Abstract
Recent discoveries suggest that a novel second messenger, bis-(3′→5′)-cyclic di-GMP (c-diGMP), is extensively used by bacteria to control multicellular behavior. Condensation of two GTP to the dinucleotide is catalyzed by the widely distributed diguanylate cyclase (DGC or GGDEF) domain that occurs in various combinations with sensory and/or regulatory modules. The crystal structure of the unorthodox response regulator PleD from Caulobacter crescentus, which consists of two CheY-like receiver domains and a DGC domain, has been solved in complex with the product c-diGMP. PleD forms a dimer with the CheY-like domains (the stem) mediating weak monomer–monomer interactions. The fold of the DGC domain is similar to adenylate cyclase, but the nucleotide-binding mode is substantially different. The guanine base is H-bonded to Asn-335 and Asp-344, whereas the ribosyl and α-phosphate moieties extend over the β2-β3-hairpin that carries the GGEEF signature motif. In the crystal, c-diGMP molecules are crosslinking active sites of adjacent dimers. It is inferred that, in solution, the two DGC domains of a dimer align in a two-fold symmetric way to catalyze c-diGMP synthesis. Two mutually intercalated c-diGMP molecules are found tightly bound at the stem–DGC interface. This allosteric site explains the observed noncompetitive product inhibition. We propose that product inhibition is due to domain immobilization and sets an upper limit for the concentration of this second messenger in the cell.
Keywords: allosteric regulation, signal transduction, x-ray crystallography
Cyclic nucleotides like cAMP or cGMP have been recognized as important low-molecular-weight signaling molecules. Although bacterial pathogens can interfere with cGMP signaling of their eukaryotic host cells (1), prokaryotes in general do not seem to use cGMP for signaling. In contrast, the cyclic dinucleotide bis-(3′→5′)-cyclic diGMP (c-diGMP) has been shown to regulate cell surface-associated traits and community behaviorlike biofilm formation in a number of bacterial species (2–5). The general importance of c-diGMP is underscored by the omnipresence of the diguanylate cyclase (DGC) domain (hitherto named GGDEF or DUF1) in bacterial genomes, where it occurs in various combinations with other sensory and/or regulatory modules (6, 7). Despite the wide distribution and obvious regulatory relevance of DGC proteins, structural and functional information about this class of regulators is largely missing.
In Caulobacter crescentus, pole remodeling during development is regulated by several polar sensor histidine kinases, which control the DGC activity of the response regulator PleD (8, 9). PleD is composed of a CheY-like receiver domain (D1), a CheY-like adapter domain (D2), and a DGC domain. Upon phosphorylation of D1, activated PleD sequesters to the differentiating pole, where it catalyzes the conversion of two molecules of GTP to c-diGMP (10). Thus, by coupling of activity and subcellular localization, the PleD readout is controlled in time and space. The conformational changes invoked by phosphorylation of response regulator receiver (RRR) domains have been studied thoroughly on CheY (11) and other single domain receiver proteins (for a review, see ref. 12). Little, however, is known about how these alterations are transmitted to the effector protein or domain, and only few intact multidomain response regulators have been analyzed structurally. Distinct mechanisms of activation, such as relief of active site obstruction [CheB (11, 13)], dimerization [DrrB, DrrD (14)], or both [NarL (15)], have been proposed.
Here, we report the structure of full-length PleD, which allows conclusions to be drawn regarding its mechanism of activation and product inhibition. Also, by providing a detailed view on the enzyme–product interactions, our study offers insight into the catalytic mechanism of this widespread family of regulatory proteins.
Materials and Methods
Overexpression and Purification. Full-length PleD with a C-terminal His-6 tag was overexpressed in Escherichia coli strain BL21 (DE3) pLysS. Purification was performed on a NiSO4-charged HiTrap chelating HP column (Amersham Pharmacia Biosciences) with elution at ≈200 mM imidazole. The pooled fractions were dialyzed overnight in 20 mM Tris·HCl (pH 8.0)/100 mM NaCl/1 mM DTT at 4°C. The protein solution was concentrated and, after clarification, loaded on a Superdex 200 HR 10/30 column(Amersham Pharmacia Biosciences), preequilibrated with the same buffer. PleD appeared as monomer, and fractions contributing to the peak were pooled. Selenomethionine-substituted PleD was expressed by using the metabolic inhibition pathway. The purification procedure was the same as for native PleD.
