Abstract
Autophagy requires diverse membrane sources and involves membrane trafficking of mATG9, the only membrane protein in the ATG family. However, the molecular regulation of mATG9 trafficking for autophagy initiation remains unclear. Here we identified two conserved classic adaptor protein sorting signals within the cytosolic N-terminus of mATG9, which mediate trafficking of mATG9 from the plasma membrane and trans-Golgi network (TGN) via interaction with the AP1/2 complex. Src phosphorylates mATG9 at Tyr8 to maintain its endocytic and constitutive trafficking in unstressed conditions. In response to starvation, phosphorylation of mATG9 at Tyr8 by Src and at Ser14 by ULK1 functionally cooperate to promote interactions between mATG9 and the AP1/2 complex, leading to redistribution of mATG9 from the plasma membrane and juxta-nuclear region to the peripheral pool for autophagy initiation. Our findings uncover novel mechanisms of mATG9 trafficking and suggest a coordination of basal and stress-induced autophagy.
Keywords: autophagy, mATG9 trafficking, Src kinase, ULK1 kinase, basal autophagy
Introduction
Individual cells constantly monitor environmental and cellular cues in order to maintain cellular homeostasis and survive under different conditions. In response to sub-lethal stresses, cells undergo rapid adaptive changes in their metabolism to protect themselves against potential damage. This is orchestrated through a multifaceted cellular program, which involves the concerted action of diverse general and stress response pathways. One of the key pathways that mediate normal cellular homeostasis and stress-induced adaptation is macroautophagy (hereafter called autophagy). Autophagy is a cell survival process in which portions of the cytosol and damaged or unwanted organelles are engulfed into a double-membrane autophagosome and delivered to the lysosomes for degradation and recycling1,2. The major autophagy pathway involves the activation of the ULK1/ATG13/FIP200 complex, the phosphatidylinositol 3-kinase complex and the mATG9 cycling machinery to initiate the formation of a phagophore/isolation membrane, leading to subsequent expansion and maturation of the autophagosome3. Under normal physiological conditions, autophagy is maintained at a basal homeostatic level, which is responsible for protein quality control and turnover of intracellular organelles4. In response to nutrient deprivation or other sub-lethal stresses, autophagy is geared up to a high flux to efficiently degrade and recycle cytoplasmic components for homeostasis and cell survival. Defects in autophagy have been causally linked with degenerative, inflammatory, metabolic and neoplastic diseases5,6.
Multiple membrane sources, including mitochondria7, endoplasmic reticulum (ER)8,9,10,11, Golgi apparatus12,13 and plasma membrane14 contribute to nascent autophagosome formation under stress conditions. Mobilization of these diverse membrane sources requires dynamic membrane trafficking events, including the budding of vesicles from donor membrane structures, membrane remodeling during phagophore formation, directional vesicle movement, and homotypic/heterotypic fusion of vesicles during autophagosome formation and maturation. Of note, mATG9, the only multi-spanning membrane protein in the ATG family, was found to traffic through the plasma membrane15, the trans-Golgi network (TGN)13,16,17,18, early endosomes, late endosomes and recycling endosomes. During starvation-induced autophagy, mATG9 moves from the TGN to the peripheral pool and co-localizes with endosome markers and the autophagosome marker LC3. The redistribution of mATG9 appears to be mediated by ULK1, as it is inhibited by ULK1 knockdown17. In addition, it has been reported that a sub-population of mATG9 resides on the plasma membrane and can interact with the AP2 complex for internalization via clathrin-mediated endocytosis. Internalized mATG9 traffics from early to recycling endosomes for fusion with ATG16L1-positive structures15. It has been shown that two TBC domain-containing RABGAPs, TBC1D5 and TBC1D14, are involved in regulating mATG9 trafficking and autophagosome formation19,20. TBC1D5 interacts with LC3, the AP2 complex and mATG9 during autophagy, and this interaction may function to recruit mATG9 vesicles to autophagic membranes. TBC1D14, previously identified as a negative regulator of autophagy, binds to RAB11 and inhibits vesicular transport from recycling endosomes and autophagosome formation21. A recent report indicated that mATG9 trafficking is regulated by TBC1D14 and the TRAPPIII complex independently of ULK120. Recruitment of the TRAPPIII complex to tubulated recycling endosomes by TBC1D14 activates RAB1 to promote endosome-Golgi trafficking, which may recycle mATG9 to maintain autophagy flux. Despite significant progress, the exact molecular mechanisms that regulate mATG9 trafficking and coordinate upstream nutrient sensing signals with mATG9 trafficking to fine-tune autophagy flux under normal or starvation conditions remain elusive. Here, we identified two adaptor protein sorting signals within the N-terminus of mATG9 which can be recognized by the AP1 and AP2 complexes to mediate trafficking of mATG9 from the plasma membrane and TGN. Src kinase promotes the interaction between mATG9 and AP1/2 complex by directly phosphorylating mATG9 at Tyr8, and this phosphorylation is required for constitutive mATG9 trafficking. We also found that in response to starvation, phosphorylation of mATG9 at Tyr8 by Src functionally cooperates with Ser14 phosphorylation by ULK1 to promote redistribution of mATG9 from the plasma membrane and juxta-nuclear region to the peripheral pool for autophagy initiation.
Results
The mATG9 N-terminus contains two conserved adaptor protein sorting signals
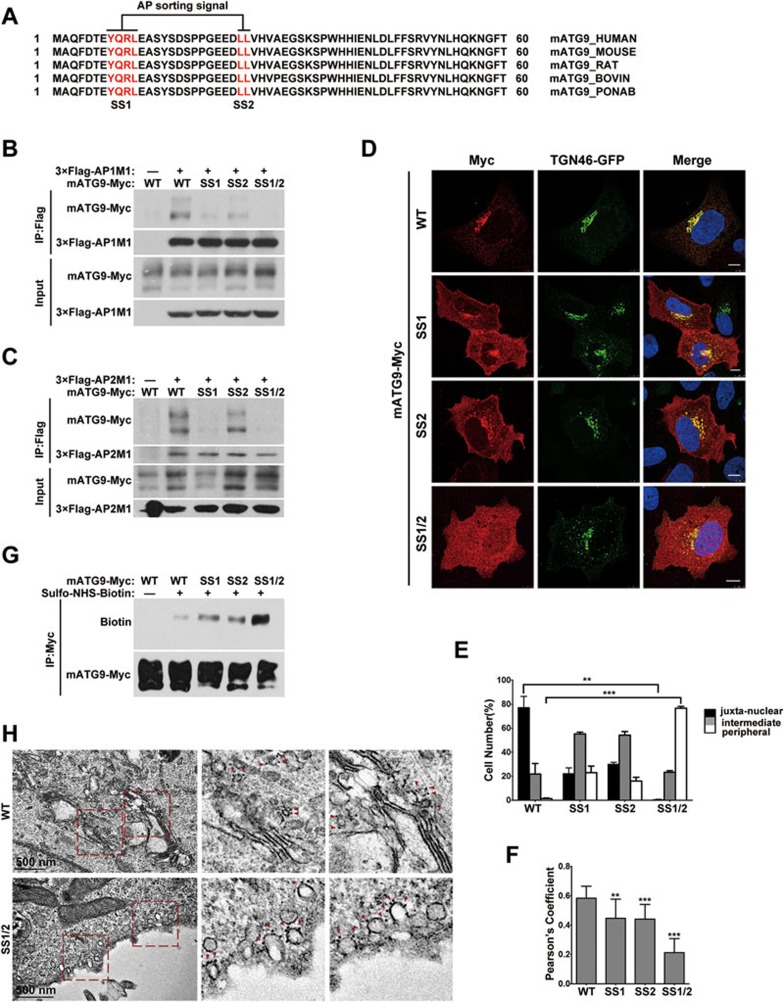
To understand the mechanism underlying mATG9 trafficking, we hypothesized that mATG9 may contain specific sorting signals responsible for trafficking. We thus searched the N- and C-termini of mATG9, which are both cytosolic, and identified two putative conserved adaptor protein (AP) sorting signals in the N-terminus (Figure 1A). These are a tyrosine-based sorting signal Y(8)QRL(11) and a dileucine-based signal EEDL(25)L(26). Tyrosine-based sorting signals are recognized by the mu subunit of the AP1/2 complexes, while dileucine-based sorting signals are recognized by multiple subunits of the complexes22,23,24. We therefore created mATG9 proteins with Y8/L11 mutated to alanine (SS1), L25/L26 mutated to alanine (SS2), or both mutations (SS1/2), and examined the interaction of mATG9 with the mu subunits from the AP1 and AP2 complexes (Figure 1B and 1C). Indeed, the single mutation of these two sorting signals weakened the mATG9-AP1/2 complex interaction and the double mutation completely abolished the interaction. In vitro pull-down assays using a purified His-tagged N-terminal fragment of mATG9 (His-ATG9N-HA, aa1-66), wild-type or carrying a Y8A mutation, further verified the physical interactions with the mu subunits (Supplementary information, Figure S1A and S1B). Immunofluorescent staining showed that mutations of the sorting signals weakened or completely abolished the juxta-nuclear distribution of mATG9 and its co-localization with TGN46, a marker for TGN (Figure 1D-1F; Supplementary information, Figure S1C and S1D). Sorting signal mutations also impaired the co-localization of mATG9 with RAB11-positive recycling endosomes (Supplementary information, Figure S1E and S1F). Hypothesizing that this dispersed localization of the SS1/2 mutant may occur in early endosome, we co-expressed mATG9 with a Rab5 GTPase-deficient mutant (Rab5 Q79L), which stimulates aberrant fusion of early endosomes25. The result showed that the Rab5 mutant co-localized only with wild-type mATG9, but not the SS1/2 mutant, ruling out the possibility of early endosomal localization (Supplementary information, Figure S1G and S1H). To further investigate whether these sorting signal mutations led to cytoplasmic localization or plasma membrane retention of mATG9, we carried out cell surface biotinylation assays to measure the proportion of plasma membrane-localized mATG9 (Figure 1G). The biotinylation of mATG9 was moderately enhanced by the single mutations and very strongly enhanced by the double mutation, suggesting that the mutants are retained at the plasma membrane. Consistent with this, immunogold-labeled EM analysis revealed that in cells harboring the SS1/2 mutation, mATG9 was retained at the plasma membrane due to a severe endocytosis defect (Figure 1H and Supplementary information, Figure S1I). Thus, we identified two classic AP sorting signals in the mATG9 N-terminus and we showed that mutation of these two sorting signals leads to decreased interaction of mATG9 with AP complexes and increased retention of mATG9 at the plasma membrane.
