Pre-mRNA splicing in eukaryotes requires joining together the nucleotides of the various mRNA-coding regions (exons) after recognizing them from the normally vastly superior number of non-mRNA-coding sequences (introns). For three excellent reviews on general splicing and its regulation, refer to references 14, 62, and 70. In eukaryotes, the vast majority of splicing processes are catalyzed by the spliceosome, a very complex RNA-protein aggregate which has been estimated to contain several hundred different proteins in addition to five spliceosomal snRNAs (1, 54, 62, 63, 81, 109). These factors are responsible for the accurate positioning of the spliceosome on the 5′ and 3′ splice site sequences. The reason why so many factors are needed reflects the observation that exon recognition can be affected by many pre-mRNA features such as exon length (5, 97), the presence of enhancer and silencer elements (8, 62), the strength of splicing signals (45), the promoter architecture (29, 55), and the rate of RNA processivity (86). In addition, the general cellular environment also exerts an effect, as recent observations suggest the existence of extensive coupling between splicing and many other gene expression steps (69) and even its modification by external stimuli (96).
In the midst of all this complexity, it has also been proposed that pre-mRNA secondary structures can potentially influence splicing activity. However, despite a steady increase of reports invoking their effects on splicing regulation, the last specific review on this subject is now more than 10 years old (3). Here, we propose to address again this specific issue in the current perspective of the general field. Before we do this, however, we have to answer a basic question.
DO PRE-mRNAS PRESENT SECONDARY STRUCTURE IN VIVO?
Two properties of RNA molecules cannot be denied: their natural tendency to form highly stable secondary and tertiary structures in vitro and in vivo (9, 27, 39) and the observation that alterations in these structures represent a well-known regulatory mechanism for many RNA cellular processes (60).
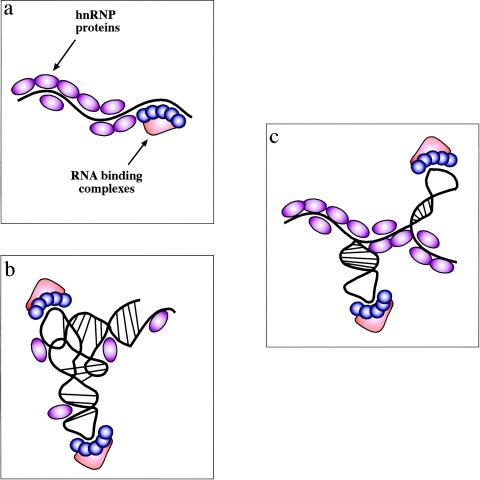
In this particular respect, however, a question that still remains to be addressed conclusively regards the presence of secondary structures in pre-mRNAs in vivo. That this existence may not simply be taken for granted comes from early experimental evidence. In fact, it was suggested that in vitro evidence regarding the possible influence of RNA structure on splicing (94) could not be accurately reproduced in vivo (95). The reason why this should be so goes back to the classical concept that RNA is coated in vivo by proteins. In fact, heterogeneous ribonucleoprotein particles have been known since early studies and the major protein family involved, the hnRNP proteins, are very abundant in mammalian cells. These RNA-protein interactions may well prevent mRNAs from folding in stable secondary structures (34) (Fig. 1a). For this reason, it was hypothesized that, following transcription, pre-mRNA may be allowed only a very limited timespan to fold (36). Consistent with this view, studies with artificial constructs used for the quantification of enhancer activities yielded results which supported the hypothesis that these pre-mRNA molecules behaved largely as a linear structure (44).
FIG. 1.
Experimental models of RNA secondary structures in mRNAs. There are three possible experimental models of RNA secondary structures in mRNAs. (a) The first one is represented by the view that hnRNP proteins bind the mRNA as it gets transcribed by RNA polymerase II and keep it in a largely linear conformation. In this case, binding of specific factors is regulated only by the competitive advantage provided by sequence-specific interactions over the generic RNA binding affinities of all hnRNP proteins. (b) The opposite situation is one where the drive to form RNA secondary and tertiary structures is stronger than the ability of RNA-binding proteins to prevent it (and maybe even stabilized by these proteins). In this case, the role played by generic RNA-binding proteins is severely reduced and specific complexes can bind through a mix of sequence-specific and structure-specific recognition. (c) Between these two models is a situation that should encompass many cellular mRNAs. In this case, the potential “ironing” of the mRNA by its weak or aspecific interactions with hnRNPs can indeed maintain the mRNA in a largely linear conformation. However, in particular regions the mRNA is still able to form localized RNA structures which might represent, together with the nucleotide sequence, preferential binding sites for specific nuclear complexes. Of course, given the enormous variety of mRNAs produced by the cell these models cannot be considered mutually exclusive, although there is probably a distinct preference for the model in panel c.
