Cytoskeletal motor proteins are ATPases that use the energy released from ATP hydrolysis to move along the cytoskeletal elements of microtubules and actin microfilaments. Found among all eukaryotic organisms, kinesins are microtubule-based motor proteins with a conserved kinesin motor domain, and myosins are actin microfilament-based motor proteins with a conserved myosin motor domain. Cytoskeletal motor proteins directly contribute to the organization of various cytoskeletal arrays during cell division and cell growth in plant tissues. They are also responsible for the motility of molecules and organelles, and the segregation of genetic materials during mitosis and meiosis. In the genome of the model plant Arabidopsis (Arabidopsis thaliana), there are at least 61 genes encoding kinesins and 17 genes encoding myosins. Most Arabidopsis kinesins and all myosins are evolutionarily divergent from their counterparts in animals and fungi. Little is known about the functions of most plant kinesins and myosins. Arabidopsis kinesins form a number of subfamilies. The mitotic kinesins in the BIMC/Kinesin-5 and the NCD/Kinesin-14 subfamilies appear to be similar to those in fungi and animals. Others, however, are very divergent, as their nonmotor sequences are unique to plants. Some of Arabidopsis kinesins are associated with microtubules, mitochondria, Golgi stacks, and vesicles. They affect microtubule organization, organelle distribution, and vesicle transport, respectively. Ultimately, Arabidopsis kinesins contribute directly or indirectly to cell division and cell growth in various tissues. Arabidopsis myosins are classified into two subfamilies: class VIII and class XI. The class XI myosins are associated with various organelles/vesicles. Functions of Arabidopsis myosins are still elusive. Future efforts will be devoted to deciphering not only the functions of these motors by molecular genetics but also the molecular mechanisms underlying how these roles are played.
INTRODUCTION OF CYTOSKELETAL MOTORS
Cytoskeletal motors use the energy released from ATP hydrolysis to move unidirectionally along tracks of microtubules and actin microfilaments. To date, cytoskeletal motors have been found in all studied eukaryotic organisms. Common structural features of cytoskeletal motors have been vividly depicted in a recent review article (Vale, 2003).
Kinesins
Members of the kinesin superfamily are one of the two families of microtubule-based motors that share a catalytic core of about 350 amino acids containing an ATP-binding site and a microtubule-binding site. The catalytic core is often juxtaposed with an α-helical domain of smaller than 50 amino acids, which is called the neck region (Endow, 1999). The catalytic core plus the neck form a kinesin motor domain. To date, it has been found that the budding yeast Saccharomyces cerevisiae has the fewest kinesin genes with six, and flowering plants have the most, with Arabidopsis having 61 kinesin genes (Reddy and Day, 2001b; Vale, 2003).
Kinesins are grouped into more than a dozen subfamilies by phylogenetic analyses of their motor domains (Schoch et al., 2003; Dagenbach and Endow, 2004). A recent effort by the kinesin research community has introduced a new nomenclature system to name established kinesin subfamilies from Kinesin-1 to Kinesin-14 (Lawrence et al., 2004).
Kinesins transport various vesicles and organelles along microtubules in different cell types of a wide range of organisms (Vale, 2003; Wozniak et al., 2004). Recently, a number of studies indicate that kinesins participate in RNA distribution in Drosophila (Tekotte and Davis, 2002). Several lines of evidence also point out that kinesins interact with scaffolding linkers of signaling modules to allow the modules to be localized at particular intracellular sites (Schnapp, 2003). Multifaceted roles of different kinesins are also found during mitosis, i.e. for centrosome separation, chromosome attachment to microtubules, chromosome aggregation to the metaphase plate, sister chromatid segregation, maintenance of bipolar spindle, and spindle elongation (Sharp et al., 2000).
A kinesin Web site has been launched to summarize the knowledge from the discovery of kinesins to the latest findings (http://www.proweb.org/kinesin/).
Myosins
Myosins are motors that travel along actin microfilaments. To date, members of the myosin superfamily are classified into at least 18 subfamilies (Berg et al., 2001). Almost all myosins travel toward the barbed (+) end of actin microfilaments (Berg et al., 2001). Myosin VI is the only one known to move toward the pointed (−) end (Buss et al., 2002).