Figure 1.
The mATG9 N-terminus contains two conserved adaptor protein sorting signals. (A) Alignment of mATG9 N-terminus sequences from different mammalian species reveals that two putative AP sorting signals (red color letters) are highly conserved. (B, C) HEK293T cells were transiently co-transfected with wild-type (WT) mATG9-Myc or the indicated mutants and 3×Flag-AP1/2M1. 24 h after transfection, cells were collected for immunoprecipitation with anti-Flag antibody. M1 is the mu subunit of the AP1 and AP2 complexes. (D) U2OS cells were co-transfected with WT mATG9-Myc or the indicated mutants (red) and TGN46-GFP (green). 24 h after transfection, cells were fixed and immunostained with anti-Myc antibody. Cells were counterstained with DAPI (blue). Scale bar, 10 μm. (E) The distribution of mATG9 or the indicated mutants in cells from D was assessed and quantified in a blind fashion (mean ± SEM; n = 100 cells from three independent experiments, **P < 0.01, ***P < 0.001). (F) Pearson's coefficient was determined for the co-localization between mATG9 or the indicated mutants and TGN46 in cells from D. Data were analyzed by ImageJ (means ± SEM; n = 30 cells from three independent experiments, **P < 0.01, ***P < 0.001). (G) HeLa cells were transfected with mATG9-Myc or the indicated mutants for 24 h, and then incubated at 4 °C with ice-cold Sulfo-NHS-Biotin solution for 30 min, rinsed and lysed. Total mATG9 was immunoprecipitated with anti-Myc antibody and the proportion of biotinylated mATG9 was assessed by immunoblotting with anti-Biotin antibody. (H) HeLa cells were transfected with mATG9-Myc or the SS1/2 mutant for 24 h, and then processed for immunogold electron microscopy with anti-Myc antibody. Boxed areas in the left image are shown enlarged on the right. Gold nanoparticles are indicated by red color arrows. Scale bar, 500 nm. See also Supplementary information, Figure S1.
Src directly phosphorylates the mATG9 N-terminus at Tyr8
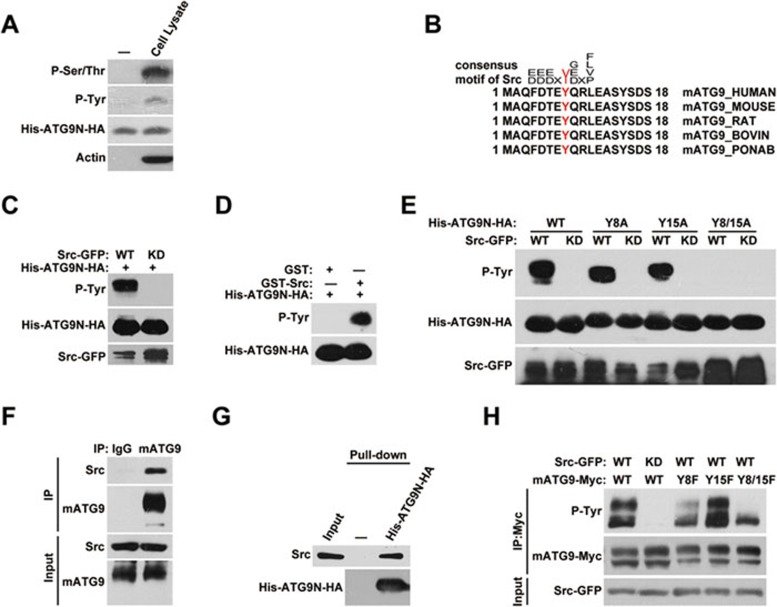
Given that mATG9 disperses from the juxta-nuclear region under starvation stress17, we postulated that there must be a regulatory mechanism underlying this mATG9 redistribution process. Noticing multiple phosphorylatable residues near the AP sorting signals, we hypothesized that phosphorylation may participate in this regulation. In vitro kinase assays using the purified N-terminus of mATG9 and HEK293T cell lysates revealed that the mATG9 N-terminus can be phosphorylated at both serine/threonine and tyrosine residues (Figure 2A). We also subjected the samples to mass spectrometry analysis and confirmed that several residues near the AP sorting signals were phosphorylated (Supplementary information, Figure S2). To determine which kinases are responsible for phosphorylation of the mATG9 N-terminus, we performed sequence alignments and found that the Y8 residue of mATG9 is within a typical consensus phosphorylation motif for Src kinase (Figure 2B)26. Incubation of the purified mATG9 N-terminal peptide with immunoprecipitated wild-type Src-GFP and purified recombinant GST-Src kinase from Sigma, but not kinase-dead Src-GFP, confirms that Src kinase can directly phosphorylate the mATG9 N-terminus in vitro (Figure 2C and 2D). Point mutation analysis revealed that both Y8 and Y15 of mATG9 can be phosphorylated by Src in vitro (Figure 2E). Immunoprecipitation analysis showed that Src interacts with mATG9 (Figure 2F and Supplementary information, Figure S3A). Pull-down experiments using immobilized mATG9 N-terminal peptide revealed that the N-terminal region of mATG9 directly interacts with Src (Figure 2G). To confirm that Y8 and Y15 are authentic Src kinase phosphorylation sites in vivo, we introduced the point mutations of Y8F, Y15F and Y8/15F into mATG9 to measure the phospho-signal when Src-GFP is overexpressed (Figure 2H). The signal is much reduced in the Y8F and Y8/15F mutants, while the Y15F signal is similar to that of wild-type mATG9. These data suggested that mATG9 is mainly phosphorylated by Src at Y8, although additional Src phosphorylation sites may exist.
Figure 2.
Src directly phosphorylates the mATG9 N-terminus at Y8. (A) Purified His-ATG9N-HA peptide (aa1-66) was incubated with control lysis buffer or HEK293T cell lysate and incubated at 30 °C for 30 min in kinase buffer. The reaction products were subjected to western blotting using anti-phosphotyrosine and anti-phosphoserine/threonine antibodies to analyze the phosphorylation status of the mATG9 N-terminus. (B) Alignment of mATG9 N-terminal sequences from different mammalian species reveals the conserved consensus motif for Src kinase. The phosphorylated tyrosine (Y) is in red. (C) In vitro Src kinase assay. Immunoprecipitated wild-type (WT) or kinase-dead (KD) Src was incubated with purified His-ATG9N-HA peptide at 30 °C for 30 min in kinase buffer. (D) GST or purified recombinant GST-Src (Sigma) protein was incubated with purified His-ATG9N-HA peptide at 30 °C for 30 min in kinase buffer to assay Src kinase activity in vitro. (E) In vitro Src kinase assay was performed as described in C using His-ATG9N-HA or the indicated point-mutated peptides. (F) HeLa cells were harvested and lysed by NP-40 lysis buffer for immunoprecipitation with anti-mATG9 antibody. (G) HEK293T cell lysate was incubated with immobilized His-ATG9N-HA peptide at 4 °C overnight. (H) HEK293T cells were co-transfected with WT mATG9-Myc or the indicated mutants and Src-GFP or KD Src for 24 h. Total mATG9 was immunoprecipitated with anti-Myc antibody and the phosphotyrosine level was assessed by immunoblotting with anti-phosphotyrosine antibody. See also Supplementary information, Figures S2 and S3A.
