SUMMARY
Chromosomal translocations of the mixed-lineage leukemia (MLL) gene with various partner genes result in aggressive leukemia with dismal outcomes. Despite similar expression at the mRNA level from the wild-type and chimeric MLL alleles, the chimeric protein is more stable. We report that UBE2O functions in regulating the stability of wild-type MLL in response to interleukin-1 signaling. Targeting wild-type MLL degradation impedes MLL leukemia cell proliferation and down-regulates a specific group of target genes of the MLL chimeras and their oncogenic cofactor, the Super Elongation Complex. Pharmacologically inhibiting this pathway substantially delays progression and improves survival of murine leukemia through stabilizing wild-type MLL protein, which displaces the MLL chimera from some of its target genes and therefore relieves the cellular oncogenic addiction to MLL chimeras. Stabilization of MLL provides us with a paradigm in the development of therapies for aggressive MLL leukemia and perhaps for other cancers caused by translocations.
Keywords: MLL leukemia, Wild-type MLL, MLL chimera, UBE2O, Interleukin 1, IRAK4, IRAK1, Super Elongation Complex (SEC)
Graphical Abstract
Etoc
Stabilizing wild-type MLL proteins may be a therapeutic approach for leukemia’s resulting from MLL translocations
INTRODUCTION
Mixed-lineage leukemia (MLL) including acute myeloid (AML) and acute lymphoblastic leukemia (ALL) are very aggressive blood cancers with unique clinical and biological characteristics, and are often lethal due to the development of resistance and high relapse rates with established therapies including hematopoietic stem cell transplantation (Pigneux et al., 2015; Tomizawa et al., 2007). MLL leukemia are characterized and driven by the recurrent translocations of an allele of the MLL gene (MLL, KMT2A) with a variety of other chromosomes (Mohan et al., 2010b). These MLL rearrangements predominantly occur in pediatric patients, including 69–79% of infant leukemia, and are associated with the poorest prognosis of any acute leukemia subset (Meyer et al., 2013; Mohan et al., 2010b). MLL leukemia remains a clinical challenge with an urgent need for the development of more effective targeted therapies for these aggressive cancers.
The MLL gene encodes a member of the Complex of Proteins Associated with Set1 (COMPASS) family of enzymes that are conserved from yeast to human and catalyze methylation of histone H3 lysine 4 (Miller et al., 2001; Shilatifard, 2012; Wang et al., 2009). MLL is a large protein comprised of about 4000 amino acids, but due to proteolytic processing by Taspase I, primarily exists in cells as two fragments, an N-terminal 320 KDa fragment (N320) and a C-terminal 180 KDa (C180) fragment (Yokoyama et al., 2011). MLL plays an essential and non-redundant role in development and hematopoiesis (Jones et al., 2012; Shilatifard, 2012). Homozygous Mll knockout mice are embryonic lethal, while Mll heterozygous mice are haploinsufficient with segmentation defects and hematological abnormalities (Yu et al., 1995).
The MLL gene has been characterized in translocations with over 70 different translocation partner genes that generate diverse oncogenic MLL chimeras possessing an MLL N-terminal fragment fused in-frame to a C-terminal portion of the partner (Meyer et al., 2013). Some of the most common fusion partners of MLL in leukemia are subunits of the Super Elongation Complex (SEC) (Lin et al., 2010) and/or the DOT1L complex (DotCom) (Mohan et al., 2010a; Nguyen et al., 2011). The inclusion of MLL’s chromatin-targeting domains in the MLL chimeras, together with their fusion to SEC and DotCom subunits, could result in aberrant recruitment of SEC and/or DotCom to deregulate MLL target genes, leading to increased proliferation and defects in differentiation (Mohan et al., 2010b; Shilatifard, 2012; Smith et al., 2011).
Gene disruption of the remaining wild-type copy of MLL, but not loss of histone methyltransferase activity, has been reported to jeopardize the initiation of MLL-AF9 leukemia in hematopoietic stem and progenitor cells (HSPCs) (Mishra et al., 2014; Thiel et al., 2010), consistent with the general requirement of MLL for embryonic hematopoiesis and the specification of postnatal HSPCs (Gan et al., 2010; Jude et al., 2007; McMahon et al., 2007). However, the existence of patient-derived MLL leukemia cell lines (such as ML2) with a deletion of the entire wild-type MLL locus suggests that wild-type MLL may not be required for the maintenance of MLL leukemia. Interestingly, in AML patients with partial tandem duplications of the MLL gene (MLL-PTD), wild-type MLL transcription is generally silenced and contributes to MLL-PTD mediated leukemia (Whitman et al., 2005). However, the interplay between wild-type MLL and the MLL chimeras in MLL-rearranged leukemia remain elusive given the fact that chromatin-interacting domains from the N-terminus are shared between oncogenic MLL chimeras and wild-type MLL.
Here, we found that wild-type MLL protein is much less abundant than the MLL chimeras in MLL leukemia cells. Therefore, we reasoned that the stabilization of the wild-type copy of the MLL protein could displace MLL chimeras from chromatin and therefore evade the oncogenic addition of these cells to MLL chimeras. To test this hypothesis, we established biochemical approaches and a genome-wide shRNA screen to identify factors regulating MLL protein degradation. These studies identified UBE2O and the interleukin 1 (IL-1) pathway in regulating MLL stability. Disruption of the balance between wild-type MLL and MLL chimeras through chemical inhibition of the interleukin-1 receptor-associated kinases (IRAK) impedes MLL leukemia cell proliferation both in vitro and in vivo. Together, our study reveals that targeting MLL/COMPASS degradation pathways is a promising strategy for treating the aggressive and otherwise refractory MLL leukemia, while also and providing a new paradigm in the development of therapies through protein stabilization that perhaps can be applied for the treatment of other cancers resulting from translocations.
RESULTS
UBE2O interacts with an internal region of MLL and promotes wild-type MLL degradation
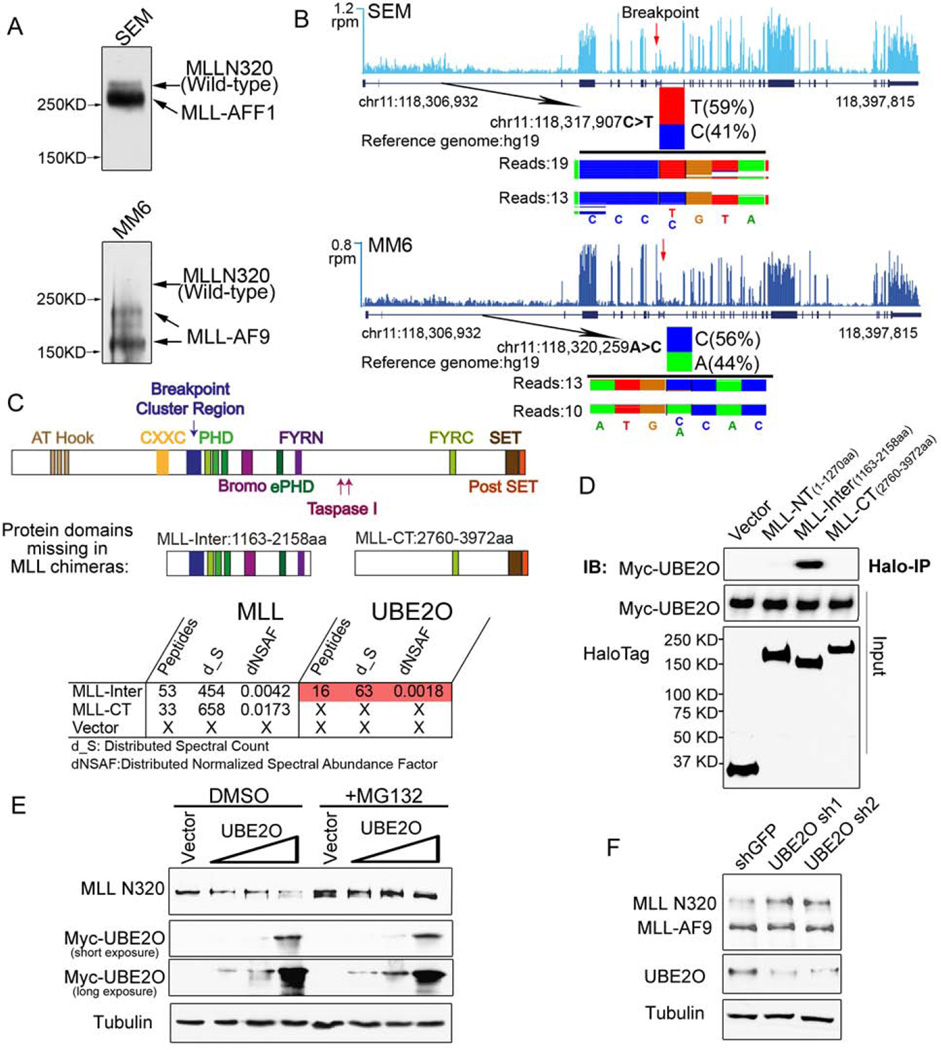
We first validated the specificity of the MLL antibodies using wild-type and MLL null mouse embryonic fibroblast (MEF) cells (Figure S1A–B and data not shown???). To determine the stability of the MLL chimera and the wild-type MLL, we analyzed MLL levels in multiple leukemia cell lines, and found that the wild-type MLL protein is much less abundant than the corresponding MLL chimera when assayed by western blotting (Figure 1A and S1C). To determine if this observation was a consequence of lower MLL mRNA levels, we used total RNA-seq with SEM (MLL-AFF1, also called MLL-AF4) and MM6 (MLL-AF9) leukemia cells to identify MLL allele-specific single-nucleotide polymorphisms (SNP) that are in regions N-terminal to the breakpoints. Each SNP has similar levels of RNA expression (Figure 1B), suggesting that wild-type MLL protein could be less stable than the MLL chimeras and that stabilization of MLL by targeting MLL degradation factors might be a potential strategy for MLL leukemia treatment.
Figure 1. UBE2O interacts with an MLL-internal region and promotes wild-type MLL protein degradation.
(A) MLL-rearranged leukemia cell lines SEM (MLL-AFF1), MM6 (MLL-AF9) have relatively low levels of wild-type MLL protein compared to the MLL chimeras. Immunoblotting of MLL N320 with the D2M7U monoclonal antibody was performed with total cell lysates of SEM and MM6 cells.
(B) Wild-type MLL and MLL fusion alleles are transcribed at similar levels in SEM and MM6 cells. Total RNA-seq was performed with SEM and MM6 cells. Genome browser tracks of MLL RNA are shown, with reads per million (r.p.m.) indicated on the y-axis. The single nucleotide polymorphisms N-terminal to the breakpoint of the MLL gene at chr11:118,317,907 (C>T) seen in SEM cells and at chr11:118,320,259 (A>C) in MM6 cells are shown below each track, with the number of sequencing reads from each allele indicated.
(C–D) Identification of UBE2O as a MLL-Inter specific interacting protein. Halo-tagged MLL-internal region (MLL-Inter) and C-terminal region (MLL-CT), both of which are missing in the MLL chimeras, were transiently transfected into HEK293 cells. MudPIT analysis identified the UBE2O as the most abundant protein specifically interacting with MLL-Inter (Table S1). Interaction between UBE2O and MLL-Inter was confirmed by co-immunoprecipitation (D).
(E) Ectopic expression of UBE2O induces MLL degradation. HEK293 cells were transfected with increasing amounts of Myc-UBE2O expression plasmid. 24 h later, cells were treated with DMSO or MG132 for 12 h.
(F) Depletion of UBE2O specifically stabilizes wild-type MLL, but not the MLL chimera MLL-AF9. UBE2O was depleted with two independent shRNAs in Flag-MLL-AF9 HEK293 cells. 4 days after infection, the MLL N320 and MLL-AF9 levels were determined with the D2M7U antibody.
See also Figures S1 and Table S1.
MLL chimeras share the same N-terminus with wild-type MLL but lose internal regions C-terminal of the breakpoint (Figure 1C). In order to identify proteins that associate with these missing regions, we expressed and purified the Halo-tagged portions of an internal region (MLL-Inter) and a C-terminal region (MLL-CT) in HEK293 cells. Multidimensional protein identification technology (MudPIT) analysis of the co-eluted proteins identified the Ubiquitin-Conjugating Enzyme E2O (UBE2O, an E2/E3 ubiquitin ligase) to be the most abundant protein specifically interacting with MLL-Inter (Table S1), which was confirmed by co-immunoprecipitation (Figure 1C–D). In agreement, UBE2O does not interact with the most common MLL chimeras (MLL-AF9, MLL-AFF1, MLL-ENL, MLL-ELL) (Figure S1D) (Lin et al., 2010). Further truncation of MLL-Inter demonstrates that the region spanning the MLL breakpoint region and the first PHD finger domain (1163–1482aa) is required and sufficient for the MLL-UBE2O interaction (Figure S1E–F and Table S1).
