Abstract
The bacterial pathogen Xanthomonas campestris pv. vesicatoria (Xcv) uses a type III secretion system (TTSS) to translocate effector proteins into host plant cells. The TTSS is required for Xcv colonization, yet the identity of many proteins translocated through this apparatus is not known. We used a genetic screen to functionally identify Xcv TTSS effectors. A transposon 5 (Tn5)-based transposon construct including the coding sequence for the Xcv AvrBs2 effector devoid of its TTSS signal was randomly inserted into the Xcv genome. Insertion of the avrBs2 reporter gene into Xcv genes coding for proteins containing a functional TTSS signal peptide resulted in the creation of chimeric TTSS effector::AvrBs2 fusion proteins. Xcv strains containing these fusions translocated the AvrBs2 reporter in a TTSS-dependent manner into resistant BS2 pepper cells during infection, activating the avrBs2-dependent hypersensitive response (HR). We isolated seven chimeric fusion proteins and designated the identified TTSS effectors as Xanthomonas outer proteins (Xops). Translocation of each Xop was confirmed by using the calmodulin-dependent adenylate cydase reporter assay. Three xop genes are Xanthomonas spp.-specific, whereas homologs for the rest are found in other phytopathogenic bacteria. XopF1 and XopF2 define an effector gene family in Xcv. XopN contains a eukaryotic protein fold repeat and is required for full Xcv pathogenicity in pepper and tomato. The translocated effectors identified in this work expand our knowledge of the diversity of proteins that Xcv uses to manipulate its hosts.
Keywords: bacterial plant pathogenesis, virulence proteins
Many phytopathogenic bacteria use a conserved type III secretion system (TTSS) to infect plant cells (1). The structural components of the TTSS apparatus are encoded by the hypersensitive response and pathogenicity (hrp) gene cluster, a pathogenicity island required for these bacteria to colonize susceptible plants and to elicit the hypersensitive response (HR) in resistant plants (2). TTSSs facilitate the translocation of a group of proteins, called type III or TTSS effectors, into plant and animal cells (1). The precise function of phytopathogenic TTSS effectors is not known; however, their actions within the plant cell enable bacteria to grow outside plant cells (3).
We are interested in elucidating how Xanthomonas campestris pv. vesicatoria (Xcv) uses TTSS effectors to colonize its pepper and tomato hosts. Xcv is a Gram-negative bacterium that infects leaves and fruit, causing necrotic lesions and chlorosis, resulting in leaf abscission and fruit loss (4). Several Xcv effectors have been identified (5), and molecular analysis has revealed new insight into the role of these proteins in Xcv pathogenesis. AvrBs2, AvrBs3, HpaA, and XopA are required for full virulence of Xcv (6–9). XopD and AvrXv4 encode cysteine proteases that cleave the small ubiquitin-like modifier (SUMO) from plant SUMO-protein conjugates (10, 11), potentially disrupting eukaryotic processes regulated by SUMO modification. AvrBs3 is translocated into the plant nucleus and binds DNA, inducing transcription of several genes, including auxin-induced proteins, α-expansins, and pectate lyases (9).
These studies provide only a glimpse of the role of TTSS effectors in Xcv pathogenesis because this pathogen may contain many more effectors. Computer-based analyses of Pseudomonas syringae pv. tomato (Pst) and Ralstonia solanacearum have identified 40–50 effectors in each organism by using two criteria: (i) proximity to hrp-dependent promoter elements, suggesting co-regulation with the TTSS, and (ii) N-terminal export-associated signal peptides defined by known TTSS effectors (12–14). In Xcv, the expression of hrp genes and some TTSS effectors is under the control of the response regulator HrpG (15). Several Xcv effectors, including XopA, XopB, XopC, XopD, and XopJ were discovered by using cDNA-amplified fragment length polymorphism (AFLP) analysis of Xcv 85*, which contains a constitutively active HrpG (16). The existence of effectors regulated independently of HrpG indicates that alternative searches for Xcv effectors are necessary.
In this study, we performed a genetic screen to functionally identify Xcv TTSS effectors by exploiting the modular nature of these proteins. Generally, the N terminus of effectors is sufficient to target protein reporters into plant cells, whereas the C terminus is sufficient to activate HR in resistant plants (17, 18). Using this information, Guttman et al. (19) randomly integrated a gene reporter encoding the C terminus of the AvrRpt2 effector lacking its TTSS signal sequence throughout the Pseudomonas pv. maculicola (Psm) genome (19). By creating TTSS effector::AvrRpt2 chimeric fusion proteins that activate an AvrRpt2-dependent HR in plants, they functionally identified 13 new effectors and predicted another 38 effectors by means of bioinformatics (19).
Using a similar strategy, we show that the Xcv AvrBs2 effector can be used as a reporter protein to identify Xcv effectors. AvrBs2 was selected because its TTSS signal has been identified (20) and it elicits a strong HR in resistant BS2 pepper cells (6, 20). We have defined a minimal domain of AvrBs2 capable of triggering HR in resistant BS2 pepper. This domain, devoid of known TTSS signal sequences, was used as a reporter and randomly inserted into the Xcv genome by means of transposon 5 (Tn5). We have identified seven translocated effectors by looking for Xcv insertion strains that induce an avrBs2-dependent HR in BS2 pepper. The identified TTSS effectors are designated as Xanthomonas outer proteins (Xops). Here, we describe the initial characterization of these proteins and discuss their potential roles in pathogenesis.
Materials and Methods
Bacterial Strains, Growth, and Matings. Escherichia coli, Agrobacterium tumefaciens, and Xcv strains were grown as described (10). Bacterial strains used in this study are listed in Table 3, which is published as supporting information on the PNAS web site.