Src phosphorylates mATG9 at Y8 to promote its constitutive trafficking
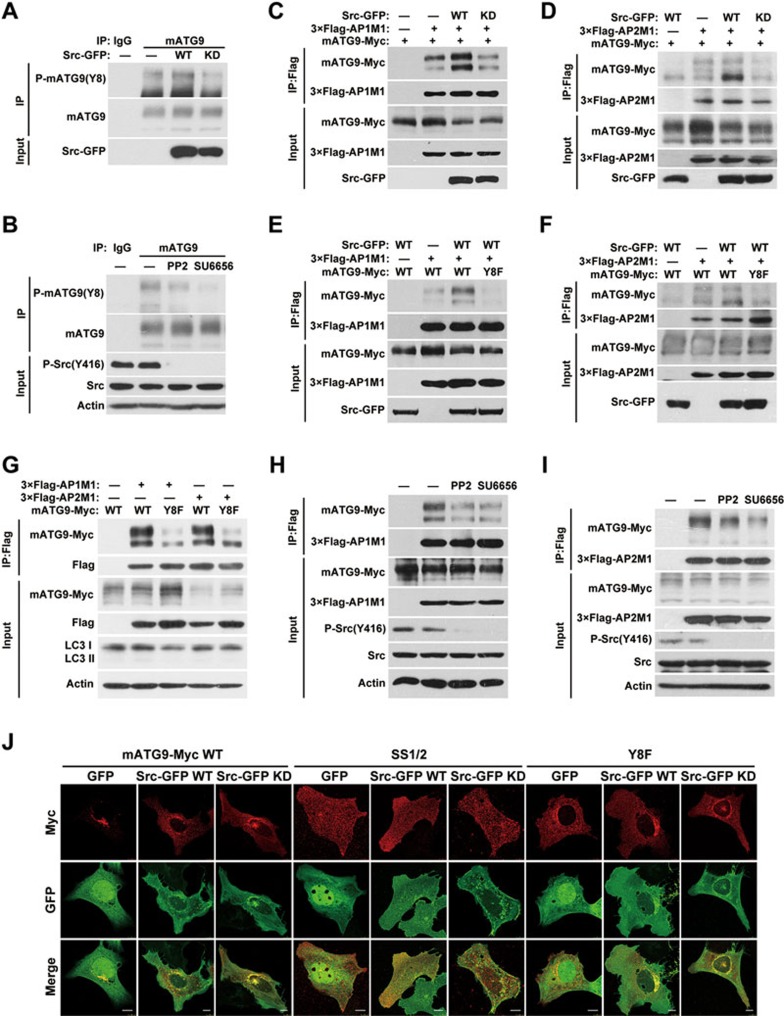
We next examined whether phosphorylation of Y8 by Src kinase is of functional importance in regulating the trafficking of mATG9. We first generated and verified a specific antibody recognizing the phosphorylated state of Y8 (Supplementary information, Figure S3B). Y8 of mATG9 is phosphorylated under normal unstressed conditions in vivo (Figure 3A), and this phosphorylation is decreased upon treatment with the Src inhibitors or Src siRNA (Figure 3B and Supplementary information, Figure S3C). Ectopic expression of wild-type Src further enhances the phosphorylation of Y8, whereas kinase-dead Src inhibits the phosphorylation (Figure 3A). We then asked whether this phosphorylation of mATG9 affects its interaction with the AP1/2 complexes and subsequent trafficking. As shown in Figure 3C and 3D, ectopic expression of wild-type Src, but not kinase-dead Src, was able to enhance the interaction between mATG9 and the AP1/2 complexes. More importantly, this enhancement was blocked by the Y8F mutation of mATG9 (Figure 3E and 3F). As Y8 is highly phosphorylated under normal conditions, we checked the interactions between the AP1/2 complex mu subunits and wild-type mATG9 or the Y8F mutant under normal culture conditions. We found that Y8F weakened the interaction (Figure 3G). To rule out the possibility that the Y8F mutation of mATG9 may change the conformation of the sorting signal, we used synthesized peptides of mATG9 (aa1-18) phosphorylated or unphosphorylated at Y8 for competitive binding of AP1/2 complex mu subunits in vitro (Supplementary information, Figure S3D and S3E). The results revealed that phosphorylated peptide at Y8 has a stronger affinity with AP1/2 complex mu subunits compared with unphosphorylated peptide. Similarly, the Src inhibitors PP2 or SU6656 reduced the interactions (Figure 3H and 3I). However, the Y8F mutant of mATG9 was not sensitive to PP2 or SU6656 treatment (Supplementary information, Figure S3F), suggesting that the phosphorylation at Y8 by Src promotes the interaction of mATG9 with AP1/2 complex. Immunostaining analysis showed that ectopic expression of Src drives the spreading of wild-type mATG9, but not the SS1/2 or Y8F mutants, from the juxta-nuclear region, while ectopic expression of kinase-dead Src caused partial retention of wild-type mATG9 on the plasma membrane (Figure 3J). Taken together, these data showed that Src can phosphorylate mATG9 at Y8 under normal conditions and promote constitutive mATG9 trafficking.
Figure 3.
Src phosphorylates mATG9 at Y8 to promote constitutive trafficking of mATG9. (A) HeLa cells were transfected with Src-GFP or kinase-dead (KD) Src for 24 h, and collected for immunoprecipitation with anti-mATG9 antibody. (B) HeLa cells were treated with the Src kinase inhibitors PP2 (10 μM) or SU6656 (10 μM) for 12 h and collected for immunoprecipitation with anti-mATG9 antibody. (C, D) HEK293T cells were co-transfected with mATG9-Myc, 3×Flag-AP1/2M1 and GFP vector, Src-GFP or KD Src for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (E, F) HEK293T cells were co-transfected with WT mATG9 or the Y8F mutant, 3×Flag-AP1/2M1 and Src-GFP for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (G) HeLa cells were co-transfected with WT mATG9-Myc or the Y8F mutant and 3×Flag-AP1/2M1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (H, I) HeLa cells were co-transfected with mATG9 and 3×Flag-AP1/2M1 for 24 h, and then treated with vehicle or the Src kinase inhibitors PP2 (10 μM) or SU6656 (10 μM) for 12 h. Cells were collected for immunoprecipitation with anti-Flag antibody. (J) U2OS cells were co-transfected with WT mATG9-Myc or the indicated mutants (red) and GFP vector, Src-GFP or KD Src (green) for 24 h, and then fixed and immunostained with anti-Myc antibody. Scale bar, 10 μm. See also Supplementary information, Figure S3B-S3C.
Stimulation of Src kinase activity by EGF enhances mATG9 retrograde trafficking
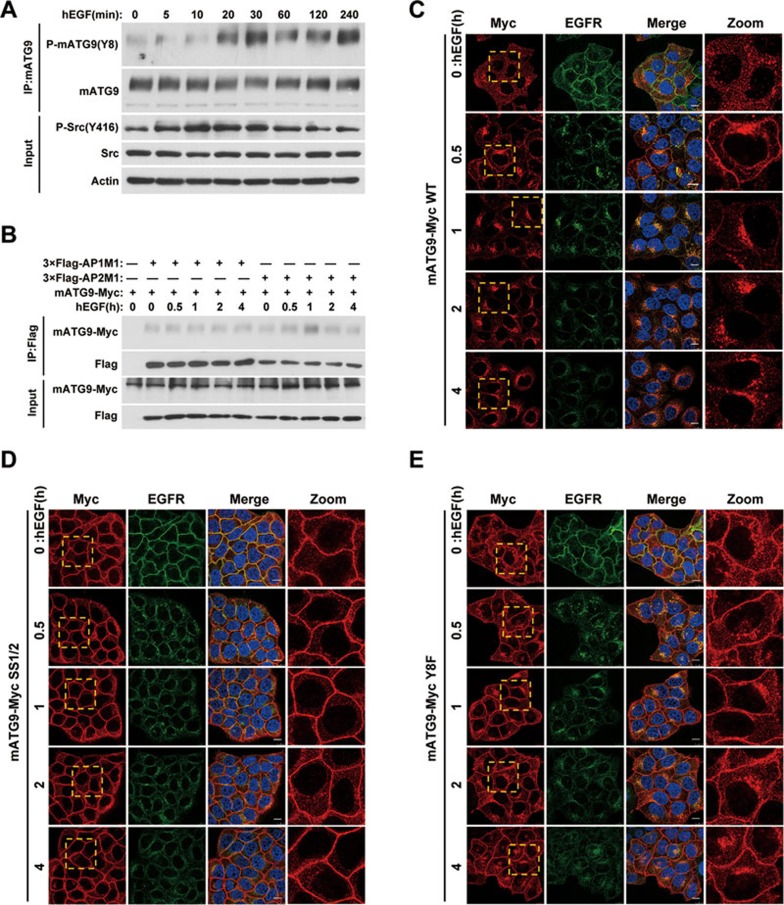
Src can be activated in response to stimulation of various cell surface receptor-mediated signaling pathways including the epidermal growth factor receptor (EGFR) pathway27. We next tested whether hEGF stimulation regulates mATG9 phosphorylation and trafficking. In response to hEGF stimulation, phosphorylation of mATG9 at Y8 was significantly increased immediately after the activation of Src (Figure 4A and Supplementary information, Figure S3G). hEGF treatment rapidly enhanced the interaction of mATG9 with the AP2 complex, but not the AP1 complex, suggesting that mATG9 is trafficked from the plasma membrane by endocytic transport in response to hEGF stimulation (Figure 4B and Supplementary information, Figure S3H). We further hypothesized that stimulation of Src kinase with hEGF treatment may affect mATG9 trafficking and distribution by phosphorylation of Y8. To address this question, we generated an Atg9a-knockout (KO) HeLa cell line using the CRISPR/Cas9 system, and stably reintroduced wild-type mATG9 or the Y8F and SS1/2 mutants into the KO cells (Supplementary information, Figure S3I-S3K). After serum-starving the cells for 24 h and stimulating them with hEGF for different lengths of time, we monitored mATG9 trafficking and distribution. As shown in Figure 4C, mATG9 was dispersed in serum-starved Atg9a KO HeLa cells stably reconstituted with mATG9-Myc. In response to hEGF stimulation, mATG9 was partially localized to the plasma membrane, and was then internalized together with EGFR before finally recovering its juxta-nuclear localization. In the case of the SS1/2 mutant, neither serum starvation nor hEGF treatment affected the plasma membrane retention of mATG9 (Figure 4D). Interestingly, cells reconstituted with the Y8F mutant showed an intermediate phenotype between the cells reconstituted with wild-type mATG9 and SS1/2; they were partially resistant to hEGF stimulation (Figure 4E), whereas the internalization of EGFR was not affected. These findings indicated that the phosphorylation of Y8 by Src kinase is essential for retrograde trafficking and juxta-nuclear localization of mATG9 following EGF stimulation or under unstressed condition.