Ectopic expression of UBE2O in HEK293 cells induces proteasome-mediated MLL degradation in a dose-dependent manner (Figure 1E). Knockdown of UBE2O by two independent shRNAs has no effect on MLL mRNA expression (Figure S1G) but increases wild-type MLL protein levels (but not MLL-AF9 protein levels) in Flag-MLL-AF9 HEK293 cells (Figure 1F). Together, these data demonstrate that UBE2O specifically interacts with the MLL internal region and induces degradation of wild-type MLL protein.
UBE2O mediates Interleukin-1 pathway-induced MLL degradation
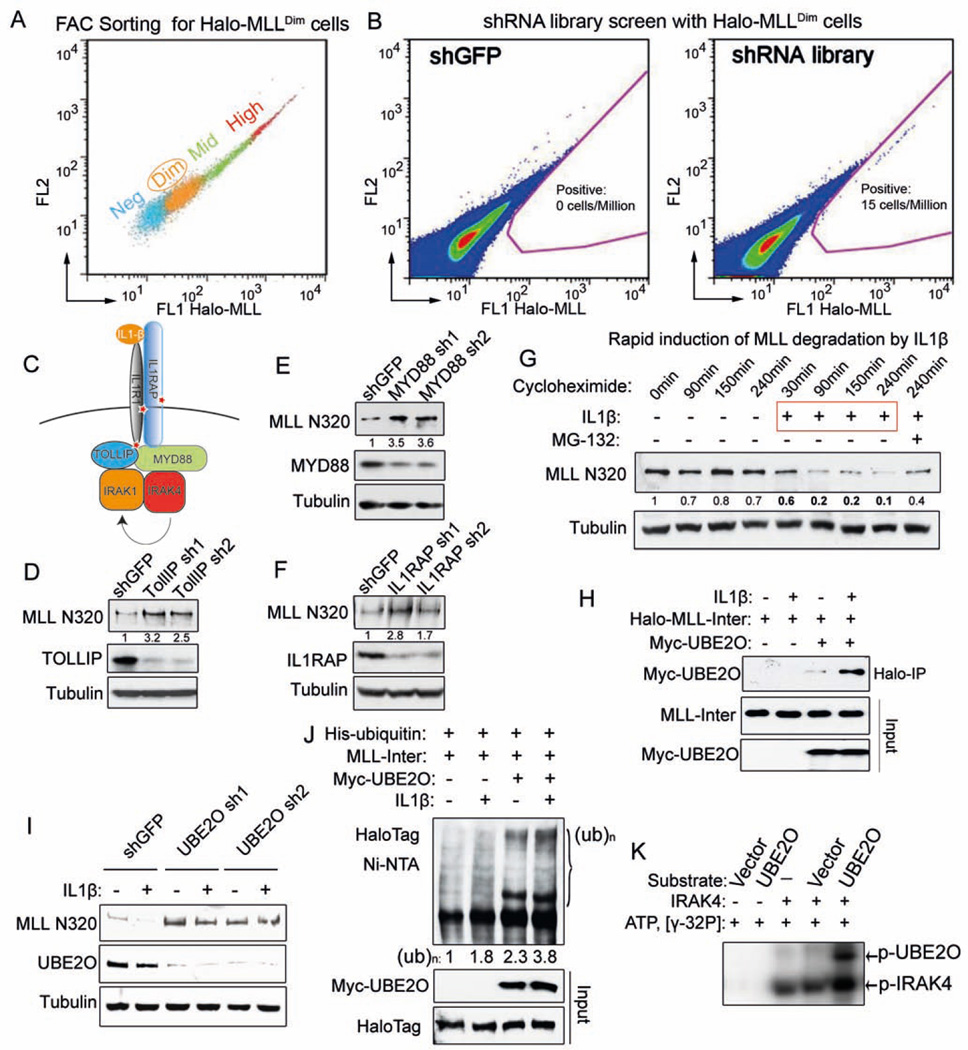
To identify the molecular pathways involved in MLL degradation, we employed a genome-wide shRNA screen. We first generated a stable cell line with random integration of Halo-MLL that can fully reconstitute the MLL complex (Figure S2A–B). Cells stained with HaloTag ligand were sorted into Halo-MLLNeg, Halo-MLLDim, Halo-MLLMid and Halo-MLLHigh (Figure 2A and S2C). The Halo-MLLDim cells were transduced with TRC lentiviral libraries and selected with puromycin for 1–2 weeks before performing flow cytometry to sort cells with increased Halo-MLL protein levels (Figure S2D). We reproducibly (4 out of 4 times) sorted a population of cells with elevated Halo-MLL signal in the shRNA library-transduced cells compared to negative control (shGFP) transduced cells (Figure 2B). Sequencing of the shRNA sequences from these sorted cells identified 303 gene targets (enriched in at least two different sortings) (Table S2).
Figure 2. Genome-wide shRNA screen identifies the IL-1 pathway in promoting MLL degradation through an MLL-UBE2O interaction.
(A) Stably-transfected, randomly integrated Halo-MLL HEK293 cells were stained with HaloTag R110 ligand and sorted by flow cytometry to get the low-expressing (Halo-MLLDim Halo-MLL cells.
(B) Representative sort for shGFP and TRC lentiviral libraries infected Halo-MLLDim cells. Halo-MLLDim cells were infected with lentiviral shRNA libraries or shGFP. Flow cytometry sorting was performed to obtain cells with increased Halo-MLL expression. Gating for cells with increased HaloTag R110 signal is indicated in pink.
(C) Identification of the IL-1 pathway in the shRNA screen. Components of the IL-1 pathway, including IL1R1, IL1RAP and TOLLIP were represented in the 303 enriched genes and are indicated with a red star (Table S2).
(D–F) Knockdown of IL-1 pathway components increases endogenous MLL protein. Knockdown of IL-1 pathway components and changes in MLL N320 protein levels were determined by western blotting. Fold changes of MLL N320 protein relative to shGFP are indicated. Tubulin serves as a loading control.
(G) IL-1β rapidly induces MLL N320 degradation in 293C6 cells, which have ectopically expressed IL-1 receptors IL1R and IL1RAP. 293C6 cells were stimulated with PBS or 50 ng/ml IL-1β for the indicated time. MG-132 was added prior to IL-1β induction. Fold changes relative to 0 minutes are indicated.
(H) IL-1β increases MLL-UBE2O interaction. Halo-MLL-Inter and Myc-UBE2O plasmids were cotransfected into 293C6 cells. 24 h later, these cells were stimulated with IL-1β for 30 min in the presence of MG-132 before MLL purification with HaloLink resin. Myc-UBE2O was detected with anti-Myc antibody and the inputs were blotted with anti-HaloTag and anti-Myc antibodies.
(I) UBE2O depletion disrupts IL-1β-induced MLL degradation. After UBE2O depletion for 4 days, 293C6 cells were stimulated with 50 ng/ml IL-1β for 90 min.
(J) IL-1β stimulates UBE2O–mediated MLL-Inter ubiquitination. His-tagged ubiquitin, Myc-UBE2O, and Halo-MLL-Inter plasmids were cotransfected into 293C6 as indicated. 24 h later, these cells were stimulated with IL-1β for 45 min in the presence of MG-132. His-tagged ubiquitinated proteins were purified with Ni-NTA agarose and blotted with anti-HaloTag antibody.
(K) IRAK4 directly phosphorylates UBE2O in vitro. 100 ng IRAK4 was incubated with eluates (purified from either vector or Flag-UBE2O transfected HEK293 cells) in the presence of γ-32P ATP. The phosphorylated IRAK4 and UBE2O proteins were visualized by autoradiography.
See also Figures S2 and Table S2.
PANTHER pathway analysis (Mi et al., 2013) of these targets showed that the interleukin 1 (IL-1) and cytokine receptor activity terms were significantly enriched (Figure S2E). In the shRNA screening, IL1R1, IL1RAP and TOLLIP were enriched targets (Figure 2C and Table S2). To determine if the IL-1 pathway regulates endogenous MLL protein, we depleted TOLLIP, MYD88 and IL1RAP within the IL1 pathway (Figure 2C shown by *) and observed increased levels of endogenous MLL protein (Figure 2E–F), with no obvious effect on MLL mRNA expression (Figure S2F).
We further stimulated HEK-293C6 cells, which ectopically express IL1R1 and IL1RAP (Lu et al., 2009), with IL-1β in the presence of the protein synthesis inhibitor cycloheximide. We found that IL-1β rapidly induces MLL degradation and increases the MLL-Inter-UBE2O interaction (Figure 2G–H). Depletion of UBE2O diminishes IL-1-induced endogenous MLL protein degradation (Figure 2I) while ectopic expression of UBE2O increases MLL-Inter ubiquitination, which could be further elevated by IL-1β stimulation (Figure 2J). Furthermore, IRAK4 could directly phosphorylate UBE2O in the in vitro kinase assay (Figure 2K), raising the possibility that phosphorylation of UBE2O by IRAK4 could be a regulatory signal for the enhanced MLL-UBE2O interaction and subsequent MLL degradation induced by IL-1β.
IRAK kinase inhibition increases the stability and chromatin occupancy of wild-type MLL
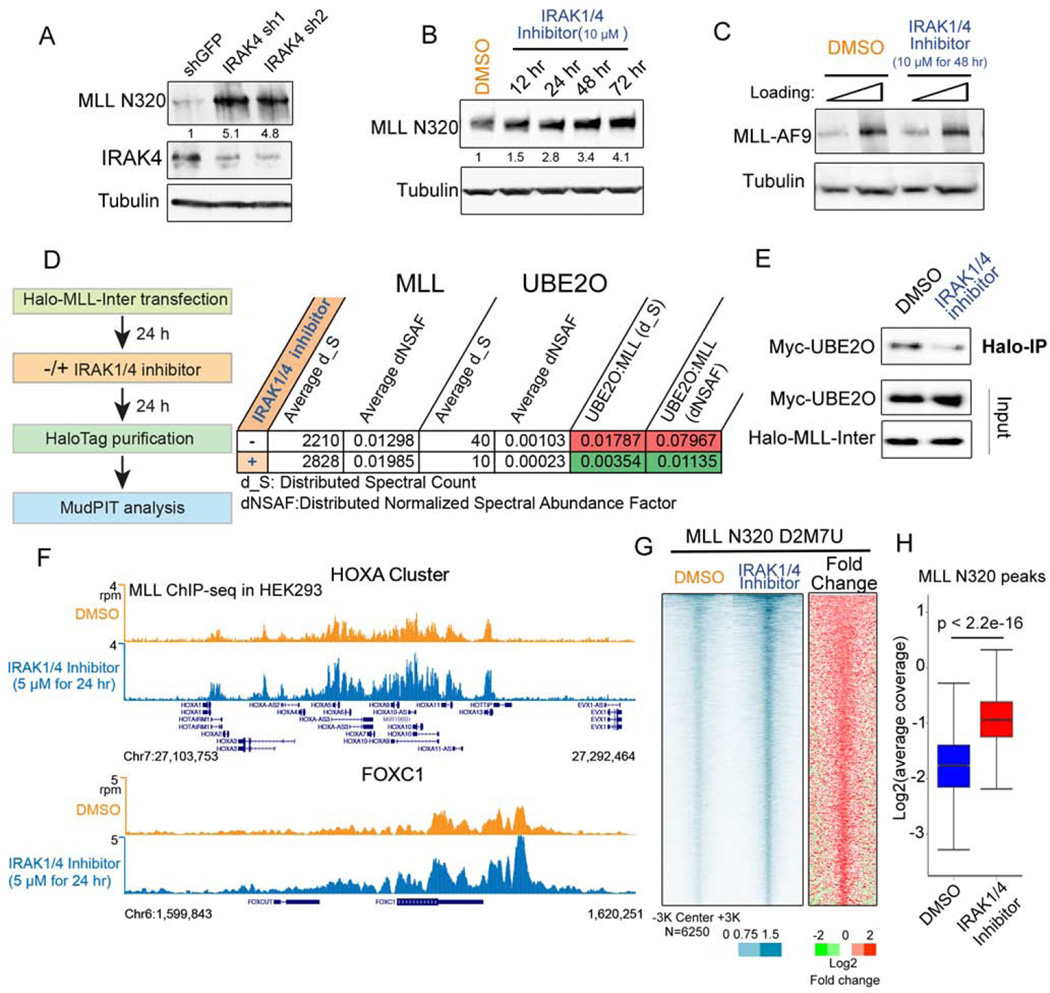
Since depleting IRAK4 protein levels can increase MLL protein levels (Figure 3A), we asked whether IRAK4’s kinase activity was required for MLL degradation. Treating cells with a small-molecule inhibitor of IRAK1/4 (IRAK1/4 inhibitor I), which has been developed to inhibit the IRAK1 and IRAK4 kinase activities (Powers et al., 2006), led to increased levels of MLL protein in a time- and dose-dependent manner (Figure 3B and S3A). A cycloheximide chase assay also demonstrates that the IRAK1/4 inhibitor increases MLL protein stability (Figure S3B). Consistent with the role of the IL-1 pathway and UBE2O in MLL degradation, IRAK inhibition does not affect MLL-AF9 and MLL-AFF1 protein levels (Figure 3C and S3C), suggesting that the IRAK kinase activity specifically signals for the wild-type MLL degradation but not for the MLL chimeras. Furthermore, we measured the MLL-UBE2O interaction with or without IRAK1/4 inhibitor treatment using MudPIT analysis and found that IRAK inhibition substantially decreased the MLL-UBE2O interaction (Figure 3D and Table S1), which was confirmed by co-immunoprecipitation (Figure 3E).
