Abstract
5S rRNA is a ribosomal core component, transcribed from many gene copies organized in genomic repeats. Some eukaryotic species have two 5S rRNA types defined by their predominant expression in oogenesis or adult tissue. Our next-generation sequencing study on zebrafish egg, embryo, and adult tissue identified maternal-type 5S rRNA that is exclusively accumulated during oogenesis, replaced throughout the embryogenesis by a somatic-type, and thus virtually absent in adult somatic tissue. The maternal-type 5S rDNA contains several thousands of gene copies on chromosome 4 in tandem repeats with small intergenic regions, whereas the somatic-type is present in only 12 gene copies on chromosome 18 with large intergenic regions. The nine-nucleotide variation between the two 5S rRNA types likely affects TFIII binding and riboprotein L5 binding, probably leading to storage of maternal-type rRNA. Remarkably, these sequence differences are located exactly at the sequence-specific target site for genome integration by the 5S rRNA-specific Mutsu retrotransposon family. Thus, we could define maternal- and somatic-type MutsuDr subfamilies. Furthermore, we identified four additional maternal-type and two new somatic-type MutsuDr subfamilies, each with their own target sequence. This target-site specificity, frequently intact maternal-type retrotransposon elements, plus specific presence of Mutsu retrotransposon RNA and piRNA in egg and adult tissue, suggest an involvement of retrotransposons in achieving the differential copy number of the two types of 5S rDNA loci.
Keywords: 5S ribosomal RNA, maternal rRNA, zebrafish, embryogenesis, target-specific retrotransposon
INTRODUCTION
5S ribosomal RNA (rRNA) is a small (∼120 nucleotide [nt]) RNA component of the ribosome complex in most domains of life (Szymanski et al. 2002; Dinman 2005). Together with two larger rRNAs, 5.8S and 28S rRNAs, it forms in eukaryotes the scaffold of the large ribosomal unit, while the fourth type of rRNA, 18S, does so for the small ribosomal subunit. Counterintuitively, in most eukaryotes the 18S, 5.8S, and 28S rRNA genes are transcribed as a single transcriptional unit (45S) with the rRNA sequences separated by internal-transcribed spacers (ITS), whereas 5S rRNA genes exist as individual genes. All rRNA genes occur in units of tandem repeats that are interspersed with variable nontranscribed spacers (NTSs) (Long and Dawid 1980). However, while 5S rRNA genes are individually transcribed by RNA polymerase III outside the nucleolus (Ciganda and Williams 2011), the 45S rRNA transcriptional units occur in so-called nucleolus organizer regions (NORs) and are transcribed inside the nucleolus by RNA polymerase I (Pederson 2011). The 45S rRNA precursor is subsequently processed into the three individual rRNA types by a complex and well-regulated mechanism (Henras et al. 2015). Remarkably, 45S rRNA production can be boosted by additional amplification at the DNA level (rDNA). Especially during oogenesis this results, for example, in Xenopus laevis, in the unique production of large extra-chromosomal DNA circles containing hundreds to thousands of rDNA repeating units (Cohen et al. 1999).
While at first it would seem that the numerous rRNA gene copies throughout the genome reflect the large amount of rRNA needed for cell functioning (“dosage repetition”), it turns out that the 5S rRNA gene also exhibits so-called “variant repetition” (Long and Dawid 1980). Indeed, it has been shown in some eukaryotic species that there are at least two families of 5S rRNA (Wegnez et al. 1972). These two families differ by just a few nucleotides and have been defined as oocyte- and somatic-type, for they are expressed during different developmental stages: The somatic-type 5S rRNA is transcribed during oogenesis, late embryogenesis, and adult form (Denis and Wegnez 1977), whereas the oocyte-type 5S rRNA is abundantly transcribed in developing oocytes, scarcely in early embryogenesis and switched off in somatic cells (Guinta et al. 1986). Interestingly, the two 5S rRNA variants also show a different genomic organization: In Xenopus, the 5S rRNA somatic type gene has about 400 copies in the genome, while the oocyte gene type has about 20,000 copies (Peterson et al. 1980).
Such variant repetition of 5S rRNA genes was first shown in X. laevis (Wegnez et al. 1972) and later in a few other organisms (Komiya et al. 1986; Martins and Wasko 2004; Dimarco et al. 2012). However, poorly represented or even entirely absent rDNA sequences seem to be a common issue in many currently assembled genomes (Treangen and Salzberg 2012), which might hamper the discovery of 5S rRNA variants. A recent review emphasized our lack of knowledge regarding full rRNA sequences in higher eukaryotes, because of the imperfection of modern technology for highly repetitive sequence analysis, and doubted whether all the rDNA repeats are intact functional genes or what other functional sequences are interspersed with them (Shaw and Brown 2012). For example, the 5S rRNA of zebrafish (Danio rerio) has not been thoroughly described, despite being one of the most versatile model organisms (Dooley 2000; Briggs 2002; Langheinrich et al. 2002; Veldman and Lin 2008; Norton and Bally-Cuif 2010). In fish, 5S rRNA seems to also have an oocyte- and somatic-type (Martins et al. 2002; Martins and Wasko 2004), but their expression pattern has never been investigated, yet.
Usually, 5S rDNA loci are, with the exception of interspersing retrotransposon elements, quite strictly, repetitively organized. 5S rRNA genes in zebrafish are insertion targets for a specific family of retrotransposons called Mutsu (Kojima and Fujiwara 2004). The DNA sequence of Mutsu retrotransposon elements is approximately 5500 base pairs (bp) long and consists of two open reading frames (ORFs) flanked by untranslated regions (UTRs). The first ORF encodes an RNA-binding protein that functions as a chaperone and the second ORF encodes a protein complex that has endonuclease and reverse transcriptase activity. Mutsu retrotransposons belong to the group of long interspersed nuclear elements (LINE) that have a distinct way of replicating and integrating themselves into the genome (Swergold 1990; Ostertag and Kazazian 2001; Elbarbary et al. 2016). Thus far, three subfamilies of Mutsu have been identified: MutsuDr1-3. Their sequence-specific insertion targets are located ∼20 nt downstream from the 5′ terminus of 5S rRNA genes, which results in disruption of the 5S rRNA genes. It is believed that Mutsu retrotransposons could still be active in the zebrafish genome, as their ORFs can encode functional proteins (Kojima and Fujiwara 2004).
In this study, we first characterize all the putative 5S rRNA sequences present in the zebrafish genome and then, by using a wide range of biological samples, demonstrate for the first time the existence of maternal- (oocyte-) and somatic-type 5S rRNA variants in this model organism. Also, many available 5S rRNA genes, although being organized quite strictly, seemed not to be expressed in all samples tested. Even more exciting is the observation that the part of the 5S rRNA gene that varies between the two families might be linked to the functioning of the Mutsu family of retrotransposons and consequently suggest involvement of retrotransposons in the copy-number difference of the two 5S rDNA loci.
