and was labeled at the 3′ end with Cy3 fluorescent dye. Glycosylase activity on the undamaged DNA substrate and the cisplatin ICL substrate were assessed by incubating with purified bacterial UDG and APE1 and separating the products on a DNA sequencing gel. Lanes 2 and 7 of Figure 1C show the formation of a 19-mer product when UDG and APE1 are incubated with the undamaged and ICL DNA substrates, respectively. This is consistent with an appropriate incision occurring at an abasic site generated by removal of the uracil. This product served as a positive control for the reaction catalyzed by glycosylases from cell extracts. The uracil containing DNA substrates were also incubated with HeLa cell extract, which contains UNG and other active glycosylases. Upon incubation of the DNA substrates with HeLa cell extracts, the 19-mer product was observed (Figure 1C, lanes 4 and 9, respectively). To determine if the uracil adjacent to the ICL is removed by UNG, we used a highly specific inhibitor of UNG, uracil glycosylase inhibitor (UGI) and gauged the effect on the incised product. UGI blocked the activity of the purified bacterial UDG on both damaged and undamaged substrates (Figure 1C, lanes 3 and 8 respectively). After incubation of HeLa cell extracts with UGI, the 19-mer product formation was still observed on the undamaged substrate (lane 5), supporting the notion that redundant uracil glycosylases can actively remove uracil from an undamaged DNA substrate. However, when the UGI pre-incubated HeLa cell extract was incubated with the cisplatin ICL DNA substrate, the product was not detected (Figure 1C, lane 10). This indicates a highly specific requirement for UNG in the removal of uracil adjacent to cisplatin ICLs.
These data support the results from the colony survival studies, further indicating that UNG processes the uracil adjacent to cisplatin ICLs and this processing is essential to activate BER and maintain cisplatin sensitivity. We repeated the experiment with WT and UNG null MEF cell extracts for both undamaged and cisplatin ICL DNA substrates. As expected, when undamaged and ICL DNA substrates were incubated with WT cell extract the 19-mer product was detected, indicating uracil removal and APE1 incision at the AP site (Supplementary Figure S1D, lanes 4 and 9, respectively). The 19-mer product was detected when WT MEF extract was incubated with UGI and undamaged DNA, consistent with the results obtained with HeLa cell extracts (Supplementary Figure S1D, lane 5). The 19-mer product was also detected with UNG null cell extract (Supplementary Figure S1E, lane 4) which highlights the redundancy of uracil glycosylases to catalyze the removal of uracil from an undamaged duplex DNA substrate. Furthermore, when UNG null cell extract was supplemented with purified UDG, there was no increase in product formation (Supplementary Figure S1E, lane 5) suggesting that the efficiency of removal of uracil from undamaged duplex DNA by other uracil glycosylases is as efficient as UNG. When the ICL uracil DNA substrate was incubated with the UGI, consistent with the results obtained with HeLa cell extract, we did not observe product with extracts from WT MEFS or UNG−/− MEFs (Supplementary Figures S1D and E, lanes 10 and 9, respectively). Product formation was restored when the UNG−/− extracts were supplemented with purified bacterial UDG (Supplementary Figure S1E, lane 10) suggesting a specific requirement for UNG in removing uracil adjacent to cisplatin ICLs. Furthermore, previously we have shown that UNG −/− MEFs are resistant to cisplatin as compared to WT MEFS. The resistant phenotype in these cells can be correlated with increased ICL DNA repair. Moreover, when we assessed the rate of repair of cisplatin intrastrand adducts, we did not see any difference between UNG proficient and deficient cells. These data indicate that intrastrand adduct repair remains unaffected and BER protein levels do not influence intrastrand adduct repair after cisplatin treatment [5]. Based on these results, we conclude that UNG is the primary glycosylase responsible for removing uracil adjacent to cisplatin ICLs.
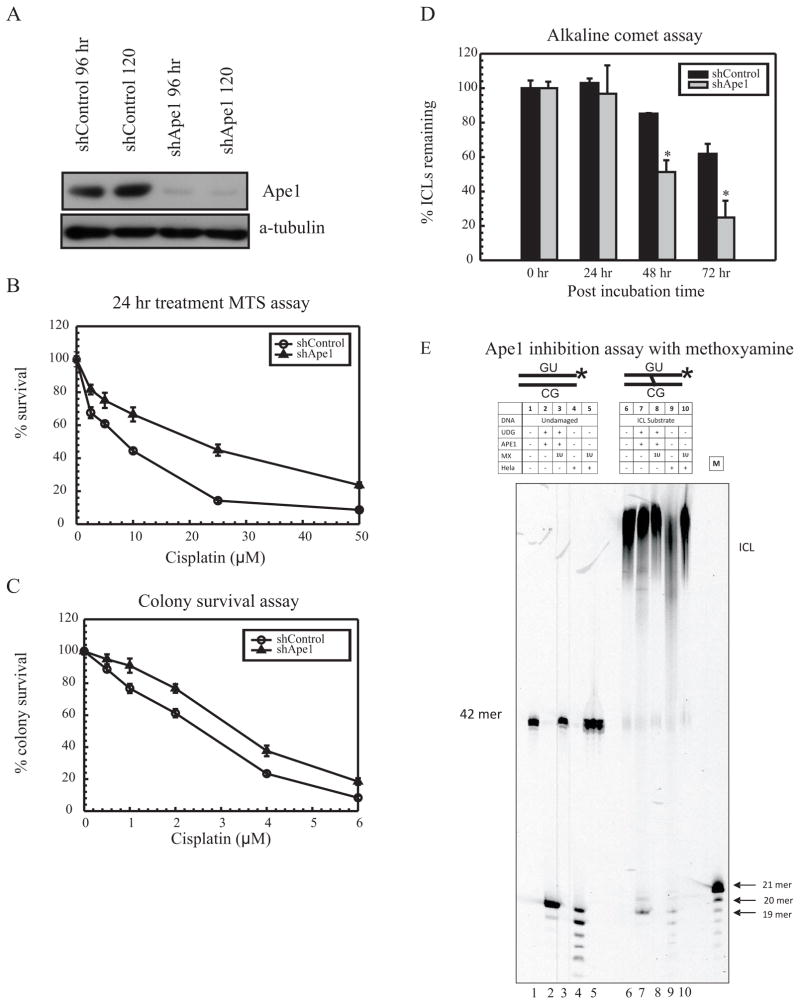
APE1 knockdown gives rise to cisplatin resistance
