Abstract
Long non-coding RNAs (lncRNAs) have emerged as important in cancer development and progression. The impact of diet on lncRNA expression is largely unknown. Sulforaphane (SFN), obtained from vegetables like broccoli, can prevent and suppress cancer formation. Here we tested the hypothesis that SFN attenuates the expression of cancer-associated lncRNAs. We analyzed whole genome RNA-sequencing data of normal human prostate epithelial cells and prostate cancer cells treated with 15 μM SFN or DMSO. SFN significantly altered expression of ~100 lncRNAs in each cell type, and normalized the expression of some lncRNAs that were differentially expressed in cancer cells. SFN-mediated alterations in lncRNA expression correlated with genes that regulate cell cycle, signal transduction, and metabolism. LINC01116 was functionally investigated because it was overexpressed in several cancers, and was transcriptionally repressed after SFN treatment. Knockdown of LINC01116 with siRNA decreased proliferation of prostate cancer cells, and significantly upregulated several genes including GAPDH (regulates glycolysis), MAP1LC3B2 (autophagy) and H2AFY (chromatin structure). A 4-fold decrease in the ability of the cancer cells to form colonies was found when the LINC01116 gene was disrupted through a CRISPR/CAS9 method, further supporting an oncogenic function for LINC01116 in PC-3 cells.. We identified a novel isoform of LINC01116 and bioinformatically investigated the possibility that LINC01116 could interact with target genes via ssRNA:dsDNA triplexes. Our data reveal that chemicals from the diet can influence the expression of functionally important lncRNAs, and suggest a novel mechanism by which SFN may prevent and suppress prostate cancer.
Keywords: chemoprevention, long non-coding RNA, LINC01116, prostate cancer, sulforaphane, diet-gene interactions
1 Introduction
While it has long been known that thousands of long noncoding RNAs (lncRNAs) are produced in human cells, only recently have their myriad epigenetic regulatory functions come to light. LncRNAs are defined as RNAs longer than 200 nucleotides that do not encode proteins [1]. Like mRNAs, lncRNAs can be regulated by transcription factors, undergo post-transcriptional processing, form complex secondary and tertiary structures and exhibit tissue- and development-specific expression [2, 3]. LncRNAs differ from mRNAs in that their primary structures are generally poorly conserved between species and they are expressed at very low to moderate levels, relative to mRNAs [2, 3]. While the molecular functions of most lncRNAs remain largely unknown, there is growing evidence that lncRNAs have functional roles in cell biology and development [4, 5]. Additionally, their dysregulation can contribute to multiple disease processes [1]. Mechanistically, lncRNAs have been shown to modulate gene expression at the level of transcription by associating with DNA and chromatin modifying complexes, thereby mediating alteration of the local epigenetic landscape [1, 6]. LncRNAs have also been shown to act as decoys for transcription factors [7], regulate protein activity [2, 8], and function as structural components in subcellular structures like nuclear paraspeckles [3, 9]. A growing list of lncRNAs are being identified as dysregulated in cancer cells, and several studies have shown that lncRNAs can contribute to the etiology of diseases like prostate cancer [10–12].
Among men, prostate cancer is the second most frequently diagnosed cancer globally, and is the second leading cause of cancer-related deaths in the United States [13, 14]. This high prevalence translates to a significant societal and financial burden and highlights the great need to prevent and suppress prostate cancer formation [14]. Importantly, previous studies have shown that increased consumption of cruciferous vegetables reduces the risk of developing prostate cancer [15, 16]. When cruciferous vegetables, such as broccoli and broccoli sprouts, are chopped or chewed, glucoraphanin interacts with the enzyme myrosinase, producing the phytochemical sulforaphane (SFN) [16]. SFN has been shown to have both chemopreventive and cancer suppressive properties in carcinogen-induced and genetic models of cancer including a model of prostate cancer [16, 17]. Importantly, SFN also exhibits cancer-specific cytotoxic and anti-proliferative effects in human prostate cells [18–20]. Little is known about the interaction of lncRNAs and dietary factors. We hypothesized that lncRNAs could be a chemopreventive target of the phytochemical SFN. Using RNA-sequencing (RNA-seq) we determined genome-wide expression levels for lncRNAs, and investigated whether these changes have functional importance.
2 Methods
2.1 Culturing and Treatment of Cells
Normal prostate epithelium cells (PREC) were obtained from Lonza (Basel, Switzerland) and cultured as recommended (Lonza) [18, 21]. Androgen-dependent (LNCaP) and androgen-independent (PC-3) prostate cancer cells were obtained from American Type Tissue Collection (Manassas, VA) and cultured as previously described [18]. All cells were confirmed to be mycoplasma free and have the expected allelic composition (Idexx Radil, Columbia, MO). Each cell line was treated at 50–70% confluent with SFN (LKT Laboratories, St. Paul, MN) dissolved in dimethylsulfoxide (DMSO) at 5 or 15 μM concentration, or with an equal volume of DMSO (vehicle control) that was 0.03% of the total media volume. Adherent cells were harvested at 6 and 24 hours post-treatment. The 15 μM concentration of SFN was chosen because ~15 μM of total SFN metabolites has been observed in the plasma of mice orally dosed with 20 μmoles of SFN, and prostates have one of the highest accumulations of SFN among tissues examined [22]. This concentration is achievable from consuming 1 to 2 cups (106 – 212 g) of broccoli sprouts. The three different prostate cell lines were used because they represent different states of prostate cancer progression and based on previous reports showing SFN-induced HDAC inhibition, cell cycle arrest (G1 arrest in LNCaP cells, G2/M in PC-3 cells), and apoptosis in prostate cancer cells, but not in normal prostate epithelial cells [18–20].
2.2 RNA Sequencing and LncRNA Identification
Cells were treated in triplicate, and total RNA was isolated and subsequently sequenced on an Illumina HiSeq 2000 machine at the Center for Genome Research and Biocomputing core facility at Oregon State University as previously described [21]. Sequencing data were processed in two separate pipelines. The Tuxedo Suite was used in the first pipeline [23]; RNA-seq reads were aligned to the human Illumina iGenome (GRCh37/hg19) using TopHat version v2.0.14 with the hg19 iGenome annotation guide and default parameters plus --no-coverage-search and –no-novel-juncs. TopHat was also run permitting novel junctions for purposes of novel transcript assembly. Cufflinks version v2.2.1 was used to assemble transcript models and compare these to the reference hg19 iGenome annotation. In the second pipeline LncRNA annotations were downloaded from Ensembl.org using Biomart. Differentially expressed lncRNAs were identified in the GENE-counter pipeline based on the criterion of being significantly altered in the experimental versus control condition (p < 0.05) according to the NBPSeq software package using default parameters [24]. Differentially expressed genes were identified based on the criteria of being significantly differentially expressed and exceeding a threshold of 20 normalized reads in at least one treatment group. Log2 fold changes that approached infinity were reassigned as 20. The raw RNA-seq reads and differential expression data have been previously deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE48812 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48812) [21].