Gene Manipulation and Plasmid Construction. Gene constructs generated by PCR were verified by restriction analysis and sequencing. For expression in Xcv, genes were subcloned into the broad host range vector pVSP61 and expressed under the control of native promoters. For transient expression using A. tumefaciens, genes were subcloned into the binary vector pMDD1 and expressed by a CaMV 35S promoter. Relevant plasmids are listed in Table 3. Information regarding constructs is available on request.
Construction of AvrBs2 Reporters. avrBs262–574::HA was amplified from pMDD1(avrBs21–574::HA) (courtesy of D. Dahlbeck, University of California, Berkeley) and subcloned into pMDD1. To make avrXv41–100::avrBs262–574::HA, the avrXv41–100 BglII fragment from pMS107 (10) was ligated to pBS(BglII::avrBs262–574::HA), creating pMS107(avrXv41–100:: avrBs262–574::HA). This gene was moved into pVSP61 and pMDD1. BS2 peppers were inoculated with 6 × 108 colony-forming units (CFU)/ml A. tumefaciens carrying pMDD1 clones for transient expression or 2 × 108 CFU/ml Xcv carrying pVSP61 clones as described (10).
Construction of pTn5(avrBs262–574::HA) Transposon. A pTn5cat plasmid derivative was constructed by exchanging the cat gene with the avrBs262–574::HA reporter gene inserted in frame adjacent to the left inverted repeat (IR) of Tn5 (21). Digestion of pUIRM504 by EcoRI and ScaI removed the cat gene and the left IR. A SpeI linker was ligated into this vector to create a unique site at the 5′ end of the Tn5 transposon. PCR amplification and subcloning of the N terminus of avrBs2 added a SpeI site and a 27-bp Tn5 IR upstream of the avrBs262–574 coding sequence, forming pBS(SpeI::IR::avrBs262–574::HA). This chimeric gene was inserted into the SpeI site of the modified pTn5 vector to create pTn5(avrBs262–574::HA), which is referred to as pTn5(avrBs2) hereafter.
Construction of Xcv 85-10 ΔavrBs2 Strain. p815avrBs2::Gm, a cosmid encompassing a 20-kb genomic region surrounding avrBs2, contains a replacement of the NdeI/ClaI fragment of the avrBs2 coding sequence with a gentamycin resistance cassette (22). p815avrBs2::Gm was introduced into Xcv 85-10 by triparental mating to exchange the Gm cassette with the chromosomal avrBs2 gene by homologous recombination.
Xcv Effector Screen. pTn5(avrBs2) was introduced into Xcv 85-10 ΔavrBs2 by triparental mating. Mutagenized strains were replica plated onto NYGA (0.5% peptone/0.3% yeast extract/2% glycerol/1.5% agar) rifampicin/kanamycin/tetracycline to identify tetracycline-sensitive strains, which contain a true Tn5(avrBs2) insertion rather than genomic integration of the plasmid. These strains were grown overnight in NYGB (0.5% peptone/0.3% yeast extract/2% glycerol) rifampicin at 28°C. Pools of eight strains, containing ≈1 × 108 CFU per strain, were combined, centrifuged, and resuspended in 1 mM MgCl2 to 1 × 109 CFU/ml per pool. Pools were hand-inoculated into BS2 pepper leaves and kept under continuous light for 2–3 days. Each Xcv strain from a pool that showed an avrBs2-dependent HR was independently inoculated at 2 × 108 CFU/ml to identify the HR-positive insertion strain. Genes disrupted by the transposon were identified by plasmid rescue and sequencing. National Center for Biotechnology Information blast algorithms were used to identify homologs (23).
Southern Blot Analysis. We transferred 5 μg of genomic DNA digested with BamHI or EcoRV to nylon membranes, and they were visualized by using the CDP-Star chemiluminescent detection system (Amersham Pharmacia). A 1,000-bp fragment of avrBs2 was used to confirm single Tn5(avrBs2) insertions. Whole or partial xop ORFs were hybridized to DNA isolated from Xanthomonas spp. to probe for xop homologs. avrBs2 and xop hybridizations were done at 65°C and 55°C, respectively.
Xop::Calmodulin-Dependent Adenylate Cyclase (Cya) Assays. The promoter and coding regions of each xop gene 5′ to the Tn5 insertion site were amplified by PCR and subcloned into the BglII site in pMS107 (24) to create translational Cya fusions. Each xop::cya construct was subcloned into pVSP61. E. coli strains carrying the pVSP61(xop::cya) plasmids were triparentally mated into Xcv 85-10 hrpG* (25) and Xcv 85-10 hrpG* ΔhrpF (26). The expression and activity of Xop::Cya proteins in bacteria and plants were assessed by immunoblot analysis and cAMP ELISA as described (10).
Signal Sequence Analysis. The first 55 aa of each protein and the remaining C-terminal residues were compared for amino acid composition. The statistical significance of amino acid biases between these groups was measured by using the Kolmogorov–Smirnov test. Analyzed effectors included Xops, identified here, and the following, with GenBank accession numbers given in parentheses: Xcv 81-23 effector AvrBs1 (P19520); Xcv 75-3 effectors AvrBs2 (AAD11434), AvrBsT (AAD39255), and Avr-Rxv (Q08678); Xcv 71-21 effector AvrBs3 (P14727); Xcv 85-10 effectors XopA (AAL78294), XopB (AAK72487), XopC (AAR23832), XopD (AAL78292), XopJ (AAK72486), HpaA (AAD21323), and XopX (AAT39020); and Xcv 91-118 effectors AvrXv3 (AAG18480) and AvrXv4 (AAG39033).