UNG removal of uracils that flank cisplatin ICLs results in abasic sites. These abasic sites are recognized by APE1, and following cleavage of the phosphodiester bond, facilitate recruitment and DNA synthesis by Polβ. To further understand the involvement of APE1 in mediating cisplatin cytotoxicity, APE1 was knocked down in MDA-MB-231 cells using gene specific lentiviral shRNA infection. 85–90% knockdown (KD) efficiency was achieved, checked at both the protein and transcript level by using western blot analysis (Figure 2A) and qRT-PCR (Data not shown). APE1 KD increases cell survival/resistance (~2 fold) after cisplatin treatment (Figure 2B and 2C). APE1 KD in HeLa cells resulted in similar survival trends as MDA-MB-231 (Supplementary Figures S2AB). Resistance to oxaliplatin was not observed, which is consistent with the structural differences in ICLs formed by oxaliplatin and cisplatin (Supplementary Figure S2C). These data indicate that decreased amounts of APE1 give rise to increased cellular survival in response to cisplatin. In order to understand the mechanism for increased cisplatin-specific resistance in APE1 KD cells, we performed modified alkaline comet assays to study the rate of ICL DNA repair in these cells. At 48- and 72- hour time points, increased ICL DNA repair was observed as compared to the control cells (Figure 2D). This increased ICL DNA repair correlates with the increased resistance seen in APE1 KD cells. Our previous studies have shown that methoxyamine pretreatment before addition of cisplatin gives rise to a similar increased rate of repair of cisplatin ICLs as compared to cisplatin alone treated cells [5]. Furthermore, the rate of intrastrand adduct repair remained unaffected indicating that BER proteins are not involved in the repair of cisplatin intrastrand adducts. In addition, the rate of oxaliplatin ICL repair remained unchanged suggesting that the effects observed are specific to cisplatin [5]. These results are consistent with our previous observations where inhibition of BER endonuclease cleavage gives rise to increased repair of cisplatin ICLs leading to cisplatin resistance [5].
Figure 2.
Effect of APE1 on cisplatin resistance (A) Western blot analysis in WT MDA-MB-231 cells following APE1 knockdown. (B) MTS assay with shControl (open circles) shAPE1 (closed triangles). (C) Colony survival assay with shControl (open circles) shAPE1 (closed triangles). (D) Modified alkaline comet assay with shControl and shAPE1 cells at 0, 24, 48 and 72 hour time points. Results are represented as mean ± SE from 3 independent experiments. (* p < 0.05) (E) In vitro APE1 inhibition assay: Lane 1, undamaged DNA alone; Lane 2, undamaged DNA treated with UDG and APE1 to generate a 19 mer product; Lane 3, reaction of the undamaged DNA substrate treated with UDG, APE1 and 1 unit of 200 μM Methoxyamine; Lane 4, DNA incubated with HeLa extract; Lane 5, DNA incubated with HeLa extract and methoxyamine. Lanes 6–10 follow the same set up as lanes 1–5, but with ICL substrate. Both undamaged and ICL substrates contain a central uracil and a 3′ Cy3 label. M is a 21-nt marker.
The downstream processing events that occur upon the removal of uracil were assessed using synthetic DNA substrates. In Figure 2E, purified APE1 cleaves at an abasic site in both undamaged and cisplatin ICL DNA following uracil removal (Lanes 2 and 7). To confirm the abasic site (formed through uracil removal by UDG) was indeed further processed by APE1 cleavage in cancer cell extracts, methoxyamine was used to inhibit APE1 cleavage. Methoxyamine binds to an abasic site and blocks the APE1 induced cleavage reaction [25]. The addition of methoxyamine blocked cleavage by purified APE1 at the abasic site, indicating that methoxyamine is effectively binding at the abasic site (Figure 2E, lane 3). Cleavage of undamaged DNA by APE1 was also inhibited when methoxyamine was added to HeLa extracts (Figure 2E, lane 5). Consistent with undamaged DNA, when methoxyamine was added along with purified UDG and APE1 to the ICL substrate, 19mer product was not detected (Figure 2E, lane 8). Similarly, the product formation was inhibited when methoxyamine was added to the HeLa extract (Figure 2E, lane 10). These results demonstrate that APE1 is the DNA endonuclease involved in the processing of uracils adjacent to cisplatin ICLs.
