Abstract
Human cells have twenty-three pairs of chromosomes but in cancer, genes can be amplified in chromosomes or in circular extrachromosomal DNA (ECDNA), whose frequency and functional significance are not understood1–4. We performed whole genome sequencing, structural modeling and cytogenetic analyses of 17 different cancer types, including 2572 metaphases, and developed ECdetect to conduct unbiased integrated ECDNA detection and analysis. ECDNA was found in nearly half of human cancers varying by tumor type, but almost never in normal cells. Driver oncogenes were amplified most commonly on ECDNA, elevating transcript level. Mathematical modeling predicted that ECDNA amplification elevates oncogene copy number and increases intratumoral heterogeneity more effectively than chromosomal amplification, which we validated by quantitative analyses of cancer samples. These results suggest that ECDNA contributes to accelerated evolution in cancer.
Cancers evolve in rapidly changing environments from single cells into genetically heterogeneous masses. Darwinian evolution selects for those cells better fit to their environment. Heterogeneity provides a pool of mutations upon which selection can act1, 5–9. Cells that acquire fitness-enhancing mutations are more likely to pass these mutations on to daughter cells, driving neoplastic progression and therapeutic resistance10, 11. One common type of cancer mutation, oncogene amplification, can be found either in chromosomes or nuclear ECDNA elements, including double minutes (DMs)2–4, 12–14. Relative to chromosomal amplicons, ECDNA is less stable, segregating unequally to daughter cells15, 16. DMs are reported to occur in 1.4% of cancers with a maximum of 31.7% in neuroblastoma, based on the Mitelman database4, 17. However, the scope of ECDNA in cancer has not been accurately quantified, the oncogenes contained therein have not been systematically examined, and the impact of ECDNA on tumor evolution has yet to be determined.
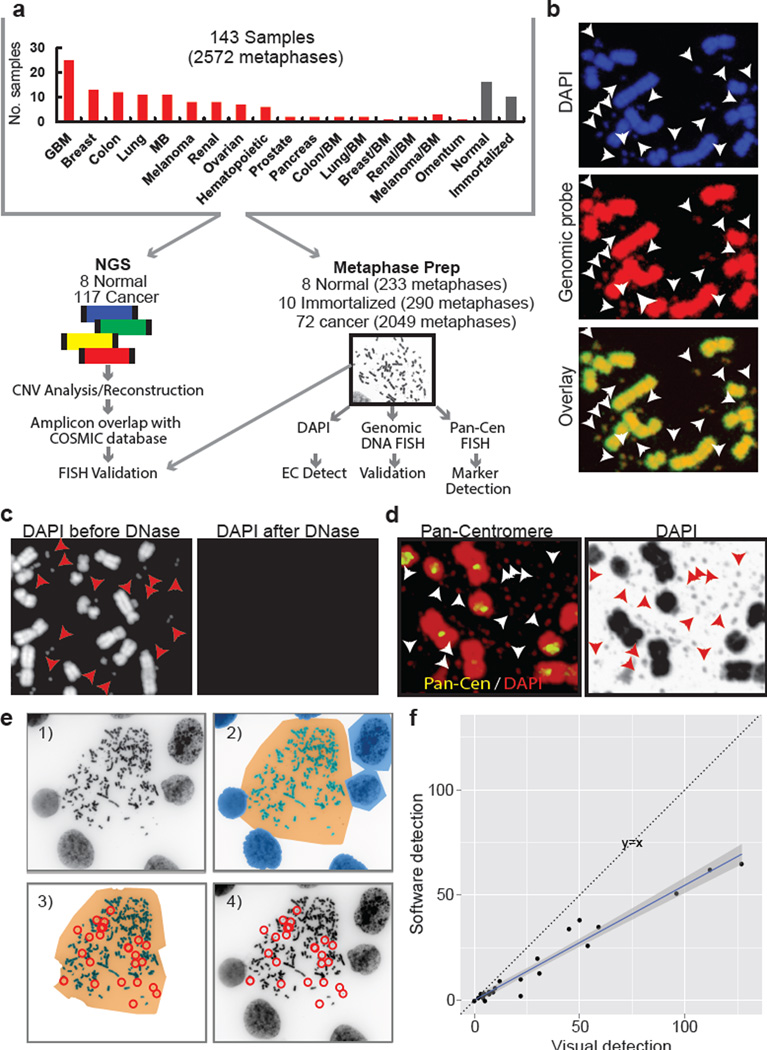
DNA sequencing permits unbiased analysis of cancer genomes, but it cannot spatially resolve amplicons to specific chromosomal or EC regions. Bioinformatic analyses can potentially infer DNA circularity18, but EC amplicons may vary from cell to cell. Consequently, ECDNA oncogene amplification may be greatly underestimated. Cytogenetic analysis of tumor cell metaphases can localize amplicons, but this technique does not permit unbiased discovery. To quantify the spectrum of ECDNA in human cancer and systematically interrogate its contents, we integrated whole genome sequencing (WGS) of 117 cancer cell lines, patient-derived tumor cell cultures and tumor tissues from a range of cancer types (Fig. 1A), with bioinformatic and cytogenetic analysis of 2049 metaphases from 72 cancer cell samples for which metaphases could be obtained. Additionally, 290 metaphases from 10 immortalized cell cultures, and 233 metaphases from 8 normal tissue cultures were analyzed, for a total of 2572 metaphases (Source Data Table S1 and Methods).
Fig. 1. Integrated next-generation DNA sequencing and cytogenetic analysis of ECDNA.
a, Schematic diagram of experimental flow. b, Representative metaphases stained with DAPI and a genomic DNA FISH probe (ECDNA, arrows). c, DNase treatment abolishes DAPI staining of chromosomal and ECDNA (arrows). d, Pan-centromeric FISH reveals absence of a centromere in ECDNAs (arrows). e, Schematic illustration of ECdetect. e.1) DAPI-stained metaphase as input. e.2) Semi-automated identification of ECDNA search region via segmentation. e.3) Conservative filtering, removing non-ECDNA components. e.4) ECDNA detection and visualization. (F). Pearson correlation between software-detected and manual calls of ECDNA (R: 0.98, p < 2.2 × 10−16.
The fluorescent dye DAPI, 4', 6-diamidino-2-phenylindole, permits ECDNA detection (Fig. 1B), as confirmed using genomic DNA and centromeric FISH probes (Fig. 1B–D; Extended Data Fig. E1). We developed an image analysis software package ECdetect (Fig. 1E; Methods), providing a robust, reproducible and highly accurate method for quantifying ECDNA from DAPI-stained metaphases in an unbiased, semi-automated fashion. ECdetect accurately detected ECDNA and was highly correlated with visual detection (r=0.98, p < 2.2 × 10−16, Fig. 1F), permitting quantification in 2572 metaphases, including at least 20 metaphases from each sample.
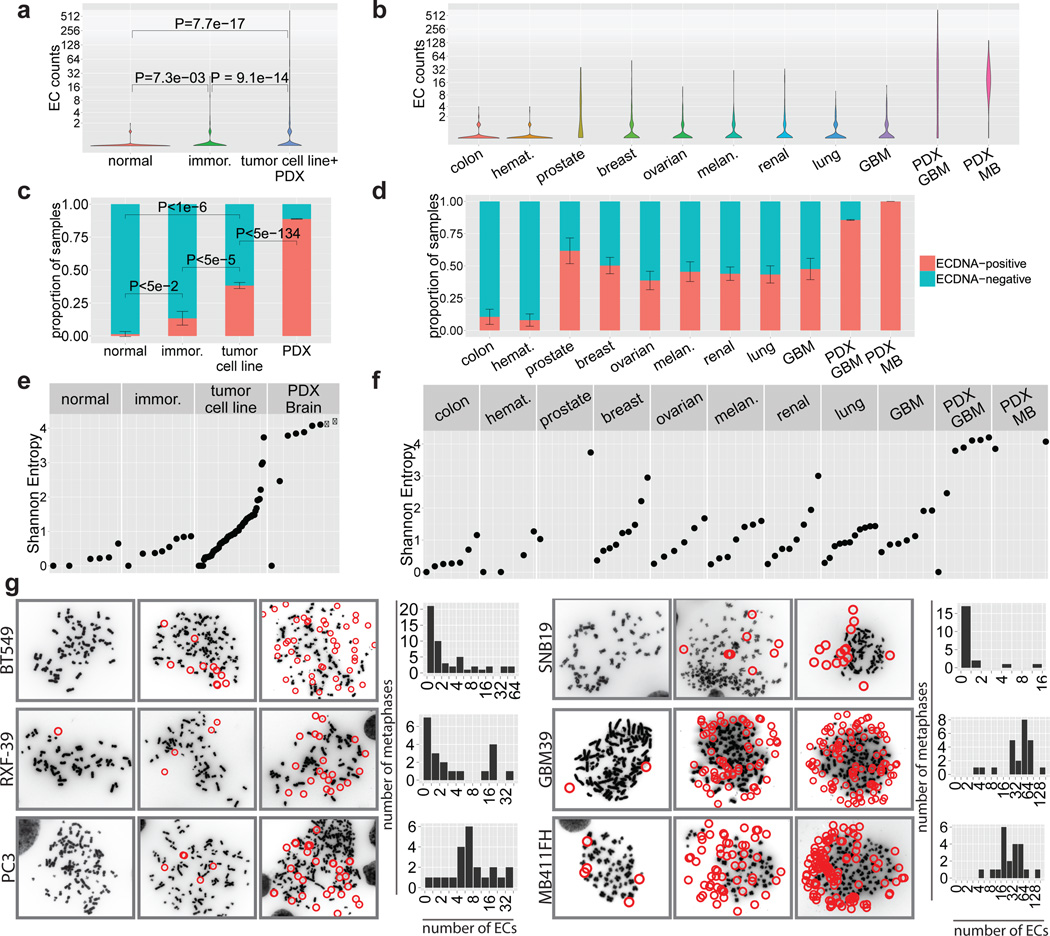
ECDNA was abundant in the cancer samples (Fig. 2A), but was rarely found in normal cells. Approximately 30% of the ECDNAs were paired DMs (Source Data Table S2). ECDNA levels varied among tumor types, with substantially higher levels in patient-derived cultures (Fig. 2B). Using the conservative metric of at least 2 ECDNAs in ≥ 10% (2 of 20) metaphases, ECDNA was detected in nearly 40% of tumor cell lines and nearly 90% of patient-derived brain tumor models (Fig. 2C–D; Methods; Extended Data Fig. E2). No significant associations between ECDNA level and either: a) primary vs. metastatic status; b) untreated vs. treated samples or c) un-irradiated vs. post-irradiated tumors were detected (Source Data Table S2). The diverse array of treatments relative to sample size limited our ability to definitively determine the impact of specific therapies on ECDNA levels. ECDNA number varied greatly from cell to cell within a tumor culture (Fig. 2E–G; Extended Data Fig. E3; Supplementary Section 2.3), as quantified by the Shannon Index19. These data demonstrate that ECDNA is common in cancer, varies greatly from cell to cell, and is very rare in normal tissue.