2.3 Quantitative Real-Time PCR
Total RNA was collected from indicated cells using a standard Trizol extraction method (Life Technologies). cDNA was synthesized using 1 μg of total RNA and SuperScript III First-Strand Synthesis SuperMix (Life Technologies). Real time PCR was done using primers that amplify all known transcript isoforms of each gene as a single product of expected size, between 140 and 300 bp, with the exception of LINC01116 isoforms where the primers where designed to only amplify the designated isoform. Primer sequences were as indicated (Supplemental Table 1, all supplemental material is attached in a zip file). Reactions were performed using Fast SYBR Green Mastermix (Life Technologies) on 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). PCR conditions were programmed as follows: 95°C for 20 s, followed by 40 cycles of denaturing at 95°C for 1 s, annealing and extension at 58°C for 20 s, followed by a dissociation curve at 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. A dilution series of 103, 104, 105, 106, and 107 copies of template DNA served as internal standard for quantification [25]. Data represent the copy number of the gene of interest normalized to the copy number of the indicated housekeeping gene. GAPDH was used as a housekeeping gene in SFN experiments as previously described [21]. For knockdown experiments β-actin was used as housekeeping due to consistent expression in all knockdown conditions.
2.4 Guilt By Association
In order to form hypotheses about the function of the identified lncRNAs, we performed a correlation-based, guilt-by-association analysis to identify significantly associated pathways. Each of the putative SFN-repressed and cancer-associated lncRNAs was investigated by identifying significantly correlated and anti-correlated genes. Each set of significantly correlated genes was compared to gene subsets annotated in Reactome pathway terms downloaded from mSigDB (CP:Reactome). Significant overlap between correlated gene sets and Reactome pathways were identified by computing a hypergeometric p-value for each set of correlated genes followed by a Benjamini-Hochberg procedure to compute false discovery rate (FDR). We imposed an FDR threshold of 0.05. Significantly associated pathways are represented in a heat map where 6 out of the 7 lncRNAs of interest showed significant association with pathways that are plausibly relevant to cancer.
2.5 Manipulation of LINC01116 Expression
Growing PC-3 cells were transfected with siRNA or plasmids with the TransIT-X2 reagent (Mirus, Madison, WI) following the manufacturer’s protocol. For LINC01116 knockdown custom Silencer Select siRNA for LINC01116 was designed, synthesized (Ambion), and transfected into cells using the standard manufactures protocol. Knockdown of LINC01116 RNA was validated 48 h post transfection. The effect of knockdown on PC-3 cell death and proliferation was quantified using trypan blue and a hemocytometer as previous described [26]. The CRISPR/CAS9 system was utilized to knock out or mutate the LINC01116 gene. Cells were transfected with a plasmid (DNA 2.0, Menlo, CA) that encoded GFP, Nickase Cas9 protein, and two guide RNAs that targeted DNA mutations specifically to the LINC01116 promoter region just upstream of isoform 2 and 3, and in the first exon of isoform 1. Alternatively, negative control cells were transfected with the same plasmid except the guide RNAs targeted DNA mutations to a region of DNA on chromosome 13 where no genes are present. Twenty-four hours following transfection, cell sorting was performed on a MoFlo XDP cytometer (Beckman Coulter, Inc.). Fluorescence was excited using a blue (488 nm) laser; cells were identified in a bivariate scatter plot gated to exclude debris. Cells exhibiting the highest fluorescence intensity in the green channel (510–550 nm) were sorted, using purify mode and a drop envelope of 1, either into 12 × 75 mm culture tubes or 96-well plates on a CyCLONE robotic collector. Cells were verified to be GFP positive and immediately reseeded at 10,000 cells per 25 cm2 flask and cultured for 15 days to determine clonogenic survival where colonies of more than 60 cells were counted. Cell lines were generated through standard culturing techniques from single cells contained in the 96 well plates. Negative control and possible LINC01116 mutant cell lines were assessed for expression of LINC01116 RNA (as described above). PCR was also performed using standard protocols and 5′-GTTCAAGTGCGTCCGGGTTT-3′ (forward) and 5′-CGGACTTCTTTTCCAGGCGG-3′ (reverse) primers, which flank the DNA sequence around the location where CAS9 protein acted in the LINC01116 gene.
2.6 LINC01116 Correlation Analysis
We used custom Perl and R scripts to compute Pearson’s correlation values between each LINC01116 isoform and all transcripts in the human transcriptome, or between the LINC01116 gene and all genes in the genome, using FPKM expression values reported by Cuffdiff version 2.2.1 [27]. P-values were calculated from the Z-score of the Pearson’s correlation values using a two-tailed test. The Benjamini-Hochberg procedure was used to compute q-values and control the false discovery rate (FDR). Positive or negative correlations between genes/transcripts and LINC01116 or one of its isoforms at FDR 0.05 were considered to be significant.
2.7 RNA Structure Prediction
Minimum free energy secondary structures were predicted using RNAfold version 2.1.9 with no isolated base pairs, and with dangling energies on both sides of a helix in any case (-d 1). Base pair probabilities were color coded using relplot.pl which is part of the Vienna RNA package (version 2.1.9) [28]
2.8 Identification of Putative LINC01116 Regulatory Targets
To identify putative regulatory targets for LINC01116 transcripts, we assessed capacity for ssRNA:dsDNA triplex formation between each LINC01116 transcript and the promoters of all anti-correlated transcripts, where promoters were defined as 2 or 4 kb of DNA centered at the transcription start site. Potential for triplex formation was computed with Triplexator using the option –fr off with a permitted error rate of 8 percent and a minimum length requirement of 14 bases [29]. Although we ran without filtering repeats, none of the identified TFOs were repetitive.