Inhibition of either repair or redox domain of APE1 gives rise to cisplatin resistance
APE1 has two major functions: one involved in DNA repair that is important for BER and the other a redox function responsible for regulating the ROS levels by influencing transcription (7). In order to understand the role of these two domains in mediating cisplatin efficacy, myricetin and E3330 were used to inhibit the DNA repair function and redox activity of APE1, respectively [26, 27]. BER proteins primarily repair the DNA damage caused by alkylating agents. Therefore, inhibition of the DNA repair domain by myricetin or methoxyamine treatment should sensitize the cells to alkylating agents like methyl methanesulfonate (MMS). MDA-MB-231 breast cancer cells were treated with MMS in an MTS assay after pretreatment with myricetin and methoxyamine. Expectedly, the use of both inhibitors sensitized the cells to MMS treatment (Supplementary Figure S3). The same concentration of myricetin was used for pretreatment in combination with cisplatin. Myricetin treatment resulted in increased resistance to cisplatin as compared to cisplatin alone control cells in both colony survival and MTS assays (Figure 3A and 3B, respectively). These results are consistent with our previous studies where we used methoxyamine to block abasic sites and showed that pretreatment with methoxyamine gave rise to resistance to cisplatin [5]. Similarly, when the cells were pretreated with increasing concentrations of E3330, a corresponding increase in cisplatin resistance was observed (Figure 3C). These results suggested that inhibition of either the DNA repair or redox domain of APE1 causes cisplatin resistance. The APE1 redox activity has been shown to regulate transcription factors associated with various DNA repair genes [27]. APE1 is also involved in redox-based co-activation and DNA binding of several transcription factors including nuclear factor- κβ (NF- κβ), p53, and activator protein -1 (AP-1). AP-1 and p53 have been shown to regulate DNA repair proteins involved in BER, MMR and NER pathways. To assess the effect of APE1 redox inhibition on these repair pathways, the expression levels of MMR and NER proteins were monitored following E3330 treatment. APE1 redox inhibition led to decreased expression of MSH2, MSH6, MLH1 and ERCC1 (Figure 3D). A similar effect is observed with shRNA-mediated KD of APE1 (Figure 3D). Despite having shown that APE1 incision activity is important for processing at a cisplatin ICL site, the resistant phenotype seen in E3330 treated cells and APE1 KD cells could also be attributed to decreased expression of MMR proteins. The lack of MMR proteins have been previously shown to enhance ICL DNA repair and mediate cisplatin resistance [20].
Figure 3.
Knockdown of APE1 in MDA-MB-231 breast cancer cells (A) Colony survival assay with E3330 (closed circles) or Myricetin (closed triangles) pretreatment followed by cisplatin treatment compared with cisplatin alone (open circles). (B) MTS assay with 35 μM Myricetin pretreatment followed by cisplatin treatment (closed triangles) compared with cisplatin alone (open circles). (C) MTS assay with 10 μM (closed triangles), 25 μM (closed circles) and 50 μM (open squares) of E3330 pretreatment followed by cisplatin treatment compared with cisplatin alone (open circles). (D) Western blot analysis for MSH2, MSH6, MLH1, ERCC1, APE1 and α-tubulin after E3330 or myricetin pretreatment or APE1 knockdown +/− cisplatin treatment. Results are represented as mean ± SE from 3 independent experiments.
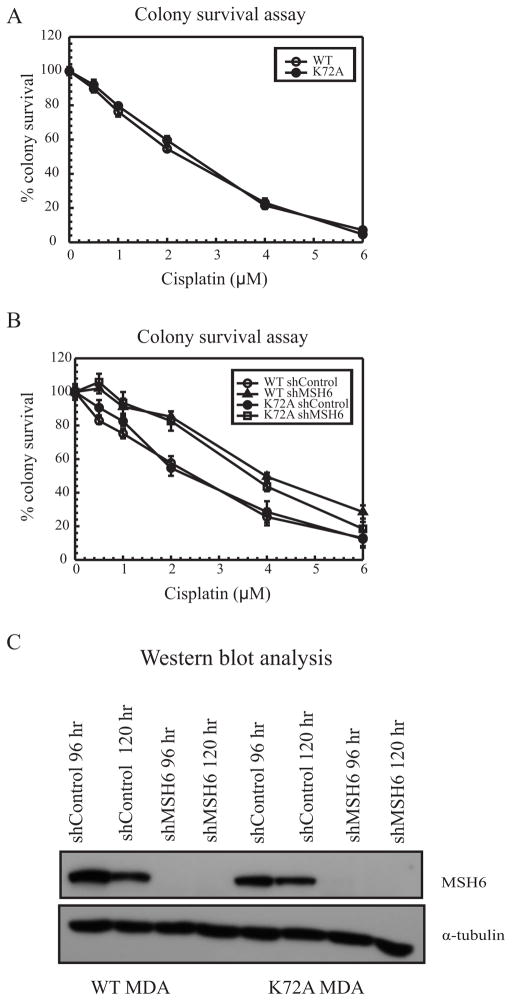
Mutation in the lyase domain of Polβ does not confer cisplatin resistance
In previous studies, we have shown that the polymerase activity of Polβ is essential for mediating cisplatin sensitivity [12]. The Polβ KD cells showed a cisplatin resistant phenotype, whereas a Polβ polymerase dead D256A mutant showed cisplatin sensitivity [12]. It was observed that the polymerase domain of Polβ is essential for MMR recruitment and activity in this mechanism. To further evaluate the activities of Polβ that are critical for mediating cisplatin efficacy, we assessed the role of the lyase domain in cisplatin cytotoxicity. The dRP lyase activity of Polβ has been shown to be required for the reversal of hypersensitivity seen in Polβ null cells in response to MMS treatment [28, 29]. Lysine 72, important in the dRP lyase beta-elimination catalytic reaction, was mutated to alanine because it has been shown to decrease lyase activity by more than 90% without affecting its DNA binding ability [29]. The removal of the dRP moiety is essential to inhibit activation of cell death and development of drug resistance. Cell survival was tested in MDA-MB-231 cells with the K72A Polβ mutation using increasing concentrations of MMS in MTS assays. The lyase dead K72A cells were more sensitive to MMS treatment as compared to WT cells; validating the requirement of dRP lyase activity of Polβ for resistance to MMS induced DNA damage (Supplementary Figure S4A). When treated with increasing concentration of cisplatin in a colony survival assay, no difference was observed in cisplatin cytotoxicity for the K72A cells compared with control cells (Figure 4A). These results suggest that the lyase activity of Polβ does not play a role in mediating cisplatin cytotoxicity.