In Planta Growth of Xcv Δxop Strains. To create xop knockouts, 300-bp fragments internal to each xop gene were subcloned into the suicide vector pLVC18-RfC (courtesy of C. Morales, University of California, Berkeley). pLVC18(xop) plasmids were moved into Xcv 85-10 by triparental mating, and selected by growth on NYGA rifampicin/tetracycline. Sequencing confirmed homologous recombination within the gene of interest. Then, 1 × 105 CFU/ml suspensions of Xcv 85-10 and Xcv Δxop strains were hand-inoculated into multiple bs2 pepper or VF36 tomato leaves. Plants were grown under 10 h of light per day at 22°C. Leaf disks (total area, 1 cm2) were ground in 1 ml of MgCl2 and dilution-plated onto selective NYGA in triplicate to determine bacterial load.
Results
AvrBs2 as a Translocation Reporter. To use AvrBs2 as a TTSS reporter, we defined the minimal AvrBs2 peptide capable of activating BS2-dependent resistance in pepper. AvrBs2 is composed of 714 aa, of which amino acids 1–58 contain the TTSS secretion and translocation signals and amino acids 62–714 activate BS2 disease resistance (17). C-terminal deletion analysis of AvrBs2 shows that amino acids 575–714 are not required for BS2 recognition (D. Dahlbeck and B. Staskawicz, unpublished results). Therefore, the minimal region required for AvrBs2-specific HR activity in planta was predicted to be amino acids 62–574. Transient expression of AvrBs262–574::HA in BS2 pepper leaves induced a strong HR response (Fig. 1), similar to that observed with the mature polypeptide (data not shown). This finding shows that AvrBs262–574::HA is a functional protein reporter for avrBs2-dependent HR activity in plant cells.
Fig. 1.
avrBs2-dependent HR activity in BS2-resistant pepper. A. tumefaciens (At) strains carrying the empty vector pMDD1, pMDD1(avrBs262–574::HA), or pMDD1(avrXv41–100::avrBs262–574::HA) were inoculated into resistant BS2 pepper leaves at 6 × 108 CFU/ml. Xcv ΔavrBs2 strains carrying the empty vector pDD62, pDD62(avrBs262–574::HA), or pDD62(avrXv41–100::avrBs262–574::HA) were inoculated into BS2 leaves at 2 × 108 CFU/ml. Symptoms were recorded 24–48 h postinoculation.
We tested TTSS delivery of AvrBs262–574::HA to ascertain whether the protein can serve as a reporter to identify effectors. A useful reporter must accumulate in Xcv but cannot traverse the TTSS. To create an Xcv strain suitable for screening, we replaced the chromosomal copy of the avrBs2 gene in Xcv 85-10 with a gentamycin resistance cassette. Xcv 85-10 ΔavrBs2 did not trigger an avrBs2-dependent HR in BS2 pepper (Fig. 1) or produce AvrBs2 protein (data not shown). Moreover, Xcv 85-10 ΔavrBs2 exhibited reduced growth on susceptible plants as compared with Xcv 85-10, consistent with published results (data not shown and ref. 6). Xcv 85-10 ΔavrBs2 pVSP61(avrBs262–574::HA) expressed AvrBs262–574:: HA protein (data not shown) but failed to produce an avrBs2-dependent HR in pepper (Fig. 1). However, Xcv 85-10 ΔavrBs2 expressing the fusion protein AvrXv41–100::AvrBs262–574::HA triggered an avrBs2-dependent HR (Fig. 1). This result demonstrated that the AvrBs2 reporter fused to a functional TTSS signal peptide could be targeted through the Xcv TTSS. Furthermore, transient expression of AvrXv41–100::AvrBs262–574::HA in pepper elicited the same phenotype as AvrBs262–574::HA, indicating that modification of the reporter does not affect its recognition by BS2 (Fig. 1). These studies show that AvrBs262–574::HA can be used as a sensitive TTSS reporter in the Xcv ΔavrBs2 strain.
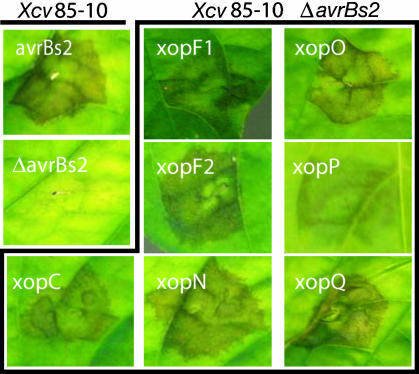
A Genetic Screen to Identify TTSS Effectors. The AvrBs262–574::HA reporter was randomly integrated into the Xcv ΔavrBs2 genome to create protein fusions, an approach previously used by Guttman et al. (19). To do this, we modified a Tn5-based transposon, pTn5cat, that randomly integrates into Xanthomonas spp. (21). In pTn5(avrBs2), the avrBs262–574::HA gene fragment is downstream of the left IR of Tn5. Approximately 14,000 Xcv Tn5 insertion strains were assayed for AvrBs2 activity on pepper. We identified seven strains that triggered HR when inoculated into BS2 pepper leaves (Fig. 2). The timing and strength of the HR elicited by each strain varied, suggesting that the stability and/or quantity of protein translocated to the host was effector-dependent. We confirmed that the HR was avrBs2-specific by inoculating both resistant BS2 and susceptible bs2 pepper leaves. All strains elicited a BS2-dependent HR (data not shown). Southern blot analysis verified that only one Tn5(avrBs2) insertion was responsible for each phenotype (data not shown).