Enzymatic Assays. The assay for the measurement of the initial velocity of c-diGMP has been described (10). In short, GTP containing [α-32P]GTP (Amersham Pharmacia Biosciences; 5 nCi/microliter) was added to 50 μl of a solution containing 12.5 μg of PleD in 50 mM Tris·HCl (pH 7.8)/100 mM NaCl/10 mM MgCl2/5 mM 2-mercaptoethanol. Aliquots were taken at regular time intervals, and the product was assayed by thin-layer chromatography. For the product inhibition measurements, PleD was preincubated with c-diGMP at 25°C for 5 min. Care was taken to remove any bound c-diGMP from PleD carried over from the purification by extensive dialysis.
DGC activity was also assayed indirectly by monitoring the production of PPi by using an enzyme coupled spectrophotometric assay (16). The reaction mixture contained PleD in 20 mM Tris·HCl (pH 8.0)/100 mM NaCl/5 mM MgCl2/100 μM GTP and pyrophosphatase (500 milliunits/ml). The reaction was stopped by transferring a 100-μl reaction mixture into the phosphate assay reagent (pH ≈ 0; 1 ml final volume) containing molybdate and malachite green. The phosphate content in commercial GTP products had to be reduced from ≈5% to <0.5% by anion exchange chromatography to reduce background absorption (17).
Crystallography. Crystals were obtained at room temperature by hanging-drop vapor diffusion. For this, PleD at a nominal concentration of 200 μM = 10 mg/ml (assuming an ε280 of 9200 M–1·cm–1) in 20 mM Tris·HCl (pH 8.0)/100 mM NaCl/1 mM DTT/2 mM MgCl2/0.8 mM synthetic (10) c-diGMP was mixed with the reservoir (1.0 M glycine, pH 9.2/2% dioxane/14.5% polyethylene glycol 20 000) at a ratio of 1:1. Selenomethione-substituted crystals were obtained in the same manner, but by using a reservoir solution containing 1.0 M N-tris[hydroxymethyl]methyl-3-aminopropanesulfonic acid (TAPS) (pH 9.0), 2% dioxane, and 11% polyethylene glycol 20000. Diffraction data were collected from a single native (≈15 μm in diameter) and a single Se-Met substituted crystal (60 μm) at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland, and were processed with mosflm/scala (18). Eighteen selenium positions were identified by using shelxd (19). Phase refinement was performed by sharp/solomon (20) and was followed by two-fold averaging and phase extension by using dm (21). The model was built at the interactive graphics and refined by using refmac (18), imposing strict noncrystallographic symmetry constraints for the two copies in the asymmetric unit except for residues 117, 164, 168, and 404. The entire main chain is defined by electron density except for residues 137–146, 282–288, and the C-terminal His-tag.
