Abstract
Telomeres are specialized nucleoprotein complexes that are essential for preserving chromosome integrity in eukaryotic cells. Several potential telomere binding proteins have recently been identified in higher plants, but nothing is known about their in vivo functions. We previously identified NgTRF1 as a double-stranded telomeric repeat binding factor in tobacco (Nicotiana tabacum) and here show that the binding of NgTRF1 to telomeric repeats inhibits telomerase-mediated telomere extension. To determine whether NgTRF1 is involved in telomere length regulation, we established transgenic tobacco BY-2 cell lines that overexpress or suppress NgTRF1. Pulsed-field gel electrophoresis showed that 35S::NgTRF1 cells exhibited significantly shortened telomeres (45 to 10 kb), whereas 35S::antisense-NgTRF1 cells contained longer telomeres (80 to 25 kb) compared with wild-type and 35S::GUS control cells (65 to 15 kb), indicating that telomere length inversely correlates with the amount of functional NgTRF1 in BY-2 cells. 35S::NgTRF1 cells with shorter telomeres displayed a progressive reduction in cell viability and stopped dividing after 25 to 40 successive rounds of 12-d batch subculture, in sharp contrast with control cells, which have an unlimited capacity for division. Internucleosomal DNA fragmentation, mitochondrial release of cytochrome c, and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling positive nuclei were detected in 35S::NgTRF1 cells during prolonged subculture, indicating that enhanced cell death was attributable to an apoptosis-like mechanism. 35S::antisense-NgTRF1 cells containing low levels of NgTRF1 also exhibited a progressive decrease in cell viability and apoptotic cell death, but less so than did 35S::NgTRF1 cells, suggesting that the level of NgTRF1 is critically associated with cell viability. Taken together, these data indicate that perturbation of NgTRF1 expression results in changes in telomere length and stability, which in turn causes apoptotic cell death in transgenic BY-2 cells. These results are discussed in light of the suggestion that NgTRF1 is involved in the mechanism by which telomere length and stability are maintained. We further suggest that the structural stability of telomeres, in addition to length maintenance, is essential for their function and for the immortality of BY-2 cells.
INTRODUCTION
Telomeres are specialized nucleoprotein complexes that protect the termini of linear eukaryotic chromosomes. This shielding function was originally deduced in classical cytogenetic studies of flies by Muller and of maize (Zea mays) by McClintock (Muller, 1938; McClintock, 1941). Their pioneering studies provided insights for defining the mechanisms that counteract chromosome shortening and instability. Subsequently, it was shown that telomeres are capped to prevent exonucleolytic degradation and end-to-end fusion with other chromosomes (Blackburn, 1991, 2001; Greider, 1996). Telomeres must also be fully regenerated when conventional DNA polymerases replicate linear DNA molecules. It appears that without a mechanism to ensure complete replication of chromosome extremities, terminal attrition of chromosomal DNA would eventually lead to the loss of genetic information and accelerate the entry of cells into senescence. In most eukaryotes, this end-replication problem has been solved by the ribonucleoprotein telomerase, which lengthens the terminal regions of telomeric DNA by the RNA-templated addition of tandemly repeated G-rich arrays (Zakian, 1995; Linger and Cech, 1998; Nugent and Lundblad, 1998; Liu, 1999). Telomeric sequences vary among species—TTAGGG in humans and all other vertebrates, TG1-3 in budding yeast (Saccharomyces cerevisiae), and TTTAGGG in higher plants—but the fundamental functions of telomeres and telomerase are very similar throughout eukaryotes (Blackburn, 1991; Shore, 2001). Telomeric repeats vary considerably in length among different species, but each species maintains a fixed average telomere length.
In addition to telomerase activity, specific proteins that bind telomere repeat sequences appear to be critical for telomere integrity and function. Telomere binding proteins can be divided into two distinct classes according to their substrate specificities. Members of the first group, which bind to the single-stranded 3′ extension at telomere termini, are necessary for chromosome capping and telomerase regulation. Cdc13 in budding yeast and Pot1 in fission yeast and human belong to this group (Nugent et al., 1996; Baumann and Cech, 2001). The second class comprises double-stranded telomeric repeat binding proteins, including yeast Rap1p, Tel2, and Taz1p, and human TRF1 and TRF2 (Shore 1994; Chong et al., 1995; Runge and Zakian, 1996; Bilaud et al., 1997; Vassetzky et al., 1999). Rap1p was reported to act as a negative regulator of telomere elongation by forming a complex with Rif1p and Rif2p (Wotton and Shore, 1997). Human TRF1, a suppressor of telomere elongation, is involved in a negative feedback mechanism that inhibits telomerase at the ends of individual telomeres (van Steensel and de Lange, 1997; Smucker and Turchi, 2001). A dominant-negative allele of TRF2 induces end-to-end chromosome fusions in metaphase and anaphase cells, indicating that TRF2 plays a key role in protecting telomeres in human cells (van Steensel et al., 1998). It has been reported that TRF2, along with TRF1, acts as a negative regulator of telomere length (Smogorzewska et al., 2000). Another telomeric protein, Pin2, has been identified in HeLa cells (Shen et al., 1997). Pin2 is identical in sequence to TRF1, except for an internal deletion of 20 amino acids, suggesting that these two proteins may be derived from the same gene, PIN2/TRF1 (Shen et al., 1997). The Pin2-interacting protein PinX1 was shown to bind the telomerase catalytic subunit hTERT and to potently inhibit its activity (Zhou and Lu, 2001).
Compared with the extensive knowledge of yeast and mammalian telomeres, an understanding of the structure and biological roles of telomeres in higher plants is still rudimentary. Recent studies have shown that telomere structure and the pattern of telomerase regulation in higher plants are very similar to that in other eukaryotes (reviewed in Shippen and McKnight, 1998; McKnight et al., 2002). As is the case for mammals, there is a tight correlation between telomerase activity and cell division capacity in higher plant tissues (Killan et al., 1995, 1998; Fitzgerald et al., 1996; Heller et al., 1996; Riha et al., 1998; Tamura et al., 1999; Yang et al., 2001, 2002). Likewise, the level of mRNA encoding a telomerase reverse transcriptase catalytic subunit is abundantly present in callus and shoot apical meristems, which possess high levels of telomerase activity, as opposed to leaf tissue, which has no detectable activity, as observed for Arabidopsis thaliana and rice (Oryza sativa) (Fitzgerald et al., 1999; Oguchi et al., 1999; Heller-Uszynska et al., 2002). Telomerase activity and telomere structure were demonstrated in Arabidopsis to be critical for plant development. Telomerase-null mutants display cytogenetic damage, which is correlated with severe developmental defects in both vegetative and reproductive organs (Riha et al., 2001).
Previously, cDNAs encoding potential telomeric DNA binding proteins were isolated from rice and Arabidopsis (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001). Like human TRF1/Pin2 and TRF2, the predicted protein sequences of rice RTBP1 and Arabidopsis AtTRP1 and AtTBP1 contain a single Myb domain at their C termini, and they exhibit sequence similarity to Myb-related double-stranded telomeric DNA binding proteins from other organisms. Gel retardation assays showed that the Myb domain of both the rice and Arabidopsis proteins is able to bind to double-stranded, but not single-stranded, plant telomeric DNA. However, these studies did not provide information about the physiological roles and developmental regulation of these telomere-related proteins. Recently, we isolated a cDNA clone, pNgTRF1, which encodes a tobacco (Nicotiana tabacum) protein that can interact specifically with double-stranded plant telomeric repeat sequences (Yang et al., 2003). The NgTRF1 transcript is present only in tissues that undergo negligible levels of cell division. During a 9-d culture period of tobacco BY-2 suspension culture cells, the NgTRF1 gene was highly activated in stationary phase cells that were not proliferating, whereas telomerase activity positively correlated with cell division (Yang et al., 2002; 2003). In synchronized BY-2 cells, NgTRF1 is selectively expressed in G1 phase, in contrast with telomerase, which is active during S phase. These findings indicate that telomerase activity and NgTRF1 expression are differentially regulated in an opposing fashion during cell division in tobacco.
We are interested in elucidating how telomere length and stability are controlled in higher plants. Because nothing is known about the physiological role of telomere binding proteins with respect to telomere function and structure, the specific aim of this study was to explore the possible relationship between telomere binding proteins and telomerase activity in BY-2 cells. For this purpose, we constructed transgenic BY-2 cell lines in which NgTRF1 was overexpressed or suppressed. We present results indicating that perturbation of the tobacco telomeric repeat binding factor NgTRF1 expression results in changes in telomere length and stability, which in turn causes apoptotic cell death. These results are discussed in light of the suggestion that NgTRF1 is involved in maintaining telomere length and stability in tobacco cells. We further suggest that the structural stability of telomeres, in addition to proper length maintenance, is essential for telomere function in tobacco cells.
