Abstract
Organismal function is, to a great extent, determined by interactions among their fundamental building blocks, the cells. In this work, we studied the cell-cell interactome of fetal placental trophoblast cells and maternal endometrial stromal cells, using single-cell transcriptomics. The placental interface mediates the interaction between two semiallogenic individuals, the mother and the fetus, and is thus the epitome of cell interactions. To study these, we inferred the cell-cell interactome by assessing the gene expression of receptor-ligand pairs across cell types. We find a highly cell-type-specific expression of G-protein-coupled receptors, implying that ligand-receptor profiles could be a reliable tool for cell type identification. Furthermore, we find that uterine decidual cells represent a cell-cell interaction hub with a large number of potential incoming and outgoing signals. Decidual cells differentiate from their precursors, the endometrial stromal fibroblasts, during uterine preparation for pregnancy. We show that decidualization (even in vitro) enhances the ability to communicate with the fetus, as most of the receptors and ligands up-regulated during decidualization have their counterpart expressed in trophoblast cells. Among the signals transmitted, growth factors and immune signals dominate, and suggest a delicate balance of enhancing and suppressive signals. Finally, this study provides a rich resource of gene expression profiles of term intravillous and extravillous trophoblasts, including the transcriptome of the multinucleated syncytiotrophoblast.
The long duration of eutherian fetal development requires a considerable level of negotiation between fetal and maternal needs and capacities. The key locus of this negotiation is between trophoblast cells of the placenta and the endometrium of the maternal uterus, in addition to hormonal signals. Eutherian trophoblasts are ancestrally invasive (Wildman et al. 2006), and it is likely that even secondarily noninvasive placentation, such as that of hoofed animals or lemurs, is due to the evolution of a less permissive uterus, rather than the evolution of less invasive trophoblasts (D'Souza and Wagner 2014). Differentiation of endometrial stromal fibroblasts to form the decidua, which accepts the implanting conceptus, is an additional evolutionary novelty to accommodate trophoblast invasiveness also found in humans (Wagner et al. 2014).
Maternal-fetal interactions are also central in human pregnancy. In addition to standard communication between adjacent cells within an individual, the maternal-fetal interface also integrates two semiallogenic individuals, the fetus and the mother. The manifold functions in negotiating maternal and fetal interests (e.g., nutrient and gas exchange, anchoring, immunity) are reflected in heterogeneous placental structure, encompassing many distinct cell types. Placental cell fate is determined in early eutherian development, as the outer layer of the blastocyst, the trophectoderm, is the precursor of placental tissue. Enveloped by the trophectoderm is the inner cell mass, which gives rise to the embryo proper and further extraembryonic tissues, like the yolk sac, the amnion, and the allantois. Following the implantation of the blastocyst into decidualized endometrium, a specialized population of placental trophoblasts, extravillous trophoblasts (EVTs), invade the maternal decidua and vessels and thereby generate lacunas filled with maternal blood in which the developing surface-enlarging fetal villi become bathed. EVTs migrate out of the anchoring villi into maternal endometrium and partially into myometrium. A subset of EVTs is involved in remodeling maternal spiral arteries, thereby acquiring endothelial character, while another subset fuse to form the placental bed, and yet another set is involved in the uterine gland remodeling (Ji et al. 2013; Maltepe and Fisher 2015). The placental chorionic villi, which are bathed in maternal blood, contain fetal blood vessels and are covered by a continuous multinucleated layer of syncytiotrophoblast. This layer arises and is maintained through pregnancy by the fusion of the underlying cytotrophoblasts and represents an alternative differentiation fate to EVT. The syncytiotrophoblast is in direct contact with maternal blood and is the main interface between maternal and fetal circulation.
Implantation outside of the decidualized uterine region is deeper, often reaching the myometrium (i.e., placenta accreta) and can be fatal to the mother at birth (Hannon et al. 2012). In addition to forming a maternal barrier against invasive trophoblast, decidualization has been suggested to actively control trophoblasts. For example, the invasiveness and growth of cultured trophoblast cells is decreased in conditioned medium from decidual cells (Lewis et al. 1993; Zhu et al. 2009; Godbole et al. 2011). Decidual effects are not uniformly suppressive; rather they involve fine-tuned interactions via numerous pathways (Knofler 2010). For example, a number of decidual growth factors secreted starting at implantation likely support trophoblast invasion. Similarly, the maternal immune system is not simply suppressed at the introduction of the allograft but rather modulated to add active support (Knofler and Pollheimer 2012; Erlebacher 2013; Fock et al. 2013). Given the pervasive evidence for mutual interaction between maternal and fetal tissues, the resulting utero-placental phenotype is best conceptualized as a unit resulting from a rich network of developmental interactions.
Identification of cell-cell crosstalk requires examining maternal and fetal cells jointly. This cannot be done in healthy human pregnancy, and therefore most questions are addressed either on the term placenta alone, or in maternal or fetal cell culture. The utility of animal models for this purpose is often questioned, as the evolutionary relationships, and therefore homology between placental cell lineages across species, are not well understood (Mess 2014; Maltepe and Fisher 2015). Here, we approach this question by inferring the cell interactome of trophoblast cells and maternal decidua from single-cell transcriptomes, complemented with laser microdissected syncytiotrophoblast and cell culture of endometrial stromal cells. While the inferred cell communication network remains hypothetical, the coexpression of ligands and receptors in adjacent cells is stronger evidence for functional importance than the gene expression of a single cell type. In combination with downstream gene expression effects of the signaling, this information promotes rich functional insights, because cell interactions are part of the core biological function of these tissues (Semrau and van Oudenaarden 2015). In addition, the high level of resolution in the single-cell transcriptome allows identification of potentially important rare cell types, whose signal is too diluted to be detected in tissue-level transcriptomes (Buettner et al. 2015; Crosetto et al. 2015; Stegle et al. 2015).
Results
Sampling the villous tissue of two human term placentas resulted in 87 single-cell transcriptomes, which grouped into five distinct clusters (Fig. 1; Supplemental Fig. S6). Of these, three large clusters consisted of 23, 28, and 28 cells, respectively, while sets of three and five cells represent the remaining two small clusters. We identified the three large and one small cluster as belonging to different types of placental trophoblasts and the remaining small cluster as maternal immune cells. These data were complemented by two transcriptomes of syncytiotrophoblast, collected from a single placenta by laser microdissection, the transcriptomes of primary undifferentiated endometrial stromal fibroblast (ESF), and the transcriptome of in vitro differentiated primary decidual cells from two patients. The cells included in this study thus represent adjacent maternal (decidua) and fetal (syncytiotrophoblast and extravillous trophoblast) cells of utero-placental interface, as well as the precursors of decidualized cells and syncytiotrophoblast (ESF and cytotrophoblast). This approach enables identification of putative interactions and their development. Finally, we generated two tissue-level transcriptomes of human term placenta for comparison. All transcriptomes were normalized as TPM (transcripts per million) to the same set of human coding genes prior to comparison.
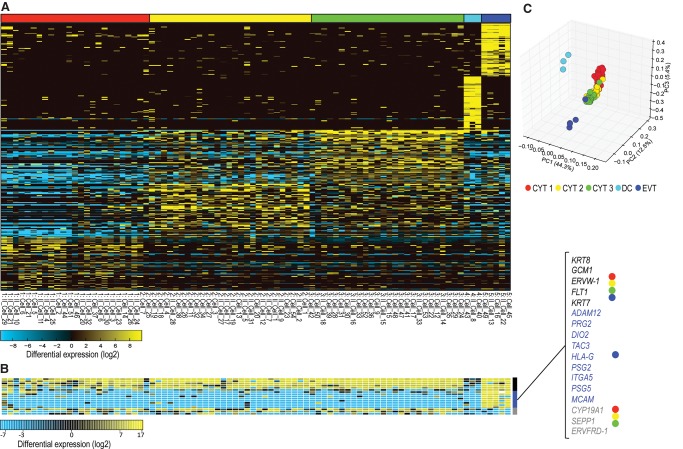
Figure 1.
The gene expression in the single-cell experiment. (A) Expression of the top 300 genes with greater than twofold differential expression between clusters that were previously established by hierarchical clustering. x-axis: The cells with roman II denote the second batch of cells. Note that the cells from both batches are represented in all clusters. (B) Known trophoblast markers are shown separately, with general trophoblast markers in black, extravillous trophoblasts in blue, and cytotrophoblast-specific expression in gray. (C) First three principal components based on 300 marker genes (full PCA in Supplemental Fig. S8). Clusters correspond to the clustering in the heat map in A.
