ABSTRACT
The near-minimal bacterium Mesoplasma florum constitutes an attractive model for systems biology and for the development of a simplified cell chassis in synthetic biology. However, the lack of genetic engineering tools for this microorganism has limited our capacity to understand its basic biology and modify its genome. To address this issue, we have evaluated the susceptibility of M. florum to common antibiotics and developed the first generation of artificial plasmids able to replicate in this bacterium. Selected regions of the predicted M. florum chromosomal origin of replication (oriC) were used to create different plasmid versions that were tested for their transformation frequency and stability. Using polyethylene glycol-mediated transformation, we observed that plasmids harboring both rpmH-dnaA and dnaA-dnaN intergenic regions, interspaced or not with a copy of the dnaA gene, resulted in a frequency of ∼4.1 × 10−6 transformants per viable cell and were stably maintained throughout multiple generations. In contrast, plasmids containing only one M. florum oriC intergenic region or the heterologous oriC region of Mycoplasma capricolum, Mycoplasma mycoides, or Spiroplasma citri failed to produce any detectable transformants. We also developed alternative transformation procedures based on electroporation and conjugation from Escherichia coli, reaching frequencies up to 7.87 × 10−6 and 8.44 × 10−7 transformants per viable cell, respectively. Finally, we demonstrated the functionality of antibiotic resistance genes active against tetracycline, puromycin, and spectinomycin/streptomycin in M. florum. Taken together, these valuable genetic tools will facilitate efforts toward building an M. florum-based near-minimal cellular chassis for synthetic biology.
IMPORTANCE Mesoplasma florum constitutes an attractive model for systems biology and for the development of a simplified cell chassis in synthetic biology. M. florum is closely related to the mycoides cluster of mycoplasmas, which has become a model for whole-genome cloning, genome transplantation, and genome minimization. However, M. florum shows higher growth rates than other Mollicutes, has no known pathogenic potential, and possesses a significantly smaller genome that positions this species among some of the simplest free-living organisms. So far, the lack of genetic engineering tools has limited our capacity to understand the basic biology of M. florum in order to modify its genome. To address this issue, we have evaluated the susceptibility of M. florum to common antibiotics and developed the first artificial plasmids and transformation methods for this bacterium. This represents a strong basis for ongoing genome engineering efforts using this near-minimal microorganism.
KEYWORDS: antibiotic markers, chromosomal origin of replication, Mesoplasma, plasmids, synthetic biology, transformation methods
INTRODUCTION
Mollicutes are a class of bacteria mainly characterized by small genome sizes (0.58 to 2.2 Mbp), small cell dimensions (∼0.2 to 0.4 μm), and the absence of a cell wall (1–3). Mollicutes are thought to have derived from low-GC-content Gram-positive bacteria through genome reduction, which resulted in a significant simplification of their metabolic pathways (1–3). Consequently, many bacteria of this class have evolved a parasitic lifestyle with the ability to infect various plants and animals, including humans (1, 2). Unlike other small-genome bacteria, such as chlamydias and rickettsias, Mollicutes can be cultured in acellular medium, except for phytoplasmas, which are obligate parasites of plants (4). The remarkable genomic simplicity of Mollicutes makes members of this class attractive candidates to develop minimal cells in which the thorough characterization of global cellular mechanisms will be more easily achievable (5, 6).
Mesoplasma florum, first described as Acholeplasma florum in 1984 (7), constitutes a particularly interesting member of the Mollicutes as a new model for systems and synthetic biology studies. M. florum is closely related to the mycoides cluster of mycoplasmas, which includes Mycoplasma mycoides and Mycoplasma capricolum; these have become model organisms for whole-genome cloning (8–10), genome transplantation (8, 11, 12), and genome minimization (13). However, M. florum shows higher growth rates (∼34 min), has no known pathogenic potential, and possesses a significantly smaller genome that positions this species among some of the simplest free-living organisms (1, 6, 14, 15). For example, M. florum L1 (RefSeq accession no. NC_006055.1), the first representative of its species, has a total genome size of only ∼793 kb, compared to ∼1.2 Mb and ∼1.0 Mb for M. mycoides and M. capricolum, respectively (1). M. florum also uses an alternative genetic code (Mycoplasma/Spiroplasma code) in which the UGA codon signals the incorporation of a tryptophan in the nascent protein rather than a stop codon, a feature that limits horizontal gene transfer from and to other microorganisms (16, 17). Despite these advantageous characteristics, practically no genetic tools are currently available to reduce and reprogram the genome of M. florum or to build artificial gene circuits.
Many Mollicutes phylogenetically related to M. florum, including M. mycoides, M. capricolum, and Spiroplasma citri, have been successfully transformed with artificial plasmids containing a chromosomal origin of replication (oriC) (18–26). oriC-based plasmids have multiple uses, such as expression of exogenous genes, inactivation of target genes by recombination, or complementation of chromosomal mutations. Since Mollicutes are naturally susceptible to tetracycline, the tetM gene derived from the Tn916 transposon of Enterococcus faecalis is often used as an antibiotic resistance marker for robust oriC-based plasmid selection (18–26). Following transformation in a recipient cell, the oriC plasmids can replicate due to specific interactions of the DnaA protein with sequences called DnaA boxes (24, 27). In Mollicutes, DnaA boxes have been shown to generally be located within the two AT-rich intergenic regions flanking the dnaA gene, with a proposed 9-bp asymmetric sequence of 5′-TT(A/T)TC(C/A)ACA-3′ (21, 24). By virtue of their sequence homology, oriC plasmids can also integrate into the oriC region of the host cell chromosome by recombination events (18–24, 26).
In this work, we evaluate the susceptibility of M. florum L1 to common antibiotics and describe the successful utilization of the predicted oriC region of M. florum L1 chromosome to generate the first replicable plasmids in this microorganism. These oriC plasmids were characterized for their transformation frequency, stability, and their propensity to recombine at the chromosomal oriC region of M. florum. We also report successful oriC plasmid transformation using electroporation or conjugation as alternative transformation methods to the more traditional polyethylene glycol (PEG)-mediated procedure and investigate the capacity of M. florum to replicate heterologous oriC plasmids. The genetic tools developed in this study will contribute to ongoing efforts toward building an M. florum-based near-minimal cellular chassis for synthetic biology.
RESULTS
Antibiotic susceptibilities of M. florum L1.
While several studies have established antibiotic susceptibilities in members of the Mollicutes class, the sensitivity of M. florum to some commonly used antibiotics was lacking. Using growth inhibition assays, we tested 12 antibiotics commonly used for genetic manipulation in bacteria (Table 1). We confirmed that some drugs were ineffective against M. florum, which could be used to eliminate contaminating bacteria when needed. As expected, M. florum L1 showed natural resistance to ampicillin, rifampin, sulfamethoxazole, and trimethoprim, displaying MICs above 100 μg/ml for each of these antibiotics (Table 1). Interestingly, M. florum was resistant to kanamycin and gentamicin but slightly susceptible to streptomycin and spectinomycin. M. florum also showed a high sensitivity to chloramphenicol, erythromycin, and puromycin, exhibiting MICs of 5 to 8.5 μg/ml, 1 to 1.5 μg/ml, and 8 to 15.5 μg/ml, respectively (Table 1). Finally, M. florum showed susceptibility to tetracycline, with an MIC of less than 10 μg/ml.
TABLE 1.
MICs of some common antibiotics against M. florum L1
Identification of putative DnaA boxes within the oriC region of M. florum.
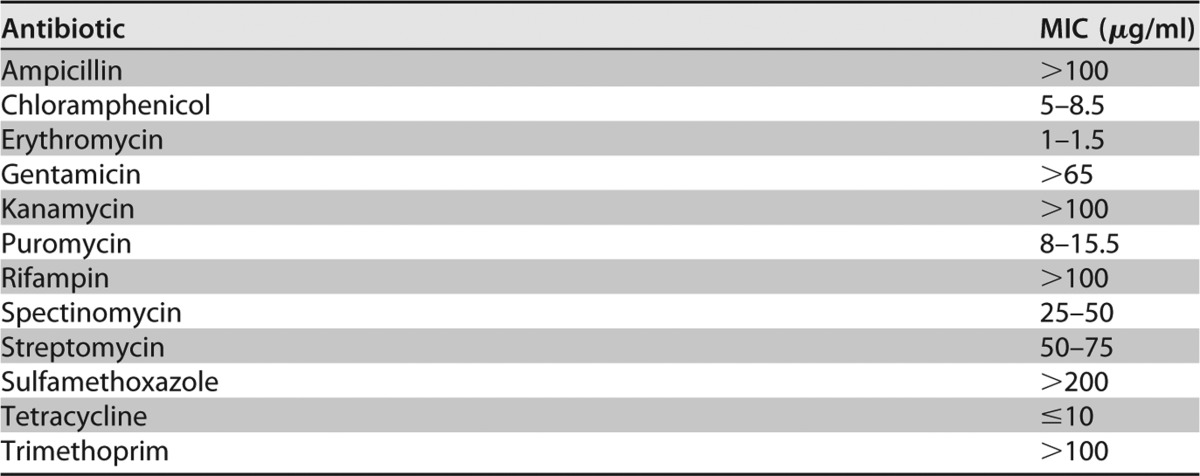
Previously, no self-replicative plasmid had been either identified in or developed for M. florum. The susceptibility of M. florum to tetracycline (Table 1) offers the possibility to take advantage of the widely used tetM resistance marker for plasmid selection. However, the localization of putative DnaA boxes in M. florum remains unknown, hindering our ability to develop plasmids based on the oriC region of the chromosome. We therefore compared the oriC regions of 11 selected representative members of the Spiroplasma group using multiple-sequence alignment (Fig. S1 and Table S4) and evaluated the phylogenetic relationships between species using sequence similarity (Fig. 1A). We observed that the differences in the oriC region sequence are consistent with the Mollicutes phylogeny based on conserved proteins (26, 28) and 16S rRNA sequences (29, 30). Mycoplasmas of the mycoides cluster (M. leachii, M. capricolum, and M. mycoides) shared an oriC region with a high percentage of nucleotide similarity (>90%), while S. citri and S. kunkelii were more phylogenetically distant and characterized by a more divergent oriC sequence (Fig. 1A and Table S4). As expected, M. florum was phylogenetically closer to the mycoides cluster than the spiroplasmas based on the oriC region sequence but remained clearly separated from all analyzed mycoplasmas (Fig. 1A and Table S4).