RESULTS
NgTRF1 Inhibits in Vitro Telomerase Activity
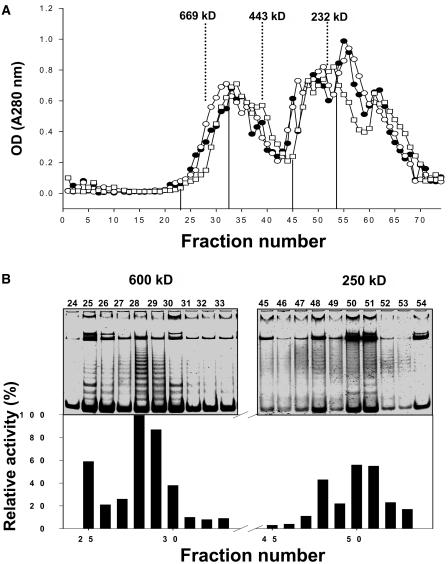
Telomerase activity is positively correlated with the cell division program, but by contrast, NgTRF1 gene activity was inversely associated with cell division during a 9-d culture period of BY-2 cells (Yang et al., 2003). In synchronized BY-2 cells, NgTRF1 is selectively expressed in G1 phase, whereas telomerase activity peaks in S phase. These patterns prompted us to investigate the biological function of NgTRF1 with regard to its possible relationship with telomerase activity. As a first step, we partially isolated telomerase activity from BY-2 cells. Nuclear lysates were extracted from actively dividing BY-2 cells, and telomerase activity was fractionated by gel filtration. As shown in Figure 1, telomerase activity, which was evaluated by means of the telomere repeat amplification protocol (TRAP) assay, eluted as a single major peak with an apparent molecular mass of ∼600 kD and an additional minor peak of ∼250 kD. Previously, Greene and Shippen (1998) showed that telomerase is present as a 280-kD complex in the vegetative stage of the ciliate Euplotes crassus, whereas upon the initiation of macronuclear development, it assembles into larger complexes of 550 kD, 1600 kD, and 5 MDa. Given that the predicted molecular sizes of the telomerase reverse transcriptase (TERT) subunits of Arabidopsis and rice are 131 and 137 kD, respectively, tobacco telomerase is expected to be assembled into large complexes (Oguchi et al., 1999; Heller-Uszynska et al., 2002).
Figure 1.
Fractionation of Telomerase Complexes by Gel Filtration.
(A) Tobacco BY-2 cell extracts were fractionated by gel filtration and the concentration of protein for each fraction was determined. Numbers at the top of the figure indicate the sizes of molecular mass markers in kD (thyroglobulin, 669 kD; ferritin, 440 kD; catalase, 232 kD).
(B) Telomerase activity elutes as a major 600-kD complex and an additional lower molecular weight complex (250 kD). After gel filtration and concentration of proteins, telomerase activity was evaluated by the TRAP assay.
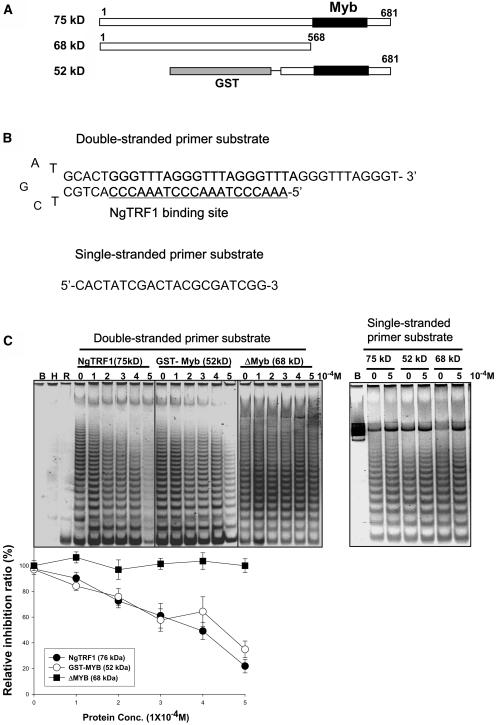
To assess if NgTRF1 could affect the ability of telomerase to extend telomeric DNA, three different bacterially expressed NgTRF1 derivatives were individually incubated with the major 600-kD telomerase fractions (Figure 2A). The degree of telomerase activity was monitored by the TRAP assay with a partial duplex DNA substrate primer containing an optimal NgTRF1 binding site [TTTA(GGGTTTA)3GGG] (Figure 2B). The results in Figure 2C show that telomerase elongation was not observed in the absence of the telomerase fraction (lane B) or in the presence of RNase- or heat-treated fraction (lanes R and H), in which telomerase activity is inactivated. On the other hand, a high level of telomerase activity was readily detected in the absence of NgTRF1, which indicates that multiple repeats were added to the double-stranded substrate primer (Figure 2C). Telomerase extension activity was markedly reduced in the presence of increasing concentrations of full-length NgTRF1. NgTRF1 at 0.2 mM substantially inhibited telomerase extension, and at 0.5 mM almost completely abolished enzyme activity (Figure 2C). A similar concentration-dependent inhibitory profile was observed when the GST-NgTRF1441-681 mutant protein, which contains the Myb-like motif and C-terminal region, was included in the telomerase fraction. As a next step, we coincubated the tobacco telomerase fraction with the NgTRF11-568 derivative, which lacks the Myb-like DNA binding domain. This mutant protein did not inhibit telomerase activity. In parallel, we performed control reactions using a single-stranded substrate primer for telomerase that lacks a binding site for NgTRF1 (Figure 2B). The enzymatic activity of telomerase remained unchanged in the presence or absence of NgTRF1 proteins, confirming the specificity of our TRAP assay with a duplex substrate primer (Figure 2C). Overall, these data argue that under our experimental conditions, NgTRF1 effectively inhibits telomerase activity in vitro in a dose-dependent manner. This result led us to hypothesize that the tobacco telomere binding protein NgTRF1 is involved in controlling telomerase activity by an as-yet unidentified mechanism.
Figure 2.
Inhibition of Telomerase-Mediated Extension by NgTRF1.
(A) Diagram of full-length NgTRF1 and two deletion derivatives used for the telomerase inhibition assay. The shaded boxes indicate the GST polypeptide, and the black boxes depict the Myb-like domain of NgTRF1. The coding region outside the Myb-like domain is shown in open boxes. The molecular mass of each polypeptide is indicated.
(B) Sequences of DNA substrates used in this assay. The NgTRF1 binding site in the double-stranded primer substrate is underlined.
(C) Telomerase extension assays were performed with BY-2 cell lysates in the presence or absence of the double-stranded and single-stranded primer substrates. NgTRF1 and derivatives at concentrations of 0, 0.1, 0.2, 0.3, 0.4, and 0.5 mM were added to the primer substrates before incubation in tobacco cell extract. Lane B, no extract; lane H, heat-treated extract; lane R, RNase A-treated extract. The extent of inhibition was quantified for NgTRF1 and the two deletion derivatives by normalizing the intensities of telomerase extension products for each concentration of protein to the intensities of extension products in the control (without added NgTRF1).
Overexpression and Suppression of NgTRF1 Cause Changes in Telomere Length in Transgenic BY-2 Cells
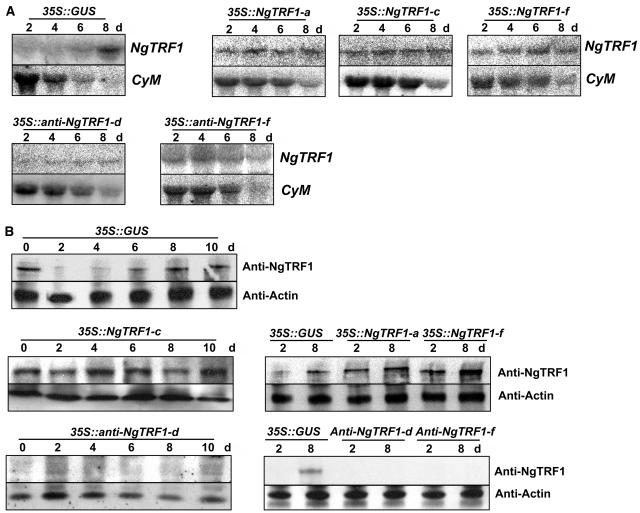
To determine whether NgTRF1 is involved in telomere length regulation in vivo, we established transgenic BY-2 cell lines that overexpressed or suppressed NgTRF1. BY-2 cells were transformed with binary vectors carrying a fusion of the 35S promoter of Cauliflower mosaic virus and pNgTRF1 in the sense or antisense orientation by use of the Agrobacterium tumefaciens–mediated transformation method (Lee and Kim, 2003). For each transgene, independent primary transformed cells were selected by resistance to kanamycin, and transgenic suspension cells were then propagated. As a control, we also generated 35S::GUS cell lines. The presence of each transgene was confirmed by genomic DNA gel blot and PCR analyses (data not shown). Among numerous transgenic lines, three independent sense lines, 35S::NgTRF1-a, 35S::NgTRF1-c, and 35S::NgTRF1-f, and two independent antisense lines, 35S::anti-NgTRF1-d and 35S::anti-NgTRF1-f, all of which exhibited markedly altered levels of NgTRF1 transcript under normal culture conditions, were chosen for further analysis. As found previously, the amount of NgTRF1 mRNA in 35S::GUS control cells was very low during the logarithmic phase of growth, began to accumulate at day 6, and attained a maximal level in the lag phase, when these cells stopped proliferating (Figure 3A). The gene encoding the mitotic cyclin CYM, a positive control of cell division, was specifically expressed during the exponential growth phase of 35S::GUS control cells. By contrast, high-level constitutive expression of NgTRF1 mRNA was clearly observed in every 35S::NgTRF1 transgenic line examined during the entire 8-d culture period (Figure 3A). Immunoblot analysis using an anti-NgTRF1 antibody also revealed that the level of NgTRF1 protein was significantly upregulated in transgenic cells in comparison with that in wild-type cells (Figure 3B). On the other hand, a negligible amount of NgTRF1 protein was detected in 35S::anti-NgTRF1 cells at all time points, although constitutive NgTRF1 mRNA was observed, which was probably because of the strongly elevated level of antisense mRNA (Figures 3A and 3B). These results indicate that the sense and antisense transgenes are functional in tobacco cells and that they effectively increase and suppress endogenous NgTRF1 expression, respectively.
