Abstract
The leucine zipper tumor suppressor 2 (LZTS2) was identified as a tumor susceptibility gene within the 10q24.3 chromosomal region, and is approximately 15Mb from the PTEN locus. This region containing the both loci is frequently deleted in a variety of human malignancies, including prostate cancer. LZTS2 is a ß-catenin-binding protein and a negative regulator of Wnt signaling. Overexpression of PTEN in prostate cancer cell lines reduces ß-catenin-mediated transcriptional activity. In this study, we examined the collaborative effect of PTEN and LZTS2 using multiple in vitro and in vivo approaches. Co-expression of PTEN and LZTS2 in prostate cancer cells shows stronger repressive effect on ß-catenin mediated transcription. Using a newly generated mouse model, we further assessed the effect of simultaneous deletion of Pten and Lzts2 in the murine prostate. We observed that mice with both Lzts2 and Pten deletion have an earlier onset of prostate carcinomas as well as an accelerated tumor progression compared to mice with Pten or Lzts2 deletion alone. Immunohistochemical analyses show that atypical and tumor cells from compound mice with both Pten and Lzts2 deletion are mainly composed of prostate luminal epithelial cells and possess higher levels of cytoplasmic and nuclear β-catenin. These cells also exhibit a higher proliferative capacity than cells isolated from single deletion mice. These data demonstrate the significance of simultaneous Pten and Lzts2 deletion in oncogenic transformation in prostate cells and implicates a new mechanism for the dysregulation of Wnt/β-catenin signaling in prostate tumorigenesis.
Introduction
The leucine zipper tumor suppressor 2 (LZTS2), also called Lapser1, was originally identified based on homology with the LZTS1 tumor suppressor [1]. Lzts2 null mice showed no obvious pre- or post-natal lethality, but a portion of the mice developed defects in the kidney and urinary tract, including renal/ureteral duplication, hydroureter, and hydronephrosis [2]. Aged Lzts2 null mice also presented with increased spontaneous tumor development [3]. When treated with N-butyl-N-(4-hydroxybutyl) nitrosamine (BBN), both homozygous and heterozygous Lzts2 deletion mice showed increased susceptibility to urinary bladder carcinoma development [3]. LZTS2 has also been shown to interact with ß-catenin [4]. A Rev-like leucine-rich, CRM1/exportin-regulated nuclear export signal (NES) sequence was identified within the carboxyl terminal region of LZTS2. Through this NES site, LZTS2 can modulate the export of nuclear ß-catenin, reducing the transcriptional activity of ß-catenin in the cell [4]. These data suggest that LZTS2 is a bona fide regulator of ß-catenin and plays critical role in development and tumorigenesis.
The tumor suppressor PTEN is a phosphoprotein/phospholipid dual-specificity phosphatase [5]. Somatic mutation of PTEN frequently occurs in a variety of human tumors, including prostate cancer [6]. It has been shown that PTEN inhibits the activity of AKT/PKB, a key effector of the phosphatidylinositol 3-kinase (PI3K) signaling pathway, and functions as a tumor suppressor [7]. Activation of AKT can phosphorylate a number of downstream substrates, including glycogen synthase kinase 3ß, GSK3ß, [8,9]. Loss of PTEN increases GSK3ß phosphorylation and results in inhibiting ß-catenin degradation through the destruction complex [10].
Deletion of the human chromosomal 10q23-24 has been frequently observed in many human tumors, including prostate cancer. PTEN was identified within 10q23.3 region [11,12], and LZTS2 is located at 10q24.3, approximately 15Mb from the PTEN locus [1]. Intriguingly, both 10q23.3 and 10q24.3 regions, containing PTEN and LZTS2, are frequently deleted in a variety of human tumors [1,13]. PTEN deletion is closely associated with prostate cancer initiation and progression [6]. LZTS2 is expressed in human testis, prostate, and ovary tissues [4], and reduced expression of LZTS2 transcripts and proteins has been observed in prostate cancer tissues [3]. Similar to humans, in the mouse, Lzts2 is located on chromosome 19, only 11Mb from the mouse Pten gene [14]. In this study, we observed that PTEN and LZTS2 collaboratively enhance the transcriptional activity of ß-catenin in prostate cancer cells. To fully investigate the collaborative role of PTEN and LZTS2 in prostate tumor development, we generated a mouse model, in which both floxed Pten and Lzts2 alleles were targeted on chromosome 19. We subsequently crossed this mouse line with Probasin-Cre4 mice [15], and generated Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice. Using these mouse models, we characterized the biological consequences of the loss of either or both Pten and Lzts2 in the mouse prostatic luminal epithelium. We detected increased cellular proliferation in the prostates of Lzts2LoxP/LoxP PtenloxP/Wt:PB-Cre4 compound mice, and observed accelerated tumor development and aggressive tumor invasion. These data elucidate a collaborative role of loss of both Pten and Lzts2 in prostate tumorigenesis, and implicate a critical role of Wnt/ß-catenin in prostate tumorigenesis.
Experimental procedures
Cell cultures and transfections
Human prostate cancer cell lines, PC3 and DU145, were maintained in DMEM supplemented with 5% fetal calf serum (FCS) (HyClone, Denver, CO). An AR-positive prostate cancer cell line, LNCaP, was maintained in T-medium (Invitrogen, Carlsbad, CA) with 5% FCS. Transient transfections were carried out using a LipofectAMINE transfection kit or LipofectAMINE 2000 (Invitrogen, Carlsbad, CA).