Fig. 2. ECDNA is found in nearly half of cancers and contributes to intra-tumoral heterogeneity.
a, Distribution of ECDNA per metaphase from 72 cancer, 10 immortalized and 8 normal cell cultures, Wilcoxon rank sum test. b, ECDNA distribution per metaphase stratified by tumor type. c, Proportion of samples with ≥ 2 ECDNAs in ≥ 2 per 20 metaphases. Data shown as mean ± SEM. (methods). d, Proportion of tumor cultures positive for ECDNA by tumor type. e, Shannon diversity index (SI). Each dot represents an individual cell line sampled with ≥ 20 metaphases. f, SI by tumor type. g, DAPI-stained metaphases with histograms.
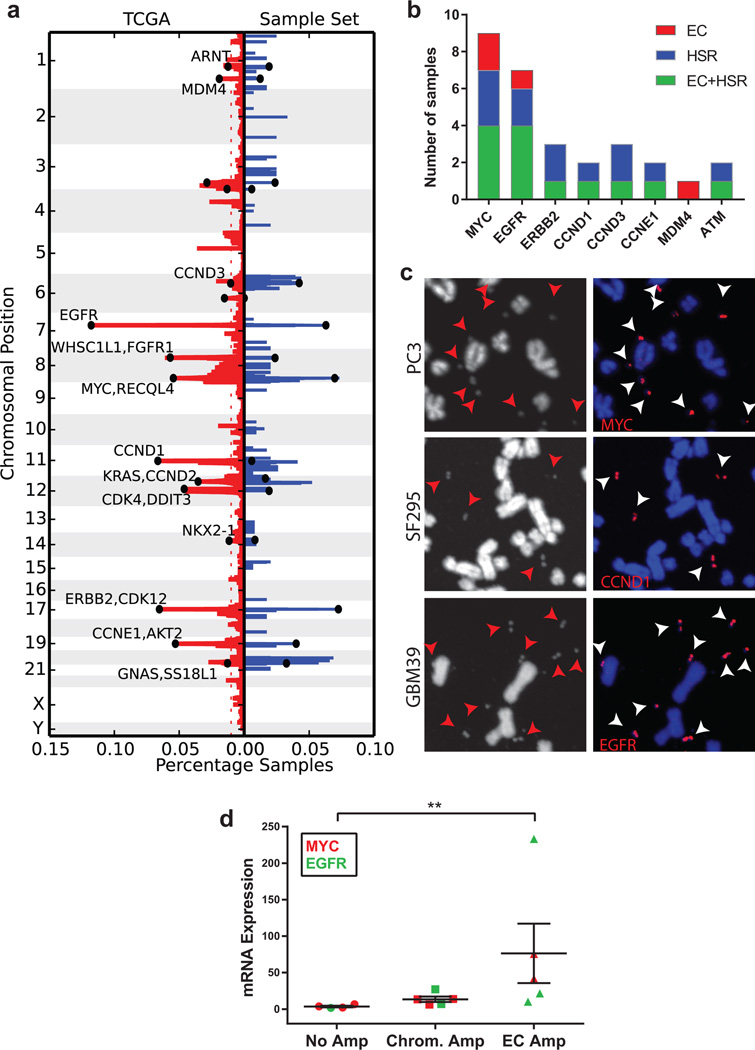
WGS with median coverage of 1.19× (Extended Data Fig. E4) revealed focal amplifications that were nearly identical to the amplifications found in the TCGA analyses of the same cancer types (Fig. 3A; Source Data Table S3), including amplified oncogenes found in a pan-cancer analysis of 13 different cancer types20. All of the amplified oncogenes tested were found solely on ECDNA, or concurrently on ECDNA and chromosomal homogenous staining regions (HSRs) (Fig. 3B–C; Extended Data Fig. E5–6). Oncogenes amplified in ECDNA expressed high levels of mRNA transcripts (Fig. 3D) and the copy number diversity of commonly amplified oncogenes in ECDNA far exceeded their copy number diversity if they were on other chromosomal loci (Extended Data Fig. E7).
Fig. 3. The most common focal amplifications in cancer are contained on ECDNA.
a, Comparison of the frequency of focal amplifications detected by next generation sequencing of 117 cancer samples studied here (blue), with those of matched tumor types in the TCGA (red), demonstrates significant overlap and representative sampling (p-value 10−6 based upon random permutations of TCGA amplicons; Methods). b, Localization of oncogenes by FISH. c, Representative FISH images of focal amplifications on ECDNA (arrows). d, EGFRvIII and c-Myc mRNA level, measured by qPCR (p < 0.001, Mann-Whitney test), mean ± SEM. n=17; each data point represents qPCR values from three technical replicates.
To determine whether extra- and intrachromosomal structures had a common origin, we developed ‘AmpliconArchitect’ to elucidate the finer genomic structure using sequencing data (Methods). To better understand the relationship between subnuclear location and amplicon structure, we took advantage of spontaneously occurring subclone of GBM39 cells in which high copy EGFRvIII shifted from ECDNA exclusively to HSRs. Independent replicates of GBM39 containing an ECDNA amplicon, revealed a consistent circular structure of 1.29 MB containing one copy of EGFRvIII (Extended Data Fig. E8). Remarkably, the GBM39 subclone harboring EGFRvIII exclusively on HSRs had an identical structure with tandem duplications containing multiple copies of EGFRvIII, indicating that the HSRs arose from reintegration of EGFRvIII-containing ECDNA elements (Extended Data Fig. E8)14. In GBM39 cells, resistance to the EGFR tyrosine kinase inhibitors is caused by reversible loss of EGFRvIII on ECDNA21. Structural analysis revealed a conservation of the fine structure of the EGFRvIII amplicon containing ECDNA in naïve cells, in treatment, and upon regrowth with discontinuation of therapy (Extended Data Fig. E9), indicating that ECDNA can dynamically relocate to chromosomal HSRs while maintaining key structural features14, 22.