Notwithstanding these results, there are also some problems with the view that this situation may be applied to the vast majority of pre-mRNA molecules. Clearly, considering the enormously diverse sequences of all processed pre-mRNAs, it would be quite over the line to propose the presence of highly stable secondary structures (Fig. 1b) that resemble those of the highly conserved tRNAs, rRNAs, IRES, or other stability-, replication-, and localization-controlling elements present in several 3′UTRs of prokaryotic and eukaryotic mRNAs, in which proteins may also play a key role in stabilizing the structure (60). However, in between these two extremes there may exist a third possibility, represented by the existence of a loose amount of RNA-specific secondary structures which might, under normal conditions, influence the splicing machinery (Fig. 1c). Significantly, several studies along this line have been reported. For example, in organisms such as Saccharomyces cerevisiae, probing of pre-mRNA structures by dimethyl sulfate in vivo has demonstrated the existence of secondary structure formation between the 5′ splice site and the branch point capable of promoting U1snRNP assembly in the early splicing stages (21). Although there is no comparable evidence for human systems, it has been reported recently that single-nucleotide polymorphisms are capable of inducing in vivo different structural folds in mRNA structures (88) (however, the effect of these single-nucleotide polymorphisms on splicing or function has not yet been tested). In addition, statistical analysis of mRNA coding sequences has revealed that the calculated mRNA folding is more stable than expected by chance, suggesting that codon bias may favor the existence of mRNA structures (87). Even though these results have been challenged using a different set of statistical tools and genes (107), considerations analogous to those of Seffens and Digby (87) have been recently reported concerning bacterial RNA (57).
An additional possibility to indirectly assess this issue is to investigate whether, and to what extent, the binding of splicing factors can be affected by or affect the RNA secondary structures. Clearly, any indications along these lines would represent a sound experimental basis for speculations regarding the role played by RNA secondary structure in splicing.
EFFECTS OF RNA SECONDARY STRUCTURE ON BINDING OF RNA SPLICING FACTORS (AND VICE VERSA)
There are several excellent reviews regarding the topic of how proteins bind sequence specifically to single-stranded RNA (2, 30, 85) and a more recent report regarding binding to double-stranded sequences (17). Indeed, several reports in the recent literature suggest that RNA secondary structure plays an important role in binding. For example, binding of proteins to RNA (CNG)n trinucleotide repeats in vivo closely matches the in vitro results that predict that these repeats are folded in a characteristic hairpin shape (93). The most recent observation is that the predictive ability in the search for novel RNA binding targets for well-known proteins can be greatly enhanced if secondary structure is taken into consideration. A recent example of this is represented by HuR (68) (Fig. 2a), a protein that binds specific mRNA subsets and is involved in the posttranscriptional regulation of gene expression (68). Considering that an increasing number of RNA binding proteins behave like HuR, that is, seem to recognize loosely defined sequence motifs, it would not be surprising if in several cases RNA secondary structures represented constraining elements capable of shaping well-defined target regions in the presence of loose sequence conservation.
FIG. 2.
Effects of RNA secondary structure on protein binding motifs. (a) Loosely conserved RNA binding sequences can be induced to display a uniform binding surface following the formation of a hairpin structure. The example chosen is the protein HuR, which binds to AU- and U-rich mRNA regions and affects their stability and translation. Identification of a large set of target sequences has shown that the they consist of two complementary 6-bp motifs (highlighted in red and violet) and a loop region which contains only a conserved U residue (highlighted in blue) (68). (b) Splicing factors are also capable of affecting RNA spatial distribution (besides being affected by it). For example, binding of SF-1, U2AF65, and U2AF35 to a small 62-nt RNA containing a branch point sequence (BPS), polypyrimidine tract (PPT), and 3′ splice site (3'ss) has been shown by hydroxyl radical iron-EDTA probing to bend the RNA in such a way as to bring together the 3′ splice site and branch point region, thus helping the formation of the splicing commitment complex (59).
With regard to specific factors capable of affecting the splicing process, it has to be noted that the binding of several positive (B52, SRp55, and NOVA-1) and negative (hnRNP A1) regulators of splicing have been shown to depend on RNA secondary structures as well as on the target nucleotide sequences (10, 31, 78, 89). Recently, the fact that most major members of the SR protein family have been observed to be potentially affected by the conformation of a target RNA may indicate that structural influences may be a widespread occurrence, at least for the components of this important family of splicing modifiers (13).
Interestingly, this relationship between splicing factors and the RNA spatial distribution may well go both ways, providing a potentially even greater level of flexibility in the control of splicing. For example, it has recently been proposed that binding of U2AF65 alone to the 3′ splice site has the result of “compacting” the RNA in such a way as to bring in close proximity to each other the 3′ splice site and the branch site region (59) (Fig. 2b). It should be noted that these studies were performed using an artificial short RNA (62 nucleotides [nt]) containing the branch region, a polypyrimidine tract, and a 3′ splice site, and thus further experiments will have to be performed in order to verify whether these effects play a wider role in vivo. Nonetheless, this finding shows that protein factors are not just passive players in the “binding and folding game,” and hopefully future studies will develop this emerging concept and its implications.
EFFECTS OF RNA SECONDARY STRUCTURE ON 5′,3′ SPLICE SITE AND BRANCH POINT ELEMENTS
For obvious reasons, the earliest and most numerous reports regarding the ability of RNA secondary structures to affect the splicing process concern conserved key regions that define an exon (i.e., 5′ splice site, 3′ splice site, and branch site). These reports include many diverse organisms and genes. For example, they include viruses such as hepatitis B virus (67), adenovirus (22, 41, 76), human immunodeficiency virus type 1 (31, 52), Rous sarcoma virus (15), yeasts (21, 33, 42, 43, 47, 80, 102), plants such as Nicotiana plumbaginifolia (66), Drosophila (23), and rats and mice (26, 28, 105). In humans, secondary structures which affect the recognition of conserved splice site consensus sequences have been proposed in the generation of human growth hormone isoforms (37), the tau gene (46, 53, 100, 101, 108), the Hprt gene (49, 98), and the hnRNPA1 gene (6).