DNA plasmids, and luciferase and ß-galactosidase assays
TOPflash (pGL3-OT) and FOPflash luciferease (pGL3-OF) reporters were obtained from Dr. Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD). A CMV-driven ß-galactosidase (ß-gal) reporter was generated by cloning the lacZ gene into the pcDNA3 vector [16]. The pcDNA-Tcf4 construct was provided by Dr. H. C. Clevers (CBG, Utrecht, The Netherlands). Expression constructs of human PTEN were generously provided by Dr. William Sellers (Dana-Farber Cancer Institute, Boston, MA). The full-length cDNA of human ß-catenin was cloned into pCDNA3 expression vector and mutants of ß-catenin with a single point mutation in the GSK3ß phosphorylation sites were generated by a PCR-based mutagenesis scheme as described previously [16]. LZTS2 expression vectors and shRNA pLentiviral vectors were generated as previously described [2,4].
Luciferase activity was measured in relative light units (RLU) as previously described [2,4,16,17]. Briefly, 50 μl of cell lysate was used for luciferase assays. The light output is measured after a 5 sec delay following injection of 50 μl luciferase buffer and 50 μl luciferin by the dual injector luminometer, according to the manufacturer’s instructions (Analytical Luminesence Lab., San Diego, CA). The RLU from individual transfections were normalized by measurement of ß-galactosidase activity expressed from a co-transfected plasmid. Individual transfection experiments were done at least three times in triplicate and the results are reported as mean luciferase/ß -galactosidase (±SD) from representative experiments.
Mouse mating and genotyping
We have previously generated a floxed allele for the mouse Lzts2 gene on chromosome 19 [2]. Mice homozygous for floxed Pten exon 5, PtenloxP/loxP, were obtained from the Jackson Laboratory (Strain#: 004597, Bar Harbor, ME). We then intercrossed Lzts2LoxP/+ with PtenloxP/LoxP mice to generate Lzts2LoxP/+-PtenloxP/LoxP compound mice through homologous recombination. To make the prostate specific conditional knockout mouse line, we bred the above mice with PB-Cre4 mice [15] to generate Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP-PtenloxP/Wt:PB-Cre4 mice in this study.
Genomic DNA samples isolated from mouse tail tips or embryo yolk sacs were used for genotyping as described in our previous reports [2,18]. Three primers were used to identify wild type and Lzts2 deleted alleles, including common forward primer, 5′-TACCATCTGAGTTGCTGATTGC-3′; wild type reverse primer, 5′-AGAGAGGAAGGAATGGGAGATC-3′; deleted reverse primer, 5′-CACAAGGAATGCTCCAACCCTG-3′. PCR was performed as follows: 5 min 94°C and then 35 cycles of 94°C for 45 sec, 60°C for 45 sec, and 72°C for 80 sec, followed by a final step at 72°C for 10 min. For Pten allele, we used the forward primer (5’-TCCCAGAGTTCATACCAGGA-3’) and the reverse primer (5’-AATCTGTGCATGAAGGGAAC -3’) to distinguish the wild type and target alleles by amplifying the flanking loxP sites. For detection of deleted exon 5, the forward primer 5’-ACTCAAGGCAGGGATGAGC-3’, and reverse primer, 5-GCTTGATATCGAATTCCTGCAGC-3’ were used [19]. The forward primer 5’GATCCTGGCAATTTCGGCTAT-3’ and reverse primer 5’GCAGGAAGCTACTCTGCACCTTG-3’ were used to detect the PB-Cre transgene. Genomic DNA fragments were amplified at 95°C for 5 min, then 95°C for 45 sec, 58°C for 40 sec, and 72°C for 60 sec for 36 cycles, then 72°C for 5 min. We made littermate controls lacking the Cre transgene in all experiments. All animal experiments performed in this study were approved by the ethics committee of the Administrative Panel on Institutional Animal Care and Use Committee at Stanford University and Beckman Research Institute/City of Hope, respectively.
Histological analyses and immunohistochemistry
In this study, we used the new guidelines recommended by The Mouse Models of Human Cancers Consortium Prostate Pathology Committee in 2013 for our pathological analyses [20]. Mouse tissues were fixed and processed as described in our previous study [21]. Slides were subsequently counterstained with 5% (w/v) Harris hematoxylin. For histological analysis, 5-μm serial sections were processed from xylene to water through a decreasing ethanol gradient, stained with hematoxylin and eosin, and processed back to xylene through an increasing ethanol gradient. For immunohistochemical assays, 5-μm sections were boiled in 0.01M citrate buffer (pH 6.0) or Tris-EDTA-Tween (pH 9.0) for 20 mins after re-dehydration from xylene to water, and blocked by 5% goat serum. Tissue sections were then incubated with 1:500 dilution of anti-mouse/human AR (Rabbit polyclonal Ab, Santa Cruz, sc-816), 1:100 anti-Pten (Rabbit mAb, Cell Signaling, 9559), 1:300 dilution of anti-p63(Rabbit polyclonal Ab, Santa Cruz, sc-8343), 1:3000 of anti-Ki67(Mouse mAb, Novacastsra, NCL-ki67), 1:300 of anti-E-cadherin(Mouse mAb, BD Transduction Laboratories, c20820), 1:1000 of anti-K5 (Rabbit polyclonal, Covance, PRB-160P), 1:1000 of anti-K8 (Mouse mAb, Covance, MMS-162P), 1:200 of synaptophysin antibody (Rabbit polyclonal, Invitrogen, 180130), 1:500 of anti-ß-catenin (Mouse mAb, BD Transduction Laboratories, 610154) or an “in-house” rabbit polyclonal anti-Lzts2 antibody [2], in 1% of goat serum at 4°C overnight. Tissues were incubated with biotinylated goat anti-mouse or goat anti-rabbit (Vector Laboratories, BA-1000 or BA-9200) at 1:1000 dilution for 1 hr at room temperature followed by a 30 min incubation with horse radish peroxidase (HRP)-conjugated streptavidin (Vector Laboratories, SA-5004). Immunostainings were visualized using DAB kit (Vector Laboratories, SK-4100). Images for all HE and immunohistochemistry experiments in this study were acquired on a Leica dissecting microscope (model MZ95) using Zeiss Axiovision software.