Fig. 2.
Xcv Xop::AvrBs2 proteins induce avrBs2-dependent HR during infection. BS2-resistant pepper leaves were independently inoculated with a 2 × 108 CFU/ml suspension of Xcv 85-10, Xcv 85-10 ΔavrBs2, or seven Xcv 85-10 ΔavrBs2 strains each containing an xop::avrBs262–574::HA gene fusion. Symptoms were recorded 75 h postinoculation.
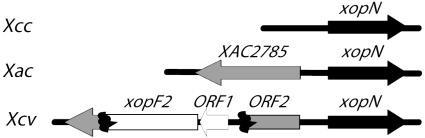
Identity of Xop Effectors. To identify the site of transposon integration, we determined the DNA sequence adjacent to Tn5 for each HR-inducing Xcv strain. Each strain contained a single insertion in a unique gene, generally near the 5′ end (Table 1). We have designated these genes as xops in accordance with current effector nomenclature. blast analysis showed that the seven Xop proteins are found only in phytopathogenic bacteria (Table 1). Three Xops are similar to known or predicted TTSS effectors. XopC is an effector previously discovered in Xcv strain 85-10 (27). XopQ shares homology with the R. solanacearum effector RipB (14) and the Pst DC3000 effector HolPtoQ (19). XopO is homologous to the Pseudomonas pv. pisi effector AvrRps4 (28). XopN and XopP have homology to hypothetical proteins from Xanthomonas spp. (29). Fragments of the xopF1 gene are located within the hrp cluster of many Xanthomonas spp., although a complete ORF is present only in the Xcv and Xanthomonas oryzae pv. oryzae (Xoo) hrp clusters. XopF2 is 59% identical and 68% similar to XopF1 when analyzed with the pairwise blast algorithm (23) although it is located adjacent to xopN (Fig. 3). Comparison of this genomic region with syntenic regions in Xanthomonas axonopodis pv. citri (Xac) and Xanthomonas campestris pv. campestris (Xcc) showed that xopF2 was inserted into ORF2, which is homologous to XAC2785 (29). Xcc does not contain orthologs of xopF2 or xAC2785. xopF2 seems to be cotranscribed with ORF1, a small gene of 148 aa with a predicted pI of 4.93 (Fig. 3). Structural analysis of ORF1 using the protean program (DNASTAR, Madison, WI) indicated that the protein may contain amphipathic α-helices. These characteristics are shared by type III chaperones (30), suggesting that ORF1 may encode an Xcv chaperone.
Table 1. Identity of Xop effectors.
| Effector | Homology (GenBank accession no.) | blastp,* bits/e-value | Predicted size, aa | Insertion site, aa | Homologs in sequenced species |
|---|---|---|---|---|---|
| XopC | Type III effector XopC (AAR23832) | 575/e-163 | 834 | 708 | Rs |
| XopF1 | Xoo conserved hypothetical protein ORF1 (BAD30000) | 879/0.0 | 670 | 151 | Xoo, Xcc† |
| XopF2 | Xoo conserved hypothetical protein ORF1 (BAD30000) | 517/e-145 | 667 | 108 | Xoo, Xcc† |
| XopN | Xac conserved hypothetical protein XAC2786 (NP _ 643095) | 1059/0.0 | 733 | 197 | Xac, Xcc |
| XopO | Psp avirulence protein AvrRps4 (AAB51082) | 119/4e-26 | 220 | 93 | Psp |
| Pst DC3000 | |||||
| XopP | Xac conserved hypothetical protein XAC1208 (NP_641544) | 865/0.0 | 658 | 321 | Xac, Xcc, Rs |
| XopQ | Xac conserved hypothetical protein XAC4333 (NP_644627) | 704/0.0 | 464 | 81 | Xac, Pst DC3000, Xcc, Rs |
blastp queries were done by using databases at the National Center for Biotechnology (23).
blastn of nucleotide sequence also yields homology with Xac, Xag, and Xcc.
Fig. 3.
Organization of the xopN/xopF2 genomic region. ORF1 and xopF2 are depicted as white arrows. XopN homologs are shown as black arrows. XAC2785 and its disrupted Xcv homolog ORF2 are depicted as gray arrows. Arrowheads indicate the predicted direction of transcription.
To gain insight into effector function, we examined the predicted secondary structure of each Xop using a three-dimensional position-specific scoring matrix (3d-pssm, www.sbg.bio.ic.ac.uk/∼3dpssm) and identified two Xop proteins with structural homologs (31). The most similar fold to XopN is found in the constant regulatory domain of protein phosphatase 2a (pr65/A) with an e-value of 0.0701. This protein contains HEAT (huntingtin, elongation factor 3, PR65/A, TOR1) repeats, which are tandem anti-parallel α-helices that stack on each other to form a solenoid and are structurally superimposable with the Armadillo (ARM) repeat family (32). 3d-pssm analysis of XopQ shows a significant match with an e-value of 1.66e–08 to the crystal structure of the inosine-uridine nucleoside N-ribohydrolase, IU-NH, from the protazoan parasite Crithidia fasciculata (33).
Xop::Cya Assays Confirm TTSS-Dependent Translocation. Although we removed known TTSS signals from the AvrBs262–574 reporter, this peptide may contain additional translocation signals. Therefore, we used the Cya domain of Bordetella pertussis cyclolysin as a biochemical reporter (24, 26) to confirm that the identified Xops are sufficient to target non-TTSS proteins to the plant cell. Xop::Cya proteins were made by fusing each xop promoter region and coding sequence upstream of the transposon insertion to the cya reporter. The xopF2::cya gene construct also contained the proposed chaperone, ORF1. Constructs were expressed in the isogenic strains Xcv hrpG*, which contains a point mutation in the HrpG response regulator leading to constitutive expression of the TTSS (15), and Xcv hrpG* ΔhrpF, which lacks the putative TTSS translocon HrpF and does not translocate proteins into plant cells (34).