Figure 4.
Stimulation of Src kinase activity by EGF promotes retrograde trafficking of mATG9. (A) HeLa cells were serum-starved for 24 h, and then stimulated with 50 ng/ml hEGF for the indicated times. Cells were collected for immunoprecipitation with anti-mATG9 antibody and phospho-mATG9 was assessed by immunoblotting with a specific antibody against Y8. (B) HeLa cells stably expressing mATG9-Myc were transfected with 3×Flag-AP1/2M1, serum-starved for 24 h, and then stimulated with hEGF for the indicated times. Cells were collected for immunoprecipitation with anti-Flag antibody. (C-E) Atg9a KO HeLa cells reconstituted with wild-type mATG9 (C) or the SS1/2 (D) or Y8F (E) mutants were starved of serum for 24 h, and then stimulated with hEGF for the indicated times. Cells were fixed and immunostained with anti-Myc (red) and anti-EGFR (green) antibodies. Nuclei are stained with DAPI (blue). Boxed areas in the left panels are enlarged in the right panels. Scale bar, 10 μm. See also Supplementary information, Figure S3D-S3F.
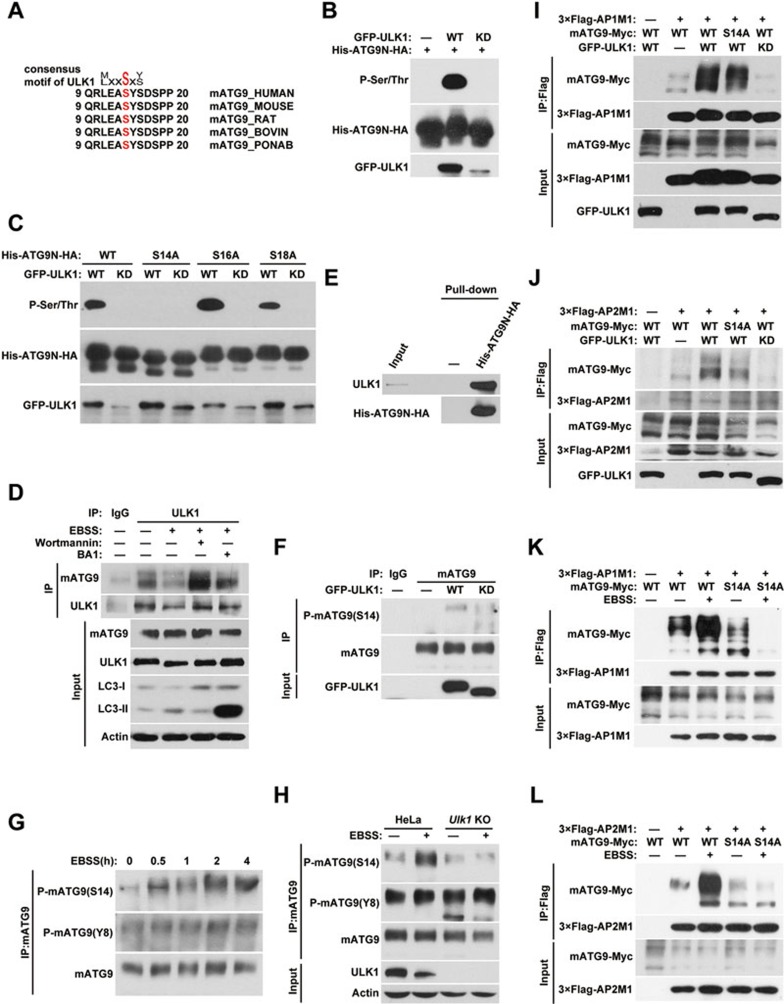
ULK1 directly phosphorylates mATG9 at S14 to promote its trafficking under starvation stress
Under starvation stress, mATG9 redistributes from the TGN to endosomes in a ULK1-dependent manner17. ULK1 is a serine/threonine kinase that is essential for initiation of autophagy. The redistribution of mATG9 is also important for autophagy initiation, although the precise regulatory mechanism is not clear. We thus wished to investigate whether ULK1 regulates mATG9 trafficking via a sorting signal-based mechanism during starvation. By sequence alignment, we found that the S14 residue of mATG9 lies within a consensus phosphorylation motif for ULK1 kinase (Figure 5A)28. In vitro kinase assays showed that wild-type ULK1 but not kinase-dead ULK1 can directly phosphorylate the purified mATG9 N-terminal peptide (Figure 5B). We individually mutated the three serine residues in the mATG9 N-terminal peptide and found that only the S14A mutation abrogates the phospho-signal, which suggests that S14 is the specific phosphorylation site for ULK1 kinase (Figure 5C). Immunoprecipitation experiments showed that ULK1 interacts with mATG9 (Figure 5D). Using two different inhibitors, Wortmannin (PI3K inhibitor) and BA1 (lysosome inhibitor), we blocked EBSS starvation-induced autophagy at the early and late stage, respectively, and found that the interaction between mATG9 and ULK1 was dramatically enhanced at the early stage, suggesting that phosphorylation of mATG9 by ULK1 is of functional importance for autophagy initiation. Pull-down experiments revealed direct interaction of in vitro translated ULK1 protein with the purified mATG9 N-terminal peptide (Figure 5E). We next investigated the functional consequence of S14 phosphorylation on mATG9 redistribution and autophagy initiation. We first generated a specific antibody recognizing the phosphorylated state of S14 (Supplementary information, Figure S4A) and showed that ectopic expression of wild-type ULK1, but not a kinase-dead mutant, enhanced the phosphorylation of S14 (Figure 5F). Unlike Y8, which is highly phosphorylated under normal unstressed conditions, S14 of mATG9 showed a relatively low level of phosphorylation (Figure 5F). Upon EBSS treatment, the phosphorylation of S14 is drastically increased, whereas the phosphorylation of Y8 is relatively stable (Figure 5G and Supplementary information, Figure S4B). Knockout of Ulk1 using the CRISPR/Cas9 system in HeLa cells abolished S14, but not Y8, phosphorylation of mATG9, further supporting our notion that ULK1 is responsible for S14 phosphorylation under starvation (Figure 5H and Supplementary information, Figure S4C). We next speculated that phosphorylation by ULK1 may promote the redistribution of mATG9 from the plasma membrane and TGN to endosomes in response to starvation. We thus introduced the non-phosphorylatable S14A mutant of mATG9 into HEK293T cells and examined the interaction of mATG9 with the AP1/2 complexes in the presence of ectopically expressed wild-type ULK1 or kinase-dead ULK1 (Figure 5I and 5J). Wild-type ULK1, but not the kinase-dead form, significantly enhanced the mATG9-AP1/2 complex interaction, whereas the enhancement was partially blocked in S14A mutant. In contrast to Y8, the S14A mutation of mATG9 did not reduce the interaction between mATG9 and the AP1/2 complex due to the relatively low level of S14 phosphorylation under normal conditions (Supplementary information, Figure S4D). Previous work reported that starvation stress caused redistribution of mATG9 from the TGN to peripheral endosome membranes in a ULK1-dependent manner17. Accordingly, we found that EBSS starvation enhanced the mATG9-AP1/2 complex interaction and the mATG9 S14A mutation impaired this enhancement (Figure 5K and 5L). We also stably reintroduced wild-type mATG9 or the S14A mutant into Atg9a KO HeLa cells and treated the cells with EBSS starvation. Wild-type mATG9 dispersed quickly in response to EBSS treatment, whereas the S14A mutant showed prolonged juxta-nuclear localization and delayed redistribution (Supplementary information, Figure S4E). Based on these results, we conclude that ULK1 phosphorylates mATG9 at S14 to promote mATG9 trafficking and redistribution from the plasma membrane and TGN to endosomes in response to starvation stress.