Figure 4.
Polβ lyase activity and cisplatin resistance (A) Colony survival assay with WT MDA-MB-231 (open circles) and K72A MDA-MB-231 (closed circles). (B) Colony survival assay with WT shControl (open circles), WT MSH6 KD (closed triangles), K72A shControl (closed circles) and K72A MSH6 KD (open squares). Results are represented as mean ± SE from 3 independent experiments. (C) Western blot analysis with MSH6 knockdown in WT and K72A MDA-MB-231 cells.
To ensure that the K72A cells still retained the ability to become resistant to cisplatin, the MMR protein MSH6 was knocked down in both WT and K72A cells to assess colony survival following cisplatin treatment (Figure 4B). Knocking down MSH6 gave rise to cisplatin resistance in WT cells, which is consistent with our previous studies, while MSH6 KD in K72A cells also gave rise to resistance to cisplatin when compared with K72A control cells (Figure 4B) [20]. The levels for MSH6 protein KD are shown in Figure 4C. These results were in contrast with MMR knockdown in D256A cells, where MSH2 deficiency did not lead to cisplatin resistance, highlighting the importance of the polymerase domain of Polβ in nucleotide misincorporation and subsequent MMR protein recruitment near cisplatin ICL sites [12]. Methoxyamine mediated inhibition of BER in D256A cells resulted in a cisplatin resistant phenotype [12]. Similarly, myricetin mediated inhibition of BER in Polβ K72A cells gave rise to increased resistance to cisplatin, indicating that BER upstream of Polβ can still mediate cisplatin sensitivity (Supplementary Figure S4B). These data strongly support a non-functional role of Polβ lyase domain in mediating cisplatin efficacy.
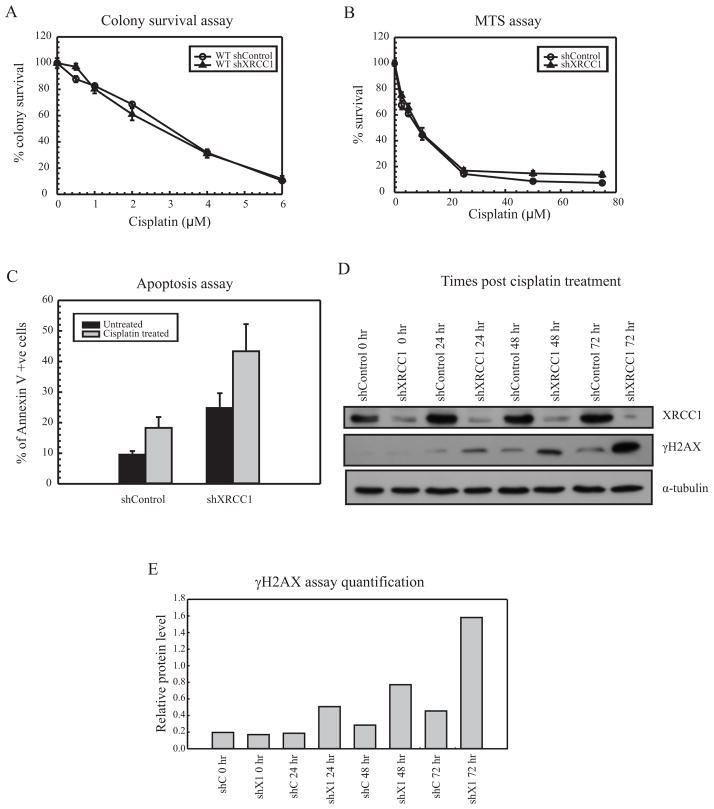
XRCC1 deficiency gives rise to cisplatin sensitive phenotype
XRCC1 acts as a scaffolding protein that interacts with multiple DNA repair enzymes. Loss of XRCC1 has been shown to give rise to cisplatin sensitivity in gastric cancer cells as well as ovarian cancer cells [16, 19]. Novel mechanisms involving ubiquitin-mediated degradation of XRCC1 in cisplatin sensitive cells have also been presented in gastric cancer cells. To understand the role of XRCC1 as a downstream BER protein involved in mediating cisplatin sensitivity, XRCC1 was knocked down in MDA-MB-231 cells and the KD efficiency was assessed using qRT-PCR (Supplementary Figure S5). The effect of this knockdown on cell survival in response to cisplatin was analyzed using colony survival and MTS assays. The raw values for untreated cells showed increased cell death in WT shXRCC1 cells when compared to WT shControl cells (not shown). However, when normalized to 100 percent individually, the XRCC1 deficient cells remained sensitive to cisplatin treatment in both survival assays (Figure 5A and 5B). In order to gain a better understanding for the enhanced cisplatin sensitivity following XRCC1 knockdown, we assessed apoptosis in these cells. The percentage of apoptotic cells in shControl and shXRCC1 showed loss of XRCC1 increased induction of apoptosis in untreated as well as cisplatin treated cells when compared to WT cells (Figure 5C). The apoptosis experiments were conducted by treating for 24 hours with cisplatin to get significantly higher percentage of apoptotic cells versus 2 hours for colony survival and MTS assays. Analysis of γH2AX levels in XRCC1 KD cells showed increased phosphorylation of H2AX from 24 to 72 hour times points following cisplatin treatment compared to control cells. In addition, we analyzed the level of γH2AX in XRCC1 knockdown cells followed by cisplatin treatment. Loss of XRCC1 led to increased phosphorylation of H2AX from 24 hr to 72 hr time point when compared with shControl cells. Increased γH2AX are indicative of increased strand breaks and enhanced DNA damage. These results suggest that cisplatin sensitivity in XRCC1 KD cells can be attributed to increased DNA damage (Figure 5D). Unlike what is observed with UNG, APE1 and Polβ, loss of XRCC1 does not lead to cisplatin resistance.