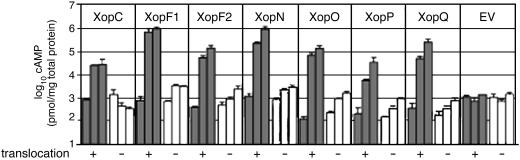
All fusion proteins were equally expressed in the translocation-competent (Xcv hrpG*) and translocation-deficient (Xcv hrpG* ΔhrpF) strains (Fig. 6, which is published as supporting information on the PNAS web site). Surprisingly, xopF1::cya and xopP::cya encoded proteins with larger molecular weights than expected given the predicted Met start codons. We identified GTG start codons with Shine–Delgarno sites (35) upstream of the predicted start that would yield the appropriate size fusion proteins. Protein extracts were then isolated from Xcv hrpG* and Xcv hrpG* ΔhrpF expressing each Xop::Cya fusion to test for endogenous Cya enzyme activity. Each Xop::Cya protein had little endogenous Cya activity, but addition of calmodulin to protein extracts from both strains restored enzyme activity (data not shown). To assess Xop-dependent TTSS translocation of the Cya reporter into plant cells, we inoculated all strains into susceptible pepper leaves and measured cAMP levels in tissue samples at 0, 8, and 27 h after inoculation. Leaves infected with Xcv hrpG* strains expressing each Xop::Cya protein accumulated 10- to 1,000-fold higher levels of cAMP compared with mock infections, in an HrpF- and time-dependent manner (Fig. 4). This finding confirms that all Xop proteins are targeted to the plant by means of the Xcv TTSS in the absence of the AvrBs2 reporter.
Fig. 4.
TTSS-dependent translocation of Xop::Cya proteins into plant cells. Susceptible bs2 pepper leaves were inoculated with a 5 × 108 CFU/ml suspension of Xcv expressing each Xop::Cya protein. Xcv 85-10 hrpG* strains are translocation competent (+, gray bars) and Xcv 85-10 hrpG* ΔhrpF strains are translocation deficient (–, white bars). cAMP levels were measured as the log10 (pmol cAMP per mg total protein). Tissue samples were taken at 0, 8, or 27 h postinoculation. Time points are shown by sequential bars.
Analysis of Xcv TTSS Signals. We compiled the sequence of our six new Xop proteins with 14 known Xcv effectors (see Materials and Methods) to determine whether the N termini of these proteins exhibit an amino acid bias similar to that reported for effectors from other phytopathogens. In Pst DC3000 and R. solanacearum, the N-terminal 50 aa of effectors have significantly higher levels of serine and proline and decreased levels of leucine as compared with proteins not associated with the TTSS (12, 14, 19). Because the genome sequence of Xcv is not available, we used the C termini of our effectors as a control for amino acid composition. We found that the levels of Ser (14.8%), Pro (11.7%), and Leu (6.9%) in the first 55 aa of 20 Xcv TTSS effectors are statistically different from levels of Ser (8.3%), Pro (5.4%), and Leu (9.9%) in the C-terminal portions of these proteins. Other rules defined for Pst signal sequences are less conserved in Xcv and R. solanacearum (12, 14). Hydrophobic residues at position 5 are rare in Pst, whereas 9 of 20 of the Xcv effectors have a Met, Ile, Leu, Val, Phe, Tyr, or Trp at this position. Similar to R. solanacearum, 8 of 20 of the Xcv effectors have an Arg or Lys at position 2, and a majority (18 of 20) contain at least one basic residue in the first 7 aa. In contrast to both Pst and R. solanacearum effectors, where acidic residues are not found in the first 12 aa, half of the Xcv effectors incorporate at least one Asp or Glu in this region.
Contribution of Xops to Xcv Pathogenicity. We disrupted each gene in Xcv 85-10 by homologous recombination and tested mutant strains for a reduction in pathogenicity in susceptible plants. None of the Xcv Δxop strains exhibited growth defects when grown in a rich NYGB medium or minimal media that mimics the conditions in the apoplast (data not shown). Each Xcv Δxop strain was inoculated into susceptible pepper and tomato leaves by hand infiltration, and in planta bacterial growth was measured. By day 8, Xcv ΔxopN bacteria grew to only 1/10 to 1/100 the level of Xcv 85-10 in pepper and tomato (Fig. 5). Thus, XopN is required for maximal Xcv growth in both pepper and tomato plants. No other Xcv Δxop mutant exhibited significant growth defects in either host (data not shown).
Fig. 5.
Growth of Xcv ΔxopN in susceptible hosts. Susceptible bs2 pepper plants (A) and VF36 tomato plants (B) were hand-inoculated with a 1 × 105 CFU/ml suspension of Xcv 85-10 or the mutant strain Xcv 85-10 ΔxopN. Data points represent the mean log10 cells per cm2 of leaf tissue ± SD of a representative growth assay. This analysis was performed three times per plant host.