Preparation of whole cell lysates and nuclear extracts, and immunoprecipitation and blotting
Different aged mouse embryos were cut into small pieces, homogenized, and then used for making both cytosolic and nuclear extracts as described previously [10,16]. The cytosolic fractions were prepared in digitonin lysis buffer (1% digitonin, 150 mM NaCl, 50 mM Tris-HCl pH 7.5, 10 mM MgCl2) or in RIPA buffer (0.5% Nonidet P-40, 0.3% Triton X-100, 15mM MgCl2, 5mM EDTA, 150mM NaCl, 50mM Tris-HCl pH 7.8), respectively [22]. Nuclear extracts were prepared as described previously [23].
Protein fractions for immunoblotting were boiled in SDS-sample buffer and then resolved on a 10% SDS-PAGE. The proteins were transferred onto a nitrocellulose membrane and probed with anti-ß-catenin antibody (Santa Cruz Biotechnology), anti-tubulin (clone DM1A, Neomarker), PCNA (PC10, Termo Fisher Scientific), or the polyclonal Lzts2 antibody [2]. Proteins were detected using the ECL kit (Amersham, Arlington Heights, IL). The antibody against tubulin (Neomarker, Fremont, CA) was used for protein loading.
Statistical analyses
We presented the data as the mean ±SD. We made comparisons between groups, using a two-sided Student’s t test. P<0.05 and P<0.01 were considered significant.
Results
PTEN expression regulates ß-catenin transcriptional activity
It has been shown that wild-type PTEN expression inhibits the enhancement of ß-catenin mediated transcriptional activity in prostate cancer cells [10]. LZTS2 has also been shown to interact with ß-catenin and modulate the export of nuclear ß-catenin, reducing the transcriptional activity of ß-catenin [4]. In addition, both human and murine Pten and Lzts2 genes are closely localized on chromosome 10 or 19, respectively [14,24]. Furthermore, the deletion of both 10q23.3 and 10q24.3 regions that contain PTEN and LZTS2 genes have been frequently observed in a variety of human tumors [1,13]. Therefore, based on these lines of evidence, we examined the collaborative effect of PTEN and LZTS2 in regulating ß-catenin activity. We performed transient transfections in several prostate cancer cell-lines using either wild-type or stabilized mutant ß-catenin to assess PTEN expression in ß-catenin mediated transcription. These ß-catenin mutants contain point mutations within the phosphorylation site of GSK3β (S33F or S37A), which prevents degradation via the ubiquitin proteasome pathway. As shown in Fig 1, co-expression of TCF4 and ß-catenin induced transcription of the TOPflash (pGL3-OT) reporter in all three prostate cancer lines, including LNCaP, PC3, and DU145 (Fig 1A, 1B and 1C). Interestingly, a significant reduction of ß-catenin mediated transcriptional activity was observed when a wild-type PTEN vector was co-transfected with the wild-type ß-catenin expression vectors in all of three different cell lines (see lines 1 versus lines 2 in Fig 1A–1C, P<0.05). In contrast, there is almost no change in samples co-transfected with either stabilized mutant ß-catenin vectors in the presence or absence of PTEN (lines 3 to 6, Fig 1A–1C). The well described ß-catenin mutants used above are impervious to degradation by the destruction complex [25,26]. Therefore, these results suggest that PTEN can negatively regulate ß-catenin-mediated transcription in a GSK3ß-dependent manner.
Fig 1. PTEN represses ß-catenin transcriptional activity in multiple prostate cancer cell lines.

(A) LNCaP cells were transfected with TOPflash (pGL3-OT) or FOPflash (pGL3-OF) luciferase reporter (100 ng), pcDNA3-ß-gal (25 ng), pcDNA3-Tcf4 (5 ng), and the wild-type or mutants of pcDNA3-Flag-ß-catenin (50 ng). Either an empty pCMV5 vector or pCMV5-PTEN were co-transfected with the above plasmids. Cell lysates were measured for luciferase and ß-gal activities. Similar experiments were performed in (B) PC-3 and (C) DU-145 cells. The data represent the mean ± S.D. of three independent samples. “*” means P<0.05.
PTEN and LZTS2 collaboratively regulate β-catenin transcriptional activity
Next, we examined the possible collaborative effect of PTEN and LZTS2 on ß-catenin-mediated transcription. Co-expression of TCF4 and ß-catenin showed a transcriptional induction of pGL3-OT in LNCaP cells (Fig 2A). Transfection of PTEN or LZTS2 alone repressed wild type ß-catenin mediated transcriptional activity (lines 2 and 3, Fig 2A), while co-transfection of both PTEN and LZTS2 displayed significantly stronger repression (p<0.01, line 4 versus line 1, Fig 2A). In contrast, LZTS2 expression showed a repression on pGL3-OT promoter/reporter mediated by both wild type and mutated ß-catenin (lines 7, 8, 11, and 12, Fig 2A). These data suggest that LZTS2 can repress ß-catenin mediated transcription collaboratively with PTEN, and its regulatory mechanism of ß-catenin is distinct from PTEN-mediated repression [4]. We then evaluated the repressive effect of endogenous LZTS2 using short hair-pin RNA (shRNA) interference. Transfection of LZTS2 shRNA, but not control shRNA, showed reduced expression of endogenous LZTS2 proteins in LNCaP cells (Fig 2C). These knockdown effects also resulted in a dosage-dependent activation of both wild type and stabilized mutant ß-catenin with mutations of the serine residues on the pGL3-OT promoter/reporter in LNCaP cells (Fig 2B). In contrast, there is no change in samples transfected with the control shRNA vector, suggesting that the above effect was due to LZTS2 knock-down. Taken together, these data demonstrate the role of LZTS2 in the regulation of ß-catenin-mediated transcription.