Figure 3.
Expression Profiles of the NgTRF1 and CYM Genes in the 35S::GUS, 35S::NgTRF1 and 35S::anti-NgTRF1 Lines.
(A) RNA gel blot analysis of NgTRF1 and CYM expression in transgenic BY-2 suspension culture cells. Total RNA was isolated from sense and antisense NgTRF1 lines at the indicated time points during the 8-d culture period and subjected to RNA gel blot analysis with a 32P-labeled pNgTRF1 probe. The CYM gene was included for comparison as a gene expressed during exponential growth phase.
(B) Expression of the NgTRF1 protein in 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 cells during the 8-d culture period. Total proteins were prepared from sense and antisense NgTRF1 lines at the indicated time points, and subjected to protein gel blot analysis using an anti-NgTRF1 antibody and an anti-actin antibody as a loading control.
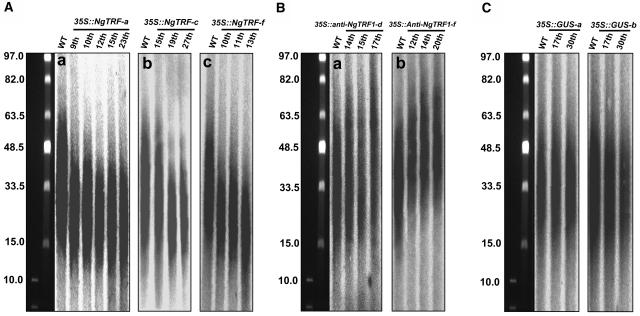
To explore whether the altered expression of NgTRF1 affects telomere metabolism in tobacco cells, we next measured the length of telomeres in wild-type, 35S::NgTRF1, 35S::anti-NgTRF1, and 35S::GUS control cells. Total genomic DNA was isolated from each transgenic cell line, digested with the restriction enzyme TaqI, and resolved by pulsed-field electrophoresis. DNA on the gel was then blotted and hybridized with a telomere repeat probe (TTTAGGG)70 (Figure 4). In our experimental conditions, samples from wild-type cells migrated as a typical telomeric smear, with fragments ranging from ∼65 to 15 kb (Fajkus et al., 1995, 1996). On the other hand, all of the 35S::NgTRF1 cell lines analyzed showed significantly shortened telomeres, whose lengths ranged between 45 and 10 kb, depending on the line (Figure 4A). Telomere length also varied with the stage of subculture. For example, the 35S::NgTRF1-c line contains telomeres ranging from 45 to 15 kb in length after the 15th successive round of a 12-d batch subculture, at a time corresponding to 183 d after gene transfer, whereas telomeres at the 27th round of subculture (318 d after gene transfer) are 35 to 12 kb in length (Figure 4A, panel b). Line f, which showed the most severe growth defect (see below), has telomeres 30 to 10 kb long after 13 rounds of subculture (panel c). In addition, we observed that longer telomeres were more severely affected than shorter telomeres and, hence, a reduction in length was more easily detected for longer telomeres. In transgenic line a, 65-kb–long telomeres were shortened to 40 kb, whereas 15-kb–long telomeres were reduced to ∼10 kb (panel a). A similar situation was observed for lines c (panel b) and f (panel c). Generally, telomere shortening was apparent at ∼100 d after gene transfer, and this length was continuously maintained for at least 200 to 300 d of growth, which corresponds to 17 to 25 successive rounds over the 12-d cell culture period.
Figure 4.
Telomere Length in Transgenic BY-2 Cells.
Total genomic DNA was isolated from wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 tobacco cells during long-term batch subculture. DNA was digested with TaqI, separated by pulsed-field gel electrophoresis, and probed with labeled (TTTAGGG)70 plant telomeric repeats. The positions of molecular weight–size markers are shown at the side of each panel. Numbers at the top of each gel indicate successive rounds of a 12-d batch subculture.
(A) Telomere shortening in 35S::NgTRF1-a (panel a), 35S::NgTRF1-c (panel b), and 35S::NgTRF1-f (panel c) cells.
(B) Enhanced telomere elongation in 35S::anti-NgTRF1-d (panel a) and 35S::anti-NgTRF1-f (panel b) cells.
(C) 35S::GUS control lines, like wild-type cells, maintained a constant telomere length (65 to 15 kb).
By contrast, overexpression of antisense NgTRF1 mRNA caused a modest enhancement of telomere elongation in two independent cell lines, resulting in telomeres 80 to 25 kb long (Figure 4B). As was observed with 35S::NgTRF1 cells, these longer telomeres were seen ∼150 d after antisense gene transfer, corresponding to the 12 to 13th round of subculture, and they were maintained at this greater length for at least 20 rounds. However, we observed that telomeric DNA tracts were maintained at a constant size in both wild-type and 35S::GUS control transgenic cells cultured for long periods (Figure 4C), suggesting that alterations in the length of telomeres was not an experimental artifact but was specific to the action of sense and antisense NgTRF1 mRNAs in transgenic lines. Thus, we interpret these results as evidence that there is an inverse correlation between the amount of functional NgTRF1 and the length of telomeres in BY-2 cells. With this in mind, we speculate that NgTRF1 is involved in telomere length regulation in tobacco cells.
Growth Defects of 35S::NgTRF1 and 35S::anti-NgTRF1 Transgenic Cells
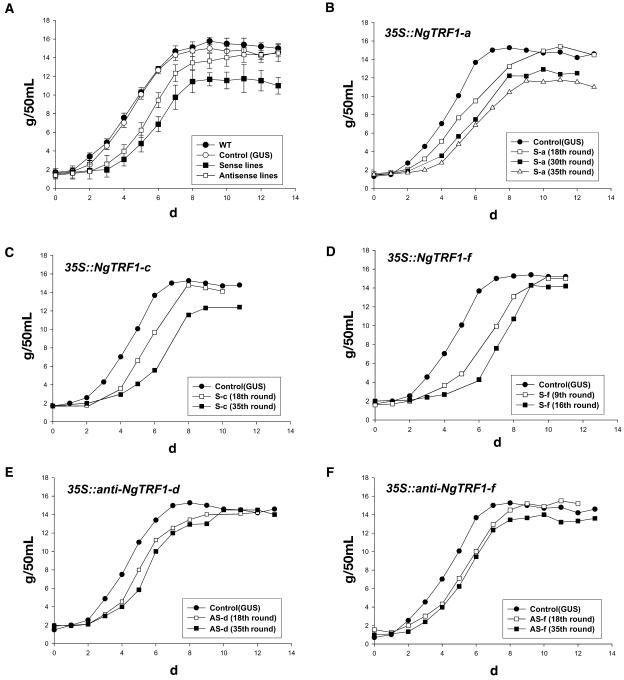
To address the mechanistic outcome of alterations in the level of NgTRF1 and changes in telomere length, we next assayed the proliferative capacity of 35S::NgTRF1 and 35S::anti-NgTRF1 transgenic cell lines. Growth curves determined by estimating the cellular fresh weight revealed that wild-type and 35S::GUS control lines displayed typical growth patterns with comparable growth rates during the 12-d batch culture (Figure 5A). Cellular proliferation began very rapidly after subculture. After a short lag phase period of ∼1 d, cells rapidly entered the logarithmic growth phase and reached the stationary stage after 7 d. On the other hand, 35S::NgTRF1 cells grew more slowly than both wild-type and 35S::GUS cells. Their slow growth rate was most easily observed at early stages because they exhibited a long (2 to 3 d) lag phase. Consequently, these transgenic lines slowly progressed into the logarithmic growth stage, showed a significantly lower fresh weight population during days 4 to 8, and eventually reached stationary phase 9 to 10 d after subculture (Figures 5A to 5D). We did not observe any difference in cell size between these transgenic lines and wild-type BY-2 cells, indicating that the slow growth rate did not result from changes in cell size (data not shown). More importantly, we noted that every 35S::NgTRF1 cell line examined displayed a gradual retardation in growth rate during prolonged subculture. For example, the growth rates of 35S::NgTRF1-a and 35S::NgTRF1-c lines were significantly reduced after 35 successive rounds of subculture (413 d after gene transfer) compared with that after the 18th round (209 d after gene transfer) (Figures 5B and 5C). Similarly, the proliferative ability of 35S::NgTRF1-f cells in the 16th round of subculture (185 d after gene transfer) was lower than that of ninth-round cells (104 d after gene transfer) (Figure 5D). This result indicates that 35S::NgTRF1 transgenic cells with shorter telomeres exhibit a progressive decline in cell viability. To our surprise, none of the 35S::NgTRF1 cells could divide after 300 to 450 d (depending on the cell line) of suspension culture, and eventually died. Among the three 35S::NgTRF1 cell lines, line f showed the most severe growth defect and died after 25 successive rounds of subculture, in sharp contrast with wild-type and 35S::GUS control cells, which had an unlimited capacity for cell division.
Figure 5.
Growth Curves for Transgenic BY-2 Cells.
Growth curves of wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 tobacco cells were determined by estimating the fresh weight of the cells during prolonged culture periods.
(A) Comparison of growth curves for wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 tobacco cells.
(B) to (D) A gradual retardation in growth rate was found for 35S::NgTRF1 transgenic tobacco cells during 18 to 35 successive rounds of batch subculture (lines a and c) and during 9 to 16 successive rounds of batch subculture (line f).
(E) and (F) Decreased cell viability was also found for 35S::anti-NgTRF1 transgenic cells during 18 to 35 successive rounds of batch subculture (lines d and f).