Figure 3. IRAK inhibition stabilizes MLL protein and increases genome-wide MLL occupancy.
(A) IRAK4 knockdown leads to increased levels of endogenous MLL protein in HEK293 cells. Fold changes of MLL N320 protein relative to shGFP are indicated.
(B) IRAK1/4 inhibitor stabilizes endogenous MLL protein. HEK293 cells were treated with 10 µM IRAK1/4 inhibitor for the indicated times. Fold changes of MLL N320 protein relative to vehicle (DMSO) are indicated.
(C) IRAK1/4 inhibitor has no obvious effect on MLL-AF9 stabilization. Flag-MLL-AF9 HEK293 cells were treated with 10 µM IRAK1/4 inhibitor for 2 days and subjected to western blotting with the FLAG monoclonal antibody.
(D–E) IRAK1/4 inhibitor decreases MLL-UBE2O interaction. Halo-MLL-Inter transfected HEK293 cells were treated with 10 µM IRAK1/4 inhibitor 24 h, purified with HaloLink resin and subjected to MudPIT analysis.
(F) Genome browser tracks of MLL-N320 D2M7U ChIP-seq at HOXA and FOXC1 loci after DMSO or IRAK1/4 inhibitor treatment for 24 h. IRAK1/4 inhibitor increases MLL occupancy at HOXA and HOXC loci.
(G) Heat map analysis of MLL occupancy after IRAK1/4 inhibitor treatment in HEK293 cells. Each row represents a peak of MLL occupancy (N=6250), with rows ordered by decreasing MLL occupancy in the inhibitor-treated condition.
(H) MLL occupancy is significantly increased after IRAK1/4 inhibitor treatment. Box plots depict the read coverage for all MLL peaks. The p value was calculated with the Wilcoxon signed-rank test.
See also Figures S3 and Table S1.
To investigate the consequences of MLL stabilization and its association on chromatin, we performed ChIP-seq of MLL in HEK293 cells with the presence or absence of the IRAK1/4 inhibitor. IRAK1/4 inhibitor enhances MLL occupancy at the well-characterized MLL target genes, HOXA, HOXC, and FOXC1, as revealed by two different MLL N320 antibodies (CST D2M7U and Bethyl NT86) (Figure 3F and S3D). Genome-wide analysis demonstrates that IRAK1/4 inhibition results in significant increases in MLL occupancy (Figure 3G–H and S3D–F), demonstrating that the MLL protein stabilized by IRAK inhibition can access chromatin.
Stabilization of MLL through IRAK inhibition and UBE2O depletion regulates a specific gene regulatory network in MLL leukemia
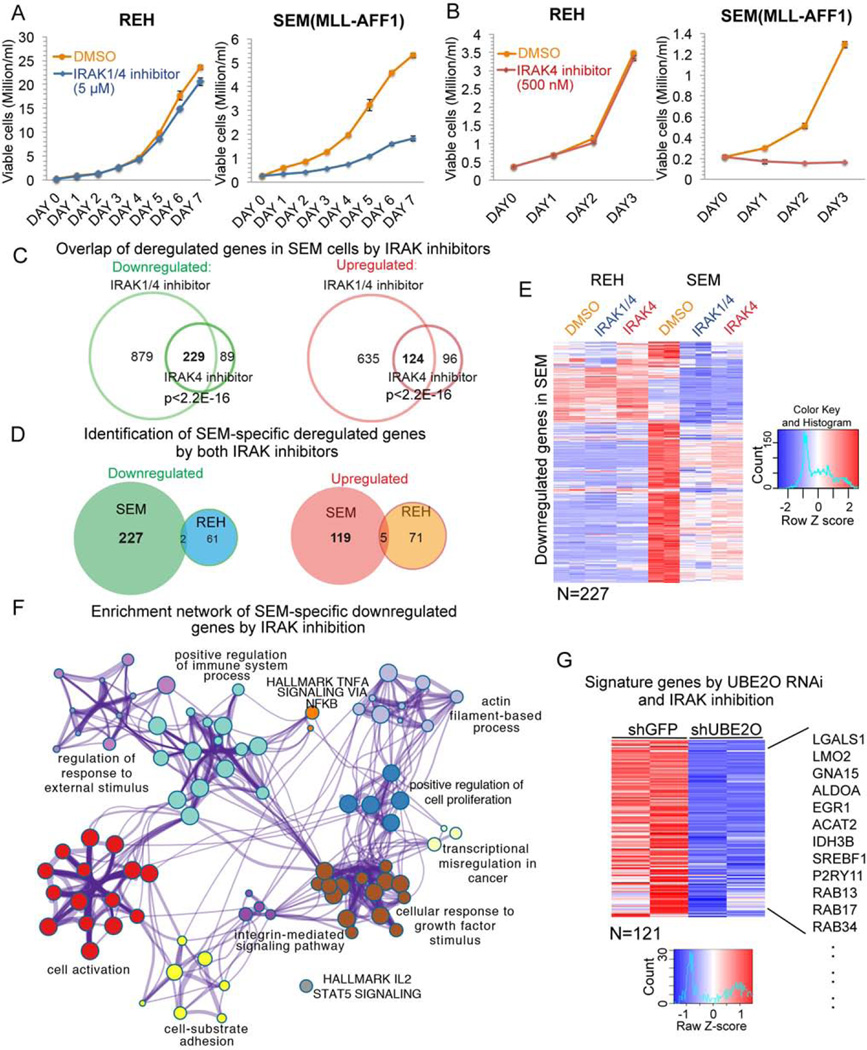
To measure the consequence of stabilizing MLL in MLL leukemia cells, we determined the effects of IRAK inhibition on cell proliferation of REH (MLL-germline leukemia) and SEM (MLL-AFF1) cells. Both of these cell lines are derived from precursor B-cell ALL patient blast cells and have therefore been studied when comparing non-MLL (REH) and MLL-dependent (SEM) leukemia gene expression (Guenther et al., 2008). Treatment with 5 µM IRAK1/4 inhibitor results in decreased proliferation of SEM cells, but not REH cells (Figure 4A). To avoid potential off-target effects of the IRAK1/4 inhibitor, we also used a second inhibitor named IRAK4 inhibitor compound 26, which was characterized as a more specific inhibitor of IRAK4 (Tumey et al., 2014). Treatment with 500 nM of this IRAK4 inhibitor led to an even greater inhibition of SEM cell proliferation and decreased cell viability with no detectable effect on the REH cells (Figure 4B).
Figure 4. Stabilization of MLL through IRAK inhibition or UBE2O depletion impedes cell proliferation and deregulates a gene regulatory network in MLL leukemia.
(A) IRAK1/4 inhibitor treatment results in slower growth of MLL leukemia SEM cells but has no effect on non-MLL leukemia REH cells. Viable cells were seeded at 0.2 million/ml, and were monitored by trypan blue exclusion staining and counted using a Vi-CELL XR cell counter. Data are represented as Mean ± SD (n=3).
(B) 500 nM IRAK4 inhibitor treatment blocks SEM cell growth, but not REH cell growth. Data are represented as Mean ± SD (n=3).
(C) Venn diagram of deregulated genes in SEM cells by IRAK1/4 and IRAK4 inhibitors. 229 genes were downregulated and 124 genes were upregulated by both inhibitors.
(D) Venn diagram of deregulated genes in SEM and REH cells by both inhibitors. Little overlap was observed between REH and SEM cells.
(E) Hierarchical clustering of 227 genes specifically downregulated in SEM cells but not REH cells by both inhibitors. Heat maps of Z score-normalized values are displayed.
(F) Network enrichment analysis by Metascape (Tripathi et al., 2015) of the 227 genes downregulated only in SEM cells (Table S3). Each cluster is represented by different colors and a circle node represents each enriched term.
(G) UBE2O depletion and IRAK inhibition affect a common subset of genes in SEM cells. 121 out of 227 genes downregulated by IRAK inhibition are also decreased after UBE2O knockdown. Some examples of common downregulated genes are indicated to the right. Heat maps represent Z score-normalized values.
See also Figures S4.
Total RNA-seq of SEM and REH cells after 2 days of IRAK1/4 or IRAK4 inhibitor treatment was performed to characterize gene expression profile changes. We found that expression of the IKK/NF-κB downstream targets HOXA9 and MEIS1 (Kuo et al., 2013) was not reduced by IRAK inhibition in SEM cells, suggesting that IKK/NF-κB signaling may not be the major target of IRAK inhibition in MLL leukemia cells. However, we found that in the presence of the IRAK1/4 inhibitor, 238 genes were downregulated and 186 upregulated genes in both REH and SEM cells (Figure S4A), with the gene ontology analysis of these genes consistent with a previous report in Myelodysplastic Syndrome (MDS) cells (Figure S4B) (Rhyasen et al., 2013). However, this set of deregulated genes would not explain the different response to IRAK inhibition by the MLL leukemic SEM cells.
To determine the set of genes specifically deregulated in SEM cells, we compared gene expression changes in SEM and REH cells with both IRAK inhibitors (Figure 4C). We found 227 downregulated and 119 upregulated genes in SEM cells but not REH cells (Figure 4D–E and S4C). Gene ontology analysis (Tripathi et al., 2015) of the SEM-specific downregulated genes shows that the cell activation, cellular response to growth factor stimulus, positive regulation of cell proliferation, and integrin-mediated signaling pathway are among the top enriched terms (Figure 4F, S4D and Table S3), while no significantly enriched terms were reported for the SEM-specific upregulated genes (Table S4).
Similar to IRAK inhibition, depletion of UBE2O led to a greater defect in SEM cell proliferation compared to REH cells (Figure S4E–F). Furthermore, ectopic expression of the MLL N-terminus (1–1250 aa), which cannot interact with UBE2O, but possesses chromatin binding domains (Figure S4G), results in a substantial reduction of SEM cell proliferation, indicating that destabilization of wild-type MLL is required for MLL leukemia cell proliferation (Figure S4H). Our RNA-seq analysis of SEM cells after UBE2O knockdown found that 121 of the 227 genes that were downregulated by the IRAK inhibitors were also decreased by UBE2O depletion (Figure 4G). These genes include genes related to cell activation (LGALS1, GNA15, ALDOA and EGR1) and cellular response to growth factor stimulus (P2RY11, SREBF1, RAB13, RAB17, and RAB34) (Figure 4G). Together, these results demonstrate that targeting MLL degradation through either IRAK inhibition or UBE2O depletion decreases cell proliferation and downregulates a specific gene regulatory network in MLL leukemia cells.
Determinants of the increased sensitivity of MLL leukemia cells to IRAK inhibition
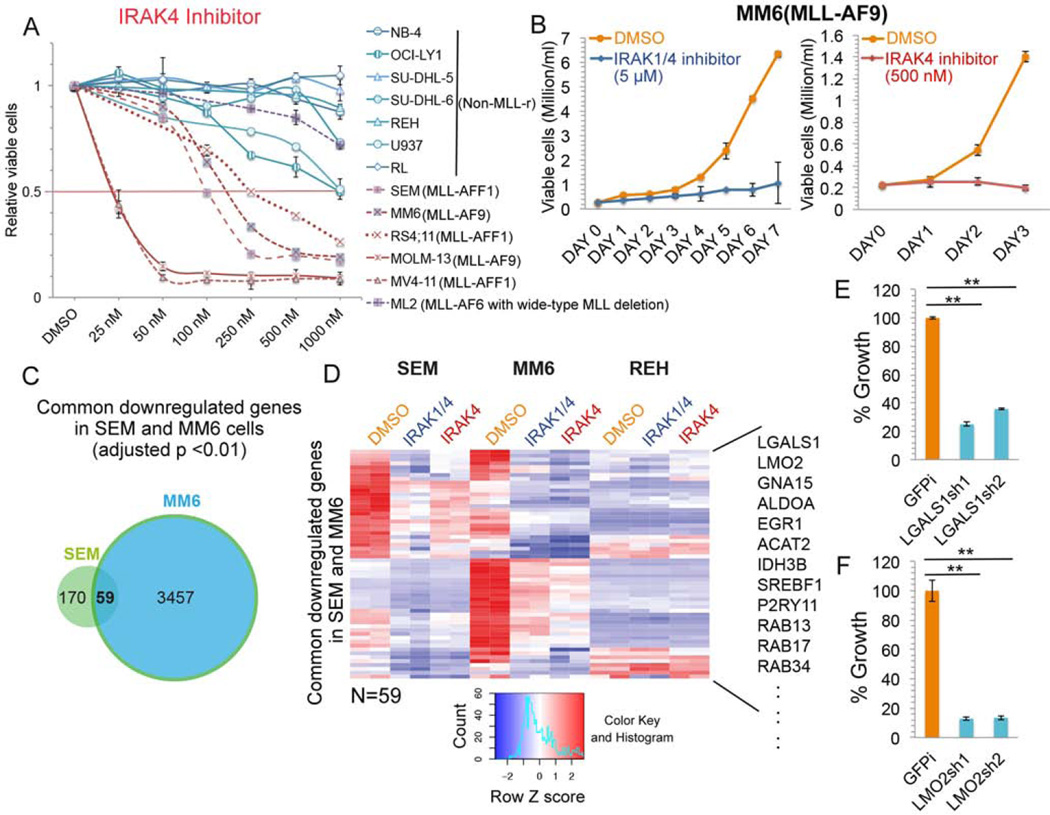
To further determine the effectiveness of IRAK inhibition for MLL leukemia cell growth, we performed dose-dependent studies with multiple patient-derived leukemia cell lines including MLL leukemia and non-MLL leukemia or lymphoma cells. We found that IRAK4 inhibitor treatment preferentially impedes cell growth of MLL-rearranged AML and ALL leukemia cells (Figure 5A), including the AML MM6 cells for which we demonstrated similar mRNA expression from the wild-type and translocated alleles (Figure 1A–B and Figure 5B). Interestingly, the MLL-AF6 positive ML2 leukemia cells, which have a deletion of the wild-type MLL allele, are not sensitive to the IRAK4 inhibitor (Figure 5A). Consistent with the lower sensitivity of ML2 leukemia cells, we found that depleting wild-type MLL in MM6 cells with a shRNA targeting the MLL C-terminus reduces the sensitivity of MM6 cells to IRAK inhibition (Figure S5A). These results suggest that wild-type MLL is required for the preferentially sensitivity of MLL leukemia cells to IRAK inhibition.