RESULTS AND DISCUSSION
5S rRNA variants in zebrafish
To make an inventory of the zebrafish 5S rRNA variation, we first established all potential 5S rRNA gene sequences present in the zebrafish genome. For this, we grouped the 5S rDNA sequences as annotated by Rfam (Griffiths-Jones et al. 2003) together with similar sequences in the zebrafish genome (assembly GRCz10). A total of 758 putative 5S rRNA gene sequences plus genome location are annotated by the Rfam bioinformatics prediction algorithms. As some of these putative 5S rRNA gene sequences are identical, i.e., gene copies, we collapsed them to 734 unique sequences, i.e., 5S rRNA variants. Furthermore, since the Rfam approach might not have detected all 5S rRNA genes, we leniently searched the genome for sequences similar to the 734 Rfam-predicted unique 5S rRNA variant sequences. In this way we detected 9081 5S rRNA genes in the zebrafish genome (Supplemental Table S1) that consisted of 6309 unique sequences of putative 5S rRNA variants. Often gene copies of these 5S rRNA variants are present as well-organized tandem gene repeats dispersed throughout the 25 chromosomes, with a strong preference for chromosome 4 (Supplemental Figs. S1, S2).
Because collectively these 5S rRNA gene copies including NTSs represent ∼0.1% of the complete zebrafish genome, it is obvious that they are not all actively expressed. To identify those 5S rRNA variants that are expressed, we analyzed the expression of each putative 5S rRNA variant in a wide variety of tissues. For this we applied an adapted small-RNA-seq approach on an egg pool (51 M reads), an embryonic time series (49 M reads), a whole-body adult male sample (40 M reads), and a female adult tail sample (7.7 M reads). The latter sample was added to include somatic female tissues without ovaries and developing oocytes. Mapping the small-RNA-seq reads to the sequences of the 6309 unique putative 5S rRNA variants revealed that only 83 (Supplemental Tables S1, S2) were expressed above the set threshold in the analyzed zebrafish tissues. These 5S rRNA gene variants are collectively present in 2513 gene copies in the genome, with three frequently occurring variants; 5Sseq1 (1663 copies), 5Sseq22 (533 copies), and 5Sseq31 (44 copies) (Supplemental Fig. S3). All other variants have less than 15 copies in the genome (Supplemental Fig. S3). The reduction from 9081 putative to 2513 functional 5S rRNA genes is in line with the known fact that there are large numbers of 5S rRNA pseudogenes (Griffiths-Jones et al. 2005; Pinhal et al. 2011). It has to be noted though that it is difficult to sequence 5S rRNA with regular next-generation sequencing protocols. It may, therefore, be that the expression of some 5S rRNA variants is underestimated. Also, it is impossible to determine which of the gene copies are expressed for each 5S rRNA variant.
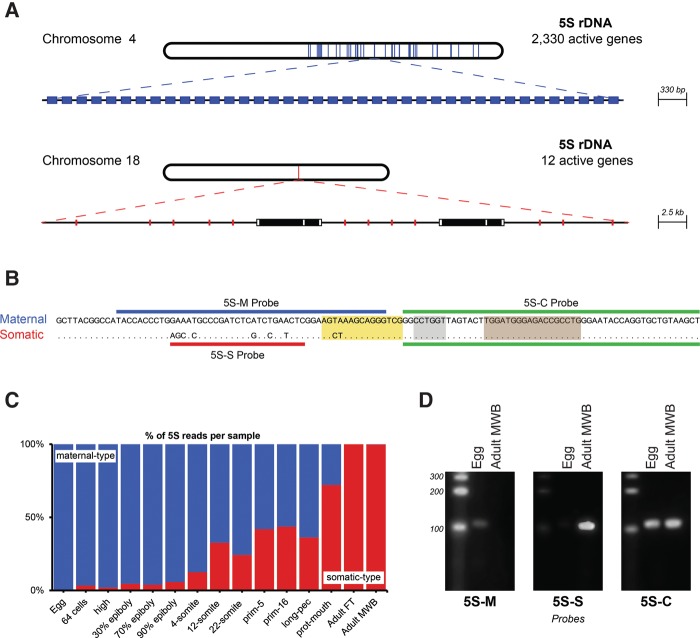
The 5S rRNA variants are quite similar, as all but two variants (5Sseq86 and 5Sseq87) showed only 1- or 2-nt differences compared to the 5S rRNA consensus sequence (Supplemental Fig. S3). 5Sseq86 and 5Sseq87 showed eight and nine sequence differences, respectively, seven of which are identical (Supplemental Figs. S3, S4). Where 5Sseq86 and 5Sseq87 are located on chromosome 8 (one copy) and 18 (12 copies), respectively (Fig. 1A), all other 5S rRNA gene variants are primarily located on chromosome 4 (Fig. 1A), sometimes chromosome 22, or nonassembled genomic scaffolds (Supplemental Fig. S3). The presence of 5S rRNA repeats on the long arm of chromosome 4 has also been shown by fluorescence in situ hybridization (Gornung et al. 2000; Phillips and Reed 2000).
FIGURE 1.
5S rRNA variant types and their expression in zebrafish. (A) Chromosomal localization and organization of the maternal-type (blue) and somatic-type 5S (red) rRNA main clusters in the zebrafish genome. The colored parts on the chromosomes indicate the areas with 5S rRNA genes. In the zoomed-in parts, each colored rectangle is an individual 5S rRNA gene and each white rectangle is a MutsuDrS1 retrotransposon with two potential ORFs (black). (B) Sequence comparison of the 5S rRNA variant types. Maternal, Seq1 sequence; somatic, Seq87 sequence. Internal control regions are shown as colored boxes. A box (yellow); internal element (gray); C Box (light brown) (Schramm and Hernandez 2002; Martins and Wasko 2004). The probes used for Northern blotting are indicated with colored lines. (C) Expression of the 5S rRNA maternal and somatic types indicated by percentage of total 5S rRNA sequencing reads. (Adult FT) Adult female tail; (Adult MWB) adult male whole-body. (D) Northern blot analyses with total RNA from zebrafish eggs and adult (male) tissue and probes as indicated in B. Probe 5S-M recognizes a sequence specific for the maternal-type 5S RNA; probe 5S-S a sequence specific for somatic-type 5S RNA; and probe 5S-C a common sequence between the two 5S RNA variants. Each panel contains lanes from the same gel. In the second panel, the contrast was adjusted as the LNA 5S-S probe gave a very bright signal.
This genomic organization seems to be concordant with that found in Xenopus (Nietfeld et al. 1988) and other fish (Martins and Wasko 2004), like Oreochromis niloticus (Martins et al. 2002). Remarkably, neither variants 5Sseq86 and 5Sseq87 are present in the Rfam database, probably due to their sequence differences compared to the consensus sequence, combined with the fact that their NTSs are significantly longer (several kb) than most NTSs (57 nt) between the other zebrafish 5s rRNA genes (Fig. 1A). This genomic organization is also observed in Oreochromis niloticus, where there are also two 5S rRNA gene variants: one with a short spacer and the other with a long one (Martins et al. 2002). However, 5Sseq86 and 5Sseq87 are present in recently developed 5SRNAdb version 2 (Szymanski et al. 2016) as variant sequences E02726 and E02725, respectively. In this database for zebrafish two other 5S rRNA sequences are present, E02727 and E02728, which seem to be 5Sseq87 variants, as they are different by 1 nt. However, we were not able to detect these gene variants in the zebrafish genome. All other 5S rRNA variants we characterized are not present in the 5SRNAdb, despite occurring most frequently in the genome.