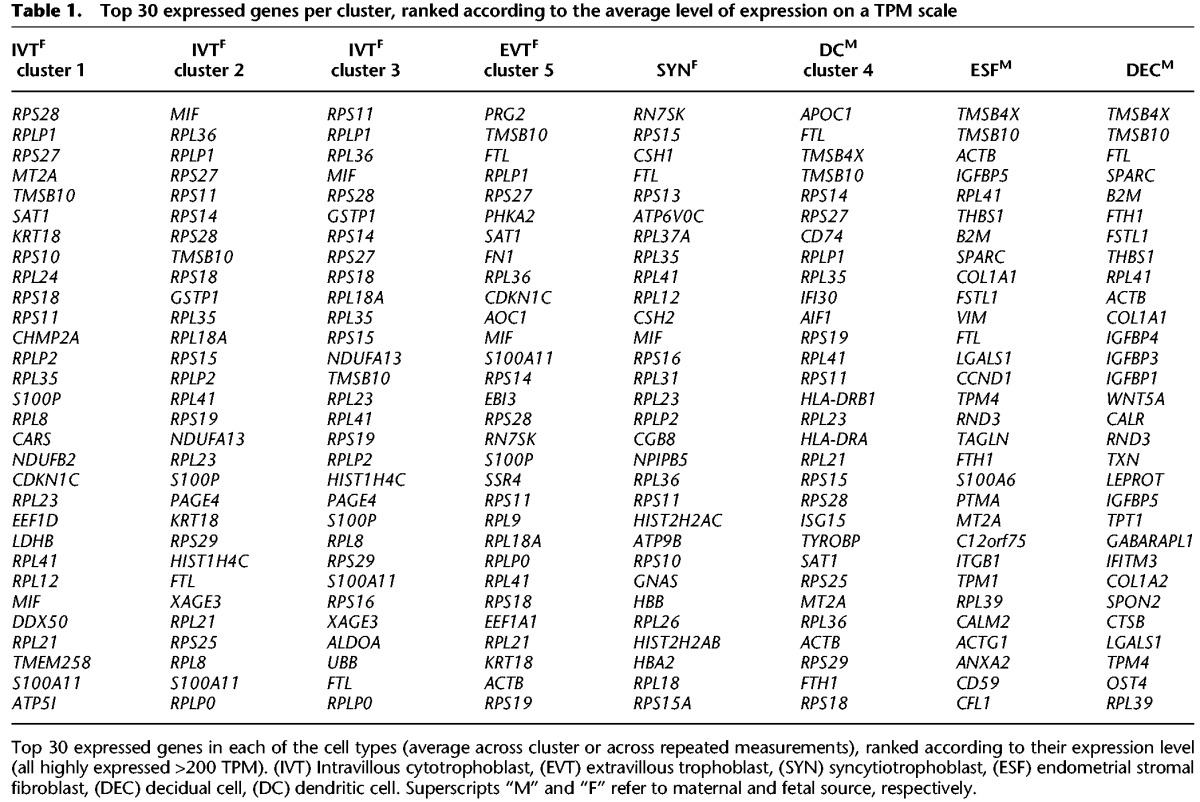
In the following, we first present the main expression patterns starting with comparison of tissue-level transcriptomes to the sum total of single-cell transcriptomes and then characterize the clusters by their diagnostic signatures relative to the particular assembly of cells. The description is limited to diagnostic gene expression, with the noteworthy observations of potential interest for experimental follow-up described in greater depth in Supplemental Information. The 30 most highly expressed genes in each cluster relative to other clusters are listed in Table 1 (averaged values) (full normalized expression levels in Supplemental Table S1). The second part of the study focuses on the cell communication network, its structure, and maternal and fetal contributions to pathways known to play major roles at the maternal-fetal interface. To our knowledge, this is the first characterization of the cell communication network at the human maternal-fetal interface.
Table 1.
Top 30 expressed genes per cluster, ranked according to the average level of expression on a TPM scale
Expression profiles
Tissue-level villous placenta RNA-seq
Various factors may result in cells acquired by the pipeline not being represented in proportion to their frequency in the starting tissue. For example, the cells’ adhesive properties or size may bias their passage through the Fluidigm cell, as is most apparent in the large multinucleated syncytiotrophoblast, which we targeted separately. In addition, rare cell types will evade limited sampling. Our tissue preparation enriched for trophoblasts, which is the likely reason that the endothelial cells of placental vasculature or the villous mesenchymal cells are not represented. To estimate what portion of placental gene expression is covered by the single-cell transcriptomes, we compared the single-cell data with the tissue-level transcriptomes, generated from different individuals but the same type of sample (term Cesarean section, not in labor).
The top 25% (2108) most highly expressed genes in the whole placental transcriptome comprise 80% of the total aligned placental mRNA. Only 20 of these genes were not detected at all in the combined single-cell transcriptomes, and a further 34 genes were detected in less than three cells (syncytiotrophoblast-specific genes excluded) (Supplemental Table S2). The list includes several highly expressed RNA-genes with placental function (e.g., H19 and MIRLET7-family); however, it is not clear what missing cell population may be implicated. These genes do not show high expression in the decidual cells, which is the cell type commonly included in preparations of whole placenta. Given this high level of correspondence between tissue-level transcriptomes and the single-cell data, we conclude that the trophoblast cells at term are by far the most abundant cells, whereas the missing cells are rare and/or overlap greatly with respect to their expression with the cells captured in our samples. Consistent with the view that single-cell analysis may reveal expression profiles of rare cells, our analysis recovers expression of over 4000 genes, which are expressed at levels >2 TPM in five or more cells but are not detected in the tissue-level transcriptome at levels ≥1TPM. In this comparison, we chose a lower threshold for the tissue-level transcriptome, as this transcriptome has been sequenced at greater depth than the single-cell transcriptomes. We report the top 1000 of these genes in the Supplemental Table S3, ranked by the number of expressing cells.
Placental trophoblast cells
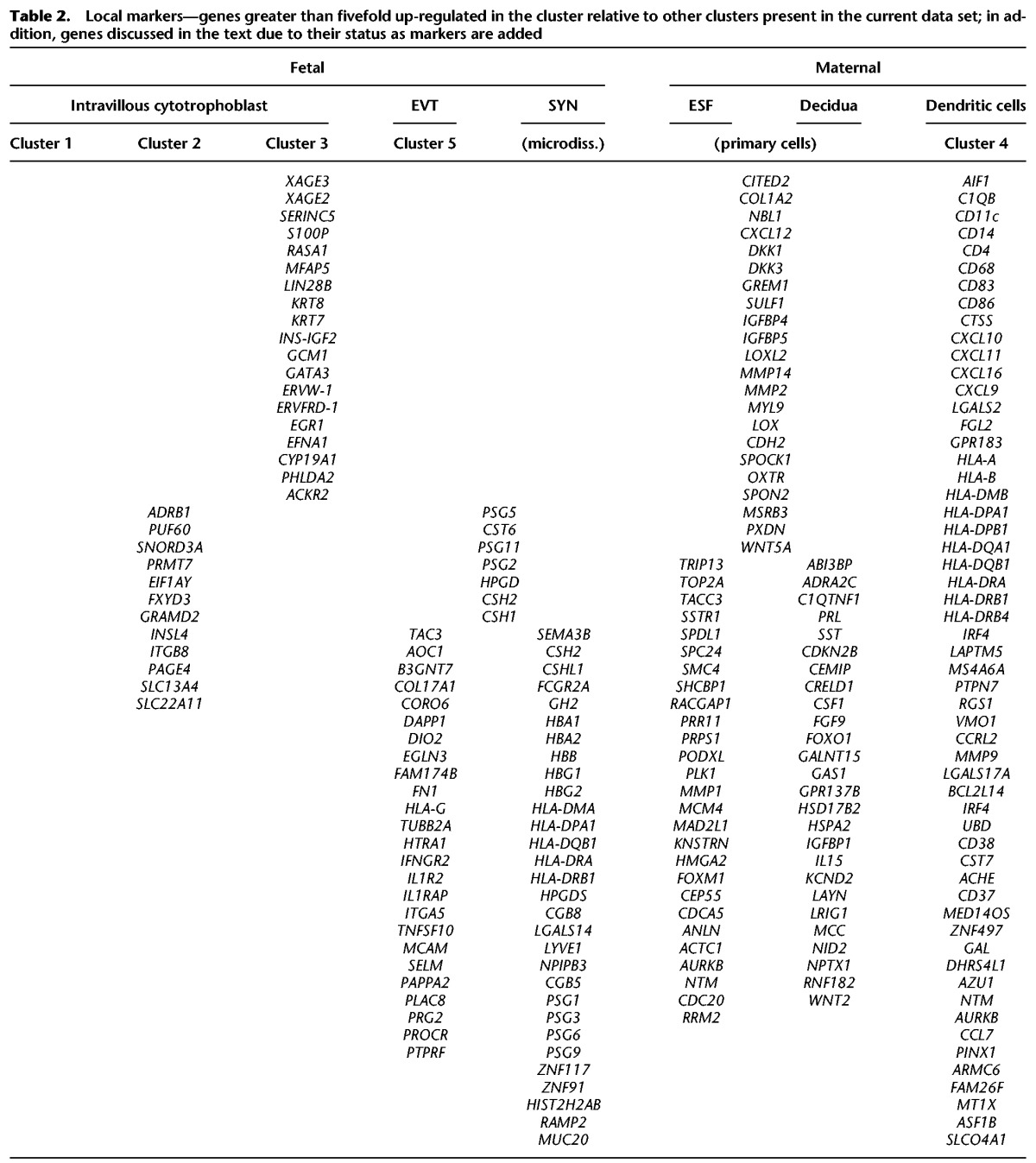
We clustered cells by their single-cell gene expression profiles into five clusters. A combination of known trophoblast markers (KRT7, KRT8, GCM1, CYP19A1) distinguishes clusters 1, 2, 3, and 5 from cluster 4 and from endometrial stromal cells (Fig. 1B). Further diagnostic genes are listed in Table 2, showing genes with >10-fold higher expression in trophoblast than in uterine and immune cells. Among the trophoblast clusters, clusters 1 and 5 show lower expression of ribosomal proteins, a signature typical of later stages of differentiation in cell lineages leading toward extravillous trophoblast (Ji et al. 2013) and the syncytiotrophoblast (Handwerger and Aronow 2003). The large trophoblast clusters 1–3 were identified as intravillous cytotrophoblast (see Supplemental Text) and the smaller cluster 5 as extravillous trophoblast.