FIG 1.
Sequence analysis of the predicted oriC region of 11 selected Mollicutes of the Spiroplasma group. (A) Phylogenetic tree based on the oriC region sequence of the chromosome using maximum likelihood. The number on each node indicates the percentage with which each branch topology was supported. The tree is drawn to scale, with branch lengths representing the number of substitutions per site. (B) Putative DnaA binding motif found using MEME. (C) Localization of putative DnaA boxes within the intergenic regions upstream and downstream of dnaA. Putative DnaA boxes on positive and negative DNA strands are indicated by red rectangles positioned above and below the chromosomal line, respectively. Regions are drawn to scale. S. citri and S. kunkelii guaB-dnaA intergenic region is cut for presentation purposes, as well as represented genes.
We next hypothesized that the conservation property of the oriC region in the Spiroplasma group could be used to identify putative DnaA boxes in M. florum. We submitted the DNA sequence of the two intergenic regions flanking the dnaA gene found in representative species of the Spiroplasma group to the de novo motif discovery tool MEME (31) and detected a motif that is highly consistent with the previously proposed putative DnaA box consensus of Mollicutes [TT(A/T)TC(C/A)ACA] (21, 24) (Fig. 1B). We then searched the precise localization of putative DnaA boxes within the two oriC intergenic regions using MAST (32) and observed that the number of DnaA boxes and their organization were reminiscent of the species phylogenetical relationships (Fig. 1A and C, Fig. S1, and Table S5). For instance, members of the mycoides cluster all shared the same four putative DnaA boxes located at approximately 6 bp, 47 bp, 144 bp, and 185 bp upstream of dnaA, with the exception of M. leachii, in which the box 185 bp upstream of dnaA was not detected due to a transversion mutation (C→A) at position 6 of the consensus sequence. Species of the mycoides cluster also shared a unique putative DnaA box located ∼1,391 bp downstream of the start codon of dnaA (Fig. 1C and S1 and Table S5). Interestingly, this box is shared and highly conserved (7 out of 10 bp) between all 11 selected Mollicutes.
Mycoplasma yeatsii, Mycoplasma putrefaciens, M. florum, and spiroplasmas were distinguished from the mycoides cluster mostly by the number and position of putative DnaA boxes located upstream of dnaA. For example, putative DnaA boxes located ∼6 bp and ∼185 bp before the dnaA gene in the mycoides cluster were not detected in other analyzed Mollicutes (Fig. 1C and S1 and Table S5).
Development of M. florum oriC-based plasmids.
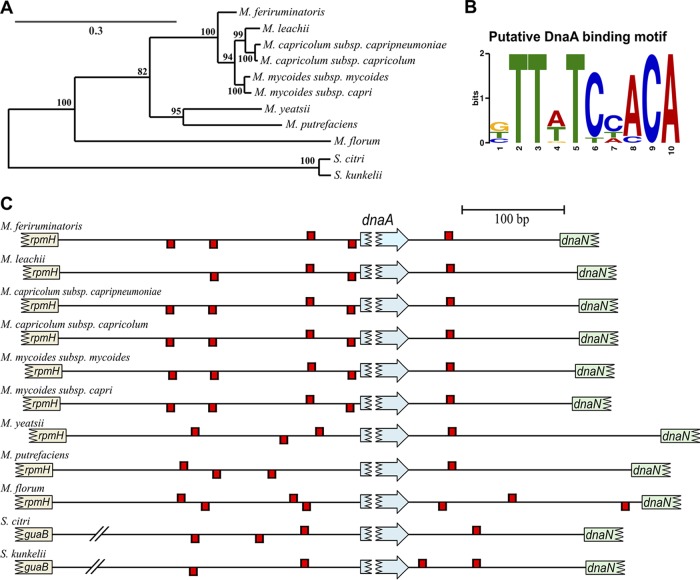
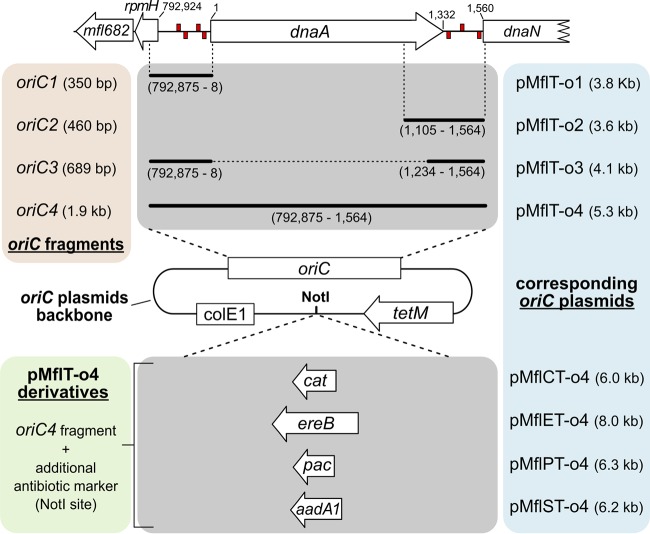
In total, seven putative DnaA boxes were identified within the oriC region of M. florum. Four of them were located in the intergenic region between rpmH and dnaA, whereas three boxes were found in the intergenic region between dnaA and dnaN (Fig. 1C and S1 and Table S5). Except for the two boxes located ∼1,363 bp and ∼1,545 bp downstream of the dnaA start codon, all DnaA boxes found in M. florum coincided with boxes found in one or many Mollicutes analyzed here. However, the importance of both intergenic regions for plasmid replication, as well as the presence of a copy of the dnaA gene, remained to be established in M. florum. We therefore developed four different plasmids based on the localization of predicted DnaA boxes within the oriC region of the M. florum chromosome: two plasmids containing either the rpmH-dnaA or the dnaA-dnaN intergenic region (pMflT-o1 and pMflT-o2, respectively), one plasmid containing both regions but lacking the dnaA gene (pMflT-o3), and another plasmid including the whole oriC-dnaA locus (pMflT-o4) (Fig. 2). The tetM gene, coding for a tetracycline ribosomal protection protein, was chosen as a selectable marker in the oriC plasmids and was specifically recoded to be functional in both Escherichia coli and M. florum. Following assembly in E. coli, oriC plasmids were transformed in M. florum L1 by a PEG-mediated procedure (3, 33). Intriguingly, pMflT-o1 and pMflT-o2 failed to produce any detectable tetracycline-resistant transformant, while pMflT-o3 and pMflT-o4 transformation resulted in several hundreds to thousands of colonies on solid medium, with overall frequencies of 4.06 × 10−6 and 4.16 × 10−6 transformants per viable cell, respectively (Fig. 3A).
FIG 2.
Schematic representation of M. florum oriC-based plasmids. M. florum oriC plasmids contain various oriC fragments, a ColE1 replication origin, and a tetracycline resistance cassette (tetM). oriC fragments were based on the predicted oriC region of M. florum L1 chromosome, and their respective coordinates and sizes are indicated in brackets. Coordinates of the start codon of rpmH and dnaN, as well as the start and stop codons of dnaA, are also indicated. Putative DnaA boxes found in the rpmH-dnaA and dnaA-dnaN intergenic regions (see Fig. 1C) are represented by red rectangles on positive and negative DNA strands. oriC fragments were assembled with tetM and ColE1 fragments to produce pMflT-o1, pMflT-o2, pMflT-o3, and pMflT-o4 plasmids. Additional antimicrobial resistance gene cassettes were cloned in the NotI site of pMflT-o4 or a derivative plasmid to generate pMflCT-o4 (cat), pMflET-o4 (ereB), pMflPT-o4 (pac), or pMflST-o4 (aadA1).
FIG 3.
