Abstract
Introduction
This study was designed to investigate the combined influence of ATM and Ki67 on clinical outcome in early stage hormone receptor positive breast cancer (ES-HPBC), particularly in patients with smaller tumors (< 4 cm) and fewer than four positive lymph nodes.
Methods
532 formalin-fixed paraffin-embedded specimens of resected primary breast tumors were used to construct a tissue microarray. Samples from 297 patients were suitable for final statistical analysis. We detected ATM and Ki67 proteins using fluorescence and brightfield immunohistochemistry respectively, and quantified their expression with digital image analysis. Data on expression levels were subsequently correlated with clinical outcome.
Results
Remarkably, ATM expression was useful to stratify the low Ki67 group into subgroups with better or poorer prognosis. Specifically, in the low Ki67 subgroup defined as having smaller tumors and no positive nodes, patients with high ATM expression showed better outcome than those with low ATM, with estimated survival rates of 96% and 89% respectively at 15 years follow up (p = 0.04). Similarly, low-Ki67 patients with smaller tumors, 1-3 positive nodes and high ATM also had significantly better outcomes than their low ATM counterparts, with estimated survival rates of 88% and 46% respectively (p = 0.03) at 15 years follow up. Multivariable analysis indicated that the combination of high ATM and low Ki67 is prognostic of improved survival, independent of tumor size, grade, and lymph node status (p = 0.02).
Conclusions
These data suggest that the prognostic value of Ki67 can be improved by analyzing ATM expression in ES-HPBC.
Keywords: ATM, Ki67, early stage hormone receptor positive breast cancer, automated quantitative immunofluorescence analysis, disease specific overall survival
INTRODUCTION
The incidence of early stage breast cancer (stage I-III) has been increasing over the last 20 years largely due to the introduction of nation-wide mammogram screening program (Canadian Cancer Statistics 2015). At the time of the collection of these tissues (and excluding the 20% of in-situ cases) there would likely have been around 50% of patients with stags I, with around 40% with stage II and III, the remainder being stage IV or unknown. Although it is interesting that we do not have population data readily available for treatments given, approximately 60% of the patients with early stage breast cancer would have been given only adjuvant endocrine therapy.
Early stage hormone receptor-positive breast cancer (ES-HPBC) has a relatively good prognosis; it is, however, molecularly diverse and distinctive for late relapse. Consequently, a certain subset of patients with ES-HPBC has a poor prognosis. ES-HPBC can be broadly divided into luminal A [estrogen receptor (ER)/progesterone receptor (PR) strong positive, human epidermal growth factor receptor 2 (HER2) negative, well differentiated and less proliferative] and luminal B (ER/PR low to moderate positive, poorly differentiated and highly proliferative) subtypes with a 10-year breast cancer-specific overall survival (DSOS) of 92% (95% CI 90-94) and 79% (95% CI 74-85) respectively (p < 0.001) [1].
Ki67, a protein expressed exclusively in proliferating cells [2], is commonly used to determine luminal subtypes in ES-HPBC [1], and is adopted as a prognostic marker in breast cancer in general [3, 4]. However, the prognostic value of Ki67 in ES-HPBC is a matter of controversy [5–8]. A wide variety of assay methods and significant inter-observer variability [9–12] have made it difficult to clearly and consistently determine the value of Ki67 as a standalone prognostic marker. Nevertheless, Ki67 is commonly used in clinical decision-making, particularly to select which patients with supposedly good prognosis could avoid chemotherapy. While the weight of evidence is against such use [5–8], one cannot ignore the value of Ki67 as part of a prognostic index. It is notable that Ki67 is among the genes carrying the greatest weight in the recurrence score-calculating algorithm of Oncotype Dx, which is a validated prognostic and predictive assay with extensive use in ES-HPBC [13, 14]. Such use suggests that Ki67 is best used along with other biomarkers, which would improve its prognostic utility while avoiding pitfalls that emerge from variability in assay methods.
We and others previously showed that ataxia telangiectasia mutated (ATM), a protein that is critical in maintaining genomic stability [15], is significantly reduced in breast cancer as compared to normal breast tissue. We also showed that reduced ATM expression is associated with poor clinical outcome in early stage breast cancer [16–18]. Moreover, low ATM is strongly related to known poor prognostic factors such as larger tumor size, positive lymph nodes (LN), and high grade in ES-HPBC [18]. ATM is, however, not an independent prognostic factor in a multivariable analysis in ES-HPBC [18].
In this study, we used immunohistochemistry (IHC) and automated semi-quantitative digital analysis to detect and quantify ATM and Ki67 in resected primary tumors from patients with ES-HPBC. We hypothesized that the combination of both biomarkers would be a better prognostic indicator than either biomarker alone. Specifically, we assessed the breast cancer specific survival as an outcome using both biomarkers in LN negative or positive (LN = 1-3) ES-HPBC. We further asked if ATM status was useful to stratify the outcome of ES-HPBC patients, particularly in the subgroup with low Ki67 expression.
RESULTS
Patient characteristics
Our study adhered to the REMARK criteria for the study of biomarkers [19]. The clinical and tumor characteristics of the patients in the initial clinical cohort (n = 819), the selected TMA cohort (n = 532) and the final cohort (n = 297) are shown in Appendix 1B. It is notable that the patients’ characteristics and clinico-pathological features including age, tumor size, grade, LVI and LN in these three cohorts are similar (insignificant p value in Appendix 1B), suggesting that the final cohort could be representative of our initial clinical cohort. For the final cohort, we only included ES-HPBC (stage I-III) patients with Ki67/ATM data and confirmed ER/PR positivity and HER2 negativity (Appendix 1A). There were thus differences between the final cohort and the other two cohorts (the clinical and TMA cohorts) with regard to stage, ER/PR, and HER2 status (Appendix 1B).
The clinical and tumor characteristics of these patients in the final cohort, which was subdivided into four groups based on ATM and Ki67 expression levels, is shown in Table 1. Briefly, mean age at diagnosis was 65 years. The majority of cases had smaller tumors (96%; T1/T2), and were non-high grade (80%; grades 1/2), LN negative (74%) and stages I and II (93%), indicative of the early staging of this cohort. Sixty percent of patients had adjuvant radiation treatment (RT). All patients (100%) received adjuvant endocrine therapy in the form of tamoxifen and 19% of patients also received aromatase inhibitors as adjuvant endocrine treatment. No patients in this cohort received adjuvant chemotherapy.