Developmental-stage-specific expression of 5S rRNA variants
Once all 5S rRNA gene variants were defined, it became possible to determine their expression during development in the sequenced RNA content from egg until adult tissue samples. It became quickly clear that there are two distinct types of 5S rRNA genes: Of the 83 expressed zebrafish 5S rRNA variants, only two, 5Sseq86 and 5Sseq87, were virtually absent in egg, but they made up almost 100% of all 5S rRNA in adult tissue (Supplemental Table S2). Hence there are maternal-type 5S rRNA genes that produce virtually all 5S rRNA in eggs and somatic-type 5S rRNA genes that produce virtually all 5S rRNA in adult somatic tissues (Fig. 1C). This is similar to observations in Xenopus, even though the expression of rRNA types in zebrafish is almost mutually exclusive for the mentioned developmental stages (Supplemental Table S2), whereas in Xenopus this was reported to be less absolute (Wolffe and Brown 1988). Given that one 5S rRNA type is replaced by another 5S rRNA type, which resembles the maternal mRNA clearance during early embryogenesis (Schier 2007; Lee et al. 2014), we named the egg-dominant type “maternal” instead of “oocyte” type like in Xenopus, as mentioned earlier (Wegnez et al. 1972). Analyses of 12 intermediate embryonic-development stages showed that the maternal-type 5S rRNA is steadily replaced by the somatic-type 5S rRNA during embryogenesis (Fig. 1C). This is likely caused by the continuous degradation of the maternal-type rRNA accumulated in an oocyte, combined with the continuous expression of somatic-type rRNA during embryogenesis.
rRNA genes are not expressed during the first stages of Xenopus embryogenesis and the onset of 5S RNA synthesis is found at the blastula stage (Wormington and Brown 1983). By the end of gastrulation, the Xenopus embryo is synthesizing almost exclusively the somatic-type 5S rRNA. Although we did not quantitatively measure zebrafish 5S rRNA, the relative read counts show that there probably is somatic-type 5S rRNA expression at an earlier embryonic stage in zebrafish: At the egg stage, only 0.1% of the reads originate from somatic-type 5S rRNA, whereas in the first three embryonic stages this is already 3.3% (Fig. 1C; Supplemental Table S2). This is in line with the findings in Xenopus and sea urchins, where 5S rRNA synthesis can be detected in the early blastula stage (Miller 1974). The regulation of the transcription of the 5S rRNA types in zebrafish is likely regulated epigenetically or by chromatin arrangement, as is shown for Xenopus (Bouvet et al. 1994) and other organisms (Mathieu et al. 2003; Douet and Tourmente 2007; Bellavia et al. 2013). For instance, the long arm of chromosome 4 in zebrafish, where the maternal-type 5S rDNA clusters are located, has very few protein-coding genes and high heterochromatin content (Howe et al. 2013), which could be a way to suppress the maternal-type 5S rRNA expression during embryogenesis and in adult tissues.
Confirmation of 5S rRNA types expression
To confirm the observed developmental-stage-specific expression of 5S rRNA variants, we performed qPCR analysis with primers specific for the two expressed 5S rRNA types. The results clearly confirmed the RNA-seq findings (Supplemental Fig. S5). However, because both approaches rely on cDNA synthesis, we also checked our sequencing results by Northern blot analysis. This is feasible because the differential expression of the 5S rRNA variants is rather absolute. We designed three probes that were either specific for one variant type or could detect them both (Fig. 1B). These Northern blot analyses confirmed our sequencing results (Fig. 1D).
To verify the validity of our RNA-seq approach for 5S rRNA, we applied it also to Xenopus laevis, as it is the most studied organism for the 5S rRNA heterogeneity during development (Brown 2004). It has been reported that X. laevis has three main 5S rRNA gene types: X. laevis oocyte-type (Xlo), X. laevis trace oocyte-type (Xlt), and X. laevis somatic-type (Xls) (Wolffe and Brown 1988). RNA from X. laevis egg and adult tissue (kidney) was sequenced and the results (Supplemental Table S3) confirmed the data present in the literature (Wolffe and Brown 1988): Somatic-type 5S rRNA constitutes over 93% of the 5S rRNA present in adult tissue, while in the egg almost all (∼99%) 5S rRNA present is of oocyte origin (Supplemental Table S3). Confirming the presence of the Xenopus 5S rRNA types gives us confidence that our next-generation sequencing approach, in which we sequence full-length 5S rRNA molecules, can be used to discriminate between the expressions of closely related 5S rRNA gene variants.
Interpreting the differences between the 5S rRNA types
The zebrafish major maternal- and somatic-type 5S rRNA variant sequences differ by 9 nt (8%), which are all located in the 5′ half of the rRNA sequence (Fig. 1B). These differences are located in 5S rRNA residues that are moderately conserved between organisms (Supplemental Fig. S6B,C). It is worth noting that phylogenetic analyses suggest that this differentiation of maternal- and somatic-type in 5S rRNA occurred independently in the lineages of lamprey, bony fishes, and amphibians (Komiya et al. 1986). In X. laevis, the sequences of the two 5S rRNA families differ in 6 nt yet only one is at the same position as in zebrafish (Supplemental Fig. S6A,C). Remarkably, this was also a differential position in Misgurnos fossilis and Tilapia (Supplemental Fig. S6C). Perhaps more relevant is the fact that in both zebrafish and X. laevis most differential nucleotides are in the B-loop of the rRNA molecule (Supplemental Fig. S6A).
The sequence differences between the 5S rRNA types obviously may have an effect at the DNA level and/or RNA level. For instance, although TFIIIA binds to the internal element of the 5S rRNA gene promoter (Fig. 1B), which has no nucleotide difference between the two 5S rRNA types, the close-by upstream nucleotide differences in the internal control region still might affect transcription. In X. laevis, TFIIIA and ribosomal protein L5 bind differently to the 5S RNA types. During oogenesis the maternal-type 5S rRNA is preferentially stored as 7S ribonucleoprotein particle (7S RNP) (= 5S rRNA + TFIIIA), or 42S RNP (= 5S rRNA + Thesaurin b/p43) in the cytoplasm whereas the somatic-type is rapidly integrated in the ribosome complex via the binding of the L5 ribosomal protein (5S RNP) (Allison et al. 1991, 1995): 5S rRNAs, maternal- and somatic-type genes, are transcribed during the early stages of the oogenesis before assembly and storage of the ribosomes (Mairy and Denis 1971). While during these stages the TFIIIA amount is at its maximum (Dixon and Ford 1982), it binds both 5S types and stores them in the cytoplasm. During vitellogenesis, the synthesis of the ribosome's components and their assembly starts (Scheer et al. 1976), while the L5 protein is also produced. The Xenopus somatic-type 5S rRNA binds to L5 because of its higher affinity to L5, and this RNP complex is then transported into the nucleus to be integrated into ribosomes. The Xenopus maternal-type 5S is also recruited by L5, but at a slower rate. It has then been proposed that the somatic-type 5S rRNA may be suitable for short-term use in protein synthesis in the developing oocyte, whereas the maternal-type could be used in the ribosomes that need to be stored for many months in the mature oocyte (Miller 1974). Xenopus somatic-type 5S rRNA has not been detected in the pool of stored ribosomes, suggesting that it may result in lower metabolic stability of its ribosomes (Denis and Wegnez 1977). Such a process could also exist in zebrafish because of the nine maternal- versus somatic-type differential nucleotides; three reside in the 5S region where the L5 is known to interact (Supplemental Fig. S6A; Ciganda and Williams 2011). In fact, the affinity of L5 to 5S rRNA seems to be directly linked to the nucleotides in helix III and loop C (Scripture and Huber 2011). It is interesting to note that in Xenopus, a U43 → A artificial mutation in loop C, which resulted in a marked decrease in affinity to L5, is identical to the difference between the somatic (U43) and maternal (A43) types in Zebrafish (Scripture and Huber 1995).