Table 2.
Local markers—genes greater than fivefold up-regulated in the cluster relative to other clusters present in the current data set; in addition, genes discussed in the text due to their status as markers are added
Cytotrophoblast (CYT; Clusters 1–3)
These clusters likely represent a continuum in the differentiation of cytotrophoblasts. Genes specifically expressed in cytotrophoblasts are summarized in Table 2. Intravillous cytotrophoblasts are a very heterogeneous group, a finding often encountered at single-cell-level transcriptomes (Brunskill et al. 2014; Olsson et al. 2016). The expression profiles of these cells suggest that differentiation proceeds by individual cells turning on—or off—sets of genes in a different order rather than a uniform progression toward a terminal expression profile. Detailed characterization of intercellular heterogeneity and cell differentiation is beyond our present scope and will be addressed separately.
Extravillous trophoblast (EVT; Cluster 5)
This cluster of cells is unique in its expression of nonclassical human leukocyte antigen HLA-G and integrin 5 alpha ITGA5—both considered markers of extravillous trophoblast, KRT7 (Telugu et al. 2013; Borbely et al. 2014) and ADAM12 (Biadasiewicz et al. 2014). Further EVT-specific genes and other highly expressed genes are listed in Tables 1 and 2 and discussed in the Supplemental Material. These cells differ from cytotrophoblasts by expression of some endothelial genes, such as adhesion molecule MCAM (Jouve et al. 2013; Kaspi et al. 2013), lowered E-cadherin (CDH1) (Zhou et al. 1997), and the receptor for vascular endothelial growth factor FLT1. Note that MCAM may be a receptor for WNT5A, affecting cell motility (Ye et al. 2013). While EVTs are involved in invasion in early pregnancy (Bischof et al. 2000), some of the invasion-associated signature may be lacking at term (Ji et al. 2013). At term, genes up-regulated in EVT relative to cytotrophoblast are involved in modulation of extracellular matrix, vascularization, and immune pathways (Table 2; Supplemental Fig. S1B).
Syncytiotrophoblast (SYN; laser microdissection)
This cell type was collected in two samples from a single term placenta by laser microdissection and analyzed by the conventional RNA-seq pipeline. The two samples were analyzed separately and show a high correlation of expression (r = 0.99, in square root-transformed TPM), indicating a technically robust transcriptome. Among the most highly expressed syncytiotrophoblast-specific genes are hormones (CSH2, CSHL1, GH2), CGA, and pregnancy-specific glycoproteins (PSG-family) (detail in Supplemental Fig. S2). Several highly expressed genes are concomitantly restricted to syncytiotrophoblast and extravillous trophoblast: PSG2 and -5, chorionic somatotropin (CSH1 and CSH2), and the key prostaglandin catabolic enzyme, HPGD. An intriguing observation is the expression of human MHC type II antigen in syncytiotrophoblast; uniquely among trophoblasts, this cell layer expresses a profile of human leukocyte antigens similar to the profile of the uterine dendritic cell cluster (HLA-DRA, HLA-DRB1 and -5, HLA-DMA, HLA-DQB1, HLA-DPA1), albeit at levels of expression 20- to 30-fold lower than those in dendritic cells.
Maternal cells
Uterine dendritic cells (DCs; Cluster 4)
Cluster four consists of three cells lacking the trophoblast markers but expressing MHC II subunits (HLA-DRA in particular) (Table 2), characteristic of antigen-presenting cells. These cells lack expression of CD19, CD209, and CD163 but express the combination of markers ITGAX+/ CD14+/CD4+/CD83+/ CD86+. The genes CLEC4C, THBD, CD1C, CD80, IL10, and IL12B that characterize some of the decidual DCs (Kammerer et al. 2008; Talayev et al. 2010) were not found. CD14 is often considered a marker of macrophages, with which dendritic cells are closely related. It has, however, been shown that CD14 expression is retained in dendritic cells when they develop in the presence of trophoblasts (Talayev et al. 2010). These facts, together with the source of cells in this experiment, suggest that these are uterine dendritic cells and thus of maternal origin (Kammerer et al. 2000; Tagliani and Erlebacher 2011). Even if we cannot exclude that some of the markers are missing in our data as a consequence of methodological or biological variability, particularly due to low number of cells of this type in our sample (N = 3), the expression profile is nonetheless sufficiently consistent for the identification of these cells as uterine dendritic cells.
Endometrial stromal fibroblasts and decidual cells (ESFs and DECs; cell culture)
These cells stem from primary cultures from two patients at days 3 and 9 of the menstrual cycle and were cultured following standard procedures. Decidual cells in this study have been differentiated in vitro (see Methods; for associated gene expression changes, see Brar et al. 2001). For the most significantly up- and down-regulated genes, see Table 3; a portion of these will be discussed in the context of interactions below. Differentiation marks a transition from the proliferative into decidual state. Correspondingly, the bulk of down-regulated genes in decidua relative to the endometrial stromal fibroblasts are involved in cell cycle, replication, and DNA repair (Supplemental Fig. S3). In contrast, multiple metabolic, immune, and hormonal pathways are up-regulated in decidualization.
Table 3.
Genes up- and down-regulated during decidualization

Maternal-fetal cell-cell communication
We used public databases of ligand-receptor relationships (Methods) to infer cell interactions. While based on mRNA expression rather than function, many putative interactions are supported by the expression of downstream target genes in the particular cells or have empirical support in prior studies and have been tested here (below).
The plot in Figure 2 shows the inferred cell-cell interactions, with the cell types indicated by color, and expressed ligands and receptors listed along the outer margins. We included ligands and receptors expressed beyond 30 TPM, to allow visualization. The arrows connect expressed ligand and receptor genes, indicating an interaction, with gray arrows representing the full interactome, whereas the black arrows signify a set of interactions between adjacent maternal and fetal cells. Figure 3 illustrates the increase in the potential for interactions between uterine and adjacent trophoblast cells during decidualization as interaction plots (Fig. 3A,B) and quantitatively as the number of edges (Fig. 3C).