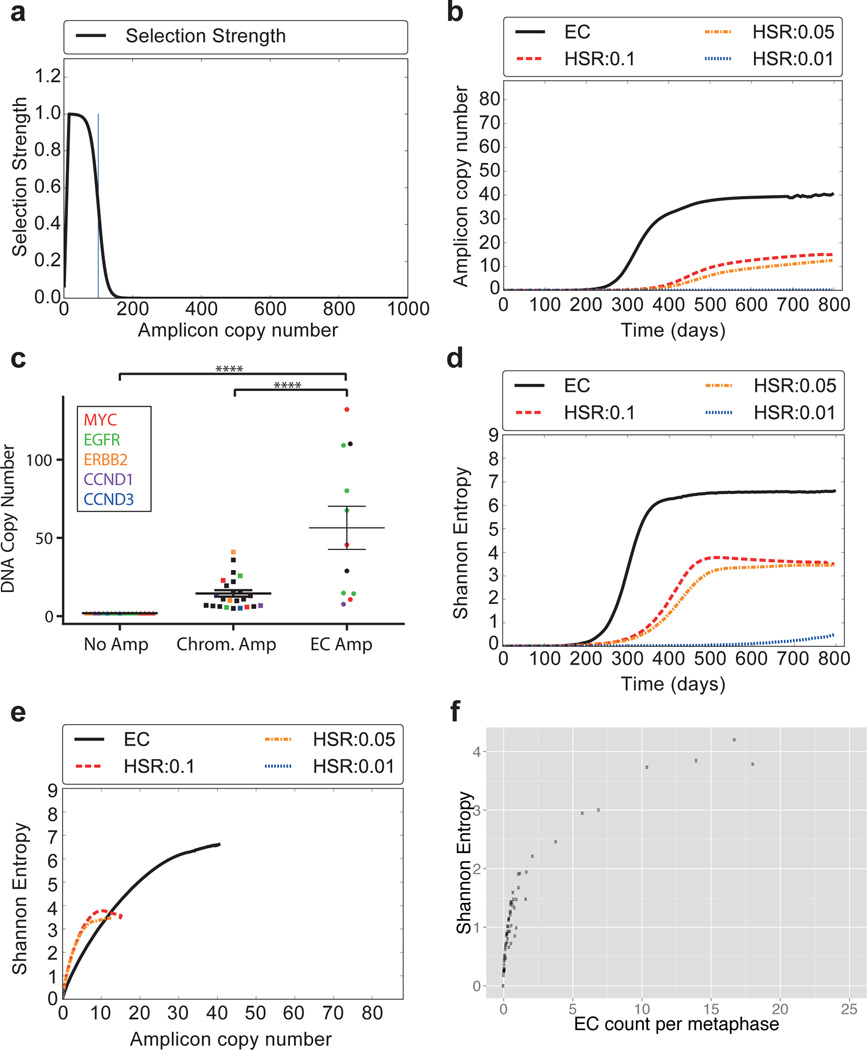
Does ECDNA localization confer any particular benefit? We hypothesized ECDNA amplification may enable an oncogene to rapidly reach higher copy number because of the unequal segregation to daughter cells15 than would be possible by intrachromosomal amplification. We used a simplified Galton-Watson branching process to model the evolution of a tumor23, where each cell in the current generation either replicates or dies to create the next generation. A cell with k copies of the amplicon is selected for replication with probability bk; bk/(1-bk) = 1 + sfm(k). We provided a positive selection bias towards cells with higher ECDNA counts by choosing s ϵ {0.5,1} along with different selection functions for f. Specifically, fm(k) increases to a maximum value fm(15) = 1, then declines in a logistic manner with fm(m) = 0.5 to reflect metabolic constraints (Methods). We allowed the amplicon copy number to grow to 1000 copies (Extended Data Fig. E10), but set bk = 0 for k ≥ 103. During cell division, the 2k copies resulting from the replication of each of the k ECDNA copies segregate independently into the two daughter cells. We contrasted this with an intrachromosomal model of duplication with identical selection constraints, but with the change in copy number affected by mitotic recombination, and achieved by incrementing or decrementing k by 1, with duplication probability pd. A range of values for pd, (0.01 ≤ pd ≤ 0.1) was used, where the upper bound reflects a change in copy number once every 5 divisions. The full assumptions of the model are explained in detail in Supplementary Material Section 4. Starting with an initial population of 105 cells, with s = 0.5 and m = 100 and a selection function f100(k) (Fig. 4A), we find that an oncogene can reach much higher copy number in a tumor if it is amplified on ECDNA, rather than on a chromosome (Fig. 4B). As predicted by the model, we detected significantly higher copy number of the most frequently amplified oncogenes EGFR (including EGFRvIII) and c-MYC, when they were contained within ECDNA instead of within chromosomes (Fig. 4C). We also reasoned that if an oncogene is amplified intra-chromosomally, the heterogeneity of the tumor (in terms of the distribution of copies of the oncogene) would stabilize at a much lower level. In contrast, unequal segregation of ECDNA would be likely to rapidly enhance heterogeneity and maintain it. Our model confirmed this prediction (Fig. 4D), consistently for a wide range of simulation parameters (Supplementary Material Section 4.3). The heterogeneity of copy number change stabilizes and even decreases over time10, 24, much as predicted in Fig. 4C–D. We also tested the validity of the model by comparing the Shannon entropy against the average number of amplicons per cell in our tumor samples. Heterogeneity of a tumor with respect to oncogene copy number would be more likely to rise relatively slowly if it is present on a chromosome, but would rise more rapidly and be maintained much longer, if that oncogene is present on ECDNA, as confirmed by a plot of Shannon entropy vs copy number (Fig. 4E). Moreover, the predicted correlation in Fig. 4E is completely recapitulated by the experimental data (Fig. 4F), thereby validating the central tenets of the model.
Fig. 4. Theoretical model for focal amplification via extrachromosomal (EC) and intrachromosomal (HSR) mechanisms.
Simulated change in copy number via random segregation (EC) or mitotic recombination (HSR), starting with 105 cells, 100 of which carry amplifications. a, The selection function f100(k) reaches maximum for k=15, then decays logistically. b, Growth in amplicon copy number over time. c, DNA copy number stratified by oncogene location. (p<0.001, ANOVA/Tukey’s multiple comparison). N=52; data points include top five amplified oncogenes, mean ± SEM. d, Change in heterogeneity (SI) over time. e, Correlation between copy number and heterogeneity. f, Experimental data showing correlation between ECDNA counts and heterogeneity matches the simulation in panel E.
There is growing evidence that genetically heterogeneous tumors are remarkably difficult to treat10. The data presented here identifies a mechanism by which tumors maintain cell-to-cell variability in the copy number and transcriptional level of oncogenes that drive tumor progression and drug resistance. We suggest that EC oncogene amplification may enable tumors to adapt more effectively to variable environmental conditions by increasing the likelihood that a subpopulation of cells will express that oncogene at a level that maximizes its proliferation and survival12, 21, 25–28, rendering tumors progressively more aggressive and difficult to treat over time. Even when using a selection function that only mildly depends on copy number, we detected a very large difference between intra-and extrachromosomal amplification mechanisms leading to higher copy number of amplicons and greater heterogeneity of copy number. Thus, even small increases in selection advantage conferred by oncogenes amplified on ECDNA would be expected to yield a very high fitness advantage (Supplementary Material Section 4.3). The strikingly high frequency of ECDNA in cancer, as shown here, coupled to the benefits to tumors of EC gene amplification relative to chromosomal inheritance, suggest that oncogene amplification on ECDNA may be a driving force in tumor evolution and the development of genetic heterogeneity in human cancer. Understanding the underlying molecular mechanisms of tumor evolution, including oncogene amplification in ECDNA, may help to identify more effective treatments that either prevent cancer progression or more effectively eradicate it.
Methods
Cytogenetics
Metaphase cells were obtained by treating cells with Karyomax (Gibco) at a final concentration of 0.01 µg/ml for 1–3 hours. Cells were collected, washed in PBS, and resuspended in 0.075 M KCl for 15–30 minutes. Carnoy’s fixative (3:1 methanol/glacial acetic acid) was added dropwise to stop the reaction. Cells were washed an additional 3 times with Carnoy’s fixative, before being dropped onto humidified glass sides for metaphase cell preparations. For ECdetect analyses, DAPI was added to the slides. Images in the main figures were captured with an Olympus FV1000 confocal microscope. All other images were captured at a magnification of 1000 with an Olympus BX43 microscope equipped with a QiClick cooled camera. FISH was performed by adding the appropriate DNA FISH probe onto the fixed metaphase spreads. A coverslip was added and sealed with rubber cement. DNA denaturation was carried out at 75 °C for 3–5 minutes and the slides were allowed to hybridize overnight at 37 °C in a humidified chamber. Slides were subsequently washed in 0.4× SSC at 50 °C for 2 minutes, followed by a final wash in 2× SSC/0.05% Tween-20. Metaphase cells and interphase nuclei were counterstained with DAPI, a coverslip was applied, and images were captured.
Cell culture
The NCI-60 cell line panel (gift from Andrew Shiau-obtained from NCI) was grown in RPMI-1640 with 10% FBS under standard culture conditions. Cell lines were not authenticated, as they were obtained from the NCI. The PDX cell lines were cultured in DMEM/F-12 media supplemented with Glutamax, B27, EGF, FGF, and Heparin. Lymphoblastoid cells (gifts from Bing Ren) were grown in RPMI-1640, supplemented with 2 mM glutamine and 15% FBS. IMR90 and ALS6-Kin4 (gift from John Ravits and Don Cleveland) cells were grown in DMEM/F-12 supplemented with 20% FBS. Normal human astrocytes (NHA) and normal human dermal fibroblasts (NHDF) were obtained from Lonza and cultured according to Lonza-specific recommendation. Cell lines were not tested for mycoplasma contamination.
Tissue samples
Tissues were obtained from the Moores Cancer Center Biorepository Tissue Shared Resource with IRB approval (#090401). All samples were de-identified and patient consent was obtained. Additional tissue samples that were obtained were approved by the UCSD IRB (#120920).
DNA library preparation
DNA was sonicated to produce 300–500bp fragments. DNA end repair was performed using End-it (Epicentre), DNA library adapters (Illumina) were ligated, and the DNA libraries were amplified. Paired-end next generation sequencing was performed and samples were run on the Illumina Hi-Seq using 100 cycles.