In Xenopus, the (long) repeats of 45S rRNA transcriptional units are amplified extra-chromosomally during the oogenesis to enable the production of the massive amount of rRNAs needed in the developing embryo (Cohen et al. 1999). For the (short) 5S rRNA genes, no DNA gene amplification is needed as they occur in multiple copies in the genome (Peterson et al. 1980). To verify if this is also the case for zebrafish 5S rRNA genes and knowing that amplified rDNA is still present in the mature egg (Thomas et al. 1977), we analyzed the copy number of maternal- and somatic-type 5S rRNA genes with real-time PCR in both zebrafish egg and adult tissue. Our results show that the 5S maternal-type rRNA genes are not amplified in the oocyte (Supplemental Fig. S7). It is interesting to notice that 12 5S rRNA copies suffice to produce the, as compared to normal genes, still quite large quantity of 5S rRNA in somatic cells. At the same time, we have to be aware that rRNA genes are notoriously poorly assembled and annotated in any genome (Treangen and Salzberg 2012), even though the current assembly of the zebrafish genome seems rather accurate with respect to rDNA sequences.
Linking 5S rRNA variants to different members of the retrotransposon Mutsu family
As we were investigating the different genomic organization (Fig. 1A) of the two 5S rRNA variants on chromosomes 4 and 18, we noticed that the rather strictly organized repeats of maternal-type 5S rRNA gene copies were regularly interrupted by DNA stretches of ∼5 kb without obvious gene copies. Upon further investigation, these spaces typically contained retrotransposon sequences. Mutsu has been reported as a family of autonomous non–long-terminal-repeat (non-LTR) retrotransposons that insert the genome via specific target sequences in zebrafish 5S rRNA genes (Kojima and Fujiwara 2004). These retrotransposons encode for two proteins: an RNA-binding protein that functions as a chaperone and a protein complex that has endonuclease and reverse transcriptase activity. There are three Mutsu subfamilies reported: MutsuDr1, MutsuDr2, and MutsuDr3, with their associated specific DNA target sequence. As it turned out, the Mutsu retrotransposon target sequences are located exactly in the region of the 5S rRNA gene where the maternal- and somatic-type rRNA genes differ the most (6 out of 9 nt differences) (Fig. 2A). As the Mutsu retrotransposons function via a specific target sequence, this means that only specific MutsuDr subfamilies can insert either in maternal- or somatic-5S rRNA genes.
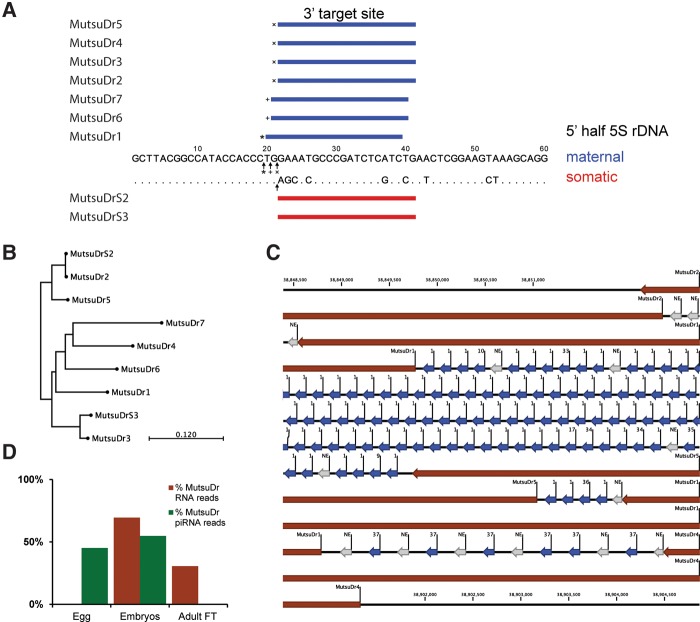
FIGURE 2.
Retrotransposon family MutsuDr linked to 5S rRNA loci. (A) 5′ half of the two rRNA types that include all nucleotide differences. Indicated are the sequence-specific MutsuDr 3′ target sites in the maternal- (blue) and somatic-type (red) 5S rDNA sequence. Small arrows show the insertion sites. (B) Tree of the different MutsuDr retrotransposon subfamilies constructed based on their genomic consensus DNA sequences. (C) An example of the maternal-type 5S rRNA genome organization on chromosome 4 (Supplemental File SF8): expressed 5S rRNA gene copies (5Sseq variant number, blue arrow), nonexpressed 5S rRNA gene copies (NE, gray arrow), and MutsuDr retrotransposon elements (brown arrow). The 5S RNA NE elements right downstream from a Mutsu element are remnants of the 5S rRNA gene that was disrupted by the insertion of the retrotransposon. (D) Relative expression of MutsuDr RNA (brown) and MutsuDr piRNAs (green) as indicated by the fraction of the sequencing reads found for each type. (Embryos) The combined samples as indicated in Figure 1C; (adult FT) adult-female tail.
To further explore the relationship between Mutsu retrotransposons and the 5S rRNA types, we searched for Mutsu elements within the 5S rRNA genomic locations at chromosomes 4 and 18 (39.6 Mb and 45.5 kb, respectively). We found 27 MutsuDr1-3 retrotransposon elements (Dr1: 11; Dr2: 11; and Dr3: 5) on chromosome 4 (Fig. 2C), mostly associated with the 5S rRNA maternal-type gene repeats (Supplemental Table S4; Supplemental Fig. S8). These however, did not explain all the observed genomics spaces within the 5S rRNA repeat regions. Closer inspection revealed four new Mutsu subfamilies, labeled MutsuDr4-7 with different numbers of elements (Dr4: 16; Dr5: 11; Dr6: 7; and Dr7: 6) (Supplemental Table S4; Supplemental Figs. S8–S10; Supplemental File S1). At the somatic-type rRNA region on chromosome 18, two copies of MutsuDr2 appear to be present. However, at closer inspection, we discovered in both copies an identical deleterious 26 nt gap in the ORF1 of the retrotransposon. Because of this and the different target sequence, we labeled these elements a new subfamily MutsuDrS2. In the same region, one other Mutsu element was found, which was similar (∼93%) to MutsuDr3, yet also with a different target sequence, hence subfamily MutsuDrS3 (Fig. 2B; Supplemental Table S4; Supplemental Figs. S8–S10; Supplemental File S1).