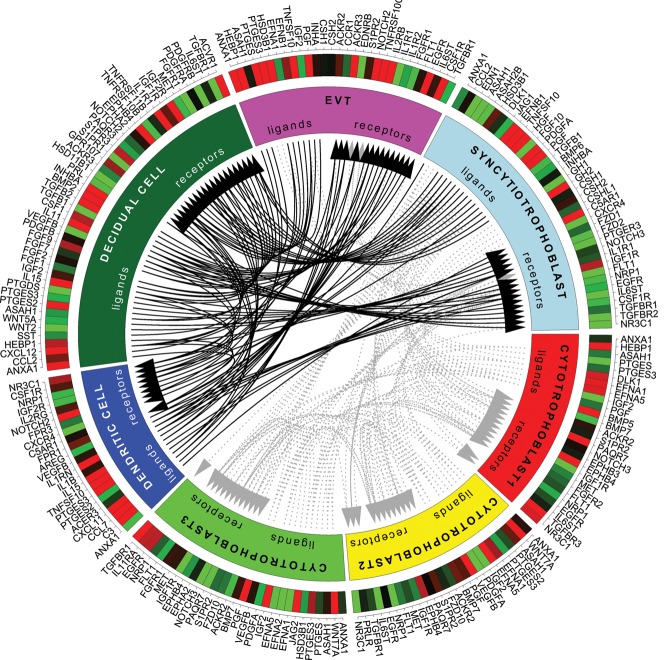
Figure 2.
Putative signaling between expressed receptors and their ligands in pregnant uterus at term. Compartments represent cell types, and their expressed ligands and receptors are noted along the outer margin. Average cluster gene expression was used, with expression threshold 30 TPM. Black arrows denote putative interactions between directly adjacent maternal and placental cells (maternal decidua and placental EVT and SYN), and dotted gray lines denote putative interactions with cytotrophoblasts.
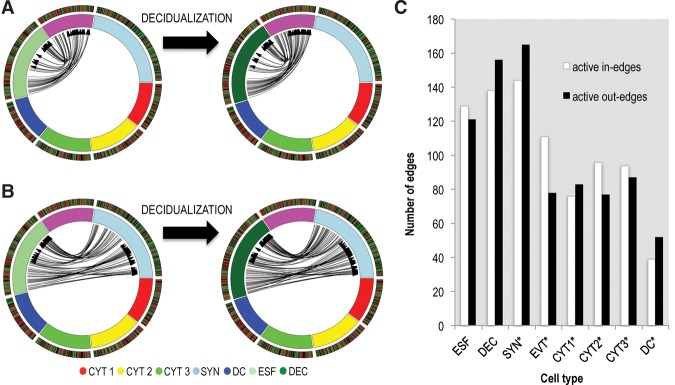
Figure 3.
Decidualization increases the cell-cell signaling potential with the adjacent EVT (purple, A) and syncytiotrophoblast (light blue, B). Average cluster gene expression is shown, with genes expressed >5 TPM included. (C) Average numbers of putative interactions per cell type, separated into in- and out-edges. Asterisks refer to the number of interactions in the context of decidua, rather than undifferentiated endometrial stromal fibroblasts.
The structure of the cell-cell communication network: DECs are an interaction hub
The most striking aspect of the interactome in Figure 2 is the large number of interactions between decidual cells and the syncytium, suggesting that the uterine decidua acts as a hub of feto-maternal cell signaling. Figure 3C summarizes the total number of outgoing and incoming edges of each cell type. The interactions with placental and dendritic cells are assessed for decidualized cells (rather than ESF), as this reflects the in vivo situation during pregnancy. Uterine cells and the syncytiotrophoblast thus lead in the potential for interactions among the cells of maternal-fetal interface. The dominance of decidual signaling is maintained if the overlap in ligands and receptors across closely related trophoblasts is removed (which could enhance the pattern). We further asked how likely the observed structure is based merely on the observed number of ligands/receptors per cell type, using a permutation test. We find that, given the observed number of ligands and receptors across cells, only 20% of the randomly generated networks have a higher number of connected decidual ligands (Supplemental Fig. S10). As this is at least in part due to the high number of ligands and receptors expressed in maternal endometrial stroma, we next asked how likely it is that decidual cells will express the number of ligands and receptors given the expectations from other cells in the data set. This is highly unlikely for the number of expressed receptors and ligands, as well as the number of those involved in putative interactions (in all cases, P < 0.05 for decidualized, nondecidualized cells, and syncytiotrophoblast), confirming the significance of the characterization as a hub. To specifically assess the contribution of decidualization to the potential for cell communication, we compared the inferred interactions of fetal cells with ESFs to those with DECs in Figure 3, A and B. The up-regulation of uterine ligands and receptors during decidualization enhances signaling to the syncytiotrophoblast and EVT, as well as from syncytiotrophoblast. In fact, a majority of receptors and ligands up-regulated in decidualization likely communicate with trophoblast cells (Supplemental Fig. S4). The involved pathways will be discussed below.
GPCR spectrum characterizes cell types
Cells exert their biological function, in large part, by interacting with other cells. This suggests that cell types with different biological functions would show distinct profiles of ligands and receptors, as these enable the cell-type–specific interactions. Therefore, the receptor/ligand profile may serve as a robust biological characterization of the cell types. In our data, this is the case in particular with respect to G-protein-coupled receptors (Fig. 4; Supplemental Table S4). Figure 4A shows the heat map of GPCR expression among cells, showing the specificity of GPCR signaling among clusters. Distinct signatures of different cell types are contrasted in our data set with very similar signatures of the three cytotrophoblast clusters. This supports the inference that these three clusters are closely related groups of cells representing the same cell type. Specificity of GPCR profiles suggests an interesting way to characterize cell-type identity; namely, by focusing specifically on the cell's interaction potential. Thereby, it may be possible to distinguish between the cell's biological role within a tissue captured in interactions from the more variable aspects of the cell's gene expression phenotype.
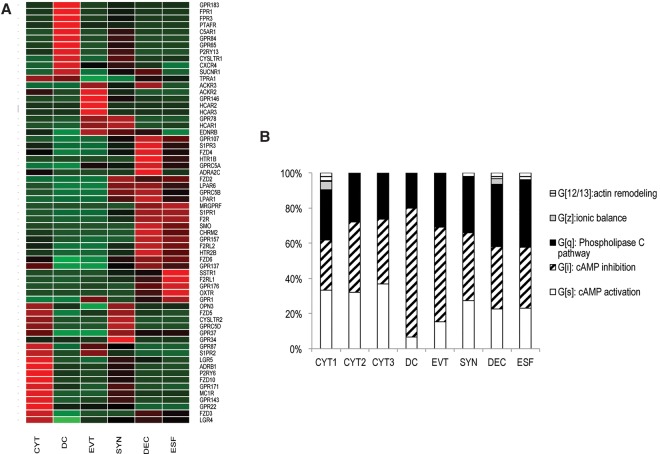
Figure 4.
Distribution of G-protein-coupled receptors among cell types. (A) The expression heat map of GPCRs across sampled cells. (B) Characterization of cell clusters by class of G-protein alpha subunit used.
Of over 400 GPCRs in the IUPHAR data set (http://www.guidetopharmacology.org), 116 are expressed >2 TPM in at least one of our cells, most in a cell-type–specific manner. For example, GPCRs expressed in cytotrophoblasts include multiple members of the WNT-pathway (mainly Frizzled and LGR families), purinergic and chemokine receptors. Dendritic cells are rich in chemokine receptors, complement system receptors, and prostaglandin E receptors. Decidua differs from endometrial stromal fibroblast in the 260-fold up-regulation of adrenergic receptor 2C and the expression of endothelin and chemokine receptors, while it shares with endometrial stromal fibroblasts the expression of serotonin and oxytocin receptors (down-regulated in decidualization), thrombin receptor-like 2 (F2RL2), and several Frizzled receptors (FZD4, -6). The downstream G-protein subunits, in particular the G-protein alpha subunits, can be associated with functional classes via their specific second messenger systems. We used this association to compare cell types with respect to the types of GPCR signaling they are involved in. The plot in Figure 4B shows that the most distinct cell type with respect to GPCR signaling is the dendritic cell, in which most GPCR signaling is associated with inhibition of cAMP signaling. Somewhat distinct is EVT, in which cAMP inhibition represents a greater proportion of GPCR signaling than in other trophoblasts or endometrium.
Uterine decidualization changes the interaction potential
The cell-cell interactions that are enabled by decidualization of the uterine stroma are of particular interest. These can be found by comparing the interactions of the decidualized with those of undifferentiated stromal fibroblasts.