3 Results
3.1 Effects of SFN treatment on lncRNA expression in human prostate cell lines
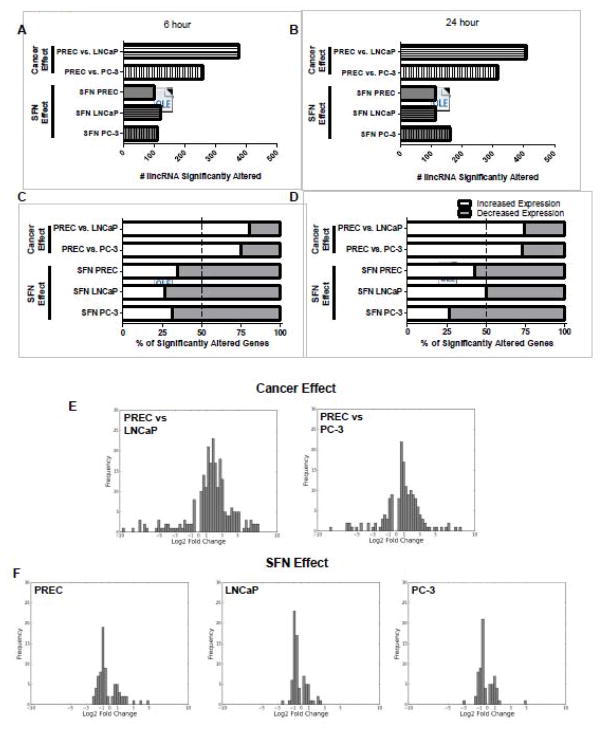
In order to investigate lncRNAs in the context of prostate cancer and SFN we mined previously published RNA-seq datasets [21]. We first used GENE-counter to identify putative lncRNAs that are differentially expressed, as a factor of cell type (normal versus prostate cancer). Pairwise comparisons were made between normal and cancerous cell lines treated with the vehicle control (DMSO). Between 257 and 407 lncRNAs were significantly differentially expressed in androgen-dependent LNCaP and androgen-independent PC-3 cells as compared to normal PREC cells (designated here and in the text as cancer effect, Fig. 1A–B and Supplemental Table 2, all supplemental material is attached in a zip file). Secondly, to determine the extent to which expression of lncRNAs is altered by SFN treatment in prostate cells, pairwise comparisons were also made within each cell line and time point between the SFN and vehicle control treatments. An average of 119 lncRNAs were significantly altered by SFN treatment in each prostate cell line at each time point (Fig. 1A and 1B, and Supplemental Table 3).
Figure 1. Genome-wide effects of cancer and SFN on lncRNA expression.
The cancer effect was determined by comparing the lncRNA levels found in LNCaP and PC-3 cells compared to normal PREC cells. The SFN effect was determined in each of the three cell lines by direct comparison of lncRNA levels of samples treated with 15 μM SFN compared to their respective vehicle control at the same time point. A–B) Bars represent the number of lncRNAs that were significantly altered by prostate cancer development or by SFN treatment at the 6 or 24-hour time points. C–D) Data represent the percentage of lncRNAs that had a significant increase (white), or decrease in expression (grey) at the C) 6 h and D) 24 h time points. E–F) Frequency plots of the amplitude of change in lncRNA expression, expressed as log2-fold, which was significantly altered under the indicated comparisons. Data are from the 6-hour time point although similar log2 fold distributions were also observed at the 24-hour time point (data not shown).
Comparisons of transcript expression in prostate cancer cells versus normal cells showed that the majority of differentially expressed lncRNAs (75.8%) were significantly upregulated in LNCaP and PC-3 prostate cancer cells as compared to normal cells (Fig. 1C–D). In contrast, the majority (64.5%) of lncRNAs significantly differentially expressed after SFN treatment decreased in expression. This trend was consistently observed, with the exception of 24h LNCaP cells, and shows that the majority of lncRNAs altered by SFN treatment undergo a decrease in gene expression (Fig 1C–D). A comparison of the cancer effect and SFN effect frequency plots shows that the extent of changes in lncRNA expression (i.e. log2 fold change) was more modest between SFN treatment and control than between normal and cancer cell lines (Fig. 1E–F). Taken together, these results show that lncRNA expression is altered in prostate cancer cells and that lncRNA expression can be altered by SFN treatment.
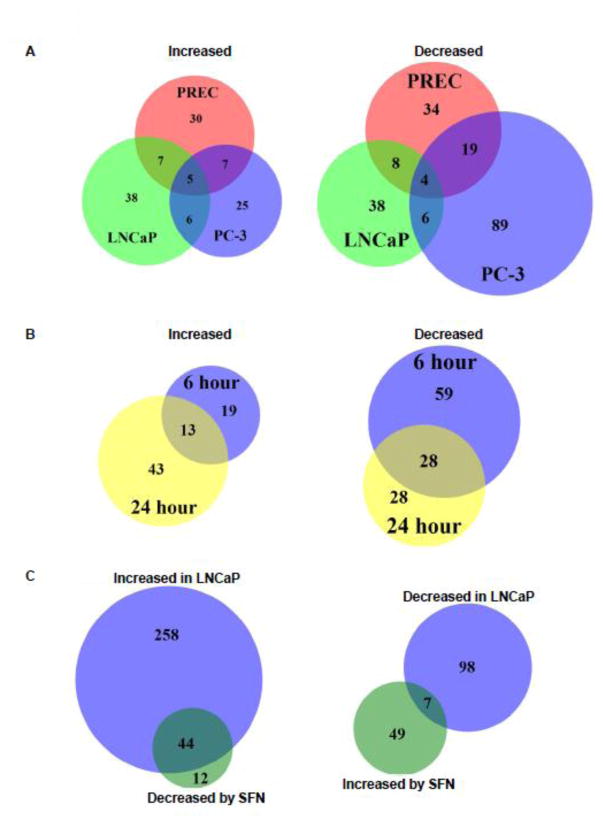
Venn diagrams were generated to evaluate how many lncRNAs were altered by SFN treatment in all three cell types. The majority of lncRNAs whose expression was altered with SFN were specific to each cell line (Figure 2A). This differential lncRNA expression was expected because we have observed a differential cellular response to SFN treatment in the same prostate cell lines [18, 21]. The effect of SFN on lncRNA expression is also dynamic over time, as the proportion of lncRNAs with SFN-induced changes in expression at both time points only exceeded 50% for one of the six conditions tested (Figure 2B and data not shown).
Figure 2. Comparison of SFN-induced changes in lncRNA expression in normal prostate epithelial cells and prostate cancer cells.