Table 1. Comparison of patients’ characteristics and clinico-pathological features of breast tumors in high and low ATM/ Ki67 groups in the final cohort.
| Baseline Characteristics | Full Cohort N=297 (100%) |
Low Ki67 Group | High Ki67 Group | Low Ki67 Group | ||
|---|---|---|---|---|---|---|
| High ATM /Low Ki67 N=145 (49%) |
Low ATM/ Low Ki67 N=105 (35%) |
High ATM/ High Ki67 n=21 (7%) |
Low ATM/ High Ki67 N=26 (9%) |
Low Ki67/high ATM vs Low ATM/ Low Ki67 p value |
||
| Age median (min-max) | 65 (38-96) | 63(38-96) | 68(38-89) | 68(43-78) | 68(50-86) | 0.150 |
| Tumor size | ||||||
| T1/T2 (<5cm) | 285 (96%) | 141 (48%) | 99 (33%) | 21 (7%) | 24 (8%) | 0.329 |
| T3/T4 (≥5cm) | 12 (4%) | 4 (1%) | 6 (2%) | 0 (0%) | 2 (1%) | |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Grade | ||||||
| 1 | 77(26%) | 48 (16%) | 26 (9%) | 1 (0.5%) | 2 (1%) | 0.090 |
| 2 | 161 (54%) | 80 (27%) | 61 (20%) | 10 (3%) | 10 (3%) | |
| 3 | 47 (16%) | 10 (4%) | 15 (5%) | 9 (3%) | 13 (4%) | |
| Missing | 12 (4%) | 7 (2%) | 3 (1%) | 1 (0.5%) | 1 (0.5%) | |
| LVI | ||||||
| Negative | 180 (61%) | 90 (30%) | 64 (21%) | 12 (4%) | 14 (5%) | 0.584 |
| Positive | 50 (17%) | 20 (7%) | 18 (6%) | 4 (1%) | 8 (3%) | |
| Missing | 67 (22%) | 35 (12%) | 23 (8%) | 5 (2%) | 4 (1%) | |
| LN | ||||||
| 0 | 220 (74%) | 114 (38%) | 82 (28%) | 13 (4%) | 11 (4%) | 1.0 |
| >0 | 77 (26%) | 31 (10%) | 23 (7%) | 8 (3%) | 15 (5%) | |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Stage | ||||||
| I | 168 (57%) | 94 (32%) | 63 (21%) | 7 (2%) | 4 (1%) | 0.012 |
| II | 108 (36%) | 47 (16%) | 29 (10%) | 12 (4%) | 20 (7%) | |
| III | 21 (7%) | 4 (1%) | 13 (4%) | 2 (1%) | 2 (1%) | |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| ER/PR status | ||||||
| Positive | 297 (100%) | 145 (49%) | 105 (35%) | 21 (7%) | 26 (9%) | |
| Negative | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| HER2 Status | ||||||
| Positive | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Negative | 297 (100%) | 145 (49%) | 105 (35%) | 21 (7%) | 26 (9%) | |
| Missing | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| RT | ||||||
| Yes | 178 (60%) | 97 (33%) | 57 (19%) | 9 (3%) | 15 (5%) | 0.107 |
| No | 113 (38%) | 47 (16%) | 44 (15%) | 11 (4%) | 11 (4%) | |
| unknown | 6 (2%) | 1 (0.5%) | 4 (1%) | 1 (0.5%) | 0 (0%) | |
| Chemo | ||||||
| Yes | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| No | 297 (100%) | 145 (49%) | 105 (35%) | 21 (7%) | 26 (9%) | |
| Endocrine | ||||||
| Tamoxifen | 297 (100%) | 145 (49%) | 105 (35%) | 21 (7%) | 26 (9%) | |
| Aromatase inhibitor | 56 (19%) | 26 (9%) | 18 (6%) | 6 (2%) | 6 (2%) | 1.0 |
ATM and Ki67 expression analysis
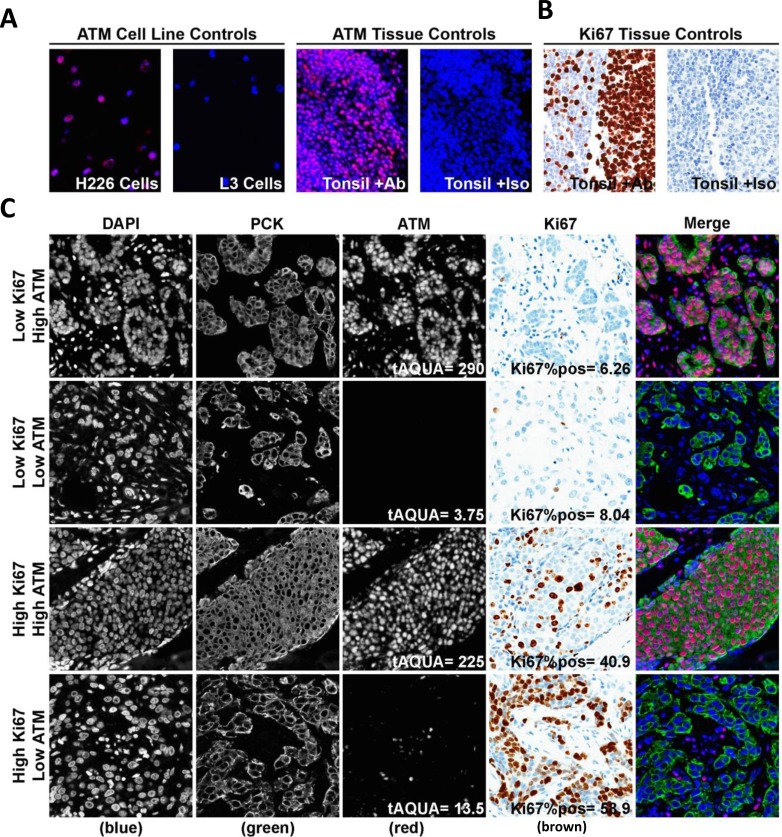
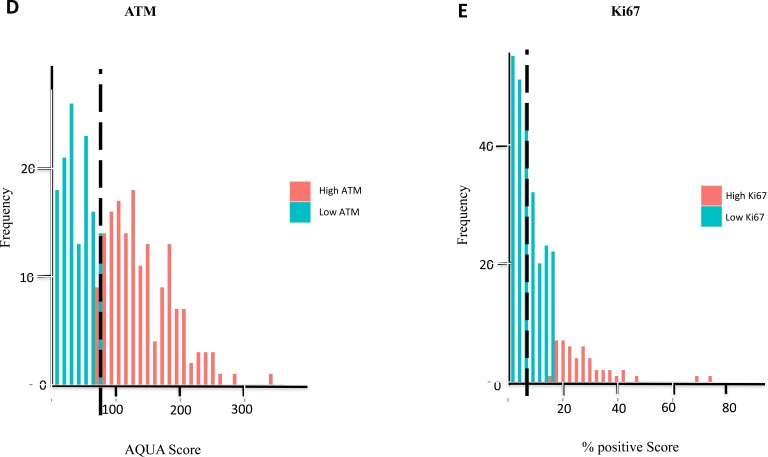
ATM protein was detected by fluorescence IHC, and quantified by digital image analysis using AQUAnalysis software [20] (Figure 1). Antibody specificity was confirmed using the L3 ATM-deficient human squamous lung carcinoma cell line and the age-matched H226 ATM-expressing human squamous lung carcinoma cell line as controls (Figure 1A, left two panels) [18]. As expected, no IHC signal was detected in the L3 cells, whereas the H226 showed clear nuclear staining. Positive and negative (by omission of the primary antibody) tonsil tissue controls were included in each run (Figure 1A, right two panels). Normal tonsil tissue stained with the Ki67 or isotype control antibodies served as positive and negative controls respectively for Ki67 (Figure 1B). Representative images of the low Ki67/high ATM, low Ki67/low ATM, high Ki67/high ATM, and high Ki67/low ATM specimens are shown in Figure 1C. ATM expression was determined within each patient tissue core as the average Cy3 pixel intensity within the pan-cytokeratin positive malignant cell area, and represented as an AQUA score (Figure 1C). For each patient sample, the average AQUA score over triplicate cores was used to define the ATM expression score. Ki67 expression was indicated as the percentage of the analyzed tumor area that contained positive (DAB) staining (Figure 1C). The histograms in Figure 1D and 1E indicate the distributions of Ki67 and ATM expression scores. The median ATM AQUA score and Ki67 percent positive score in this cohort were 85.8 (95% CI: 72.3-98.3) and 7.0 (95% CI: 6.2-8.4) respectively (Figure 1D and 1E, hashed black line).