Figure 5.
ULK1 phosphorylates mATG9 at S14 to promote its trafficking under starvation stress. (A) Alignment of the mATG9 N-terminal sequences from different mammalian species reveals the consensus motif for ULK1 kinase. (B) In vitro ULK1 kinase assay. Immunoprecipitated GFP protein, wild-type ULK1 (WT) or kinase-dead ULK1 (KD) was incubated with purified His-ATG9N-HA peptide (aa1-66) at 30 °C for 30 min in kinase buffer. (C) In vitro ULK1 kinase assay was performed as described in B using His-ATG9N-HA or the indicated point-mutated peptides. (D) HeLa cells were treated with or without EBSS for 2 h in the presence or absence of the PI3K inhibitor Wortmannin (100 nM) or the lysosome inhibitor BA1 (20 nM), and then collected for immunoprecipitation with anti-ULK1 antibody. (E) ULK1 protein produced by an in vitro transcription/translation system was incubated with immobilized His-ATG9N-HA peptide at 4 °C overnight. (F) HeLa cells were transfected with GFP-ULK1 or KD ULK1 for 24 h, and collected for immunoprecipitation with anti-mATG9 antibody. (G) HeLa cells were treated with EBSS for the indicated times, and then collected for immunoprecipitation with anti-mATG9 antibody. The proportion of phospho-mATG9 was assessed by immunoblotting with specific antibodies against Y8 or S14. (H) Parental cells or Ulk1 KO HeLa cells were treated with or without EBSS for 2 h. Cells were collected for immunoprecipitation with anti-mATG9 antibody, and phospho-mATG9 was assessed by immunoblotting with a specific antibody against Y8 or S14. (I, J) HEK293T cells were co-transfected with WT mATG9 or the indicated mutants, 3×Flag-AP1/2M1 and GFP, GFP-ULK1 or KD ULK1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (K, L) HeLa cells were co-transfected with WT mATG9 or the S14A mutant and 3×Flag-AP1/2M1 for 24 h, and then starved in EBSS for 2 h. Cells were collected for immunoprecipitation with anti-Flag antibody. See also Supplementary information, Figure S4.
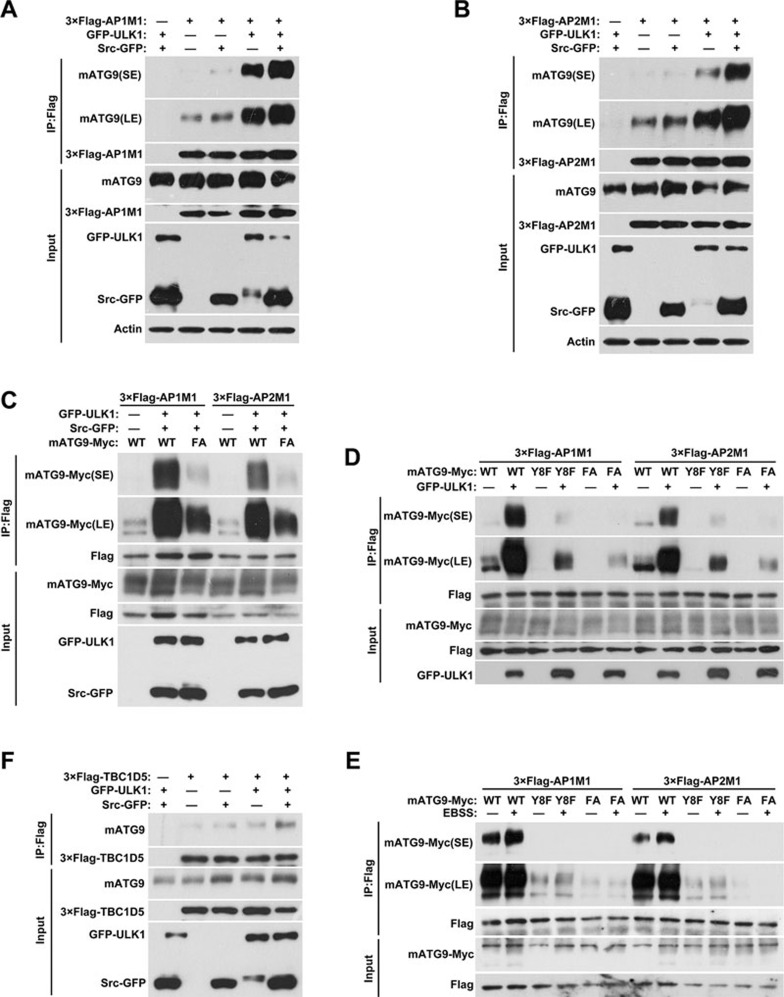
Phosphorylation of Y8 and S14 functionally cooperates to regulate mATG9 trafficking and redistribution
Since Y8 and S14 are phosphorylated by Src and ULK1, respectively, under starvation stress in vivo (Figure 5G and 5H), we next addressed how they functionally cooperate to regulate mATG9 trafficking. Ectopic expression of Src or ULK1 kinase alone enhances the mATG9-AP1/2 complex interaction, with ULK1 having a greater effect than Src (Figure 6A and 6B; Supplementary information, Figure S5A and S5B). Co-expression of Src and ULK1 kinases markedly promotes the mATG9-AP1/2 complex interaction, suggesting that they have a synergistic effect. As expected, this enhancement of the mATG9-AP1/2 complex interaction was blocked by double mutation of the phosphorylation sites Y8F/S14A (designated FA) of mATG9 (Figure 6C). As shown in Figure 5H, the phosphorylation of Y8 was not evidently altered in Ulk1 KO cells. We anticipated that these two sites are phosphorylated by separate processes, but once phosphorylated, the sites cooperate to enhance the mATG9-AP1/2 complex interaction. To further clarify this, we ectopically expressed ULK1 alone, and investigated the mATG9-AP1/2 complex interaction in the presence of wild-type mATG9, the Y8 mutant (which has a normal S14 residue) and the Y8F/S14A double mutant (Figure 6D). The result showed that the basal level of interaction was strongly reduced by Y8F and abolished by Y8F/S14A. The ULK1-induced interaction enhancement was reduced by Y8F and greatly reduced by Y8F/S14A. Similar results were observed in cells that were starved in EBSS to induce activation of ULK1 (Figure 6E). A previous report demonstrated that internalized mATG9 subsequently interacts with the RabGAP protein TBC1D5 to promote autophagy initiation19. We thus examined the interaction of mATG9 with TBC1D5 in cells with ectopic expression of Src or ULK1 (Supplementary information, Figure S5C-S5F). As expected, wild-type Src or ULK1, but not the kinase-dead forms, enhanced the interaction of mATG9 with TBC1D5, and the phosphorylation site mutants blocked their interaction. Notably, co-expression of Src and ULK1 kinases markedly increased this interaction (Figure 6F). Based on these results, we propose that mATG9 is phosphorylated at Y8 under normal conditions to sustain its constitutive trafficking between the TGN, plasma membrane and endosomes. In response to starvation, ULK1 is activated to phosphorylate mATG9 at S14. The phosphorylation at both sites of mATG9 synergistically promotes mATG9-AP1/2 complex interaction and accelerates the extrusion of mATG9 vesicles from the plasma membrane and TGN to initiate autophagy.
Figure 6.
Phosphorylation of Y8 and S14 cooperates to regulate mATG9 trafficking and redistribution. (A, B) HEK293T cells were co-transfected with 3×Flag-AP1/2M1 and GFP vector, Src-GFP and/or GFP-ULK1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (C) HEK293T cells were co-transfected with WT mATG9-Myc or the FA mutant, 3×Flag-AP1/2M1, Src-GFP and GFP-ULK1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (D) HEK293T cells were co-transfected with WT mATG9-Myc or the indicated mutants, 3×Flag-AP1/2M1, GFP vector or GFP-ULK1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. (E) HeLa cells were co-transfected with WT mATG9-Myc or the indicated mutants and 3×Flag-AP1/2M1 for 24 h, and then starved in EBSS for 2 h. Cells were collected for immunoprecipitation with anti-Flag antibody. (F) HEK293T cells were co-transfected with 3×Flag-TBC1D5 and GFP vector, Src-GFP and/or GFP-ULK1 for 24 h, and then collected for immunoprecipitation with anti-Flag antibody. See also Supplementary information, Figure S5.