Fig 2. LZTS2 and PTEN collaboratively repress β-catenin-mediated transcription.
(A) One hundred ng of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-ß-gal, 5 ng of TCF4 expression vector, and 50 ng of wild-type or mutant ß-catenin were co-transfected with either pcDNA3-FLAG-hLZTS2 or pCMV5-PTEN plasmids alone or together into LNCaP cells. Cell lysates were measured for luciferase and ß-gal activities. (B) LNCaP cells were transfected with either 100 ng of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-ß-gal, 50 ng of wild-type or mutant ß-catenin, and control or Lzts2-targeted shRNA as indicated. Luciferase and ß-gal activities were measured as described above. The data represent the mean ± S.D. of three independent samples. (C) LNCaP cells were transfected with either control or LZTS2 targeted shRNAa representative western blot with antibodies against human LZTS2 or Tubulin is shown. “*” or “**” means P<0.05 or <0.01, respectively.
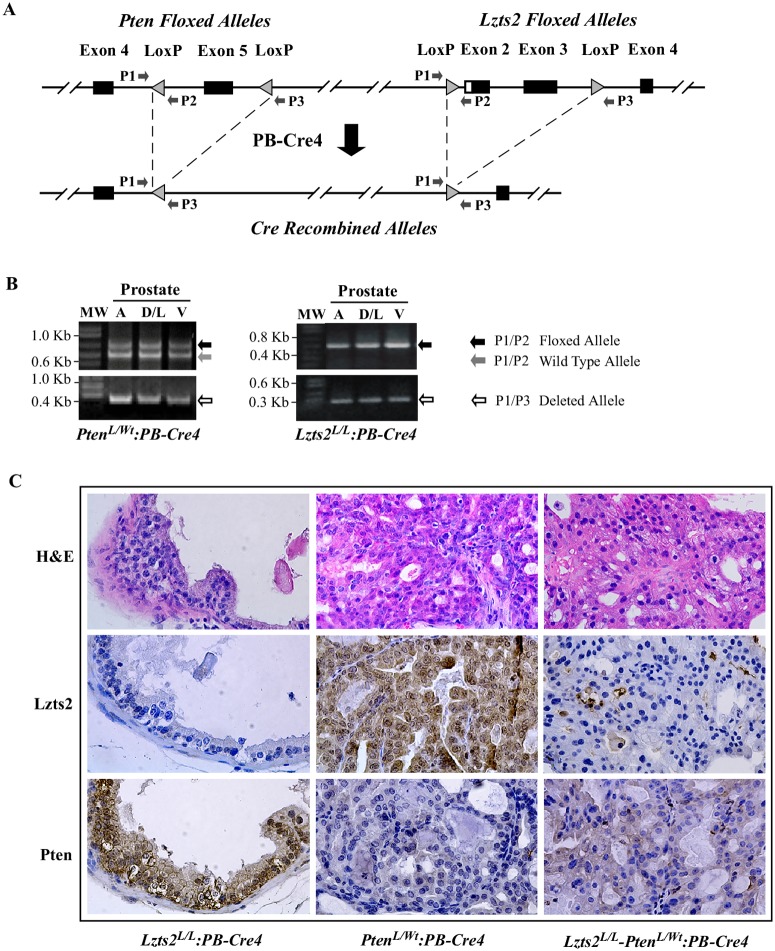
Generation of the Lzts2 and Pten compound mice
To further examine the collaborative role of PTEN and LZTS2 in vivo, we took a loss-of-function approach to directly address the biological significance of PTEN and LZTS2 in tumorigenesis using an Lzts2 and Pten deficient mouse strain. Because murine Lzts2 is located approximately 11Mb away from Pten [14], we recombined floxed Pten and Lzts2 loci into chromosome 19 by crossing Pten and Lzts2 floxed mice [2,19]. To examine the role of Pten and Lzts2 in the murine prostate, we subsequently crossed this mouse model with Probasin-Cre4 mice [15], and generated Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice (Fig 3A). Using specific primers (Fig 3A), we assessed mouse genotypes using genomic PCR analysis. We observed both appropriate floxed and deleted Pten and Lzts2 alleles in mouse prostate tissues (Fig 3B). We then evaluated Pten and Lzts2 expression in prostate tissues, which were isolated from 6–8 month old mice with different genotypes, using immunohistochemistry. As shown in Fig 3C, Lzts2 staining was observed in prostatic luminal cells of PtenloxP/Wt:PB-Cre4 mice, but very low or no staining with Lzts2 antibody was detected in samples isolated from Lzts2LoxP/LoxP:PB-Cre4 and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice. In a similar vein, decreased staining with a Pten specific antibody was observed in prostate tissue samples isolated from both PtenloxP/Wt:PB-Cre4 and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice. These data demonstrate that either or both Lzts2 and Pten are deleted in the prostate of Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice, respectively.
Fig 3. Generation of mice with prostate-specific Lzts2 and Pten deletion.
(A) Schematic representation of Lzts2 and Pten compound mice. On chromosome 19, loxP sites flank exon 5 of the pten gene and exons 2 and 3 of the lzts2 gene. Crossing with PB-Cre mice results in prostate-specific recombination of the loxP sites and removal of these exons from this locus. (B) Genomic PCR analysis of Lzts2 and Pten alleles in prostate tissues using specific primers as detailed in (A). (C) Histological and immunohistochemical analysis of 6–8 month old Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice. Top panel depicts H&E staining of prostate tissues. Immunohistochemistry of Lzts2 (middle) and Pten (lower) on sequential sections are shown below.