Figure 1. Semi-quantitative fluorescence immunohistochemistry and digital image analysis for ATM and Ki67 in ES-HPBC.
A. Positive and negative controls for ATM were ATM-expressing H226 cells and ATM-deficient L3 cells, as well as in breast cancer tissue with either rabbit IgG or primary anti-ATM antibody. B. Normal tonsil tissue stained with the Ki67 or isotype control antibodies served as positive and negative controls respectively for Ki67. C. Representative fluorescence images for low Ki67/high ATM, low Ki67/low ATM, high Ki67/high ATM and high Ki67/low ATM expression in ES-HPBC cohort. DAPI-stained nuclei are depicted in blue (first column), pan-cytokeratin-stained malignant cells are depicted in green (second column), and ATM is depicted in red (third column). Haematoxylin-stained nuclei are depicted in blue, while DAB-stained Ki67-positive nuclei are depicted in brown (fourth column). D., E. Histograms representing the distribution of ATM expression (D) and Ki67 expression (E) in ES-HPBC. The hashed black line represents median Ki67/ATM expression in this cohort.
Association between Ki67/ATM protein expression and clinical outcome
Patients were stratified into subgroups based on high and low Ki67/ATM scores, using independent cut-points identified by X-Tile software [21]. The cut-point was set at an AQUA score of 72.2 for ATM and a percent positive score of 16.9 for Ki67 respectively. As both the high Ki67 groups (high Ki67/high ATM and high Ki67/low ATM) have low study numbers (n = 21 and 26 respectively), we focused our further statistical analysis on low Ki67 groups. Patients with low Ki67/low ATM were significantly more likely than their low Ki67/high ATM counterparts to have stage III disease (p = 0.012) (Table 1). There were no associations between low Ki67/low ATM and other well-known prognostic factors including tumor size (p = 0.177), LN status (p = 1.0), or grade (p = 0.086) (Table 1), implying that low Ki67/low ATM may constitute an independent prognostic factor.
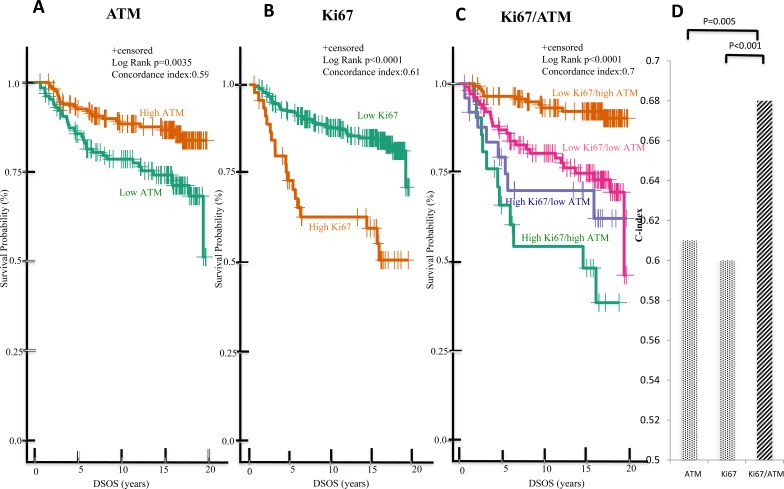
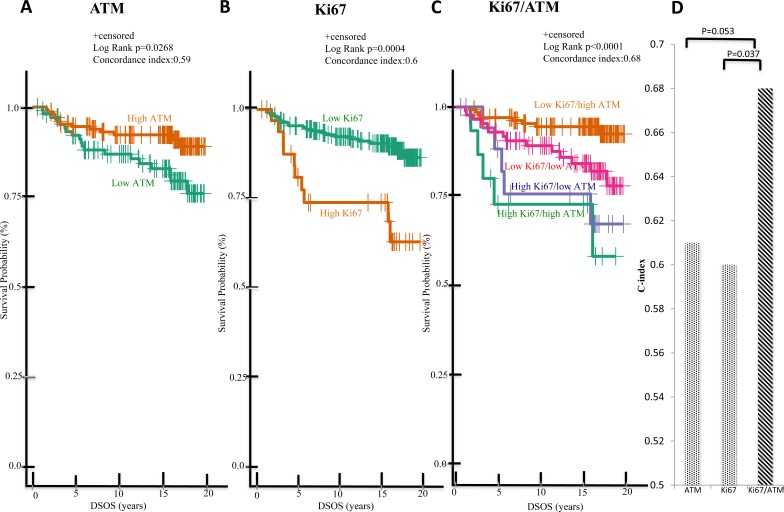
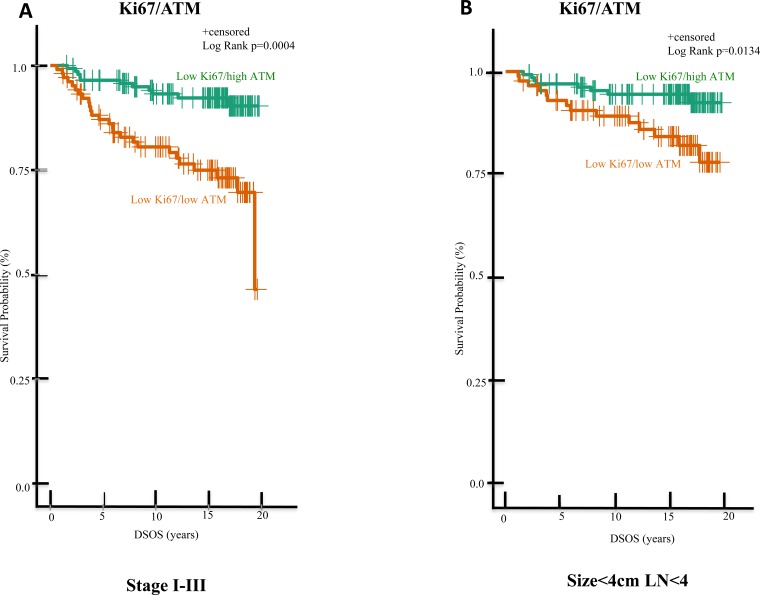
To further investigate the prognostic value of these biomarkers, patients were separated into sub-groups based on high or low ATM or Ki67 expression, and the survival data were subjected to further analysis using the Kaplan-Meier method. Consistent with previous studies [3, 4, 18], DSOS significantly differed based on ATM or Ki67 alone. For patients with stage I-III HPBC, the estimated 15-year DSOS rates were: 87% and 74% for high and low ATM group respectively (p = 0.0035); and 85% and 60% for low and high Ki67 group respectively (p < 0.001) (Figure 2A and 2B). For ES-HPBC patients with smaller tumors (size < 4cm) and fewer LN (LN < 4), the estimated 15-year DSOS rates were: 92% and 79% for high and low ATM group respectively (p = 0.0268); and 90% and 74% for low and high Ki67 group respectively (p = 0.0004) (Figure 3A and 3B). More importantly, the combination of Ki67 and ATM was a stronger predictor of DSOS than either biomarker alone, as indicated by the higher C-index in the combination (Ki67/ATM: 0.7; Ki67 alone: 0.61; ATM alone: 0.59; p < 0.001 for combined Ki67/ATM vs Ki67 alone and p = 0.005 for combined Ki67/ATM vs ATM alone) (Figure 2D). Similarly, in the subgroup of ES-HPBC patients defined as tumor size < 4cm and LN < 4, combined biomarkers seem to have a better prognostic performance than either biomarker alone. (Ki67/ATM: 0.68; Ki67 alone: 0.60; ATM alone: 0.59; p = 0.037 for combined Ki67/ATM vs Ki67 alone and p = 0.053 for combined Ki67/ATM vs ATM alone) (Figure 3D). Overall, patients with low Ki67 and high ATM scores showed the most favorable clinical outcome with estimated 15-year DSOS rates of greater than 92% (Figure 2C & 3C).