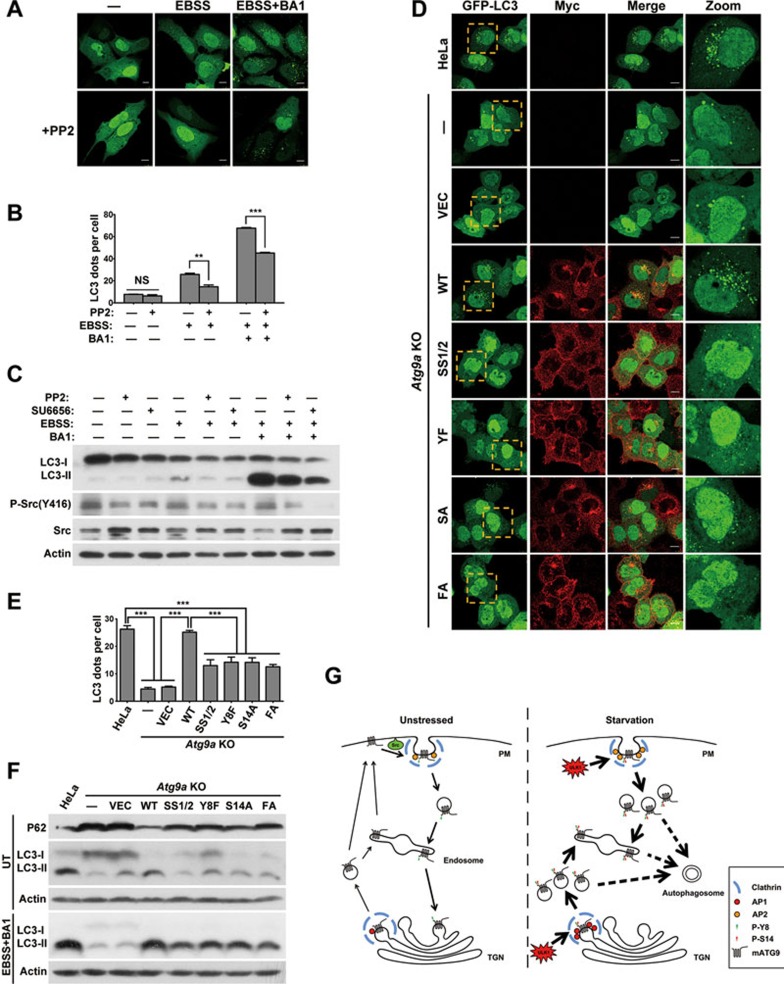
Phosphorylation of both Y8 and S14 is essential for autophagy initiation
It has been assumed that mATG9 may play a role in lipid or protein delivery for nascent autophagosome formation during autophagy initiation29. We thus examined whether the phosphorylation of these two sites is required for autophagy regulation. Cells pretreated with the Src inhibitor PP2 showed fewer LC3 dots and lower autophagy flux in response to EBSS starvation (Figure 7A and 7B). Consistent with this, western blotting analysis also confirmed that the Src inhibitors PP2 or SU6656 reduced LC3-II conversion and autophagy flux during starvation-induced autophagy (Figure 7C and Supplementary information, Figure S6A). To confirm the role of Src kinase in autophagy, we transfected HeLa cells with Src siRNA and measured autophagy flux. The results showed that knockdown of Src kinase led to inhibition of autophagy induction and flux (Supplementary information, Figure S6B). To further study the specific function of the mATG9 phosphorylation, we reintroduced the phosphorylation site mutants into Atg9a KO HeLa cells and measured autophagy flux when the cells were starved in EBSS in the presence of the lysosome inhibitor BA1 (Figure 7D and 7E). As expected, the number of LC3 dots markedly decreased in Atg9a KO cells compared with wild-type cells. The formation of LC3 dots was fully rescued in Atg9a KO cells by reconstitution with wild-type mATG9, but not the AP sorting signal mutant SS1/2 or the phosphorylation site mutants Y8F, S14A and FA (Y8F/S14A). In addition, western blotting analysis revealed that compared with the parental HeLa cells, LC3-I and the autophagy substrate P62 accumulated in Atg9a KO HeLa cells under normal conditions, suggesting that basal autophagy is defective (Figure 7F, Supplementary information, Figure S6C and S6E). The mRNA level of P62 showed no significant alteration (Supplementary information, Figure S6D). The basal autophagy defect was rescued by reconstitution with wild-type mATG9, but not the AP sorting signal mutant SS1/2, or the phosphorylation site mutants Y8F, S14A and FA. In the presence of BA1, Atg9a KO HeLa cells reconstituted with the mATG9 mutants SS1/2, Y8F, S14A and FA exhibited a decrease in LC3-II conversion and autophagy flux compared with the parental HeLa cells or wild-type mATG9-reconstituted KO cells upon EBSS treatment (Figure 7F and Supplementary information, S6F). We also treated these cells with mTORC1 inhibitors, Torin1 or Rapamycin. Similar results were obtained with EBSS treatment, suggesting that these stimuli share the same mechanism for mATG9 regulation and autophagy induction (Supplementary information, Figure S6G). Together, these data suggested that the phosphorylation of mATG9 at Y8 and S14A are both essential for basal and starvation-induced autophagy.
Figure 7.
Phosphorylation of both Y8 and S14 is essential for autophagy initiation. (A) U2OS cells were transfected with GFP-LC3 for 24 h, then treated with or without EBSS for 2 h in the presence or absence of the Src inhibitor PP2 (10 μM) and the lysosome inhibitor BA1 (Bafilomycin A1, 20 nM). Scale bar, 10 μm. (B) The number of LC3 dots in cells from A was assessed and quantified in a blind fashion by ImageJ (mean ± SEM; n = 100 cells from three independent experiments, NS, not significant, **P < 0.01, ***P < 0.001). (C) HeLa cells were pretreated with Src inhibitor PP2 (10 μM) or SU6656 (10 μM) for 10 h, and then treated with or without EBSS for 2 h in the presence or absence of Src inhibitor PP2 (10 μM) or SU6656 (10 μM) and the lysosome inhibitor BA1 (20 nM). (D) Atg9a KO HeLa cells stably reconstituted with wild-type mATG9 or the indicated mutants were transfected with GFP-LC3 for 24 h, and then starved in EBSS for 2 h in the presence of the lysosome inhibitor BA1 (20 nM). Cells were fixed and immunostained with anti-Myc antibody. Scale bar, 10 μm. (E) The number of LC3 dots in cells from D was assessed and quantified in a blind fashion by ImageJ (mean ± SEM; n = 100 cells from three independent experiments, ***P < 0.001). (F) Atg9a KO HeLa cells stablely reconstituted with wild-type mATG9 or the indicated mutants were starved in EBSS for 2 h in the presence of the lysosome inhibitor BA1 (20 nM). (G) Schematic diagram showing the regulation of mATG9 trafficking and autophagy initiation under normal unstressed and starvation conditions. Under normal conditions, mATG9 is phosphorylated by Src at Y8 to sustain its constitutive trafficking and juxta-nuclear localization. In response to starvation stress, mATG9 is phosphorylated at Y8 by Src and at S14 by ULK1 to synergistically promote redistribution of mATG9 from the plasma membrane and TGN to endosomes for autophagy initiation.
Discussion
Sorting signals and mATG9 trafficking
One of our major findings is the identification of two sorting signals which are critical for mATG9 trafficking. These signals are recognized by the AP1/2 complexes for directional movement of mATG9 from the plasma membrane and TGN. Double mutation of the sorting signals led to plasma membrane, but not TGN, retention of mATG9, suggesting that the interactions of the sorting signals with the AP1/2 complexes are essential for endocytic trafficking. Our results also support the idea that the plasma membrane is one of the primary membrane sources for autophagy, thus helping to answer the longstanding question of how diverse membrane sources contribute to autophagosome formation. Recently, Imai et al.30 also identified the mAtg9 sorting motif at its N-terminus, which mediated mATG9 traffic by interaction with AP2, consistent with our results. Our results also extend a recent report showing that TBC1D5 and the AP2 complex directly interact with ATG9 to facilitate its endocytic trafficking19. Sequence alignments show that the sorting signals and the phosphorylation sites are highly conserved in mammals, suggesting that our newly characterized regulatory mechanism is likely to be widely used for mATG9 trafficking and autophagy initiation. It is also possible that ATG9 in yeast is regulated by the same mechanisms, and further experimental work is warranted to investigate this.