Venn diagrams showing the number and overlap of A) the lncRNAs whose expression levels were significantly altered in PREC, LNCaP, and PC-3 cells with 24 hours SFN treatment. B) Venn diagrams showing the number and overlap of lncRNAs in LNCaP cells that were significantly increased or decreased with either 6 or 24-hour treatment of SFN. C) lncRNAs that were either significantly increased or decreased in expression in LNCaP cells, relative to normal cells, and significantly altered by SFN in the opposing direction in LNCaP cells at the 24-hour time point. A–C) Data are from a single condition (ie. time point or cell line) but similar plots were observed at the A) 6 h time point, B) with PREC and PC-3 cell lines, and C) PC-3 cells (data not shown).
To determine which lncRNAs may contribute to chemopreventive effects of SFN, we next identified the set of lncRNAs that were significantly upregulated in cancer cells compared to control PRECs, and significantly downregulated by SFN treatment in the same cancer cell type (Figure 2C). We also identified the set of lncRNAs that were downregulated in cancer cells compared to PRECs, and increased by SFN in the same cancer cell type. While there was significant overlap between these lncRNA sets, SFN was more likely to attenuate a cancer effect if the lncRNA was increased in the cancer cell line and decreased by SFN treatment (Figure 2C). The lncRNAs with prostate-cancer-associated expression changes that were also mitigated by SFN treatment were considered for further analysis. A subset of these lincRNAs were verified by qPCR to independently verify the SFN effect on lncRNA expression (Figure 3) and their gene expression in the RNA-seq data set was graphed (Supplemental Figure 1). Positive controls Malat1 and Neat1 were upregulated in cancer cells relative to PRECs. The abundance and significant changes in expression of our short list of lncRNAs of interest are shown (Supplemental Figure 1).
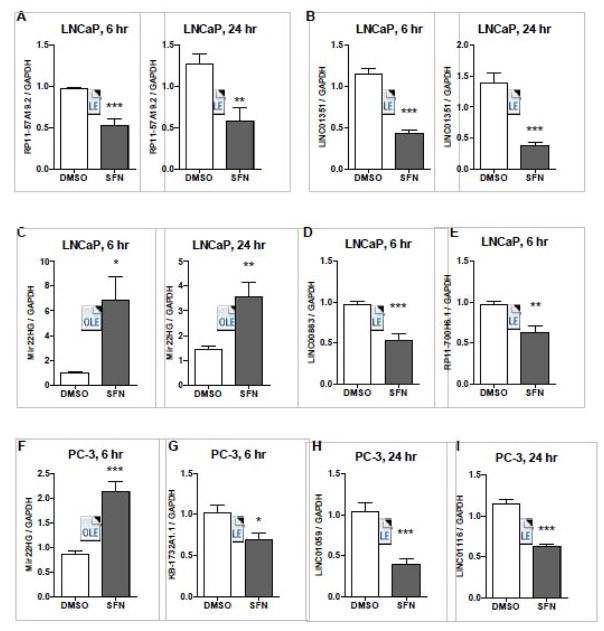
Figure 3. Validation of SFN’s capacity to normalize the expression of lncRNAs that were differentially expressed in prostate cancer cells.
LNCaP and PC-3 cells were treated with 15 μM SFN or vehicle control (DMSO) for 6 or 24 hours, and qPCR was performed. Bars are representative of the mean expression levels of the indicated lncRNA normalized to GAPDH and expressed relative to the mean expression levels in cells treated with DMSO. Samples were collected in at least triplicate, in two independent experiments (n=6–7) and stars indicates significant differences between the control and treatment groups as calculated by an unpaired, two-tailed t test where * p < 0.05, ** p < 0.01, *** p < 0.001.
Among all the lncRNAs and conditions examined by qPCR, ~86% of the tests confirmed results identified by RNA-seq (Figure 3). RP11-57A19.2 was overexpressed in LNCaP cells and its expression decreased by 6 and 24-hour after treatment with SFN (Figure 3A). LINC01351 (also called RP11-547I7.2) was overexpressed in LNCaP cells and decreased with SFN treatment at both 6 and 24-hour time points (Figure 3B). LINC00883 (also called RP11-446H18.3) was overexpressed in LNCaP cells and decreased with SFN treatment in PREC, LNCaP and PC-3 cells (Supplemental Figure 1 and Figure 3D). RP11-700H6.1 was overexpressed in LNCaP cells and decreased by SFN at the 6h time point (Figure 3E). MIR22HG (also called C17orf91) was suppressed in both LNCaP and PC-3 cells, and increased by SFN treatment in PREC, LNCaP and PC-3 cells (Supplemental Figure 1 and Figure 3C and F). KB-1732A1.1 was overexpressed in PC-3 cells and decreased by SFN in PrEc and PC-3 cells (Supplemental figure 1 and Figure 3G). LINC01059 (also called AP000783.2) was overexpressed in PC-3 cells and decreased by SFN treatment in PC-3 cells (Supplemental Figure 1 and Figure 3H). LINC01116 (also called AC017048.3) was overexpressed in PC-3 cells and decreased in PREC and PC-3 cells with SFN (Supplemental Figure 1 and Figure 3I). We also confirmed that expression of a subset of lncRNAs was significantly altered with a lower concentration of SFN (5 μM SFN, Supplemental Figure 2). With the lower dose in SFN the fold change in lncRNA expression was smaller, but the SFN-induced change in expression was significantly altered in at least one time point examined, and was altered in the same direction (up- or down-regulated) as was found with 15 μM SFN.
To investigate possible links between these lncRNAs and cancer, we performed guilt-by-association analysis; after identifying the genes strongly correlated or anticorrelated with our lncRNAs of interest based on their expression in the RNA-seq dataset, we then cross-referenced these gene lists with Reactome pathways. This analysis revealed that most of the lncRNAs on our short list were associated with metabolism, gene expression, disease, signal transduction, and the immune system (Supplemental Figure 3). In addition, some of the lncRNAs were associated with cell cycle, developmental biology, extracellular matrix organization and cell-cell communication, which are highly relevant to the etiology of cancer.