Figure 2. Low Ki67/high ATM expression in malignant tumors predicts favorable DSOS in ES-HPBC (stage I-III).
Kaplan-Meier analysis of DSOS between patients with high and low ATM expression alone A., high and low Ki67 expression alone B., and of patients in these four groups (low Ki67/high ATM, low Ki67/low ATM, high Ki67/high ATM, high Ki67/low ATM expression) C. C-index was used to evaluate the model prediction accuracy of single (ATM or Ki67) versus combined biomarkers (Ki67/ATM) D.
Figure 3. Low Ki67/high ATM expression in malignant tumors predicts favorable DSOS in ES-HPBC (size < 4cm, LN < 4).
A subgroup of patients with tumor size < 4cm and LN < 4 were selected for further analysis. Kaplan-Meier analysis of DSOS between patients with high and low ATM expression alone A., high and low Ki67 expression alone B., and of patients in these four groups (low Ki67/high ATM, low Ki67/low ATM, high Ki67/high ATM, high Ki67/low ATM expression) C. C-index was used to evaluate the model prediction accuracy of single (ATM or Ki67) versus combined biomarkers (Ki67/ATM) D.
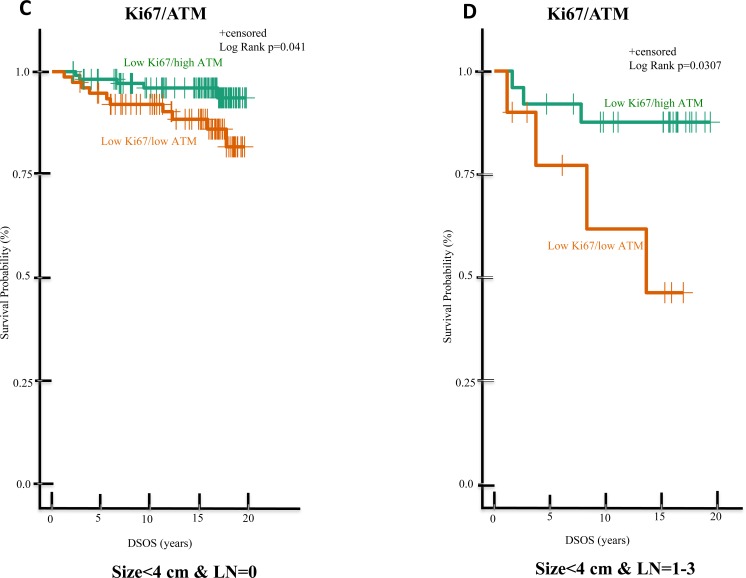
We focused the majority of our subsequent statistical analyses on the low Ki67 groups, due to the low numbers of high Ki67 tumors in our final cohort. Low Ki67/low ATM patients had significantly lower 15-year DSOS rates (75%) than the low Ki67/high ATM group (92%; p = 0.004; Figure 4A). Among patients with smaller tumors (size < 4cm) and fewer positive nodes (LN < 4), the low Ki67/low ATM cases also exhibited poorer DSOS rates (84%) than those with low Ki67/high ATM (94%; p = 0.0134; Figure 4B). Similar trends were evident in patients with node-negative patients with smaller tumors (tumor size < 4cm and LN = 0; 96% vs 89%; p = 0.0411; Figure 4C) and in patients with tumor size < 4cm and LN = 1-3 (88% vs 46%; p = 0.0307; Figure 4D).
Figure 4. In the low Ki67 subgroup of ES-HPBC, patients with high ATM vs low ATM expression significantly differ in DSOS.
Shown are Kaplan-Meier analysis of DSOS of ES-HPBC patients with A. stage I-III tumors and low Ki67 expression; B. low Ki67 expression, smaller tumors (size < 4 cm) and fewer positive nodes (LN < 4), C. no positive nodes, low Ki67, and smaller tumors (size < 4 cm), D. low Ki67, smaller tumors (size < 4 cm) and LN = 1-3.
To determine if the combined expression score for Ki67 and ATM was an independent prognostic factor for DSOS in the low Ki67 group, we used Cox proportional hazards regression models. The multivariable model consisted of known clinical prognostic factors such as tumor size, grade, LN status, LVI and age. As expected, tumor size and grade were significantly associated with survival (Table 2). LN status was trending but not significantly associated with survival, likely due to the small sample size of positive LN in this low Ki67 ES-HPBC group. In this subgroup of ES-HPBC with low Ki67 expression, the significant association between combined biomarkers of Ki67/ATM and DSOS persisted in the multivariable model, adjusted for known prognostic variables (p = 0.02) (Table 2).
Table 2. Low Ki67/high ATM expression independently predicts favorable disease survival in a multivariate model in low Ki67 subgroup of ES-HPBC (Stage I-III).
| Variables | pvalue | HR (95%CI) |
|---|---|---|
| Low Ki67/high ATM vs Low Ki67/low ATM | 0.02 | 0.36 (0.15-0.88) |
| Tumor size (T1/2 vs T3/4) | 0.01 | 0.16 (0.04-0.69) |
| LN status (- vs +) | 0.14 | 0.49 (0.20-1.25) |
| LVI (_- vs +) | 0.16 | 0.51 (0.20-1.31) |
| Grade (1/2 vs 3) | <0.0001 | 0.28 (0.11-0.67) |
| Age (<65 vs >65) | 0.01 | 0.26 (0.10-0.74) |
Stage I-III patients with low Ki67 expression were selected for this analysis. A multivariable model was created using selected known prognostic factors such as tumor size, lymph node (LN) status, lymphovascular invasion (LVI), grade and age. Cox proportional hazards regression methods were used to calculate p values, hazard ratio (HR) and confidence interval (CI) of each variable in the model.
DISCUSSION
In this study, we showed that the combined Ki67/ATM expression index is a stronger prognostic factor of DSOS than either biomarker alone. When patients were grouped by Ki67 and ATM expression, the ES-HPBC patients with low Ki67 and high ATM scores had the most favorable prognosis. Most importantly, ATM expression identified a subset of patients who had poor prognosis, even with low Ki67 scores. This observation held true across the spectrum of ES-HPBC cases, regardless of stage, tumor size, or node status. To our knowledge, this is the first study to examine the prognostic value of both biomarkers together in breast cancer.