Distribution of Xops among Xanthomonas spp. We determined the presence of each xop gene in a diverse group of Xanthomonas spp. using Southern hybridization (Table 2). We hybridized xop-specific probes to Xanthomonas strains with different characteristics. Xanthomonas spp. have been grouped according to the following traits: phenotype on different plant cultivars (36), amylolitic activity designated as serovars A–D (37), and 16S rRNA nucleotide homology (38). Xcv 75-3 and Xcv 81-18 are closely related to Xcv 85-10, because all three strains are pepper race 2, tomato race 1, and serovar A. Xcv 81-23, Xcv 81-18, Xcv 597, and Xcv 82-8 are also serovar A strains, but they are different pepper or tomato races. Xcv 71-4 and Xcv 81-6 are both in serovar B, Xv 938 in serovar C, and Xv 444 in serovar D (38). X. axonopodis pv. glycines (Xag), Xac, Xanthomonas campestris pv. armoraciae (Xca), Xcc, and X. oryzae pv. oryzae (Xoo) are pathogenic on different host plants.
Table 2. Distribution of xop genes in phytopathogenic bacteria.
| Serovar A
|
B
|
C
|
D
|
Xanthomonas spp.
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 85-10 | 75-3 | 81-23 | 81-18 | 597 | 82-8 | 71-4 | 81-6 | Xv938 | Xv444 | Xcg | Xac | Xca | Xcc | Xoo | |
| XopC | X | X | X | X | X | X | X | ||||||||
| XopF1 | X | X | X | X | X | X | X | W | X | X | X | X | X | W | X |
| XopF2 | X | X | X | X | X | X | X | W | X | ||||||
| XopN | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| XopO | X | X | X | X | X | X | |||||||||
| XopP | X | X | X | X | X | X | X | X | X | X | X | X | |||
| XopQ | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X |
Genomic DNA was probed with each xop gene by Southern hybridization. X, strong hybridization; W, weak hybridization; blank, no hybridization. All numbered strains are different serovars of Xcv. Other strain abbreviations are as follows: Xv, X. vesicatoria; Xcg, X. campestris pv. glycines; Xac, X. axonopodis pv. citri; Xca, X. c. pv. armoraciae; Xcc, X. c. pv. campestris; Xoo, X. oryzae pv. oryzae.
All seven xop genes tested hybridized to DNA from each Xcv serovar A strain (Table 2). Consistent with blast results (Table 1), xopF1, xopN, xopP, and xopQ are conserved across most Xanthomonas spp., whereas xopC, xopF2, and xopO are present only in closely related Xcv strains. Protein homologs of XopO and XopQ are present in Pseudomonas spp.; however, we did not observe hybridization of xopO and xopQ probes to Pseudomonas spp. DNA (data not shown). This result was not unexpected, because nucleotide homology for the respective genes is low (50% identity for Xcv xopQ and Pst holPtoQ) or undetectable (Xcv xopO and Psp avrRps4).
Discussion
In this work, we used AvrBs2 as a sensitive reporter (19) to isolate new Xcv effectors. The AvrBs2/Bs2 reporter system is a tool that can be used to identify translocated effectors in bacterial pathogens that infect other naturally occurring or transgenic BS2 plant lines. We identified four TTSS effectors of unknown function (XopN, XopF1, XopP, and XopF2), one previously identified Xcv effector (XopC), one effector with homology to a known P. syringae effector (XopO = AvrRps4), and one that is widely conserved among phytopathogens (XopQ = RipB, HolPtoQ). The fact that we did not detect all known Xcv 85-10 effectors is not surprising. We screened 14,000 insertion strains, but only one of six permutations of transposon insertion in an effector gene will be detected because recognition of AvrBs2 relies on the creation of a translational fusion. Many more insertion strains must be tested for this screen to reach saturation.
Understanding acquisition and evolution of type III effectors in Xcv may help us deduce the roles of these proteins in pathogenicity. As a first step, we examined Xop acquisition by using the three criteria used for analysis of P. syringae effectors: GC content, strain distribution, and genomic context (39). xopN, xopQ, xopP, and the C terminus of xopF1 are conserved across Xanthomonas spp. (Table 2), and their GC content is comparable with that of the Xcv hrp cluster (8). These xop genes may be members of a “core” group of Xanthomonas spp. effectors. xopC and xopO did not hybridize to many distant Xanthomonas spp., but have homologs in two tomato phytopathogens, Pst and R. solanacearum (Tables 1 and 2), suggesting that these genes may have been introduced into Xcv by horizontal gene transfer. In fact, the xopC gene is flanked by 62-bp IRs and multiple ORFs predicted to encode transposases and integrases (8). A peptide fragment with homology to the Xcc IS1479 transposase (29) was identified upstream of xopO. Both xopC and xopO have GC contents 10–15% lower than the Xcv hrp cluster (8). These genes may be recent additions to the Xanthomonas spp. effector cadre considering their uneven species distribution, low GC content, and association with mobile elements. As Xop homologs are identified in other bacterial species, we can more fully explore the evolution of these effector genes among diverse pathogens.
Recent articles report a bias in the amino acid composition of TTSS signal sequences from P. syringae spp. and R. solanacearum as compared with N termini of nonsecreted proteins (12, 14, 19). We found that Xcv TTSS signal sequences share attributes with both R. solanacearum and Pst, but Xcv elements defining these signals are more similar to R. solanacearum. This finding is consistent with the phylogenetic distribution of the TTSS machinery: Xanthomonas and Ralstonia TTSS genes are similar, whereas Pseudomonas sequences are more divergent (40). Thus, it is likely that subtle differences in the TTSS signal sequences of each organism may provide specificity to the structures they traverse. However, the fact that Xcv can secrete R. solanacearum PopA, Pseudomonas pv. glycinea AvrB, and Yersinia pseudotuberculosis YopE effectors (25) suggests that there may be redundancy or promiscuity for effector targeting. The field has yet to discover the precise combination of factors that defines a type III signal peptide.