We next monitored the cell division capacity of 35S::anti-NgTRF1 cells. Because 35S::NgTRF1 cells, which contain shorter telomeres, displayed a growth defect, we expected that 35S:anti-NgTRF1 cells, which have longer telomeres, would not. However, as presented in Figures 5A, 5E, and 5F, the antisense transgenic lines also showed moderate reductions in growth rate with a 2-d-long lag phase when compared with wild-type and 35S::GUS control cells. As were the cases for the 35S:NgTRF1 lines, the growth rates of antisense line d and line f cells progressively decreased with prolonged subculture (Figures 5E and 5F). Thus, it appears that proper level of NgTRF1 is required for the maintenance of cell viability. These reductions, however, were less pronounced than those seen for the sense transgenic lines and, hence, 35S::anti-NgTRF1 lines exhibited the phenotype intermediate between that of wild-type and 35S::NgTRF1 cells (Figure 5A). Unlike sense lines, 35S:anti-NgTRF1 cells were still alive after 40 successive rounds of subculture, although their proliferative capacities were significantly lowered. These results reveal that the amount of functional NgTRF1 is critically associated with cell viability: growth is progressively reduced in response to an elevated level of NgTRF1, which induces telomere shortening, and also in response to a decreased level of the protein during long-term culture. Taken together, our transgenic analyses support the notion that NgTRF1 plays a role in telomere length regulation (Figure 4) and cell viability (Figure 5) in BY-2 cells.
35S::NgTRF1 Cells Undergo Apoptosis
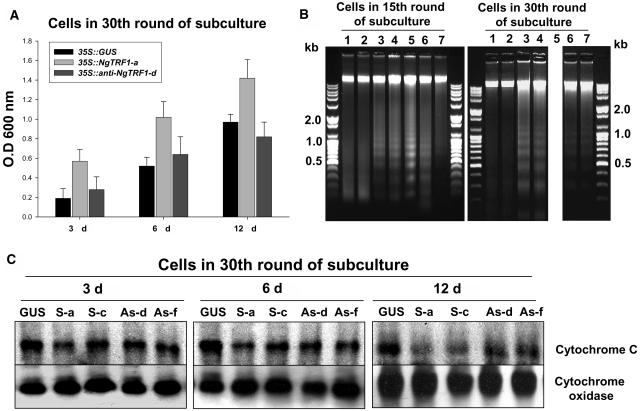
To further define the physiological relevance of the changes in telomere length and the decrease in viability of transgenic BY-2 cells, both of which resulted from perturbation of the NgTRF1 level, we assessed the extent of cell death using the Evans blue assay. To make experimental conditions consistent, we used transgenic cells from the 30th round of batch subculture. During the 12-d culture, sense and antisense lines were scored for cell death as indicated by the intensity of Evans blue staining (Figure 6A). In 35S::GUS control cells, Evans blue staining increased concomitantly with the progression of culture and attained a maximal level at day 12, indicating that the percentage of dead cells peaked at stationary phase. Notably, 35S::NgTRF1 cells showed a significantly higher degree of Evans blue uptake throughout the entire period, suggesting that the percentage of living cells declined in sense lines. As determined by growth-curve analysis, the differential degree of Evans blue staining was most readily seen in early growth phase. In 3-d-old cells, the intensity of staining in the sense line was ∼3 times higher than in control cells, whereas there was a 1.5-fold induction of staining at day 12 (Figure 6A). On the other hand, the staining patterns of 35S::anti-NgTRF1 cells were intermediate between those of the GUS control and sense cells at day 3 and 6 of subculture, indicating that a low level of NgTRF1 also caused increased death of tobacco cells. These results, in concert with the growth-curve data, are consistent with the view that cell death was more frequent in 35S::NgTRF1 lines, and also in 35S::anti-NgTRF1 lines, than in the GUS control line. Thus, cell death appears to be sensitive to the level of functional NgTRF1 in BY-2 cells.
Figure 6.
Effect of Altered NgTRF1 Expression on Viability of Transgenic BY-2 Cells.
(A) Evaluation of cell death in 35::GUS, 35::NgTRF1-a, and 35S::anti-NgTRF1-d cells. Transgenic cells at the 30th round of batch subculture were stained with Evans blue as described in Methods. Cellular uptake of Evans blue was quantified by spectrophotometry. Data are expressed as the means ± sd of three experiments.
(B) Oligonucleosomal DNA fragmentation in 35::NgTRF1 and 35S::anti-NgTRF1 cells. Total genomic DNA was isolated from wild-type, 35::GUS, 35::NgTRF1, and 35S::anti-NgTRF1 cell lines at the 15th or 30th round of batch subculture. DNA was analyzed by 1.2% agarose gel electrophoresis with ethidium-bromide staining. Lane 1, wild type; lane 2, 35::GUS; lane 3, 35::NgTRF1-a; lane 4, 35::NgTRF1-c; lane 5, 35::NgTRF1-f; lane 6, 35S::anti-NgTRF1-d; lane 7, 35S::anti-NgTRF1-f. Positions of molecular weight–size markers are shown at the side of each panel. Note that a DNA sample from 35::NgTRF1-f cells at 30th round of batch subculture could not be obtained (lane 5) because these cells died after 25 rounds.
(C) Immunoblot analysis of cytochrome c release from mitochondria of transgenic BY-2 cells in the 30th round of batch subculture. Proteins were isolated from mitochondrial fractions, separated on 12.5% (w/v) SDS-PAGE, and analyzed by protein gel blotting with an antibody against cytochrome c. The level of cytochrome oxidase was also monitored as a protein loading control. GUS, 35S::GUS; S-a, 35S::NgTRF1-a; S-c, 35S::NgTRF1-c; As-d, 35S::anti-NgTRF1-d; As-f, 35S::anti-NgTRF1-f.
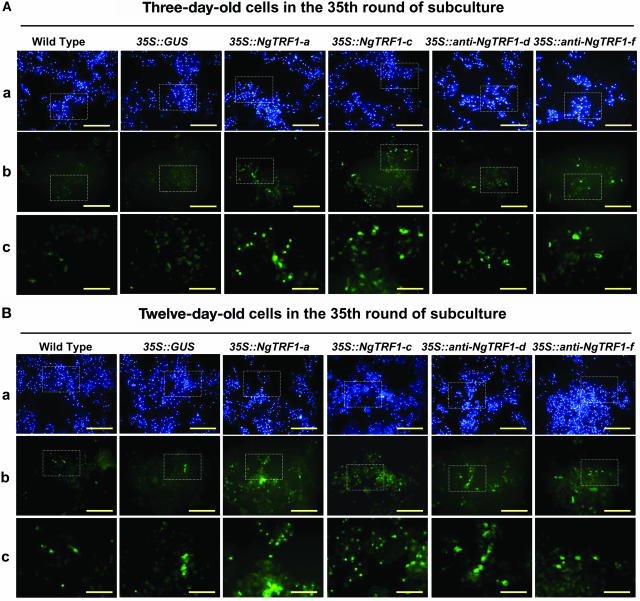
These results prompted us to investigate the mechanism by which cell death occurred. We first used 4′,6-diamidino-2-phenylindole (DAPI), a UV-excitable dye that can specifically bind double-stranded DNA, to assess nuclear structure. Wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 cells were fixed, stained with DAPI, and observed by fluorescence microscopy. As shown in Figures 7A and 7B, most of the transgenic cells in the 35th round of subculture were DAPI positive, indicating that nuclear degradation did not occur to an appreciable extent in these cells during the 12-d batch culture. We next employed the terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) technique, an in situ assay that measures the fragmentation of DNA in individual cells by the incorporation of fluorescein 12-dUTP at free 3′-OH ends (Gavrieli et al., 1992; Mittler and Lam, 1995). The results of the TUNEL assay are summarized in Figure 7 and Table 1. Nearly all of the 3-d-old wild-type and GUS control cells exhibited TUNEL-negative nuclei although there was a slight increase in the number of TUNEL-positive nuclei in cells in the stationary growth phase (Figure 7). We observed that ∼2% of the DAPI-positive signals in control cells were TUNEL-positive at day 12 (Table 1). Notably, there was a marked elevation in the number of TUNEL-positive 35S::NgTRF1 nuclei in the 35th round of subculture. The percentage of TUNEL-positive cells was estimated to be 9.5 to 12.0% at day 3 and 14.0 to 17.6% at day 12, depending on the transgenic line (Table 1). This suggests that the lower cell viability detected by Evans blue staining may account for the higher percentage of TUNEL-positive cells in 35S::NgTRF1 lines. Likewise, there were TUNEL-positive cells in both the 35S::anti-NgTRF1-d and -f lines, but the frequency of these cells was lower (4.1 to 7.3%) compared with the 35S::NgTRF1 cells (Figure 7, Table 1), a result that is consistent with the growth curve (Figure 5) and Evans blue staining (Figure 6A) data.
Figure 7.
Nuclear Staining and in Situ Detection of DNA Fragmentation in Wild-Type and Transgenic Tobacco BY-2 Cells at Different Stages of Subculture.
(A) Three-day-old wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 transgenic cells in the 35th round of batch subculture were stained with DAPI (panel a) and assayed with the TUNEL technique (panel b) as described in Methods to detect nuclei and fragmented nuclear DNA, respectively. Panel c shows a magnified view of the boxed portion in panels a and b. Bars = 25 μm in panels a and b; bars = 100 μm in panel c.