Results and Discussion
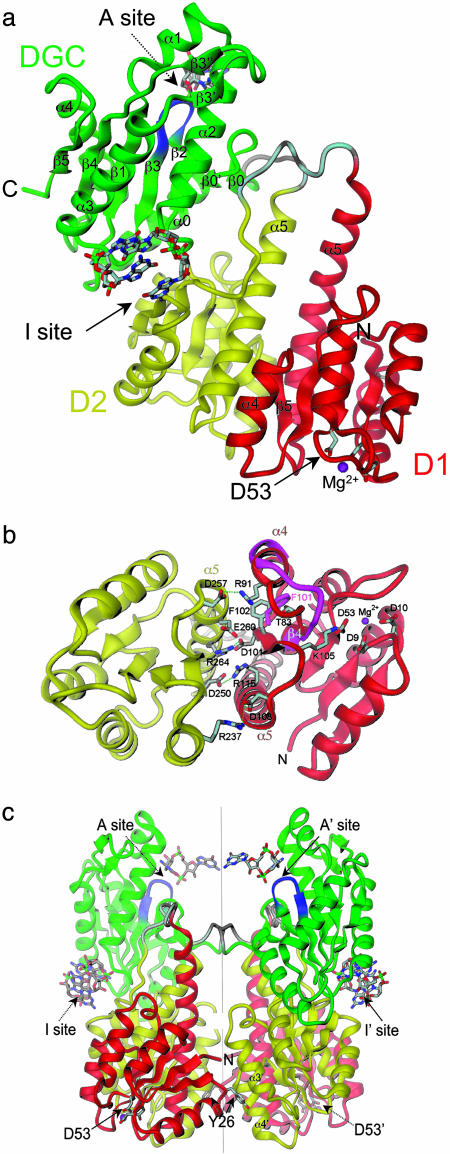
Crystal Structure. The crystal structure of nonphosphorylated PleD has been solved by multiwavelength anomalous diffraction phasing on Se-Met substituted protein to 2.7 Å (Tables 1 and 2). The structure shows a linear arrangement of three structural domains (D1, D2, and DGC; Fig. 1a) that are connected by single disordered loops. As anticipated, domains D1 and D2 show the typical RRR fold with (βα)5 topology. However, in both domains, the C-terminal helix α5 is considerably extended and protrudes from the globular domain, apparently enhancing the D1-D2 contact. Both domains closely resemble each other (rms deviation = 1.2 Å for 119 Cα). Quasiisologous contacts are formed between helix α5 of one domain and α4, β5, and α5 of the other domain. The C-terminal extensions of the α5 helices form apolar contacts, whereas their N-terminal parts are involved in several ionic interactions with α4 and β5 (Fig. 1b).
Table 1. Crystallographic data.
| SeMet
|
||||
|---|---|---|---|---|
| Native | Peak | Inflection | Remote | |
| Data collection | ||||
| Wavelength, Å | 0.97950 | 0.97950 | 0.97970 | 0.97700 |
| Resolution, Å | 2.7 | 3.2 | 3.0 | 3.0 |
| Highest shell, Å | 2.85-2.70 | 3.27-3.20 | 3.16-3.00 | 3.16-3.00 |
| Space group | P42212 | |||
| a = b, Å | 135.9 | 134.9 | ||
| c, Å | 169.2 | 168.1 | ||
| No. of unique reflections | 42,707 | 26,249 | 32,080 | 32,140 |
| Redundancy | 5.7 (5.5) | 4.1 (3.9) | 4.8 (4.9) | 4.6 (3.6) |
| I/σ | 7.2 (1.9) | 6.2 (2.1) | 7.2 (1.8) | 7.3 (1.1) |
| Completeness, % | 99.3 (98.7) | 99.9 (100) | 99.9 (100) | 99.6 (97.6) |
| Anom. completeness, % | — | 99.7 | 99.9 | 97.5 |
| Rmerge, % | 8.3 (36) | 10.5 (33) | 9.4 (39) | 11.0 (63) |
| Phasing | ||||
| Resolution shell | 13.12 7.69 | 5.98 5.06 | 4.47 3.72 | 3.46 3.16 |
| Figure of merit | 0.74 0.67 | 0.62 0.55 | 0.50 0.34 | 0.27 0.13 |
| Mean figure of merit | 0.39 | |||
Values in parentheses are for the highest-resolution shell.
Table 2. Model statistics.
| No. of molecules in a.u. | 2 |
| Resolution, Å | 50.0-2.7 |
| R/Rfree, % | 21.0/23.9 |
| rms deviation | |
| Bond lengths, Å* | 0.008 |
| Bond angles, °* | 1.2 |
| rms ΔB of bonded atoms, Å2 | |
| Main chain | 1.13 |
| Side chain | 2.42 |
| Protein atoms | 6883 |
| Water molecules | 15 |
| Ligand atoms | 233 |
| Residues in Ramachandran core, % | 92.8 |
| Residues in disallowed region, % | 0.0 |
| Average B, Å2 | 25.6 |
rms deviation from ideal stereochemistry.
Fig. 1.