For a variety of reasons, the use of Ki67 for clinical decision-making has been mired in controversy. One such reason surrounds its use as an independent prognostic tool, which is a concern we address in this paper. Several years ago, two extensive meta-analyses were conducted on the use of Ki67 in this context [3, 4]. One was based on 46 studies involving 12,155 patients and the other was based on 43 studies involving 15,790 patients. Both analyses suggested that Ki67 is a useful prognostic tool in both node-positive and node-negative early stage breast cancer. Nevertheless, the independent prognostic value of Ki67 has been frequently challenged [5–8]. In a recent review, Andre and colleagues challenged the use of Ki67 in making clinical decisions for patients with early stage breast cancer and one to three positive axillary nodes [8]. They argue that Ki67 fails to identify a sizable proportion of patients who may have poor prognosis [8]. Our current work highlights the value of secondary biomarkers, such as ATM, in identifying patients whose prognosis is significantly poorer than expected in that low Ki67 subgroup. Fully 35% of patients in our final cohort (n = 105/297; Table 1) were categorized as low Ki67/low ATM, and the poorer prognosis of this cohort highlights a significant deficiency in the application of Ki67 alone as a prognostic factor.
One of the common shortcomings with the use of Ki67 as a biomarker is the poor reliability of manual analysis methods. Even with centralized staining, inter-observer concordance with manual scoring is often quite low [10, 22]. For this study, we used an automated digital image analysis platform to score Ki67, which can potentially reduce the effect of human bias (inter- and intra- observer variability) and improve analytical validity. This is an approach that has been recommended by several other studies [11, 12]. The ACIS analysis platform we used in this study offers high concordance with well-trained manual scorers, as documented in similar applications [23–26]. Additionally, utilizing a computer-generated algorithm provides consistent scoring between samples, and minimizes run-to-run variation. One limitation with the Ki67 scoring method in our study is the potential influence from contributing stromal components and tumor-infiltrating intra-epthelial immune cells. Manually selecting tumor regions minimizes potential influence from stromal components; however, it doesn't completely eliminate this potential bias. The addition of a stain for cytokeratin to identify tumor areas may improve the specificity of this biomarker testing, by enhancing definition of tumor areas. Additionally, utilizing multi-staining IHC approaches combined with multispectral imaging that allows for several target antibodies to be used and analyzed on a single slide, biomarkers such as CD68 could also be used to identify, and remove, intratumoral infiltrating leukocytes such as macrophages from Ki67 calculations.
Another source of inconsistency with Ki67 as a prognostic marker in prospective-retrospective clinical studies [3, 4] is the possible qualitative interaction of Ki67 with adjuvant chemotherapy. Qualitative interaction of a biomarker with treatment is defined as patients with a positive biomarker (ie. high Ki67) who are likely to benefit from adjuvant chemotherapy, whereas patients with a negative biomarker (ie. low Ki67) likely derive no benefit or may even be harmed by adjuvant chemotherapy [27]. Therefore, high Ki67 would not necessarily result in a poor prognosis if patients received effective chemotherapy. Similarly, low Ki67 would not necessarily result in a good outcome if patients received harmful chemotherapy. In addition, high Ki67 could predict good or poor outcome depending on if a tumor will respond and be sensitive to a given chemotherapy [28]. In our study, none of patients in the TMA cohort received adjuvant chemotherapy and all patients received tamoxifen as adjuvant endocrine therapy indicating a relatively homogenous study population. Therefore, it is unlikely that the biomarkers investigated in this cohort would succumb to the treatment effect. Biologically, however, the prognostic impact of low Ki67/high ATM is easily justified. A low Ki67 score is indicative of a relatively slow-proliferating, less aggressive tumor; while there were no associations between Ki67/ATM indices and standard clinico-pathological features of the tumors (Table 1), such a correlation would naturally be biased by the various stages at which patients present with disease. As suggested by our previous publication [18], low ATM expression would highlight tumors that are more genetically unstable. The combination of high proliferative rate and low DNA stability is thus clearly a harbinger of relapse, which would, as our data again show, is independently prognostic of DSOS (Table 2 and Appendix 3).
In this study, we identified a significant difference in survival for patients based on Ki67 status. These data contradict those of some large-scale clinical trial studies (BIG 1-98 [29], IBCSGVIII-IX [30], ATAC [31]) which showed that the statistically significant prognostic value of Ki67 was relatively low. For example, the absolute difference of 4-year disease free survival (DFS) in patients with high vs low Ki67 expression was only 3% in the BIG 1-98 trial (93% vs 90%) [29]. In our study, the survival difference was larger between high and low Ki67 expression alone group, which is about 20% difference at 5 years follow up (93% vs 73% respectively, Figure 2B). Although this is likely the effect of sample size and the different treatment regimens in these studies, we believe that this difference is also likely a result from different Ki67 cut-points. Since Ki67 data in our study constitutes a continuous variable generated automatically by the analysis software, we chose to use the X-tile program to define an optimal cut-point. X-tile program is a validated methodology to define biomarker cut-point based on the survival outcome (DSOS) [21]. An additional important observation from our study is that the DSOS difference was more pronounced after five to ten years suggesting that these biomarkers are useful to detect late breast cancer associated mortality in ES-HPBC (Figure 2A-C). We observed a similar trend in DFS using the combined biomarkers in the final cohort as well as the subgroup patients with tumor size <4cm and LN<4 with a significant p value of <0.0001 and 0.0026 respectively (data not shown).
A number of highly reproducible RNA-based multigene analyses have been recently developed and validated for prognostic and predictive purposes of ES-HPBC [13, 32–34]. One of which is Oncotype Dx (Genome Health 21-gene recurrence score), which has been approved in use as a prognostic and predictive biomarker for adjuvant chemotherapy in node negative ES-HPBC [34]. It is notable that proliferation genes, including Ki67, are heavily weighted in the Oncotype Dx algorithm to calculate recurrence scores. However, the multigene assays can be logistically difficult and cost-prohibitive for routine clinical use in much of the world. A simplified assay, such as IHC 4 score consisting of standard IHC breast biomarkers such as ER, PR and HER2 as well as Ki67, may have a similar prognostic value when compared to Oncotype Dx in ES-HPBC [31]. Therefore, these observations highlight not only the importance of prognostic value of Ki67 in ES-HPBC but also the fact that when Ki67 is used in the mixture of multigene analysis such as in Oncotype Dx assay and IHC4 score, the prognostic value is significantly enhanced. This supports our finding that the prognostic performance of Ki67 biomarker is significantly improved and refined by ATM status. (C-index in Figure 2D/3D and multivariable analysis in Appendix 3). These two molecular biomarkers represent two hallmarks of cancer—uncontrolled proliferation and genome instability [35]. The combination of these two biomarkers may dictate the distinct luminal tumors’ behavior thereby predicting the prognosis. It would be interesting to further investigate the prognostic value of the combined biomarkers Ki67/ATM when ER/PR intensity scores are taken into consideration in ES-HPBC. The combined IHC biomarkers (Ki67/ATM) with the incorporation of ER/PR in the future could be a much simpler and cheaper tool than RNA based multigene assays if this could be further validated in a larger cohort and prospective clinical studies.
Currently, the predictive validity of Oncotype Dx in ES-HPBC patients with 1-3 LN remains unknown. While breast oncologists are eagerly awaiting the results from three ongoing randomized clinical trials (Rxponder [36], MINDACT [37], OPTIMA [38]) to address this question, Ki67 IHC is intended to guide clinicians to weigh risk-benefit ratio of adjuvant chemotherapy as this is widely available and financially acceptable in most laboratories. Our study revealed that low Ki67 alone may not provide enough prognostic information for clinicians to abort adjuvant chemotherapy in node negative or node positive (LN=1-3) ES-HPBC. In Figures 4A-D, we identified certain high-risk patients with a poorer prognosis if ATM protein expression is low despite a low Ki67 who may warrant further intervention in hopes of reducing their risk of relapse and improving DSOS. This data is consistent with the view presented in a recent review paper cautioning the use of Ki67 in guiding treatment decisions for node positive ES-HPBC [8]. The survival difference between patients with low Ki67/high ATM and low Ki67/low ATM should be, however, interpreted cautiously as our study numbers in these subgroups of node positive (LN-1-3) ES-HPBC are low (n=25 and 10 in low Ki67/high ATM and low Ki67/low ATM groups respectively).