A Web site has been designated for myosins: http://www.mrc-lmb.cam.ac.uk/myosin/myosin.html (U.S. version: http://www.proweb.org/myosin/index.html).
Compared to what has been known on animal and fungal cytoskeletal motors, we know much less about their counterparts in angiosperms. This difference is largely because there are only a few laboratories that study plant motors. A recent comprehensive review article is devoted to summarizing early findings of plant cytoskeletal motors (Reddy, 2001).
SIXTY-ONE ARABIDOPSIS KINESINS
The completed Arabidopsis genome contains at least 61 genes encoding polypeptides with the kinesin catalytic core (Reddy and Day, 2001b). The amino acid sequences of most of these kinesins were deduced from the annotation of genomic sequences. A few of them have their cDNA sequences determined. Arabidopsis kinesins vary dramatically regarding to the motor domain position within the polypeptides. Coiled-coil domains are found among most of them. Phylogenetic analysis of their motor domains has placed most of them into established kinesin subfamilies (Reddy and Day, 2001b). When an Arabidopsis kinesin and an animal or fungal one are grouped into the same subfamily, however, one should not simply interpret that they share a similar function. This is not only because their nonmotor sequences often show no common features, but also because bootstrap values are often less than 50% in phylogenetic analyses of the motor domains (Dagenbach and Endow, 2004).
To date, kinesins identified from angiosperms other than Arabidopsis all have homologs in this small model plant. In this Update article, we summarize recent findings on Arabidopsis kinesins from in vitro and/or in vivo studies. A few examples were brought in from other angiosperms because their Arabidopsis homologs have not been studied yet.
Microtubule Minus End-Directed Kinesins
Although the conventional kinesin and most other kinesins are microtubule plus end-directed motors, a number of C-terminal motor kinesins do the opposite (Ovechkina and Wordeman, 2003). It has been determined by serendipity that a 14-amino acid neck peptide immediately next to the catalytic core is the directionality determinant (Endow, 1999).
Based on the presence of the signature neck sequence, there are 21 Arabidopsis genes encoding minus end-directed kinesins (Reddy and Day, 2001b). Phylogenetic analyses of the motor domains of these kinesins also indicate that they are more closely related to each other than to other Arabidopsis kinesins (Reddy and Day, 2001b; Dagenbach and Endow, 2004). Among these predicted minus end-directed kinesins, however, some have the motor domain located at the N terminus, while some others have it in the middle (Reddy and Day, 2001b).
KATA/ATK1 and Close Relatives
Kinesins encoded by the KATA/B/C and At4g05190 genes are closely related to the NCD (non-claret disjunction) kinesin from Drosophila melanogaster and the Kar3p kinesin from S. cerevisiae (Reddy and Day, 2001b), so they belong to the NCD/Kinesin-14 subfamily. KATA/ATK1 has been shown as a nonprocessive microtubule minus end-directed motor (Marcus et al., 2002).
The Arabidopsis KATA/ATK1 protein plays a critical role in microtubule organization at the spindle pole and the spindle midzone during meiosis, and loss-of-function mutations consequently cause abnormal chromosome segregation during microsporogenesis (Chen et al., 2002). The atk1-1 mutation also prevents microtubule accumulation at mitotic spindle poles at early stages of spindle assembly (Marcus et al., 2003). Nevertheless, such an effect on microtubule organization during prometaphase does not affect the ultimate outcome of mitosis (Marcus et al., 2003). The atk1-1 phenotype on microtubule organization could be interpreted by the lack of microtubule-bundling activities in the spindle. Similar to what has been found in NCD, KATA/ATK1 and three other kinesins may contain a microtubule-binding site at their N terminus due to sequence similarity. The genetic data also suggest that some nonoverlapping functions of KATA/ATK1 and At4g05190 are expected despite the high degree of sequence identity.