Although many of these cases contain individual peculiarities, there seem to be two rather intuitive and unifying mechanisms involved. The most common one is represented by the presence of structural elements which may hinder the accessibility of selected sequences by basic splicing factors. In this way they have been proposed to hinder intron processivity and promote skipping of the exon both in an artificial context (43) (Fig. 3a) and in the context of a pathological defect involving the human tau gene (100) (see Fig. 6 and below). Depending on the system analyzed this inhibition has been observed to target only the acceptor site, the donor site, or both. With special regard to the 3′ splice site, however, it should be noted that recent attempts to correlate the presence of loosely defined secondary structures in 3′ splice site definition have resulted in a small (5 to 10%) but significant improvement in predictive ability (84), indicating that this region may be particularly sensitive to the presence of structured RNA.
FIG. 3.
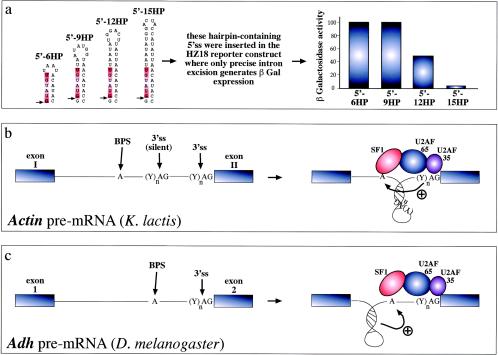
RNA structural elements and splicing efficiency. (a) Series of short artificial hairpins (HP) of increasing length containing a 5′ splice site (indicated by an arrow, while the consensus sequence is highlighted in red). These constructs were engineered in the yeast RP51A intron and assayed for functionality in a reporter construct which contains a wild-type intron inserted in the lacZ gene. In this construct, only precise excision of the intron generates β-galactosidase expression. Following transfection in yeast cells, it was determined that longer hairpins have an increasing ability to sequester the donor site and inhibit the early steps of spliceosome assembly (43). Hairpins can also change the relative distances of splicing regulatory elements and thus affect the final outcome. For example, in the yeast actin intron (b) the branch-point sequence (BPS) is located far away from the 3′splice site (3'ss), which is also preceded by a silent 3'ss (silent) that is not normally used by the splicing machinery. The reason for this lies in the proposed folding of the region between the BPS and the correct 3'ss in a hairpin structure. The function of this structure would be twofold: to bring the BPS into working range of the correct 3'ss and to sequester the silent 3'ss, preventing its use by the splicing machinery (33). Finally, hairpin structure formation near the branch point of the Adh gene intron 1 (3c) has been recently proposed to play an active role in splicing through a distinct mechanism. In this case, hairpin formation has been proposed to force the branch point sequence (BPS) into an unpaired conformation that would be better recognized by the splicing machinery (23).
FIG. 6.
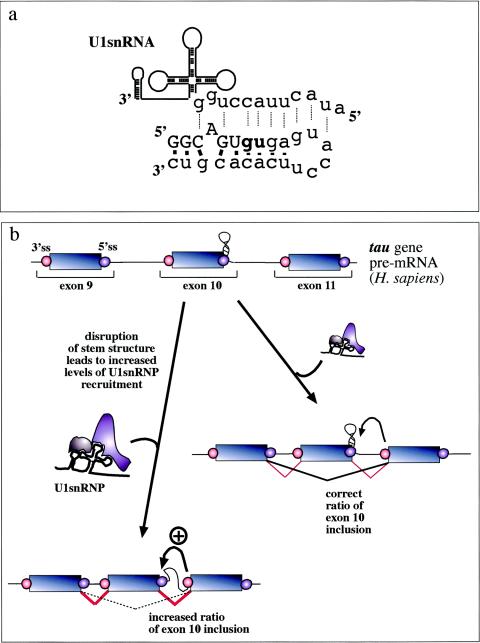
Involvement of RNA structural elements in human disease. In the alternatively spliced tau exon 10, a stem-loop element regulates the accessibility of the 5′ splice site by U1snRNP. (a) What NMR studies have uncovered (100). This element consists of a stem-loop structure involving the 5′ exon/intron junction (uppercase/lowercase) from −5 to +19 which is capped by a flexible loop of 6 bp. Base pairings between the nucleotides in this region (solid bars) prevent recognition by U1snRNA (dotted lines). (b) Mutations which disrupt the structural element result in an increased accessibility of this region and an increased binding of U1snRNP to the donor site. As a consequence, exon recognition is improved and the resulting alterations in the balance between exon 10+ and exon 10− transcripts inside the cell have been proposed to lead to frontotemporal dementia and parkinsonism linked to chromosome 17 (46, 53, 100, 101, 108).
The second mechanism involves a more indirect effect, whereby RNA secondary structures that do not involve the conserved splicing sequences can nonetheless vary the relative distance between these elements. These changes can then determine considerable variation in splice site usage or efficiency. An example of such an event has been seen to occur in the yeast Kluyveromyces lactis actin pre-mRNA, where varying the distance between the branch point element and two potential 3′ splice sites determines efficient use of the distal acceptor site (33) (Fig. 3b). Alternatively, structural constraints may also have the effect of indirectly promoting branch site use by keeping it in a single-stranded accessible configuration, such as was described for the Drosophila Adh gene (23) (Fig. 3c).
EFFECTS OF RNA SECONDARY STRUCTURE ON EXONIC/INTRONIC ENHANCER OR SILENCER ELEMENTS
In addition to splicing consensus sequences, there is also a smaller (but ever increasing) number of cases where structural constraints have been described to affect less-defined cis-acting sequences such as exonic/intronic splicing enhancers (ESE/ISE) or silencer elements (ESS/ISS) (8, 92).