Crystal structure of PleD. (a) The monomer consists of three domains connected by disordered linker peptides (light gray). Domains D1 (residues 2–140, red) and D2 (residues 141–285, yellow) show the CheY-like fold. D1 carries the phosphoacceptor D53. The catalytic DGC domain (286–454) is shown in green. The GGEEF signature motif is located on the β-hairpin (blue) and constitutes part of the active site (A-site) to which a c-diGMP molecule is bound. Two c-diGMP molecules are found at the D2/DGC interface (I-site). (b) The D1(red)/D2(yellow) interface as viewed along the quasi-two-fold axis. Compared with the view in a, the structure has been rotated by 90° approximately around the horizontal. Ionic residues in the interface and residues implicated in activation (phosphoacceptor D53, K105, T83, and F102) are shown. The trace of the β4-α4 loop and F101 of phosphorylated FixJ [magenta; PDB ID code 1d5w (23)] is shown superimposed on D1. (c) The two monomers of the asymmetric unit form a two-fold dimer. The view is related by a –60° rotation about the dimer (vertical) axis with respect to the view in a. The c-diGMP molecules that are bound to sites A and A′ crosslink to another dimer above (see Fig. 2a).
Only D1 carries all of the residues required for RRR activation (11). These are primarily aspartates D9, D10, and the phosphoacceptor D53 of the “acidic pocket”, but also residues K105, T83, and F102 that have been shown to undergo large structural changes upon activation (11). Strong density is bridging the three carboxylates of the acidic pocket and main-chain carbonyl 55. In line with its coordination geometry, this has been modeled as a Mg2+ ion. Comparison of D1 with other known RRR structures confirms that D1 is in the nonactivated conformation with the β4–α4 loop in an “outward” orientation. The structural changes accompanying activation in RRRs are known (22). In Fig. 1b, (nonactivated) PleD and activated FixJ (23) are superimposed. From this, one would predict that the largest conformational change upon PleD activation would occur in the β4–α4 loop with a concomitant shift (1–2 Å) in α4 that participates in the D1–D2 interaction. Another prominent change would be a rotameric switch of F102. Aromatic residues in this position have been shown to swing from a half-buried location into a pocket between α4 and the β-sheet that opens up on activation (11, 23). Notably and similar to CheB (24), F102 is located at a central position of the interface (Fig. 1b). This argues for a repacking of the D1–D2 interface upon phosphorylation. How this may tie in with a global model of PleD activation is discussed below.
The DGC (GGDEF) domain consists of a five-stranded central β-sheet surrounded by helices (Fig. 1a). Discounting two additional short β-hairpins (β0, β0′ and β3′, β3′′) and the N-terminal helix α0, the succession of secondary structure elements is (βααββαβαβ), for which the topology is identical and the arrangement closely similar to that of the catalytic core of adenylate cyclase (AC) (25) as well as to the “palm” domain of DNA polymerases (26) (Fig. 5, which is published as supporting information on the PNAS web site). Structural relatedness with AC has been proposed earlier by sequence threading (27). Obviously, the DGC domain is functionally related to AC and DNA polymerases in that it also catalyzes 3′–5′ phosphodiester formation. The GG(D/E)EF signature motif locates to the central β-hairpin (Fig. 1a), which, as in AC and DNA polymerases, constitutes part of the active site.
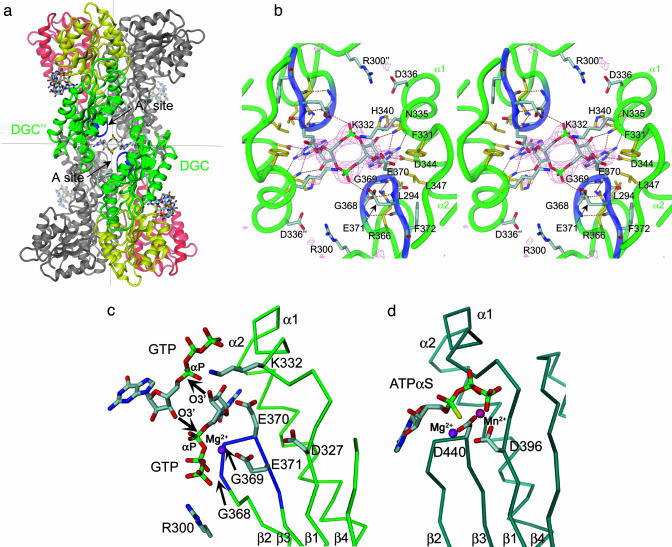
In the crystal, PleD monomers are arranged to local dimers (Fig. 1c). Isologous contacts are formed, with Y26 of D1 being tucked between helices α3 and α4 of D2 and the N terminus contacting α3. The small contact area (2 × 425 Å2) is consistent with the monomeric state of PleD in solution. Along the direction of their symmetry axis, dimers are associated head to head to form tetramers of 222 symmetry. Intriguingly, this interaction does not involve protein–protein contacts. Instead, as shown in Fig. 2a, c-diGMP is bound with its two guanine bases in a two-fold symmetric fashion to the active site formed by the adjacent monomers. This crosslinking of adjacent dimers represents a serendipitous crystallization artifact, because it provides comprehensive information about the product–enzyme interactions that appears relevant also for the situation in solution where the product would bridge the catalytic domains within a dimer (see below).