The Calmodulin-Binding KCBP/ZWI Kinesin
KCBP/ZWI is a single gene in the Arabidopsis genome that encodes a unique calmodulin-binding kinesin (Reddy and Day, 2001b). The only noticeable phenotype of the loss-of-function zwi mutations is the reduction of branch formation in leaf trichomes (Oppenheimer et al., 1997). KCBP/ZWI may act as a microtubule stabilization factor during cell morphogenesis, as the zwi mutant phenotype can be suppressed by the application of taxol, a microtubule-stabilizing agent (Mathur and Chua, 2000). Recent studies indicate that the cotton (Gossypium hirsutum) GhKCBP kinesin decorates cortical microtubules in cotton fibers, thus suggesting that KCBP/ZWI may stabilize microtubules in the interphase cell cortex by directly binding to them (Preuss et al., 2003).
The search for KCBP/ZWI-interacting proteins has revealed several novel ones (Day et al., 2000; Folkers et al., 2002; Reddy et al., 2004). Two of them play roles in trichome branching as well (Folkers et al., 2002; Reddy et al., 2004). However, it is not known whether KCBP/ZWI affects the functions of these interacting proteins, or vice versa.
The Actin-Binding KCH Kinesins
Among 21 putative Arabidopsis minus end-directed kinesins, seven distinguish themselves from others by having a unique calponin-homology (CH) domain at the N terminus (Reddy and Day, 2001b). Because these kinesins have only been reported in organisms of the kingdom Plantae, they are named KCH for kinesins with a CH domain in order to be distinguished from other kinesins (Preuss et al., 2004). The presence of the neck sequence of minus end-directed kinesins suggests that they are probably also minus end-directed motors. Functions on these Arabidopsis KCHs, however, are not understood.
The presence of a CH domain in a kinesin is intriguing as it is typically found in actin-binding proteins like calponin and fimbrin (Gimona et al., 2002; Korenbaum and Rivero, 2002). The cotton GhKCH1 has been tested for actin binding (Preuss et al., 2004). The N-terminal region of GhKCH1, including its CH domain, cosediments with actin microfilaments in vitro. GhKCH1 decorates cortical microtubules in cotton fibers and is occasionally associated with actin microfilaments as well (Preuss et al., 2004). It has been suggested that GhKCH1 and other KCHs may act as dynamic linkers between microtubules and actin microfilaments, and such a link may be important for spatial organization of either or both cytoskeletal elements.
Other Minus End-Directed Kinesins
There are nine other Arabidopsis kinesins in the category of minus end-directed motors. We do not have any clue about their functions. Among them, four have the motor domain located at the N terminus. Again, such a feature has never been reported for kinesins from animals and fungi. One of the minus end-directed kinesins with an N-terminal motor domain, AtGRIMP/KCA1, is a substrate of cyclin-dependent kinase (Vanstraelen et al., 2004). It interacts with the geminivirus protein AL1 and is probably involved in geminivirus-induced cell division (Kong and Hanley-Bowdoin, 2002). It will be very interesting to explore whether some of these Arabidopsis kinesins play similar roles as cytoplasmic dyneins in animal and fungal cells in, for example, nuclear migration.
The N-Terminal Motor Kinesin AtFRA1 (Kinesin-4)
The AtFRA1 kinesin, which belongs to the Kinesin-4 or KIF4/chromokinesin subfamily, was identified by an elegant screen for fragile fiber mutants (Zhong et al., 2002). Animal kinesins in this subfamily typically play roles in chromatid motility and chromosome condensation, activities associated with mitosis (Wang and Adler, 1995; Kwon et al., 2004; Mazumdar et al., 2004). The fra1 mutant, however, does not show defects in cell division (Zhong et al., 2002). The only phenotype observed is that cellulose microfibril orientation was altered in fibers of the inflorescence stems (Zhong et al., 2002). Thus, AtFRA1 may directly or indirectly contribute to cellulose microfibril deposition in the cell wall. This is one of the examples that indicate sequence similarity in the motor domain does not confer any functional relationship among kinesins from different organisms.