(B) Twelve-day-old wild-type, 35S::GUS, 35S::NgTRF1, and 35S::anti-NgTRF1 transgenic cells in the 35th round of batch subculture were stained with DAPI (panel a) and assayed with the TUNEL technique (panel b). Panel c shows a magnified view of the boxed portion in panels a and b. Bars = 25 μm in panels a and b; bars = 100 μm in panel c.
Table 1.
Percentage of Apoptotic Cells of Tobacco BY-2 Cells in the 35th Round of Subculture
| Days in Subculture | Cell Line | Total Cell No. Counted | Apoptotic Cell No. | Ratio |
|---|---|---|---|---|
| Three-day-old cells | Wild type | 1950 | 4 | 0.02% |
| 35S::GUS | 1840 | 5 | 0.02% | |
| 35S::NgTRF1-a | 1752 | 211 | 12.0% | |
| 35S::NgTRF1-c | 1356 | 129 | 9.5% | |
| 35S::Anti-NgTRF1-d | 1950 | 89 | 4.6% | |
| 35S::Anti-NgTRF1-f | 1940 | 80 | 4.1% | |
| Twelve-day-old cells | Wild type | 1900 | 44 | 2.3% |
| 35S::GUS | 1840 | 37 | 2.0% | |
| 35S::NgTRF1-a | 1645 | 291 | 17.6% | |
| 35S::NgTRF1-c | 1657 | 232 | 14.0% | |
| 35S::Anti-NgTRF1-d | 1570 | 115 | 7.3% | |
| 35S::Anti-NgTRF1-f | 1640 | 85 | 5.2% |
Nuclear DNA fragmentation is typical of apoptotic cell death (Ellis et al., 1991; Gavrieli et al., 1992; Vaux and Strasser, 1996; Lam et al., 1999). Because the in situ TUNEL assay is a sensitive method to detect chromosomal degradation, we hypothesized that the enhanced cell death of NgTRF1 transgenic lines was mediated by an apoptosis-like mechanism. To address this possibility, we extracted total genomic DNA from wild-type and transgenic BY-2 cells and analyzed it by agarose gel electrophoresis with ethidium-bromide staining. Interestingly, the results shown in Figure 6B revealed apoptosis-specific DNA ladders in 35S::NgTRF1 cells. This DNA profile appeared in the 15th round of subculture, and became more pronounced at later stages of batch subculture (the 30th round of subculture), which correlates well with the progressive DNA fragmentation observed using the TUNEL assay (Figure 6B). The ladders are composed of multiples of ∼180 bp, the approximate length of DNA contained within a nucleosome, and are indicative of internucleosomal DNA cleavage. Similarly, ladders were also detected in 35S::anti-NgTRF1 lines, albeit to a lower degree than in 35S::NgTRF1 lines (Figure 6B). By contrast, we failed to detect the induction of DNA cleavage in wild-type and GUS control cells during the 12-d culture period. Thus, nuclear DNA fragmentation is not an experimental artifact but is specific to NgTRF1 transgenic cells.
In mammalian cells, mitochondria play a pivotal role in the induction of apoptosis by releasing intermembrane space components into the cytosol, such as cytochrome c, a key enzyme of the electron transport chain (Cai et al., 1998; Green and Reed, 1998). Cytochrome c then activates caspase-mediated proteolysis, leading to the internucleosomal cleavage of chromosomal DNA (Enari et al., 1998). Recently, translocation of cytochrome c from mitochondria to the cytosolic fraction has been reported during apoptosis and programmed cell death (PCD) in higher plants (Stein and Hansen, 1999; Sun et al., 1999; Balk and Leaver, 2001; Tiwari et al., 2002). These studies indicate that cytochrome c release is an essential step in plant cell PCD as in animal PCD. Thus, we determined the localization of cytochrome c in control and NgTRF1 transgenic cells. Mitochondrial proteins extracted from GUS and NgTRF1 transgenic cells were separated by SDS-PAGE and probed with a specific antibody. In control cells, the amount of cytochrome c in the mitochondrial fraction was virtually constant during the entire period of suspension subculture, indicating that there was only negligible release of cytochrome c (Figure 6C). However, in 35S::NgTRF1 lines, the abundance of mitochondrial cytochrome c protein varied depending on the growth phase. The level gradually decreased during the progression of subculture, with a very low level being detectable at day 12, when transgenic cells exhibited maximal TUNEL-positive signals and DNA fragmentation (Figure 6C). Notably, a difference in the amount of cytochrome c in 35S::GUS and 35S::NgTRF1 lines was apparent in 3- and 6-d-old culture cells, suggesting the early release of cytochrome c from mitochondria in NgTRF1 sense cells. Cytochrome c oxidase, a membrane-bound enzyme involved in the electron transport chain, was assayed by immunoblot analysis as a negative control. The level of this protein remained unchanged in all stages of culture in every transgenic line tested (Figure 6C). The loss of cytochrome c from mitochondria was also detected in 35S::anti-NgTRF1 cells, but, as found previously, fluctuations in the level of protein were less pronounced than in 35S::NgTRF1 cells. Therefore, cell death in 35S::NgTRF1 and 35S::anti-NgTRF1 transgenic lines is accompanied by the release of cytochrome c from mitochondria, raising the possibility that an apoptosis-like mechanism is responsible for the high frequency of death in these cells. Taken together, our results show that tobacco cells containing a high level of NgTRF1 and short telomeres lose their immortality through apoptotic cell death. Furthermore, a specific level of NgTRF1 might be essential for telomere functions, and hence, cells lacking NgTRF1 are subject to apoptosis. Based on these results, we hypothesize that perturbation of NgTRF1 expression results in changes in telomere length and stability, which, in turn, triggers apoptosis-like cell death in transgenic cells.
DISCUSSION
In most species investigated to date, telomeres are highly conserved structures that consist of tandemly repeated G-rich arrays of DNA that are synthesized and maintained by the action of telomerase. Mammalian telomeres and telomerase have attracted much interest, as their structures and activities are intimately linked to cellular proliferation, differentiation, aging, and tumor growth (Zakian, 1995; Linger and Cech, 1998; Nugent and Lundblad, 1998; Liu, 1999). Although much less is known about plant telomeres and telomerase in comparison with mammalian systems, it is increasingly likely that the function and structure of telomeres and also the patterns of telomerase control are largely conserved in higher plants and animals (Shippen and McKnight, 1998; McKnight et al., 2002). The primer extension/nick translation assay was used to show that telomere architecture is developmentally regulated in the dicot plants Silene latifolia and Arabidopsis; the fraction of telomeres with detectable G-overhang structures changes substantially, from 50% in seedlings to 30% in leaves (Riha et al., 2000). These results, along with the growth- and cell division–dependent expression of NgTRF1 and telomerase activity (Yang et al., 2002, 2003), suggest that plant telomeres are developmentally dynamic. Arabidopsis telomerase-null mutants generated by a T-DNA insertion exhibit a decrease in telomere length by ∼500 bp per generation (Fitzgerald et al., 1999). Although Arabidopsis can survive for up to 10 generations without telomerase, the last five generations of telomerase-deficient mutants show severe cytogenetic damage with numerous developmental anomalies, indicating that telomerase activity is critical for the maintenance of telomere integrity in higher plants (Riha et al., 2001). Thus, as was found for mammals, the structure and function of telomeres are essential for the plant life cycle.
Studies on the organization of telomeric chromatin indicate that telomeres are packaged into specialized nucleoprotein complexes and that nonhistone proteins, an integral component of these complexes, are responsible for telomere integrity and function (Collins, 2000; Blackburn, 2001; Shore, 2001). Telomere binding proteins have been extensively characterized in different eukaryotes, including ciliates, yeast, and humans. These proteins have been shown to suppress telomere elongation and to be involved in the regulation of telomere length in vivo. A negative feedback function that regulates telomere length has been identified for yeast Rap1p and human TRF1/Pin2 and TRF2, which inhibit the action of telomerase at the ends of telomeres (van Steensel and de Lange, 1997; Wotton and Shore, 1997; Smogorzewska et al., 2000; Smucker and Turchi, 2001).
We recently identified NgTRF1 as a double-stranded telomeric repeat binding factor in tobacco plants (Yang et al., 2003). Expression analysis showed that NgTRF1 gene activity was negatively associated with the cell division capacity of tobacco BY-2 suspension cells, whereas telomerase activity was positively correlated with cell division, indicating that telomerase activity and NgTRF1 expression are regulated in an opposing fashion during growth (Yang et al., 2002, 2003). In this study, we obtained biochemical data that indicate that NgTRF1 effectively inhibits telomerase activity in vitro (Figures 1 and 2). These and previous results raise the tantalizing possibility that NgTRF1 is involved in the control of telomerase activity and thereby mediates telomere length in tobacco cells. This hypothesis is particularly crucial because virtually nothing has been known about the in vivo function of telomere binding proteins in higher plants. To address this proposal, we constructed transgenic BY-2 cells that overexpressed or suppressed NgTRF1. By pulsed-field gel electrophoresis, we detected typical telomeric smears consisting of fragments 65- to 15-kb long, whose length was maintained during prolonged culture periods in both wild-type and 35S::GUS control cells (Figure 4C). However, we found that 35S::NgTRF1 tobacco cells had significantly shortened telomeres (45 to 10 kb), which varied in length in different transgenic lines and at different stages of subculture (Figure 4A). Closer inspection revealed that 65- to 60-kb–long telomeres were shortened by up to 25 kb, whereas shorter telomeres, 20- to 15-kb long, were shortened by only 5 kb, suggesting that longer telomeres are more sensitive to an elevated level of NgTRF1. Another striking phenotype of NgTRF1-overexpressing cells was a progressive reduction of cell viability (Figures 5 and 6A). Consequently, all 35S::NgTRF1 cells with short telomeres died after 25 to 40 successive rounds of 12-d batch subculture. The enhanced death of 35S::NgTRF1 cells appeared to be attributable to an apoptosis-like mechanism. This view is supported by the fragmentation of internucleosomal DNA (Figure 6B), the mitochondrial release of cytochrome c (Figure 6C), and the presence of TUNEL-positive nuclei (Figure 7) in 35S::NgTRF1 cells, all of which are typical phenomena associated with apoptosis.