Transformation frequencies of M. florum oriC plasmids and recombination with the chromosome. (A) Transformation frequencies of M. florum oriC plasmids using polyethylene glycol (PEG)-mediated transformation procedure. o1, pMflT-o1; o2, pMflT-o2; o3, pMflT-o3; o4, pMflT-o4. Error bars indicate the standard deviations calculated from the results of six independent biological replicates. Asterisks indicate transformation frequencies below the detection limit. (B and C) Southern blot analysis of pMflT-o3 (B) and pMflT-o4 (C) recombination with the M. florum chromosome. Fragment sizes corresponding to the integrated and extrachromosomal forms of each plasmid are indicated. I, plasmid integrated at the oriC region of the chromosome; IA, plasmid integrated at the rpmH-dnaA intergenic region; IB, plasmid integrated at the dnaA-dnaN intergenic region; E, plasmid as an extrachromosomal element. Twelve isolated M. florum clones were analyzed for each plasmid (clone number indicated above each well). P, purified plasmid control; N, M. florum L1 wild-type (WT) genomic DNA (negative control).
Growth analysis revealed that pMflT-o4 transformants were not affected by tetracycline concentrations considerably higher than those tolerated by M. florum L1 (Fig. S2A). In fact, the tetM gene conferred resistance to tetracycline concentrations exceeding 100 μg/ml (Table 2), a concentration at least 10 times higher than the MIC of the M. florum wild-type strain (Table 1). Similar results were also obtained for M. florum carrying pMflT-o3 (data not shown). Because additional selectable markers would offer a broader range of possibilities, genes conferring resistance to chloramphenicol (cat), erythromycin (ereB), puromycin (pac), and spectinomycin/streptomycin (aadA1) were introduced into pMflT-o4 to generate pMflCT-o4, pMflET-o4, pMflPT-o4, and pMflST-o4 plasmids, respectively (Fig. 2). We observed that the pac gene included in pMflPT-o4 conferred a protection against >200 μg/ml puromycin (Table 2 and Fig. S2B), a concentration 20 times higher than the MIC of the wild-type L1 strain (Table 1). Similar results were obtained with M. florum carrying pMflST-o4 growing in medium with or without spectinomycin or streptomycin (Fig. S2C and D and Table 2). For pMflET-o4, growth inhibition assays suggested very weak protection against erythromycin that is not sufficient to be exploited robustly (data not shown). Similarly, our data suggest that the cat gene of the pMflCT-o4 plasmid is not functional in M. florum, since no protection against chloramphenicol was observed (data not shown).
TABLE 2.
MICs of M. florum carrying different antibiotic resistance markers
| Plasmid | Antibiotic | Gene conferring resistance | MIC (μg/ml) |
|---|---|---|---|
| pMflT-o4 | Tetracycline | tetM | >100 |
| pMflPT-o4 | Puromycin | pac | >200 |
| pMflST-o4 | Spectinomycin | aadA1 | >200 |
| Streptomycin | aadA1 | >200 |
Homologous recombination with the host chromosome.
Since oriC-based plasmids are known to frequently recombine at the oriC region of the chromosome due to sequence homology (18–24, 26), 12 M. florum isolated clones carrying pMflT-o3 or pMflT-o4 were analyzed by Southern blotting using a radiolabeled probe targeting a region of the tetM gene to discriminate between the integrated and extrachromosomal forms of the plasmids (Fig. S3). Interestingly, all pMflT-o3 and pMflT-o4 tested clones showed the presence of recombination events with the host chromosome after overnight growth with selective antibiotics (Fig. 3B and C). More specifically, the majority of pMflT-o3 tested clones exhibited a recombined form of the plasmid at the dnaA-dnaN intergenic region (10 out of 12), while only 2 clones showed a band corresponding to the recombined element at the rpmH-dnaA region (Fig. 3B). In addition, a total of 17 out of 24 analyzed clones were found to carry the oriC plasmids as extrachromosomal elements (9/12 clones for pMflT-o3 and 8/12 clones for pMflT-o4). All clones that presented a band corresponding to the extrachromosomal form of the elements also showed a recombination event with the oriC region of M. florum chromosome (17/17 clones), suggesting the presence of heterogeneous populations of cells deriving from the same initial colony (Fig. 3B and C). Taken together, these results indicate that plasmids based on the oriC of M. florum have a strong tendency to recombine with oriC region of the chromosome, regardless of the presence of a copy of the dnaA gene.
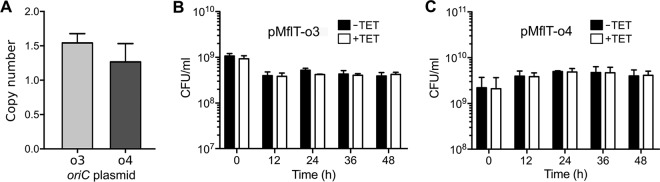
oriC plasmid copy number and stability.
Using quantitative PCR (qPCR) analysis, we next quantified the number of pMflT-o3 and pMflT-o4 copies per cell relative to that of the M. florum chromosome. Plasmid copy number was determined by comparing the relative abundance of the tetM gene of individual pMflT-o3 and pMflT-o4 clones to the control strain M. florum L1 clone 3632 containing one copy of tetM integrated in the chromosome. We observed that the overall copy numbers of pMflT-o3 and pMflT-o4 were between 1 and 2 copies per M. florum genome (Fig. 4A). We then sought to determine if these oriC plasmids were stable over several generations by maintaining M. florum L1 carrying either pMflT-o3 or pMflT-o4 under continuous culture conditions for 48 h without tetracycline. Colony counts revealed no significant reduction in tetracycline-resistant colonies during and after continuous growth without selective pressure (Fig. 4B and C). Considering that M. florum has a doubling time of ∼34 min in ATCC 1161 medium (14, 15), this indicates that pMflT-o3 and pMflT-o4 plasmids can be stably maintained for at least 85 generations without detectable loss.
FIG 4.
M. florum oriC plasmid copy number and stability. (A) Number of oriC plasmids per M. florum cell obtained by quantitative PCR targeted on the tetM gene. o3, pMflT-o3; o4, pMflT-o4. (B and C) Evaluation of pMflT-o3 (B) and pMflT-o4 (C) stability in M. florum under continuous culture conditions for up to 48 h. For each represented time point, cells were plated on ATCC 1161 solid medium with (+TET, white bars) or without (−TET, black bars) tetracycline, and CFU per milliliter were quantified. Error bars represent standard deviations from the results of three independent biological replicates.
Alternative transformation methods.
Transformation by electroporation is generally successful with most cell types and was previously reported for S. citri and Mycoplasma genitalium (3, 34). This method requires fewer steps than PEG-mediated transformation and might offer higher transformation frequencies. However, we are not aware of any report documenting the successful transformation of M. florum using electroporation. We therefore optimized the electroporation procedure with M. florum using the pMflT-o4 plasmid and observed drastic effects of electroporation voltage on transformation frequency (Fig. 5A). Indeed, the transformation frequency was just above the detection limit of approximately 1 × 10−9 transformants per viable cell when 0.5 kV was used (2.18 × 10−9 transformants per viable cell), while using 2.5 kV yielded more than 70,000 transformants per ml of M. florum culture (7.87 × 10−6 transformants per viable cell), which is comparable to the frequency observed for PEG-mediated transformation (Fig. 3A).
FIG 5.
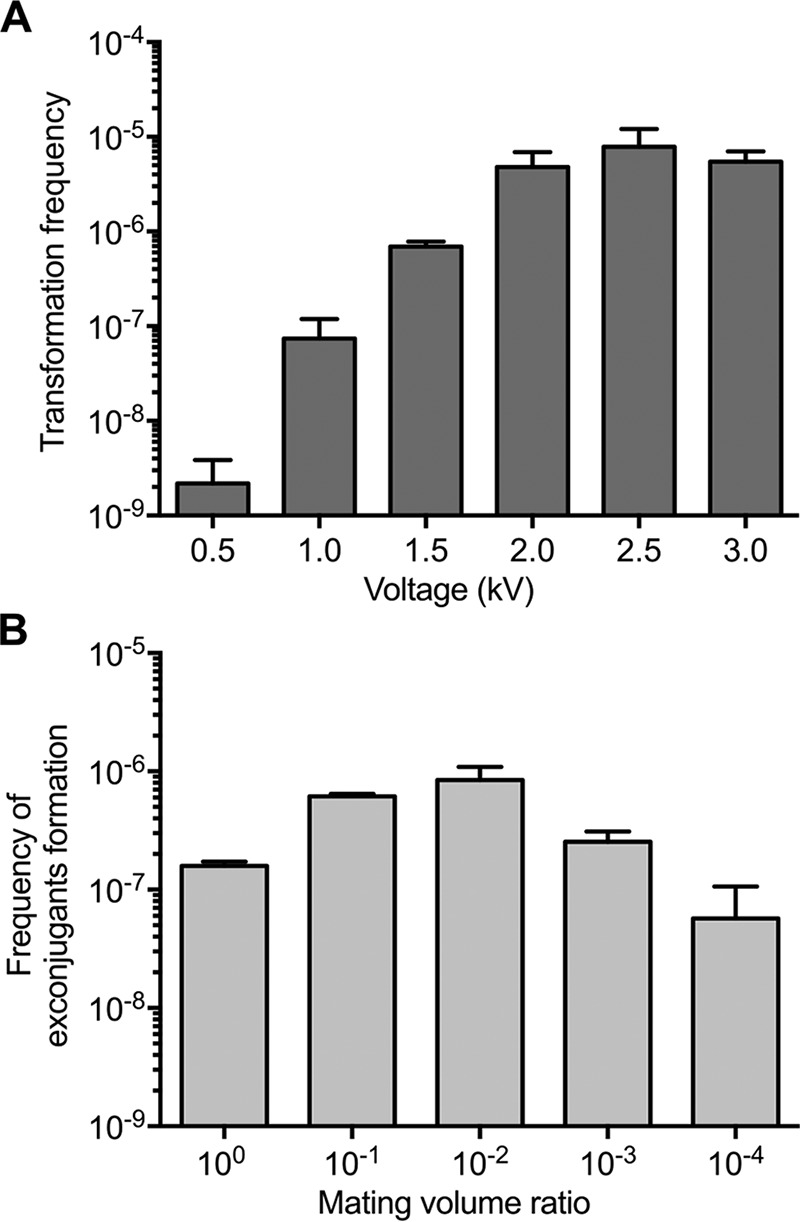
Frequencies of plasmid introduction in M. florum by electroporation or conjugation. (A) Transformation frequencies of pMflT-o4 in M. florum L1 using the electroporation procedure with 1-mm cuvettes and different voltage values. Error bars indicate the standard deviations from the results of three independent biological replicates. (B) pMflT-o4 transfer rates by conjugation using different mating volume ratios of donor (E. coli MFDpir) and recipient cells (M. florum L1). Indicated mating volume ratios are calculated by dividing the volume of M. florum culture by the volume of E. coli culture mixed during the conjugation process (see Table S3). pMflT-o4 transfer frequency is expressed as the number of exconjugants per viable recipient CFU. Error bars indicate the standard deviations from the results of three independent biological replicates.