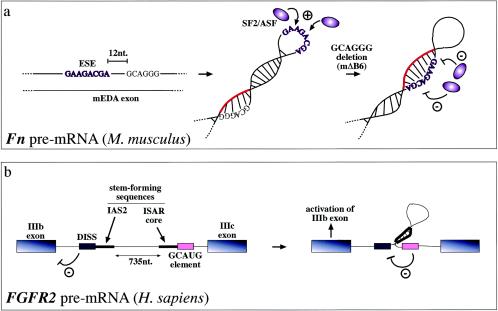
For example, a human-specific ESS sequence in the fibronectin EDA exon has been shown to affect the binding of SR proteins to an ESE sequence which lies 13 nt upstream in the primary RNA sequence. Under normal conditions, the function of this ESS sequence has been proposed to stabilize the secondary structure of the ESE sequence in such a way as to allow binding of SR proteins (77). Additional characterization of the ESS/ESE system in the mouse and human EDA exons showed that while human and mouse ESE sequences behaved in an identical fashion, mutations introduced in the mouse ESS sequence (putatively identified by sequence homology) had no effect on exon splicing. Structural analysis of the mouse EDA exon showed that regardless of its few nucleotide changes in sequence (8 of 270) from the human exon the two RNA secondary structures differed considerably (13). By comparing how the mouse structure responded to homologous deletions in its putative ESS sequences, it was thus finally demonstrated that changes in splicing behavior with respect to the human ESS sequence could be accounted for by a conformational shift from a loop to a stem in the ESE structure (Fig. 4a). This shift prevented binding of SF2/ASF and resulted in exon skipping without modifying directly the SF2/ASF binding motif (13). Therefore, different structural constraints in mice and humans could thus account for what appeared to be an apparently contradictory splicing behavior.
FIG. 4.
Steric hindrance has also been found to occur in ESE sequences, which promote recognition of the correct 3′ splice sites and 5′ splice sites. In the case of the mouse (and human) fibronectin EDA exons, secondary structural elements can stabilize the conformation of the ESE sequence and enhance its SR protein binding capabilities (13, 77). For example, mutations that do not directly affect the mouse ESE sequence have been demonstrated to cause a conformational change in this region (from a loop to a stem) which hinders SF2/ASF protein binding and in this way abolishes exon recognition (a). Highlighted in red is the RNA region which, following the mΔB6 deletion, blocks the ESE sequence. Alternatively, RNA secondary structures can also function on ESE/ESS regulatory regions indirectly. For example, in the FGFR2 gene they contribute to regulating the inclusion of the mutually exclusive IIIb (expressed in epithelial cells) and IIIc (expressed in mesenchymal cells) exons. In this case, the function of the stem structure formed by the intronic activating sequence 2 (IAS2) and intronic splicing activator and repressor (ISAR) element would be that of approximating an inhibitory intronic sequence (a GCAUG-rich sequence) relative to the distal intronic splicing silencer (DISS) element, which would normally repress exon IIIb inclusion (4). Inactivation of this element would then lead to activation of exon IIIb splicing in epithelial cells (b).
A somewhat analogous situation has also been recently described for the SMN1/SMN2 genes; Miyaso et al. (74) have identified an ISE element consisting of a conserved 24-nt stem-loop structure in intron 7. Disruption of this secondary structure leads to loss of binding of an as-yet-unidentified trans-acting factor, and this can influence the splicing process (but only in the presence of the C-to-T transition which occurs in position 6 of exon 7). Considering that this transition has been shown to involve directly several protein-binding signatures such as SF2/ASF (19) and hnRNP A1 (56) and is close to a Tra2-β1 (51) binding site, it will be interesting to analyze the potential interplay between all these factors and the identified ISE element. Significantly, an in silico search made by Miyaso et al. has shown that this element seems to be present in a variety of intron sequences from several genes, raising the possibility that this structurally defined ISE may play a wider role in the general splicing field (74). Finally, from a structural point of view it has to be noted that this exon may also harbor a stem-loop element near its 3′ splice site region (90), although the effect of this structure on exon 7 splicing still remains to be determined.
A different mechanism from the ones presented above has been recently proposed for the human FGFR2 gene. In this case, the formation of a double-stranded RNA created by the joining of two single-stranded elements (creating a loop of 735 nt) was initially observed to regulate splicing of the mutually exclusive IIIb and IIIc exons (32, 75). Mutational studies have demonstrated that the fundamental feature is in the double-stranded structure and not in the FGFR2-specific sequences. Further work on the subject has recently suggested that the function of this structure would be to approximate an intronic control element that inactivates a previously mapped ISS sequence localized near the IIIb exonic sequence (4). In fact, as shown in Fig. 4b, in linear conditions this novel intronic control element would be too far away to have any effect on the functioning of the ISS element. Interestingly, a phylogenetic analysis of this structure from sea urchin to humans has demonstrated that functional conservation of this structure has been maintained for over 600 million years, highlighting the resilience of mRNA secondary structures during evolution independently of the specific nucleotide sequences (73).