The Internal Motor Kinesin: Kinesin-13
Arabidopsis has two kinesins, AtKinesin-13A (At3g16630) and AtKinesin-13B (At3g16060) that belong to the Kinesin-13 subfamily (Reddy and Day, 2001b), previously known as MCAK or KIN I. The similarity of Kinesin-13A and -13B to kinesins of the same subfamily from other kingdoms, however, is only limited to the catalytic core (L. Lu, Y.-R.J. Lee, R. Pan, and B. Liu, unpublished data). They lack a Lys-rich neck motif that is commonly found in animal Kinesin-13s (Ovechkina et al., 2002), implying that they are probably different from animal Kinesin-13s. AtKinesin-13A is specifically associated with Golgi stacks. Loss-of-function mutations only induce an additional branching event in leaf trichomes, but do not affect cell division or other aspects of growth (L. Lu, Y.-R.J. Lee, R. Pan, and B. Liu, unpublished data).
Animal internal motor kinesins in the Kinesin-13 subfamily are not motors. Instead, they are microtubule depolymerases activated by microtubule end binding (Walczak, 2003). They are required for mitotic spindle assembly and chromosome segregation (Ovechkina and Wordeman, 2003; Rogers et al., 2004). Therefore, this again demonstrates that sequence homology in the motor domain alone does not guarantee functional similarity for kinesins from different organisms.
Kinesins in the BIMC (Kinesin-5) Subfamily
Four Arabidopsis genes, At2g28620, At2g36200, At2g37420, and 3g45850, encode kinesins in the BIMC/Kinesin-5 subfamily (Reddy and Day, 2001b). Besides the sequence conservation in the motor domain of these four kinesins and their animal and fungal counterparts, all Kinesin-5s contain a conserved phosphorylation site for the key cell-cycle kinase p34cdc2 (Liu and Lee, 2001). It would be critical to examine localization patterns and functions of individual isoforms of plant Kinesin-5s. Another urgent task is to functionally characterize these four Arabidopsis Kinesin-5s using molecular genetics approaches. All four loci have been hit by T-DNA-mediated mutagenesis. It is only a matter of time before these mutations are characterized.
Kinesins for Cytokinesis
Plant cytokinesis is mechanistically different from that in animals and fungi (Mayer and Jurgens, 2004). One of the distinctions is that microtubules play a leading role during the assembly of the cell plate via the phragmoplast in plants. The plus end of phragmoplast microtubules is facing the division site, while the minus end is facing the reforming daughter nuclei (Liu and Lee, 2001). The assembly of the cell plate involves the delivery of Golgi-derived vesicles toward the plus end of phragmoplast microtubules and the fusion of these vesicles. During the course of cell-plate assembly, more microtubules are continuously assembled toward the periphery of the phragmoplast, while in the central region microtubules are disassembled concomitantly. Several lines of evidence have suggested that a number of N-terminal motor kinesins are involved in different aspects of cytokinesis. These kinesins all localize to the division site, but in distinct fashions.
AtNACK1/HIK Kinesins and a MAP Kinase Cascade
AtNACK1/HIK was independently identified as an activator of a mitogen-activated protein (MAP) kinase cascade and as an essential player for cytokinesis (Nishihama et al., 2002; Strompen et al., 2002). AtNACK1/HIK is in a unique plant subfamily tentatively classified as At1 (Dagenbach and Endow, 2004). The tobacco (Nicotiana tabacum) homolog NACK1 physically interacts with a MAP kinase kinase kinase called NPK1, and they form a complex during cell division (Nishihama et al., 2002). Both NACK1 and NPK1 localize to the midline in the phragmoplast of tobacco BY-2 cells (Nishihama et al., 2002). AtNACK1/HIK is essential for the completion of cell-plate formation during vegetative growth (Nishihama et al., 2002; Strompen et al., 2002). Three Arabidopsis homologs of the tobacco NPK1, ANP1 to ANP3, show redundant function(s) during cytokinesis, as loss-of-function mutations in individual genes do not have a noticeable phenotype (Krysan et al., 2002). The anp2 anp3 mutation combination leads to incomplete cell-plate formation (Krysan et al., 2002). Likewise, the MAP kinase kinase ANQ1, downstream of ANP1 to ANP3 in the kinase cascade, is also essential for cytokinesis (Soyano et al., 2003). The fact that NACK1 is required for NPK1 localization (Nishihama et al., 2002) implies that NACK1 may act as an active transporter for NPK1.