Decidualization up-regulates growth factors FGF9, BMP2, and PDGFB, for which the receptors are expressed in syncytiotrophoblast and EVT, and the receptors are also up-regulated in decidua itself. In addition, decidual up-regulation of IL15 and CSF1 introduces potential to signal to the receptors in EVT and dendritic cells. Decidual cells also likely communicate with dendritic cells via IL1B, which is produced by dendritic cell and signals to decidual cell receptor IL1R1. Further potential for interaction is the decidual up-regulation of CCL2, which binds to atypical chemokine receptor ACKR2 expressed in trophoblast and dendritic cells and is associated with termination of inflammatory response by chemokine scavenging. Up-regulated autocrine signaling includes somatostatin (SST) and likely neurostatin signaling via down-regulated SSTR1 and up-regulated GPR107 receptor (Samson et al. 2008; Yosten et al. 2012, 2015). WNT4 expression is initiated, with its receptors expressed in trophoblast and decidua (below). Decidualization down-regulates uterine neuropilin (NRP1), reducing its potential inhibition of VEGFB in trophoblast.
Signaling changes in trophoblast differentiation
The comparison of receptor/ligand spectra expressed in differentiated extravillous trophoblasts and syncytiotrophoblasts to that of cytotrophoblasts also suggests the signaling differences that arise during trophoblast differentiation in the two lineages. The signaling interactions likely gained in EVT involve interleukin receptors IL1R1 and IL1R2 potentially interacting with maternal decidua and dendritic cells, EFNB1 and TNFSF10, as well as growth factor receptors of decidual growth factors. Similarly, the differentiation toward syncytiotrophoblast involves gain of chemokine and growth factor receptors (IL1R1, TGFBR1 and -2, PDGFRA, BMPR2) but in addition adds growth signaling toward decidua by of IGF1, HGF, and DLK1.
Maternal-fetal interactions in main signaling pathways
Here, we characterize the fetal and maternal contributions to the main signaling pathways and their change upon decidualization (see Supplemental Material for IGF, GH and prolactin pathways).
Wnt signaling
Decidualization entails differentiation toward epithelial-like character, reflected in up-regulation of several Wnt ligands, above all, the induction of WNT4 and up-regulation of WNT2 and WNT5A. Intravillous cytotrophoblasts show moderate expression of Wnt ligands (WNT3, WNT7A), and WNT6 is expressed in syncytiotrophoblast. Expression of Wnt receptors (Frizzled family) in fetal and maternal cells suggests autocrine as well as paracrine signaling. Specifically, FZD2, -4, and -6 are expressed in decidualized cells, and FZD3, -4, -5, and -6 are expressed in cyto- and syncytiotrophoblast. WNT4, a marker of decidualization, likely binds to the FZD6 receptor, possibly similar to signaling during mesenchymal-to-epithelial transition in kidney development (Lyons et al. 2004). Canonical and noncanonical pathways have been suggested for WNT4 signaling in endometrium (Hou et al. 2004; Knofler and Pollheimer 2013). The up-regulation of WNT2 and putative binding to the FZD4 receptors (Klein et al. 2008) in cytotrophoblast and decidua may augment canonical WNT4 signaling. The expression of downstream transcription factors TCF7L2 in trophoblast and decidua, as well as LEF1 in syncytiotrophoblast, further supports activation of canonical Wnt signaling in maternal and placental cells via autocrine and paracrine signals.
Prostaglandins
Short-distance signaling of prostaglandins makes their expression highly informative. The enzyme-expression of the prostaglandin biosynthesis pathway is summarized in Supplemental Figure S5. Briefly, the phospholipase A (PLA2G16) catalyzing the early step is broadly expressed, and cyclooxygenase 1 (PTGS1) is up-regulated in decidualization. The enzymes active in the later stages of the pathway seem complementarily present in maternal and placental term cells: The prostaglandin D synthetase PTGDS increased fourfold in decidualization; HPGDS is expressed in syncytiotrophoblast; prostaglandin E synthetase PTGES in EVT and cytotrophoblast, and moderately in decidua; and the oxidoreductase FAM213B catalyzing the biosynthesis of prostaglandin F2alpha (PGF2a) is expressed in decidua. Interestingly, while prostaglandin F receptor is weakly expressed in syncytiotrophoblast and decidua, prostaglandin F receptor antagonist, PTGFRN, is highly expressed in decidua. In addition, the main prostaglandin catabolic enzyme, 15-hydroxy dehydrogenase (HPGD), is highly expressed in all term trophoblast cell types (in EVT > 2000 TPM) and absent in maternal cells. Collectively, the data suggest prostaglandin E2 synthesis in decidua and trophoblast and prostaglandin F2alpha predominantly in decidua. Receptor expression is generally weak, and prostaglandin F receptor antagonist and the catabolic enzyme HPGD may further inhibit the prostaglandin signaling at the maternal-fetal interface. These patterns suggest a uterine autocrine signaling of PGF2a, whereas the trophoblast PGE2 signaling may be directed toward decidua. In vivo analysis will reveal whether low levels of decidual PGE-receptors are artifacts of decidualization in the absence of trophoblast.
Interleukin and TNF signaling
The interleukin expression among the collected cells is highest in dendritic cells and is comprised of IL1B, IL15, and IL32. IL1B expression is matched by high expression of its receptor IL1R1 in decidua, up-regulated >40-fold during decidualization. The adjacent fetal EVT and the syncytiotrophoblast express IL1R1, and EVT also highly expresses IL1R2, a decoy receptor for both IL1B and IL1R1. Interestingly, maternal dendritic cells also express anti-inflammatory IL1RN, the IL1-receptor antagonist. Thus, while maternal immune cells produce pro-inflammatory signals and decidua and trophoblasts are capable of perceiving them, maternal immune cells and placental cells concurrently express strong multifaceted anti-inflammatory signals, potentially inducing a sustained anti-inflammatory state (see details in Supplemental Material).
Tumor necrosis factor alpha and beta (TNF, LTA) are not expressed in our cells; however dendritic cells express genes of the tumor necrosis factor ligand family, TNFSF. In addition, EVT and syncytiotrophoblast highly express the apoptosis-inducing TNFSF10. This is a gene for the ligand of death receptor TNFRSF10B, a gene which is up-regulated in decidualization. Decidualization also increases expression of TNFRSF11B, coding for a decoy receptor for TNFSF11 and TNFSF10. These data suggest that active signaling from the trophoblast to decidua via TNFSF10 and TNFRSF10B is modulated by decidual TNFRSF11B expression. The downstream targets are described in the Supplemental Material.
Steroid hormones progesterone, estrogen, and glucocorticoids
Steroid hormone biosynthesis is well represented at the feto-maternal interface (Supplemental Fig. S6). The upstream enzymes catalyzing steroid and specifically progesterone synthesis (CYP11A1 and HSD3B1) are expressed in all trophoblast cells. On the maternal side, decidualization is accompanied by a strong up-regulation of the enzyme with a primary role in converting estrone to estradiol (HSD17B2) but also catabolizing progesterone. Decidua expresses a series of progesterone-catabolizing enzymes: AKR1C1, SRD5A1, and SRD5A3. The conversion of progesterone to estrogen via androgens is inhibited in human pregnancy by the lack of CYP17A1, the enzyme catalyzing the first step of the conversion. In contrast, aromatase (CYP19A1), the enzyme converting the second step, is highly expressed in term syncytio- and cytotrophoblast. Whereas progesterone- and estrogen nuclear receptors PGR and ESR1 are weakly expressed in decidual or placental cells, the less-understood membrane-associated receptors PGRMC1 and -2 are highly expressed in all trophoblasts and up-regulated in decidualization (PGRMC2: fourfold to reach >800 TPM). Collectively, data suggest high progesterone (P4) production in the fetal compartment, a moderate increase in P4-catabolism in decidualization, and high overall expression of membrane-bound P4 receptors.
The progesterone signal may also be mediated via the glucocorticoid receptor (GR; gene NR3C1) (Karalis et al. 1996), which is generally expressed most highly in dendritic cells and is down-regulated in uterine decidualization. Signaling via GR could explain P4's modulation of maternal immune cells (for review, see Butts et al. 2010). Dendritic cells are a candidate mediator of maternal tolerance for fetal antigens (Tagliani and Erlebacher 2011), with P4 proposed as a signal reducing DCs’ capacity to stimulate T-cells (Ivanova et al. 2005; Hughes et al. 2008; Butts et al. 2010). Previous work using PGR antagonist RU486 to reverse the effect of progesterone concluded that the effect is mediated by PGR (Butts et al. 2010). However, the fact that RU486 also antagonizes GR, the absence of PGR, and high expression of GR in uterine dendritic cells suggest that the observed effect on dendritic cells is mediated by P4 binding to GR. In accordance, P4 signaling via GR has been recently shown to modulate IL1B-induced COX2 expression in myometrium (Lei et al. 2012).