LONG-RANGE EFFECTS OF RNA SECONDARY STRUCTURES ON AVAILABILITY OF PRE-mRNA REGIONS
A still very much obscure mechanism through which RNA secondary structure has been proposed to influence the splicing process is by affecting higher-order structures of the pre-mRNA molecule. The first evidence that alterations of extensive secondary structural elements involving both exonic and intronic sequences were responsible for splicing alterations came from the analysis of the chicken β-tropomyosin gene (25, 64) and the dystrophin gene (71). A somewhat related concept has been recently taken up again concerning the presence of conserved polypurinic and polypyrimidinic sequences in the intronic regions of a variety of genes (72) which might be able to pair off with each other and thus exclude determinate exons from the splicing “queue.” It remains unclear, however, how these structures might be responsible for exon skipping, although for the chicken β-tropomyosin gene recent work indicates that under conditions that favor RNA structure formation there is a generalized interference with U1-U6 snRNP interactions (91) (Fig. 5a). Nonetheless, further experimentation will hopefully allow us to provide some information on this. For example, in the rp51b intron of S. cerevisiae the efficient splicing of a 325-nt intron requires the pairing of two short interacting sequences which are normally 200 nt apart, an event which probably facilitates cooperative interactions between intron-spanning factors (65).
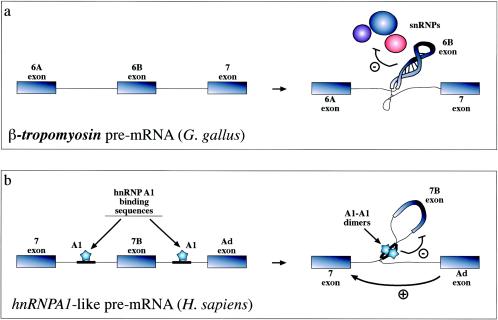
FIG. 5.
Recognition of exons can also be inhibited by formation of extensive RNA secondary structures such as the one shown (a) for the chicken β-tropomyosin gene. In this case, the 6B exonic sequence and its surrounding intronic sequences fold upon themselves in a complex structure that has the ability to affect the interaction of all snRNPs (U1 to U6) with the pre-mRNA (91). Formation of RNA structures that can “loop out” an entire pre-mRNA has also been described to occur in the hnRNPA1 pre-mRNA. However, in this case inhibition is mediated by proteins (hnRNP A1 itself) binding on either side of the exon and interacting with each other (b) (79). The way these structures have been proposed to act is through steric hindrance of the looped-out splice sites (which even if it does not hinder U1snRNP binding may be incompatible with later splicing events). Alternatively, or in addition, these structures might function by simply providing a competitive advantage for distal 5′ splice sites which are moved closer to the acceptor sequence.
An analogous mechanism which has received extended experimental testing in recent years has also been described to occur in the hnRNPA1 gene (7, 79). In this case, the hnRNP A1 factor itself has been shown to bind on either side of an exon and directly promote exclusion through a “looping out” mechanism (Fig. 5b). At present, the proposed mechanisms of action involve an active hindrance of the looped-out 5′ splice site (probably in postcommitment processing steps because U1snRNP binding did not seem to be altered in the looped-out exon) (20) and/or approximation of the distal 5′ splice site, potentially providing a competitive advantage. Interestingly, a similar situation may also occur in the splicing regulation of the neuron-specific c-src exon N1 by the polypyrimidine tract binding protein. Also in this case, polypyrimidine tract binding protein binding on either side of this exon and looping out the N1-containing RNA may contribute to its suppression and prevent its inclusion in nonneuronal cells (24, 103).
CAN ALTERATIONS IN PRE-mRNA SECONDARY STRUCTURE BE INVOLVED IN HUMAN DISEASE?
Considering the evident complexity involved in correct pre-mRNA processing it is not surprising that splicing alterations have been increasingly reported as being involved in many genetic diseases. On purpose, this review does not intend to be an exhaustive analysis of the ever-growing connections between mutations, splicing, and disease, as this has been the subject of several excellent recent reviews (16, 18, 38, 40, 82, 83). However, among this ever-increasing body of evidence that links splicing with disease it is worthwhile to point out that changes in RNA structure have also been invoked to play a role in pathogenic processes involving the dystrophin gene (71), the NF-1 gene (50, 58), and, more recently, the CFTR gene (48). In these cases, however, there is no evidence by experimental probing that the proposed structures follow the in silico predictions. Furthermore, in IVS8 of the CFTR gene it is still unclear how the predicted structures relate to splicing factors binding in the same position (11, 12, 104). At this stage, however, a word of caution is warranted regarding the fact that these described examples are principally based on association studies between splicing activity and in silico predictions of pre-mRNA structures such as those obtainable by Mfold (110) or Pfold (61). The drawback of these approaches is represented by the fact that computer algorithms provide a folding prediction (and often more than one) for virtually any RNA sequence and are strongly biased by the length of the RNA sequence examined. For this reason, although in silico predictions represent an invaluable tool for the researcher in this field, special care should be exercised when predicted pre-mRNA structures are correlated with splicing behavior. As an example, in silico studies of NF-1 gene transcripts (50, 58), which are implicated in the generation of human tumors, have been challenged by successive reports (99, 106). In fact, these studies have shown that the analyses reporting correlations between in silico predicted changes in secondary structure and splicing in these systems are heavily dependent on the RNA window taken into consideration, making it very difficult to assign significance to the suggested correlations. An analogous situation has occurred concerning the splicing control of exon 2 in the human hprt gene, where the proposed role of RNA secondary structure based on in silico evidence (49) has not received any support in a more recent analysis performed using updated parameters and including part of the flanking intron sequences (98).