Fig. 2.
Ligand binding to the active site of PleD. (a) Dimers are packed head-to-head to form 222 tetramers. The view is rotated by 135° about the vertical axis with respect to Fig. 1c. The dimers are held together by two c-diGMP ligands that are located on a local two-fold axis of the tetramer (the viewing direction), the ligand molecule in the back has been omitted for clarity. (b) Stereographic close-up view of a. The G368GEEF signature motif comprises residues important for substrate binding (G368, G369, and E371) and catalysis (E370). The omit map of the ligand is contoured at 4 σ. The side chains of E370, E371, R300, K332, and D336 are partly disordered. (c) Complex of DGC with substrate GTP-Mg as modeled on the basis of the product complex shown in b. The positions of the guanine, ribose, and α-phosphoryl moieties are the same as in the product complex structure. The upper substrate, which has been shifted arbitrarily by ≈2 Å to the upper right, would be bound to another two-fold related DGC domain (not shown). The side-chain conformations of E370 and K332 have been adjusted to bring the functional groups into catalytically competent positions. The arrows indicate the nucleophilic attack of the 3′-oxygens on the α-phosphates. (d) AC in complex with substrate analog ATP-α-S [light-blue, PDB ID code 1cjk (25)], view corresponding to that in c. The Cα-trace corresponds to that part of the α-chain, which is structurally homologous to DGC. The β-chain, which provides specific interactions with the adenine base, has been omitted for clarity.
Active Site and Catalytic Mechanism. Fig. 2b shows that the guanine base is inserted in a pocket formed mainly by helices α1 and α2 of DGC. All of its polar atoms except O6 make specific interactions, explaining the observed GTP versus ATP discrimination (10). The N335 side chain forms H-bonds with N3 and N2, whereas N1 is recognized by the D344 carboxylate. Both residues are highly conserved among DGC domains. Furthermore, the base abuts the apolar side chains of L294, F331, and L347. The ribosyl and the α-phosphate are extending over the turn of the β2-β3-hairpin, with G369 being completely covered, explaining glycine conservation at this position. The 2′-hydroxyl is H-bonded to N335, and a phosphate nonester oxygen is in the vicinity of the main-chain amide 369.
On the basis of the observed complex structure, extrapolations regarding the binding mode of the GTP substrate to an individual DGC domain as well as the catalytic mechanism can be proposed (Fig. 2c). Considering the specific interactions with the protein, it is most likely that the guanyl base does not change its position during catalysis. Furthermore, we assume that the ribosyl and α-phosphate moieties largely keep their positions. In this case, the β- and γ-phosphates of the substrate would be easily accommodated close to G368 of the β-hairpin and would probably be bound via a Mg2+ ion to E371, with R300 conferring further stabilization. For catalysis to occur, two substrate-loaded DGC domains have to arrange themselves in a similar way like the interdimer crosslink in the crystal (Fig. 2 b and c). This might be facilitated by charge complementarity between D336 and R300 (Fig. 2b). Subsequently, the 3′-OH group of the GTP substrate has to be deprotonated to allow an intermolecular nucleophilic attack onto the α-phosphate. E370 appears well poised for acting as a general base, whereas K332 would stabilize the charge of the developing pentacoordinated phosphoryl transition state and the pyrophosphate leaving group. All of the residues proposed to be of functional importance are highly conserved among DGC sequences.