3.2 Role of LINC01116 in an aggressive prostate cancer cell line
In order to address the functional importance of the lncRNAs, we manipulated the expression of the lncRNAs in cancer cells. Suppression of LINC01116 produced the strongest phenotype (Figure 4A). ENCODE also showed that LINC01116 has high levels of histone 3 lysine 27 acetylation (H3K27Ac) in its first exon and transcription factors previously implicated in cancer (E2F1, RAD21, EGR1 and MYC) have been found near the transcription start; these are additional lines of evidence that LINC01116 is transcribed and may be regulated by transcription factors that are often dysregulated in cancer. For these reasons we pursued LINC01116 to determine if it is functionally significant in PC-3 cells. Using siRNA and transient transfections we knocked down LINC01116 expression by 60–70% (Figure 4B). This knockdown did not affect the number of cells that stained positive for trypan blue, a dye that indicates cell death, but it significantly decreased proliferation of PC-3 cells (Figure 4C and D). To independently verify this pro-proliferative role of LINC01116, we attempted to knockout LINC01116 in PC-3 cells using the CRISPR/CAS9 system. Cells were transfected with a plasmid that encoded GFP, Nickase Cas9 protein, and two guide RNAs that together were designed to cause base substitutions to the promoter and first exon of LINC01116 to disrupt gene expression. Over three experiments, we tried to generate LINC01116 knockout cell lines from over 4,320 GFP positive PC-3 cells. Survival and proliferation were severely impaired in these cells, and no surviving cell lines had a complete LINC01116 knockout. One cell line was generated that contained a heterozygous mutation for LINC01116, which consisted of a 23 base pair insertion, confirming that the CRISPR system mutated the DNA at the LINC01116 loci (Figure 4E). As a positive control we generated an Nrf2 mutant using the same technology (Supplemental Figure 4). Since we could not create a LINC01116 knockout cell line, we next sought to determine if mutation of LINC01116 in PC-3 cells would alter the potential of the cancer cells to form colonies. PC-3 cells transfected with the plasmid engineered to cause disruption of LINC01116 expression caused a significant 5-fold reduction in clonogenic survival (Figure 4F).
Figure 4. LINC01116 enhances proliferation and clonogenic survival in PC-3 cells.
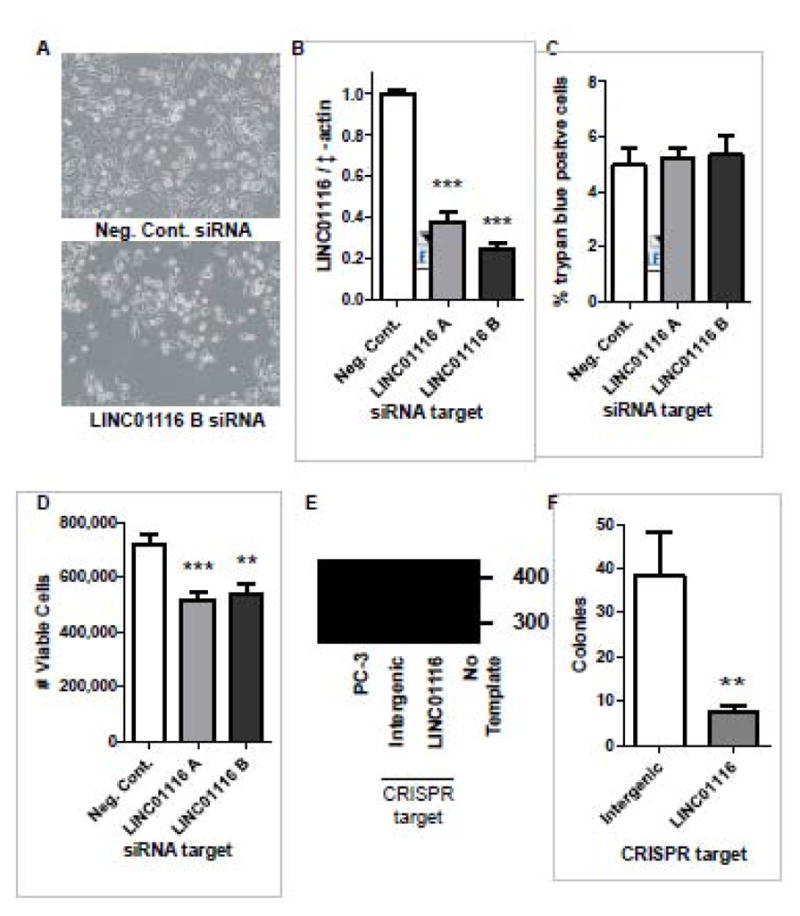
A–D) PC-3 prostate cancer cells were transfected with negative control siRNA or two different siRNAs to knock down LINC01116, and assayed 48 hours post transfection for (A) cellular appearance (10x magnification), (B) LINC01116 expression levels, (C) number of cells staining positive for trypan blue, and (D) number of viable cells as an indicator of cell proliferation. B–D) Data represent an average of at least 10 replicates that were obtained from at least 3 independent experiments. E–F) PC-3 cells were transfected with GFP labeled CRISPR/CAS9 plasmids designed to create mutations in the LINC01116 gene (in the LINC01116 promoter/first exon), or a negative control plasmid that was designed to mutate DNA in a region with no genes (Intergenic). GFP positive cells were sorted and (E) cell lines were generated from single cells or (D) clonogenic survival was completed. E) Representative DNA gel obtained from a PCR with DNA isolated from indicated cell lines and primers that flanked the location where the CAS9 nickase enzyme was directed to create mutations. Sanger sequencing confirmed the bands are PCR products from LINC01116 and the larger band in the LINC01116 well was LINC01116 sequence with a 23 bp insertion. B–D, F) Bars are indicative of the mean ± SEM and **, and *** indicates significant differences between the control and treatment group as calculated by an unpaired, two-tailed t test, where p < 0.01 and p <0.001 respectively.
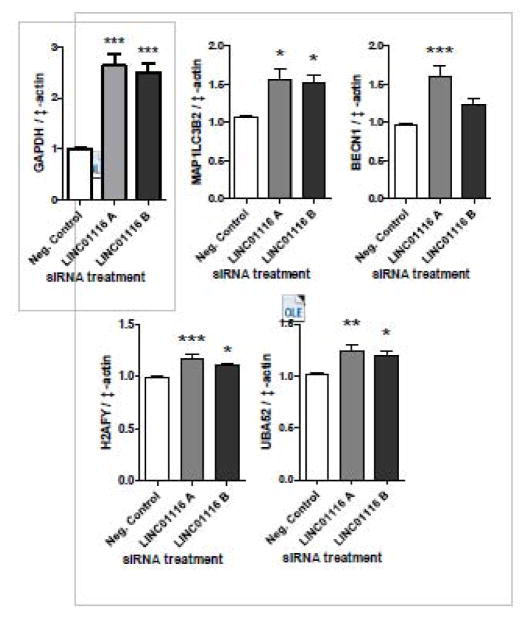
LncRNAs have been shown to regulate expression of target genes through multiple mechanisms. To learn more about the possible functions of LINC01116, we used our RNA-seq data to identify genes whose expression patterns anti-correlated with the expression patterns of LINC01116 (Supplemental Table 4). These gene lists contained Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Microtubule-Associated Protein 1 Light Chain 3 Beta 2 (MAP1LC3B2), Beclin 1, Autophagy Related (BECN1), H2A Histone Family, Member Y (H2AFY), and Ubiquitin A-52 Residue Ribosomal Protein Fusion Product 1 (UBA52) which have been previously linked to glycolysis, autophagy, regulation of chromatin structure and ubiquitination respectively. To determine whether LINC01116 levels may be causally related to the expression of these genes, we evaluated their expression in PC-3 cells where LINC01116 was knocked down with siRNA (Figure 4B). Knockdown of LINC01116 consistently resulted in a significant increase in GAPDH, MAP1LC3B2, H2AFY and UBA52 expression at the mRNA level (Figure 5). BECN1 mRNA was also upregulated with LINC01116 suppression but this change was only significant with one of the siRNAs tested.