Figure 5. Determinants of the increased sensitivity of MLL leukemia cells to IRAK inhibition.
(A) IRAK4 inhibitor-specific inhibition of MLL leukemia cell proliferation. Multiple MLL leukemia and non-MLL leukemia or lymphoma cell lines were cultured with different doses of IRAK4 inhibitor for 3 days. Data are represented as Mean ± SD (n=3).
(B) Cell growth of MLL-AF9 positive AML MM6 cells is inhibited by IRAK inhibition. Data are represented as Mean ± SD (n=3).
(C) Venn diagram analysis identifies 59 common downregulated genes by both IRAK inhibitors in MLL-AFF1 SEM and MLL-AF9 MM6 cells. P value was determined with the hypergeometric test.
(D) Hierarchical clustering of the 59 common genes downregulated in SEM and MM6 cells after IRAK inhibition. Some examples of common downregulated genes are indicated to the right.
(E–F) Depletion of LGALS1 and LMO2 results in reduced growth of MM6 cells. MM6 cells were transduced with shGFP control (GFPi) or two different lentiviral shRNA constructs. After selection with puromycin for 4 days, viable cells were seeded at 0.2 million/ml and cultured for 3 more days before cell viability counting. Data represent the Mean ± SD (n=3). **, p<0.005, One-Way ANOVA.
See also Figures S5.
Comparing the differentially expressed genes in SEM and MM6 cells after IRAK inhibition, we found 59 downregulated genes and 28 upregulated genes that are shared between both cell lines (Figure 5C and S5B). These common downregulated genes contain genes related to cell activation and cellular response to growth factor stimulus (Figure 5D). Among these genes, LGALS1 and LMO2, were previously identified to be highly expressed in MLL leukemia (Armstrong et al., 2002). Depletion of LGALS1 or LMO2 also reduces MM6 cell proliferation (Figure 5E–F), indicating that IRAK inhibition could prevent MLL leukemia cell proliferation, at least partially, through downregulation of LGALS1 and LMO2 expression.
Using the UCSC cancer genome database (Children’s Oncology Group [COG], POG 9906), we found that IL-1 pathway components MYD88, IRAK1 and IRAK4 are expressed at a higher level in MLL-rearranged ALL patients than non-MLL ALL patients (Figure S5C). Furthermore, enhanced expression of Irak1 and Irak4 was found in primary and secondary leukemia in an MLL-AF9 mouse model (Figure S5D) (Liu et al., 2014). Together, these findings indicate a dependence of MLL leukemia on IL-1 signaling and suggest that targeting wild-type MLL degradation by IRAK inhibition is a potential therapeutic approach for the MLL translocation-based leukemia.
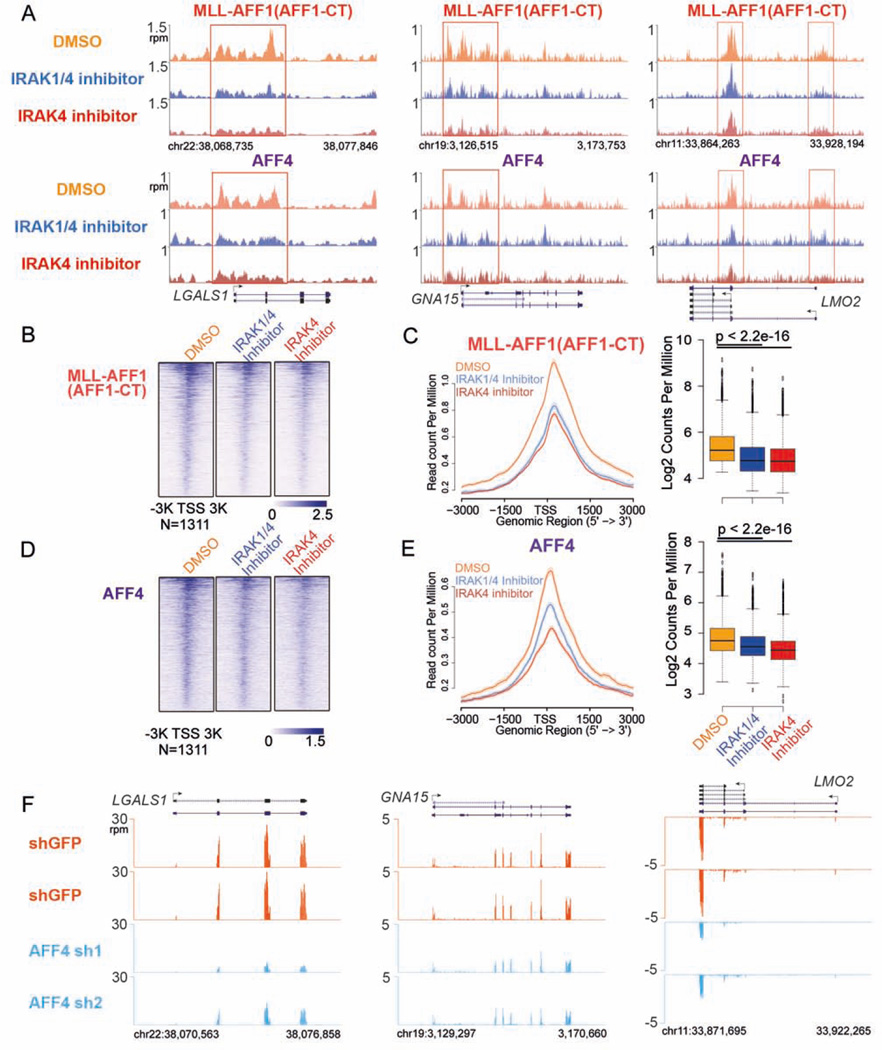
IRAK inhibition displaces the MLL chimera and subunits of SEC at a subset of target genes
As described above, IRAK inhibition significantly increases MLL chromatin occupancy and preferentially impedes MLL leukemia cell proliferation. We sought to profile the chromatin occupancy of the MLL chimera to determine the mechanistic basis of IRAK inhibition abrogating the oncogenic potential of MLL chimeras. However, our MLL antibodies recognize the MLL N-terminal epitopes shared by the wild-type MLL and the MLL chimeras. Therefore, we employed a strategy based on comparing co-occupancy of the MLL fusion partner AFF1 (AFF1 C-terminal) and the SEC subunits that are recruited by the MLL chimeras (Lin et al., 2010; Yokoyama et al., 2010). We tested this strategy in Flag-MLL-AFF1 HEK293 cells and observed increased occupancy of MLL-NT at the sites of normal MLL occupancy in HEK293 cells (Figure S6A–B). The increase of MLL-AFF1 chimera occupancy was also detected by the co-enrichment of AFF1 (AFF1-CT antibody), which is generally not enriched at most of these genes in HEK293 cells (Figure S6A–B). Furthermore, SEC subunits recruitment to these MLL-AFF1 binding sites was demonstrated by AFF4 ChIP-seq (Figure S6A–B).
MLL-AFF1 (AFF1-CT) and AFF4 ChIP-seq in SEM cells demonstrates that MLL-AFF1 and AFF4 occupancies are both reduced in the promoter-proximal regions of LGALS1, GNA15 and LMO2 genes after IRAK inhibitors treatment (Figure 6A), consistent with the downregulation of these genes by IRAK inhibition. Genome-wide analysis further reveals that MLL-AFF1 occupancy is significantly decreased (more than 25%) around the promoter-proximal regions of 1,311 genes (Figure 6B–C), while AFF4 occupancy is decreased at these regions as well (Figure 6D–E), demonstrating that IRAK inhibition displaces MLL chimeras and SEC from these chromatin regions.
Figure 6. IRAK inhibition displaces MLL chimera and SEC occupancy at a subset of MLL chimera and SEC target genes.
(A) IRAK inhibitors decrease MLL-AFF1 and SEC occupancy at the LGALS1, GNA15 and LMO2 genes in SEM cells. Genome browser views of MLL-AFF1 (AFF1-CT) and SEC component AFF4 occupancy at the LGALS1, GNA15 and LMO2 genes are shown. Red boxes indicate the promoter-proximal regions with decreased MLL-AFF1 and AFF4 occupancy.
(B–C) We identified 1311 promoter regions (± 3kb of the TSS) in which MLL-AFF1 occupancy was altered by IRAK inhibitors. These regions are plotted as heatmaps (B) and metagene plots, and the Wilcoxon signed-rank test was used to show that MLL-AFF1 occupancy is significantly decreased after IRAK inhibition (C).
(D–E) Heatmap, metagene and statistical analysis of AFF4 occupancy at the 1311 promoter regions.
(F) RNA-seq genome browser track examples indicating that AFF4 knockdown reduces the expression of the LGALS1, GNA15 and LMO2 genes in SEM cells.
See also Figures S6.
To further determine if LGALS1, LMO2 and GNA15 are direct target genes of MLL chimeras and SEC, we performed AFF4 knockdown in SEM cells (Figure S6C) and found that AFF4 depletion results in decreased mRNA expression of LGALS1, LMO2 and GNA15 genes as well (Figure 6F and S6D). These results demonstrate that IRAK inhibition displaces MLL chimeras and its oncogenic cofactor SEC at a subset of MLL chimera target genes.
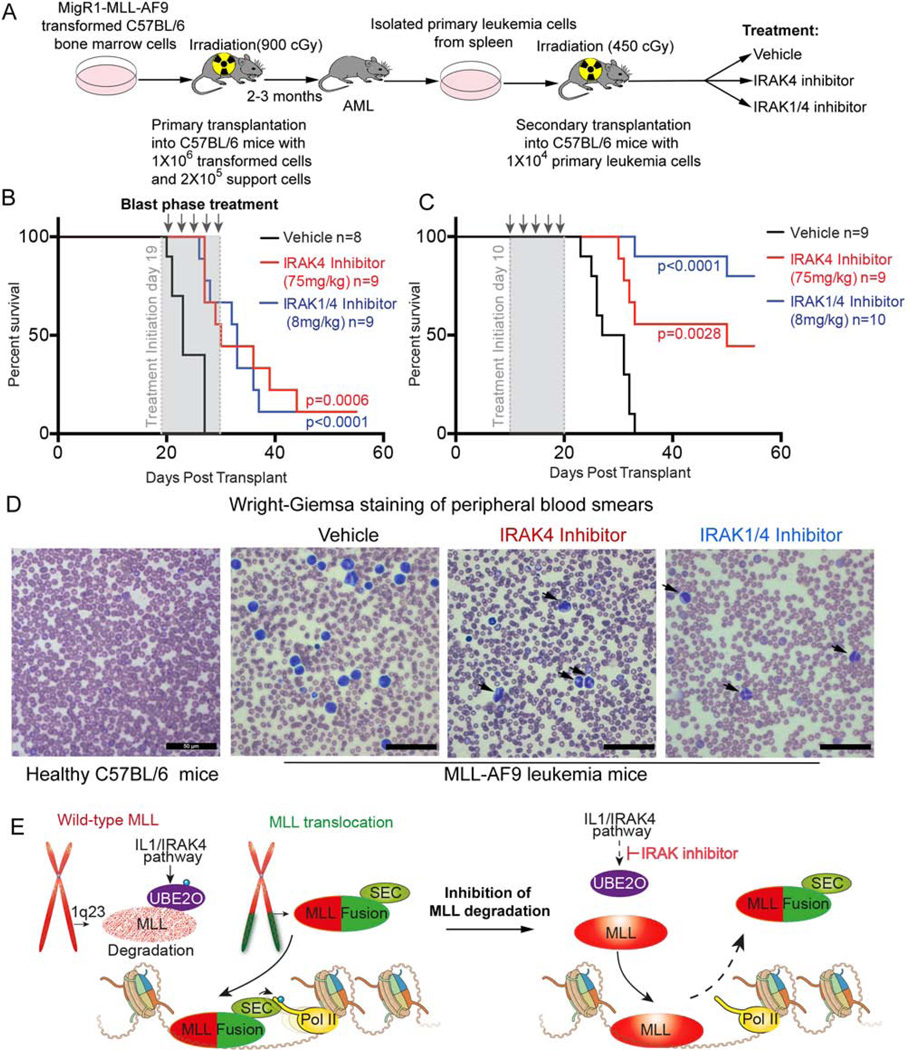
IRAK inhibitors substantially delay the progression and improve the survival of murine MLL-AF9 leukemia in vivo
We further assessed the effects of the IRAK inhibitors in vivo using the murine MLL-AF9 leukemia transplantation model (Figure 7A) (Volk et al., 2014). Primary MLL-AF9 leukemia cells are sensitive to IRAK inhibition based on the decreased colony formation and cell proliferation in vitro (Figure S7A–B). To measure the potential of IRAK inhibitors as first-line treatment, we initiated the injection of the animals with IRAK inhibitors on day 19 after transplantation, just before they succumb to leukemia. Intraperitoneal injection with IRAK1/4 inhibitor (8 mg/kg), IRAK4 inhibitor (75 mg/kg) (Tumey et al., 2014) or vehicle was performed every other day for 10 days. At sacrifice, the leukemia was confirmed by enlarged spleen, liver weights, elevated white blood cell counts (Figure S7C–E), and histological analysis. Both IRAK inhibitors significantly extended survival of the recipients beyond the 27 days when all of the vehicle treated mice succumbed to the disease, and extended the life of the AML mice to more than 55 days, with one mouse from each IRAK inhibitor-treated group still alive at day 55 (Figure 7B).