There is evidence that telomere shortening leads to apoptosis in human cells. For instance, cells with inducible dominant-negative mutants of hTERT, which have a markedly reduced level of endogenous telomerase activity, exhibit chromosomal damage and short telomeres, which in turn are associated with apoptotic cell death in human tumor A431 cells (Zhang et al., 1999). It has been suggested that the perpetuation of tumor and other immortal cells requires telomerase activity and telomere length maintenance, and tumors with short telomeres might be effectively and rapidly killed (Zhang et al., 1999). Overexpression of TRF1/Pin2 in human breast tumors induces apoptosis in cells with short telomeres, but not in cells containing long telomeres, indicating that apoptotic cell death is telomere-length dependent (Kishi et al., 2001). Thus, upregulation of TRF1/Pin2 can induce apoptosis in cancer cells. Those results, along with the data in this study, lead us to consider that overexpression of NgTRF1 inhibits telomerase activity and reduces telomere length during long-term subculture, which in turn induces apoptotic cell death in transgenic BY-2 cells. Thus, it is tempting to propose that a balance between telomerase activity and the level of functional NgTRF1 acts as a molecular switch to maintain the immortality of BY-2 cells. There may be possible antagonistic function between NgTRF1 and telomerase activity in the control of telomere length. We currently do not know the mechanism by which NgTRF1 inhibits telomerase activity. In this regard, it is worth noting that the purified Myb domain of NgTRF1 inhibited telomerase activity in vitro to an extent comparable to that of full-length NgTRF1 (Figure 2C). In addition, NgTRF1 failed to inhibit telomerase activity in the presence of a single-stranded primer substrate; an inhibitory effect was found only with a double-stranded primer substrate that contains the NgTRF1 binding site (Figure 2C). This result suggests that NgTRF1 might not directly interact with the telomerase complex. In human HeLa cells, the Pin2 binding protein PinX1, but not Pin2, was shown to bind hTERT, thereby inhibiting telomerase activity and influencing tumorigenicity (Zhou and Lu, 2001). It was previously reported that tobacco nuclei contain telomeric proteins that inhibit telomerase activity and hence might be involved in telomere length regulation (Fulneckova and Fajkus, 2000). Thus, it is possible that an as-yet unidentified NgTRF1-associating factor(s) may control telomerase activity. Alternatively, NgTRF1 may function as a major structural component of telomeres and may simply hinder the accessibility of telomeric ends to telomerase complexes, resulting in the nonspecific inhibition of telomerase activity.
In contrast with what we observed in 35S::NgTRF1 lines, 35S::anti-NgTRF1 cells contained a very low level of NgTRF1 protein (Figure 3B) and exhibited significantly longer telomeres during long-term subculture (Figure 4B). Thus, the downregulation of NgTRF1 results in longer telomeres, as opposed to upregulation, which causes telomere shortening in transgenic BY-2 cells, indicating that NgTRF1 is indeed functional in tobacco cells. Recently, it was reported that Caenorhabditis elegans with longer telomeres lived longer (Joeng et al., 2004). The extension of lifespan of worms, which was achieved by the overexpression of HRP1, a single-stranded telomeric binding protein, was found to be attributed to increased telomere length and not to the everexpression of HRP-1 per se (Joeng et al., 2004). However, we clearly observed that antisense-NgTRF1 transgenic lines with longer telomeres displayed an intermediate phenotype between that of 35S::GUS control cells and 35S::NgTRF1 cells in terms of cell viability and apoptosis. The 35S::anti-NgTRF1 lines showed a progressive reduction in cell viability and increased apoptosis during batch subculture, but to a lesser extent than did NgTRF1-overexpressing lines (Figures 5, 6, and 7). Several lines of evidence indicate that telomere function critically depends on structural aspects of telomeres in addition to their length, and that formation of a functional telomere requires the expression of telomere binding proteins (Blackburn, 2001). In various mammalian cell types, a dominant negative version of TRF2 induces apoptosis that is mediated by the p53- and ataxia telangiectasia mutated kinase-dependent pathway. This indicates that telomeres lacking TRF2 directly trigger apoptosis, perhaps because they resemble damaged DNA (Karlseder et al., 1999). Karlseder et al. (2002) recently proposed that the cellular senescence of primary human culture cell lines is induced by a change in the protected status of telomeres rather than by a loss of telomeric DNA. Furthermore, deletion of TRF1 in mouse embryonic stem cells induces growth defects and chromosomal instability. However, shortening or elongation of telomeres in these cells was not observed in short-term culture, which indicates that TRF1 has a crucial function in telomere structure in addition to length regulation (Iwano et al., 2004). Taken together, it is attractive to speculate that the growth defect and apoptosis of 35S::anti-NgTRF1 tobacco cells is not because of longer telomeres per se, but possibly results from the destabilization of telomeric architecture caused by a low level of NgTRF1. The cellular level of NgTRF1 may serve as a regulatory marker in the cell division program, and telomeres lacking NgTRF1 may signal cells to cease proliferation and to enter apoptosis. Thus, the structural integrity of telomeres, in addition to their proper length, could be a critical factor for telomere functions in tobacco cells. Overall, this proposal is consistent with the notion that NgTRF1 has a dual nature in BY-2 cells: it is involved in the control of telomerase activity, and it functions as a structural component of telomeric ends.
Among the numerous transgenic BY-2 cell lines constructed, the 35S::NgTRF1-f line exhibited the most severe phenotype. These cells died after 25 successive rounds of batch subculture. It should be noted that although all NgTRF1-overexpressing cells have short but similarly sized telomeres, 35S::NgTRF1-f cells contain the shortest telomeres observed, 35 to 10 kb (Figure 4A, panel c). Interestingly, Hemann et al. (2001) observed that it is not the average but rather the shortest telomeres that signal telomere dysfunction and limit the survival of mouse cells. It would be intriguing to determine whether the severe phenotype of the 35S::NgTRF1-f line is indeed because of shortened telomeres.
Although NgTRF1 transgenic cells had marked growth defects, which were possibly caused by telomere shortening or telomere dysfunction, they died only after long-term batch subculture (25 to 45 successive rounds of 12-d culture). Riha et al. (2001) found that later generations of telomerase-deficient Arabidopsis mutants have an unexpectedly extended lifespan and remained metabolically active, which is consistent with the more plastic nature of plant development and genome organization. The long-term survival of NgTRF1 tobacco cells may reflect a greater tolerance for telomeric dysfunction, in comparison with animal cells. We are now constructing transgenic tobacco plants that overexpress or suppress the NgTRF1 gene. The detailed characterization of these plants will help us to more precisely define the biochemical and physiological functions of NgTRF1 in tobacco cells. Further functional studies of NgTRF1 and characteristics of the proteins it interacts with will be required to further the understanding of the relationship between cell viability and the dynamic functions of telomeres in higher plants.
METHODS
Tobacco Suspension Cell Culture
The tobacco BY-2 (Nicotiana tabacum cv bright yellow-2) suspension cell line was maintained in Murashige and Skoog salt medium (Wako Pure Chemical, Osaka, Japan) on a rotary shaker (150 rpm) at 25°C in the dark. Every 8 to 12 d, 10 mL of stationary phase cells were transferred to 90 mL of fresh medium.
Cloning and Expression of Full-Length NgTRF1 and Deletion Mutants
The plasmids pGEX4T-1 (Pharmacia, Uppsala, Sweden) and pProEX (Invitrogen, Groningen, The Netherlands) were used for the expression of the GST-NgTRF1 and 6X His-NgTRF1 fusion proteins. Escherichia coli BL21 (DE3) strains containing the plasmids were grown at 37°C in 100 mL 2× LB medium (10 g Tryptone, 10 g yeast extract, 5 g NaCl per liter) supplemented with 1% glycerol as an additional carbon source and 70 μg/mL ampicillin. Cells at OD600 0.6 to 1.0 were induced with 1 mM IPTG, grown for an additional 4 h, and centrifuged. The pellet was resuspended in phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride. The suspension was sonicated on ice with Vibracell sonicator (Sonics and Materials, Danbury, CT), and Triton X-100 was added to a final concentration of 1%. Fusion proteins were purified by affinity chromatography using glutathione Sepharose 4B from GST purification modules (Pharmacia) or Ni-NTA agarose column chromatography (QIAGEN, Valencia, CA). The GST tags were removed by cleavage with thrombin.
Fractionation of the Telomerase Complex by Gel Filtration
Tobacco cell extracts were obtained using the protocol described by Fitzgerald et al. (1996) with modifications. Briefly, a portion (0.5 g) of BY-2 cells was ground with a mortar and pestle in liquid nitrogen and suspended in 2 mL ice-cold extraction buffer W (50 mM Tris-acetate, pH 7.5, 5 mM MgCl2, 100 mM potassium glutamate, 20 mM EGTA, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 0.6 mM vanadyl ribonucleoside complex, 1.5% [w/v] polyvinylpyrrolidone, and 10% glycerol). The extract was centrifuged at 13,000 rpm for 15 min at 4°C, PEG 8000 (Sigma, St. Louis, MO) was added to the supernatant to a final concentration of 10%, and the mixture was stirred for 30 min at 4°C and centrifuged at 13,000 rpm for 5 min. The resulting pellet was resuspended in 0.5 mL buffer W and centrifuged at 13,000 rpm for 5 min at 4°C. The supernatant was used immediately for telomerase activity assay or stored at −80°C. The protein concentration of the cell extracts was determined by the method of Bradford using a Bio-Rad (Hercules, CA) protein assay kit with BSA as a standard.