Bacterial conjugation is another common method to deliver plasmids in several species. Conjugation allows the mobilization of large DNA molecules, can reach high transfer frequencies, and is possible between phylogenetically distant organisms. However, we are not aware of any report of plasmid delivery between E. coli and Mollicutes. To investigate conjugation as another alternative transformation method for M. florum, we included the transfer origin of broad-host-range plasmid RP4 (oriTRP4) in the backbone of our oriC plasmids and tested different mating ratios using the E. coli MFDpir strain (35) as a donor (Table S3). Our results indicate that plasmid conjugation can generate more than 400 colonies per experiment, reaching a frequency of 8.44 × 10−7 transformants per viable cell (Fig. 5B). This frequency is slightly lower than those observed for PEG-mediated transformation and electroporation (Fig. 3A and 5A). No colony was observed for controls lacking the donor or recipient cells. Coincubation of M. florum cells with 1 μg of purified pMflT-o4 plasmid yielded only two tetracycline-resistant colonies in a single replicate out of three independent experiments, which sits right at the detection limit of our assay. PCR amplifications performed on exconjugants confirmed that the resulting clones truly harbored the pMflT-o4 plasmid and were not spontaneous mutants or contaminants (data not shown).
Transformation of M. florum with heterologous oriC plasmids.
We previously observed that the oriC regions of closely related Mollicutes shared some similarities relatively to their sequence and DnaA box organization (Fig. 1 and S1 and Tables S4 and S5). However, it is still unclear what degree of sequence divergence the replication machinery of Mollicutes can tolerate, and more specifically for M. florum. To better define these parameters, we investigated the capacity of M. florum to replicate heterologous oriC plasmids containing the oriC region and dnaA gene of closely related Mollicutes. Using PEG-mediated transformation, we first attempted to transform M. florum with oriC plasmids previously developed in M. mycoides (pMYCO1 and pMYSO1), M. capricolum (pMCO3), and S. citri (pSD4) (21, 24, 25) (Fig. S4A and Table 3). Unfortunately, these plasmids failed to yield any transformants, since their tetracycline resistance cassette was not properly expressed from the spiralin promoter in M. florum (data not shown) (19, 25). We therefore constructed four pMflT-o4 derivative plasmids in which the oriC region of M. florum was replaced by the oriC region of M. mycoides (pMmcT and pMmmT), M. capricolum (pMcapT), and S. citri (pSciT-o4) (Fig. S4B and Table 3). These new heterologous oriC plasmids were all shown to confer tetracycline resistance in E. coli. However, in contrast to the M. florum oriC-based pMflT-o4 plasmid, none of the new heterologous oriC constructs yielded any tetracycline-resistant colony when transformed in M. florum.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| EC100D pir+ | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG pir+ (DHFR) (Smr) | Epicentre |
| MFDpir | MG1655 RP4-2-Tc::[ΔMu1::aac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA (Aprar Zeor Emr) | 35 |
| MM294 | F− glnX44(AS) λ− endA1 spoT1 thiE1 hsdR17 creC510 | E. coli Genetic Stock Center (strain 6315) |
| Mesoplasma florum | ||
| L1 | ATCC 33453 | |
| L1 clone 3632 | mfl169::Tn-tetM | This study |
| Plasmids | ||
| pMflT-o1 | ColE1 oriTRP4 M. florum oriC1 tetM (Tcr) | This study |
| pMflT-o2 | ColE1 M. florum oriC2 tetM (Tcr) | This study |
| pMflT-o3 | ColE1 oriTRP4 M. florum oriC3 tetM (Tcr) | This study |
| pMflT-o4 | ColE1 oriTRP4 M. florum oriC4 tetM (Tcr) | This study |
| pMflCT-o4 | ColE1 oriTRP4 M. florum oriC4 tetM cat (Tcr Cmr) | This study |
| pMflET-o4 | ColE1 oriTRP4 M. florum oriC4 tetM ereB (Tcr Emr) | This study |
| pMflPT-o4 | ColE1 oriTRP4 M. florum oriC4 tetM pac (Tcr Pur) | This study |
| pMflST-o4 | ColE1 oriTRP4 M. florum oriC4 tetM aadA1 (Tcr Spr Smr) | This study |
| pMYCO1 | ColE1 M. mycoides subsp. capri oriC tetM bla (Tcr Apr) | 24 |
| pMYSO1 | ColE1 M. mycoides subsp. mycoides oriC tetM bla (Tcr Apr) | 24 |
| pMCO3 | ColE1 M. capricolum subsp. capricolum oriC tetM bla (Tcr Apr) | 24 |
| pSD4 | ColE1 S. citri oriC tetM bla (Tcr Apr) | 25 |
| pMmcT | ColE1 oriTRP4 M. mycoides subsp. capri oriC tetM (Tcr) | This study |
| pMmmT | ColE1 oriTRP4 M. mycoides subsp. mycoides oriC tetM (Tcr) | This study |
| pMcapT | ColE1 oriTRP4 M. capricolum subsp. capricolum oriC tetM (Tcr) | This study |
| pSciT-o4 | ColE1 oriTRP4 S. citri oriC tetM (Tcr) | This study |
| ereB-pUC57 | ColE1 bla ereB (Apr Emr) | This study |
| pTT01 | ColE1 tetM (Tcr) | This study |
| pUC19 | ColE1 bla (Apr) | 66 |
| pSW23T | R6K oriTRP4 cat (Cmr) | 67 |
DHFR, dihydrofolate reductase gene; Smr, streptomycin resistant; Aprar, apramycin resistant; Zeor, zeocin resistant; Emr, erythromycin resistant; Tcr, tetracycline resistant; Cmr, chloramphenicol resistant; Pur, puromycin resistant; Spr, spectinomycin resistant; Apr, ampicillin resistant.
DISCUSSION
In order to develop new genetic manipulation tools for the near-minimal bacterium M. florum, we investigated antibiotic susceptibility and oriC replication in this organism. We first validated that M. florum was indeed resistant to ampicillin, rifampin, sulfamethoxazole, and trimethoprim (Table 1), which are class-specific resistances shared among members of the Mollicutes (36–40). We also observed that M. florum was resistant to kanamycin and gentamicin (Table 1). Interestingly, the sensitivity of Mollicutes to aminoglycosides has been reported to vary among strains and isolates (37, 39–44). Similarly to the rifampin resistance in Mollicutes (37, 38), it is likely that M. florum resistance to kanamycin and gentamicin depends on variations in the targeted gene products, e.g., the 16S rRNA of the 30S ribosome subunit. More importantly, we showed that M. florum was sensitive to antibiotics generally effective against Mollicutes (36, 37, 40, 45), i.e., tetracycline, chloramphenicol, erythromycin, and puromycin (Table 1). M. florum was also found to be relatively sensitive to streptomycin and spectinomycin (Table 1).
The evaluation of M. florum antibiotic susceptibilities allowed us to investigate the functionality of different markers frequently used in bacteria. As expected, tetM and pac genes conferred M. florum resistance to high concentrations of tetracycline and puromycin (Table 2 and Fig. S2A and B). These markers were previously shown to be functional in several Mollicutes, including M. capricolum and M. mycoides (18–26, 45). On the other hand, the functionality of the aadA1 gene in M. florum was interesting since it is, to our knowledge, the first time that this genetic marker has been artificially introduced in a bacterium of the Mollicutes class (Table 2 and Fig. S2C and D). The cat and ereB genes did not confer protection against their cognate antibiotics in M. florum. However, these markers were functional in E. coli carrying pMflCT-o4 and pMflET-o4 plasmids and have been employed in other Mollicutes (46–50). Since cat and ereB were recoded to be functional in E. coli and in M. florum, it remains possible that they were not properly or sufficiently expressed in M. florum to confer a resistance phenotype.