Figure 5.
XRCC1 activity and cisplatin resistance (A) Colony survival assay with WT MDA-MB-231 cells shControl (open circles) and shXRCC1 (closed triangles) treated with cisplatin. (B) MTS assay with WT shControl (open circles) and shXRCC1 (closed triangles) treated with cisplatin. (C) Apoptosis assay with WT shControl and shXRCC1 cells treated with cisplatin for 24 hours. Percentage of Annexin V +ve cells were measured using flow cytometry. Results are represented as mean ± SE from 3 independent experiments. (* p < 0.05) (D) Western blot analysis in WT shControl and shXRCC1 cells treated with cisplatin for 2 hours, harvested at 0, 24, 48 and 72 hour time points and probed for XRCC1, γH2AX and α-tubulin. (E) Quantification of γH2AX levels following shControl (shC) or shXRCC1 (shX1) at different time points. The γH2AX assay followed by XRCC1 knockdown was repeated twice and the quantification of relative protein levels was done using imageJ software.
Discussion
In this report, we present evidence that the BER pathway participates in the removal of uracils adjacent to cisplatin ICL (summarized in the model Figure 6). Due to the structural distortion, cytosines adjacent to the cisplatin-specific ICLs become extra-helical and preferentially undergo oxidative deamination generating a uracil. Whether this deamination is solely due to spontaneous deamination or protein induced deamination is currently under investigation. An earlier study from our lab demonstrated that treatment of cells with cisplatin leads to an increase in the number of abasic sites [5]. It has also been observed that a cytosine adjacent to the cisplatin specific ICL is more readily converted to a uracil upon treatment with sodium bisulfite than a cytosine in undamaged duplex DNA. A uracil adjacent to cisplatin ICL activates the BER pathway in the vicinity of the crosslink. The results reported here establish that the uracil is removed by a UNG dependent process leading to subsequent BER pathway activation (Figure 6). HeLa cell extracts have proven to be great tools for determining the mechanistic aspects of DNA repair as they are highly enriched for DNA repair proteins [30]. Loss of the BER intermediate upon incubation with UGI, a highly specific inhibitor of UNG, has been helpful in establishing the importance of this glycosylase in cisplatin ICL processing. Cells have redundant pathways for the removal of G/U mismatches that arise due to deamination or incorrect incorporation of dUMP by DNA polymerases. Currently, four different glycosylases UNG, SMUG1, TDG and MBD4 have all been shown to catalyze the removal of uracils from DNA [31]. Apart from these glycosylases in BER, MMR has also been shown to efficiently remove G/U mismatches [32]. The choice of the specific pathway is dependent on the relative expression of each protein as well as the efficiency of catalysis. UNG has robust activity for removing uracils occurring after DNA deamination. Knockdown of UNG in breast cancer cells (MDA-MB-231) gave rise to a cisplatin resistant phenotype, whereas SMUG1 KD did not have any effect on cisplatin sensitivity. Moreover, MBD4 KD did not have any effect on cisplatin cytotoxicity. The in vitro experiments, using a specific UNG small molecule inhibitor UGI and UNG deficient MEF extracts, clearly established UNG as the primary glycosylase responsible for the removal of uracil adjacent to a cisplatin ICL containing DNA. The crystal structure of UNG bound to uracil on a DNA substrate has been solved [33] and showed uracil flipped out from the duplex DNA into the catalytic center where the glycosidic bond is cleaved [34]. The presence of uracil adjacent to cisplatin ICL in an already extra-helical conformation should allow for highly efficient recognition by UNG. In the case of an undamaged duplex DNA substrate, even though UNG is abundant and predominant other redundant glycosylases can still function to remove the uracil.
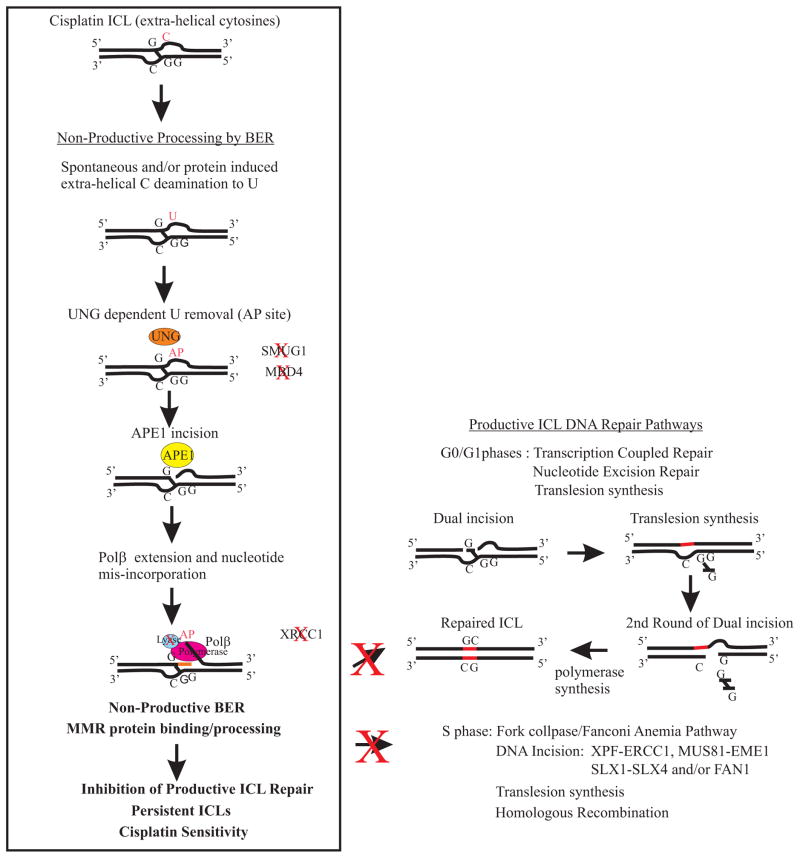
Figure 6.