We previously showed that ATM protein expression level in malignant tumors independently predicts DSOS in early stage hormone negative breast cancer. ATM expression was not prognostic in ES-HPBC in a multivariable analysis, however, because ATM protein expression level is strongly associated with known clinico-pathological prognostic factors such as tumor size, grade and LN status in ES-HPBC [18]. In the current study, we confirmed this observation that ATM biomarker alone was not an independent prognostic marker in the multivariable model (p=0.15; Appendix 3); however, the combination of Ki67/ATM biomarkers was an independent prognostic factor (p=0.01; Appendix 3). This further supports our hypothesis that the prognostic performance is significantly strengthened by the combined biomarkers. With regard to ATM staining and analysis, we used fluorescent IHC, cytokeratin masking and automated AQUA image analysis, which can provide greater sensitivity and broader range of protein quantification than the brightfield equivalent. It should be noted, however, that fluorescence IHC and the associated analysis platform are not currently used in clinical practice; therefore some data presented in this study may not be directly applicable. It is however possible that they could be translated into clinically relevant assays upon identification of the levels of protein expression that offer prognostic value.
In spite of the promising nature of our results, there were practical constraints that can only be addressed in a follow-up study. Out of the 532 primary breast tumors in our eligible cohort, only 297 patient samples were suitable for final statistical analysis. Other samples were removed due to inconclusive hormone receptor status, poor tissue quality, or inadequate staining. Consequently, whereas the high Ki67 group showed clearly poorer prognosis than their low Ki67 counterparts (Figure 2B), we were unable to perform extensive sub-group analyses due to the resulting small numbers (n=21 and 26 in ATM high/Ki67 high and ATM low/Ki67 high groups respectively). This may explain the overlap of survival curves for these two subgroups (Figure 2C and Figure 3C and Appendix 2). A larger validation cohort is warranted to see if there is any significant survival difference between patients with ATM high/Ki67 high and ATM low/Ki67 high expression.
In summary, we demonstrated that the combined biomarker analysis (Ki67/ATM) significantly increased the strength of predicting clinical outcome compared to either biomarker (Ki67 or ATM) alone. We identified a subgroup of ES-HPBC patients with low Ki67/high ATM expression who have the most favourable clinical outcome. More importantly, we revealed the significant survival difference in low Ki67 group based on high or low ATM protein expression level. The latter suggests that breast oncologists should not rely on Ki67 expression alone to decide on adjuvant chemotherapy as this may mislead clinicians to undertreat certain patients with ES-HPBC.
MATERIALS AND METHODS
Case Selection and Clinical Data Collection
This study was approved by the University of Calgary Conjoint Faculties Research Ethics Board (Ethic ID # REB15-1284), in accordance with the Tri-Council Policy Statement on Research with Human Subjects. Consents directly from patients were not required in this study as per Ethics. The clinical data were collected retrospectively through chart review. The initial clinical cohort consisted of 819 patients diagnosed with breast cancer at the Tom Baker Cancer Centre, as previously described [39]. Within this cohort, 532 patients diagnosed between 1985 and 2000 had resected primary tumor samples available in formalin-fixed paraffin-embedded (FFPE) tissue blocks for construction of a tissue microarray (TMA). Patients who had stage I to III breast cancer, with no evidence of metastatic disease at diagnosis, and had undergone either lumpectomy or mastectomy were included for the final analysis. Estrogen receptor (ER) and progesterone receptor (PR) statuses were assessed by local breast pathologists at the time of diagnosis and later evaluated on TMA using an independent staining method [39]. Samples with discordant results between initial pathological diagnosis and later evaluation on TMA were excluded. HER2 expression was not routinely assessed at the time of diagnosis for this cohort but was tested retrospectively by IHC [39]. Cases found to be HER2 positive were excluded. In the end, 297 cases that were successfully stained with both biomarkers of ATM and Ki67, and with confirmed ER/PR positivity and HER2 negativity were available for statistical analysis (Appendix 1A).
Tissue Microarray Generation
All archived FFPE blocks were retrieved from Calgary Laboratory Services and reviewed by a pathologist (AMM). TMAs were constructed as previously described [18]. Appropriate controls for the protein targets were available either as tissue controls on the TMA slides or as separate slides within the same staining runs.
Fluorescence IHC for ATM Detection and Automated Semi-Quantitative Digital Analysis
Fluorescence IHC against ATM was performed on TMA sections as previously described [18]. The following primary antibodies were applied in sequence: rabbit anti-ATM (rabbit monoclonal, clone Y170, ab32420, 1:1000, Abcam, Cambridge, MA, USA), mouse anti-vimentin to identify the stromal compartment (mouse monoclonal, clone V9, 1:5000, DAKO), and guinea pig anti-pan-cytokeratin to identify the epithelial compartment (guinea pig polyclonal, catalog number BP5069, 1:200, Acris Antibodies, San Diego, CA, USA). Automated image acquisition was performed using an Aperio Scanscope® FL (Aperio Inc., Vista, CA, USA) slide scanner. Images were acquired using filters specific for DAPI to define the nuclear compartment, fluorescein isothiocyanate (FITC) to define cytokeratin for the tumor compartment and Cy3 to define ATM. Images were then analyzed using the AQUAnalysis® program, version 2.4.4.1. Briefly, a tumor-specific binary mask was generated to distinguish the cancer cells from surrounding stromal tissue by thresholding the pan-cytokeratin images. All images were processed using optimized threshold values, and manually validated as described [18]. Antibody specificity was validated using the L3 ATM-deficient humansquamous lung carcinoma cell line and the H226 ATM-expressing human squamous cell lung carcinoma cell line as controls in addition to normal tonsil tissue (with no evidence of pathological tonsillitis) treated with either ATM or Ki67 antibody and matched isotype control slides (Figure 1A). ATM expression was calculated as the mean Cy3 pixel intensity within the tumor mask as defined by positive pan-cytokeratin (Figure 1C). For each patient sample, the average AQUA score over triplicate cores was used to define the ATM expression score.
Brightfield Ki67 Detection and Automated Digital Analysis
Following previously described deparaffinization and rehydration procedures, slides were pre-treated by immersing them in the EnVision FLEX Target Retrieval Solution, low pH (DAKO), and heating to 97°C for 20 minutes in a PT-Link pre-treatment module. Ki67 was detected using the FLEX mouse monoclonal antibody (MIB-1) and EnVision FLEX reagents following the manufacturer's recommended protocol. 3’-diaminobenzidine tetrahydrochloride (DAB) was used to visualize Ki67, while the FLEX hematoxylin counterstain allowed for visualization of the tissue and nuclei. Ki67 was quantified using the ACIS® III Automated Cellular Imaging System (DAKO) [40]. Briefly, TMA slides were scanned and tumor areas were hand selected to exclude stromal areas using the ACIS software. A unique algorithm was developed that identified distinct color thresholds for nuclear hematoxylin and DAB staining, and an overall percent positive score was calculated by taking the total Ki67 (DAB) nuclear score and dividing it by total nuclear score (DAB plus hematoxylin). The final Ki67 score for analysis was obtained by taking an average of up to three tumor cores.