Using available genomic sequences of Mollicutes closely related to M. florum, we constructed a putative DnaA binding motif and we identified putative DnaA boxes within previously uncharacterized oriC regions of members of the Spiroplasma group, such as M. leachii, M. putrefaciens, and, more importantly, M. florum (Fig. 1B and S1 and Table S5). Our predicted DnaA binding motif is highly consistent with the previously proposed putative DnaA box consensus of Mollicutes [TT(A/T)TC(C/A)ACA] and is reminiscent of the consensus sequence found in E. coli (21, 24, 27, 51). Furthermore, high-confidence putative DnaA boxes previously identified in M. mycoides, M. capricolum, and S. citri using E. coli DnaA binding consensus were successfully identified using our approach (24). Still, it is possible that more degenerate DnaA boxes exist and contribute to the chromosomal replication in these bacteria but were not detected by our motif, according to our search parameters. For example, Lartigue et al. (24) proposed a degenerate putative DnaA box located ∼30 bp from the start codon of rpmH in M. mycoides and M. capricolum that was not identified by our method (Fig. 1C and S1).
Plasmids harboring both M. florum oriC intergenic regions, with or without a copy of the dnaA gene (pMflT-o4 and pMflT-o3, respectively), were found to transform M. florum at approximately the same frequency (Fig. 3A). These results indicate that cis-expression of the DnaA protein or the spacing provided by the dnaA gene between the two clusters of DnaA boxes is probably not essential for proper plasmid replication and maintenance in M. florum. Intriguingly, even if the majority of analyzed transformants showed extrachromosomal forms of the oriC plasmids after overnight culture (Fig. 3B and C), we observed that recombination with the M. florum chromosome also occurred for all tested clones, corroborating previous observations indicating that oriC plasmids are highly recombinogenic in Mollicutes (18–24, 26). Since both the pMflT-o3 and pMflT-o4 plasmids were present in approximately one copy per cell relative to the M. florum chromosome (Fig. 4A), this suggests that a dynamic state between the circular and the integrated forms of the plasmids may exist within a clonal population of cells. Nevertheless, we showed that both constructs were maintained for at least 85 generations (48 h of continuous growth) without any selection (Fig. 4B and C). It remains to be determined if the extrachromosomal form is disfavored over time and if the long-term oriC plasmid stability is dependent on integration events. Additional experiments will also be necessary to engineer M. florum oriC plasmids to remain as extrachromosomal molecules, or conversely, to perform specific gene targeting.
Using pMflT-o4, we also demonstrated that electroporation and conjugation are viable transformation methods for M. florum (Fig. 5), thus offering alternative procedures that require less material and hands-on time than the PEG-mediated transformation protocol. Interspecies conjugation from E. faecalis to Mycoplasma gallisepticum (52), Mycoplasma arthritidis (53), or Mycoplasma hominis (54) has previously been reported to deliver a Tn916 transposon. However, our results constitute the first reported example of plasmid conjugation from E. coli to a Mollicutes species. Although the current results using the RP4 conjugation machinery showed slightly lower plasmid transfer rates than the electroporation and PEG-mediated transformation frequencies (Fig. 3A and 5), this approach might be improved with the use of alternative conjugative systems that could be better adapted for gene transfer into Mollicutes. For example, certain machineries could be better adapted for the absence of a cell wall, or specific pili could stabilize the contact between E. coli and the comparatively small M. florum cells. It will be interesting to test whether our conjugation system is also working with other Mollicutes and what factors might affect transfer frequency.
Interestingly, plasmids containing only one oriC intergenic region (pMflT-o1 and pMflT-o2) were not able to replicate in M. florum (Fig. 3A), while only the sole intergenic region located downstream of dnaA was shown to be sufficient for plasmid replication in S. citri (25). Unfortunately, minimization efforts have not been reported for M. mycoides and M. capricolum oriC plasmids, thus preventing any comparison with M. florum. We also observed that oriC plasmids containing the heterologous oriC region of M. mycoides, M. capricolum, and S. citri (Fig. S5) failed to replicate in M. florum. It is, however, unclear why M. florum failed to replicate these heterologous oriC plasmids since they contain their own heterologous dnaA gene. One possibility that could explain this host/plasmid incompatibility is that the heterologous DnaA proteins of M. mycoides, M. capricolum, and S. citri were not sufficiently expressed in the M. florum context due to differences in the dnaA gene sequence, especially in the promoter region, or simply unable to interact with other proteins responsible for the DNA replication in M. florum (e.g., helicase). If this is the case, then the M. florum DnaA protein would have to properly recognize the DnaA boxes of the heterologous oriC regions to ensure plasmid replication. This recognition could, however, be impaired by divergences observed in the oriC region sequence and DnaA box organization of the Spiroplasma group. Indeed, M. florum and the mycoides cluster share 62% to 64% nucleotide identity at this region and only 57% with S. citri (Fig. 1A and C and Table S4). However, it was shown that even closely related species with high similarity of oriC region and DnaA box organization can fail to replicate heterologous oriC plasmids (18, 22, 24, 26). For instance, plasmids harboring the oriC region of M. mycoides were shown to replicate in M. capricolum (92% nucleotide identity), whereas the reverse experiment was shown to be unsuccessful (24). Furthermore, M. capricolum was also recently shown to allow the replication of oriC plasmids developed from S. citri, M. leachii, M. putrefaciens, and, most importantly, M. florum (12). Besides oriC region similarities, it is clear that much remains to be understood about the factors allowing or limiting replication of heterologous oriC plasmids between Mollicutes species. Broad-host-range vectors based on natural plasmid replicons could circumvent this limitation while also offering the possibility of introducing more than one plasmid per bacterium, potentially allowing a wide range of copies per cell. So far, plasmids have been isolated from some Mycoplasma and Spiroplasma species (30, 55) but not from Mesoplasma species. Additional work will be needed to experimentally test plasmids of interest in M. florum.
In summary, we report the development of the first genetic tools specifically designed for the near-minimal bacterium M. florum: two oriC plasmid configurations (pMflT-o3 and pMflT-o4), three functional antibiotic resistance markers (tetM, pac, and aadA1), and three different transformation methods (PEG-mediated, electroporation, and conjugation). This initial set of genetic tools will now be available for introducing genes in M. florum and will constitute a strong basis for other genetic engineering approaches. For example, oriC plasmids could be used to insert genes required for whole bacterial chromosome cloning in Saccharomyces cerevisiae. This strategy is now possible for M. florum (12), which offers the opportunity to efficiently modify its genome using the powerful yeast genetic engineering tools. Whole-genome cloning and transplantation have notably been used for the creation of the first synthetic bacterial genome and a quasiminimal genome based on M. mycoides subsp. capri (8–11, 13), and they will offer new opportunities for the development of an M. florum simplified cell chassis.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study are described in Table 3. E. coli strains EC100D pir+ and MM294 were routinely grown in Luria-Bertani (LB) broth at 37°C. E. coli strain MFDpir was grown at 37°C in LB broth supplemented with 0.3 mM diaminopimelic acid (DAP) and 200 μg/ml erythromycin. M. florum strain L1 (ATCC 33453) was grown at 34°C in ATCC 1161 medium. All strains were grown using an orbital shaker incubator and preserved at −80°C in their respective growth medium containing 25% (vol/vol) glycerol. Unless specified, antibiotics were used at the following concentrations for E. coli: ampicillin, 100 μg/ml; chloramphenicol, 34 μg/ml; erythromycin, 200 μg/ml; streptomycin, 50 μg/ml; spectinomycin, 100 μg/ml; and puromycin, 125 μg/ml. Unless specified, tetracycline was used at 15 μg/ml for either E. coli or M. florum. Penicillin was used at 200 U/ml for M. florum.
ATCC 1161 medium preparation.
To prepare 1 liter of ATCC 1161 medium, 17.5 g of heart infusion broth, 40 g of sucrose, and 12 g of agar (for solid medium) were first mixed in 710 ml of water before being autoclaved at 121°C. After sterilization, the mixture was cooled to room temperature (broth) or to 55°C (solid), and 200 ml of horse serum (catalog no. H1138; Sigma), 90 ml of 15% (wt/vol) yeast extract, 8 ml of 0.5% (wt/vol) phenol red, and 200 U/ml penicillin G were added. The pH was then adjusted to 7.6 with sterile NaOH. The final composition of ATCC 1161 medium was heart infusion broth, 17.5 g/liter; sucrose, 40 g/liter; agar (for solid medium), 12 g/liter; horse serum, 20% (vol/vol); yeast extract, 1.35% (wt/vol); phenol red, 0.004% (wt/vol); and penicillin G, 200 U/ml.
Antimicrobial susceptibility assays.