Model of BER processing at cisplatin ICL sites and potential pathways blocked by non-productive processing. Our data support a dependence on UNG for uracil removal at sites flanking cisplatin ICLs. There is also a dependence on APE1 DNA incision and Polβ polymerase activity. Our data is consistent with the 5′ dRP lyase activity of Polβ not playing a role in cisplatin sensitivity and non-productive ICL processing. In addition, XRCC1 does not appear to influence cisplatin ICL non-productive processing. The pathways on the right side are productive ICL DNA repair pathways that have been shown to occur in both G0/G1 and S phases of the cell cycle [reviewed in (45, 46)]. We have previously shown an epistatic relationship between BER and MMR in mediating cisplatin sensitivity via specific nucleotide mis-incorporation by Polβ at cisplatin ICL sites and subsequent MMR protein binding (12). In addition, if we knockdown the productive DNA repair pathways (e.g., ERCC1-XPF) we overcome the cisplatin resistant phenotype following loss of BER/MMR (5). Our model and data predict that non-productive BER/MMR processing at cisplatin ICL sites will block productive ICL DNA repair pathways regardless of cell cycle phase and result in persistent cisplatin ICLs that enhance cisplatin efficacy.
The classical BER reaction proceeds through a channeling of the substrates, and is aptly named the “passing the baton” model [35]. APE1 was shown to increase the overall efficiency of DNA repair by directly participating in the BER reaction. It has been proposed to have a role in the displacement of a glycosylase present on the abasic site as well as the recruitment of Polβ to the cleaved abasic sites [36]. Our previous studies have shown that blocking abasic sites with methoxyamine resulted in cisplatin resistance, and similar results were seen in the in vitro studies in this paper where APE1 KD in MDA-MB-231 cells lead to cisplatin resistance. Together, this resistance correlated with increased repair of cisplatin ICLs, further supporting a role for APE1-mediated processing of DNA at cisplatin ICLs [5]. Previous studies have shown that down-regulation of APE1 in human non-small cell lung cancer cells sensitized them to cisplatin chemotherapy [8]. Cisplatin resistant CP70 ovarian cancer cells showed altered APE1 expression levels as compared to cisplatin sensitive A2780 cells. Suppression of APE1 in A2780 and CP70 gave rise to increased cellular sensitivity to cisplatin [37]. Also, in sensory neuronal cultures APE1 knockdown lead to increased sensitivity to cisplatin treatment [38]. However, our studies using human breast cancer MDA-MB-231 cells and cervical cancer HeLa cells showed increased resistance to cisplatin after APE1 knockdown. Studies have also shown that APE1 knockdown leads to increased sensitivity in response to alkylating agents like BCNU and the crosslinking drug MMC. A differential response is seen in the case of cisplatin which most likely can be attributed to the unique structure formed by cisplatin ICLs, which leads to activation and recruitment of different repair proteins (11). It was also found that the addition of methoxyamine to the glycosylase reactions blocked APE1 activity, clearly indicating that abasic sites are formed adjacent to the ICL. Targeting APE1 directly with inhibitors of the repair or redox domains by myricetin and E3330, respectively, have been shown to sensitize various cancer cells when used alone [39, 40]. We showed that inhibition of either domain by myricetin and E3330 also resulted in cisplatin resistance in MDA-MB-231 cells. The mechanism of resistance induced by myricetin might be similar to the cells treated with methoxyamine. The lack of ability to repair abasic sites may involve an increase in the repair of cisplatin ICLs and ICL induced DSBs thus leading to increased cisplatin resistance. Myricetin has been shown to sensitize BRCA1 deficient cells [40]. However, in our studies, MDA-MB-231 cells WT for BRCA1 gave a resistant phenotype in response to cisplatin. APE1 is one of the only DNA repair proteins known to have a redox function independent of its DNA repair function. Pretreatment with E3330 also gave a cisplatin resistant phenotype. The inhibition of the redox domain of APE1 has been shown to decrease the DNA binding ability of a variety of transcription factors. Some of these transcription factors, like AP-1 and p53, modulate the expression levels of a number of DNA repair proteins. We postulate that the change in expression levels of mismatch repair proteins observed (Figure 3D) may be responsible for the cisplatin resistant phenotype with E3330, as MMR deficiency has been associated with cisplatin resistance. The differences that have been observed between cell lines/types with the loss/inhibition of APE1 in response to cisplatin may be regulated by differential protein expression or pathway activation and will have to be more thoroughly investigated.
In our studies, we also addressed whether BER processing near the cisplatin ICL sites takes place via SP-BER or LP-BER pathway. The choice of repair pathway is dependent on the concentration of a number of cellular factors, e.g. concentration of ATP and ratio of repair factors near the damage site. At low ATP concentration, XRCC1 displaces Polβ from DNA stimulating the LP-BER pathway, whereas at high ATP concentration DNA ligase III binding prevents strand displacement and continuation with SP-BER. XRCC1 plays a crucial role in stabilizing DNA ligase III which is required for SP-BER [41]. In addition, increased concentration of PARP1 inhibits LP-BER, while increased concentration of APE1 stimulates LP-BER [42]. Previously, we have shown that polymerase dead Polβ D256A mutant shows a cisplatin sensitive phenotype. MSH2 KD in D256A cells does not give rise to cisplatin resistance indicating that the polymerase function of Polβ is essential for error prone DNA synthesis and generation of mismatches that activate the MMR pathway. In the current study, the lyase domain mutant of Polβ did not result in differences in cisplatin sensitivity, suggesting that DNA synthesis near cisplatin ICL sites might proceed via the long patch BER pathway. Knocking down MMR in these cells showed a similar 2-fold resistance to cisplatin, signifying the essential role of the Polβ polymerase domain in recruitment of MMR proteins and mediating cisplatin sensitivity.