Empirical validation
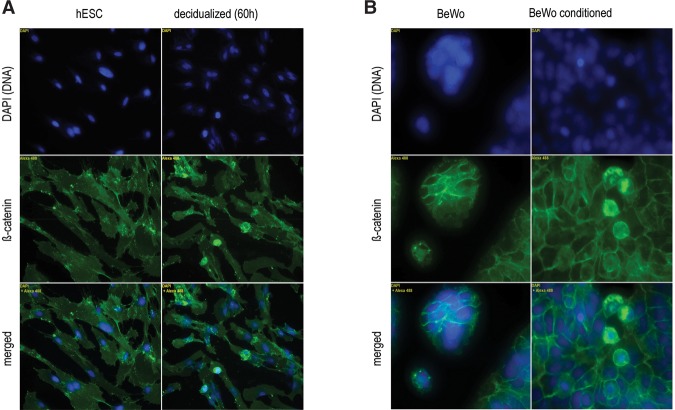
We chose Wnt-signaling to validate some of our results. Transcriptome analysis predicted induction by WNT4 and/or WNT2 of canonical Wnt pathway in maternal and fetal cells upon decidualization. The downstream effect of the canonical Wnt signal is nuclear translocation of beta-catenin. Using endometrial stromal cells (hESC) and cytotrophoblast cells (BeWo), we tested the predicted activation by conditioning the growth medium of the trophoblast cells with the supernatant of the maternal cells and determined beta-catenin localization using immunocytochemistry. As shown in Figure 5A, decidualization indeed activates the canonical pathway in endometrial stromal cells. Adding supernatant from decidualized cells to the growth medium of the trophoblast cells also activates the canonical Wnt pathway in trophoblast cells (Fig. 5B), whereas this does not occur upon adding supernatant from nondecidualized cells (or fresh decidualization medium) (Supplemental Fig. S9).
Figure 5.
Proof of concept: Canonical Wnt signaling is induced in decidualization in maternal cells, as well as in fetal cells conditioned with supernatant of maternal decidualized cells. Anti-beta-catenin antibody is shown in green, and DAPI nuclear staining in blue. The translocation of beta-catenin into the nucleus marks activation of the canonical Wnt signaling pathway. (A) Endometrial stromal fibroblasts (HESC) show beta catenin nuclear localization upon decidualization (Nikon, 20×). (B) BeWo show no nuclear localization in normal growth medium; however, the nuclear localization occurs upon addition of the supernatant from decidualized cells (Nikon, 40×).
Discussion
The data presented in this study reveal two main patterns in addition to the detailed characterization of the cells. The first is a model of the cell-cell communication network at the feto-maternal interface, with some interesting aspects that will be discussed below. The second interesting pattern is the cell-type specificity of GPCR profiles, suggesting a novel way of characterizing cell types.
When discussing our cell-cell network model, it is important to note that this model is inferred from RNA expression rather than the presence of corresponding proteins, which is dependent upon the degree of post-transcriptional regulation. Nevertheless, the expression of both interacting partners arguably offers strong suggestive evidence of actual cell-cell signaling. Maternal decidual cells and the syncytium are at the center of the cell-cell interaction network. Many (≥20) decidual signals are also directed toward and received by the extravillous trophoblast cells. Data show that the majority of receptors and particularly ligands up-regulated during decidualization have their complement ligand or receptor expressed in placental cells. Note that the decidual-trophoblast match in this study involves the portion of changes intrinsic to the decidualization program and does not depend on the presence of interaction partners, because the complementary gene expression changes were observed in vitro. Decidualization in vitro predominantly activates ligands of receptors in the adjacent trophoblast cells. This is consistent with the observations that the presence of factors from the maternal decidua affects the growth of trophoblast cells (Menkhorst et al. 2012). The potential to receive incoming trophoblast signals is similar between endometrial stromal fibroblasts and in vitro decidualized cells.
Interestingly, immune-regulatory as well as growth-regulatory interactions between maternal and placental cells show mutual concurrent expression of the enhancing and the inhibitory signals. We interpret this pattern as a signature of the fine-tuned regulation, which, as in any control circuit for maintaining homeostasis, must involve both inhibitory and enhancing effectors (Bernard 1856; Bernard et al. 1927). This principle is currently best appreciated in the immune system (Kotas and Medzhitov 2015) and is reflected in cytokine and prostaglandin pathways in these data. Yet, the principle likely also applies to regulation of another energetically costly process, namely growth during pregnancy. In the case of pregnancy, a dynamic balance must accommodate not just different cells or organs within an individual but also transient close interaction with another individual and its growing requirements. It will therefore be particularly useful to focus on the interaction dynamics occurring as new stable states are reached in pregnancy or even just anticipated in the nonpregnant cycle. The fact that the uterine cells decidualized in the absence of trophoblasts show many changes required for maternal-fetal interactions that likely also occur in the sterile menstrual cycle, strongly suggests that information collected in the nonpregnant cycle (i.e., in menstrual shedding) could be predictive of pregnancy outcomes.
An interesting result of potentially broad significance is the cell-type specificity of G-protein-coupled receptor profiles. GPCRs are used by a variety of ligands in numerous organismal contexts, from immune system to hormonal and neuronal signaling. GPCR signaling is evolutionarily old, and the diversification of receptors has been related to the increase of organismal complexity following the evolution of multicellularity and the associated diversification of cell types (Bradford et al. 2013; de Mendoza et al. 2014). Indeed, GPCRs are expressed in a cell-specific manner, with less heterogeneity of expression among cells of the same type. We interpret this as a signature of the biological role of a cell type, which is in part realized by the information transferred in cell-cell communication. If this observation can be confirmed in studies of other cell types, GPCR expression may constitute a mechanism for conveying cell-type identity within a focal species.
The novel insights from this work are enabled by the single-cell resolution of transcriptomes. However, the approach has limitations to consider. The cell collection was intentionally not guided by cell sorting using specific markers; nonetheless, the sample is biased due to morphological and chemical properties of the cells, such as cell size or cell adhesion. This is important for the comparison of single-cell data with tissue-level transcriptomes. Tissue transcriptomes are dominated by the transcription profiles of the common cells, whereas single-cell transcriptomes are biased by the physical properties of the cells. Provided that the most common cells are not also the most easily attainable in the single-cell pipeline, the two approaches provide complementary information.
We extended the analysis of sampled placental cells with in vitro decidualized primary endometrial stromal fibroblasts as a proxy for uterine lining. The transcriptome of these cells exhibits most of the known markers of decidualization. A potential limitation to these data is that in vitro cell cultures could lack some gene expression that arises due to the in vivo context or by interaction with adjacent cell types (e.g., Calabria and Shusta 2008; Gellersen and Brosens 2014; LoVerso et al. 2015). Future in vivo decidual cell transcriptomes will provide an answer to this question.
Finally, the study required combining multiple approaches to obtain transcriptomic data. We have accounted for this diversity to the extent possible in the computational part of the analysis. There are also methodological differences prior to and during the sequencing pipeline, such as tissue handling, amplification methods, and facility, which may, to some extent, influence the overall comparability of expression levels. In spite of these limitations, our results are consistent with previous experimental studies. We are thus encouraged to consider the novel insights of this study to be robust. It will be interesting to broaden this approach in the future with additional cell types, in particular myometrial cells, and earlier gestational stages, as well as to use these results as a standard against which to interpret disease-related deviations.
Methods
Generating transcriptomes
Single-cell transcriptomes
Fresh, term placental tissue from normal pregnancies was obtained following elective Cesarean section and minced finely. Tissue underwent serial trypsin and collagenase digests as previously described (Troja et al. 2014), and the resulting cell suspension was layered on a continuous Percoll gradient. Cells were collected from the 20%–60% fractions. Collected cells were filtered through a 40-µm cell filter and underwent Trypan blue exclusion to assess viability. Cell suspension at 3 × 105 cells/mL was submitted to Auto Prep System and handled according to the manufacturer's instructions. Libraries were subjected to paired-end 75-bp RNA sequencing on an Illumina HiSeq2500, resulting in a median of 3.65 million reads per cell. The reads were aligned to human assembly (hg19) (see Supplemental Fig. S7 for fragment lengths and mapped reads) using Bowtie, and assessed for abundance by eXpress (http://www.rnaexpress.org) (see also http://bio.math.berkeley.edu/eXpress/index.html; Roberts and Pachter 2013).