MIC values were determined by the growth inhibition assay, according to the broth microdilution method, in a 96-well microplate (56). The following antibiotics were tested for the M. florum L1 wild-type strain: ampicillin, chloramphenicol, erythromycin, gentamicin, kanamycin, puromycin, rifampin, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim. For M. florum L1 carrying pMflT-o4, pMflPT-o4, pMflCT-o4, and pMflET-o4, tetracycline, puromycin, chloramphenicol, and erythromycin, respectively, were tested. For M. florum L1 carrying pMflST-o4, spectinomycin and streptomycin were tested separately. Assays were conducted with three biological replicates in a final volume of 200 μl of ATCC 1161 medium supplemented with decreasing concentrations of the tested antibiotic. The medium was inoculated with ∼1.0 × 107 CFU of a log-phase batch culture for all tested strains. Microplates were next incubated at 34°C for 14 h. Bacterial growth was assessed by measuring the optical density at 560 nm every hour with a microplate reader (Synergy HT; BioTek). The metabolic activity of M. florum was previously shown to result in the acidification of the ATCC 1161 growth medium, causing changes in the absorbance of phenol red at 560 nm that correlate with the number of CFU (15). The MIC of each antibiotic was defined as the lowest tested concentration that inhibited the growth of M. florum (56).
Sequence analysis of the oriC region of the Spiroplasma group.
DNA sequence of the oriC region of selected representative members of the Spiroplasma group (M. florum L1, RefSeq accession no. NC_006055.1; M. capricolum subsp. capricolum ATCC 27343, RefSeq accession no. NC_007633.1; M. capricolum subsp. capripneumoniae 9231-Abomsa, RefSeq accession no. NZ_LM995445.1; Mycoplasma leachii PG50, RefSeq accession no. NC_014751.1; M. mycoides subsp. capri GM12, RefSeq accession no. NZ_CP001621.1; M. mycoides subsp. mycoides PG1, RefSeq accession no. NC_005364.2; Mycoplasma putrefaciens KS1, RefSeq accession no. NC_015946.1; Mycoplasma yeatsii GM274B, RefSeq NZ_CP007520.1; S. citri GII3-3x, GenBank accession numbers AM285301.1 and AM285302.1 [57]; Spiroplasma kunkelii CR2-3x, RefSeq accession no. NZ_CP010899.1; and Mycoplasma feriruminatoris G5847, GenBank accession no. ANFU01000022.1 [58]) were aligned using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) tool (3.8.31) (59). Alignments were cured using Gblocks 0.91b (60), and phylogeny was assessed using PhyML 3.1/3.0 aLRT (61), with a bootstrapping procedure repeated 1,000 times. A phylogenetic tree was drawn using TreeDyn (62).
The consensus sequence for DnaA boxes of the Spiroplasma group was generated by providing the intergenic regions upstream and downstream of the dnaA gene to the Multiple Em for Motif Elicitation (MEME) tool (31) using the “any number of repetitions” option and a maximum motif length of 15 bp. Precise locations of DnaA boxes within the oriC region of each Mollicutes chromosome were determined using the Motif Alignment and Search Tool (MAST) and the found MEME matrix (32). Positive and negative DNA strands were treated as separate strands, and only motifs with a P value below 1.0 × 10−5 were considered significant hits.
Plasmid construction.
The plasmids and oligonucleotides used in this study are listed in Tables 3 and S1, respectively. Detailed methodology of oriC plasmid construction is described in Text S1. M. florum oriC plasmids were constructed as depicted in Fig. 2. DNA fragments were amplified by PCR using VeraSeq 2.0 DNA polymerase (Enzymatics) and purified using Solid Phase Reversible Immobilization (SPRI) bead capture using Agencourt AMPure XP magnetic beads (Beckman Coulter) (63). Briefly, M. florum oriC fragments were amplified from M. florum L1 genomic DNA (gDNA), a tetM resistance cassette was amplified from pTT01, the ColE1 replication origin was amplified from pUC19 (GenBank accession no. L09137), and oriTRP4was amplified from pSW23T (GenBank accession no. AY733066). PCR fragments were assembled using the Gibson Assembly master mix (New England BioLabs) to generate pMflT-o1, pMflT-o3, and pMflT-o4 plasmids. pMflT-o2 was built by circularizing the 3.6-kb fragment of pMflT-o4 ClaI digestion. pMflPT-o4, pMflST-o4, and pMflCT-o4 plasmids were generated by cloning the pac, aadA1, and cat resistance cassettes into the NotI site of pMflT-o4, respectively. pMflET-o4 was obtained by cloning the ereB resistance cassette into a pMflT-o4 derivative plasmid. pMcapT, pMmmT, pMmcT, and pSciT-o4 plasmids were created using the pMflT-o4 backbone and the heterologous oriC fragment of M. capricolum, M. mycoides, or S. citri (Text S1 and Fig. S4). Plasmids were cloned in chemically competent E. coli strain EC100D pir+ cells, except for pMflPT-o4, which was cloned in E. coli strain MM294. Constructions were analyzed by restriction enzyme digestion, and M. florum oriC plasmid sequences were confirmed by paired-end Illumina sequencing at the Laboratoire de Génomique Fonctionnelle de l'Université de Sherbrooke (Quebec, Canada). Plasmid sequences and annotations are available in GenBank format at http://lab-rodrigue.recherche.usherbrooke.ca/m_florum_plasmids/.
Polyethylene glycol transformation.
M. florum L1 competent cells were prepared for PEG-mediated transformation by centrifuging 1 ml of a mid-logarithmic-phase bacterial culture at 21,100 × g and 10°C for 1 min. The cell pellet was washed with S/T buffer (10 mM Tris-HCl [pH 6.5], 250 mM NaCl) and centrifuged again under the same conditions. Cells were resuspended in 200 μl of 0.1 M CaCl2, incubated 30 min on ice, and then transformed using a PEG-mediated transformation procedure (3, 33). Briefly, 400 μl of modified ATCC 1161 medium (horse serum replaced by NaCl at a final concentration of 0.4% [wt/vol]) and 1 μg of plasmid DNA were added to the previously resuspended cells, and the solution was gently mixed by inverting the tube a few times. Then, one volume of 2× fusion buffer (20 mM Tris-HCl [pH 6.5], 250 mM NaCl, 20 mM MgCl2, 10% [wt/vol] PEG 8000) was immediately added, and cells were gently mixed. Cells were incubated for 50 min at 34°C and then poured into 5 ml of prewarmed ATCC 1161. The culture was gently mixed again and then incubated for 3 h at 34°C without shaking. After, cells were centrifuged at 7,900 × g and 10°C for 5 min, and the pellet was resuspended in 600 μl of ATCC 1161. Cells were serially diluted from 100 to 10−7 and plated on ATCC 1161 medium supplemented with tetracycline. To calculate the transformation frequency, 5 μl of each dilution was also spotted on ATCC 1161 medium without tetracycline. Plates were incubated at 34°C, colonies were counted, and transformation frequency was calculated according to the number of transformants obtained per recipient CFU. Assays were performed using at least three independent biological replicates.
Southern blot hybridization.
The gDNA of isolated clones of M. florum L1 carrying the pMflT-o3 or pMflT-o4 oriC plasmid was purified using the Quick-gDNA MiniPrep kit (Zymo Research), according to the manufacturer's specifications. Five hundred nanograms of gDNA was then digested at 37°C overnight using HindIII-HF restriction enzyme (New England BioLabs). After digestion, restriction fragments were separated on a 0.8% agarose gel, and DNA was depurinated and denatured by soaking the gel for 15 min in 0.25 M HCl and 0.4 M NaOH, respectively. DNA was then transferred onto a nylon membrane (Hybond-XL; Amersham Biosciences) by capillarity using 0.4 M NaOH. DNA was fixed to the membrane by UV cross-linking (700 J) and blot prehybridized for 1 h in Church buffer (0.25 M NaHPO4, 7% [wt/vol] SDS, 1× Denhardt's reagent, 1 mM EDTA). Labeled probe for tetM was synthesized by PCR from the pMflT-o4 DNA template using OneTaq DNA polymerase (New England BioLabs), the pBOT2-F/tetM-probe-R primer pair (Table S1), and 0.008 μM EasyTide-dCTP, [α-32P]-3000 Ci/mmol 10 mCi/ml (PerkinElmer). The following cycling conditions were used: (i) 30 s at 94°C; (ii) 30 cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 68°C; and (iii) 5 min at 68°C. Radiolabeled DNA probe was separated from unincorporated radioactive nucleotides using Bio-Spin columns (Bio-Rad), according to the manufacturer's recommendations. Purified tetM probe was next denatured at 95°C for 5 min, mixed with 10 ml of Church buffer, and added to the membrane for hybridization at 65°C overnight with gentle shaking. After hybridization, the membrane was washed twice for 5 min each using 2× SSC (0.3 M NaCl, 30 mM sodium citrate) containing 1% (wt/vol) SDS at 50°C and washed again using 0.2× SSC containing 1% (wt/vol) SDS at 55°C. Restriction fragments containing the tetM gene were finally visualized by autoradiography using a Typhoon FLA 9500 imaging system (GE Healthcare Life Sciences).