Most of the Xops that we isolated were not required for Xcv growth on pepper and tomato hosts. This finding is not surprising considering that the loss of a single TTSS effector does not typically affect pathogenicity. This result may reflect effector functional redundancy or simply the lack of sensitive assays available to assess effector roles in bacterial–plant interactions. However, the growth of Xcv xopN mutants was significantly impaired in both pepper and tomato (Fig. 5). xopN is present in many Xanthomonas spp. but not found in the published genomes of other bacterial pathogens containing TTSS (Tables 1 and 2). This finding suggests that XopN may be an important Xanthomonas-specific effector. XopN shares structural homology with a superfamily of proteins containing tandemly repeated α-helices, known as ARM/HEAT repeats, that are found in almost all eukaryotes but are rare in bacteria (32). XopN is the first pathogen molecule thought to mimic this eukaryotic fold. The Arabidopsis genome contains many proteins with HEAT repeats including protein phosphatase 2A PR65/A regulatory subunits (41); several homologs of the nuclear transport protein importin-β (42); and AtCAND1, which is involved in the regulation of auxin signaling (43). We speculate that XopN may use HEAT repeat structures to modulate plant signal transduction by mimicking or interfering with proteins containing these repeats. The identification of XopN plant interactors will provide clues to its role in Xcv pathogenicity.
XopQ is conserved among many plant pathogens (Tables 1 and 2). These proteins have structural homology to an inosine-uridine nucleoside N-ribohydrolase enzyme, a protein implicated in the ability of many organisms to salvage nucleotides from their environment (44). Like other homologs, XopQ contains conserved aspartate residues found in the active site of the enzyme (44). Although nucleoside hydrolase enzymes are present in Xac and Xcc and likely present in Xcv as well, these genes do not share sequence homology with XopQ (29). It is possible that XopQ functions as a scavenging hydrolase in planta, or may bind and sequester nucleosides important for plant signaling and/or metabolism.
In summary, we have identified seven TTSS effectors used by Xcv during pathogenesis of pepper and tomato. Predictive homology of two effectors points the way toward elucidating their role in disease. Additional Xops may continue to be identified, defining a repertoire of effectors used by Xcv during infection. Probing the biochemical function of the Xops would give us clues as to plant processes modified by these effectors. Moreover, it would allow us to better understand the diversity of mechanisms used by this phytopathogen to modify its host during pathogenesis.
Supplementary Material
Acknowledgments
We thank G. Minsavage and J. Jones (University of Florida, Ganesville), D. Dahlbeck, C. Morales and B. Staskawicz (University of California, Berkeley), M. Whalen (San Francisco State University, San Francisco), and A. Alvarez-Morales (University of Irapuato, Irapuato, Mexico) for strains and plasmids, N. Guiso (Institut Pasteur, Paris) for antiserum, S. Shaw for statistical assistance, and M. Nishimura, C. Starker, M. Marks, and our laboratory for critical reading of the manuscript. M.B.M. is supported by Department of Energy Grant DE-FG02-03ER15443.
Author contributions: J.A.R. and M.B.M. designed research; J.A.R., B.B., J.B.R., T.T., and J.V. performed research; J.A.R. and M.B.M. analyzed data; and J.A.R. and M.B.M. wrote the paper.
Abbreviations: Xcv, Xanthomonas campestris pv. vesicatoria; TTSS, type III secretion system; HR, hypersensitive response; Xop, Xanthomonas outer protein; hrp, HR and pathogenicity; Pst, Pseudomonas syringae pv. tomato; Xac, Xanthomonas axonopodis pv. citri; Xcc, Xanthomonas campestris pv. campestris; Xoo, Xanthomonas oryzae pv. oryzae; Cya, calmodulin-dependent adenylate cyclase; CFU, colony-forming unit; IR, inverted repeat.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY756267 (XopN/XopF2), AY756268 (XopQ), AY756269 (XopO), AY756270 (XopP), and BK005592 (XopF1)].