Conditional deletion of Lzts2 accelerates Pten-mediated oncogenic transformation in the mouse prostate
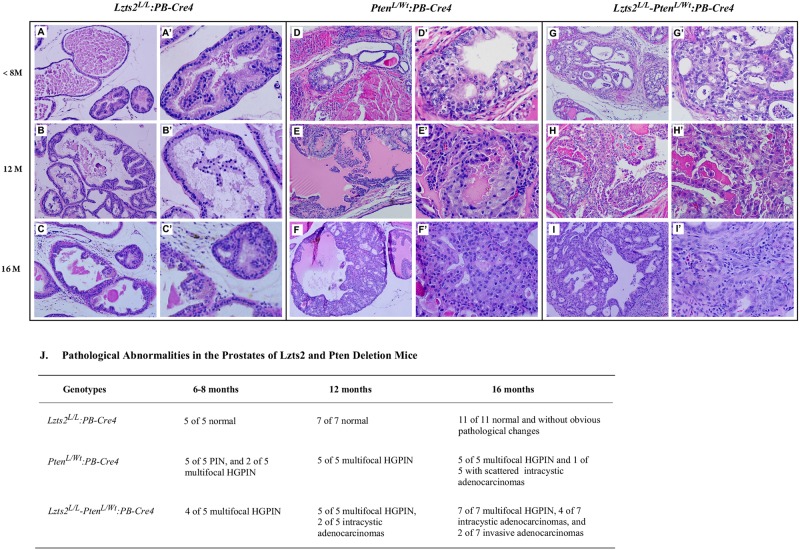
Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice as well as Lzts2LoxP/LoxP:PB-Cre4 and PtenloxP/Wt:PB-Cre4 mice were born at the expected Mendelian ratios and appeared normal with no obvious differences from their wild-type littermates at birth. We systematically examined male mice starting at 2-months of age and followed them until at least 16-months of age. We did not observe obvious abnormalities in 16 to 22-month-old Lzts2LoxP/LoxP: PB-Cre4 mice (Fig 4A–4C’). Adhering to recommendations of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee [20], we observed the development of prostatic intraepithelial neoplasia (PIN) in 6-month-old PtenloxP/Wt:PB-Cre4 mice. The PIN lesions first occurred in ventral prostate (VP), and then extended to dorsal (DP), lateral (LP), and anterior (AP) lobes. With time, these mPIN lesions progressed towards high-grade mPIN lesions or prostatic intracystic adenocarcinomas (Fig 4E–4F’). These lesions originated predominantly in the dorsal/lateral prostate (D/LP) and ventral prostate (VP) lobes, which is consistent with previous observations [19]. Notably, more Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice developed HGPIN lesions at 6-months of age than PtenloxP/Wt:PB-Cre4 mice (Fig 4G and 4G’). The compound mice also showed accelerated tumor development. At 12-months of age, half of the Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice developed prostatic intracystic adenocarcinomas, and at 16-months, almost all of the Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice progressed to prostatic intracystic adenocarcinomas (Fig 4J). Using the Fisher's exact test, we analyzed the difference in prostatic adenocarcinoma formation between PtenloxP/Wt:PB-Cre4 and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice in 12 and 16 age groups, and observed a significant difference (P<0.05). These results clearly demonstrate that deletion of Lzts2 accelerates prostate tumor progression in PtenloxP/Wt:PB-Cre4 mice.
Fig 4. Prostate-specific Lzts2 deletion accelerates Pten-mediated tumorigenesis.
Prostates from mice between the ages of 2 to 16-months were analyzed for neoplastic lesions following H&E staining as per the guidelines from the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Panels depict 10x images (A-I) and 40x images (A’-I’) of Lzts2LoxP/LoxP:PB-Cre4 (panels A-C’), PtenloxP/Wt:PB-Cre4 (panels D-F’), and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 (panels G-I’) mice. (J) Table describing pathological abnormalities in cohorts of aged Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice.
Identifying cellular origins of atypical and tumor cells
Mouse prostatic epithelium is composed of several cell types, including basal and luminal epithelial cells, as well as neuroendocrine cells. Previous studies have shown that luminal epithelial cell markers have been detected in PIN and prostatic adenocarcinoma lesions in Pten prostate conditional knockout mice with ARR2PB-Cre [19]. To determine the cellular origin of PIN in Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice, we performed comprehensive immunohistochemical analyses to examine a series of prostatic cellular markers on these high-grade PIN lesions (Fig 5). Atypical cells of PIN lesions failed to immunoreact with Lzts2 (Fig 5B1 and 5B3). Most atypical prostatic cells in the sample of Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice showed typical nuclear immunoreactivity with Ar (Fig 5C3), which is similar to the PtenloxP/Wt:PB-Cre4 mice (Fig 5C2). In samples isolated from both PtenloxP/Wt:PB-Cre4 and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice, atypical cells showed positive immunoreactivity for E-cadherin and CK8, secretory epithelial markers (Fig 5D2, 5D3, 5E2 and 5E3), but showed no immunoreactivity for the neuroendocrine cell marker, synaptophysin (Fig 5H1–5H3). Immunoreactivity for CK5 and p63, the cellular markers for prostatic basal epithelial cells, appeared mainly in the basal compartment of normal prostatic glands, but rarely in atypical cells in the above mice (Fig 5F2, 5F3, 5G2 and 5G3). Taken together, these data demonstrate that prostatic atypical cells in Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mainly contain luminal cellular markers.
Fig 5. Mouse PINs from Lzts2-Pten compound mice are composed primarily of luminal epithelial cells.