DNA extraction
Cells were collected and washed with 1× cold PBS. Cell pellets were resuspended in Buffer 1 (50 mM Tris, pH 7.5, 10 mM EDTA, 50 µg/ml RNase A), and incubated in Buffer 2 (1.2% SDS) for 5 minutes on ice. DNA was acidified by the addition of Buffer 3 (3 M CsCl, 1 M potassium acetate, 0.67 M acetic acid) and incubated for 15 minutes on ice. Samples were centrifuged at 14,000 × g for 15 minutes at 4 °C. The supernatant was added to a Qiagen column and briefly centrifuged. The column was washed (60% ethanol, 10 mM Tris pH 7.5, 50 µM EDTA, 80 mM potassium acetate) and eluted in water.
DNase treatment
Metaphase cells were dropped onto slides and visualized via DAPI. Coverslips were removed and slides washed in 2× SSC, and subsequently treated with 2.5% trypsin, and incubated at 25 °C for 3 minutes. Slides were then washed in 2× SSC, DNase solution (1 mg/ml) was applied to the slide, and cells were incubated at 37 °C for 3 hours. Slides were washed in 2× SSC and DAPI was again applied to the slide to visualize DNA.
ECDNA count statistics
In Figures 2A and 2B, the violin plots represent the distribution of ECDNA counts in different sample types. In order to compare the ECDNA counts between the different samples, we use a one-sided Wilcoxon rank sum test, where the null hypothesis assumes the mean ECDNA count ranks of the compared sample types equal.
Estimation of frequency of samples containing ECDNA
There is a wide variation in the number of ECDNA across different samples and within metaphases of the same sample. We want to estimate and compare the frequency of samples containing ECDNA for each sample type. We label a sample as being ECDNA-positive by using the pathology standard: a sample is deemed to be ECDNA-positive if we observe ≥ 2 ECDNA in ≥ 2 images out of 20 metaphase images. Therefore, we ensure that every sample contains at least 20 metaphases.
We define indicator variable Xij = 1 if metaphase image j in sample i has ≥ 2 ECDNA; Xij = 0 otherwise. Let ni be the number of metaphase images acquired from sample i. We assume that Xij is the outcome of the jth Bernoulli trial, where the probability of success pi is drawn at random from a beta distribution with parameters determined by ∑jXij. Formally,
We model the likelihood of observing k successes in n = 20 trials using the binomial density function as:
Finally, the predictive distribution p(k), is computed using the product of the Binomial likelihood and Beta prior, modeled as a “beta-binomial distribution”29.
We model the probability for sample i being ECDNA-positive with the random variable Yi such that:
The expected value of Yi is:
Let T be the set of samples belonging to a certain sample type t, e.g. immortalized samples.
We define
We estimate the frequency of samples under sample t containing ECDNA (bar heights on Figures 2C and 2D) as
and error bar heights (Figure 2C and 2D) as:
assuming independence among samples i ϵ T. For any αi or βi = 0, we assign them a sufficiently small ε. For more detail, please see Supplementary Material Section 1.
Comparison of ECDNA presence between different sample types
We construct binary ECDNA presence distributions, based on the ECDNA counts, such that an image with ≥ 2 ECDNA is represented as a 1, and 0 otherwise. In order to compare the ECDNA presence between the different samples, we use a one-sided Wilcoxon rank sum test using the binary ECDNA presence distributions, where the null hypothesis assumes the mean ranks of the compared sample types equal.
ECdetect: Software for detection of extrachromosomal DNA from DAPI staining metaphase images
The software applies an initial coarse adaptive thresholding30, 31 on the DAPI images to detect the major components in the image with a window size of 150×150 pixels, and T = 10% Components breaching 3000 pixels and 80% of solidity are masked, and small components discarded. Weakly connected components (CC) of the remaining binary image are computed to find the separate chromosomal regions. CC breaching a cumulative pixel count of 5000 are considered as candidate search regions, and their convex hull with a dilation of 100 pixels are added into the ECDNA search region. Following the manual masking and verification of the ECDNA search region, a second finer adaptive thresholding with a window size of 20×20 pixels and T = 7% is performed. Components that are greater than 75 pixels are designated as non-ECDNA structures and their 15 pixel neighborhood is removed from the ECDNA search region. Any component detected with a size less than or equal to 75 and greater than or equal to 3 pixels inside the search region is detected as ECDNA. For more detail, please see Supplementary Material Section 2.
Bioinformatic datasets
We sequenced 117 tumor samples including 63 cell lines, 19 neurospheres and 35 cancer tissues with coverage ranging from 0.6× to 3.89× and an additional 8 normal tissues as controls. See Extended Data Figure E4 for the coverage distribution across samples. We mapped the sequencing reads from each sample to hg19 (GRCh37) human reference genome32 from UCSC genome browser33 using BWA software version 0.7.9a34. We inferred an initial set of copy number variants from these mapped sequence samples using the ReadDepth CNV software35 version 0.9.8.4 with parameters FDR = 0.05 and overDispersion = 1.
We downloaded copy number variation calls (CNV) for 11079 tumor-normal samples covering 33 different tumor types from TCGA. We applied similar filtering criteria to ReadDepth output and TCGA calls to eliminate false CN amplification calls from repetitive genomic regions and hotspots for mapping artefacts.
We used the filtered set of CNV calls from ReadDepth as input probes for AmpliconArchitect which revealed the final set of amplified intervals and the architectures of the amplicons. See Supplementary Material Section 3 for more details.
Reconstruction using AmpliconArchitect
We developed a novel tool AmpliconArchitect (AA), to automatically identify connected amplified genomic regions and reconstruct plausible amplicon architectures. For each sample, AA takes as input an initial list of amplified intervals and whole genome sequencing (WGS) paired-end reads aligned to the human reference. It implements the following steps to reconstruct the one or more architectures for each amplicon present in the sample: (a) Use discordant read-pair alignments and coverage information to iteratively visit and extend connected genomic regions with high copy numbers. (b) For each set of connected amplified regions, segment the regions based on depth of coverage using a mean-shift segmentation to detect copy number changes and discordant read-pair clusters to identify genomic breaks. (c) Construct a breakpoint graph connecting segments using discordant read-pair clusters. (d) Compute a maximum likelihood network to estimate copy counts of genomic segments. (e) Report paths and cycles in the graph that identify the dominant linear and circular structures representing one. (Supplementary Material Section 3)
Comparison of CNV gains between the sequencing sample set and TCGA
We compared our sample set against TCGA samples to test the assumption that the genomic intervals amplified in our sample set are broadly representative of a pan-cancer dataset, by comparing against TCGA samples. Here, we deal with an abstract notation to represent different datasets and describe a generic procedure to compare amplified regions. Consider a set of K samples. For any k ∈ [1, …, K], let Sk denote the set of amplified intervals in sample k.
Let c be the cancer subtype for sample k. We compare Sk against TCGA samples with sub-type c. Let T denote the set of all genomic regions which are amplified in at least 1% of TCGA samples of subtype c. For each interval t ∈ T, let ft denote its frequency in TCGA samples of subtype c. We define a match score
The cumulative match score for all samples is defined as:
To compute the significance of statistic D, we do a permutation test. We generate N random permutations of the TCGA intervals for subtype c and estimate distribution of match scores of our sample set against the random permutations. We choose a random assignment of locations of all intervals in T, while retaining their frequencies. For the jth permuted set Tj, we computed the cumulative match score Dj relative to our sample set. Thus the significance of overlap between our sample set and the TCGA amplified intervals is estimated by the fraction of random permutations with Dj > D. Computing 1 million random permutations generated exactly one permutation breaching the TCGA score D, implying a p-value ≤ 10−6.
Oncogene Enrichment
We compared the rank correlation of the most frequent oncogenes in our sample set with the top oncogenes as reported by TCGA pan-cancer analysis by Zack et al20. We identified 14 oncogenes occurring in 2 or more samples of our sample set and compared these with the top 10 oncogenes from the TCGA pan-cancer analysis. We found that 7 out of the top 10 oncogenes were represented in our list of 14 oncogenes. Considering 490 oncogenes in the COSMIC database, the significance of observing 7 or more oncogenes in common in the two datasets is given by the hypergeometric probability
Amplicon structure similarity
We found high similarity between amplicon structures of biological replicates (e.g. Extended Data Figure E8). We estimate probability of common origin between two samples by measuring the pairwise similarity between amplicon structures. In reconstructing the structures (Supplementary Material Section 3), we identify a set of locations representing change in copy number and we use the locations of change in copy number to estimate the similarity in amplicon structures.