References
- 1.Buttner, D. & Bonas, U. (2002) EMBO J. 21, 5313–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonas, U., Schulte, R., Fenselau, S., Minsavage, G., Staskawicz, B. & Stall, R. (1991) Mol. Plant–Microbe Interact. 4, 81–88. [Google Scholar]
- 3.Alfano, J. R. & Collmer, A. (2004) Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- 4.Stall, R. (1995) in Pathogenesis and Host Specificity in Plant Diseases: Histopathological, Biochemical, Genetic, and Molecular Bases, eds. Singh, U. S., Singh, R. P. & Kohmoto, K., (Pergamon, Oxford), Vol. 1, pp. 167–184. [Google Scholar]
- 5.Buttner, D. & Bonas, U. (2003) Curr. Opin. Plant Biol. 6, 312–319. [DOI] [PubMed] [Google Scholar]
- 6.Kearney, B. & Staskawicz, B. J. (1990) Nature 346, 385–386. [DOI] [PubMed] [Google Scholar]
- 7.Huguet, E., Hahn, K., Wengelnik, K. & Bonas, U. (1998) Mol. Microbiol. 29, 1379–1390. [DOI] [PubMed] [Google Scholar]
- 8.Noel, L., Thieme, F., Nennstiel, D. & Bonas, U. (2002) J. Bacteriol. 184, 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marois, E., Van den Ackerveken, G. & Bonas, U. (2002) Mol. Plant–Microbe Interact. 15, 637–646. [DOI] [PubMed] [Google Scholar]
- 10.Roden, J., Eardley, L., Hotson, A., Cao, Y. & Mudgett, M. B. (2004) Mol. Plant–Microbe Interact. 17, 633–643. [DOI] [PubMed] [Google Scholar]
- 11.Hotson, A., Chosed, R., Shu, H., Orth, K. & Mudgett, M. B. (2003) Mol. Microbiol. 50, 377–389. [DOI] [PubMed] [Google Scholar]
- 12.Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., Schechter, L. M., Janes, M. D., Buell, C. R., Tang, X., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwiesler-Vollick, J., Plovanich-Jones, A. E., Nomura, K., Bandyopadhyay, S., Joardar, V., Kunkel, B. N. & He, S. Y. (2002) Mol. Microbiol. 45, 1207–1218. [DOI] [PubMed] [Google Scholar]
- 14.Cunnac, S., Occhialini, A., Barberis, P., Boucher, C. & Genin, S. (2004) Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- 15.Wengelnik, K., Rossier, O. & Bonas, U. (1999) J. Bacteriol. 181, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noel, L., Thieme, F., Nennstiel, D. & Bonas, U. (2001) Mol. Microbiol. 41, 1271–1281. [DOI] [PubMed] [Google Scholar]
- 17.Mudgett, M. B., Chesnokova, O., Dahlbeck, D., Clark, E. T., Rossier, O., Bonas, U. & Staskawicz, B. J. (2000) Proc. Natl. Acad. Sci. USA 97, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman, D. S. & Greenberg, J. T. (2001) Mol. Plant–Microbe Interact. 14, 145–155. [DOI] [PubMed] [Google Scholar]
- 19.Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G. & Greenberg, J. T. (2002) Science 295, 1722–1726. [DOI] [PubMed] [Google Scholar]
- 20.Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., Stall, R. E. & Staskawicz, B. J. (1999) Proc. Natl. Acad. Sci. USA 96, 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsch-Moreno, R., Hernandez-Guzman, G. & Alvarez-Morales, A. (1998) Plasmid 39, 205–214. [DOI] [PubMed] [Google Scholar]
- 22.Swords, K. M., Dahlbeck, D., Kearney, B., Roy, M. & Staskawicz, B. J. (1996) J. Bacteriol. 178, 4661–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul, S. F., Madden, T. L., Schèaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sory, M. P. & Cornelis, G. R. (1994) Mol. Microbiol. 14, 583–594. [DOI] [PubMed] [Google Scholar]
- 25.Rossier, O., Wengelnik, K., Hahn, K. & Bonas, U. (1999) Proc. Natl. Acad. Sci. USA 96, 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casper-Lindley, C., Dahlbeck, D., Clark, E. T. & Staskawicz, B. J. (2002) Proc. Natl. Acad. Sci. USA 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noel, L., Thieme, F., Gabler, J., Buttner, D. & Bonas, U. (2003) J. Bacteriol. 185, 7092–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinsch, M. & Staskawicz, B. (1996) Mol. Plant–Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- 29.da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature 417, 459–463. [DOI] [PubMed] [Google Scholar]
- 30.Bennett, J. C. & Hughes, C. (2000) Trends Microbiol. 8, 202–204. [DOI] [PubMed] [Google Scholar]
- 31.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 32.Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W. & Bork, P. (2001) J. Mol. Biol. 309, 1–18. [DOI] [PubMed] [Google Scholar]
- 33.Degano, M., Gopaul, D. N., Scapin, G., Schramm, V. L. & Sacchettini, J. C. (1996) Biochemistry 35, 5971–5981. [DOI] [PubMed] [Google Scholar]
- 34.Buttner, D., Nennstiel, D., Klusener, B. & Bonas, U. (2002) J. Bacteriol. 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gold, L. (1988) Annu. Rev. Biochem. 57, 199–233. [DOI] [PubMed] [Google Scholar]
- 36.Minsavage, G., Dahlbeck, D., Whalen, M. C., Kearney, B., Bonas, U., Staskawicz, B. & Stall, R. (1990) Mol. Plant–Microbe Interact. 3, 41–47. [Google Scholar]
- 37.Bouzar, H., Jones, J. B., Stall, R., Hodge, N. C., Minsavage, G., Benedict, A. A. & Alvarez, A. M. (1994) Phytopathology 84, 663–671. [Google Scholar]
- 38.Jones, J. B., Bouzar, H., Stall, R. E., Almira, E. C., Roberts, P. D., Bowen, B. W., Sudberry, J., Strickler, P. M. & Chun, J. (2000) Int. J. Syst. Evol. Microbiol. 50, 1211–1219. [DOI] [PubMed] [Google Scholar]
- 39.Rohmer, L., Guttman, D. S. & Dangl, J. L. (2004) Genetics 167, 1341–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, L., Paulsen, I. T., Tchieu, J., Hueck, C. J. & Saier, M. H., Jr. (2000) J. Mol. Microbiol. Biotechnol. 2, 125–144. [PubMed] [Google Scholar]
- 41.Slabas, A. R., Fordham-Skelton, A. P., Fletcher, D., Martinez-Rivas, J. M., Swinhoe, R., Croy, R. R. & Evans, I. M. (1994) Plant Mol. Biol. 26, 1125–1138. [DOI] [PubMed] [Google Scholar]
- 42.Cingolani, G., Petosa, C., Weis, K. & Muller, C. W. (1999) Nature 399, 221–229. [DOI] [PubMed] [Google Scholar]
- 43.Cheng, Y., Dai, X. & Zhao, Y. (2004) Plant Physiol. 135, 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellāe, R., Schramm, V. L. & Parkin, D. W. (1998) J. Biol. Chem. 273, 2118–2126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.