Figure 7. IRAK inhibition substantially delays disease progression and improves survival of MLL-AF9 leukemia mice.
(A) Schematic of the development of secondary murine MLL-AF9 leukemia. After transformation of MLL-AF9, c-Kit+ HPSCs were transplanted into lethally irradiated C57BL/6 mice with 1×106 transformed cells and 2×105 support cells. Leukemia cells from primary AML mice were isolated and transplanted into sublethally irradiated C57BL/6 mice. Drug treatments were started at day 10 or 19 after transplantation.
(B) Kaplan-Meier survival curves of secondary transplanted C57BL/6 mice after vehicle and IRAK1/4 or IRAK4 inhibitor treatment at day 19 (blast phase). Vehicle or IRAK inhibitors were administered every other day by intraperitoneal injection for a total of 5 treatments. Leukemia was confirmed at the endpoint for each transplant mouse. The number (n) indicates the number of mice in each group. The p values were calculated using the log rank test.
(C) Kaplan-Meier survival curves of vehicle and IRAK1/4 or IRAK4 inhibitors treated C57BL/6 mice transplanted with 1 × 104 primary MLL-AF9 leukemia cells. 10 days after transplantation, vehicle or IRAK inhibitors were administered every other day by intraperitoneal injection for a total of 5 treatments. The p values were calculated using the log rank test.
(D) Wright-Giemsa staining of peripheral blood smears from vehicle and IRAK inhibitor-treated leukemic mice at the endpoint. Arrows indicate partially differentiated MLL-AF9 blast cells after IRAK inhibition.
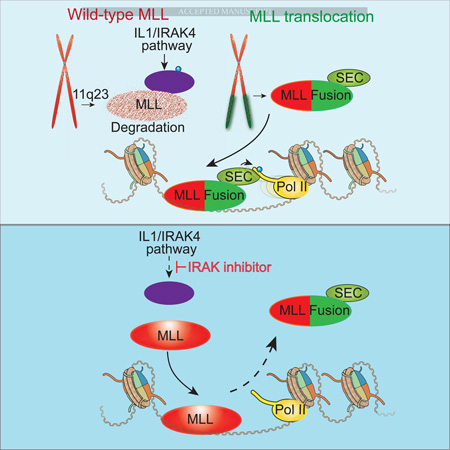
(E) In the leukemic cells, the wild-type MLL (shown in speckled red) expressed from the non-translocated chromosome (red) is less stable than the MLL chimera (shown as solid red for MLL and green for the fusion partner). Preventing wild-type MLL degradation (MLL now solid red) through inhibition of the IL1/IRAK pathway (via the IRAK inhibitor) allows wild-type MLL to compete with the MLL chimera for chromatin occupancy. The release of the MLL chimera from chromatin upon stabilization of the wild-type copy of MLL removes the oncogenic addiction of leukemic cells to MLL chimeras.
See also Figures S7.
We also treated cohorts of animals 10 days after transplantation (Figure 7C). Strikingly, 8/10 mice from the IRAK1/4 inhibitor treated group and 4/9 mice from the IRAK4 inhibitor group still did not develop MLL-AF9 leukemia as of day 55, while all of the vehicle-treated mice succumbed to the disease by 31 days after transplantation (Figure 7C). IRAK inhibitor treatment of mice led to a substantial decrease of leukemic blasts in the peripheral blood as seen by white blood cell counting (Figure S7E) or visually in blood smears from the MLL-AF9 leukemic mice at the endpoint (Figure 7D). Wright-Giemsa staining also revealed that the blasts from IRAK inhibitor treated MLL-AF9 leukemic mice are partially differentiated (Figure 7D). Together, these data suggest that pharmacologic inhibition of IRAK can delay the progression and improves survival of the aggressive MLL-AF9 leukemia in vivo.
DISCUSSION
Murine MLL chimera knock-in mice develop leukemia with a long latency, which indicates that multiple cooperating events and/or signaling pathways are required in the pathogenic process (Li and Ernst, 2014). Despite the complexity of these diseases and the remaining challenges for effective treatments, biochemical and developmental insights have led to the proposal of several therapeutic strategies, including the use of small molecules that block the Menin-MLL interaction (Borkin et al., 2015) or disrupt the chromatin binding of the bromodomain-containing protein 4 (BRD4) (Dawson et al., 2011). Inhibitors of the methyltransferase activity of DOT1L have also been tested in phase 1 clinical trials and are being explored for use in combination with other therapies (Epizyme, 2015). Recent studies (Fong et al., 2015; Rathert et al., 2015) also reported recurring development of resistance to BRD4 inhibitors.
Here, we provide a unique mechanism for the treatment of MLL-translocation based leukemia via the stabilization of the wild-type copy of MLL (Figure 7E). Through our biochemical and molecular screens, we demonstrated that the IL-1 pathway initiates the specific degradation of wild-type IRAK1/4 and UBE2O, while MLL chimeras escape UBE2O–mediated degradation for lack of the MLL-UBE2O interacting region (Figure 7E). The higher expression of IL-1 pathway components in MLL-translocated ALL leukemia patient cells, and the increased Irak1 and Irak4 expression in mouse models, correlate with the low abundance of wild-type MLL protein in MLL leukemia cells (Figure 1A–B and S1C). The IRAK1/4 inhibitor has recently been shown to sensitize a subset of MDS and T-ALL cells that exhibit high expression of IRAK1 to BCL2 inhibitor treatment, although IRAK1/4 inhibitor alone does not substantially impair these cells (Li et al., 2015; Rhyasen et al., 2013). Here, we found profound effects of IRAK inhibition alone for MLL leukemia both in vitro and in vivo. Mechanistically, we found that targeting wild-type MLL degradation through IRAK inhibition or UBE2O depletion impedes MLL leukemia cell proliferation and downregulates a common subset of MLL chimera target genes. These genes (Figure 4F) are likely to contribute to the observed antiproliferative effects in MLL leukemia cells. Indeed, knockdown of LGALS1 or LMO2 leads to decreased MLL leukemia cell proliferation (Figure 5E–F), indicating that downregulation of these genes contributes to the cell growth inhibition observed after IRAK inhibition or UBE2O knockdown. The downregulation of these genes could be explained by the decreased occupancy of the MLL chimeras and the associated SEC, a key cofactor for MLL leukemia (Smith et al., 2011). Together, these findings explain the enhanced effects of IRAK inhibition on MLL chimera-driven leukemia compared to other non-MLL rearranged leukemia and suggest that stabilization of wild-type MLL could be a potential therapeutic strategy for MLL leukemia treatment (Figure 7E).
Although a requirement for wild-type MLL in leukemogenesis has been suggested by the decreased growth of MLL leukemia cells in the presence of a small molecule that disrupts the WDR5-MLL SET domain interaction (Cao et al., 2014), a recent study demonstrated that deletion of the whole SET domain of wild-type MLL has no effect on MLL leukemogenesis, suggesting that at least the HMT activity of MLL is dispensable for MLL leukemogenesis (Mishra et al., 2014). Furthermore, ChIP-seq analysis of MLL chimera occupancy in ML2 cells, in which the entire wild-type locus is lost, demonstrated that MLL chimeras can access chromatin to mediate their oncogenic functions in the absence of wild-type MLL (Okuda et al., 2014; Wang et al., 2011). MLL fusion proteins require an open chromatin status for chromatin occupancy due to their impaired chromatin binding capability, likely due to their missing key chromatin binding modules such as PHD fingers and a bromodomain (Milne et al., 2010). These data suggest that rather than being required for the oncogenic function of MLL chimeras, wild-type MLL has the potential to outcompete the chimeras through additional chromatin binding modules. Therefore, lessening the imbalance between wild-type MLL and the more abundant oncogenic MLL chimeras could deregulate MLL chimera target gene expression and impair MLL leukemia cell proliferation.
In addition to IL-1 receptors, TOLL-like receptors can also activate the IRAK kinases, suggesting that these pathways may also have the potential to regulate MLL stability and contribute to MLL leukemia. In this study, we observed a few-fold increase of MLL occupancy after IRAK inhibition, with the MLL chimera and SEC occupancy being reduced only at a subset of genes generally associated with weak MLL chimera and SEC occupancy. Therefore, searching for additional pathways involved in regulating MLL stability could be very helpful for MLL leukemia treatment. In conclusion, our study suggests that altering the balance between wild-type MLL and MLL oncogenic fusion proteins by modulating signaling pathways is a promising approach for treating the aggressive and refractory MLL-rearranged leukemia. Furthermore, in addition to stabilization of MLL as a paradigm in the development of therapies for aggressive MLL leukemia, perhaps other cancers caused by translocations can be treated via developing similar stabilization strategies.
STAR*METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Ali Shilatifard (ASH@Northwestern.edu).
Experimental Model and Subject Details
Cell Lines
HEK293 cells, Flag-MLL-AFF1 and Flag-MLL-AF9 stable cell lines (Lin et al., 2010) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, catalog No. F6178, Sigma). The 293C6 stable cell line with the overexpression of IL1R1 and IL1RAP in HEK293 cells was a gift from George Stark’s Lab (Cleveland Clinic) and maintained in DMEM medium with 5% FBS. MONO-MAC-6 (MM6, ACC-252), RL (CRL-2261), U-937 (CRL-1593.2), RS4;11 (CRL-1873), SU-DHL-5 (CRL-2958), SU-DHL-6 (CRL-2959), MOLM13 (ACC-554), OCI-LY1 (ACC-722), THP-1 (TIB-202), ML-2 (ACC-15), NB-4 (ACC-207) and REH (ACC-22) leukemia cells were maintained in RPMI-1640 medium supplemented with 10% FBS. MV4–11 (CRL-9591) and SEM (ACC-546) leukemia cells were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM) with 10% FBS. The Mll wild-type Mll+/+ and knockout Mll−/− mouse embryonic fibroblasts (MEF) cell lines were provided by Dr. Jay Hess (University of Michigan Medical School) and cultured in DMEM with 10% FBS.
Expression Plasmids and shRNAs
Mammalian COMPASS expression constructs fused with an N-terminal HaloTag (pFENHK-Halo-MLL, pFENHK-Halo-MLL2 (KMT2B), pFENHK-Halo-SETD1A and pFENHK-Halo-MLL4 (KMT2D) plasmids) were obtained from Promega. Halo-MLL truncates were amplified by PCR with pFENHK-Halo-MLL plasmid and subcloned into the pFENHK plasmid. pCDH-CMV-MLL(1–1250)-EF1-GFP was constructed by insertion of MLL cDNA (1–1250aa) into pCDH-CMV-MCS-EF1-GreenPuro vector (SBI, Cat#: D513B-1). Myc-tagged UBE2O full-length plasmid was provided by El Bachir Affar (University of Montreal). pcDNA5-Flag-UBE2O truncate (552–1292aa) was cloned from UBE2O cDNA (CCSB Human ORFeome UBE2O Clone Without Stop Codon Accession: BC051868) (Dharmacon Inc., Clone ID: 11793) into the pcDNA5-Flag plasmid.
shRNAs for human TOLLIP (TRCN0000063693 and TRCN0000356024), IL1RAP (TRCN0000058540 and TRCN0000372626), IL1R1 (TRCN0000059260 and TRCN0000360115, MYD88 (TRCN0000008025 and TRCN0000011223) were used to knockdown the IL-1 pathway components. IRAK4 was depleted with the shRNAs: TRCN0000002064 and TRCN0000435677. LGALS1 (TRCN0000057423 and TRCN0000057424) and LMO2 (TRCN0000017128 and TRCN0000017130) were depleted with shRNAs in MM6 cells. UBE2O was depleted with shRNAs: TRCN0000004587 and TRCN0000272907. Wild-type MLL was depleted with a shRNA targeting a C-terminal region of MLL (GCCAAGCACTGTCGAAATTAC).