Statistical analysis
Summary and descriptive statistics were used to describe demographic and clinico-pathological characteristics of patients in the clinical cohort, the TMA cohort and the final cohort for statistical analysis. Histograms were used to evaluate the distribution of Ki67 and ATM expression. In the final cohort, patients were divided into four subgroups based on ATM and Ki67 expression levels. Cut-points to define low and high Ki67 and ATM expression were modeled using X-Tile software [21]. The following comparison was made in the Ki67 low group only. The Student's t-test was used to compare the continuous variable of “age” between low Ki67/high ATM and low Ki67/low ATM groups. Fisher's exact test was used to compare the categorical variables between low Ki67/low ATM and low Ki67/high ATM groups. Disease-free survival (DFS) and breast cancer- or disease-specific overall survival (DSOS) estimates were calculated using the Kaplan-Meier (KM) survival analysis, and log-rank tests were used to compare low and high Ki67/ATM groups. The C-index (concordance index) was used to evaluate the model prediction accuracy [41]. The C-index is defined as the probability of agreement for two randomly selected groups, and the agreement indicates the higher risk group predicted by the model will have shorter survival time. C-index values are ranged from 0 to 1, where 0.5 indicates random prediction, and higher values suggest better model prediction. P values for C-index comparison were obtained by 3000 bootstraps samples. Cox proportional hazards regression methods were used to assess the prognostic effect of Ki67 and ATM combined on DSOS, in a multivariable model with clinical known prognostic factors such as tumor size, grade, LN status, lymphovascular invasion (LVI) and age. The prognostic utility of each biomarker alone and in combination was tested using a backward selection approach to obtain the optimal model. All tests were two-sided, and a p-value of 0.05 was considered statistically significant. All statistical analyses were conducted using R statistical analysis software (Version 3.1.2).
APPENDIX
Acknowledgments
We thank Drs Helen Anderson and Jason Hart for tremendous institutional support.
Abbreviations
- ATM
ataxia telangiectasia mutated
- ES-HPBC
early stage hormone receptor positive breast cancer
- TMA
tissue microarray
- AQUA
automated quantitative immunofluorescence analysis
- FFPE
formalin-fixed paraffin-embedded
- ER
estrogen receptor
- PR
progesterone receptor
- HER2
human membrane epidermal growth factor receptor 2
- DSOS
disease specific overall survival
- LVI
lymphvascular invasion
- LN
axillary lymph node
Footnotes
CONFLICTS OF INTEREST
All authors declare no conflict of interests.
GRANT SUPPORT
We received financial support from Breast Cancer Society of Canada.
Authors’ contributions
XF designed the study, interpreted the results, and drafted the manuscript. HL performed the statistical analysis, helped prepare most figures and critically revised the manuscript. EK carried out the TMA staining for Ki67 and data collection and analysis. MD carried out the TMA staining for ATM and data analysis and helped prepare some figures. EE participated in optimizing antibodies and techniques for TMA staining, and helped to draft manuscript and critically revised the manuscript. AM generated HPBC cohort, participated in study design and critically revised the manuscript. DM participated in generating HPBC cohort and critically revising the manuscript. SPL-M, KR, PW, GB and AP helped conceive and design or coordinate the study, and helped to draft the manuscript. XF and AP obtained research grant for this project. All authors read and approved the final manuscript. In addition, all authors agreed to be accountable for the accuracy and integrity of the results from this study.
Authors’ information
Xiaolan Feng MD PhD FRCPC, Medical Oncologist and Research Scientist
Haocheng Li PhD, Biostatistician
Elizabeth Kornaga MSc, Quality and Data Coordinator and Research Scientist
Michelle Dean BSc, Senior Technician
Susan P Lees-Miller PhD, Senior Research Scientist
Karl Riabowol PhD, Research Laboratory Lead
Anthony M Magliocco MD, Pathologist and Senior Research Scientist
Don Morris MD PhD, Medical Oncologist and Senior Research Scientist
Peter Watson MB FRCPC, Pathologist and Senior Research Scientist
Emeka K Enwere PhD, Research Scientist
Gwyn Bebb MBMCh PhD FRCPC, Medical Oncologist and Senior Research Scientist
Alexander Paterson MD FRCPC, Medical Oncologist
Editorial note
This paper has been accepted based in part on peer-review conducted by another journal and the authors’ response and revisions as well as expedited peer-review in Oncotarget.
REFERENCES
- 1.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. Journal of the National Cancer Institute. 2009;101(10):736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast cancer research : BCR. 2006;8(6):216. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Azambuja E, Cardoso F, de Castro G, Jr., Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. British journal of cancer. 2007;96(10):1504–1513. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart-Harris R, Caldas C, Pinder SE, Pharoah P. Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast. 2008;17(4):323–334. doi: 10.1016/j.breast.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. The lancet oncology. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 6.Kontzoglou K, Palla V, Karaolanis G, Karaiskos I, Alexiou I, Pateras I, Konstantoudakis K, Stamatakos M. Correlation between Ki67 and breast cancer prognosis. Oncology. 2013;84(4):219–225. doi: 10.1159/000346475. [DOI] [PubMed] [Google Scholar]
- 7.Guiu S, Michiels S, Andre F, Cortes J, Denkert C, Di Leo A, Hennessy BT, Sorlie T, Sotiriou C, Turner N, Van de Vijver M, Viale G, Loi S, Reis-Filho JS. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- 8.Andre F, Arnedos M, Goubar A, Ghouadni A, Delaloge S. Ki67--no evidence for its use in node-positive breast cancer. Nature reviews Clinical oncology. 2015;12(5):296–301. doi: 10.1038/nrclinonc.2015.46. [DOI] [PubMed] [Google Scholar]
- 9.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, McShane L, Paik S, Penault-Llorca F, Prudkin L, Regan M, Salter J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. Journal of the National Cancer Institute. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, Viale G, Zabaglo LA, Penault-Llorca F, Bartlett JM, Gown AM, Symmans WF, Piper T, Mehl E, Enos RA, Hayes DF, et al. An international Ki67 reproducibility study. Journal of the National Cancer Institute. 2013;105(24):1897–1906. doi: 10.1093/jnci/djt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudlaugsson E, Skaland I, Janssen EA, Smaaland R, Shao Z, Malpica A, Voorhorst F, Baak JP. Comparison of the effect of different techniques for measurement of Ki67 proliferation on reproducibility and prognosis prediction accuracy in breast cancer. Histopathology. 2012;61(6):1134–1144. doi: 10.1111/j.1365-2559.2012.04329.x. [DOI] [PubMed] [Google Scholar]
- 12.Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, Besusparis J, Dasevicius D, Elie N, Iqbal Y, Bor C. A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast cancer research : BCR. 2014;16(2):R35. doi: 10.1186/bcr3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 14.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE, Sledge GW, Winer EP, Hudis C, Ingle JN, Perez EA, Pritchard KI, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. The lancet oncology. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature reviews Molecular cell biology. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 16.Bueno RC, Canevari RA, Villacis RA, Domingues MA, Caldeira JR, Rocha RM, Drigo SA, Rogatto SR. ATM down-regulation is associated with poor prognosis in sporadic breast carcinomas. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25(1):69–75. doi: 10.1093/annonc/mdt421. [DOI] [PubMed] [Google Scholar]