NACK1 appears to be required for cell-plate expansion after cytokinesis has been initiated (Nishihama et al., 2002). In order to understand how NACK1 and NPK1 regulate cytokinesis in plants, the substrates of the NPK1 MAP kinase cascade need to be identified and characterized. The substrates could be those proteins that are involved in microtubule reorganization, and those that are involved in vesicle trafficking and fusion. The MAP kinase cascade may activate these proteins for the completion of cell-plate formation.
The AtTES/NACK2 kinesin, similar to AtNACK1/HIK, however, is not required for vegetative cytokinesis (Yang et al., 2003). Instead, it is required for the establishment of the radial microtubule array prior to the meiotic cytokinesis, and thus is essential for tetrad formation during male gametogenesis (Yang et al., 2003). Whether AtTES/NACK2 interacts with ANP kinases awaits further characterization.
AtPAKRP1/AtKinesin-12A and Similar Kinesins
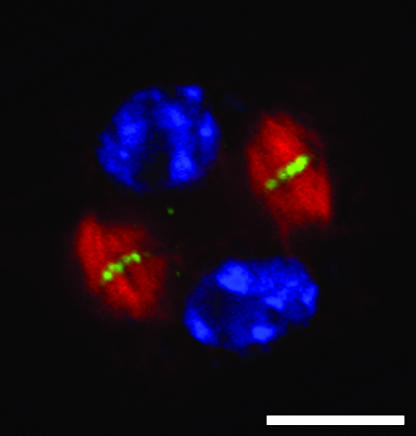
AtPAKRP1/AtKinesin-12A and two similar kinesins are specifically associated with the plus end of phragmoplast microtubules (Fig. 1; Lee and Liu, 2000; Pan et al., 2004). AtPAKRP1/AtKinesin-12A, AtPAKRP1L/AtKinesin-12B, and AtPAKRP1L′/AtKinesin-12C are predicted to be plus end-directed motors. They may push newly added microtubule segments apart in the phragmoplast in order to maintain the plus ends at the division site. Based on in vitro binding assays, AtPAKRP1/AtKinesin-12A and AtPAKRP1L/AtKinesin-12B can form homo- and/or heterodimers (Pan et al., 2004). Loss-of-function mutations in each gene do not cause a cytokinetic defect (Pan et al., 2004). Loss of AtPAKRP1/AtKinesin-12A and AtPAKRP1L/AtKinesin-12B together, however, causes frequent failure during postmeiotic cytokinesis in microspores (Y.-R.J. Lee and B. Liu, unpublished data). Therefore, these kinesins probably play synergistic roles in maintaining the organization of phragmoplast microtubules. An intriguing question is how Arabidopsis uses these three similar kinesins differently to accomplish cytokinesis at different stages of growth and development.
Figure 1.
Triple localization of AtPAKRP1L/AtKinesin-12B (green), microtubules (red), and DNA (blue) in an Arabidopsis root cell undergoing cytokinesis. Microtubules were organized into a phragmoplast array with their plus ends at or near the midline of the phragmoplast, where the cell plate would be formed. Note that AtPAKRP1L/AtKinesin-12B only appeared at the plus end of phragmoplast microtubules. Scale bar = 5 μm.
Kinesins for Delivering Golgi-Derived Vesicles
Another phragmoplast-associated kinesin, AtPAKRP2, is specifically associated with Golgi-derived vesicles in the phragmoplast (Lee et al., 2001). At least two other Arabidopsis kinesins are also exclusively associated with vesicles in the phragmoplast (Y.-R.J. Lee and B. Liu, unpublished data). Therefore, they are predicted to be examples of plus end-directed kinesin motors that deliver Golgi-derived vesicles during cytokinesis.