Src kinase in constitutive mATG9 trafficking and basal autophagy
It has long been observed that mATG9 traffics between the plasma membrane, the TGN, endosomes, and newly formed autophagosomes in response to autophagy-induced stresses13,15,17,31,32,33,34. It is thus of fundamental importance to understand the mechanism underlying mATG9 trafficking, because it is essential for autophagosome formation, and for normal cellular functions and stress responses. The finding that Src phosphorylates mATG9 at Tyr8 to regulate constitutive endocytic mATG9 trafficking is somewhat unexpected. Tyr8 phosphorylation promotes the interaction of mATG8 with AP1/2 complexes, which are known mediators of endocytosis and membrane trafficking, to sustain recycling transport and juxta-nuclear localization of mATG9 under normal unstressed condition. Our results are consistent with a large body of literatures showing that Src plays essential roles in endocytosis and membrane trafficking35. Src is able to phosphorylate multiple endocytic molecules including clathrin heavy chain (CHC), dynamin 1/2 and cortactin for constitutive EGF or transferrin uptake36,37,38,39,40. Importantly, we found that engagement of EGF with its receptor activates Src kinase to promote mATG9 trafficking. Engagement of ligands by various receptors (including integrin receptors, G-protein-coupled receptors, antigen- and Fc-coupled receptors, cytokine receptors and steroid hormone receptors) is able to activate Src kinase, which in turn regulates the functional activity of receptors and downstream events35. This places Src at a unique and important position to receive or transmit both extracellular and intracellular signals. We found that Atg9a KO cells reconstituted with Y8F mutant of mATG9 showed accumulation of type I LC3 and autophagy substrate P62 under normal unstressed condition, indicating defective basal autophagy. As we used EGF as a treatment for activating Src and promoting the retrograde transport of mATG9, our data thus imply that these signaling pathways including EGFR pathway may also regulate constitutive and basal levels of autophagy by regulating mATG9 distribution via Src kinase. A recent elegant report from Anderson's group showed that knockdown of EGFR inhibited basal and serum starvation-induced autophagy flux, suggesting a role of EGFR in sustaining basal autophagy in a kinase-independent fashion41. It is also reported that EGFR can phosphorylate Beclin1 directly to inhibit autophagy flux42. These dual roles of EGFR may be fine-tuned by Src kinase to sustain autophagy at a low but basal level (in which high autophagy flux is inhibited) and at high flux when ULK1 is activated. Further studies are needed to understand the complex and mutual regulation of the distinct kinases in autophagy regulation in normal and stress conditions. Recent reports showed that active Src kinase forms a complex with LC3B, a known autophagy cargo receptor43. Specific autophagic degradation of active Src enables the cell to fine-tune its cellular activities. This reciprocal regulation of Src and autophagy may be important for precisely adjusting the cellular response to cope with environmental and cellular cues and stresses.
Cooperation of Src and ULK1 in mATG9 trafficking and autophagy
Both basal and stress-induced autophagy are important for the maintenance of tissue homeostasis6. Our data suggest that, while Src phosphorylation of mATG9 is important for basal and constitutive autophagy, ULK1 phosphorylation of mATG9 at S14 is critical for mATG9 trafficking and initiation of stress-induced autophagy. Phosphorylation at Y8 by Src promotes the interaction mATG9 with the AP1/2 complexes, which may be important for constitutive trafficking and basal autophagy. Importantly, Src and ULK1 collaborate to synergistically enhance mATG9 trafficking, which suggests that constitutive mATG9 trafficking and basal autophagy are orchestrated to deliver a graded response to cellular and environmental cues (Figure 7G). Studies from Tooze's group showed that mATG9 trafficking can be regulated by p38MAPK44. It will be interesting to investigate whether mATG9 is a direct substrate of these kinases. It is possible that phosphorylation of mATG9 at distinct sites by different kinases regulates its trafficking or the initiation of autophagy. ULK1 could also regulate myosin II activation by phosphorylating ZIPK, which is essential for mATG9 trafficking after budding from TGN45. It should be noted that, besides autophagy, these signaling molecules are involved in regulating numerous cellular processes including nutrient uptake, cellular metabolism, cell cycle and growth control, cell fate determination and cellular survival/death programs. Our findings thus demonstrate a close integration between signals that regulate these cellular processes and those that regulate autophagy. Taken together, our study has advanced our understanding of autophagic regulation, and its precise links to other cellular processes await further exploration.
Materials and Methods
Cell culture and transfection
HeLa, HEK293T and U2OS cells were cultured in DMEM with 10% FBS (Hyclone) at 37 °C under 5% CO2. Transfection reagent PEI (Polysciences) and Lipofectamine 2000 (Invitrogen) were used for transient transfection of plasmids according to the manufacturer's instruction. Cells were analyzed 24 h after transfection. Three Src siRNAs were synthesized by Guangzhou RiboBio Co., Ltd.
Reagents and antibodies
The following primary antibodies were used: mouse anti-GFP (1:2 000, Santa Cruz), mouse anti-Myc (1:1 000, Sant Cruz), rabbit anti-Myc (1:1 000, Sigma), mouse anti-Flag (1:2 000, Sigma), rabbit anti-Flag (1:1 000, Sigma), mouse anti-HA (1:1 000, Santa Cruz), rabbit anti-Biotin (1:1 000, Cell Signaling), rabbit anti-p-Serine/Threonine (1:1 000, Abcam), mouse anti-p-Tyrosine (1:1 000, Cell Signaling), mouse anti-actin (1:10 000, Sigma), mouse anti-Src (1:1 000, Santa Cruz), rabbit anti-Src (1:1 000, Cell Signaling), rabbit anti-p-Src(Y416) (1:1 000, Cell Signaling), rabbit anti-mATG9 (1:1 000, Abcam), rabbit anti-EGFR (1:1 000, Cell Signaling), rabbit anti-ULK1 (1:1 000, Santa Cruz), rabbit anti-LC3 (1:1 000, MBL), rabbit anti-LC3 (1:2 000, Sigma), rabbit anti-P62 (1:10 000, MBL). Anti-p-mATG9 (Y8) (1:1 000) and anti-p-mATG9 (S14) (1:1 000) polyclonal antibodies were generated by immunizing rabbits with purified mATG9 phospho-peptides and affinity purified (Abgent). For western blotting analysis, HRP-conjugated anti-mouse/rabbit secondary antibodies (KPL) were used and bands were detected with chemiluminescence kits (Engreen Biosystem and Millipore). For immunostaining assays, secondary antibodies were purchased from Invitrogen (goat anti-mouse/rabbit Alexa Fluor 488/555, 1:1 000). The mTORC1 inhibitors: Torin1 (Selleck, 1 μM) or Rapamycin (Selleck, 1 μM) were employed to treat cells for 2 h.
Construction of KO cell lines
LentiCRISPR (Addgene #49535) was used to construct the Atg9a KO (target sequence: CTTCTTCTCTCGAATATCCT) and Ulk1 KO (target sequence: GACCTGGCCGACTACCTGCA) plasmids. Plasmids were transfected into wild-type HeLa cells. After 24 h, cells were selected in DMEM with 2.5 μg/ml puromycin for 4 days. The limiting dilution method was used to acquire several cell monoclones. Cells were collected for detection of mATG9 or ULK1 expression by western blotting and for sequencing of the mutations. Validated cell lines were selected for further experiments.
In vitro kinase assay
Typically, HEK293T cell lysates or immunoprecipitated GFP-ULK1 were incubated with purified His-ATG9N-HA peptide (aa1-66) in ULK1 kinase buffer (25 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) containing 100 μM ATP at 30 °C for 30 min. Similarly, recombinant GST-Src (Sigma) or immunoprecipitated Src-GFP were incubated with purified His-ATG9N-HA peptide in Src kinase buffer (100 mM Tris-HCl, pH 7.2, 125 mM MgCl2, 25 mM MnCl2, 2 mM EGTA, 250 μM Na3VO4, 2 mM DTT). Reactions were stopped by the addition of sample buffer, and then analyzed by SDS-PAGE and western blotting.
Cell surface biotinylation assay
Cells cultured in dishes were washed three times with ice-cold PBS (pH 8.0) and incubated with Sulfo-NHS-Biotin reagent (Pierce) solution at 4 °C for 20 min. Cells were washed three times in 100 mM glycine with PBS to quench the reaction and remove excess reagent, and lysed in NP-40 lysis buffer for immunoprecipitation with anti-Myc antibody. Samples were subjected to SDS-PAGE, and the proportion of mATG9 retained on the plasma membrane was evaluated by western blotting with anti-Biotin antibody.
Pull-down assay
All His-ATG9N-HA peptides were expressed in Transetta (DE3) Chemically Competent Cells (Transgen Biotech) and purified with Ni-NTA beads (GE Healthcare). For pull-down experiments, Ni-NTA beads were incubated with His-ATG9N-HA peptide at 4 °C for 2 h and then washed three times with NP-40 lysis buffer to remove unbound proteins. Beads were incubated with cell lysates at 4 °C overnight and washed five times with lysis buffer. The precipitated components were boiled with sample buffer for 5 min and analyzed by western blotting.