Let L be the total length of amplified intervals. These intervals are binned into windows of size r, resulting in Nb = L/r bins. We use a segmentation algorithm that determines if there is a change in copy number in any bin, within a resolution of r = 10,000 bp. (See Meanshift in coverage: Supplementary Materials Section 3.2.) Note that this is an over-estimate, since with split-reads and high density sequencing data, we can often get the resolution down to a few base pairs. Let S1 and S2 represent the set of bins with copy number changes in the two samples, respectively. S1 and S2 are selected from a candidate set of locations Nb. Under the null hypothesis that S2 is random with respect to S1, we expect I = S1 ∩ S2 to be small. Let m = min{|S1|, |S2|}, and M = max{|S1|, |S2|}. A p-value is computed as follows:
In looking at GBM39 replicates (Extended Data Figure E8), we find that all replicates displaying EGFR ECDNA are similar to each other. Comparing replicates in row 1 and row 2 among |Nb| = 129 bins (1.29 Mbp), |S1| = 5 corresponding to row 1 (EC sample), |S2| = 6 corresponding to row 2 (EC sample) and intersection set size |I| = 5, we compute the p-value for observing such structural similarity by random chance is 2.18 × 10−8 which is the highest p-value among all EC replicate pairs. In addition, we compare the replicates displaying EGFR on ECDNA with the culture displaying EGFR on HSR. Among |Nb| = 129 bins, |S1| = 6 corresponding to row 2 (EC), |S2| = 4 corresponding to row 4 (HSR), the intersection set has size |I| = 4 intervals giving a p-value of 1.98 × 10−5 which gives the highest p-value among the 3 ECDNA replicates compared to the HSR culture, suggesting a common origin.
A branching process model for oncogene amplification
Consider an initial population of N0 cells, of which Nα cells contain a single extra copy of an oncogene. We model the population using a discrete generation Galton-Watson branching process23. In this simplified model, each cell in the current generation containing k amplicons (amplifying an oncogene) either replicates with probability bk to create the next generation, or dies with probability 1 - bk to create the next generation. We set the selective advantage
| (1) |
In other words, cells with k copies of the amplicon stop dividing after reaching a limit of Mα amplicons. Otherwise, they have a selective advantage for 0 < k ≤ Mα, where the strength of selection is described by fm(k), as follows:
| (2) |
Here, s denotes the selection-coefficient, and parameters m and α are the ‘mid-point’, and ‘steepness’ parameters of the logistic function, respectively. Initially, fm(k) grows linearly, reaching a peak value of fm(k) = 1 for k = Ms. As the viability of cells with large number of amplicons is limited by available nutrition36, fm(k) decreases logistically in value for k > Ms reaching fm(k) → 0 for k ≥ Mα. We model the decrease by a sigmoid function with a single mid-point parameter m s.t. fm(m) = 1/2. The ‘steepness’ parameter α is automatically adjusted to ensure that min{1 – fm(Ms), fm(Mα)} → 0.
The copy number change is affected by different mechanisms for extrachromosomal (EC) and intrachromosomal (HSR) models. In the EC model, the available k amplicons are on EC elements which replicate and segregate independently. We assume complete replication of EC elements so that there are 2k copies which are partitioned into the two daughter cells via independent segregation. Formally, the daughter cells end up with k1 and k2 amplicons respectively, where
| (3) |
| (4) |
In contrast, in the intrachromosomal model, the change in copy number happens via mitotic recombination, and the daughter cell of a cell with k amplicons will acquire either k + 1 amplicons or k − 1 amplicons, each with probability pd. With probability 1 – 2pd, the daughter cell retains k amplicons. See Supplementary Material Section 4 for more details.
Code and data availability
AmpliconArchitect is available for use online at: https://github.com/virajbdeshpande/AmpliconArchitect. ECdetect will be available upon request. Whole genome sequencing data is deposited to the NCBI Sequence Read Archive (SRA) under Bioproject at http://www.ncbi.nlm.nih.gov/bioproject/338012 with Accession ID PRJNA338012. DAPI and FISH metaphase images are available for download on figshare at https://figshare.com/s/ab6a214738aa43833391.
Extended Data
Figure E1.
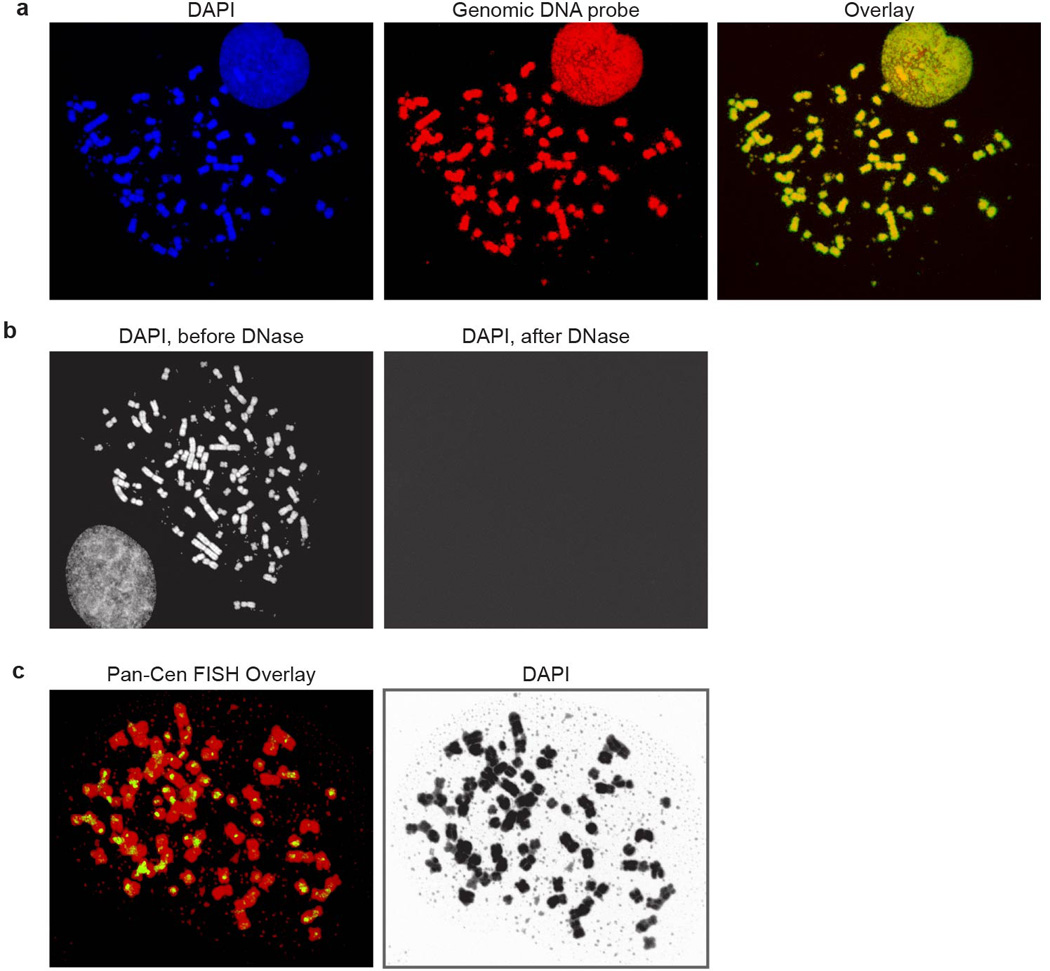
Full metaphase spreads corresponding to the partial metaphase spreads shown in Figure 1. a, Images corresponding to Fig. 1B, b, images corresponding to Fig. 1C, c, images corresponding to Fig. 1D.
Figure E2.
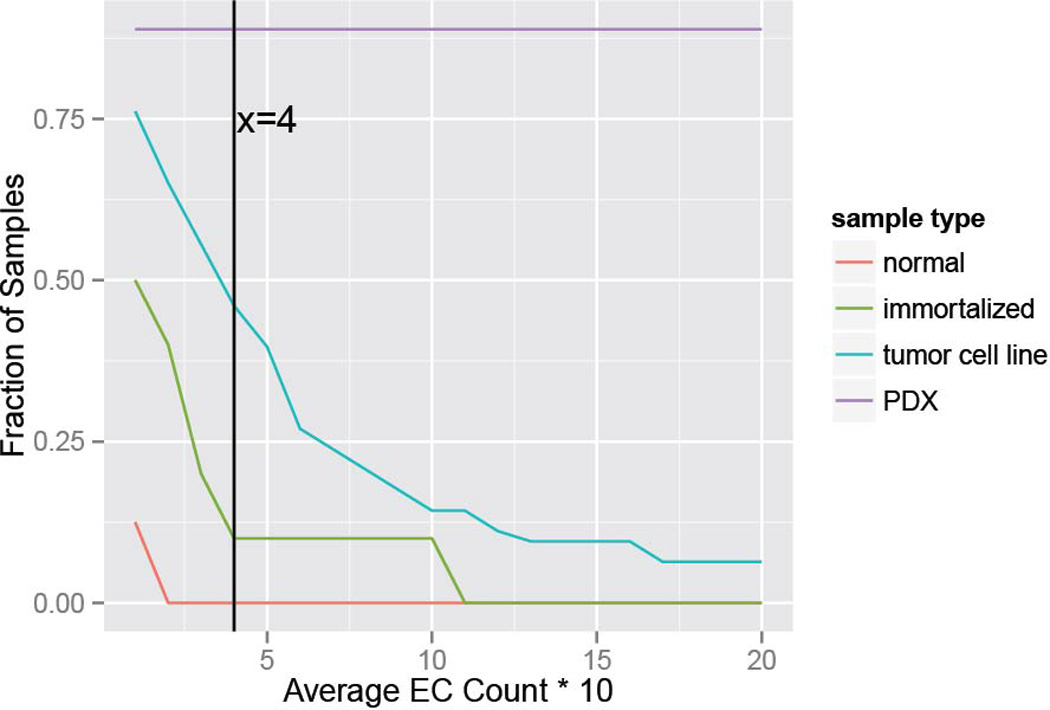
Alternative analysis of ECDNA presence according to varying criteria, stratified by sample type: Samples with a minimum number of ECDNA per 10 metaphases in average shown in x-axis are classified ECDNA-positive, and their fraction is displayed on the y-axis. The vertical line at x=4 shows that for a minimum of 4 ECDNA per 10 metaphases on average, 0% of normal, 10% of immortalized, 46% of tumor cell line and 89% of PDX samples are classified as ECDNA positive.