Immunohistochemical comparison of prostates from Lzts2LoxP/LoxP:PB-Cre4 (panel 1), PtenloxP/Wt:PB-Cre4 (panel 2), and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 (panel 3) mice. Prostates were stained with H&E (Panels A1-A3) for histological comparison and Lzts2 (B1-B3), mouse androgen receptor (C1-C3), E-cadherin (D1-D3), cytokeratin 8 (E1-E3), cytokeratin 5 (F1-F3), p63 (G1-G3), and Synaptosin (H1-H3) to characterize the atypical cells.
Conditional deletion of Lzts2 enhances prostatic cell proliferation and results in alteration of ß-catenin subcellular localization
It has been shown that deletion of Pten enhances proliferation of prostatic epithelial cells in mice [19,27,28]. In this study, we assessed whether deletion of Lzts2 enhances cell proliferation in the prostate of mice using Ki67 immunohistochemistry. We carefully quantified Ki67 immunostaining in mouse prostate tissues by counting a total of 1000 epithelial cells from five high-power fields in samples isolated from Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice in different age groups. Experiments were repeated at least three times with three different slides prepared independently in each genotype. As shown in Fig 6A–6D, we presented data prepared from 6–8 month old mice with different genotypes mice. Heterozygous deletion of Pten appears to increases cell proliferation in prostatic epithelial cells in comparison with samples isolated from Lzts2LoxP/LoxP:PB-Cre4 mice (Fig 6B1 and 6B2 versus Fig 6A1 and 6A2). Intriguingly, a significant increase was observed in Ki67 immunostaining in both mPIN and prostatic adenocarcinoma lesions in Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice when compared to those in PtenloxP/Wt:PB-Cre4 mice (Fig 6C1 and 6C2 versus Fig 6B1 and 6B2). The epithelial proliferative index increased from 80 to 240 in HGPIN lesions (P<0.01, Fig 6D). These results demonstrate that Lzts2 deletion can augment the proliferation of prostatic epithelial cells mediated by Pten deletion in the compound mice.
Fig 6. Lzts2 deletion increases cellular proliferation and nuclear ß-catenin in the mouse prostate.
(A-C). Cellular proliferation was examined by immunostaining for Ki-67. Prostate sections isolated from Lzts2LoxP/LoxP:PB-Cre4, PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice were stained for Ki-67. (D) A total of 1000 epithelial cells in each lesion from three different lesions from three mice of each genotype were evaluated for Ki-67 immunoreactivity. (E) Mouse embryonic fibroblasts (MEFs) were prepared from different genotype embryos at E10.5. Either whole cell lysates or nuclear extracts were isolated from different genotype MEFs and analyzed by Western-blotting assays for either ß-catenin (ß-cat), tubulin, or PCNA. (F-H) Representative H&E and ß-catenin staining of Prostate tissues from the three different genotype mice is shown. Boxes highlight strong nuclear ß-catenin staining observed with conditional LZTS2 deletion (F2, H2). “*” or “**” means P<0.05 or <0.01, respectively.
Previously, we have demonstrated that LZTS2 regulates the cellular level and localization of ß-catenin [4]. In this study, we also confirmed the effect of Lzts2 on cellular ß-catenin in mouse embryonic fibroblasts (MEFs). As shown in Fig 6E, both whole cell lysates and nuclear extracts prepared from different genotypes of MEFs were analyzed for levels of ß-catenin. A notable increase of nuclear ß-catenin was observed in the nuclear extract of Lzts2 null MEFs despite similar levels of total ß-catenin in whole cell lysates isolated from the same cells. We then performed immunohistochemistry to assess ß-catenin expression in prostate tissues isolated from the three genotypes of mice. We observed typical cell membrane staining of ß-catenin in prostatic luminal cells in the samples isolated from all of three different genotype mice (Fig 6F2, 6G2, and 6H2)., Slightly increased cytoplasmic ß-catenin staining was observed in some of the prostatic epithelial cells of PtenloxP/Wt:PB-Cre4 mice (Fig 6G2). Intriguingly, a clear nuclear staining of ß-catenin appears in prostatic epithelial cells of samples from Lzts2LoxP/LoxP:PB-Cre4 and Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice (Fig 6F2 and 6H2 boxed). These data further implicate the role of LZTS2 in promoting the nuclear export of ß-catenin in prostatic epithelial cells in mice.
Discussion
Human PTEN and LZTS2 are localized on the region of 10q23-24, within approximately 15Mb of each other [1]. Loss of heterozygosity (LOH) and homozygous deletions at human chromosomal region 10q23-24 are frequently found in prostate adenocarcinomas, as well as other malignancies, suggesting that multiple tumor suppressors may be present in the region [13]. Most intriguingly, approximately 10% of prostate tumor samples have been shown to possess both LZTS2 and PTEN deletion [29]. In this study, we generated a new mouse model in which both PTEN and LZTS2 were deleted simultaneously in prostatic epithelium to directly assess the biological significance and clinical relevance of PTEN and LZTS2 inactivation in prostate tumorigenesis. As we reported here, we observed accelerated oncogenic transformation and aggressive tumor phenotypes in the prostates of Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice with the deletion of both Pten and Lzts2 genes in comparison to PtenloxP/Wt:PB-Cre4 mice with Pten deletion only. Our data demonstrate the biological role of LZTS2 in tumorigenesis, and implicates the loss of both LZTS2 and PTEN as important biological and relevant events that can directly contribute to prostate cancer development and progression.