In some cases, retrotransposon copies in the genome are nonfunctional because of distorted ORFs and are considered “molecular fossils” (Richardson et al. 2015). However, analyzing the Mutsu retrotransposon elements in the maternal-type 5S rRNA region on chromosome 4, we found that overall in a surprising 42%, both ORFs were intact (Supplemental Table S4). In contrast, none of the three Mutsu retrotransposons in the somatic-type 5S rRNA region on chromosome 18 has two intact ORFs. If we assume that two intact ORFs equals a functional retrotransposon element, it is an intriguing observation that apparently in the locus with many (maternal-type) 5S rRNA genes, several potentially active retrotransposons reside, whereas in the small (somatic-type) locus there is none. It raises the question whether these retrotransposons were evolutionarily involved in achieving and maintaining the needed high maternal-type 5S rDNA gene copy number, as well as the limited somatic-type 5S rRNA gene copy (Cioffi et al. 2010; Rebordinos et al. 2013). In this scenario, one function of the sequence difference between maternal- and somatic-type 5S rRNA genes could be that the different target sequence in the somatic-type genes obstructs new integration of retrotransposons, thus stabilizing the somatic-type 5S rRNA locus.
As a first attempt to investigate this, we searched for Mutsu retrotransposon RNA in an RNA-seq experiment on all mentioned zebrafish samples, excluding the adult male whole-body sample because of the presence of testis material (Houwing et al. 2007). Mutsu RNA presence could provide us with insight into the timing of retrotransposon activity. We did detect very low, yet unmistakably Mutsu retrotransposon RNA in all but the egg and the 64-cell single embryo samples (Fig. 2D; Supplemental Table S5A). Since we sequenced about 48 million reads in the egg sample, the absence of Mutsu RNA is quite convincing. Retrotransposons are restrained by associated 30-nt piRNAs via a so-called “Ping-Pong” mechanism (Houwing et al. 2007; Iwasaki et al. 2015). We did find piRNAs specific for Mutsu retrotransposon RNA in all samples, yet in the adult female tissue sample it was virtually absent (Fig. 2D; Supplemental Table S6A). Overall, Mutsu piRNAs showed a relative read count at an average of 6500 times higher than that of Mutsu RNA. Although we tried, the low read count, combined with the significant homology between the MutsuDr retrotransposon subfamilies, hampered the detection of subfamily specific (piRNA) expression (Supplemental Tables S5B, S6B). The overall distribution of Mutsu RNA over MutsuDr subfamilies was similar to that of piRNA, hinting at a connection. A final interesting observation was that the presence of sense/antisense piRNAs appeared nonrandom over the MutsuDr elements, with each subfamily showing another pattern (Supplemental Fig. S11), which implies an intricate regulation. Altogether, the piRNAs might represent an echo of a suppression Mutsu retrotransposon activity during oogenesis and/or embryogenesis.
Conclusion
Evidently in zebrafish, much like Xenopus, a specific ribosomal system exists that is dedicated to transcription during embryogenesis and potentially oogenesis. We found a maternal-type 5S rRNA that is accumulated during oogenesis, replaced throughout the embryogenesis by a somatic-type, and thus virtually absent in adult somatic tissue. As there are only a few nucleotide differences between the two 5S rRNA types, one can speculate on the rationale why such a double system for translation exists. Storage for later use in embryogenesis rather than directly forming a functional ribosome seems the most obvious reason. This is supported by the finding in Xenopus that a 2-nt difference in the oocyte-type 5S rRNA sequence affects the riboprotein L5 binding site, which in turn leads to storage of the oocyte-type rRNA in an RNP complex instead of immediate incorporation into a ribosome complex. This might also be true for zebrafish, as the maternal-type 5S rRNA sequence differs 3 nt from the somatic-type sequence at the riboprotein L5 binding site.
Typically, sequence differences in coding regions of genes are primarily interpreted by their effect in the associated RNA or protein molecules. However, as the promoter of the 5S rRNA genes is located in the coding part of this small gene, it is to be expected that effects also occur at the DNA level. Some sequence differences between the two types of 5S rRNA genes are located at the TFIII binding site, likely resulting in altered 5S rRNA gene expression. Not only the sequence differences, but also the huge difference in gene copies, as well as genomic location at different chromosomes, offer mechanisms to organize this complex gene locus with its extreme demand for expression at a precise phase in the zebrafish life cycle. Another mechanism might be in functional coexistence of these 5S rRNA genes with retrotransposons. We observed many retrotransposon elements in the 5S rRNA loci that belong to known and new MutsuDr subfamilies. Functional involvement in the maternal-type 5S rRNA locus might be indicated by the fact that many of the present retrotransposon elements contain two intact ORFs, whereas this is not the case in the somatic-type 5S rRNA locus. However, the most compelling indication is the observation that the target site of this sequence-specific retrotransposon family is exactly at the position in the 5S rRNA gene where the most sequence differences between the two 5S rRNA types reside. This means that due to their specific integration target site, functional Mutsu retrotransposon elements in the maternal-type 5S rRNA locus are restricted to this locus. This obviously also applies to the Mutsu retrotransposon elements in the somatic-type 5S rRNA locus, where we did not find any intact retrotransposon element. Additionally, the absence and presence of Mutsu retrotransposon RNA and piRNA in eggs, respectively, together with the reversed situation in adult tissue, fuel the need to investigate the obvious link between 5S rRNA and these sequence-specific non-LTR retrotransposons. All of this sparks the notion that the retrotransposon elements are involved in the way the copy numbers evolved in both maternal- and somatic-type 5S rDNA loci (Rebordinos et al. 2013), similar to that reported in 45S (Symonová et al. 2013).
Thus, several effects at the DNA and RNA level result from the sequence differences between the two types of 5S rRNA. There will undoubtedly be more variation at the level of gene-copy number regulation, gene-expression regulation, rRNA storage, and ribosomal functioning, which is not entirely unexpected given that it happens at the core of one of the most basic cellular mechanisms.
MATERIALS AND METHODS
Biological materials
Zebrafish
Adult zebrafish (strain ABTL) were handled in compliance with local animal welfare regulations and maintained according to standard protocols (http://zfin.org). The breeding of adult fish was approved by the local animal welfare committee (DEC) of the University of Leiden, the Netherlands. All protocols adhered to the international guidelines specified by the EU Animal Protection Directive 86/609/EEC. Unfertilized eggs (oocytes) were collected by squeezing the abdomen of three spawning females and further stored as three corresponding egg pools. Whole-body male adult zebrafish samples, female adult tail samples, and egg pools were flash-frozen in liquid nitrogen and stored at −80°C. Before freezing, fish were put under anesthesia using 0.02% buffered 3-aminobenzoic acid ethyl ester (Tricaine).