Figure 5. Knockdown of LINC01116 significantly increases expression of genes that anti-correlate with LINC01116.
PC-3 prostate cancer cells were transfected with negative control siRNA or two different siRNAs to knock down LINC01116 expression. qPCR was performed from RNA collected 48 hours post-transfection. Bars are representative of the mean expression levels of the indicated gene normalized to β-actin and expressed relative to the mean expression levels in cells treated with negative control siRNA. Data represent 6–14 individual samples obtained from at least 2 independent experiments. Stars indicate significant differences between the control and treatment groups as calculated by an unpaired, two-tailed t test where * p < 0.05, ** p < 0.01, *** p < 0.001.
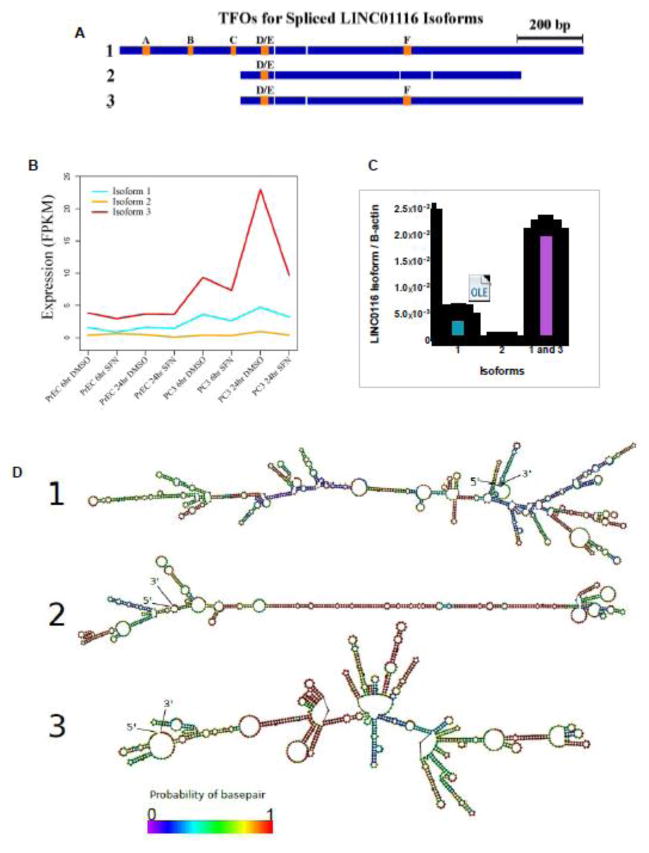
To further characterize LINC01116, we next evaluated its gene structure. The hg19 and hg38 genome annotations list two isoforms for this gene (Figure 6A, Isoforms 1 and 2). Visualization in Gbrowse of the density of mapped reads around LINC01116 indicated the existence of an alternate, unannotated isoform. The predominate abundance of this third isoform was supported by TopHat and Cufflinks analysis in all three cell lines tested (Figure 6A and B). We also independently verified the expression of all three isoforms by qPCR using primers that were specific to only one or two different isoforms (Figure 6B). Isoform 3 of LINC01116 was confirmed to be the most abundant in PC-3 cells, followed by isoform 1 and then isoform 2. We predicted secondary structures for each LINC01116 isoform using RNAfold from the Vienna RNA package (Figure 6D) [28]. Isoform 1 and 3 were both predicted to have some areas with lower probability of base pairing while isoform 2 was predicted to form a long hairpin with a high probability of base pairing.
Figure 6. LINC01116 structure and identification of novel isoform.
A) Schematic of the three spliced LINC01116 isoforms with exons (blue) and predicted triplex forming oligonucleotide (TFO) regions (orange). B) Line graph indicates the mean expression levels of each isoform of LINC01116 in PrEC and PC-3 cells, treated with 15 μM SFN or vehicle control (DMSO), as determined by RNA-Seq. Expression is calculated as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). C) qPCR results obtained with primers specific to the indicated isoform or isoforms, and RNA from PC-3 cells. Bars are representative of the mean expression levels of the indicated isoforms, relative to a standard curve of known quantity, and normalized to β-actin (n=6). D) Predicted secondary structures of each LINC01116 isoform.
Several lncRNAs have previously been reported to regulate gene expression via a tethering mechanism, wherein the lncRNA interacts with the promoter DNA of a target gene through an ssRNA:dsDNA triple helix (triplex) and physically recruits histone modifying enzymes such as Polycomb Repressive Complex 2 to mediate chromatin remodeling. Such examples include MEG3 [30], Fendrr [31], Khps1 [32], and PARTICLE [33]. To explore a possible mechanism for LINC01116, we used the program Triplexator to evaluate the potential of the lncRNA in forming a triplex. Six distinct triplex-forming oligonucleotide regions (TFOs) were predicted among the LINC01116 isoforms (Figure 6A, orange squares, and Table 1). Isoform 1 had the highest number of predicted TFOs, followed by isoform 3 and then isoform 2. These TFOs ranged from 15 to 29 nucleotides in length.