Interestingly, similar to humans, both murine Pten and Lzts2 are localized on Chromosome 19, only 11Mb apart from each other [14]. Homozygous deletion of Pten in the mouse embryo is lethal and characterized by developmental defects in the mesoderm, endoderm and ectoderm [30]. Heterozygous Pten mice develop multiple neoplasia in a wide spectrum of tissues including prostate, thyroid, colon, lymphatic system, mammary gland, and endometrium [30–32]. Conditional inactivation of Pten in the murine prostate results in PIN and invasive prostate cancer [19], suggesting a critical role between PTEN inactivation and prostate tumorigenesis. LZTS2 is expressed in testis, prostate, and ovary tissues [4], and reduced expression of LZTS2 transcripts and proteins has been observed in prostate cancer samples [3]. An increase in spontaneous tumor development has been observed in both aged Lzts2 heterozygous and homozygous knockout mice in comparison to wild type littermates [3]. These heterozygous or homozygous mice also showed an increase of BBN, a carcinogen, induced urinary bladder carcinoma development [3]. These lines of evidence suggest that both PTEN and LZTS2 play critical roles in tumorigenesis, and inactivation of both proteins may have a collaborative effect in oncogenic transformation. Our data presented in this report provide a line of evidence demonstrating combined loss of LZTS2 and PTEN as an important biological event in prostate cancer development and progression.
Multiple lines of evidence suggest that the Lzts2 gene is a tumor susceptibility gene [3]. Our previous data also showed a potential role of Lzts2 in prostate tumorigenesis. In this study, we also generated mice with conditional inactivation of Lzts2 in prostatic luminal epithelial cells using PB-Cre transgenic mice to directly examine Lzts2 in prostate tumorigenesis, [15]. We did not observe significant pathological changes in the prostate of both Lzts2LoxP/wt:PB-Cre4 and Lzts2LoxP/LoxP:PB-Cre4 mice up to 20-months of age (data not shown). These results imply that selective inactivation of Lzts2 in prostatic luminal epithelial cells by the ARR2PB promoter is insufficient to induce oncogenic transformation in prostatic luminal epithelial cells [15]. Homozygous deletion of Pten in the murine prostate results in invasive prostate cancer and metastatic prostate cancer of the lymph nodes and lung as early as ages of 2-months [19]. However, conditional heterozygous inactivation of Pten in the mouse prostate showed slow and moderate PIN and prostatic adenocarcinomas development [19]. Therefore, we used Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice to further evaluate the combined effect of Lzts2 and Pten inactivation in the prostate of mice. As detailed in this study, homozygous inactivation of Lzts2 in the mouse prostate accelerates the oncogenic transformation mediated by heterozygous loss of Pten in prostatic luminal epithelial cells. Given that PTEN loss of heterozygosity has been frequently observed in human tumors, Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 mice may mimic what occurs during the course of human prostate cancer development, and can be used to characterize this mechanism of prostate cancer initiation and progression. Specifically, identification of possible pathways and molecules that are involved in Lzts2 and Pten mediated tumorigenesis using the above mouse models would be biologically significant and clinical relevant.
Dysregulation of Wnt and ß-catenin mediated signaling pathways events in the pathogenesis of variety of human malignancies, including prostate cancer [33,34]. It has been shown that tumor cells contain high levels of nuclear ß-catenin through different regulatory mechanisms [35]. LZTS2 has been demonstrated to regulate ß-catenin nuclear export and modulate its cellular distribution and activity [4]. In this study, using Lzts2-deleted MEFs, we also assessed the effect of Lzts2 on the cellular localization of ß-catenin. Although we observed almost equal levels of ß-catenin in whole cell lysates prepared from either wild type or heterozygous and homozygous Lzts2 deletion MEFs, a significant increase of nuclear ß-catenin appears in Lzts2 null MEFs. This observation is consistent with previous data and demonstrates an important role of Lzts2 in regulating ß-catenin nuclear export [4]. PTEN exerts its function as a tumor suppressor through negative regulation of PI3K/AKT signaling pathways [5]. PI3K/Akt increases the stability of nuclear ß-catenin by phosphorylation and inactivation of the downstream substrate, GSK3ß, in prostate cancer cells, and PTEN deletion can augment PI3K/AKT action and increase cellular ß-catenin [10]. As shown in this study, prostate cancer cells co-transfected with both wild type PTEN and LZTS2 expression vectors showed less transcriptional activity of Tcf/ß-catenin than those transfected with either PTEN or LZTS2 alone. Interestingly, PTEN expression showed a much stronger inhibitory effect on wild type of ß-catenin than mutated ß-catenin. In contrast, LZTS2 expression inhibits both wild type and mutated ß-catenin activity. Through these distinct mechanisms, PTEN and LZTS2 collaboratively regulate cellular levels of ß-catenin and act as tumor suppressors to inhibit Wnt/ß-catenin-mediated oncogenic transformation in cells. In addition, we observed an increase in PIN and prostatic tumor development in Lzts2LoxP/LoxP -PtenloxP/Wt:PB-Cre4 compound mice in comparison to PtenloxP/Wt:PB-Cre4 mice. Most atypical and tumor cells in Lzts2LoxP/LoxP-PtenloxP/Wt:PB-Cre4 mice appear to be E-cadherin and CK8 positive, suggesting that they are of luminal epithelial cellular origin. In this study, we also measured cell proliferation in samples isolated from different mice. Prostatic luminal cells isolated from Lzts2LoxP/LoxP-PtenloxP/Wt:PB-Cre4 compound mice appear more proliferative than those from other genotypes of mice. We also observed more cellular ß-catenin expression in atypical and tumor cells in the prostate of PtenloxP/Wt:PB-Cre4, and Lzts2LoxP/LoxP-PtenloxP/Wt:PB-Cre4 mice. Interestingly, deletion of Lzts2 alone showed more nuclear ß-catenin expression than the other genotypes in the above samples. These data provide a link between increased cellular ß-catenin and oncogenic transformation in prostatic luminal epithelial cells. Validation of PTEN and LZTS2 loss, as well as cellular ß-catenin expression and localization within human tumor samples will provide useful information about the roles of PTEN and LZTS2 in human tumorigenesis; this knowledge may lead to the development of new therapeutic strategies for prostate cancer and other human malignancies.