Figure E3.
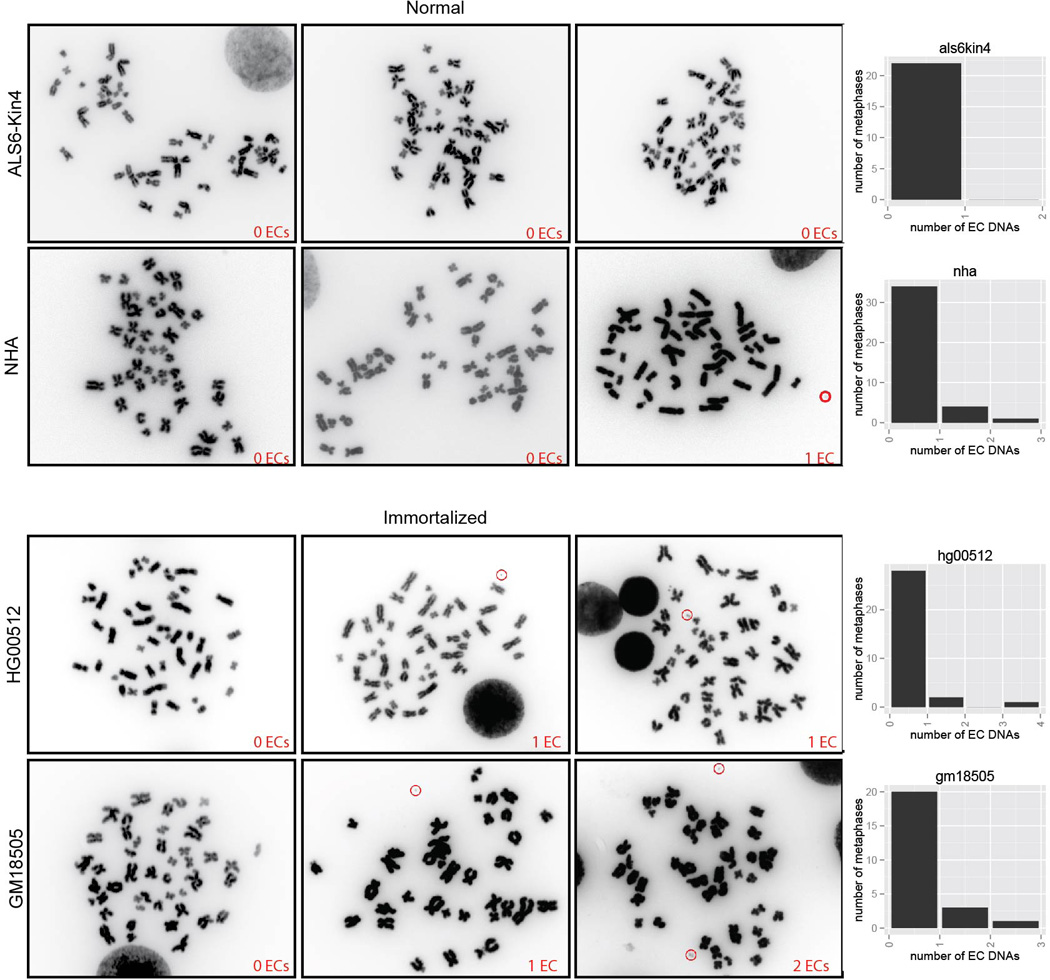
ECDNA counts in normal and immortalized cells.
Figure E4.
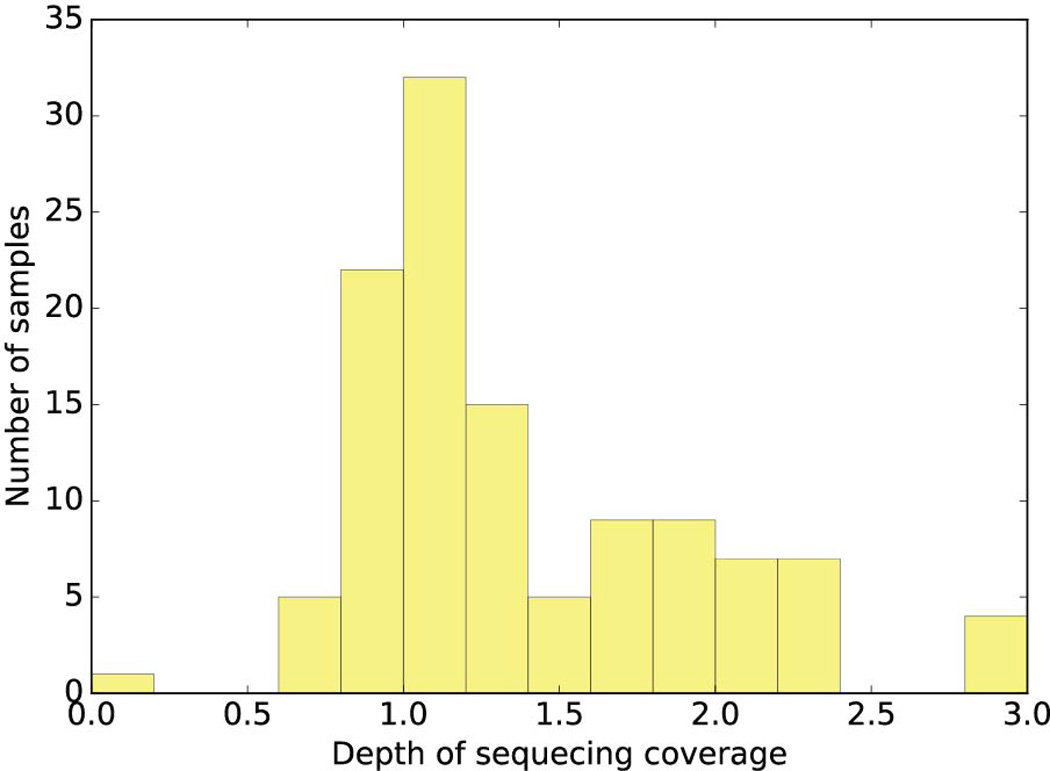
Histogram of depth of coverage for next-generation sequencing of tumor samples. We sequenced 117 tumor samples including 63 cell lines, 19 neurospheres (PDX) and 35 cancer tissues with coverage ranging from 0.6× to 3.89× (excluding one sample with 0.06 × coverage) with median coverage of 1.19×.
Figure E5.
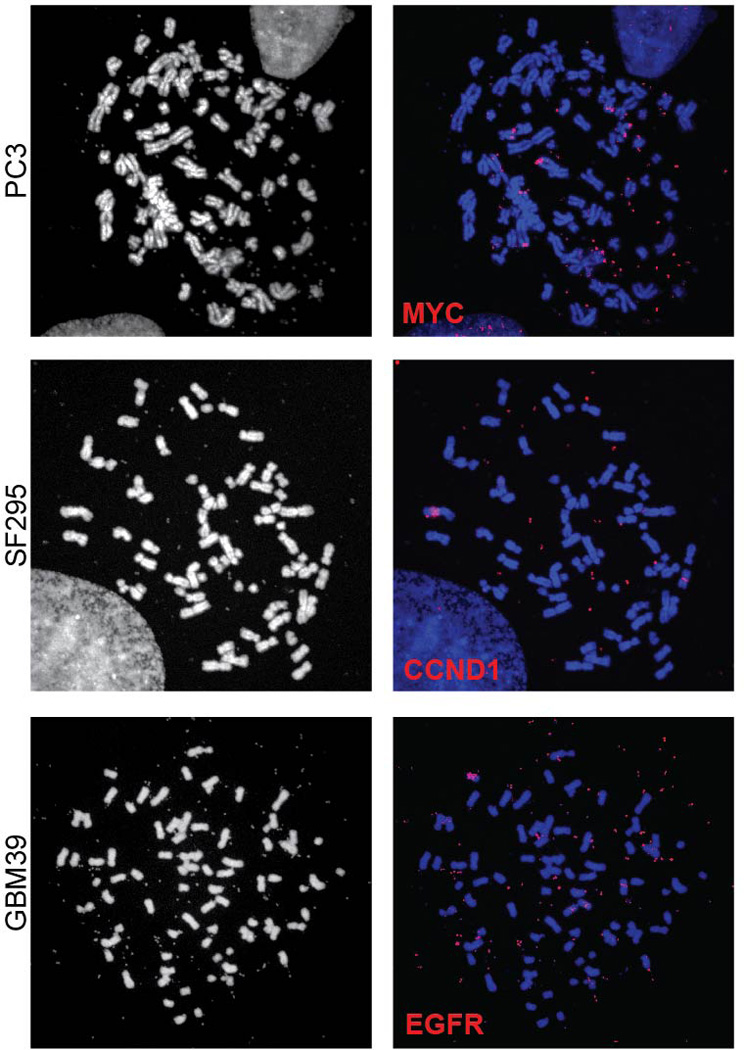
Full metaphase spreads corresponding to the partial metaphase spreads shown in Figure 3C.