Table 1.
| TFO | TFO Location | Length | Motif | Error Rate | Guanine Rate | Sequence | Isoforms | |
|---|---|---|---|---|---|---|---|---|
| TFOA | 2:177502567..177502592 | - | 27 | Y | 0.077 | 0.65 | CTCTCTCTCCTGCCCATCCCCTCCCC | 1 |
| TFOB | 2:177502434..177502447 | - | 15 | Y | 0.071 | 0.43 | TTCCCGTTTTTCCC | 1 |
| TFOC | 2:177502307..177502321 | - | 16 | R | 0.067 | 0.6 | GAGGGGAAT GGGGAA | 1 |
| TFOD | 2:177502210..177502223 - | 15 | Y | 0.071 | 0.5 | TCTTGCCTCCCCTT | 1,2,3 | |
| TFOE | 2:177502205..177502218 | - | 15 | Y | 0.071 | 0.71 | CCTCCCCTTACCCC | 1,2,3 |
| TFOF | 2:177494842..177494869 | :- | 29 | R | 0.071 | 0.29 | GGAGAAAGAAAAAAATAAGTGAAAAGGA | 1,3 |
To gain insights into the possible biological role of LINC01116, we next sought to computationally identify putative direct regulatory targets. Ideal candidates were defined as genes whose expression levels significantly anti-correlated with LINC01116 and having at least one transcript capable of forming a triplex with LINC01116 isoform 1 or 3 within 1 kb of the transcript’s transcription start site (isoform 2 was excluded from this analysis due to its low abundance). Using Triplexator, 189 such transcripts were found to possess putative triplex target sites (TTSs) in their promoters corresponding to a predicted TFO from LINC01116 isoform 1 (Supplemental Table 5), while only 3 anti-correlated genes had TTSs corresponding to an isoform 3 TFO (Supplemental Table 6). For this reason, we expanded this search up to +/− 2 kb from the transcription start site. In this case 21 different genes are predicted targets of LINC01116 isoform 3 (Supplemental Table 7). All together, these gene lists contained several genes relevant to cancer, including CNBP, MAP2K3, ANAPC16, RAD50. They also contained BECN1 and GAPDH, which we found in this study to be upregulated after LINC01116 knockdown (Figure 5).
4 Discussion
The field of lncRNAs is evolving, with a growing number of lncRNAs being identified, while their functions remain largely enigmatic [34]. In particular, very little is known about the relationship between lncRNAs and dietary factors. Here we show that lncRNA expression can be altered by treatment with SFN, a phytochemical derived from the diet. When we examined SFN’s effect on lncRNAs we found that the majority of the lncRNAs that were significantly altered with SFN were decreased in expression. SFN’s effect on lncRNAs was generally cell-specific and dynamic over time; this trend was also observed for mRNAs differentially expressed in the same prostate cells after SFN treatment [21]. It is important to note that we expected SFN treatment to produce distinct lncRNA profiles in each of the cell types because they represent various states of cancer progression, and the SFN treatment produces different cellular endpoints ranging from no visible change in normal cells, to apoptosis in advanced prostate cancer cells [18, 35, 36]. Because the cellular response to SFN is diverse, the differences in SFN-induced changes in lncRNA expression between the cell types are interesting and may contribute to the phenotypes observed. Future work could determine whether the selective induction of a pro-apoptotic response in PC-3 cells is dependent on any of the lncRNAs that are only significantly changed with SFN in this cell type.
There are a few examples of dietary impacts on lncRNA expression. Several reports explored how different dietary patterns alter the expression of the lncRNA H19, which regulates growth and cell proliferation [37–39]. Also, Li et. al. showed changes in the lncRNA expression profiles of the kidneys in the progeny of mothers were fed a low protein diet [40]. Five lncRNAs were shown to be differentially expressed in a rat model of hypertension that involved a high salt diet [41], andan independent study examined the expression of the lncRNA sONE and eNOS in relation to high salt diets and hypertension [42]. Knockout of the gene that encodes the lncRNA SRA in mice conferred resistance to high fat diet-induced obesity [43]. While this result was complicated by the fact that SRA also codes for a protein, ithighlights the exciting potential of lncRNAs in diet-based research. Taken together, this literature and our own study begin to paint a picture of the important and previously unappreciated role of lncRNAs in the body’s response to diet.
Several studies have demonstrated that lncRNAs are involved in the development and progression of multiple types of cancers, including prostate cancer [10–12]. These discoveries illustrate that lncRNAs can play important roles in cancer development and may be useful targets for cancer prevention, detection, and treatment. While it was not the focus of our study, we did confirm that hundreds of lncRNAs were differentially expressed in prostate cancer cells compared to normal prostate cells. Given SFN’s cancer chemopreventive properties, one of our goals was to understand whether lncRNAs dysregulated in cancer could be good targets for cancer prevention or suppression in response to SFN. In exploring this possibility, we identified eight lncRNAs for which SFN treatment mitigated the prostate cancer associated dysregulation of expression: RP11-57A19.2, LINC01351, LINC00883, RP11-700H6.1, Mir22HG, KB-1732A1.1, LINC01059, and LINC01116.. While each of the lncRNAs are worthy of continued investigation, it is worth noting that expression of both LINC00883 and MIR22HG was significantly altered by SFN in all three cell types. Furthermore, examination of the EMBLI-EBI expression atlas showed that LINC00883 is expressed in various types of cancers and MIR22HG was significantly downregulated in several cancer studies including prostate and lung cancer [44]. Interestingly, NRF2 has been associated with MIR22HG in a chromatin immunoprecipitation study and thus MIR22HG is likely regulated by this transcription factor [45]. NRF2 is a well-known transcription factor that has long been implicated in SFN’s chemopreventive properties [46, 47]. The expression atlas also revealed that KB-1732A1.1 was expressed in various types of cancers but little is known about its differential expression compared to normal cells, and little is known about LINC01059 [44]. In comparison, RP11-700H6.1 was expressed in prostate, breast, stomach, and lung cancers, as well as chronic lymphocytic leukemia, and RP11-57A19.2 was detected in several different types of cancer cells including prostate adenocarcinoma, lung carcinoma, and colon carcinoma [44]. LINC01351 and LINC01116 are also of particular interest because they were overexpressed in our prostate cancer cells relative to normal cells, and mining of the database revealed that LINC01351 was significantly increased in pituitary cancer and adenocarcinomas of the lung. LINC01116 was upregulated in several previous cancer studies including glioblastomas, and lung, colon and prostate cancer [44].