There is no doubt that the list of kinesins required for cytokinesis is going to grow in the near future. Plant cytokinesis requires concerted forces generated by different kinesins present in the phragmoplast.
The above discussion has only covered a fraction of Arabidopsis kinesins. There are others of which we know very little or nothing about their structure and function. For example, two N-terminal motor kinesins contain a mitochondria-targeting peptide that would allow them to be associated with the organelle (Itoh et al., 2001). Whether the endogenous proteins are indeed associated with the organelle is not known.
SEVENTEEN ARABIDOPSIS MYOSINS
The Arabidopsis myosin gene family is much simpler compared to its kinesin family (Reddy and Day, 2001a). Phylogenetic analysis has placed these myosins into two plant-specific subfamilies, Myosin VIII with four members and Myosin XI with 13 members (Berg et al., 2001; Reddy and Day, 2001a), which are more closely related to animal and fungal Myosin Vs than to other myosins. Several review articles have been devoted to summarizing early discoveries of plant myosins (Yamamoto et al., 1999; Shimmen et al., 2000; Shimmen and Yokota, 2004). Here, we only summarize recent findings on Myosin XI.
Myosin XI is probably more abundantly expressed in cells than Myosin VIII. Biochemically purified plant myosins with molecular mass at 165 to 175 kD turn out to be class XI myosins (Ma and Yen, 1989; Yokota and Shimmen, 1994; Tominaga et al., 2003). Myosin XIs have four to six IQ motifs following their motor domain (Reddy and Day, 2001a). Only three Arabidopsis Myosin XIs, MYA1, MYA2, and MYA3 (partial), have their cDNA sequences determined (Kinkema et al., 1994). Very little, if any, information can be extracted from their sequences outside the motor domain and the IQ motif, indicating that these myosins probably fulfill plant-specific functions.
A tobacco 175-kD myosin, probably the homolog of MYA1, demonstrates a high processive velocity of 7 μm/s at 35 nm steps, probably the fastest among known processive myosins (Tominaga et al., 2003). It remains to be tested whether this myosin is responsible for cytoplasmic streaming in plant cells.
One of the questions about these 13 Myosin XIs is whether each one interacts with a particular organelle or structure. Immunolocalization studies indicate that Myosin XI is associated with particles of various sizes in tobacco pollen tubes and suspension cells (Yokota et al., 1995). Antibodies against a conserved Myosin XI peptide label the plasma membrane and maybe mitochondria (Liu et al., 2001). Recently, a subclass of Myosin XI has been shown to be associated with mitochondria and plastids (Wang and Pesacreta, 2004). Obviously, polypeptide-specific probes are needed to distinguish individual Myosin XIs in order to reveal their specific intracellular localization patterns.
Functions of MYA2 have been further investigated using an Arabidopsis T-DNA insertional mutant (Holweg and Nick, 2004). This study suggests that MYA2 plays a role in regular vesicle flow, which is probably responsible for basipetal auxin transport. The homozygous mutant line demonstrates pleiotropic phenotypes on growth, i.e. bushy and dwarf plants, lack of trichome branching, and defects in tip growth of root hairs and pollen tubes (Holweg and Nick, 2004). This mya2-1 mutant also demonstrates an alteration of the division plane in root cells, which further supports a potential role of myosin in plant cytokinesis suggested by an earlier pharmacological study (Molchan et al., 2002).
CONCLUSIONS AND PERSPECTIVES
Current knowledge indicates that most kinesins and myosins of Arabidopsis and other plants are very different motors from those in other kingdoms. This is reflected by the sequence divergence in the motor domains, and more significantly by novel sequences in the nonmotor regions whose significance is largely unknown. What we have learned about plant cytoskeletal motors is probably just the tip of the iceberg. The tail sequences suggest that many tales of these Arabidopsis cytoskeletal motors are yet to be told.