Figure E6.
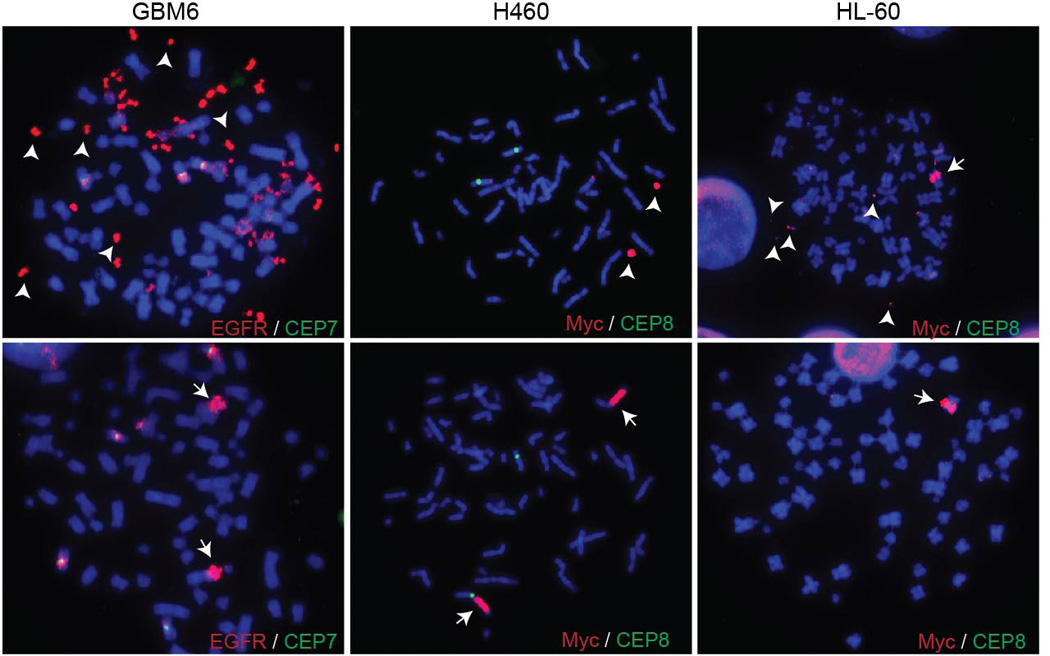
FISH images displaying both ECDNAs and HSRs in cells from the same sample.
Figure E7.
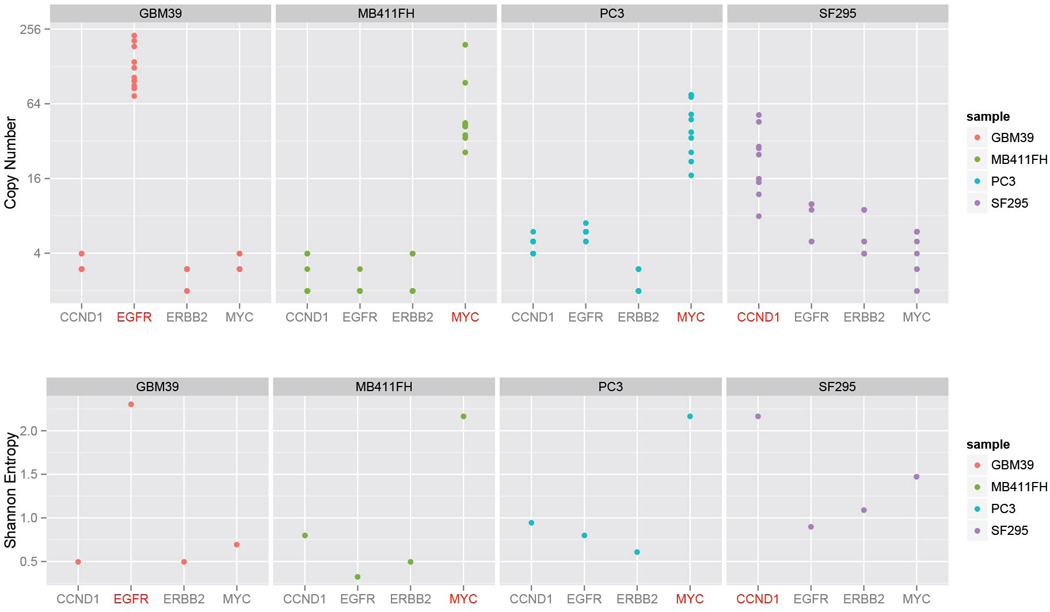
Copy number amplification and diversity due to ECDNA. To test how much of the copy number and diversity could be attributed to ECDNA, we chose FISH probes that bind to four of the most commonly amplified oncogenes in our sample set, EGFR, MYC, CCND1 or ERBB2, and quantified the cell-to-cell variability in their DNA copy number in metaphase spreads, from four tumor cell lines: GBM39, MB411FH, SF295 and PC3 cancer cells. For each cell line, only the target oncogene marked in red is known to be amplified on ECDNA (EGFR in GBM39; MYC in MB411FH and PC3, and CCND1 in SF295). The other 3 genes reside on chromosomal loci. The target oncogene shows consistently higher copy numbers (Top Panel) and diversity (Bottom Panel).
Figure E8.
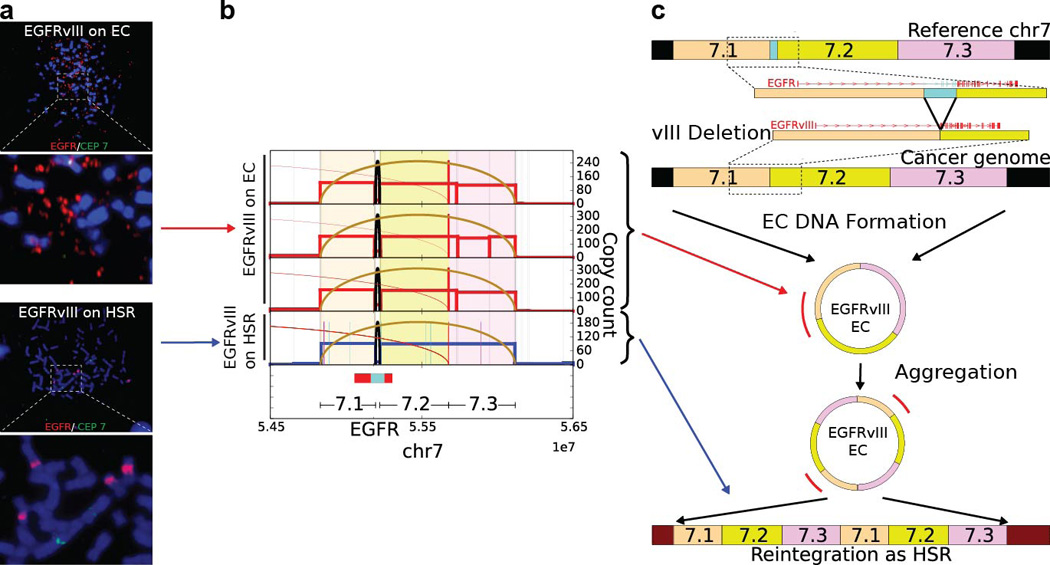
Fine structure analysis of EGFRvIII Amplification in Extrachromosomal or Chromosomal DNA in GBM39 Cells: a., FISH images revealed EGFR gene on ECDNAs (top) and HSRs (bottom) on different passes of the GBM39 cell line. Analysis of the HSR FISH images shows evidence of multiple integration sites on different chromosomes. b., Next generation sequencing of DNA from 4 independent cultures of GBM39 was used to analyze the fine structure of amplifications (Supplementary Material Section 4.3). In 3 biological replicates (rows 1 to 3) of these cultures, EGFRvIII was exclusively on ECDNA, while one of the later passage cultures (row 4) was found to contain EGFRvIII entirely on HSRs, with no detectable ECDNA. The DNA derived from different ECDNA cultures shows identical structure with some heterogeneity (p < 2.18 × 10−8 for all pairs), suggesting common origin. However, DNA derived from HSRs reveals a conserved structure that is identical to ECDNA structure (p < 1.98 × 10−5, Supplementary Material Section 2.4), possibly with tandem duplications. c., A possible progression of normal genome to cancer genome with EGFRvIII ECDNAs and amplification to a copy count of around 100 copies. The EGFRvIII ECDNAs possibly aggregate into tandem duplications and reintegrate into multiple chromosomes as HSRs such that 5–6 HSRs accommodate around 100 copies of EGFRvIII.