Murine MLL-AF9 Leukemia Model
To generate the MLL-AF9 leukemic mice, bone marrow transplantation with MigR1-MLL-AF9 was performed as previously described (Volk et al., 2014). C-Kit+ HSPCs isolated from the bone marrow of female C57BL/6 mice (8–10 weeks old) were spinoculated with MigR1-MLL-AF9 and selected with G418 for one week. Primary recipient female C57BL/6 mice (Age range from 8–10 weeks) were irradiated (900 cGy) and transplanted by tail vein intravenous injection with 1×106 MLL-AF9 transduced cells along with 2×105 wild-type support cells. The mice were monitored for signs of acute leukemia for 2–3 months and euthanized when leukemia was evident (weight loss below 16.75g, reduced mobility, malaise, palpable spleen, and hunched back). Spleens were isolated from leukemic mice and the homogenate was grown in suspension culture (RPMI-1640 supplemented with penicillin/streptomycin, 10% FBS, 100 ng/mL SCF, 50 ng/mL IL6, and 20 ng/mL IL3) for one week. 1×104 of the resulting leukemia cells were transplanted into sublethally irradiated (450 cGy) female C57BL/6 mice (8–10 weeks) via tail vein injection. Animal studies were approved by the Loyola University Chicago and Northwestern University Institutional Animal Care and Use Committees.
Method Details
HaloTag Purification and MudPIT Analysis
Plasmids encoding Halo-tagged proteins were transiently transfected into HEK293 cells. 2 days later, HEK293 cells were harvested and lysed with mammalian lysis buffer (Promega). For IRAK1/4 inhibitor treatment, 10 µM IRAK1/4 inhibitor was added 24 hours before harvest. Halo-tagged proteins were purified with HaloLink resin in the presence of Benzonase (Sigma) and eluted with TEV protease. The eluates were precipitated with TCA. After washing with acetone, the protein mixtures were digested with endoproteinase Lys-C and trypsin (Roche) and analyzed by MudPIT as previously described (Liang et al., 2015a). Original mass spectrometry data can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1090.
Generation of Halo-MLLDim HEK293 Cells
To generate Halo-tagged MLLDim cells, HEK293 cells were transfected with pFENHK-MLL plasmid with Polyethylenimine. 2 days later, the transfected cells were selected with 400 ng/ml G418 for 3 weeks and stained with HaloTag R110Direct ligand (Promega). Cells were then sorted according to the Halo-tag signals with a MoFlo sorter (Beckman Coulter) as MLLNegative, MLLDim, MLLMid, and MLLHigh. The sorted cells were grown in DMEM with 10% FBS and selected with 400 ng/ml G418. 2 weeks later, the cells were resorted according to HaloTag signals. The MLLDim were further sorted two more times.
IRAK4 Kinase Assay
HEK293 cells were transient transfected with either vector or pcDNA5-Flag-UBE2O (552–1292aa). 2 days later, Flag-UBE2O was purified from these HEK293 cells with ANTI-FLAG M2 affinity gel and eluted with FLAG peptides. Eluates from Flag-UBE2O and vector control purifications were heat-inactivated and used as substrates in radioactive kinase assays. The phosphorylation of UBE2O were measured in the presence of 100 ng His-tagged human IRAK4 (Sino Biological), 2 µCi γ-32P ATP in 20 µl kinase buffer (20 mM HEPES [pH 7.9], 8 mM MgCl2, 0.5% glycerol, 0.1% Triton X-100, 1 mM DTT). After incubation at 37 °C for 6 hours, reactions were stopped by adding 5×SDS loading buffer, and the phosphorylated proteins were visualized by SDS-PAGE and autoradiography.
Genome-wide shRNA Library Screening
A pooled TRC lentiviral library (Mi et al., 2013), which contains TRC1 81041 shRNAs and TRC1.5 17563 shRNAs, was used for lentiviral packaging with packaging vectors pΔ8.9 and pCMV-VSV-G. Lentiviral particles were harvested after 4 days and used to transduce MLLDim HEK293 cells. The lentiviral particles were titered to transduce 30%-50% of the MLLDim cells. The infected cells were further selected with 2.0 ng/ml puromycin for 1–2 weeks and stained with HaloTag R110 direct ligand (Promega). Flow cytometry sorting with MoFlo was performed to sort for cells with increased Halo-MLL protein levels (Based on the HaloTag signal intensity).
To deconvolve the shRNA composition, the sorted cells were lysed with DirectPCR Lysis Reagent (Viagen Biotech) and shRNA sequences were PCR amplified as a single mixture using vector-backbone directed universal primers from extracted genomic DNA. 4 independent cell sortings were performed representing 4 different biological replicates. The first 3 cell sortings were also amplified with a different set of primers to represent technical replicates. PCR products were cleaned up with Qiagen’s PCR purification kit and digested with Xho I. The 103 bp fragments that contain half-hairpin sequences of the shRNAs were gel purified and ligated with barcoded linkers. Illumina adapters were added to PCR products to generate libraries for next-generation sequencing.
The sequence reads were aligned to the TRC reference shRNA library using Bowtie (Langmead et al., 2009) and allowing for 2 mismatches (bowtie -p 2 -f -v 2 --best --strata -m 1). Only uniquely aligned reads were counted and ambiguous reads were excluded. The abundance of each shRNA in the sorted cells was based on the number of aligned reads. A shRNA was a “hit” in a sort if its abundance was ranked in the top 2x sorted cell number (e.g. if 400 cells were sorted, a shRNA needed to be among the 800 most abundant shRNAs). Since each gene in the shRNA library has multiple independent shRNAs, different shRNAs targeting the same gene were considered in calculating the overall enrichment of a gene. Enriched genes had shRNA hits in at least 3 of the 7 sorts to ensure that a hit occurred in at least two biological replicates.
Chromatin Immunoprecipitation Sequencing (ChIP-seq)
5×107 cells were used per ChIP assay according to a published protocol (Liang et al., 2015b). Briefly, cells were crosslinked with 1% paraformaldehyde for 15 minutes and were quenched with glycine for 5 minutes at room temperature. Fixed chromatin was sonicated with a Covaris Focused-ultrasonicator and immunoprecipitated with the indicated antibody. Libraries were prepared with the high-throughput Library preparation kit standard PCR amp module (KAPA Biosystems) for next-generation sequencing. ChIP-seq reads were aligned to the mouse (UCSC mm9) or human genome (UCSC hg19). Alignments were processed with Bowtie version 1.1.2, allowing only uniquely mapping reads with up to two mismatches within the 50 bp read. The resulting reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million, r.p.m.). For MLL ChIP-seq in 293 cells in Figure 3G–H and S3E–F, peak detection was performed with MACS (model-based analysis of ChIP-Seq) version 1.4.2 (Zhang et al., 2008) using default parameters. D2M7U and NT86 peaks that overlapped between the IRAK1/4 inhibitor treated and non-treated cells were combined and collapsed to give 6,250 MLL-occupied regions. The average coverage (calculated using r.p.m. tracks described above) across the entire region is shown in the boxplots where p values were calculated with the Wilcoxon signed-rank test. Heatmaps depict log2 fold change of coverage profiles in a 6 kb window around the merged peak center in 25 bp binned averages and sorted by total coverage in this window.
For Figure 6B–E, heatmaps and metagene plots of the genes with decreased AFF1-CT read coverage around the TSS (± 3kb) after both IRAK inhibitor treatment were plotted with ngs.plot 2.47 and ranked by read intensity. The AFF4 read coverage at these sites was plotted with the same order. In Figure S6A–B, all genes with MLL occupancy (12,300 genes) were plotted in the heatmaps ranked by read intensity. The AFF1-CT and AFF4 signals were plotted with the same order. Metagene analysis of MLL, AFF1-CT and AFF4 were calculated based on all of the genes.
Total RNA Sequencing (RNA-seq)
After IRAK inhibitors treatment for 2 days, REH, SEM and MM6 cells were collected and lysed with Trizol reagent. Total RNA was extracted from Trizol according to the manufacturer’s instructions. The RNA was treated with DNase I (NEB) and cleaned with the Qiagen RNeasy mini kit. 500 ng RNA was used for library preparation with TruSeq Stranded Total RNA with Ribo-Zero Gold kit (Illumina, RS-123–2201). The sequenced reads were aligned to the human genome (UCSC hg19) with TopHat 2.1.0 using gene annotations from Ensembl 72. Differential gene expression was performed with EdgeR (Empirical analysis of digital gene expression data in R) version 3.08 (Robinson et al., 2010). Adjusted p values were computed using the Benjamini-Hochburg method. Protein coding genes, long non-coding RNA and pseudogenes with adjusted p values less than 0.01 were used for the downstream analysis with Metascape. P values for Venn diagrams were performed with the hypergeometric test.
In vitro MLL-AF9 Assays
Primary murine MLL-AF9 cells were seeded in M3434 methylcellulose (Stemcell Technologies) at 1×103/10mm per well with indicated doses of IRAK1/4, IRAK4 inhibitor, or vehicle and incubated at 37 degrees. Colonies were enumerated after 7 days, and defined clusters with >100 cells were counted as colonies. For the liquid culture assay, primary MLL-AF9 cells were cultured in media (RPMI-1640 supplemented with penicillin/streptomycin, 10% FBS, 100 ng/mL SCF, 50 ng/mL IL6, and 20 ng/mL IL3), and stained with trypan blue. The viable cells were counted by a Vi-CELL XR cell counter (Beckman Coulter).
Animal Treatment
10 or 19 days after transplantation, mice were randomized and treated with 8 mg/kg IRAK1/4 inhibitor or 75 mg/kg IRAK4 inhibitor, or vehicle (10% DMSO and 90% PBS) every other day for 10 days. Mice were monitored for leukemia development and leukemia was verified after the mice were sacrificed upon signs of illness (weight loss below 16.75g, partial paralysis, malaise, palpable spleen, and hunched back).
Quantification and Statistical Analyses
Data are presented as Mean ± SD. The sample sizes (n) indicate the number of replicates or number of mice in each experiment and are provided in the corresponding figure legends. The peak or gene size (N) in the heatmaps indicates the number of peaks or genes included. For Figure 5E, 5F, and Figure S1G, Figure S2F, Figure S7, One-Way ANOVA tests were performed with Prism 6 (GraphPad Software, La Jolla, CA) to determine the statistical significance. P value < 0.005 (**) was considered as highly significantly different, p value < 0.05 was considered as significantly different, n.s, not significantly different, p ≥ 0.05. For Figure 3H, Figure 6C, Figure 6E, and Figure S3F, Figure S5C, the statistical significance was determined by the Wilcoxon signed-rank test using R 3.2.1 package with the p values provided in each figure. For Figure 7B and 7C, the Kaplan-Meier survival curves were plotted with GraphPad Prism 6 and the p values were calculated using the log rank test. P values for Venn diagrams in Figure 4C were calculated with the hypergeometric test in R 3.2.1. For the western blot results, representative figures of at least three biological replicates were shown. After background subtraction, the densitometric analysis of MLL bands were performed with ImageJ and normalized to the loading control Tubulin. Fold changes of MLL N320 protein were calculated based on the negative controls as indicated in the figure legends.
Data Availability
ChIP-seq and RNA-seq data have been deposited at NCBI GEO under the accession number GSE89485. The proteomics data have been deposited in the MassIVE (MSV000080298) and ProteomeXChange (PXD005233) repositories.
Supplementary Material
(A-B) Verification of the specificity of the MLL D2M7U antibody with ChIP-seq with Mll−/− and Mll+/+ MEF cells. Genome browser views of the Hoxa and Hoxc clusters are shown. ChIP-seq in the Mll wild-type MEFs with a second antibody, Bethyl NT86, is shown. ChIP-seq with Pol II N20 antibody in these MEF cells are also shown at these MLL target genes.
(C) Low abundance of wild-type MLL protein in MLL-translocated leukemia cells. MLL-rearranged leukemia cell lines RS4;11 and THP1 have relatively low levels of wild-type MLL protein compared to the MLL chimera proteins. Immunoblotting with the MLL D2M7U antibody was performed with various total leukemia cell lysates. REH and RL are leukemia cell lines that do not have an MLL translocation and serve as controls.
(D) UBE2O does not interact with the MLL N-terminal region shared with the MLL chimeras or with some of the most common MLL chimeras. Flag-MLL-NT, Flag-MLL-AF9, Flag-MLL-AFF1, Flag-MLL-ELL and Flag-MLL-ENL were purified with Anti-Flag M2 beads and subjected to MudPIT analysis (Lin et al., 2010). Distributed normalized spectral abundance factors (dNSAF) are shown. X indicates protein not found.