At present, direct experimental evidence for a role played by secondary structure in the generation of human disease is best represented only by the work performed on the mutations that affect inclusion of exon 10 in the tau gene (46, 53, 100, 101, 108), although it should be noted that one mutational study does not support these conclusions (35). Mutations in the tau gene have been associated with frontotemporal dementia and parkinsonism. In particular, mutations in the intronic region near the 5′ splice site of exon 10 correlate closely with alterations in a characteristic stem-loop structure which has been determined by nuclear magnetic resonance spectroscopy (100) (Fig. 6a). Extensive mutational analyses (46, 53) of this region and functional binding studies to monitor U1snRNP binding to the splice site (53) have shown that mutations which destabilize the helix result in an increased splice site usage owing to an increase in U1snRNP binding (53) (see the schematic diagram in Fig. 6b). Notably, the fact that a small antibiotic, neomycin, can bind to this region and stabilize the stem-loop configuration represents a promising start in the search for therapeutic agents that exploit structural motifs (101).
CONCLUSIONS
In conclusion, the picture that is beginning to emerge clearly favors the possibility that many (if not most) pre-mRNA sequences are quite capable of harboring selected regions which can fold in well-defined secondary structures in vivo. Evolutionarily this is probably not a chance occurrence, as lack of structure would certainly deprive the splicing process of an additional regulating mechanism while too much structure would end up interfering with later complex assembly steps and other layers of regulation. The functional mechanisms investigated so far mostly involve two kinds of mechanistic explanations: the occlusion/exposure of key cis-acting regulatory elements or the spatial modification of the distance between these elements. At present, the principal limitation in identifying these events is that our predictive abilities are still rather limited and safe judgement can be made only through implementation of robust functional studies and experimental probing of proposed RNA structures.
Acknowledgments
This work was supported by grants from the Telethon Onlus Foundation (Italy) (grant no. GGP02453) and FIRB (grant no. RBNE01W9PM).
REFERENCES
- 1.Adams, M. D., D. Z. Rudner, and D. C. Rio. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8:331-339. [DOI] [PubMed] [Google Scholar]
- 2.Antson, A. A. 2000. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10:87-94. [DOI] [PubMed] [Google Scholar]
- 3.Balvay, L., D. Libri, and M. Y. Fiszman. 1993. Pre-mRNA secondary structure and the regulation of splicing. Bioessays 15:165-169. [DOI] [PubMed] [Google Scholar]
- 4.Baraniak, A. P., E. L. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23:9327-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. L. 1991. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5:389-402. [DOI] [PubMed] [Google Scholar]
- 6.Blanchette, M., and B. Chabot. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3:405-419. [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 9.Brion, P., and E. Westhof. 1997. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26:113-137. [DOI] [PubMed] [Google Scholar]
- 10.Buckanovich, R. J., and R. B. Darnell. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buratti, E., A. Brindisi, F. Pagani, and F. E. Baralle. 2004. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 74:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buratti, E., T. Dork, E. Zuccato, F. Pagani, M. Romano, and F. E. Baralle. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20:1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti, E., A. F. Muro, M. Giombi, D. Gherbassi, A. Iaconcig, and F. E. Baralle. 2004. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 24:1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome. Cold Spring Laboratory Harbor Press, Cold Spring Harbor, N.Y.
- 15.Cabello-Villegas, J., K. E. Giles, A. M. Soto, P. Yu, A. Mougin, K. L. Beemon, and Y. X. Wang. 2004. Solution structure of the pseudo-5′ splice site of a retroviral splicing suppressor. RNA 10:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 17.Carlson, C. B., O. M. Stephens, and P. A. Beal. 2003. Recognition of double-stranded RNA by proteins and small molecules. Biopolymers 70:86-102. [DOI] [PubMed] [Google Scholar]
- 18.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 19.Cartegni, L., and A. R. Krainer. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30:377-384. [DOI] [PubMed] [Google Scholar]
- 20.Chabot, B., M. Blanchette, I. Lapierre, and H. La Branche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charpentier, B., and M. Rosbash. 1996. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA 2:509-522. [PMC free article] [PubMed] [Google Scholar]
- 22.Chebli, K., R. Gattoni, P. Schmitt, G. Hildwein, and J. Stevenin. 1989. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol. Cell. Biol. 9:4852-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, Y., and W. Stephan. 2003. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc. Natl. Acad. Sci. USA 100:11499-11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949-957. [DOI] [PubMed] [Google Scholar]
- 25.Clouet d'Orval, B., Y. d'Aubenton Carafa, P. Sirand-Pugnet, M. Gallego, E. Brody, and J. Marie. 1991. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science 252:1823-1828. [DOI] [PubMed] [Google Scholar]