Zebrafish embryos time course
Zebrafish embryos were collected immediately after fertilization, maintained at 28.5°C, and staged using standard morphological criteria (Kimmel et al. 1995). One embryo was collected at 12 embryonic development points: (1) 64 cells (2 hpf), (2) high stage (3.3 hpf), (3) 30% epiboly stage (4.7 hpf), (4) 70% epiboly stage (7 hpf), (5) 90% epiboly stage (9 hpf), (6) 4-somite stage (11.3 hpf), (7) 12-somite stage (15 hpf), (8) 22-somite stage (20 hpf), (9) prim-5 stage (24 hpf), (10) prim-16 (31 hpf), (11) long-pec stage (48 hpf), and (12) protruding-mouth stage (72 hpf). After collection, the embryos were snap-frozen in liquid nitrogen, and stored at −80°C. In order to maintain a uniform genetic background, all embryos were collected from the same batch of fish stock.
Xenopus
Frogs (Xenopus laevis) were housed and maintained in a temperature-controlled aquarium in accordance with institutional guidelines. For egg collection, one female frog was first injected with 50 units of human chorionic gonadotropin (hCG) 1 wk before induction of ovulation. An additional 500 units of hCG were then injected into the dorsal lymph sac 1 d before unfertilized egg collection to induce full ovulation. One pool containing 20–30 eggs was collected into 1× MBS (0.1 M CaCl2 7 mL, 10× MBS 100 mL, 5 M NaCl 4 mL, and ddH2O 888 mL). After snap-freezing in liquid nitrogen, the samples were stored at −80°C for further RNA isolation. For isolation of adult tissues, one male frog was euthanized in tricaine methane sulfonate solution (MS222) at a concentration of 5 g/L. The animal was immediately anatomized after death and kidney samples were collected for RNA isolation.
DNA isolation
Zebrafish samples were pulverized in liquid nitrogen with a mortar and pestle, and genomic DNA isolation was performed using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) following the provided protocol. DNA concentration was measured on a NanoDrop ND-2000 (Thermo Fisher Scientific) and DNA integrity was examined using the 2200 TapeStation System with Genomic DNA ScreenTapes (Agilent Technologies).
RNA isolation
Four zebrafish samples (three egg pools further pooled together, one whole-body male adult, and one female adult tail) and two X. laevis samples (one egg pool and one male kidney) were pulverized in liquid nitrogen with a mortar and pestle, and total RNA isolation was performed using the E.Z.N.A. MicroElute RNA Clean Up Kit (Omega Bio-Tek). In short, powdered tissue was homogenized in TRIzol Reagent (Thermo Fisher Scientific) and chloroform was added. After centrifugation, RNA partitioned to the upper aqueous phase, which was carefully removed and subsequently combined with 1.5× volume EtOH. This mixture (containing total RNA) was subjected to column-based RNA isolation according to the protocol of E.Z.N.A. MicroElute RNA Clean Up Kit (Omega Bio-Tek). Eluted RNA (larger than 200 nt) concentration was measured on a NanoDrop ND-2000 (Thermo Fisher Scientific) and RNA integrity was verified using a 2200 TapeStation System with Agilent RNA ScreenTapes (Agilent Technologies). Small RNA (shorter than 200 nt) was then purified from the flow-through by adding ethanol to 65%, followed by loading onto an E.Z.N.A. MicroElute RNA column (Omega Bio-Tek). The column was washed using RWT once and RPE wash buffer once (QIAGEN). The concentration of small RNA was measured on a NanoDrop ND-2000 (Thermo Fisher Scientific) and RNA integrity was examined using a 2200 TapeStation System with Agilent RNA ScreenTapes (Agilent Technologies).
Next-generation sequencing
Bar-coded large-RNA-seq and small-RNA-seq libraries for the zebrafish samples (three egg pools further pooled together, one whole-body male adult, one female adult tail, and a 12-sample embryo time course) and two X. laevis samples (one egg pool and one male kidney) were generated according to the manufacturers’ protocols using the Ion Total RNA-Seq Kit v2 and the Ion Xpress RNA-seq bar coding kit (Thermo Fisher Scientific). A few modifications were made during purification steps in the small-RNA-seq to allow sequencing of longer RNAs (up to 200 nt) than with standard sRNA protocols; namely, 153 µL ethanol instead of 120 µL during purification of cDNA and 134 µL ethanol instead of 110 µL in protocol. The size distribution and yield of the barcoded libraries were assessed using a 2200 TapeStation System with Agilent D1K ScreenTapes (Agilent Technologies). Sequencing templates were prepared by an Ion Chef System using the Ion PI Hi-Q Chef Kit (Thermo Fisher Scientific). Sequencing was performed on an Ion Proton System using Ion PI Chips v3 (Thermo Fisher Scientific) according to the instructions of the manufacturer.
Northern blotting
Probe design
Three different DNA 5′-biotinylated probes were designed: one common to both maternal-type and somatic-type (-C), one specific for maternal-type (-M), and one specific for somatic-type (-S) (Supplemental Table S7). The probes were ordered from Integrated DNA Technologies or Exiqon. Upon delivery they were immediately rehydrated with LowTE to 100 µM and further stored at −20°C.
Electrophoresis
One microgram zebrafish egg or whole-body male adult RNA and 1 µL 0.1× Biotinylated sRNA Ladder (Kerafast) were mixed with Novex TBE-Urea Sample Buffer and heated at 70°C for 3 min. They were loaded on Novex TBE-Urea Gel 6% (Thermo Fisher Scientific) following the manufacturer's protocol. Gels were run at 70 V for 105 min.
Blotting and detection
After electrophoresis, the gel was soaked in 20× SSC for 10 min. RNA was subsequently transferred to an Amersham Hybond-N+ (GE Healthcare) membrane by capillary blotting overnight. Then the membrane was exposed to short-wave UV light (254 nm) for 1 min to cross-link the RNA and immediately prehybridized with ULTRAhyb Ultrasensitive Hybridization Buffer (Thermo Fisher Scientific) for 2 h at 55°C. Of note, 5 pM of the selected probe was overnight hybridized to the prehybridized blot at 55°C. Blots were washed twice 5 min in 2× SSC, 0.1% SDS at 55°C and twice 15 min in 0.1× SSC, 0.1% SDS at 55°C. Detection was performed using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific) following the manufacturer's protocol. Pictures were made with an Odyssey Fc (LI-COR Biosciences) adjusting brightness and contrast when needed.
RT-qPCR
Primer design
Forward and reverse PCR primers were designed for the zebrafish maternal-type 5S rRNA gene, the somatic-type 5S rRNA gene, the p53 gene, and the 18S rRNA gene (Supplemental Table S7).
Reverse transcription
SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) was used to generate 5S and 18S cDNA from total RNA of a zebrafish whole-body male adult and egg pool, following the manufacturer's protocol.