(E) Mapping of MLL-UBE2O interaction domains identifies a region spanning the MLL breakpoint cluster and the first PHD finger as being required for the MLL-UBE2O interaction. The MLL-internal region (MLL-Inter), comprising the breakpoint cluster region through the FYRN domain (T1), was further truncated to remove additional PHD fingers (T2 and T3). Halo constructs were transfected in HEK293 cells and purified for MudPIT analysis (Table S1).
(F) MLL-Inter (T1) was further truncated and Halo-MLL truncations were transiently co-transfected with Myc-tagged UBE2O for immunoprecipitation and western blotting with the Myc-tag antibody.
(G) Knockdown of UBE2O has no significant effect on MLL mRNA expression. Data are represented as Mean ± SD (n=3). n.s, no significant difference with the One-Way AVONA test.
(A-B) Halo-MLL can fully reconstitute MLL/COMPASS in HEK293 cells. Biochemical purification of different COMPASS family members with HaloLink resin from transiently transfected HEK293 cells. Halo-purified SETD1A, MLL1, MLL2 (KMT2B) and MLL4 (KMT2D) were subjected to SDS-PAGE, silver staining and western blotting. Non-transfected cells (HEK293) and vector only (Halo Vector) transfected cells were used as negative controls. Antibodies recognizing the common COMPASS subunit RBBP5 were used to demonstrate that core COMPASS subunits were present in all COMPASS purifications. In contrast, Menin was only found in the MLL1 and MLL2 purifications. The composition of MLL/COMPASS is also confirmed by MudPIT analysis (B).
(C) Flow chart for the generation of Halo-tagged MLLDim cells. After transient transfection of HEK293 cells with Halo-MLL plasmid, cells were selected with G418 for 3 weeks before staining with HaloTag R110 ligand for FACS sorting for low expressing (Halo-MLLDim).
(D) Workflow for pooled lentiviral shRNA screening. Halo-MLLDim cells were infected with lentiviral shRNA libraries or shGFP and selected for 1–2 weeks with puromycin. After HaloTag R110 staining, flow cytometry sorting was performed to obtain cells with increased Halo-MLL expression. shRNAs from the sorted cells were amplified for high-throughput sequencing.
(E) Pathway analysis of the enriched 303 genes (Table S2) from the shRNA library screen identifies the interleukin 1 (IL-1) and cytokine receptor activity as significantly enriched molecular function terms. Immune response and regulation are also enriched in biological process terms. Pathway analysis was performed with PANTHER and the fold enrichments and p values are shown.
(F) Depletion of IL-1 pathway components does not affect MLL mRNA levels as determined by RT-qPCR. Data are represented as Mean ± SD (n=3). n.s, no significant difference with the One-Way AVONA test.
(A) IRAK1/4 inhibitor stabilizes MLL protein from proteasomal degradation in a dose-dependent manner. HEK293 cells were first treated with different concentrations of IRAK1/4 inhibitor for 24 h followed by treatment with DMSO or the proteasome inhibitor MG132 for 12 h.
(B) IRAK1/4 inhibitor increases the stability of endogenous MLL. After DMSO or IRAK1/4 inhibitor treatment for 24 h, HEK293 cells were treated with the protein biosynthesis inhibitor cycloheximide for 5 and 10 h.
(C) IRAK1/4 inhibition does not stabilize MLL-AFF1 chimeras. Flag-MLL-AFF1 HEK293 cells were treated with the IRAK1/4 inhibitor at 10 µM for 2 days and subjected to western blotting with anti-FLAG.
(D) IRAK1/4 inhibition increases MLL occupancy as revealed by ChIP-seq. Track examples of ChIP-seq with anti-MLL N320 (Bethyl NT86) at HOXA, HOXC, and FOXC1 loci reveal increased MLL occupancy in the presence of the IRAK1/4 inhibitor.
(E-F) Heatmap of MLL occupancy in the presence of DMSO or the IRAK1/4 inhibitor. Occupancy in reads per million (r.p.m.) ± 3kb around the center of peak is shown (N=6250). Genes are ordered by MLL occupancy in the inhibitor treatment. The log 2 fold change heat map indicates a general increase in MLL occupancy after IRAK1/4 inhibitor treatment (E). The read counts of all the MLL peaks were plotted and the Wilcoxon signed-rank test was used to calculate the p value (F).
(A-B) Venn diagram and gene ontology analysis of common deregulated genes in REH and SEM cells after IRAK 1/4 inhibitor treatment. 238 genes were downregulated and 186 genes were upregulated in both REH and SEM cells.
(C) Hierarchical clustering of the 119 genes specifically upregulated in SEM cells by both the IRAK1/4 and IRAK4 inhibitor treatments.
(D) Gene ontology analysis of SEM-specific deregulated genes by both IRAK inhibitors. The enriched terms are shown with p values and FDR-adjusted q-values.
(E-F) Depletion of UBE2O preferentially inhibits the MLL leukemic SEM cell growth compared to non-MLL REH cells. REH and SEM cells were infected with UBE2O shRNA virus and selected with puromycin for 4 days. Viable cells were seeded and monitored for 4 more days. Data represent the Mean ± SD (n=3). **, p<0.005, One-Way ANOVA.
(G-H) Ectopic expression of an MLL-N-terminal region (1-1250aa) blocks MLL leukemic SEM cell proliferation. SEM cells were infected with lentivirus expressing MLL-NT (1-1250 aa). 2 days after lentivirus infection, the SEM cells were selected with puromycin for 4 days and monitored for GFP expression (G). The viable cells were seeded at day 0, and counted by trypan blue exclusion staining with a cell viability analyzer (H). Data are represented as Mean ± SD (n=3). **, p<0.005, One-Way ANOVA.
(A) Depletion of wild-type MLL reduces the sensitivity of MM6 cells to the IRAK1/4 inhibitor. MM6 cells were depleted with a shRNA targeting the C-terminus of MLL for 3 days. Viable cells were seeded at 0.2 million/ml and cultured for 4 days in the presence or absence of 5 µM IRAK1/4 inhibitor before cell counting. Data represent the Mean ± SD (n=3).
(B) Hierarchical clustering of the 28 genes that are upregulated by the IRAK1/4 and IRAK4 inhibitor treatments in SEM and MM6 cells.
(C) MYD88IRAK1 and IRAK4 have higher expression in MLL ALL patient cells compared to other ALL leukemia patient cells (TARGET ALL microarray data from Cancer UCSC genome database. Project: COG, POG 9906). P values were calculated with the Wilcoxon signed-rank test.
(D) Irak1 and Irak4 expression profiles in iPS cells, HPSC and MLL-AF9 transformed 1st and 2nd leukemia cells from a previously published study (Liu et al., 2014).
(A) MLL-AFF1 chimera shares the same chromatin binding sites with wild-type MLL and recruits SEC. Heatmap analysis of MLL N320, AFF1-CT and AFF4 occupancy in HEK293 and Flag-MLL-AFF1 HEK293 cells. In MLL-AFF1 HEK293 cells, the D2M7U antibody could detect both wild-type MLL and MLL-AFF1. Ectopically expressed MLL-AFF1 in HEK293 cells targets the same sites as wild-type MLL as revealed by increased occupancy of both MLL N320 and AFF1-CT. AFF4, a subunit of SEC, is also recruited to these MLL-AFF1 binding sites. All protein coding genes with MLL occupancy in either HEK293 or MLL-AFF1 HEK293 cells are included in the heatmap. Each row represents a gene with MLL occupancy around the TSS site (N=12300).
(B) Metagene plot of MLL, AFF1-CT and AFF4 occupancies in HEK293 and Flag-MLL-AFF1 HEK293 cells.
(C) Genome browser RNA-seq tracks at the AFF4 gene with and without AFF4 knockdown in SEM cells.
(D) Decreased expression of 127 genes in SEM cells by both IRAK inhibition and AFF4 knockdown. Some examples of downregulated genes are indicated on the right.
(A) IRAK inhibitors suppress growth of primary murine MLL-AF9 cells. Different dosages of IRAK inhibitors were added to the isolated primary MLL-AF9 leukemia cells in the presence of IL3, IL6 and SCF. Two days later, the viable leukemia cells were monitored by trypan blue exclusion. **, p<0.005 (One-Way ANOVA).
(B) IRAK inhibitors decrease the colony formation ability of primary murine MLL-AF9 leukemia cells in vitro. 1000 primary MLL-AF9 leukemia cells were seeded and treated with DMSO or IRAK inhibitors at various dosages for the methylcellulose colony formation assay. **, p<0.005 (One-Way ANOVA).
(C-D) Spleen and liver weights of vehicle and IRAK inhibitor-treated groups at sacrifice.
(E) White blood cells counts of vehicle and IRAK inhibitor-treated groups at the endpoint. **, p<0.005 (One-Way ANOVA).
HIGHLIGHTS.
UBE2O acts downstream of the Interleukin-1 pathway to regulate MLL/COMPASS stability
Stabilizing wild-type MLL protein inhibits MLL leukemia cell proliferation
UBE2O and IRAK inhibition alter a common set of MLL chimera target genes
Targeting the IL-1 pathway is a potential therapeutic strategy for MLL leukemia
Acknowledgments
We thank the Molecular Biology core facility at the Stowers Institute for creating and sequencing libraries for next-generation sequencing. We thank Dr. El Bachir Affar (University of Montreal) and Dr. George R. Stark (Cleveland Clinic) for sharing reagents. We thank Malcolm Cook for aligning shRNA library sequences, and Laura Shilatifard for editorial assistance. This work was performed to fulfill, in part, requirements for the Ph.D. thesis research of K.L. as a student with the Open University. This study was supported by the Samuel Waxman Cancer Research Foundation to J.D.C and National Institute of Health grants, CA117907 to J.M.E., CA101774 to J.D.C., CA211428 to E.R.S., and R35CA197569 to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
K.L. and A.S. designed the experiments and K. L. conducted most of the cell and biochemical experiments. J.Z. and A.G.V. generated the murine leukemia cells with assistance from J. Cannova. A.G.V performed the animal experiments. A.R.W., E.T.B. and K.L. performed bioinformatics analyses. J.S.H. performed flow cytometry sorting. J.M.G., L.F. and M.P.W. conducted the MudPIT analysis and L.F. advised on MudPIT data interpretation. S.A.M. generated shRNA plasmids and performed NGS sequencing. K.D.S. and J.M.E. amplified and pooled the shRNA libraries. K.L., E.R.S., A.G.V., A.S. and J.D.C. analyzed and interpreted results and wrote the paper with input from all authors.