XRCC1 plays a role in BER as well as in the single strand break repair (SSBR) pathway. Lack of XRCC1 reduces the ligation efficiency in SP-BER. XRCC1 KD in HepG2 hepatocarcinoma cell lines has been shown to give rise to cisplatin sensitivity. This sensitivity has been shown to be associated with increased DNA damage and reduced DNA repair capacity [16]. Similar to this, we observed increased phosphorylation of H2AX protein, indicative of replication stress and elevated DNA DSB signaling. XRCC1 KD also increased the percentage of apoptotic cells in both untreated and cisplatin treated cells when compared with the control cells. These data suggest that XRCC1 plays a role in the repair of endogenous DNA damage and repair of damage from ROS produced during cisplatin treatment. XRCC1 has been shown to be one of the nuclear proteins that bind to duplex DNA containing cisplatin ICLs [43]. PARP1 has been shown to recognize both cisplatin intrastrand as well as interstrand cross-links [43, 44]. These proteins function in single strand break repair (SSBR) as well as BER pathways. However, the role XRCC1 plays in the processing of cisplatin ICLs is difficult to determine because of the global effects of XRCC1 knockdown. Future work using XRCC1 deficient extracts and in vitro site-specific ICL DNA substrates with purified proteins will be important to fully understand a role (if any) of XRCC1 in processing DNA at cisplatin ICL sites.
In conclusion, we demonstrated that both the APE1 repair and redox domains are required to mediate cisplatin sensitivity. Inhibition of APE1 showed increased repair of cisplatin ICLs in breast cancer cells. We also show that these mechanisms of BER processing follow a UNG mediated removal of uracil near cisplatin ICL sites and are required for maintaining cisplatin cytotoxicity. The mechanistic understanding of pathways of ICL DNA repair is essential for the improvement of existing crosslinking chemotherapy agents. Understanding how multiple repair pathways are required for repairing ICLs is crucial to understand why cancer cells develop resistance to these agents. As ICL repair can be correlated with tumor cell survival, assessing the role of different DNA repair proteins in this mechanism is clinically important. Our studies confirm UNG as the glycosylase, which removes uracil bases adjacent to cisplatin ICLs. Our studies also confirm that inhibition of the redox domain of APE1 changes the expression levels of a number of DNA repair proteins. These altered expression levels might be responsible for the differences seen in the rate of repair of cisplatin adducts, thus ultimately influencing cisplatin cytotoxicity. In addition, we did not observe any difference in cytotoxicity in response to oxaliplatin which further suggests that different ICL inducing agents activate different DNA repair pathways via their unique structural differences. In accordance with other studies, we detected increased DSB breaks and enhanced apoptosis in XRCC1 deficient cells in response to cisplatin. Overall, as cisplatin is still an integral part of various cancer treatments, unraveling the role each type of DNA lesion plays in drug efficacy under varying conditions, understanding the mechanisms of ICL DNA repair as well as understanding the role different repair proteins play in mediating drug response are all critical areas that could lead to prognostic implications for better clinical outcomes.
Supplementary Material
Acknowledgments
Financial support: The National Institutes of Health (GM088249) to SMP and (CA148629) to RWS and the Abraham A. Mitchell Distinguished Investigator fund to RWS.
The authors thank Dr. Jessica Back and Eric Van Buren from Flow cytometry core at Karmanos cancer Institute for helping with the apoptosis assays. The authors also thank members of the Patrick lab for carefully reading the manuscript.