Data Availability
All relevant data are within the paper only.
Funding Statement
This work was supported by Public Health Service grants, R01CA070297, R01CA151623, R01CA166894, R21CA190021, and R01DK104941 from the National Cancer Institute and the National Institute of Diabetes and Digestive and Kidney Diseases, as well as a Ruth L. Kirschstein Nation Research Service Award NIH/NCI T32 CA009523 (for Daniel Johnson). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cabeza-Arvelaiz Y, Thompson TC, Sepulveda JL, Chinault AC. LAPSER1: a novel candidate tumor suppressor gene from 10q24.3. Oncogene. 2001;20(46):6707–17. 10.1038/sj.onc.1204866 [DOI] [PubMed] [Google Scholar]
- 2.Peng Y, Clark C, Luong R, Tu WH, Lee J, Johnson DT, et al. The leucine zipper putative tumor suppressor 2 protein LZTS2 regulates kidney development. J Biol Chem. 2011;286(46):40331–42. 10.1074/jbc.M111.302059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson DT, Luong R, Lee SH, Peng Y, Shaltouki A, Lee JT, et al. Deletion of leucine zipper tumor suppressor 2 (lzts2) increases susceptibility to tumor development. J Biol Chem. 2013;288(6):3727–38. 10.1074/jbc.M112.417568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thyssen G, Li TH, Lehmann L, Zhuo M, Sharma M, Sun Z. LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin. Mol Cell Biol. 2006;26(23):8857–67. 10.1128/MCB.01031-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96(8):4240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–7. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95(26):15587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13(22):2905–27. [DOI] [PubMed] [Google Scholar]
- 9.Desbois-Mouthon C, Cadoret A, Blivet-Van Eggelpoel MJ, Bertrand F, Cherqui G, Perret C, et al. Insulin and IGF-1 stimulate the beta-catenin pathway through two signalling cascades involving GSK-3beta inhibition and Ras activation. Oncogene. 2001;20(2):252–9. 10.1038/sj.onc.1204064 [DOI] [PubMed] [Google Scholar]
- 10.Sharma M, Chuang WW, Sun Z. Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through GSK3beta inhibition and nuclear beta-catenin accumulation. J Biol Chem. 2002;277(34):30935–41. 10.1074/jbc.M201919200 [DOI] [PubMed] [Google Scholar]
- 11.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997;16(13):3797–804. 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62. 10.1038/ng0497-356 [DOI] [PubMed] [Google Scholar]
- 13.Rasheed BK, McLendon RE, Friedman HS, Friedman AH, Fuchs HE, Bigner DD, et al. Chromosome 10 deletion mapping in human gliomas: a common deletion region in 10q25. Oncogene. 1995;10(11):2243–6. [PubMed] [Google Scholar]
- 14.Hansen GM, Justice MJ. Pten, a candidate tumor suppressor gene, maps to mouse chromosome 19. Mammalian genome. 1998;9(1):88–90. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101(1–2):61–9. [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, et al. Linking beta-catenin to androgen signaling pathway. J Biol Chem. 2002;277(13):11336–44. 10.1074/jbc.M111962200 [DOI] [PubMed] [Google Scholar]
- 17.Verras M, Sun Z. Beta-catenin is involved in insulin-like growth factor 1-mediated transactivation of the androgen receptor. Mol Endocrinol. 2005;19(2):391–8. 10.1210/me.2004-0208 [DOI] [PubMed] [Google Scholar]
- 18.Beliakoff J, Lee J, Ueno H, Aiyer A, Weissman IL, Barsh GS, et al. The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol Cell Biol. 2008;28(1):282–92. 10.1128/MCB.00771-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–21. [DOI] [PubMed] [Google Scholar]
- 20.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73(9):2718–36. 10.1158/0008-5472.CAN-12-4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu C, Luong R, Zhuo M, Johnson DT, McKenney JK, Cunha GR, et al. Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. J Biol Chem. 2011;286(38):33478–88. 10.1074/jbc.M111.269894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Playford MP, Bicknell D, Bodmer WF, Macaulay VM. Insulin-like growth factor 1 regulates the location, stability, and transcriptional activity of beta-catenin. Proc Natl Acad Sci U S A. 2000;97(22):12103–8. 10.1073/pnas.210394297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabeza-Arvelaiz Y, Sepulveda JL, Lebovitz RM, Thompson TC, Chinault AC. Functional identification of LZTS1 as a candidate prostate tumor suppressor gene on human chromosome 8p22. Oncogene. 2001;20(31):4169–79. 10.1038/sj.onc.1204539 [DOI] [PubMed] [Google Scholar]
- 25.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–80. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 26.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 27.Kwak MK, Johnson DT, Zhu C, Lee SH, Ye DW, Luong R, et al. Conditional deletion of the Pten gene in the mouse prostate induces prostatic intraepithelial neoplasms at early ages but a slow progression to prostate tumors. PLoS One. 2013;8(1):e53476 10.1371/journal.pone.0053476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X, Ziel-van der Made AC, Autar B, van der Korput HA, Vermeij M, van Duijn P, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65(13):5730–9. 10.1158/0008-5472.CAN-04-4519 [DOI] [PubMed] [Google Scholar]
- 29.Network CGAR. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–25. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19(4):348–55. 10.1038/1235 [DOI] [PubMed] [Google Scholar]
- 31.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96(4):1563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58(2):204–9. [PubMed] [Google Scholar]
- 33.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21(12):1021–30. [DOI] [PubMed] [Google Scholar]
- 34.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7. 10.1126/science.1094291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Povelones M, Nusse R. Wnt signalling sees spots. Nat Cell Biol. 2002;4(11):E249–50. 10.1038/ncb1102-e249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper only.