Intriguingly, the proposed nucleotide-binding mode is different from that observed in AC (Fig. 2d) and DNA polymerases. In both enzymes, the phosphate-binding site is provided by the short P-loop between β1 and α1. In AC, the base is lying on the β-hairpin and is recognized by residues of the second subunit of the heterodimer, whereas in DNA polymerases the base adopts even another position, which allows base pairing with the template strand (28). Is it conceivable that DGC can bind the substrate in a similar way as AC? This appears unlikely, because there would be no interactions with the base conferring specificity and contributing to binding affinity. An alternative model with the base bound to the DGC guanine-binding pocket and the terminal phosphates bound to the P-loop appears sterically unfeasible. Thus, there seems to be no reasonable alternative to the proposed model of the substrate–DGC complex, which nevertheless has to be confirmed experimentally. In AC, a magnesium ion coordinated by two aspartates (D396 and D440; Fig. 2d) has been proposed to assist in the nucleophilic attack of the 3′-OH group onto the α-phosphate (25). Conspicuously, the DGC domain exhibits two invariant acidic residues (D327, E370) at very similar positions, but there is no indication for a bound metal, which is in line with the disorder of E370. In fact, a magnesium ion coordinated by these residues would be too distant from the substrate to be effective. E370 is proposed to act as a general base (see above), but the role of D327, if any, remains to be elucidated.
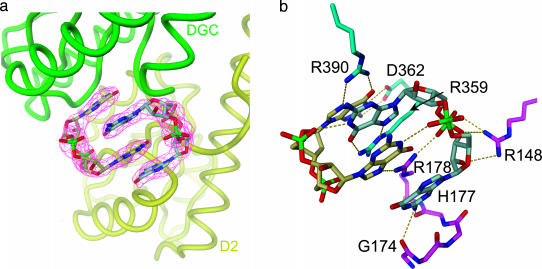
Allosteric Site and Product Inhibition. Unexpectedly, the crystal structure shows two product molecules bound to the D2/DGC interface (I-site; Figs. 1 and 3). The dimeric ligand is of compact roundish shape with mutually intercalated purine bases. Each of the two central guanyl moieties forms an intermolecular H-bond with a phosphate group. A similar structure has been observed for several crystal forms of c-diGMP (29, 30) and may be present also in solution. The ligand is bound to both D2 and DGC by a multitude of specific interactions. Hereby the common base–arginine pairing motif involving O6 and N7 of the guanyl (31) occurs three times (with R390, R359, and R178; Fig. 3b). Tight binding is consistent with the observed copurification of c-diGMP during protein preparation. Intriguingly, kinetic data reveal strong product inhibition, with a Ki (0.5 μM; Fig. 6, which is published as supporting information on the PNAS web site) about an order of magnitude lower than estimated for the cellular concentration of c-diGMP (32). Clearly, inhibition is noncompetitive (Fig. 6), i.e., independent of substrate concentration, and can thus be attributed to an allosteric effect of I-site binding.
Fig. 3.
Product binding to the allosteric inhibitory I-site. (a) A close-up view of the two mutually intercalated c-diGMP molecules (khaki and gray carbon atoms) bound at the D2 (yellow)–DGC (green) interface. The omit map of the ligand is contoured at 4 σ.(b) The ligand is tightly bound to both domains [carbons are colored in magenta (D2) and cyan (DGC)] by a multitude of specific interactions, including a recurrent arginine–guanine-binding motif. Figures were generated by dino (A. Philippsen, www.dino3d.org).
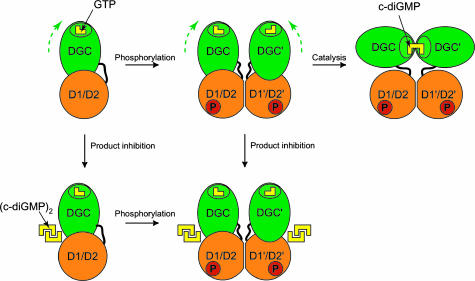
Mechanistic Model of Regulation. PleD catalyzes a condensation reaction of two identical substrates (that are bound to two identical DGC domains) to yield a C2-symmetric product molecule. In solution, nonactivated PleD appears to be monomeric, as determined by analytical ultracentrifugation and gel filtration (not shown). For the enzyme to be efficient, dimerization appears necessary, because a reaction catalyzed by monomeric PleD would be severely limited by the macromolecular diffusion rate. Based on this reasoning, a simple mechanistic model of activation and product inhibition can be proposed (Fig. 4). Phosphorylation at D53 of D1 would induce repacking of the D1/D2 interface and reorientation of D1 with respect to D2. This in turn will enhance dimer formation mediated by isologous D1-D2 contacts across the interface. In fact, the dimeric structure of nonactivated PleD in the crystal may well resemble the active dimer, although with a suboptimal interface. This view is supported by the lack of activity of N-terminally His-tagged PleD (data not shown), which would be due to interference of the tag with the interface (Fig. 1c). The low constitutive activity observed in vitro (10) would thus be due to a small fraction of nondissociated dimers. Consistent with this, the specific activity shows a strong dependence on the enzyme concentration (unpublished data).