Figure E9.
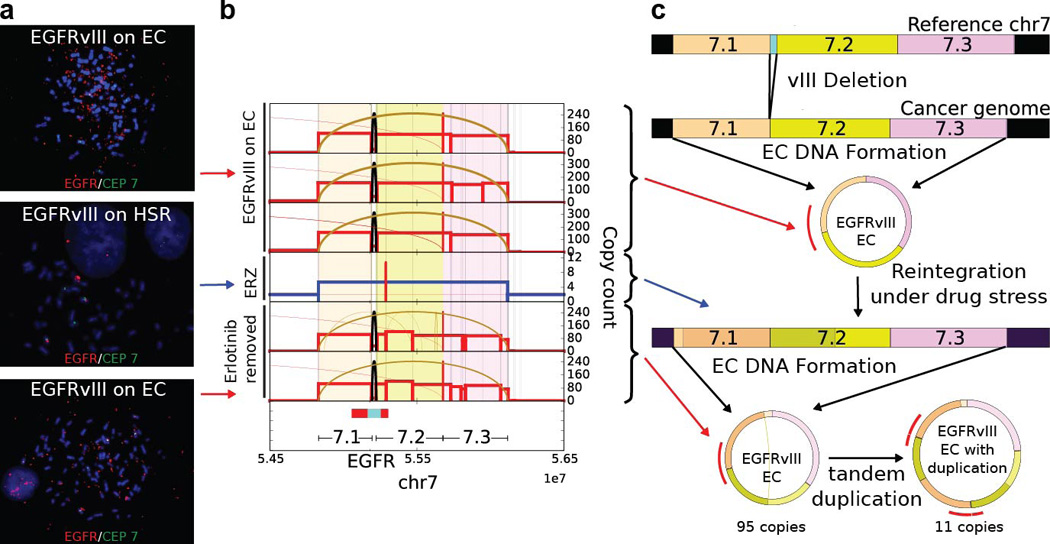
Fine structure analysis of EGFRvIII Amplification in Extrachromosomal or Chromosomal DNA in naive GBM39 cells and in response to Erlotinib Treatment (ERZ) and Drug Withdrawal: a., FISH images of naive GBM39 cells, in response to Erlotinib Treatment (ERZ) and Drug Withdrawal displayed EC amplification, HSR amplification and EC amplification respectively (top to bottom). b., Next generation sequencing of DNA from 6 independent cultures of GBM39 was used to analyze the fine structure of amplifications (Supplementary Material Section 4.3). Average copy numbers of amplified intervals as determined from sequencing analysis in naive samples (biological replicates in rows 1 to 3): 110 to 150, ERZ sample (row 4): 5.4 and Erlotinib removed (biological replicates in rows 5 and 6): 100–105. All three categories show similar fine structure indicating common origin (Methods). Erlotinib removed replicates show additional rearrangements and heterogeneity as compared to naive samples. c., Cytogenetic and sequencing progression suggests the EGFRvIII ECDNAs in naive cells get reintegrated into HSRs after drug application and the copies in the HSRs break off from the chromosomes again to form ECDNAs with copy count similar to naive cells. Drug removed samples also show additional heterogeneity in structure.
Figure E10.
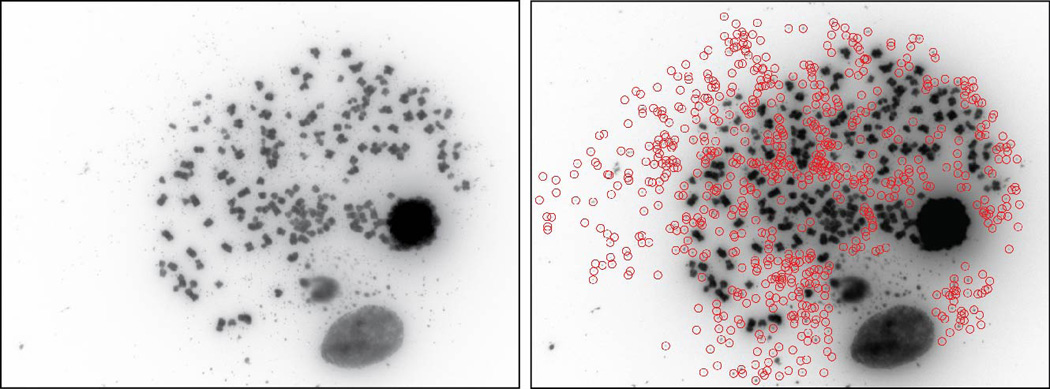
A GBM metaphase spread with large ECDNA counts (> 600), as determined by manual counting and ECdetect.
Supplementary Material
Acknowledgments
We thank Richard Kolodner, Walter Mischel, Dan Geschwind, members of the Mischel laboratory, Ali Akbari, Arya Iranmehr and Anand Patel for helpful comments. This work was supported by the Ludwig Institute for Cancer Research (PSM, BR, KA, WKC, FF), Defeat GBM Program of the National Brain Tumor Society (PSM, FF), The Ben and Catherine Ivy Foundation (PSM), generous donations from the Ziering Family Foundation in memory of Sigi Ziering (PSM); The Susan G. Komen Foundation (SAC110036), The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002) and The Breast Cancer Research Foundation (BCRF) to GMW; CureSearch for Children’s Cancer and a Leadership Award from the California Institute for Regenerative Medicine to RWR. This work was also supported by the following NIH grants: NS73831 (PSM), GM114362 (VB, VD, DB), NS80939 (FF), CA014195 and CA159859 (GMW) and CA151819 (DAN) and T32CA121938 (KMT) and NSF grants: NSF-IIS-1318386 and NSF-DBI-1458557 (VB, VD, DB).
Footnotes
Author information:
K.M.T., V.D., D.B., V. B. and P.S.M. conceived and designed the study. K.M.T,, V.D., D.B., T.K. performed experiments. D.B., V.D., and V.B. developed the ECdetect software and performed mathematical modeling and simulations.. V.D. and V.B. developed the AmpliconArchitect software. K. A, B.R., D.N., F.B.F., W.K.C., P.N.R. and G.M.W. provided analytic support. H.I.K., M.D.T., K.S., R.W., S.R.V. provided additional clinical samples and analytic support. K.M.T., V.D., D.B., V.B. and P.S.M. wrote the manuscript with feedback from all authors.
Dr. Vineet Bafna is a co-founder, has an equity interest, and receives income from Digital Proteomics, LLC. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. DP was not involved in the research presented here.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Debatisse M, Giulotto E, Wahl GM. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989;57:901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- 3.Schimke RT. Gene amplification in cultured animal cells. Cell. 1984;37:705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- 4.Fan Y, et al. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J Appl Genet. 2011;52:53–59. doi: 10.1007/s13353-010-0007-z. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 6.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 8.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12:487–493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Von Hoff DD, Needham-VanDevanter DR, Yucel J, Windle BE, Wahl GM. Amplified human MYC oncogenes localized to replicating submicroscopic circular DNA molecules. Proc Natl Acad Sci U S A. 1988;85:4804–4808. doi: 10.1073/pnas.85.13.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garsed DW, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Carroll SM, et al. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol. 1988;8:1525–1533. doi: 10.1128/mcb.8.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Windle B, Draper BW, Yin YX, O’Gorman S, Wahl GM. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 16.Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- 17.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2016 < http://cgap.nci.nih.gov/Chromosomes/Mitelman>.
- 18.Sanborn JZ, et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–6045. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almendro V, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storlazzi CT, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–1206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett’s esophagus. Cancer Prev Res (Phila) 2014;7:114–127. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra S, Whetstine JR. Different Facets of Copy Number Changes: Permanent, Transient, and Adaptive. Mol Cell Biol. 2016;36:1050–1063. doi: 10.1128/MCB.00652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schimke RT, Kaufman RJ, Alt FW, Kellems RF. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–1055. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- 27.Nikolaev S, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat Commun. 2014;5:5690. doi: 10.1038/ncomms6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biedler JL, Schrecker AW, Hutchison DJ. Selection of chromosomal variant in amethopterin-resistant sublines of leukemia L1210 with increased levels of dihydrofolate reductase. J Natl Cancer Inst. 1963;31:575–601. [PubMed] [Google Scholar]
Online Methods References
- 29.Lee PM. Bayesian statistics: an introduction. 4th. John Wiley & Sons; 2012. [Google Scholar]
- 30.Motl J. < https://www.mathworks.com/matlabcentral/fileexchange/40854>.
- 31.Bradley D, Roth G. Adaptive thresholding using the integral image. Journal of graphics, gpu, and game tools. 2007;12:13–21. [Google Scholar]
- 32.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 33.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller CA, Hampton O, Coarfa C, Milosavljevic A. ReadDepth: a parallel R package for detecting copy number alterations from short sequencing reads. PLoS One. 2011;6:e16327. doi: 10.1371/journal.pone.0016327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.