- 26.Coleman, T. P., and J. R. Roesser. 1998. RNA secondary structure: an important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry 37:15941-15950. [DOI] [PubMed] [Google Scholar]
- 27.Conn, G. L., and D. E. Draper. 1998. RNA structure. Curr. Opin. Struct. Biol. 8:278-285. [DOI] [PubMed] [Google Scholar]
- 28.Cote, J., and B. Chabot. 1997. Natural base-pairing interactions between 5′ splice site and branch site sequences affect mammalian 5′ splice site selection. RNA 3:1248-1261. [PMC free article] [PubMed] [Google Scholar]
- 29.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 30.Cusack, S. 1999. RNA-protein complexes. Curr. Opin. Struct. Biol. 9:66-73. [DOI] [PubMed] [Google Scholar]
- 31.Damgaard, C. K., T. O. Tange, and J. Kjems. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshler, J. O., and J. J. Rossi. 1991. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 5:1252-1263. [DOI] [PubMed] [Google Scholar]
- 34.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 35.D'Souza, I., P. Poorkaj, M. Hong, D. Nochlin, V. M. Lee, T. D. Bird, and G. D. Schellenberg. 1999. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl. Acad. Sci. USA 96:5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eperon, L. P., I. R. Graham, A. D. Griffiths, and I. C. Eperon. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54:393-401. [DOI] [PubMed] [Google Scholar]
- 37.Estes, P. A., N. E. Cooke, and S. A. Liebhaber. 1992. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 267:14902-14908. [PubMed] [Google Scholar]
- 38.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 39.Fontana, W., D. A. Konings, P. F. Stadler, and P. Schuster. 1993. Statistics of RNA secondary structures. Biopolymers 33:1389-1404. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Blanco, M. A., A. P. Baraniak, and E. L. Lasda. 2004. Alternative splicing in disease and therapy. Nat. Biotechnol. 22:535-546. [DOI] [PubMed] [Google Scholar]
- 41.Gattoni, R., P. Schmitt, and J. Stevenin. 1988. In vitro splicing of adenovirus E1A transcripts: characterization of novel reactions and of multiple branch points abnormally far from the 3′ splice site. Nucleic Acids Res. 16:2389-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goguel, V., and M. Rosbash. 1993. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell 72:893-901. [DOI] [PubMed] [Google Scholar]
- 43.Goguel, V., Y. Wang, and M. Rosbash. 1993. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 13:6841-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 17:6747-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green, M. R. 1991. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell Biol. 7:559-599. [DOI] [PubMed] [Google Scholar]
- 46.Grover, A., H. Houlden, M. Baker, J. Adamson, J. Lewis, G. Prihar, S. Pickering-Brown, K. Duff, and M. Hutton. 1999. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274:15134-15143. [DOI] [PubMed] [Google Scholar]
- 47.Halfter, H., and D. Gallwitz. 1988. Impairment of yeast pre-mRNA splicing by potential secondary structure-forming sequences near the conserved branchpoint sequence. Nucleic Acids Res. 16:10413-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hefferon, T. W., J. D. Groman, C. E. Yurk, and G. R. Cutting. 2004. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl. Acad. Sci. USA 101:3504-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hennig, E. E., A. H. Conney, and S. J. Wei. 1995. Characterization of hprt splicing mutations induced by the ultimate carcinogenic metabolite of benzo[a]pyrene in Chinese hamster V-79 cells. Cancer Res. 55:1550-1558. [PubMed] [Google Scholar]
- 50.Hoffmeyer, S., P. Nurnberg, H. Ritter, R. Fahsold, W. Leistner, D. Kaufmann, and W. Krone. 1998. Nearby stop codons in exons of the neurofibromatosis type 1 gene are disparate splice effectors. Am. J. Hum. Genet. 62:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofmann, Y., C. L. Lorson, S. Stamm, E. J. Androphy, and B. Wirth. 2000. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl. Acad. Sci. USA 97:9618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang, Z., J. Cote, J. M. Kwon, A. M. Goate, and J. Y. Wu. 2000. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20:4036-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 55.Kadener, S., J. P. Fededa, M. Rosbash, and A. R. Kornblihtt. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. USA 99:8185-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34:460-463. [DOI] [PubMed] [Google Scholar]
- 57.Katz, L., and C. B. Burge. 2003. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 13:2042-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufmann, D., W. Leistner, P. Kruse, O. Kenner, S. Hoffmeyer, C. Hein, W. Vogel, L. Messiaen, and B. Bartelt. 2002. Aberrant splicing in several human tumors in the tumor suppressor genes neurofibromatosis type 1, neurofibromatosis type 2, and tuberous sclerosis 2. Cancer Res. 62:1503-1509. [PubMed] [Google Scholar]
- 59.Kent, O. A., A. Reayi, L. Foong, K. A. Chilibeck, and A. M. MacMillan. 2003. Structuring of the 3′ splice site by U2AF65. J. Biol. Chem. 278:50572-50577. [DOI] [PubMed] [Google Scholar]
- 60.Klaff, P., D. Riesner, and G. Steger. 1996. RNA structure and the regulation of gene expression. Plant Mol. Biol. 32:89-106. [DOI] [PubMed] [Google Scholar]
- 61.Knudsen, B., and J. Hein. 1999. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics 15:446-454. [DOI] [PubMed] [Google Scholar]
- 62.Krainer, A. R. 1997. Eukaryotic mRNA processing. Oxford University Press Inc., New York, N.Y.