Fig. 4.
Mechanistic model of PleD regulation. The catalytic DGC domain (green) is tethered via a flexible linker peptide to the D1/D2 stem. The DGC domain is postulated to be mobile with respect to the stem, as indicated by the curved broken arrow. (Upper) PleD is activated by phosphorylation at the D1 domain, which induces dimerization mediated by the stems and allows the two substrate-binding sites (with bound GTP substrate in yellow) to approach each other and the condensation reaction (2 GTP → c-diGMP + 2 PPi) to occur. (Lower) Allosteric product inhibition occurs by binding of (c-diGMP)2 to the I-site at the stem–DGC interface, whereby the DGC domain is immobilized with respect to the stem and barred from approaching its counterpart in the dimer.
The large distance between the substrate-binding sites of the crystal dimer (Fig. 1c) would not permit catalysis. However, allowing for flexibility of the DGC domains relative to the stem (a realistic assumption considering the small D2/DGC interface area of 690 Å2), a “closed” dimer conformation competent for catalysis can be readily modeled (schematically shown in Fig. 4), in which a complete two-fold symmetric active site is formed as observed for the DGC/DGC′′ pair (Fig. 2b). In the context of this activation-by-dimerization model, the observed allosteric product inhibition can easily be explained (Fig. 4). Stabilization of the D2/DGC interface by product binding to the I-site would simply prevent encounter of the substrate-binding sites (inhibition by domain immobilization).
The tight feedback regulation of PleD by product inhibition probably demonstrates the importance of imposing an upper limit on the concentration of the second messenger c-diGMP. It remains to be shown that, as predicted by the model, inhibition indeed constitutes an overriding principle, i.e., that inhibition is independent of the phosphorylation state of the enzyme. The structure of PleD provides insight into the molecular interactions with c-diGMP that may recur in the recognition of this ligand by other proteins. The quest for downstream targets of this messenger that appears to be ubiquitous in prokaryotes (6) has started.
Supplementary Material
Acknowledgments
We thank T. Bickle, Z. Markovic-Housley, O. Mayans, and C. Peneff for critical reading of the manuscript; J. Saito for crystallization optimization; and D.V. Palanivelu and the staff of beamline X06SA of the Swiss Light Source synchrotron (Villigen, Switzerland) for assistance with data acquisition. The work was supported by Swiss National Science Foundation Grant 31-59050.99 (to U.J.).
Author contributions: B.G., U.J., and T.S. designed research; C.C., R.P., D.S., N.C.A., and T.S. performed research; D.S. and N.C.A. contributed new reagents/analytic tools; C.C., R.P., U.J., and T.S. analyzed data; and C.C., U.J., and T.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: c-diGMP, bis-(3′→5′)-cyclic di-GMP; DGC, diguanylate cyclase; RRR, response regulator receiver; AC, adenylate cyclase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1W25).