- 17.Abdel-Fatah TM, Arora A, Alsubhi N, Agarwal D, Moseley PM, Perry C, Doherty R, Chan SY, Green AR, Rakha E, Ball G, Ellis IO, Madhusudan S. Clinicopathological Significance of ATM-Chk2 Expression in Sporadic Breast Cancers: a Comprehensive Analysis in Large Cohorts. Neoplasia. 2014;16(11):982–991. doi: 10.1016/j.neo.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Li H, Dean M, Wilson HE, Kornaga E, Enwere EK, Tang P, Paterson A, Lees-Miller SP, Magliocco AM, Bebb G. Low ATM protein expression in malignant tumor as well as cancer-associated stroma are independent prognostic factors in a retrospective study of early-stage hormone-negative breast cancer. Breast cancer research : BCR. 2015;17:65. doi: 10.1186/s13058-015-0575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of NCIEWGoCD Reporting recommendations for tumor MARKer prognostic studies (REMARK) Breast cancer research and treatment. 2006;100(2):229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 20.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nature medicine. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 21.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 22.Mikami Y, Ueno T, Yoshimura K, Tsuda H, Kurosumi M, Masuda S, Horii R, Toi M, Sasano H. Interobserver concordance of Ki67 labeling index in breast cancer: Japan Breast Cancer Research Group Ki67 ring study. Cancer science. 2013;104(11):1539–1543. doi: 10.1111/cas.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciampa A, Xu B, Ayata G, Baiyee D, Wallace J, Wertheimer M, Edmiston K, Khan A. HER-2 status in breast cancer: correlation of gene amplification by FISH with immunohistochemistry expression using advanced cellular imaging system. Appl Immunohistochem Mol Morphol. 2006;14(2):132–137. doi: 10.1097/01.pai.0000150516.75567.13. [DOI] [PubMed] [Google Scholar]
- 24.Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, Duffy MJ, Jirstrom K, Gallagher WM. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res. 2008;10(5):R89. doi: 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slodkowska J, Filas V, Buszkiewicz E, Trzeciak P, Wojciechowski M, Koktysz R, Staniszewski W, Breborowicz J, Rojo MG. Study on breast carcinoma Her2/neu and hormonal receptors status assessed by automated images analysis systems: ACIS III (Dako) and ScanScope (Aperio) Folia Histochem Cytobiol. 2010;48(1):19–25. doi: 10.2478/v10042-010-0015-1. [DOI] [PubMed] [Google Scholar]
- 26.Ormenisan C, Wang J, Lawson D, Cohen C. Image cytometric HER2 in gastric carcinoma: is a new algorithm needed? Appl Immunohistochem Mol Morphol. 2013;21(5):414–419. doi: 10.1097/PAI.0b013e31827955c8. [DOI] [PubMed] [Google Scholar]
- 27.Polley MY, Freidlin B, Korn EL, Conley BA, Abrams JS, McShane LM. Statistical and practical considerations for clinical evaluation of predictive biomarkers. Journal of the National Cancer Institute. 2013;105(22):1677–1683. doi: 10.1093/jnci/djt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denkert C, Budczies J, von Minckwitz G, Wienert S, Loibl S, Klauschen F. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast. 2015;24(Suppl 2):S67–72. doi: 10.1016/j.breast.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Viale G, Giobbie-Hurder A, Regan MM, Coates AS, Mastropasqua MG, Dell’Orto P, Maiorano E, MacGrogan G, Braye SG, Ohlschlegel C, Neven P, Orosz Z, Olszewski WP, Knox F, Thurlimann B, Price KN, et al. Prognostic and predictive value of centrally reviewed Ki-67 labeling index in postmenopausal women with endocrine-responsive breast cancer: results from Breast International Group Trial 1-98 comparing adjuvant tamoxifen with letrozole. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(34):5569–5575. doi: 10.1200/JCO.2008.17.0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regan MM, Viale G, Mastropasqua MG, Maiorano E, Golouh R, Carbone A, Brown B, Suurkula M, Langman G, Mazzucchelli L, Braye S, Grigolato P, Gelber RD, Castiglione-Gertsch M, Price KN, Coates AS, et al. Re-evaluating adjuvant breast cancer trials: assessing hormone receptor status by immunohistochemical versus extraction assays. Journal of the National Cancer Institute. 2006;98(21):1571–1581. doi: 10.1093/jnci/djj415. [DOI] [PubMed] [Google Scholar]
- 31.Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, Zabaglo L, Mallon E, Green AR, Ellis IO, Howell A, Buzdar AU, Forbes JF. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(32):4273–4278. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 32.Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, Mlineritsch B, Kwasny W, Knauer M, Singer C, Jakesz R, Dubsky P, Fitzal F, Bartsch R, Steger G, Balic M, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25(2):339–345. doi: 10.1093/annonc/mdt494. [DOI] [PubMed] [Google Scholar]
- 33.Mook S, Schmidt MK, Weigelt B, Kreike B, Eekhout I, van de Vijver MJ, Glas AM, Floore A, Rutgers EJ, van ‘t Veer LJ. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2010;21(4):717–722. doi: 10.1093/annonc/mdp388. [DOI] [PubMed] [Google Scholar]
- 34.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr., Dees EC, Perez EA, Olson JA, Jr., Zujewski J, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. The New England journal of medicine. 2015;373(21):2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.A Phase III, Randomized Clinical Trial of Standard Adjuvant Endocrine Therapy +/− Chemotherapy in Patients With 1-3 Positive Nodes, Hormone Receptor-Positive and HER2-Negative Breast Cancer With Recurrence Score (RS) of 25 or Less. RxPONDER: A Clinical Trial Rx for Positive Node, Endocrine Responsive Breast Cancer. US National Library of Medicine. ClinicalTrials.gov
- 37.MINDACT (Microarray In Node-Negative and 1 to 3 Positive Lymph Node Disease May Avoid Chemotherapy): A Prospective, Randomized Study Comparing the 70-Gene Signature With the Common Clinical-Pathological Criteria in Selecting Patients for Adjuvant Chemotherapy in Breast Cancer With 0 to 3 Positive Nodes. US National Library of Medicine. ClinicalTrials.gov
- 38.Bartlett J, Canney P, Campbell A, Cameron D, Donovan J, Dunn J, Earl H, Francis A, Hall P, Harmer V, Higgins H, Hillier L, Hulme C, Hughes-Davies L, Makris A, Morgan A, et al. Selecting breast cancer patients for chemotherapy: the opening of the UK OPTIMA trial. Clinical oncology. 2013;25(2):109–116. doi: 10.1016/j.clon.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Elizabeth N, Kornaga ACK, Guggisberg Natalia, Ogilvie Travis, Morris Don G, Webster Marc, Magliocco Anthony M. A systematic comparison of three commercial estrogen receptor assays in a single clinical outcome breast cancer cohort. Submitted to Modern Pathology. 2016 doi: 10.1038/modpathol.2016.74. [DOI] [PubMed] [Google Scholar]
- 40.Magliocco A, Kornaga EN, Klimowicz AC, Sp Petrillo, Konno M, Yau A. Using automated image analysis to validate Ki67 as a clinical prognostic biomarker for estrogen-receptor positive breast cancers. 33rd San Antonio Breast Cancer Symposium (SABCS), Annual Meeting; San Antonio, Texas, USA. 2010. [Google Scholar]
- 41.Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in medicine. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.