Telomerase complexes were fractionated by gel filtration at 4°C on a Sephacryl S-400 HR column (2.5 × 100 cm; Pharmacia) equilibrated with buffer W. Tobacco lysates (15 mL) were loaded and separated at a flow rate of 1 mL/min. Fractions were tested by the TRAP assay (below) to estimate the level of telomerase enzyme activity.
Telomerase Activity Assay
The level of telomerase enzyme activity was monitored according to a modified version of the TRAP as described previously (Yang et al., 2002). The TRAP assay was performed in 40 μL of a reaction mixture containing 50 mM Tris-acetate, pH. 8.3, 50 mM potassium glutamate, 0.1% Triton X-100, 1 mM spermidine, 1 mM DTT, 50 μM each deoxynucleotide triphosphate, 5 mM MgCl2, 10 mM EGTA, 100 ng/μL BSA, and 0.5 unit Taq polymerase (Promega, Madison, WI). After the addition of partially purified tobacco cell extracts containing 1 μg total protein and 50 ng single-stranded forward primer (5′-CACTATCGACTACGCGATCGG-3′) or double-stranded forward primer (5′-CCCTAAACCCTAAACCCACTGCTCGATGCACTGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGT-3′) (Figure 2B), the telomerase reaction was allowed to proceed for 45 min at 24°C. As a negative control, some samples were pretreated with 10 ng RNase A (Sigma) or heated at 95°C for 10 min. Telomerase reaction products were amplified by 30 cycles of PCR with a reverse primer (50 ng; 5′-CCCTAAACCCTAAACCCTAAA-3′), each consisting of 30 s at 94°C, 30 s at 65°C, and 90 s at 72°C, with an additional 5 min at 72°C in an automatic thermal cycler (Perkin-Elmer/Cetus, Norwalk, CT). PCR products were separated on a 10% nondenaturing polyacrylamide gel, and the gel was stained in 0.01% SYBR Green I for 45 min and scanned on a FluorImager (Fuji, Tokyo, Japan). The activity of telomerase was calculated by the ratio of the intensity of TRAP ladders. For the inhibition assay, various concentrations (1 to 5 × 10−4 M) of the NgTRF1, GST-NgTRF1441-681, or NgTRF11-567 deletion mutant was added to the TRAP reaction buffer before the addition of partially purified telomerase super complex and then incubated for 10 min. After incubation, the TRAP assay was performed as described above.
Construction of Sense and Antisense Transgenic BY-2 Cell Lines
The vector pBI121, which contains the 35S promoter of Cauliflower mosaic virus and NOS terminator, was used for tobacco transformation. The full-length NgTRF1 cDNA (2.15 kb) with the complete open reading frame was generated by PCR using high-fidelity Ex-Tag polymerase (Takara, Kyoto, Japan). To facilitate the subcloning of PCR products, forward and reverse primers were tagged with restriction enzyme sites (BamHI and SacI). The GUS reporter gene of pBI121 was eliminated, and the tagged pNgTRF1 cDNAs were inserted into the corresponding sites of pBI121 in the sense and antisense orientations. The fusion gene constructs were transferred to Agrobacterium tumefaciens LBA4404 by electroporation according to Lee and Kim (2003). Agrobacterium was then cocultured with 2-d-old fresh tobacco BY-2 cells for 48 h. Transgenic tobacco cells were selected on MS medium supplemented with 100 μg mL−1 kanamycin and 100 μg mL−1 carbenicillin. The presence of sense and antisense transgenes in the transformants was confirmed by genomic DNA gel blot and PCR analyses. The 35S::NgTRF1 and 35S::anti-NgTRF1 transgenic tobacco cells and 35S::GUS control cells were subsequently maintained in Murashige-Skoog salt medium on a rotary shaker (150 rpm) at 25°C in the dark. Stationary phase cells (10 mL) were transferred at 12-d intervals to 90 mL of fresh medium.
Genomic DNA Isolation and Pulsed-Field Gel Electrophoresis
Total genomic DNA of tobacco BY-2 cells was isolated as described previously (Lee and Kim, 2003) and digested with TaqI restriction enzyme. The DNA fragments (10 μg per lane) were separated in a 1.0% agarose gel by pulsed-field gel electrophoresis using a CHEF-DRIII system (Bio-Rad) at 6 V/cm for 18 h at an angle of 120°, with switching times ramped from 1 to 10 s at 14°C. The gel was blotted onto a nylon membrane filter (Bio-Rad) and then hybridized to a 32P-labeled (TTTAGGG)70 fragment under stringent hybridization and washing conditions. The blots were washed and visualized by autoradiography at −80°C. Hybridization signals were quantified with a phosphorimager (Fuji).
Total RNA Isolation and RNA Gel Blot Analysis
Total RNA was prepared from wild-type and transgenic BY-2 suspension culture cells as described previously (Yang et al., 2003). RNA (20 μg per lane) was fractionated by electrophoresis in a 1% (w/v) formaldehyde-agarose gel and blotted onto a nylon membrane filter (Bio-Rad). Equal loading was confirmed by visualizing the ethidium bromide-stained rRNA content under UV light after electrophoresis. The filter was probed with a 32P-labeled EcoRI/BamHI fragment of pNgTRF1 under high-stringency hybridization and washing conditions. The blots were washed and visualized by autoradiography at −80°C. Hybridization signals were quantified with a phosphorimager (Fuji).
Protein Gel Blot Analysis
Protein samples stored at −80°C were separated by 12.5% SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked against nonspecific binding by incubation in PBS blocking buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.01% [w/v] Tween 20, and 5% [w/w] dried nonfat milk) for 1 h at room temperature and then incubated with anti-NgTRF1 antibody (1:10,000 dilution) or anti-actin antibody (1:1000 dilution; Sigma) as a loading control. The membrane was washed three times with PBS blocking buffer, incubated with horseradish peroxidase-conjugated anti-rabbit IgG (Amersham Life Science, Buckinghamshire, UK) for 1 h, and washed extensively with PBS buffer. Proteins were visualized with the ECL1 Plus Protein gel-blotting detection system (Amersham Life Science) and exposure to Kodak BioMax ML film.
Detection of Cell Death by Evans Blue Staining
Cells were stained for 5 min with a 1% solution of Evans blue and then washed five times with distilled water to remove excess unbound dye. Dye bound to dead cells was solubilized in a solution of 50% methanol and 1% SDS for 30 min at 50°C and quantified by absorbance at 600 nm.
Analysis of Cytochrome c Release
BY-2 cells were ground in a mortar and pestle on ice in grinding buffer [0.4 M mannitol, 25 mM 3-(N-morpholino)-propanesulfonic acid-KOH, pH 7.8, 1 mM EGTA, 0.1% (w/v) BSA, and 40 mM β-mercaptoethanol]. Cell debris was removed by centrifugation at 6000g for 5 min. The supernatant was recentrifuged at 12,000g for 20 min to pellet mitochondria. The crude mitochondrial pellet was resuspended in washing buffer [0.4 M mannitol, 5 mM 3-(N-morpholino)-propanesulfonic acid-KOH, pH 7.5, 1 mM EGTA, and 0.1% (w/v) BSA], layered on a Percoll density gradient (0.25 M sucrose, 5 mM EDTA, 1 mM EGTA, 0.1% [w/v] BSA, 10 mM Hepes-Tris, pH 7.5, and 15 to 28% [v/v] Percoll), and centrifuged at 40,000g for 40 min. The layer with purified mitochondria was removed using a pipette and diluted with resuspension buffer (0.4 M mannitol, 10 mM Hepes-Tris, pH 7.5, 1 mM EGTA), and the protein concentration was determined by the method of Bradford using a Bio-Rad protein assay kit with BSA as a standard.
Mitochondrial protein samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane filter. The membrane was blocked against nonspecific binding as described above and incubated with anti-cytochrome c monoclonal antibody (1:1000 dilution; Pharmingen Bioscience, San Diego, CA) or anti-cytochrome c oxidase antibody (1:1000 dilution). Each protein was visualized with the ECL1 Plus Protein gel blotting detection system (Amersham Life Science) and exposure to Kodak BioMax ML film as described previously.
Nuclear Staining and Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling
Wild-type and transgenic BY-2 cells were harvested and washed with PBS buffer at 4°C. Cells were fixed in 1% (v/v) methanol-free formaldehyde for 20 min on ice and washed twice with PBS buffer. Fixed cells were permeabilized by immersing in 0.2% (v/v) Triton X-100 solution in PBS buffer for 10 min. An apoptosis detection kit (DeadEnd Fluorometric TUNEL System; Promega) was used to detect nuclear DNA fragmentation according to the manufacturer's instructions. BY-2 cells were incubated at 37°C for 1.5 h in a reaction mixture containing terminal deoxynucleotidyltransferase and fluorescein-labeled dUTP and rinsed extensively with PBS. For detecting nuclei, cells were additionally stained with DAPI. The fluorescence signal was viewed with an Axiophot microscope (Zeiss, Jena, Germany) and photographed using the supplied camera (Zeiss) and Provia Fujichrome 400 film (Fuji).