References
- 1.Uzzau, S. & Fasano, A. (2000) Cell Microbiol. 2, 83–89. [DOI] [PubMed] [Google Scholar]
- 2.Ross, P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., Braun, S., De Vroom, E., Van der Marel, G. A., Van Boom, J. H., et al. (1987) Nature 325, 279–281. [DOI] [PubMed] [Google Scholar]
- 3.Jenal, U. (2004) Curr. Opin. Microbiol. 7, 185–191. [DOI] [PubMed] [Google Scholar]
- 4.Simm, R., Morr, M., Kader, A., Nimtz, M. & Romling, U. (2004) Mol. Microbiol. 53, 1123–1134. [DOI] [PubMed] [Google Scholar]
- 5.Tischler, A. D. & Camilli, A. (2004) Mol. Microbiol. 53, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galperin, M. Y. (2004) Environ. Microbiol. 6, 552–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galperin, M. Y., Nikolskaya, A. N. & Koonin, E. V. (2001) FEMS Microbiol. Lett. 203, 11–21. [DOI] [PubMed] [Google Scholar]
- 8.Hecht, G. B. & Newton, A. (1995) J. Bacteriol. 177, 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldridge, P., Paul, R., Goymer, P., Rainey, P. & Jenal, U. (2003) Mol. Microbiol. 47, 1695–1708. [DOI] [PubMed] [Google Scholar]
- 10.Paul, R., Weiser, S., Amiot, N. C., Chan, C., Schirmer, T., Giese, B. & Jenal, U. (2004) Genes Dev. 18, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. Y., Cho, H. S., Pelton, J. G., Yan, D., Berry, E. A. & Wemmer, D. E. (2001) J. Biol. Chem. 276, 16425–16431. [DOI] [PubMed] [Google Scholar]
- 12.Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000) Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed] [Google Scholar]
- 13.Djordjevic, S. & Stock, A. M. (1998) J. Struct. Biol. 124, 189–200. [DOI] [PubMed] [Google Scholar]
- 14.Robinson, V. L., Wu, T. & Stock, A. M. (2003) J. Bacteriol. 185, 4186–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maris, A. E., Sawaya, M. R., Kaczor-Grzeskowiak, M., Jarvis, M. R., Bearson, S. M., Kopka, M. L., Schroder, I., Gunsalus, R. P. & Dickerson, R. E. (2002) Nat. Struct. Biol 9, 771–778. [DOI] [PubMed] [Google Scholar]
- 16.Baykov, A. A., Evtushenko, O. A. & Avaeva, S. M. (1988) Anal. Biochem. 171, 266–270. [DOI] [PubMed] [Google Scholar]
- 17.Birnbaumer, L., Torres, H. N., Flawia, M. M. & Fricke, R. F. (1979) Anal. Biochem. 93, 124–133. [PubMed] [Google Scholar]
- 18.CCP4 (1994) Acta Crystallogr. D 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 19.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 20.De La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–493. [DOI] [PubMed] [Google Scholar]
- 21.Cowtan, K. D. & Zhang, K. Y. (1999) Prog. Biophys. Mol. Biol. 72, 245–270. [DOI] [PubMed] [Google Scholar]
- 22.Robinson, V. L., Buckler, D. R. & Stock, A. M. (2000) Nat. Struct. Biol 7, 626–633. [DOI] [PubMed] [Google Scholar]
- 23.Birck, C., Mourey, L., Gouet, P., Fabry, B., Schumacher, J., Rousseau, P., Kahn, D. & Samama, J. P. (1999) Struct. Folding Des. 7, 1505–1515. [DOI] [PubMed] [Google Scholar]
- 24.Djordjevic, S., Goudreau, P. N., Xu, Q., Stock, A. M. & West, A. H. (1998) Proc. Natl. Acad. Sci. USA 95, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tesmer, J. J., Sunahara, R. K., Johnson, R. A., Gosselin, G., Gilman, A. G. & Sprang, S. R. (1999) Science 285, 756–760. [DOI] [PubMed] [Google Scholar]
- 26.Doublie, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. (1998) Nature 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 27.Pei, J. & Grishin, N. V. (2001) Proteins 42, 210–216. [DOI] [PubMed] [Google Scholar]
- 28.Tesmer, J. J. & Sprang, S. R. (1998) Curr. Opin. Struct. Biol. 8, 713–719. [DOI] [PubMed] [Google Scholar]
- 29.Liaw, Y. C., Gao, Y. G., Robinson, H., Sheldrick, G. M., Sliedregt, L. A., van der Marel, G. A., van Boom, J. H. & Wang, A. H. (1990) FEBS Lett. 264, 223–227. [DOI] [PubMed] [Google Scholar]
- 30.Egli, M., Gessner, R. V., Williams, L. D., Quigley, G. J., van der Marel, G. A., van Boom, J. H., Rich, A. & Frederick, C. A. (1990) Proc. Natl. Acad. Sci. USA 87, 3235–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luscombe, N. M., Laskowski, R. A. & Thornton, J. M. (2001) Nucleic Acids Res. 29, 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, P., Mayer, R. & Benziman, M. (1991) Microbiol. Rev. 55, 35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.