PCR reaction
The complete genomic DNA content from one zebrafish egg or 3.5 ng of DNA from a whole-body male adult was used as template for copy-number determination (with p53 primers as control), while 5S cDNA was used for RT-qPCR (with 18S primers as control). SYBR Green real-time qPCR was undertaken using a 7300 Applied Biosystems Thermocycler (Applied Biosystems). The 25 µL PCR reaction consisted of 12.5 µL of 2× Platinum SYBR Green qPCR SuperMix-UDG (Thermo Fisher Scientific), 1 µL of appropriate forward and reverse primers diluted in RNase-free water, 0.5 µL of ROX Reference Dye, 6 µL of water, and 5 µL of DNA template. No-template controls were performed also for each master mix prepared. The cycling program included the following steps: 50°C for 2 min, 95°C for 2 min, followed by 45 cycles of 95°C, 15 sec, 60°C 30 sec. Results were analyzed using a 7300 System v1.4.0 (Applied Biosystems).
Bioinformatics analyses
Known 5S rRNA reference sequences
Known 5S rRNA sequences of D. rerio were downloaded from Biomart (Smedley et al. 2015) on June 1, 2016. Sequences were selected upon annotation as 5S rRNA applying a minimal-length cutoff of 100 nt. Unique 5S rRNA sequences of this first set were then aligned (Madden 2002) to the reference Danio rerio genome (GRCz10) using BLAST (Altschul et al. 1990). Positive hits were selected using minimal cutoffs of 80% for identity and 100 nt for length. Unique sequences of the intersection between the unique downloaded 5S rRNA sequences and the selected BLAST hits were selected. The obtained sequences were subsequently flanked by a stretch of 5 Ns on both the 5′ and 3′ ends, resulting in a list of Zebrafish 5S rRNA reference sequences suited for mapping analysis. Known 5S rRNA sequences of X. laevis (oocyte-type [Xlo], trace oocyte-type [Xlt] and somatic-type [Xls]) were retrieved from GenBank (Xlo: X05089.1; Xls: J01899.1; Xlt: J01012.1) and subsequently flanked by a stretch of 5 Ns on both the 5′ and 3′ ends, resulting in a list of X. laevis 5S rRNA reference sequences suited for mapping analysis.
Zebrafish 5S rRNA similarity tree
5S rRNA reference sequences were multialigned using CLC Genomics Workbench 8.5.1 (gap open cost = 20.0; gap extension cost = 20.0; end gap cost = free). A tree was then built using the same software (building method = UPGMA; nucleotide distance = Jukes–Cantor).
Mapping NGS reads to the 5S reference
Reads longer than 100 nt from all experiments were mapped against the obtained respective lists of 5S rRNA reference sequences using Bowtie2 (Langmead and Salzberg 2012) with the following settings: -np to 0, - - score-min to L, -0, -0.06 for zebrafish and -0, -0.9 for X. laevis, - -rdg and - -rfg to 1,6 in order to limit the maximal amount of mismatches to 5% and 15% for Zebrafish and X. laevis, respectively. SAMtools v1.2 (Li et al. 2009) was used to convert the alignment to the BAM file format and to retrieve the mapped read counts per 5S rRNA sequence.
Mapping results analysis
Sequences that were represented by less than 1000 reads in all samples combined were considered to be “not-expressed” and discarded as background noise. The read counts were normalized by the total of mapped reads. The expressed sequences were clustered using hierarchical clustering (hclust function with default settings in R package “stats” v3.2.1 [R Core Team 2013]) on the Euclidean distance between the scaled and centered profiles of the normalized read counts over the 19 experiments (and not on the sequence). The 5S rRNA “subsequences” and their counts were collapsed to the 5S sequences having the most similar profile according to the clustering.
Detecting MutsuDr LINE retrotransposon sequences
MutsuDr1 (L1-2_DR), MutsuDr2 (L1-5_DR), and MutsuDr3 (L1-3_DR) consensus sequences were downloaded from Repbase (www.girinst.org/repbase) and used to search through the zebrafish rRNA loci on chromosomes 4 and 18, using BLAST with default settings. For each MutsuDr subfamily, hits with a minimal length of 4500 bp and identity >90% were annotated as complete MutsuDr genomic elements and assigned as variants to each subfamily accordingly. Hits with a length between 3000 and 4500 bp were annotated as incomplete MutsuDr genomic elements. To identify new MutsuDr subfamilies, we searched for 5S rRNA intragenic spacers with a length between 4500 and 7000 bp having at least two of the following conditions: an insertion site in a 5S gene, the presence of two ORFs, and a 3′ poly(A) stretch. The located sequences were established as elements of a new MutsuDr subfamily. BLAST was then used as described above to find and annotate complete and incomplete variants of that new subfamily.
MutsuDr family similarity tree
MutsuDr consensus sequences were multialigned using CLC Genomics Workbench 8.5.1 (gap open cost = 20.0; gap extension cost = 20.0; end gap cost = free). A tree was then built using the same software (building method = UPGMA; nucleotide distance = Jukes–Cantor).
Mapping NGS reads to the MutsuDr subfamily consensus sequences
Reads shorter than 70 nt were discarded in the (large) RNA-seq experiment and reads shorter than 25 nt were discarded from the small-RNA-seq experiments. Bowtie2 with default settings was used to map the selected reads. SAMtools was used to select reads with mapping quality higher than 15, which correspond to reads mapping uniquely and with high quality to MutsuDr retrotransposon elements. SAMtools was also used to convert the alignment to a BAM file format and to retrieve the mapped reads counts.
piRNA sense and antisense coverage
SAMtools was used to select the piRNA reads oriented sense and antisense to the MutsuDr retrotransposon sequences in separate files. BEDTools v2.17.0 (Quinlan and Hall 2010) genomecov was applied on the small-RNA-seq reads to obtain for each sample the read count per nucleotide on each MutsuDr sequence (coverage depth). In each sample, the counts were normalized by the total reads in the size range of 28–32 nt (piRNA size range). For each MutsuDr sequence, the final coverage depth values per nucleotide were summed through the samples, resulting into one total coverage depth value per nucleotide position for each MutsuDr subfamily consensus sequence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Netherlands Organisation for Scientific Research (NWO), project 834.12.003.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059642.116.
Freely available online through the RNA Open Access option.