Footnotes
Conflict of interest: None declared. RWS is a scientific consultant for Trevigen, Inc
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nature Reviews Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Jung YW, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chemical Reviews. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 3.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 4.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 angstrom resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kothandapani A, Dangeti VSMN, Brown AR, Banze LA, Wang XH, Sobol RW, Patrick SM. Novel role of base excision repair in mediating cisplatin cytotoxicity. J Biol Chem. 2011;286:14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visnes T, Doseth B, Pettersen HS, Hagen L, Sousa MML, Akbari M, Otterlei M, Kavli B, Slupphaug G, Krokan HE. Uracil in DNA and its processing by different DNA glycosylases. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zharkov DO. Base excision DNA repair. Cellular and Molecular Life Sciences. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304. doi: 10.1016/j.lungcan.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Freitas S, Moore DH, Michael H, Kelley MR. Studies of apurinic/apyrimidinic endonuclease/ref-1 expression in epithelial ovarian cancer: Correlations with tumor progression and platinum resistance. Clin Cancer Res. 2003;9:4689–4694. [PubMed] [Google Scholar]
- 10.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kolstoe DD. The apurinic/apyrimidinic endonuclease activity of Apel/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress (vol 8, pg 3008, 2002) Clin Cancer Res. 2003;9:2877. [PubMed] [Google Scholar]
- 11.Kothandapani A, Patrick SM. Evidence for base excision repair processing of DNA interstrand crosslinks. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2013;743:44–52. doi: 10.1016/j.mrfmmm.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kothandapani A, Sawant A, Dangeti VSMN, Sobol RW, Patrick SM. Epistatic role of base excision repair and mismatch repair pathways in mediating cisplatin cytotoxicity. Nucleic Acids Res. 2013;41:7332–7343. doi: 10.1093/nar/gkt479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno SI, Asagoshi K, Hou EW, Khodyreva SN, Lavrik OI, Tomer KB, Yasui A, Wilson SH. HMGB1 is a cofactor in mammalian base excision repair. Molecular Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson AB, Klungland A, Rognes T, Leiros I. DNA Repair in mammalian cells. Cellular and Molecular Life Sciences. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemec AA, Wallace SS, Sweasy JB. Variant base excision repair proteins: Contributors to genomic instability. Seminars in Cancer Biology. 2010;20:320–328. doi: 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Niu YJ, Zhou YK. Increase the cisplatin cytotoxicity and cisplatin-induced DNA damage in HepG2 cells by XRCC1 abrogation related mechanisms. Toxicology Letters. 2010;192:108–114. doi: 10.1016/j.toxlet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Xu W, Wang S, Chen Q, Zhang Y, Ni P, Wu X, Zhang J, Qiang F, Li A, Roe OD, Xu S, Wang M, Zhang R, Zhou J. TXNL1-XRCC1 pathway regulates cisplatin-induced cell death and contributes to resistance in human gastric cancer. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, Chen Q, Wang Q, Sun Y, Wang S, Li A, Xu S, Roe OD, Wang M, Zhang R, Yang L, Zhou J. JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, Seedhouse C, Chan S, Madhusudan S. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. Plos One. 2013;8 doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawant A, Kothandapani A, Zhitkovich A, Sobol RW, Patrick SM. Role of mismatch repair proteins in the processing of cisplatin interstrand cross-links. DNA Repair. 2015;35:126–136. doi: 10.1016/j.dnarep.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofr C, Brabec V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J Biol Chem. 2001;276:9655–9661. doi: 10.1074/jbc.M010205200. [DOI] [PubMed] [Google Scholar]
- 22.Patrick SM, Tillison K, Horn JM. Recognition of cisplatin-DNA interstrand cross-links by replication protein A. Biochemistry. 2008;47(38):10188–10196. doi: 10.1021/bi800460d. [DOI] [PubMed] [Google Scholar]
- 23.Wood RD, Robins P, Lindahl T. Complementation of the Xeroderma Pigmentosum DNA-repair defect in cell-free-extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 24.Arora S, Kothandapani A, Tillison K, Kalman-Maltese V, Patrick SM. Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair. 2010;9:745–753. doi: 10.1016/j.dnarep.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu LL, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 26.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM. Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. Plos One. 2009;4 doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, Nyland RL, Borch RF, Qiao X, Georgiadis MM, Kelley MR. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: Small-molecule inhibition of the redox function of Ape1. Antioxidants & Redox Signaling. 2008;10:1853–1867. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis - DNA binding and 5 ′-deoxyribose phosphate lyase activities. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 29.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 30.Lyons DM, O’Brien PJ. Human base excision repair creates a bias toward-1 frameshift mutations. J Biol Chem. 2010;285:25203–25212. doi: 10.1074/jbc.M110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen HS, Visnes T, Vagbo CB, Svaasand EK, Doseth B, Slupphaug G, Kavli B, Krokan HE. UNG-initiated base excision repair is the major repair route for 5-fluorouracil in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA incorporation. Nucleic Acids Res. 2011;39:8430–8444. doi: 10.1093/nar/gkr563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena-Diaz J, Bregenhorn S, Ghodgaonkar M, Follonier C, Artola-Boran M, Castor D, Lopes M, Sartori AA, Jiricny J. Noncanonical mismatch repair as a source of genomic instability in human cells. Molecular Cell. 2012;47:669–680. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donny W, Demple B. Modulation of the 5′-deoxyribose-5-phosphate lyase and DNA synthesis activities of mammalian DNA polymerase beta by apurinic/apyrimidinic endonuclease 1. J Biol Chem. 2004;279:25268–25275. doi: 10.1074/jbc.M400804200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang J, Xiang DB, Wang D, Xin XY. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE1/Ref-1) in human ovarian cancer and indentification of the therapeutic potential of APE1/Ref-1 inhibitor. International Journal of Oncology. 2009;35:1069–1079. [PubMed] [Google Scholar]
- 38.Jiang YL, Guo CL, Vasko MR, Kelley MR. Implications of apurinic/apyrimidinic endonuclease in reactive oxygen signaling response after cisplatin treatment of dorsal root ganglion neurons. Cancer Res. 2008;68:6425–6434. doi: 10.1158/0008-5472.CAN-08-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, Maitra A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J Cell Physiol. 2009;219:209–218. doi: 10.1002/jcp.21666. [DOI] [PubMed] [Google Scholar]
- 40.Sultana R, McNeill DR, Abbotts R, Mohammed MZ, Zdzienicka MZ, Qutob H, Seedhouse C, Laughton CA, Fischer PM, Patel PM, Wilson DM, Madhusudan S. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int J Cancer. 2012;131:2433–2444. doi: 10.1002/ijc.27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caldecott KW, Mckeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA-repair protein XRCC1 and DNA ligase-III. Molecular and Cellular Biology. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sukhanova MV, Khodyreva SN, Lebedeva NA, Prasad R, Wilson SH, Lavrik OI. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005;33:1222–1229. doi: 10.1093/nar/gki266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu GY, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–4925. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang CX, Chang PV, Lippard SJ. Identification of nuclear proteins that interact with platinum-modified DNA by photoaffinity labeling. Journal of the American Chemical Society. 2004;126:6536–6537. doi: 10.1021/ja049533o. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair. 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto S, Anai H, Hanada K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes and Environment. 2016;38(9):1–8. doi: 10.1186/s41021-016-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.