Placental tissue-level transcriptome
Two human placentas (separate from above) were collected at term C-section without labor (IRB: CCHMC IRB 2013–2243), flash-frozen, and kept at −80°C until RNA extraction using TRIzol. All samples passed quality tests using Bioanalyzer 2100 (Agilent), assuring RIN > 9. Libraries were prepared with TruSeq Stranded mRNA HT (Illumina) and subjected to paired-end 50-bp RNA sequencing on an Illumina HiSeq2500, at the depth of 30 million reads per sample.
Syncytiotrophoblast transcriptome
We used a Leica LMD 7000 Laser Microdissection system to collect syncytiotrophoblast from fresh-frozen sectioned tissue. Microdissected material was collected directly into the lysis buffer of the Qiagen RNasy Plus Micro kit, and RNA was extracted subsequently. Two samples were collected from the same placenta. Due to low concentration, the Ovation RNA Amplification System (Ribo-SPIA technology, NuGen) was used to amplify cDNA. Libraries were sequenced using Illumina HiSeq2500, to obtain a minimum of 30 million 75-bp paired-end reads per sample. Reads were aligned using Bowtie, counted and normalized using RNA-eXpress.
Primary human endometrial stromal fibroblast culture transcriptome
Two primary human endometrial stromal fibroblast lines (HsESC_217S and HsESC_218S) were obtained from two patients undergoing a polypectomy. The cells were grown in DMEM-based medium and decidualized in vitro. Total RNA was extracted from undifferentiated and differentiated cells using the RNeasy Mini kit (Qiagen), followed by on-column DNase I treatment. RNA quality was assayed with the Bioanalyzer 2100 (Agilent) and samples with RIN > 9.5 sequenced using the Illumina HiSeq2500, to a minimum of 36 million paired-end reads per sample. Reads were aligned with TopHat2 to the human Ensembl cDNA build, counted using HTSeq, and normalized as transcripts per million. Two technical repeats of each of the two primary cell cultures were processed independently and the expression values averaged over four transcriptomes.
For greater detail on tissue/cell preparation as well as sequencing, see Supplemental Methods.
Immunocytochemistry
We used human trophoblast cell line BeWo (ATCC CCL98) and endometrial stromal fibroblasts (T HESC, Mor lab, Yale University, corresponding to ATCC CRL-4003). Uterine cells were decidualized in vitro by addition of cAMP and progesterone. Trophoblast cells were conditioned with decidualized cell-supernatant for 30 min, upon which the cells were fixed. Uterine nondecidualized and decidualized cells were also fixed and treated the same way. Immunocytochemistry was performed on all cells using anti-beta catenin primary antibody (Abcam, ab16051) and secondary fluorescent antibody (Abcam, ab150077).
Analysis
Single-cell transcriptomes
The resulting transcriptomes were hierarchically clustered and subsequently analyzed for specific marker expression using AltAnalyze with default settings (http://www.altanalyze.org; Emig et al. 2010). Downstream analyses included the characterization of the cluster-specific gene expression with GO-terms (using GO Elite from Gladstone Institutes: http://www.genmapp.org/go_elite/) to determine cell-type identities of the clusters. The cells from both batches are represented in all clusters, enabling us to combine the two batches in common analyses.
Combining transcriptomes to explore cell communication
For the cell-interaction analyses, the expression levels were recalculated relative to the total reads mapping to the same set of coding genes in all transcriptomes, on a TPM scale (Wagner et al. 2012). The expression values were averaged within each single-cell cluster/cell sample (Supplemental Table S1). The ligand-receptor relationships are based on data from two databases (Supplemental Table S5; http://www.guidetopharmacology.org; http://dip.doe-mbi.ucla.edu/dip/DLRP.cgi). Ligands and receptors above the cut-off threshold of 10 TPM were used to construct lists of putative interactions discussed, but only those beyond 30 TPM are shown in Figure 2, to allow better visualization. We used the freely available R package Circlize (Gu et al. 2014; https://cran.r-project.org/web/packages/circlize/) to script interaction plots with arrows connecting the ligands and corresponding receptors.
Data access
Raw and processed data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE87692 (uterine cells) and GSE87726 (placental tissue-level transcriptome, single-cell transcriptomes, and syncytiotrophoblast transcriptome).
Supplementary Material
Acknowledgments
This work was supported by the March of Dimes Prematurity Research Center Ohio Collaborative (#22-FY14-470) to L.M. and the John Templeton Foundation (grant #54860) to G.P.W. H.J. is supported by the National Institutes of Health (#R00HD068504). We thank Phillip Dexheimer from Biomedical Informatics at CCHMC for assistance in transcriptome handling, Kris McGary from Vanderbilt University for handling the data deposition in GeneBank, and the Microscopy Core of the Yale Systems Biology Institute for access to the laser microdissection system. G.P.W. thanks Hugh Taylor, Yale School of Medicine, for providing primary isolates of human endometrial fibroblasts. M.P. thanks Gil Mor, Yale School of Medicine, for providing the immortalized cell line of human endometrial fibroblast. We thank Emily DeFranco, DO and Chris DeArmond, RN from the University Hospital of the University of Cincinnati for providing placenta tissues.
Author contributions: L.M. and H.J. initiated the project; H.J. and K.O. collected and prepared placental tissue for single-cell analysis; S.G.K. collected tissue for bulk analysis; C.D.-F. extracted RNA from tissue samples; G.P.W. and A.R.C. performed the uterine cell culture experiments; A.R.C. and M.P. performed laser microdissection of syncytiotrophoblast; J.M. collected the data from the endometrial stromal fibroblasts and decidual cells; M.P. and G.P.W. analyzed the data and drafted the manuscript; all authors contributed to writing the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.207597.116.
References
- Bernard C. 1856. Introduction à l’étude de la médecine expérimentale. J.B. Baillière, Paris. [Google Scholar]
- Bernard C, Greene HC, Henderson LJ. 1927. An introduction to the study of experimental medicine. Macmillan, New York. [Google Scholar]
- Biadasiewicz K, Fock V, Dekan S, Proestling K, Velicky P, Haider S, Knofler M, Frohlich C, Pollheimer J. 2014. Extravillous trophoblast-associated ADAM12 exerts pro-invasive properties, including induction of integrin β 1-mediated cellular spreading. Biol Reprod 90: 101. [DOI] [PubMed] [Google Scholar]
- Bischof P, Meisser A, Campana A. 2000. Paracrine and autocrine regulators of trophoblast invasion—a review. Placenta 21: S55–S60. [DOI] [PubMed] [Google Scholar]
- Borbely AU, Sandri S, Fernandes IR, Prado KM, Cardoso EC, Correa-Silva S, Albuquerque R, Knofler M, Beltrao-Braga P, Campa A, et al. 2014. The term basal plate of the human placenta as a source of functional extravillous trophoblast cells. Reprod Biol Endocrinol 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford W, Buckholz A, Morton J, Price C, Jones AM, Urano D. 2013. Eukaryotic G protein signaling evolved to require G protein-coupled receptors for activation. Sci Signal 6: ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar AK, Handwerger S, Kessler CA, Aronow BJ. 2001. Gene induction and categorical reprogramming during in vitro human endometrial fibroblast decidualization. Physiol Genomics 7: 135–148. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Park JS, Chung E, Chen F, Magella B, Potter SS. 2014. Single cell dissection of early kidney development: multilineage priming. Development 141: 3093–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. 2015. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33: 155–160. [DOI] [PubMed] [Google Scholar]
- Butts CL, Candando KM, Warfel J, Belyavskaya E, D'Agnillo F, Sternberg EM. 2010. Progesterone regulation of uterine dendritic cell function in rodents is dependent on the stage of estrous cycle. Mucosal Immunol 3: 496–505. [DOI] [PubMed] [Google Scholar]
- Calabria AR, Shusta EV. 2008. A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. J Cereb Blood Flow Metab 28: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, van Oudenaarden A. 2015. Spatially resolved transcriptomics and beyond. Nat Rev Genet 16: 57–66. [DOI] [PubMed] [Google Scholar]
- de Mendoza A, Sebé-Pedrós A, Ruiz-Trillo I. 2014. The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol Evol 6: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza AW, Wagner GP. 2014. Malignant cancer and invasive placentation: a case for positive pleiotropy between endometrial and malignancy phenotypes. Evol Med Public Health 2014: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. 2010. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res 38: W755–W762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A. 2013. Immunology of the maternal-fetal interface. Annu Rev Immunol 31: 387–411. [DOI] [PubMed] [Google Scholar]