One of the frequently asked questions is why a small plant like Arabidopsis needs so many kinesins and myosins. Our answer is that besides bearing various cell types like guard cells, trichomes, pollen tubes, root hairs, and different vascular cells that may require different motors for their morphogenesis, land plants also need to meet many environmental challenges, as they cannot run away from them. Some cytoskeletal motors might participate in defense responses when a plant is challenged by environmental stresses or pathogen attacks.
We believe that in the next few years we will need to address a number of issues about these motors. The first task is to determine the intracellular localization patterns of each motor. The next demanding job is to find out the proteins they interact with. It will help us figure out what their cargoes are. Then we will need to find out when and how the motors are activated, and where their destinations are. Genetic studies are needed to address the functions of these motors. We would also like to suggest that activities of these motors are probably coordinated at a certain stage in a given cell. There must be some mechanisms for coordination between kinesins and myosins and between microtubules and actin microfilaments in plant cells.
This work was supported by grants from the U.S. Department of Energy, Division of Energy Biosciences, and by the U.S. Department of Agriculture, Cooperative State Research, Education, and Extension Service.
References
- Berg J, Powell B, Cheney R (2001) A millennial myosin census. Mol Biol Cell 12: 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Luzio J, Kendrick-Jones J (2002) Myosin VI, an actin motor for membrane traffic and cell migration. Traffic 3: 851–858 [DOI] [PubMed] [Google Scholar]
- Chen C, Marcus A, Li W, Hu Y, Calzada J, Grossniklaus U, Cyr R, Ma H (2002) The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409 [DOI] [PubMed] [Google Scholar]
- Dagenbach EM, Endow SA (2004) A new kinesin tree. J Cell Sci 117: 3–7 [DOI] [PubMed] [Google Scholar]
- Day IS, Miller C, Golovkin M, Reddy AS (2000) Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J Biol Chem 275: 13737–13745 [DOI] [PubMed] [Google Scholar]
- Endow SA (1999) Determinants of molecular motor directionality. Nat Cell Biol 1: E163–E167 [DOI] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schobinger U, Falk S, Krishnakumar S, Pollock M, Oppenheimer D, Day I, Reddy A, Jurgens G, et al (2002) The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J 21: 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ (2002) Functional plasticity of CH domains. FEBS Lett 513: 98–106 [DOI] [PubMed] [Google Scholar]
- Holweg C, Nick P (2004) Arabidopsis myosin XI mutant is defective in organelle movement and polar auxin transport. Proc Natl Acad Sci USA 101: 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Itoh R, Fujiwara M, Yoshida S (2001) Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol 127: 724–726 [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Wang H, Schiefelbein J (1994) Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant Mol Biol 26: 1139–1153 [DOI] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbaum E, Rivero F (2002) Calponin homology domains at a glance. J Cell Sci 115: 3543–3545 [DOI] [PubMed] [Google Scholar]
- Krysan P, Jester P, Gottwald J, Sussman M (2002) An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Morales-Mulia S, Rogers GC, Sharp D, Scholey JM (2004) The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol Biol Cell 15: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, et al (2004) A standardized kinesin nomenclature. J Cell Biol 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Giang HM, Liu B (2001) A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13: 2427–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B (2000) Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr Biol 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Liu B, Lee YRJ (2001) Kinesin-related proteins in plant cytokinesis. J Plant Growth Regul 20: 141–150 [Google Scholar]
- Liu L, Zhou J, Pesacreta T (2001) Maize myosins: diversity, localization, and function. Cell Motil Cytoskeleton 48: 130–148 [DOI] [PubMed] [Google Scholar]
- Ma Y-Z, Yen L-F (1989) Actin and myosin in pea tendrils. Plant Physiol 89: 586–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A, Ambrose J, Blickley L, Hancock W, Cyr R (2002) Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil Cytoskeleton 52: 144–150 [DOI] [PubMed] [Google Scholar]