Autophagy flux assay
Cells were washed three times with pre-warmed PBS and incubated with EBSS for 2 h in the presence or absence of the lysosome inhibitor Bafilomycin A1 (20 nM). Autophagy flux was evaluated by LC3-I to LC3-II conversion or the number of GFP-LC3 dots.
hEGF treatment
Cells were washed three times with pre-warmed PBS and serum-starved with DMEM containing 0.1% FBS for 24 h. For hEGF stimulation, cells were incubated with DMEM containing EGF at a final concentration of 50 ng/ml for the indicated time.
Immunogold-labeled EM analysis
For immune-electron microscopy, cells were first fixed with 2% paraformaldehyde and 0.2% glutaraldehyde in Na cacodylate buffer (pH 7.4) at 37 °C for 2 h, and then dehydrated in a graded ethanol series and embedded in acrylic resin (LRWhite). 70-nm ultrathin sections were mounted on nickel grids, and then incubated in 1% BSA in PBS containing anti-Myc antibody overnight at 4 °C. Next, the grids were washed 5 times for 5 min each in 0.5% BSA/PBS and then labeled with gold-conjugated particles in 1% BSA in PBS for 2 h at room temperature. Grids were finally washed 4 times for 5 min each in 0.5% BSA/PBS, incubated for 15 min in 1% glutaraldehyde/PBS, washed 2 times for 5 min each in PBS and 3 times in distilled water, and then stained and dried at room temperature. The samples were visualized using a 120 kV Jeol electron microscope at 80 kV and images were captured using a digital camera (AMT).
Immunoprecipitation
Cells were collected and lysed in 0.5 ml lysis buffer containing protease inhibitors (Roche) for 30 min on a rotor at 4 °C. After centrifugation at 12 000× g for 15 min, the lysates were immunoprecipitated with 2 μg specific antibody overnight at 4 °C, and 30 μL protein A/G agarose beads (Santa Cruz, SC-2003) were washed and then added for additional 3 h. Thereafter, the precipitants were washed five times with lysis buffer, and the immune complexes were boiled with loading buffer for 5 min and analyzed by SDS-PAGE. The following antibodies were used for immunoprecipitation: antibodies against Myc (Santa Cruz, sc-40, clone 9E10), Flag (Sigma, F1804, clone M2), ULK1 (Santa Cruz, sc-33182) and mATG9 (Abcam, ab108338).
Immunostaining and confocal microscopy
Cells were grown to 60% confluence on a coverslip. After treatment, cells were washed twice with PBS, and fixed with freshly prepared 4% formaldehyde at 37 °C for 30 min. Antigen accessibility was increased by treatment with 0.2% Triton X-100. Cells were incubated with primary antibodies for 1 h (or overnight at 4 °C), then washed in PBS and stained with secondary antibodies for further 1 h at room temperature. Cell images were captured with a TCS SP5 Leica confocal microscope.
Mass spectrometry
The purified mATG9 N-terminal peptide was incubated with HEK293T cell lysates for in vitro kinase assays as described above. After the reaction, the samples were boiled with SDS loading buffer and subjected to SDS-PAGE. After excision from the gel, total proteins were subjected to in-gel digestion and then analyzed by LC-MS/MS.
RNA isolation and real-time PCR
Total RNA was isolated from cultured cells using Trizol reagent (Roche). cDNA was synthesized by reverse transcription using oligo (dT) and subjected to real-time PCR with Atg9a and SQSTM1 primers in the presence of Cyber green PCR-Mix (Roche). Relative abundance of mRNA was calculated by normalization to ACTB mRNA. The following primer pairs were used to detect the mRNA levels of the following genes by RT-qPCR: Atg9a (5′-TGTTTCTCAATGAATGGAGCCTC-3′ and 5′-AAGTTAGCGATGCCAATCCAC-3′); SQSTM1 (5′-AGCTGCCCTTAGCCCTCTG-3′ and 5′-GGCTTCTTTTCCCTCCGTGCT-3′) ACTB (5′-CATGTACGTTGCTATCCAGGC-3′ and 5′-CTCCTTAATGTCACGCACGAT-3′). Data were analyzed from three independent experiments and are shown as means ± SEM.
Statistical analysis
For quantitative analyses of cultured cells (data presented as histograms), values were obtained from three independent experiments and expressed as means ± SEM. Statistical analyses were performed using Student's t-test or one-way ANOVA test, with P-values < 0.05 being considered significant. All statistical data were calculated with SPSS software.
Author Contributions
CZ conceived and designed the experiments with YZ and QC. CZ performed most of the experiments and data analysis with assistance from KM. DF performed the electron microscopy analysis. RG and LC established and verified the Atg9a and Ulk1 KO cell lines. KM, CM, Q Liu and QL contributed to plasmid construction and provided technical support. CZ, QC and YZ wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Haiteng Deng and Wenhao Zhang (Tsinghua University, China) for mass spectrometry analysis. We are deeply grateful to Professor Wei Liu (Zhejiang University, China) for his constructive suggestions and critical reading of the manuscript. This study was supported by the Ministry of Science and Technology of China (2016YFA0500201) to QC, the National Natural Science Foundation of China (31671441, 301520103904 and 31471300) to YZ and QC, and the CAS Key project of Frontier Science (QYZDJ-SSW-SMC004) and Nankai University undergraduate innovation research program (21060055338) to YZ.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Sorting signal mutations lead to plasma membrane retention of mATG9.
Identification of mATG9 N-terminal phosphorylation sites.
Active Src promotes mATG9 trafficking.
S14 phosphorylation of mATG9 is essential for its redistribution under starvation stress.
Phosphorylation of Y8 and S14 promotes mATG9 trafficking and interaction with TBC1D5.
Phosphorylation of mATG9 is essential for basal and starvation-induced autophagy.
References
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931–937. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107–132. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of mammalian autophagy in protein metabolism. Proc Jpn Acad B Phys 2007; 83:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389–393. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy 2010; 6:301–303. [DOI] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokita E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009; 5:1180–1185. [DOI] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2013; 2:e00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci 2012; 125:1706–1715. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010; 12:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013; 154:1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Reggiori F. Atg9 reservoirs, a new organelle of the yeast endomembrane system? Autophagy 2010; 6:1221–1223. [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888–3900. [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell 2010; 21:3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Dikic I. TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep 2014; 15:392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Nuhlen S, Judith D, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 2016; 35:281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. Bioessays 1999; 21:558–567. [DOI] [PubMed] [Google Scholar]
- Kelly BT, Owen DJ. Endocytic sorting of transmembrane protein cargo. Curr Opin Cell Biol 2011; 23:404–412. [DOI] [PubMed] [Google Scholar]
- Guo Y, Sirkis DW, Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol 2014; 30:169–206. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 1994; 13:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods 2013; 10:1211–1212. [DOI] [PubMed] [Google Scholar]
- Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol 2009; 6:587–595. [DOI] [PubMed] [Google Scholar]
- Egan DF, Chun MGH, Vamos M, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell 2015; 59:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 2012; 23:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Hao F, Fujita N, et al. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci 2016; 129:3781–3791. [DOI] [PubMed] [Google Scholar]
- Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 2012; 8:1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tooze SA. Autophagy regulation through Atg9 traffic. J Cell Biol 2012; 198:151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Yamamoto H, Oikawa Y, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci USA 2015; 112:3350–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol 1997; 13:513–609. [DOI] [PubMed] [Google Scholar]
- Wilde A, Beattie EC, Lem L, et al. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell 1999; 96:677–687. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yu D, Zeng XC, Zhou K, Zhan X. Receptor-mediated endocytosis involves tyrosine phosphorylation of cortactin. J Biol Chem 2007; 282:16086–16094. [DOI] [PubMed] [Google Scholar]
- Cao H, Chen J, Krueger EW, McNiven MA. SRC-mediated phosphorylation of dynamin and cortactin regulates the “constitutive” endocytosis of transferrin. Mol Cell Biol 2010; 30:781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Kim J, Lucaveche CL, et al. Src-dependent tyrosine phosphorylation regulates dynamin self-assembly and ligand-induced endocytosis of the epidermal growth factor receptor. J Biol Chem 2002; 277:26642–26651. [DOI] [PubMed] [Google Scholar]
- Ahn S, Maudsley S, Luttrell LM, Lefkowitz RJ, Daaka Y. Src-mediated tyrosine phosphorylation of dynamin is required for beta2-adrenergic receptor internalization and mitogen-activated protein kinase signaling. J Biol Chem 1999; 274:1185–1188. [DOI] [PubMed] [Google Scholar]
- Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell 2015; 160:145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Serrels B, McEwan DG, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol 2012; 14:51–60. [DOI] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J 2010; 29:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 2011; 30:636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sorting signal mutations lead to plasma membrane retention of mATG9.
Identification of mATG9 N-terminal phosphorylation sites.
Active Src promotes mATG9 trafficking.
S14 phosphorylation of mATG9 is essential for its redistribution under starvation stress.
Phosphorylation of Y8 and S14 promotes mATG9 trafficking and interaction with TBC1D5.
Phosphorylation of mATG9 is essential for basal and starvation-induced autophagy.