Quantification of oriC plasmid copy number.
The gDNA of isolated clones of M. florum L1 carrying pMflT-o3 or pMflT-o4 oriC plasmid, as well as wild-type M. florum L1 and M. florum L1 clone 3632 (mfl169::Tn-tetM), was purified using the Quick-gDNA MiniPrep kit (Zymo Research), according to the manufacturer's specifications. qPCR assays targeting the tetM gene were performed using the qPCR-tetM-F/qPCR-tetM-R primer pair (Table S1) and iQ SYBR green Supermix (Bio-Rad) at a final concentration of 1×. The relative abundance of the tetM gene was calculated using the ΔΔCT method (64) normalized to the rpoB (qPCR-rpoB-F/qPCR-rpoB-R) and rpoC (qPCR-rpoC-F/qPCR-rpoC-R) housekeeping genes (Table S1). qPCR amplifications were performed in triplicate under the following conditions: (i) 5 min at 95°C; (ii) 35 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C; and (iii) 5 min at 72°C. pMflT-o3 and pMflT-o4 copy numbers in M. florum were determined by measuring the relative abundance of the tetM gene in 12 individual clones for each plasmid compared to the M. florum L1 clone 3632 control strain containing a single copy of the tetM gene (65).
Plasmid stability assays.
One milliliter of an M. florum L1 log-phase culture carrying pMflT-o3 or pMflT-o4 growing in ATCC 1161 medium supplemented with tetracycline was centrifuged at 21,100 × g and 4°C for 1 min. The cell pellet was washed twice with 1 ml of ATCC 1161 medium without tetracycline and then resuspended in 200 μl of the same medium that was used to inoculate 20 ml of ATCC 1161 medium without tetracycline. The culture was next maintained in exponential-growth phase using a versatile continuous culture device (VCCD), as previously described (15). Five milliliters of culture was harvested every 12 h for 48 h, serially diluted from 100 to 10−7, and plated on nonselective ATCC 1161 medium and on ATCC 1161 supplemented with tetracycline. Plates were incubated at 34°C, colonies were counted, and plasmid stability was calculated according to the number of colonies growing on tetracycline divided by the number of colonies growing without tetracycline selection. Assays were performed using three independent biological replicates.
Conjugation assays.
E. coli MFDpir (35) carrying pMflT-o4 and wild-type M. florum L1 were grown until mid-logarithmic-growth phase, corresponding to ∼2.5 × 107 CFU/ml and ∼5.0 × 109 CFU/ml, respectively. Both cultures were centrifuged at 8,000 × g for 5 min, and cell pellets were resuspended in their original volume using fresh ATCC 1161 medium without penicillin and supplemented with 0.3 mM DAP (ATCC PEN−/DAP+). Conjugation assays were performed by mixing various volumes of resuspended M. florum recipient cells with 1 ml of resuspended E. coli donor cells to obtain different mating ratios (see Table S3). For each mating ratio, mixed cells were centrifuged at 16,000 × g for 2 min and washed twice with ATCC PEN−/DAP+. Cells were then resuspended in 30 μl of ATCC PEN−/DAP+, and the mating mixture was spotted on a 0.2-μm-pore nitrocellulose filter (25 mm; catalog no. 1214898; Maine Manufacturing) laid on top of an ATCC PEN−/DAP+ plate. Conjugation plates were incubated at 30°C for 24 h. Cells were recovered from the nitrocellulose filter using ATCC PEN−/DAP+ medium and serially diluted from 100 to 10−7 before plating. To select exconjugants, cells were plated on ATCC 1161 medium supplemented with tetracycline and 50 μg/ml ampicillin. Recipient cells were selected by spotting 5 μl of the 100 to 10−7 dilutions on an ATCC 1161 plate supplemented with 50 μg/ml ampicillin. Plates were incubated at 34°C, colonies were counted, and conjugation frequencies were calculated according to the number of exconjugants obtained per recipient CFU. Assays were performed using three independent biological replicates.
Electroporation of M. florum.
M. florum L1 cells were prepared for electroporation by centrifuging 1.0 ml of a mid-logarithmic-phase bacterial culture at 21,100 × g for 1 min at 4°C. The cell pellet was washed twice with an equal volume of electroporation buffer (272 mM sucrose, 1 mM HEPES [pH 7.4]). Cells were centrifuged again at 21,100 × g and 4°C for 1 min, and the cell pellet was resuspended in 100 μl of electroporation buffer. One microgram of plasmid DNA was added to 100 μl of previously prepared electrocompetent cells, and cells were transferred into a cold 1-mm electroporation cuvette. DNA was electroporated using a Gene Pulser Xcell electroporation system (Bio-Rad) set to 25 μF and 200 Ω, with a voltage varying from 0.5 to 3.0 kV. After electroporation, cells were recovered in 2 ml of ATCC 1161 medium and incubated at 34°C for 2 h. Recovered cells were serially diluted from 100 to 10−7 and plated on ATCC 1161 medium supplemented with tetracycline. To calculate transformation frequency, 5 μl of each dilution was also spotted on ATCC 1161 medium without tetracycline. Plates were incubated at 34°C, colonies were counted, and transformation frequency was calculated according to the number of transformants obtained per recipient CFU. Assays were performed using three independent biological replicates.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Carole Lartigue and Fabien Labroussaa for helpful discussions and for the kind gift of pMYCO1, pMYSO1, pMCO3, and pSD4 plasmids. We thank Joëlle Brodeur for technical assistance and Alain Lavigueur for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03374-16.
REFERENCES
- 1.Sirand-Pugnet P, Citti C, Barré A, Blanchard A. 2007. Evolution of mollicutes: down a bumpy road with twists and turns. Res Microbiol 158:754–766. doi: 10.1016/j.resmic.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Pettersson B, Johansson K-E. 2002. Taxonomy of Mollicutes, p 1–30. In Razin S, Herrmann R (ed), Molecular biology and pathogenicity of mycoplasmas. Springer, New York, NY. [Google Scholar]
- 3.Dybvig K, Voelker LL. 1996. Molecular biology of mycoplasmas. Annu Rev Microbiol 50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- 4.Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. doi: 10.1016/S0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 5.Mushegian A, Koonin E. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci U S A 93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson SN, Fraser CM. 2001. The complexity of simplicity. Genome Biol 2:comment2002.1. doi: 10.1186/gb-2001-2-2-comment2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCoy RE, Basham HG, Tully JG, Rose DL, Carle P, Bové JM. 1984. Acholeplasma florum, a new species isolated from plants. Int J Syst Bacteriol 34:11–15. doi: 10.1099/00207713-34-1-11. [DOI] [Google Scholar]
- 8.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi Z-Q, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 9.Benders GA, Noskov VN, Denisova EA, Lartigue C, Gibson DG, Assad-Garcia N, Chuang R-Y, Carrera W, Moodie M, Algire MA, Phan Q, Alperovich N, Vashee S, Merryman C, Venter JC, Smith HO, Glass JI, Hutchison CA. 2010. Cloning whole bacterial genomes in yeast. Nucleic Acids Res 38:2558–2569. doi: 10.1093/nar/gkq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lartigue C, Vashee S, Algire MA, Chuang R-Y, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA, Smith HO, Venter JC, Glass JI. 2009. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 11.Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA, Smith HO, Venter JC. 2007. Genome transplantation in bacteria: changing one species to another. Science 317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 12.Labroussaa F, Lebaudy A, Baby V, Gourgues G, Matteau D, Vashee S, Sirand-Pugnet P, Rodrigue S, Lartigue C. 2016. Impact of donor-recipient phylogenetic distance on bacterial genome transplantation. Nucleic Acids Res 44:8501–8511. doi: 10.1093/nar/gkw688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchison CA III, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi Z-Q, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 14.Baby V, Matteau D, Knight TF, Rodrigue S. 2013. Complete genome sequence of the Mesoplasma florum W37 strain. Genome Announc 1(6):e00879-13. doi: 10.1128/genomeA.00879-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matteau D, Baby V, Pelletier S, Rodrigue S. 2015. A small-volume, low-cost, and versatile continuous culture device. PLoS One 10:e0133384. doi: 10.1371/journal.pone.0133384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navas-Castillo J, Laigret F, Tully J, Bové JM. 1992. Mollicute Acholeplasma florum possesses a gene of phosphoenolpyruvate sugar phosphotransferase system and it uses UGA as tryptophan codon. C R Acad Sci III 315:43–48. (In French.) [PubMed] [Google Scholar]
- 17.Tully JG, Whitcomb RF, Hackett KJ, Rose DL, Henegar RB, Bové JM, Carle P, Williamson DL, Clark TB. 1994. Taxonomic descriptions of eight new non-sterol-requiring Mollicutes assigned to the genus Mesoplasma. Int J Syst Bacteriol 44:685–693. doi: 10.1099/00207713-44-4-685. [DOI] [PubMed] [Google Scholar]