- 63.Lamond, A. I. 1993. The spliceosome. Bioessays 15:595-603. [DOI] [PubMed] [Google Scholar]
- 64.Libri, D., A. Piseri, and M. Y. Fiszman. 1991. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science 252:1842-1845. [DOI] [PubMed] [Google Scholar]
- 65.Libri, D., F. Stutz, T. McCarthy, and M. Rosbash. 1995. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA 1:425-436. [PMC free article] [PubMed] [Google Scholar]
- 66.Liu, H. X., G. J. Goodall, R. Kole, and W. Filipowicz. 1995. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J. 14:377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loeb, D. D., A. A. Mack, and R. Tian. 2002. A secondary structure that contains the 5′ and 3′ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J. Virol. 76:10195-10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez De Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 70.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 71.Matsuo, M., H. Nishio, Y. Kitoh, U. Francke, and H. Nakamura. 1992. Partial deletion of a dystrophin gene leads to exon skipping and to loss of an intra-exon hairpin structure from the predicted mRNA precursor. Biochem. Biophys. Res. Commun. 182:495-500. [DOI] [PubMed] [Google Scholar]
- 72.Miriami, E., H. Margalit, and R. Sperling. 2003. Conserved sequence elements associated with exon skipping. Nucleic Acids Res. 31:1974-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mistry, N., W. Harrington, E. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA 9:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyaso, H., M. Okumura, S. Kondo, S. Higashide, H. Miyajima, and K. Imaizumi. 2003. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J. Biol. Chem. 278:15825-15831. [DOI] [PubMed] [Google Scholar]
- 75.Muh, S. J., R. H. Hovhannisyan, and R. P. Carstens. 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277:50143-50154. [DOI] [PubMed] [Google Scholar]
- 76.Munroe, S. H. 1984. Secondary structure of splice sites in adenovirus mRNA precursors. Nucleic Acids Res. 12:8437-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 19:2657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagel, R. J., A. M. Lancaster, and A. M. Zahler. 1998. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4:11-23. [PMC free article] [PubMed] [Google Scholar]
- 79.Nasim, F. U., S. Hutchison, M. Cordeau, and B. Chabot. 2002. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA 8:1078-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newman, A. 1987. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 6:3833-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25:1147-1149. [DOI] [PubMed] [Google Scholar]
- 82.Nissim-Rafinia, M., and B. Kerem. 2002. Splicing regulation as a potential genetic modifier. Trends Genet. 18:123-127. [DOI] [PubMed] [Google Scholar]
- 83.Pagani, F., and F. E. Baralle. 2004. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 5:389-395. [DOI] [PubMed] [Google Scholar]
- 84.Patterson, D. J., K. Yasuhara, and W. L. Ruzzo. 2002. Pre-mRNA secondary structure prediction aids splice site prediction. Pac. Symp. Biocomput. 7:223-234. [PubMed] [Google Scholar]
- 85.Perez-Canadillas, J. M., and G. Varani. 2001. Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 11:53-58. [DOI] [PubMed] [Google Scholar]
- 86.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 87.Seffens, W., and D. Digby. 1999. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 27:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen, L. X., J. P. Basilion, and V. P. Stanton, Jr. 1999. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl. Acad. Sci. USA 96:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi, H., B. E. Hoffman, and J. T. Lis. 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 17:2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh, N. N., E. J. Androphy, and R. N. Singh. 2004. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 315:381-388. [DOI] [PubMed] [Google Scholar]
- 91.Sirand-Pugnet, P., P. Durosay, B. C. Clouet d'Orval, E. Brody, and J. Marie. 1995. beta-tropomyosin pre-mRNA folding around a muscle-specific exon interferes with several steps of spliceosome assembly. J. Mol. Biol. 251:591-602. [DOI] [PubMed] [Google Scholar]
- 92.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 93.Sobczak, K., M. De Mezer, G. Michlewski, J. Krol, and W. J. Krzyzosiak. 2003. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 31:5469-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solnick, D. 1985. Alternative splicing caused by RNA secondary structure. Cell 43:667-676. [DOI] [PubMed] [Google Scholar]
- 95.Solnick, D., and S. I. Lee. 1987. Amount of RNA secondary structure required to induce an alternative splice. Mol. Cell. Biol. 7:3194-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stamm, S. 2002. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol. Genet. 11:2409-2416. [DOI] [PubMed] [Google Scholar]
- 97.Stamm, S., J. Zhu, K. Nakai, P. Stoilov, O. Stoss, and M. Q. Zhang. 2000. An alternative-exon database and its statistical analysis. DNA Cell Biol. 19:739-756. [DOI] [PubMed] [Google Scholar]
- 98.Tu, M., W. Tong, R. Perkins, and C. R. Valentine. 2000. Predicted changes in pre-mRNA secondary structure vary in their association with exon skipping for mutations in exons 2, 4, and 8 of the Hprt gene and exon 51 of the fibrillin gene. Mutat. Res. 432:15-32. [DOI] [PubMed] [Google Scholar]
- 99.Vandenbroucke, I., T. Callens, A. De Paepe, and L. Messiaen. 2002. Complex splicing pattern generates great diversity in human NF1 transcripts. BMC Genomics 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varani, L., M. Hasegawa, M. G. Spillantini, M. J. Smith, J. R. Murrell, B. Ghetti, A. Klug, M. Goedert, and G. Varani. 1999. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. USA 96:8229-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Varani, L., M. G. Spillantini, M. Goedert, and G. Varani. 2000. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Res. 28:710-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vilardell, J., and J. R. Warner. 1994. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 8:211-220. [DOI] [PubMed] [Google Scholar]
- 103.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang, H. Y., I. F. Wang, J. Bose, and C. K. Shen. 2004. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83:130-139. [DOI] [PubMed] [Google Scholar]
- 105.Watakabe, A., K. Inoue, H. Sakamoto, and Y. Shimura. 1989. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 17:8159-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wimmer, K., M. Eckart, P. F. Stadler, H. Rehder, and C. Fonatsch. 2000. Three different premature stop codons lead to skipping of exon 7 in neurofibromatosis type I patients. Hum. Mutat. 16:90-91. [DOI] [PubMed] [Google Scholar]
- 107.Workman, C., and A. Krogh. 1999. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 27:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yasuda, M., J. Takamatsu, I. D'Souza, R. A. Crowther, T. Kawamata, M. Hasegawa, H. Hasegawa, M. G. Spillantini, S. Tanimukai, P. Poorkaj, L. Varani, G. Varani, T. Iwatsubo, M. Goedert, D. G. Schellenberg, and C. Tanaka. 2000. A novel mutation at position +12 in the intron following exon 10 of the tau gene in familial frontotemporal dementia (FTD-Kumamoto). Ann. Neurol. 47:422-429. [PubMed] [Google Scholar]
- 109.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 110.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]