Acknowledgments
We thank Inhwan Hwang (Pohang University of Science and Technology, Pohang, Korea), Hyun-Sook Pai (Myongji University, Yongin, Korea), and members of WTK laboratory for their critical reading and valuable comments on the manuscript. This work was supported by grants from the Plant Diversity Research Center (21st Century Frontier Research Program funded by the Ministry of Science and Technology of Korean government, project number PF0330404-00) and the Plant Metabolism Research Center at Kyung Hee University (Science Research Center Program from the Korea Science and Engineering Foundation) to W.T.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Woo Taek Kim (wtkim@yonsei.ac.kr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026278
References
- Baumann, P., and Cech, T.R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud, T., Brun, C., Ancelin, K., Koering, C.E., Laroche, T., and Gilson, E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (1991). Structure and function of telomeres. Nature 350, 569–573. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Cai, J., Yang, J., and Jones, D.P. (1998). Mitochondrial control of apoptosis: The role of cytochrome c. Biochim. Biophys. Acta 1366, 139–149. [DOI] [PubMed] [Google Scholar]
- Chen, C.M., Wang, C.T., and Ho, C.H. (2001). A plant gene encoding a myb-like protein that binds telomeric GGTTTAG repeats in vitro. J. Biol. Chem. 276, 16511–16519. [DOI] [PubMed] [Google Scholar]
- Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. (1995). A human telomeric protein. Science 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Collins, K. (2000). Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12, 378–383. [DOI] [PubMed] [Google Scholar]
- Ellis, R.E., Yuan, J., and Horvitz, H.R. (1991). Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 7, 663–698. [DOI] [PubMed] [Google Scholar]
- Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., and Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43–50. [DOI] [PubMed] [Google Scholar]
- Fajkus, J., Kovarik, A., and Kralovics, R. (1996). Telomerase activity in plant cells. FEBS Lett. 391, 307–309. [DOI] [PubMed] [Google Scholar]
- Fajkus, J., Kovarik, A., Kralovics, R., and Bezdek, M. (1995). Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247, 633–638. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M.S., McKnight, T.D., and Shippen, D.E. (1996). Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. USA 93, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, M.S., Riha, K., Gao, F., Ren, S., McKnight, T.D., and Shippen, D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulneckova, J., and Fajkus, J. (2000). Inhibition of plant telomerase by telomere-binding proteins from nuclei of telomerase-negative tissues. FEBS Lett. 467, 305–310. [DOI] [PubMed] [Google Scholar]
- Gavrieli, Y., Sherman, Y., and Ben-Sasson, S.A. (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Greene, E.C., and Shippen, D.E. (1998). Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev. 12, 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C.W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Heller, K., Killan, A., Piatyszek, M.A., and Kleinhofs, A. (1996). Telomerase activity in plant cells. Mol. Gen. Genet. 252, 342–345. [DOI] [PubMed] [Google Scholar]
- Heller-Uszynska, K., Schnippenkoetter, W., and Kleinhofs, A. (2002). Cloning and characterization of rice (Oryza sativa L) telomerase reverse transcriptase, which reveals complex splicing patterns. Plant J. 31, 75–86. [DOI] [PubMed] [Google Scholar]
- Hemann, M.T., Strong, M.A., Hao, L.Y., and Greider, C.W. (2001). The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Hwang, M.G., Chung, I.K., Kang, B.G., and Cho, M.H. (2001). Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett. 503, 35–40. [DOI] [PubMed] [Google Scholar]
- Iwano, T., Tachibana, M., Reth, M., and Shinkai, Y. (2004). Importance of TRF1 for functional telomere structure. J. Biol. Chem. 279, 1442–1448. [DOI] [PubMed] [Google Scholar]
- Joeng, K.S., Song, E.J., Lee, K.J., and Lee, J. (2004). Long lifespan in worms with long telomeric DNA. Nat. Genet. 36, 607–611. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. (1999). P53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283, 1321–1325. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Smogorzewska, A., and de Lange, T. (2002). Senescence induced by altered telomere state, not telomere loss. Science 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- Killan, A., Heller, K., and Kleinhofs, A. (1998). Developmental patterns of telomerase activity in barley and maize. Plant Mol. Biol. 37, 621–628. [DOI] [PubMed] [Google Scholar]
- Killan, A., Stiff, C., and Kleinhofs, A. (1995). Barley telomeres shorten during differentiation but grow in callus culture. Proc. Natl. Acad. Sci. USA 92, 9555–9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi, S., Wulf, G., Nakamura, M., and Lu, K.P. (2001). Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is down-regulated in human breast tumors. Oncogene 20, 1497–1508. [DOI] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and del Pozo, O. (1999). Die and let live: Programmed cell death in plants. Curr. Opin. Plant Biol. 2, 502–507. [DOI] [PubMed] [Google Scholar]
- Lee, J.-H., and Kim, W.T. (2003). Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol. 132, 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger, J., and Cech, T.R. (1998). Telomerase and chromosome end maintenance. Curr. Opin. Genet. Dev. 8, 226–232. [DOI] [PubMed] [Google Scholar]
- Liu, J.P. (1999). Studies of the molecular mechanisms in the regulation of telomerase activity. FASEB J. 13, 2091–2104. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1941). The stability of broken ends of chromosomes in Zea mays. Genetics 26, 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, T.D., Riha, K., and Shippen, D.E. (2002). Telomeres, telomerase, and stability of the plant genome. Plant Mol. Biol. 48, 331–337. [DOI] [PubMed] [Google Scholar]
- Mittler, R., and Lam, E. (1995). Identification, characterization and purification of a tobacco endonuclease activity induced upon hypersensitive response cell death. Plant Cell 7, 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H.J. (1938). The remaking of chromosomes. Collecting Net. 8, 182–198. [Google Scholar]
- Nugent, C.I., Hughes, T.R., Lue, N.F., and Lundblad, V. (1996). Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274, 249–252. [DOI] [PubMed] [Google Scholar]
- Nugent, C.I., and Lundblad, V. (1998). The telomerase reverse transcriptase: Components and regulation. Genes Dev. 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Oguchi, K., Liu, H., Tamura, K., and Takahashi, H. (1999). Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 457, 465–469. [DOI] [PubMed] [Google Scholar]
- Riha, K., Fajkus, J., Siroky, J., and Vyskot, B. (1998). Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10, 1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Fajkus, J., Vyskot, B., and Shippen, D.E. (2000). Analysis of the G-overhang structures on plant telomeres: Evidence for two distinct telomere architectures. Plant J. 23, 633–641. [DOI] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant response to telomere dysfunction. Science 291, 1797–1800. [DOI] [PubMed] [Google Scholar]
- Runge, K.W., and Zakian, V.A. (1996). TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M., Haggblom, C., Vogt, M., Hunter, T., and Lu, K.P. (1997). Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc. Natl. Acad. Sci. USA 94, 13618–13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen, D.E., and McKnight, T.D. (1998). Telomeres, telomerase and plant development. Trends Plant Sci. 4, 126–130. [Google Scholar]
- Shore, D. (1994). Rap1: A protean regulator in yeast. Trends Genet. 10, 408–412. [DOI] [PubMed] [Google Scholar]
- Shore, D. (2001). Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11, 189–198. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smucker, E.J., and Turchi, J.J. (2001). TRF1 inhibits telomere C-strand DNA synthesis in vitro. Biochemistry 40, 2426–2432. [DOI] [PubMed] [Google Scholar]
- Stein, J.C., and Hansen, G. (1999). Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol. 121, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.-L., Zhao, Y., Hong, X., and Zhai, Z.-H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462, 317–321. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Liu, H., and Takahashi, H. (1999). Auxin induction of cell cycle regulated activity of tobacco telomerase. J. Biol. Chem. 274, 20997–21002. [DOI] [PubMed] [Google Scholar]
- Tiwari, B.S., Belenghi, B., and Levine, A. (2002). Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 128, 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Smogorzewska, A., and de Lange, T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Vassetzky, N.S., Gaden, F., Brun, C., Gasser, S.M., and Gilson, E. (1999). Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res. 27, 4687–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, D.L., and Strasser, A. (1996). The molecular biology of apoptosis. Proc. Natl. Acad. Sci. USA 93, 2239–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. [DOI] [PubMed] [Google Scholar]
- Yang, S.W., Jin, E.S., Chung, I.K., and Kim, W.T. (2001). Expression of telomerase activity is closely correlated with the capacity for cell division in tobacco plants. J. Plant Biol. 44, 168–171. [Google Scholar]
- Yang, S.W., Jin, E.S., Chung, I.K., and Kim, W.T. (2002). Cell cycle-dependent regulation of telomerase activity by auxin, ABA and protein phosphorylation in tobacco BY-2 suspension culture cells. Plant J. 29, 617–626. [DOI] [PubMed] [Google Scholar]
- Yang, S.W., Kim, D.H., Lee, J.J., Chun, Y.J., Lee, J.-H., Kim, Y.J., Chung, I.K., and Kim, W.T. (2003). Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells. J. Biol. Chem. 278, 21395–21407. [DOI] [PubMed] [Google Scholar]
- Yu, E.Y., Kim, S.E., Kim, J.H., Ko, J.H., Cho, M.H., and Chung, I.K. (2000). Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J. Biol. Chem. 275, 24208–24214. [DOI] [PubMed] [Google Scholar]
- Zakian, A.V. (1995). Telomeres: Beginning to understand the end. Science 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Mar, V., Zhou, W., Harrington, L., and Robinson, M.O. (1999). Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 13, 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.Z., and Lu, K.P. (2001). The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107, 347–359. [DOI] [PubMed] [Google Scholar]