- 18.Maglennon GA, Cook BS, Matthews D, Deeney AS, Bossé JT, Langford PR, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, BRaDP1T Consortium . 2013. Development of a self-replicating plasmid system for Mycoplasma hyopneumoniae. Vet Res 44:1–10. doi: 10.1186/1297-9716-44-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renaudin J, Marais A, Verdin E, Duret S, Foissac X, Laigret F, Bové JM. 1995. Integrative and free Spiroplasma citri oriC plasmids: expression of the Spiroplasma phoeniceum spiralin in Spiroplasma citri. J Bacteriol 177:2870–2877. doi: 10.1128/jb.177.10.2870-2877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopra-Dewasthaly R, Marenda M, Rosengarten R, Jechlinger W, Citti C. 2005. Construction of the first shuttle vectors for gene cloning and homologous recombination in Mycoplasma agalactiae. FEMS Microbiol Lett 253:89–94. doi: 10.1016/j.femsle.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordova CMM, Lartigue C, Sirand-Pugnet P, Renaudin J, Cunha RAF, Blanchard A. 2002. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J Bacteriol 184:5426–5435. doi: 10.1128/JB.184.19.5426-5435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-W, Browning GF, Markham PF. 2008. Development of a replicable oriC plasmid for Mycoplasma gallisepticum and Mycoplasma imitans, and gene disruption through homologous recombination in M. gallisepticum. Microbiology 154:2571–2580. doi: 10.1099/mic.0.2008/019208-0. [DOI] [PubMed] [Google Scholar]
- 23.Janis C, Lartigue C, Frey J, Wróblewski H, Thiaucourt F, Blanchard A, Sirand-Pugnet P. 2005. Versatile use of oriC plasmids for functional genomics of Mycoplasma capricolum subsp. capricolum. Appl Environ Microbiol 71:2888–2893. doi: 10.1128/AEM.71.6.2888-2893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lartigue C, Blanchard A, Renaudin J, Thiaucourt F, Sirand-Pugnet P. 2003. Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res 31:6610–6618. doi: 10.1093/nar/gkg848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lartigue C, Duret S, Garnier M, Renaudin J. 2002. New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid 48:149–159. doi: 10.1016/S0147-619X(02)00121-X. [DOI] [PubMed] [Google Scholar]
- 26.Sharma S, Citti C, Sagné E, Marenda MS, Markham PF, Browning GF. 2015. Development and host compatibility of plasmids for two important ruminant pathogens, Mycoplasma bovis and Mycoplasma agalactiae. PLoS One 10:e0119000. doi: 10.1371/journal.pone.0119000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messer W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev 26:355–374. [DOI] [PubMed] [Google Scholar]
- 28.Bolaños LM, Servin-Garciduenas LE, Martinez-Romero E. 2015. Arthropod-Spiroplasma relationship in the genomic era. FEMS Microbiol Ecol 91:1–8. doi: 10.1093/femsec/fiu008. [DOI] [PubMed] [Google Scholar]
- 29.Sirand-Pugnet P, Lartigue C, Marenda M, Jacob D, Barré A, Barbe V, Schenowitz C, Mangenot S, Couloux A, Segurens B, De Daruvar A, Blanchard A, Citti C. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet 3:744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breton M, Tardy F, Dordet-Frisoni E, Sagne E, Mick V, Renaudin J, Sirand-Pugnet P, Citti C, Blanchard A. 2012. Distribution and diversity of mycoplasma plasmids: lessons from cryptic genetic elements. BMC Microbiol 12:257. doi: 10.1186/1471-2180-12-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 32.Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 33.King KW, Dybvig K. 1991. Plasmid transformation of Mycoplasma mycoides subspecies mycoides is promoted by high concentrations of polyethylene glycol. Plasmid 26:108–115. doi: 10.1016/0147-619X(91)90050-7. [DOI] [PubMed] [Google Scholar]
- 34.Renaudin J, Breton M, Citti C. 2014. Molecular genetic tools of Mollicutes, p 55–76. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Wymondham, England. [Google Scholar]
- 35.Ferrières L, Hémery G, Nham T, Guérout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor-Robinson D, Bébéar C. 1997. Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J Antimicrob Chemother 40:622–630. doi: 10.1093/jac/40.5.622. [DOI] [PubMed] [Google Scholar]
- 37.Bébéar CM, Bébéar C. 2002. Antimycoplasmal agents, p 545–566. In Razin S, Herrmann R (ed), Molecular biology and pathogenicity of mycoplasmas. Springer, New York, NY. [Google Scholar]
- 38.Gaurivaud P, Laigret F, Bové JM. 1996. Insusceptibility of members of the class Mollicutes to rifampin: studies of the Spiroplasma citri RNA polymerase B-subunit gene. Antimicrob Agents Chemother 40:858–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as human pathogen. Clin Microbiol Rev 17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waites KB, Lysnyansky I, Bébéar CM. 2014. Emerging antimicrobial resistance in mycoplasmas of humans and animals, p 289–322. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Wymondham, England. [Google Scholar]
- 41.Uemura R, Sueyoshi M, Nagatomo H. 2010. Antimicrobial susceptibilities of four species of Mycoplasma isolated in 2008 and 2009 from cattle in Japan. J Vet Med Sci 72:1661–1663. doi: 10.1292/jvms.10-0165. [DOI] [PubMed] [Google Scholar]
- 42.Davis JW, Hanna BA. 1981. Antimicrobial susceptibility of Ureaplasma urealyticum. J Clin Microbiol 13:320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannan PCT. 1995. Antibiotic susceptibility of Mycoplasma fermentans strains from various sources and the development of resistance to aminoglycosides in vitro. J Med Microbiol 42:421–428. doi: 10.1099/00222615-42-6-421. [DOI] [PubMed] [Google Scholar]
- 44.Hannan PCT. 1997. Observations on the possible origin of Mycoplasma fermentans incognitus strain based on antibiotic sensitivity tests. J Antimicrob Chemother 39:25–30. doi: 10.1093/jac/39.1.25. [DOI] [PubMed] [Google Scholar]
- 45.Algire MA, Lartigue C, Thomas DW, Assad-Garcia N, Glass JI, Merryman C. 2009. New selectable marker for manipulating the simple genomes of Mycoplasma species. Antimicrob Agents Chemother 53:4429–4432. doi: 10.1128/AAC.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duret S, André A, Renaudin J. 2005. Specific gene targeting in Spiroplasma citri: improved vectors and production of unmarked mutations using site-specific recombination. Microbiology 151:2793–2803. doi: 10.1099/mic.0.28123-0. [DOI] [PubMed] [Google Scholar]
- 47.Dybvig K. 1989. Transformation of Acholeplasma laidlawii with streptococcal plasmids pVA868 and pVA920. Plasmid 21:155–160. doi: 10.1016/0147-619X(89)90061-9. [DOI] [PubMed] [Google Scholar]
- 48.Hahn TW, Mothershed EA, Waldo RH III, Krause DC. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120–124. doi: 10.1006/plas.1998.1387. [DOI] [PubMed] [Google Scholar]
- 49.Dybvig K, French CT, Voelker LL. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J Bacteriol 182:4343–4347. doi: 10.1128/JB.182.15.4343-4347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King KW, Dybvig K. 1994. Mycoplasmal cloning vector derived from plasmid pKMK1. Plasmid 31:49–59. doi: 10.1006/plas.1994.1006. [DOI] [PubMed] [Google Scholar]
- 51.Robison K, McGuire AM, Church GM. 1998. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J Mol Biol 284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 52.Ruffin DC, van Santen VL, Zhang Y, Voelker LL, Panangala VS, Dybvig K. 2000. Transposon mutagenesis of Mycoplasma gallisepticum by conjugation with enterococcus faecalis and determination of insertion site by direct genomic sequencing. Plasmid 44:191–195. doi: 10.1006/plas.2000.1485. [DOI] [PubMed] [Google Scholar]
- 53.Voelker LL, Dybvig K. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J Bacteriol 178:6078–6081. doi: 10.1128/jb.178.20.6078-6081.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts MC, Kenny GE. 1987. Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J Bacteriol 169:3836–3839. doi: 10.1128/jb.169.8.3836-3839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marenda MS. 2014. Genomic mosaics, p 15–54. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Wymondham, England. [Google Scholar]
- 56.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):S5–S16. [DOI] [PubMed] [Google Scholar]
- 57.Carle P, Saillard C, Carrère N, Carrère S, Duret S, Eveillard S, Gaurivaud P, Gourgues G, Gouzy J, Salar P, Verdin E, Breton M, Blanchard A, Laigret F, Bové JM, Renaudin J, Foissac X. 2010. Partial chromosome sequence of Spiroplasma citri reveals extensive viral invasion and important gene decay. Appl Environ Microbiol 76:3420–3426. doi: 10.1128/AEM.02954-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer A, Santana-Cruz I, Giglio M, Nadendla S, Drabek E, Vilei EM, Frey J, Jores J. 2013. Genome sequence of Mycoplasma feriruminatoris sp. nov., a fast-growing Mycoplasma species. Genome Announc 1(1):e00216-12. doi: 10.1128/genomeA.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 61.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 62.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ, Chisholm SW. 2010. Unlocking short read sequencing for metagenomics. PLoS One 5:e11840. doi: 10.1371/journal.pone.0011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Goryshin IY, Jendrisak J, Hoffman LM, Meis R, Reznikoff WS. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 66.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 67.Demarre G, Guérout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marlière P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.