- Marcus AI, Li W, Ma H, Cyr RJ (2003) A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol Biol Cell 14: 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua N (2000) Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Jurgens G (2004) Cytokinesis: lines of division taking shape. Curr Opin Plant Biol 7: 599–604 [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Sundareshan S, Misteli T (2004) Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol 166: 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchan TM, Valster AH, Hepler PK (2002) Actomyosin promotes cell plate alignment and late lateral expansion in Tradescantia stamen hair cells. Planta 214: 683–693 [DOI] [PubMed] [Google Scholar]
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, et al (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109: 87–99 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD (1997) Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA 94: 6261–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, Wordeman L (2002) K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol 159: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y, Wordeman L (2003) Unconventional motoring: an overview of the Kin C and Kin I kinesins. Traffic 4: 367–375 [DOI] [PubMed] [Google Scholar]
- Pan R, Lee YRJ, Liu B (2004) Localization of two homologous kinesin-related proteins in the phragmoplast in Arabidopsis thaliana. Planta 220: 156–164 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Delmer DP, Liu B (2003) The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol 132: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee Y-RJ, Staiger CJ, Delmer DP, Liu B (2004) A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol 136: 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN (2001) Molecular motors and their functions in plants. Int Rev Cytol 204: 97–178 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS (2001. a) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol 2: RESEARCH0024 [DOI] [PMC free article] [PubMed]
- Reddy ASN, Day IS (2001. b) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Day IS, Thomas T, Reddy AS (2004) KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G, Rogers S, Schwimmer T, Ems-McClung S, Walczak C, Vale R, Scholey J, Sharp D (2004) Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427: 364–370 [DOI] [PubMed] [Google Scholar]
- Schnapp B (2003) Trafficking of signaling modules by kinesin motors. J Cell Sci 116: 2125–2135 [DOI] [PubMed] [Google Scholar]
- Schoch C, Aist J, Yoder O, Turgeon B (2003) A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet Biol 39: 1–15 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM (2000) Microtubule motors in mitosis. Nature 407: 41–47 [DOI] [PubMed] [Google Scholar]
- Shimmen T, Ridge RW, Lambiris I, Plazinski J, Yokota E, Williamson RE (2000) Plant myosins. Protoplasma 214: 1–10 [Google Scholar]
- Shimmen T, Yokota E (2004) Cytoplasmic streaming in plants. Curr Opin Cell Biol 16: 68–72 [DOI] [PubMed] [Google Scholar]
- Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 17: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jurgens G, Mayer U (2002) The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12: 153–158 [DOI] [PubMed] [Google Scholar]
- Tekotte H, Davis I (2002) Intracellular mRNA localization: motors move messages. Trends Genet 18: 636–642 [DOI] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K (2003) Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J 22: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Torres Acosta J, De Veylder L, Inze D, Geelen D (2004) A plant-specific subclass of C-terminal kinesins contains a conserved a-type cyclin-dependent kinase site implicated in folding and dimerization. Plant Physiol 135: 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE (2003) The Kin I kinesins are microtubule end-stimulated ATPases. Mol Cell 11: 286–288 [DOI] [PubMed] [Google Scholar]
- Wang SZ, Adler R (1995) Chromokinesin-a DNA-binding, kinesin-like nuclear protein. J Cell Biol 128: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pesacreta TC (2004) A subclass of myosin XI is associated with mitochondria, plastids, and the molecular chaperone subunit TCP-1alpha in maize. Cell Motil Cytoskeleton 57: 218–232 [DOI] [PubMed] [Google Scholar]
- Wozniak M, Milner R, Allan V (2004) N-terminal kinesins: many and various. Traffic 5: 400–410 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hamada S, Kashiyama T (1999) Myosins from plants. Cell Mol Life Sci 56: 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Spielman M, Coles J, Li Y, Ghelani S, Bourdon V, Brown R, Lemmon B, Scott R, Dickinson H (2003) TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J 34: 229–240 [DOI] [PubMed] [Google Scholar]
- Yokota E, McDonald AR, Liu B, Shimmen T, Palevitz BA (1995) Localization of a 170 kDa myosin heavy chain in plant cells. Protoplasma 185: 178–187 [Google Scholar]
- Yokota E, Shimmen T (1994) Isolation and characterization of plant myosin from pollen tubes of lily. Protoplasma 177: 153–162 [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye Z-H (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]