DATA DEPOSITION
All sequencing results are accessible through the BioProject database under the project accession number PRJNA347637 (https://www.ncbi.nlm.nih.gov/bioproject).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
REFERENCES
- Allison LA, Romaniuk PJ, Bakken AH. 1991. RNA-protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev Biol 144: 129–144. [DOI] [PubMed] [Google Scholar]
- Allison LA, North MT, Neville LA. 1995. Differential binding of oocyte-type and somatic-type 5S rRNA to TFIIIA and ribosomal protein L5 in Xenopus oocytes: specialization for storage versus mobilization. Dev Biol 168: 284–295. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bellavia D, Dimarco E, Naselli F, Caradonna F. 2013. DNA-methylation dependent regulation of embryo-specific 5S ribosomal DNA cluster transcription in adult tissues of sea urchin Paracentrotus lividus. Genomics 102: 397–402. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Dimitrov S, Wolffe AP. 1994. Specific regulation of Xenopus chromosomal 5s rRNA gene transcription in vivo by histone H1. Genes Dev 8: 1147–1159. [DOI] [PubMed] [Google Scholar]
- Briggs JP. 2002. The zebrafish: a new model organism for integrative physiology. Am J Physiol Regul Integr Comp Physiol 282: R3–R9. [DOI] [PubMed] [Google Scholar]
- Brown DD. 2004. A tribute to the Xenopus laevis oocyte and egg. J Biol Chem 279: 45291–45299. [DOI] [PubMed] [Google Scholar]
- Ciganda M, Williams N. 2011. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi MB, Martins C, Bertollo LAC. 2010. Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol Biol 10: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Menut S, Méchali M. 1999. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol 19: 6682–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H, Wegnez M. 1977. Oocytes of Xenopus laevis synthesize but do not accumulate 5s RNA of somatic type. Dev Biol 58: 212–217. [DOI] [PubMed] [Google Scholar]
- Dimarco E, Cascone E, Bellavia D, Caradonna F. 2012. Functional variants of 5S rRNA in the ribosomes of common sea urchin Paracentrotus lividus. Gene 508: 21–25. [DOI] [PubMed] [Google Scholar]
- Dinman JD. 2005. 5S rRNA: structure and function from head to toe. Int J Biomed Sci 1: 2–7. [PMC free article] [PubMed] [Google Scholar]
- Dixon LK, Ford PJ. 1982. Regulation of protein synthesis and accumulation during oogenesis in Xenopus laevis. Dev Biol 93: 478–497. [DOI] [PubMed] [Google Scholar]
- Dooley K. 2000. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev 10: 252–256. [DOI] [PubMed] [Google Scholar]
- Douet J, Tourmente S. 2007. Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity (Edinb) 99: 5–13. [DOI] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351: aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornung E, De Innocentiis S, Annesi F, Sola L. 2000. Zebrafish 5S rRNA genes map to the long arms of chromosome 3. Chromosom Res 8: 362. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res 31: 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33: 121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinta DR, Tso JY, Narayanswami S, Hamkalo BA, Korn LJ. 1986. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc Natl Acad Sci 83: 5150–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. 2015. PIWI-Interacting RNA: its biogenesis and functions. Annu Rev Biochem 84: 405–433. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. 2004. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol 21: 207–217. [DOI] [PubMed] [Google Scholar]
- Komiya H, Hasegawa M, Takemura S. 1986. Differentiation of oocyte- and somatic-type 5S rRNAs in animals. J Biochem 100: 369–374. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Hennen E, Stott G, Vacun G. 2002. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol 12: 2023–2028. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to- zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB. 1980. Repeated genes in eukaryotes. Annu Rev Biochem 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Madden T. 2002. The BLAST Sequence Analysis Tool. In The NCBI Handbook (ed. McEntyre J, Ostell J), Chap. 16. National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- Mairy M, Denis H. 1971. [Biochemical studies on oogenesis. I. RNA synthesis and accumulation during oogenesis of the South African toad Xenopus laevis]. Dev Biol 24: 143–165. [DOI] [PubMed] [Google Scholar]
- Martins C, Wasko A. 2004. Organization and evolution of 5S ribosomal DNA in the fish genome. Focus Genome Res 1: 335–363. [Google Scholar]
- Martins C, Wasko AP, Oliveira C, Porto-Foresti F, Parise-Maltempi PP, Wright JM, Foresti F. 2002. Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenet Genome Res 98: 78–85. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Jasencakova Z, Vaillant I, Gendrel AV, Colot V, Schubert I, Tourmente S. 2003. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15: 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. 1974. Metabolism of 5s RNA in the absence of ribosome production. Cell 3: 275–281. [DOI] [PubMed] [Google Scholar]
- Nietfeld W, Digweed M, Mentzel H, Meyerhof W, Köster M, Knöchel W, Erdmann VA, Pieler T. 1988. Oocyte and somatic 5S ribosomal RNA and 5S RNA encoding genes in Xenopus tropicalis. Nucleic Acids Res 16: 8803–8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton W, Bally-Cuif L. 2010. Adult zebrafish as a model organism for behavioural genetics. BMC Neurosci 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HHJ. 2001. Biology of mammalian L1 retrotransposons. Annu Rev Genet 35: 501–538. [DOI] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3: a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RC, Doering JL, Brown DD. 1980. Characterization of two Xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell 20: 131–141. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Reed KM. 2000. Localization of repetitive DNAs to zebrafish (Danio rerio) chromosomes by fluorescence in situ hybridization (FISH). Chromosom Res 8: 27–35. [DOI] [PubMed] [Google Scholar]
- Pinhal D, Yoshimura TS, Araki CS, Martins C. 2011. The 5S rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays. BMC Evol Biol 11: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rebordinos L, Cross I, Merlo A. 2013. High evolutionary dynamism in 5s rDNA of fish: state of the art. Cytogenet Genome Res 141: 103–113. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr 3: MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Trendelenburg MF, Franke WW. 1976. Regulation of transcription of genes of ribosomal RNA during amphibian oogenesis. A biochemical and morphological study. J Cell Biol 69: 465–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF. 2007. The maternal-zygotic transition: death and birth of RNAs. Science 316: 406–407. [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620. [DOI] [PubMed] [Google Scholar]
- Scripture J, Huber P. 1995. Analysis of the binding of Xenopus ribosomal protein L5 to oocyte 5 S rRNA. J Biol Chem 270: 27358–27365. [DOI] [PubMed] [Google Scholar]
- Scripture JB, Huber PW. 2011. Binding site for Xenopus ribosomal protein L5 and accompanying structural changes in 5S rRNA. Biochemistry 50: 3827–3839. [DOI] [PubMed] [Google Scholar]
- Shaw P, Brown J. 2012. Nucleoli: composition, function, and dynamics. Plant Physiol 158: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10: 6718–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonová R, Majtánová Z, Sember A, Staaks GBO, Bohlen J, Freyhof J, Rábová M, Ráb P. 2013. Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol Biol 13: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 2002. 5S ribosomal RNA database. Nucleic Acids Res 30: 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Zielezinski A, Barciszewski J, Erdmann VA, Karlowski WM. 2016. 5SRNAdb: an information resource for 5S ribosomal RNAs. Nucleic Acids Res 44: D180–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Hanocq F, Heilporn V. 1977. Persistence of oocyte amplified rDNA during early development of Xenopus laevis eggs. Dev Biol 57: 226–229. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Lin S. 2008. Zebrafish as a developmental model organism for pediatric research. Pediatr Res 64: 470–476. [DOI] [PubMed] [Google Scholar]
- Wegnez M, Monier R, Denis H. 1972. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett 25: 13–20. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Brown DD. 1988. Developmental regulation of two 5S ribosomal RNA genes. Science 241: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Wormington WM, Brown DD. 1983. Onset of 5 S RNA gene regulation during Xenopus embryogenesis. Dev Biol 99: 248–257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.