REFERENCES
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53:247–261. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epizyme. Epizyme Announces Second Quarter 2015 Financial Results and Provides Corporate Update. 2015 ( https://www.sec.gov/Archives/edgar/data/1571498/000119312515279839/d23584dex991.htm)
- Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan T, Jude CD, Zaffuto K, Ernst P. Developmentally induced Mll1 loss reveals defects in postnatal haematopoiesis. Leukemia. 2010;24:1732–1741. doi: 10.1038/leu.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci U S A. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HP, Wang Z, Lee DF, Iwasaki M, Duque-Afonso J, Wong SH, Lin CH, Figueroa ME, Su J, Lemischka IR, et al. Epigenetic roles of MLL oncoproteins are dependent on NF-kappaB. Cancer Cell. 2013;24:423–437. doi: 10.1016/j.ccr.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BE, Ernst P. Two decades of leukemia oncoprotein epistasis: the MLL1 paradigm for epigenetic deregulation in leukemia. Exp Hematol. 2014;42:995–1012. doi: 10.1016/j.exphem.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Younger K, Gartenhaus R, Joseph AM, Hu F, Baer MR, Brown P, Davila E. Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J Clin Invest. 2015;125:1081–1097. doi: 10.1172/JCI75821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Gao X, Gilmore JM, Florens L, Washburn MP, Smith E, Shilatifard A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Molecular and cellular biology. 2015a;35:928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Woodfin AR, Slaughter BD, Unruh JR, Box AC, Rickels RA, Gao X, Haug JS, Jaspersen SL, Shilatifard A. Mitotic Transcriptional Activation: Clearance of Actively Engaged Pol II via Transcriptional Elongation Control in Mitosis. Mol Cell. 2015b;60:435–445. doi: 10.1016/j.molcel.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cheng H, Gao S, Lu X, He F, Hu L, Hou D, Zou Z, Li Y, Zhang H, et al. Reprogramming of MLL-AF9 leukemia cells into pluripotent stem cells. Leukemia. 2014;28:1071–1080. doi: 10.1038/leu.2013.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A. 2009;106:16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashtalir N, Daou S, Barbour H, Sen NN, Gagnon J, Hammond-Martel I, Dar HH, Therrien M, Affar el B. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014;54:392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Meyer C, Hofmann J, Burmeister T, Groger D, Park TS, Emerenciano M, Pombo de Oliveira M, Renneville A, Villarese P, Macintyre E, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27:2165–2176. doi: 10.1038/leu.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BP, Zaffuto KM, Artinger EL, Org T, Mikkola HK, Cheng C, Djabali M, Ernst P. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 2014;7:1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010a;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010b;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H, Kawaguchi M, Kanai A, Matsui H, Kawamura T, Inaba T, Kitabayashi I, Yokoyama A. MLL fusion proteins link transcriptional coactivators to previously active CpG-rich promoters. Nucleic Acids Res. 2014;42:4241–4256. doi: 10.1093/nar/gkt1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigneux A, Labopin M, Maertens J, Cordonnier C, Volin L, Socie G, Blaise D, Craddock C, Milpied N, Bacher U, et al. Outcome of allogeneic hematopoietic stem-cell transplantation for adult patients with AML and 11q23/MLL rearrangement (MLL-r AML) Leukemia. 2015;29:2375–2381. doi: 10.1038/leu.2015.143. [DOI] [PubMed] [Google Scholar]
- Powers JP, Li S, Jaen JC, Liu J, Walker NP, Wang Z, Wesche H. Discovery and initial SAR of inhibitors of interleukin-1 receptor-associated kinase-4. Bioorg Med Chem Lett. 2006;16:2842–2845. doi: 10.1016/j.bmcl.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyasen GW, Bolanos L, Fang J, Jerez A, Wunderlich M, Rigolino C, Mathews L, Ferrer M, Southall N, Guha R, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 2013;24:90–104. doi: 10.1016/j.ccr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa D, Koh K, Sato T, Kinukawa N, Morimoto A, Isoyama K, Kosaka Y, Oda T, Oda M, Hayashi Y, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21:2258–2263. doi: 10.1038/sj.leu.2404903. [DOI] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumey LN, Boschelli DH, Bhagirath N, Shim J, Murphy EA, Goodwin D, Bennett EM, Wang M, Lin LL, Press B, et al. Identification and optimization of indolo[2,3-c]quinoline inhibitors of IRAK4. Bioorg Med Chem Lett. 2014;24:2066–2072. doi: 10.1016/j.bmcl.2014.03.056. [DOI] [PubMed] [Google Scholar]
- Volk A, Li J, Xin J, You D, Zhang J, Liu X, Xiao Y, Breslin P, Li Z, Wei W, et al. Co-inhibition of NF-kappaB and JNK is synergistic in TNF-expressing human AML. J Exp Med. 2014;211:1093–1108. doi: 10.1084/jem.20130990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman SP, Liu S, Vukosavljevic T, Rush LJ, Yu L, Liu C, Klisovic MI, Maharry K, Guimond M, Strout MP, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood. 2005;106:345–352. doi: 10.1182/blood-2005-01-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Ficara F, Murphy MJ, Meisel C, Naresh A, Kitabayashi I, Cleary ML. Proteolytically cleaved MLL subunits are susceptible to distinct degradation pathways. J Cell Sci. 2011;124:2208–2219. doi: 10.1242/jcs.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-B) Verification of the specificity of the MLL D2M7U antibody with ChIP-seq with Mll−/− and Mll+/+ MEF cells. Genome browser views of the Hoxa and Hoxc clusters are shown. ChIP-seq in the Mll wild-type MEFs with a second antibody, Bethyl NT86, is shown. ChIP-seq with Pol II N20 antibody in these MEF cells are also shown at these MLL target genes.
(C) Low abundance of wild-type MLL protein in MLL-translocated leukemia cells. MLL-rearranged leukemia cell lines RS4;11 and THP1 have relatively low levels of wild-type MLL protein compared to the MLL chimera proteins. Immunoblotting with the MLL D2M7U antibody was performed with various total leukemia cell lysates. REH and RL are leukemia cell lines that do not have an MLL translocation and serve as controls.
(D) UBE2O does not interact with the MLL N-terminal region shared with the MLL chimeras or with some of the most common MLL chimeras. Flag-MLL-NT, Flag-MLL-AF9, Flag-MLL-AFF1, Flag-MLL-ELL and Flag-MLL-ENL were purified with Anti-Flag M2 beads and subjected to MudPIT analysis (Lin et al., 2010). Distributed normalized spectral abundance factors (dNSAF) are shown. X indicates protein not found.
(E) Mapping of MLL-UBE2O interaction domains identifies a region spanning the MLL breakpoint cluster and the first PHD finger as being required for the MLL-UBE2O interaction. The MLL-internal region (MLL-Inter), comprising the breakpoint cluster region through the FYRN domain (T1), was further truncated to remove additional PHD fingers (T2 and T3). Halo constructs were transfected in HEK293 cells and purified for MudPIT analysis (Table S1).
(F) MLL-Inter (T1) was further truncated and Halo-MLL truncations were transiently co-transfected with Myc-tagged UBE2O for immunoprecipitation and western blotting with the Myc-tag antibody.
(G) Knockdown of UBE2O has no significant effect on MLL mRNA expression. Data are represented as Mean ± SD (n=3). n.s, no significant difference with the One-Way AVONA test.
(A-B) Halo-MLL can fully reconstitute MLL/COMPASS in HEK293 cells. Biochemical purification of different COMPASS family members with HaloLink resin from transiently transfected HEK293 cells. Halo-purified SETD1A, MLL1, MLL2 (KMT2B) and MLL4 (KMT2D) were subjected to SDS-PAGE, silver staining and western blotting. Non-transfected cells (HEK293) and vector only (Halo Vector) transfected cells were used as negative controls. Antibodies recognizing the common COMPASS subunit RBBP5 were used to demonstrate that core COMPASS subunits were present in all COMPASS purifications. In contrast, Menin was only found in the MLL1 and MLL2 purifications. The composition of MLL/COMPASS is also confirmed by MudPIT analysis (B).
(C) Flow chart for the generation of Halo-tagged MLLDim cells. After transient transfection of HEK293 cells with Halo-MLL plasmid, cells were selected with G418 for 3 weeks before staining with HaloTag R110 ligand for FACS sorting for low expressing (Halo-MLLDim).
(D) Workflow for pooled lentiviral shRNA screening. Halo-MLLDim cells were infected with lentiviral shRNA libraries or shGFP and selected for 1–2 weeks with puromycin. After HaloTag R110 staining, flow cytometry sorting was performed to obtain cells with increased Halo-MLL expression. shRNAs from the sorted cells were amplified for high-throughput sequencing.
(E) Pathway analysis of the enriched 303 genes (Table S2) from the shRNA library screen identifies the interleukin 1 (IL-1) and cytokine receptor activity as significantly enriched molecular function terms. Immune response and regulation are also enriched in biological process terms. Pathway analysis was performed with PANTHER and the fold enrichments and p values are shown.
(F) Depletion of IL-1 pathway components does not affect MLL mRNA levels as determined by RT-qPCR. Data are represented as Mean ± SD (n=3). n.s, no significant difference with the One-Way AVONA test.
(A) IRAK1/4 inhibitor stabilizes MLL protein from proteasomal degradation in a dose-dependent manner. HEK293 cells were first treated with different concentrations of IRAK1/4 inhibitor for 24 h followed by treatment with DMSO or the proteasome inhibitor MG132 for 12 h.
(B) IRAK1/4 inhibitor increases the stability of endogenous MLL. After DMSO or IRAK1/4 inhibitor treatment for 24 h, HEK293 cells were treated with the protein biosynthesis inhibitor cycloheximide for 5 and 10 h.
(C) IRAK1/4 inhibition does not stabilize MLL-AFF1 chimeras. Flag-MLL-AFF1 HEK293 cells were treated with the IRAK1/4 inhibitor at 10 µM for 2 days and subjected to western blotting with anti-FLAG.
(D) IRAK1/4 inhibition increases MLL occupancy as revealed by ChIP-seq. Track examples of ChIP-seq with anti-MLL N320 (Bethyl NT86) at HOXA, HOXC, and FOXC1 loci reveal increased MLL occupancy in the presence of the IRAK1/4 inhibitor.
(E-F) Heatmap of MLL occupancy in the presence of DMSO or the IRAK1/4 inhibitor. Occupancy in reads per million (r.p.m.) ± 3kb around the center of peak is shown (N=6250). Genes are ordered by MLL occupancy in the inhibitor treatment. The log 2 fold change heat map indicates a general increase in MLL occupancy after IRAK1/4 inhibitor treatment (E). The read counts of all the MLL peaks were plotted and the Wilcoxon signed-rank test was used to calculate the p value (F).
(A-B) Venn diagram and gene ontology analysis of common deregulated genes in REH and SEM cells after IRAK 1/4 inhibitor treatment. 238 genes were downregulated and 186 genes were upregulated in both REH and SEM cells.
(C) Hierarchical clustering of the 119 genes specifically upregulated in SEM cells by both the IRAK1/4 and IRAK4 inhibitor treatments.
(D) Gene ontology analysis of SEM-specific deregulated genes by both IRAK inhibitors. The enriched terms are shown with p values and FDR-adjusted q-values.
(E-F) Depletion of UBE2O preferentially inhibits the MLL leukemic SEM cell growth compared to non-MLL REH cells. REH and SEM cells were infected with UBE2O shRNA virus and selected with puromycin for 4 days. Viable cells were seeded and monitored for 4 more days. Data represent the Mean ± SD (n=3). **, p<0.005, One-Way ANOVA.
(G-H) Ectopic expression of an MLL-N-terminal region (1-1250aa) blocks MLL leukemic SEM cell proliferation. SEM cells were infected with lentivirus expressing MLL-NT (1-1250 aa). 2 days after lentivirus infection, the SEM cells were selected with puromycin for 4 days and monitored for GFP expression (G). The viable cells were seeded at day 0, and counted by trypan blue exclusion staining with a cell viability analyzer (H). Data are represented as Mean ± SD (n=3). **, p<0.005, One-Way ANOVA.
(A) Depletion of wild-type MLL reduces the sensitivity of MM6 cells to the IRAK1/4 inhibitor. MM6 cells were depleted with a shRNA targeting the C-terminus of MLL for 3 days. Viable cells were seeded at 0.2 million/ml and cultured for 4 days in the presence or absence of 5 µM IRAK1/4 inhibitor before cell counting. Data represent the Mean ± SD (n=3).
(B) Hierarchical clustering of the 28 genes that are upregulated by the IRAK1/4 and IRAK4 inhibitor treatments in SEM and MM6 cells.
(C) MYD88IRAK1 and IRAK4 have higher expression in MLL ALL patient cells compared to other ALL leukemia patient cells (TARGET ALL microarray data from Cancer UCSC genome database. Project: COG, POG 9906). P values were calculated with the Wilcoxon signed-rank test.
(D) Irak1 and Irak4 expression profiles in iPS cells, HPSC and MLL-AF9 transformed 1st and 2nd leukemia cells from a previously published study (Liu et al., 2014).
(A) MLL-AFF1 chimera shares the same chromatin binding sites with wild-type MLL and recruits SEC. Heatmap analysis of MLL N320, AFF1-CT and AFF4 occupancy in HEK293 and Flag-MLL-AFF1 HEK293 cells. In MLL-AFF1 HEK293 cells, the D2M7U antibody could detect both wild-type MLL and MLL-AFF1. Ectopically expressed MLL-AFF1 in HEK293 cells targets the same sites as wild-type MLL as revealed by increased occupancy of both MLL N320 and AFF1-CT. AFF4, a subunit of SEC, is also recruited to these MLL-AFF1 binding sites. All protein coding genes with MLL occupancy in either HEK293 or MLL-AFF1 HEK293 cells are included in the heatmap. Each row represents a gene with MLL occupancy around the TSS site (N=12300).
(B) Metagene plot of MLL, AFF1-CT and AFF4 occupancies in HEK293 and Flag-MLL-AFF1 HEK293 cells.
(C) Genome browser RNA-seq tracks at the AFF4 gene with and without AFF4 knockdown in SEM cells.
(D) Decreased expression of 127 genes in SEM cells by both IRAK inhibition and AFF4 knockdown. Some examples of downregulated genes are indicated on the right.
(A) IRAK inhibitors suppress growth of primary murine MLL-AF9 cells. Different dosages of IRAK inhibitors were added to the isolated primary MLL-AF9 leukemia cells in the presence of IL3, IL6 and SCF. Two days later, the viable leukemia cells were monitored by trypan blue exclusion. **, p<0.005 (One-Way ANOVA).
(B) IRAK inhibitors decrease the colony formation ability of primary murine MLL-AF9 leukemia cells in vitro. 1000 primary MLL-AF9 leukemia cells were seeded and treated with DMSO or IRAK inhibitors at various dosages for the methylcellulose colony formation assay. **, p<0.005 (One-Way ANOVA).
(C-D) Spleen and liver weights of vehicle and IRAK inhibitor-treated groups at sacrifice.
(E) White blood cells counts of vehicle and IRAK inhibitor-treated groups at the endpoint. **, p<0.005 (One-Way ANOVA).
Data Availability Statement
ChIP-seq and RNA-seq data have been deposited at NCBI GEO under the accession number GSE89485. The proteomics data have been deposited in the MassIVE (MSV000080298) and ProteomeXChange (PXD005233) repositories.