- Fock V, Mairhofer M, Otti GR, Hiden U, Spittler A, Zeisler H, Fiala C, Knofler M, Pollheimer J. 2013. Macrophage-derived IL-33 is a critical factor for placental growth. J Immunol 191: 3734–3743. [DOI] [PubMed] [Google Scholar]
- Gellersen B, Brosens JJ. 2014. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev 35: 851–905. [DOI] [PubMed] [Google Scholar]
- Godbole G, Suman P, Gupta SK, Modi D. 2011. Decidualized endometrial stromal cell derived factors promote trophoblast invasion. Fertil Steril 95: 1278–1283. [DOI] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30: 2811–2812. [DOI] [PubMed] [Google Scholar]
- Handwerger S, Aronow B. 2003. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog Horm Res 58: 263–281. [DOI] [PubMed] [Google Scholar]
- Hannon T, Innes BA, Lash GE, Bulmer JN, Robson SC. 2012. Effects of local decidua on trophoblast invasion and spiral artery remodeling in focal placenta creta—an immunohistochemical study. Placenta 33: 998–1004. [DOI] [PubMed] [Google Scholar]
- Hou X, Tan Y, Li M, Dey SK, Das SK. 2004. Canonical Wnt signaling is critical to estrogen-mediated uterine growth. Mol Endocrinol 18: 3035–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GC, Thomas S, Li C, Kaja MK, Clark EA. 2008. Cutting edge: Progesterone regulates IFN-α production by plasmacytoid dendritic cells. J Immunol 180: 2029–2033. [DOI] [PubMed] [Google Scholar]
- Ivanova E, Kyurkchiev D, Altankova I, Dimitrov J, Binakova E, Kyurkchiev S. 2005. CD83+ monocyte-derived dendritic cells are present in human decidua and progesterone induces their differentiation in vitro. Am J Reprod Immunol 53: 199–205. [DOI] [PubMed] [Google Scholar]
- Ji L, Brkic J, Liu M, Fu G, Peng C, Wang YL. 2013. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol Aspects Med 34: 981–1023. [DOI] [PubMed] [Google Scholar]
- Jouve N, Despoix N, Espeli M, Gauthier L, Cypowyj S, Fallague K, Schiff C, Dignat-George F, Vely F, Leroyer AS. 2013. The involvement of CD146 and its novel ligand Galectin-1 in apoptotic regulation of endothelial cells. J Biol Chem 288: 2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer U, Schoppet M, McLellan AD, Kapp M, Huppertz HI, Kampgen E, Dietl J. 2000. Human decidua contains potent immunostimulatory CD83+ dendritic cells. Am J Pathol 157: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer U, Kruse A, Barrientos G, Arck PC, Blois SM. 2008. Role of dendritic cells in the regulation of maternal immune responses to the fetus during mammalian gestation. Immunol Invest 37: 499–533. [DOI] [PubMed] [Google Scholar]
- Karalis K, Goodwin G, Majzoub JA. 1996. Cortisol blockade of progesterone: a possible molecular mechanism involved in the initiation of human labor. Nat Med 2: 556–560. [DOI] [PubMed] [Google Scholar]
- Kaspi E, Guillet B, Piercecchi-Marti MD, Alfaidy N, Bretelle F, Bertaud-Foucault A, Stalin J, Rambeloson L, Lacroix O, Blot-Chabaud M, et al. 2013. Identification of soluble CD146 as a regulator of trophoblast migration: potential role in placental vascular development. Angiogenesis 16: 329–342. [DOI] [PubMed] [Google Scholar]
- Klein D, Demory A, Peyre F, Kroll J, Augustin HG, Helfrich W, Kzhyshkowska J, Schledzewski K, Arnold B, Goerdt S. 2008. Wnt2 acts as a cell type–specific, autocrine growth factor in rat hepatic sinusoidal endothelial cells cross-stimulating the VEGF pathway. Hepatology 47: 1018–1031. [DOI] [PubMed] [Google Scholar]
- Knofler M. 2010. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int J Dev Biol 54: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knofler M, Pollheimer J. 2012. IFPA Award in Placentology lecture: molecular regulation of human trophoblast invasion. Placenta 33: S55–S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knofler M, Pollheimer J. 2013. Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet 4: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotas ME, Medzhitov R. 2015. Homeostasis, inflammation, and disease susceptibility. Cell 160: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Chen L, Georgiou EX, Sooranna SR, Khanjani S, Brosens JJ, Bennett PR, Johnson MR. 2012. Progesterone acts via the nuclear glucocorticoid receptor to suppress IL-1β-induced COX-2 expression in human term myometrial cells. PLoS One 7: e50167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MP, Morlese JF, Sullivan MH, Elder MG. 1993. Evidence for decidua–trophoblast interactions in early human pregnancy. Hum Reprod 8: 965–968. [DOI] [PubMed] [Google Scholar]
- LoVerso PR, Wachter CM, Cui F. 2015. Cross-species transcriptomic comparison of in vitro and in vivo mammalian neural cells. Bioinform Biol Insights 9: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JP, Mueller UW, Ji H, Everett C, Fang X, Hsieh JC, Barth AM, McCrea PD. 2004. Wnt-4 activates the canonical β-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: functional implications of Wnt/β-catenin activity in kidney epithelial cells. Exp Cell Res 298: 369–387. [DOI] [PubMed] [Google Scholar]
- Maltepe E, Fisher SJ. 2015. Placenta: the forgotten organ. Annu Rev Cell Dev Biol 31: 523–552. [DOI] [PubMed] [Google Scholar]
- Menkhorst EM, Lane N, Winship AL, Li P, Yap J, Meehan K, Rainczuk A, Stephens A, Dimitriadis E. 2012. Decidual-secreted factors alter invasive trophoblast membrane and secreted proteins implying a role for decidual cell regulation of placentation. PLoS One 7: e31418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mess A. 2014. Placental evolution within the supraordinal clades of eutheria with the perspective of alternative animal models for human placentation. Adv Biol 2014: 1–21. [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. 2016. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537: 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10: 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson WK, Zhang JV, Avsian-Kretchmer O, Cui K, Yosten GL, Klein C, Lyu RM, Wang YX, Chen XQ, Yang J, et al. 2008. Neuronostatin encoded by the somatostatin gene regulates neuronal, cardiovascular, and metabolic functions. J Biol Chem 283: 31949–31959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau S, van Oudenaarden A. 2015. Studying lineage decision-making in vitro: emerging concepts and novel tools. Annu Rev Cell Dev Biol 31: 317–345. [DOI] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16: 133–145. [DOI] [PubMed] [Google Scholar]
- Tagliani E, Erlebacher A. 2011. Dendritic cell function at the maternal–fetal interface. Expert Rev Clin Immunol 7: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talayev VY, Matveichev AV, Lomunova MA, Talayeva MV, Tsaturov ME, Zaichenko IY, Babaykina ON. 2010. The effect of human placenta cytotrophoblast cells on the maturation and T cell stimulating ability of dendritic cells in vitro. Clin Exp Immunol 162: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telugu BP, Adachi K, Schlitt JM, Ezashi T, Schust DJ, Roberts RM, Schulz LC. 2013. Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta 34: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troja W, Kil K, Klanke C, Jones HN. 2014. Interaction between human placental microvascular endothelial cells and a model of human trophoblasts: effects on growth cycle and angiogenic profile. Physiol Rep 2: e00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Muglia L, Pavlicev M. 2014. Evolution of mammalian pregnancy and the origin of the decidual stromal cell. Int J Dev Biol 58: 117–126. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Chen C, Erez O, Grossman LI, Goodman M, Romero R. 2006. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci 103: 3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhang C, Tu T, Sun M, Liu D, Lu D, Feng J, Yang D, Liu F, Yan X. 2013. Wnt5a uses CD146 as a receptor to regulate cell motility and convergent extension. Nat Commun 4: 2803. [DOI] [PubMed] [Google Scholar]
- Yosten GL, Redlinger LJ, Samson WK. 2012. Evidence for an interaction of neuronostatin with the orphan G protein-coupled receptor, GPR107. Am J Physiol Regul Integr Comp Physiol 303: R941–R949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosten GL, Elrick MM, Salvatori A, Stein LM, Kolar GR, Ren J, Corbett JA, Samson WK. 2015. Understanding peptide biology: the discovery and characterization of the novel hormone, neuronostatin. Peptides 72: 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, Damsky CH. 1997. Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest 99: 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XM, Han T, Sargent IL, Wang YL, Yao YQ. 2009. Conditioned medium from human decidual stromal cells has a concentration-dependent effect on trophoblast cell invasion. Placenta 30: 74–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.