Because suppression of LINC01116 with siRNA produced the strongest phenotype in our cancer cell lines, we focused on this gene for further experiments investigating its functional importance in prostate cancer cells. We showed that LINC01116 promotes the growth of the aggressive prostate cancer cell line PC-3. Our multiple attempts to generate a LINC01116 knockout cell line were unsuccessful; however, this negative result suggests that LINC01116 may be essential for PC-3 cell survival. Knockdown of LINC01116 expression was associated with the upregulation of genes that significantly anti-correlated with it in prostate cells, including GAPDH, MAP1LC3B2, and H2AFY. This result supports a role for the lncRNA in prostate cancer wherein it contributes to suppression of key genes known to regulate glycolysis, autophagy, and chromatin structure [48–50]. However, it is important to note that knockdown of LINC01116 did not phenocopy SFN exposure; for example, its knockdown produced a strong increase in GAPDH expression, which we did not observe with SFN treatment.
Here we describe a new and abundant isoform for LINC01116. We also show isoform 2 is expressed at a much lower level than the othersand has a long and stable hairpin structure, suggesting the possibility of dsRNA degrading RNase III enzymes as a cause for its reduced expression [51]. It is possible that LINC01116 functions to repress target gene expression, and that this may occur via a mechanism involving formation of ssRNA:dsDNA triplexes between a LINC01116 TFO and gene promoter DNA. This potential epigenetic mechanism would be in keeping with previously identified effects of SFN on epigenetics including altered histone acetylation and DNA methylation [18, 52–57]. While extensive future experiments are needed to determine whether any of our predicted target genes are directly regulated by LINC01116 in vivo, GAPDH and BECN1 are promising targets because they are significantly anti-correlated with LINC01116, their promoters possess triplex target sites corresponding to a putative LINC01116 TFO, and they are upregulated when LINC01116 is knocked down by siRNA.
In conclusion, we established that the lncRNA LINC01116 is upregulated in a human prostate cancer cell line, is decreased by SFN treatment, and promotes cell proliferation in a human cancer cell line. We also identified a novel isoform of LINC01116 and suggested a mechanism of action. Finally, we presented data that support SFN as a promising dietary anti-cancer agent and indicate that it may exert its chemopreventive effects through multiple mechanisms, including regulation of lncRNA expression. More broadly, our data reinforce the idea that lncRNAs are an exciting new avenue for chemoprevention research, and chemicals derived from diet can alter their expression.
Supplementary Material
Acknowledgments
Funding sources: Work conducted in the authors’ laboratory is supported by NIH grants CA90890, CA65525, CA122906, CA122959, CA80176, R01GM104977, and by NIEHS Center grant P30 ES00210 and the OSU general research fund.
We thank Rong Wang, Elizabeth I. Sokolowski, Jason Cumbie, Mark Dasenko, Christopher M. Sullivan, and Matthew Peterson for technical assistance and helpful conversations.
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallory AC, Shkumatava A. LncRNAs in vertebrates: advances and challenges. Biochimie. 2015;117:3–14. doi: 10.1016/j.biochi.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–11. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff LA, Groff AF, Sauvageau M, Trayes-Gibson Z, Sanchez-Gomez DB, Morse M, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2015;112:6855–62. doi: 10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 9.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss M, Plass C, Gerhauser C. Role of lncRNAs in prostate cancer development and progression. Biol Chem. 2014;395:1275–90. doi: 10.1515/hsz-2014-0201. [DOI] [PubMed] [Google Scholar]
- 11.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Ramalho-Carvalho J, Fromm B, Henrique R, Jeronimo C. Deciphering the function of non-coding RNAs in prostate cancer. Cancer Metastasis Rev. 2016;35:235–62. doi: 10.1007/s10555-016-9628-y. [DOI] [PubMed] [Google Scholar]
- 13.International Agency for Research on Cancer. GLOBOCAN Cancer Fact Sheet: Prostate Cancer. 2010. [Google Scholar]
- 14.American Cancer Society. Cancer Facts & Figures 2012. American Cancer Society Press; Atlanta: 2012. [Google Scholar]
- 15.Liu B, Mao Q, Cao M, Xie L. Cruciferous vegetables intake and risk of prostate cancer: a meta-analysis. Int J Urol. 2012;19:134–41. doi: 10.1111/j.1442-2042.2011.02906.x. [DOI] [PubMed] [Google Scholar]
- 16.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmaco Res. 2007;55:224–36. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SV, Warin R, Xiao D, Powolny AA, Stan SD, Arlotti JA, et al. Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res. 2009;69:2117–25. doi: 10.1158/0008-5472.CAN-08-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol Nutr Food Res. 2011;55:999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27:811–9. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–74. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 21.Beaver LM, Buchanan A, Sokolowski EI, Riscoe AN, Wong CP, Chang JH, et al. Transcriptome analysis reveals a dynamic and differential transcriptional response to sulforaphane in normal and prostate cancer cells and suggests a role for Sp1 in chemoprevention. Mol Nutr Food Res. 2014;58:2001–13. doi: 10.1002/mnfr.201400269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke JD, Hsu A, Williams DE, Dashwood RH, Stevens JF, Yamamoto M, et al. Metabolism and tissue distribution of sulforaphane in Nrf2 knockout and wild-type mice. Pharm Res. 2011;28:3171–9. doi: 10.1007/s11095-011-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cumbie JS, Kimbrel JA, Di Y, Schafer DW, Wilhelm LJ, Fox SE, et al. GENE-counter: a computational pipeline for the analysis of RNA-Seq data for gene expression differences. PLoS One. 2011;6:e25279. doi: 10.1371/journal.pone.0025279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu A, Wong CP, Yu Z, Williams DE, Dashwood RH, Ho E. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin Epigenetics. 2011;3:3. doi: 10.1186/1868-7083-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaver LM, Yu TW, Sokolowski EI, Williams DE, Dashwood RH, Ho E. 3,3′-diindolylmethane, but not indole-3-carbinol, inhibits histone deacetylase activity in prostate cancer cells. Toxicol Appl Pharmacol. 2012;263:345–51. doi: 10.1016/j.taap.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–81. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell. 2015;60:626–36. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 33.O’Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, et al. PARTICLE, a triplex-forming Long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11:474–85. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 34.Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–51. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melchini A, Costa C, Traka M, Miceli N, Mithen R, De Pasquale R, et al. Erucin, a new promising cancer chemopreventive agent from rocket salads, shows anti-proliferative activity on human lung carcinoma A549 cells. Food Chem Toxicol. 2009;47:1430–6. doi: 10.1016/j.fct.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Traka MH, Spinks CA, Doleman JF, Melchini A, Ball RY, Mills RD, et al. The dietary isothiocyanate sulforaphane modulates gene expression and alternative gene splicing in a PTEN null preclinical murine model of prostate cancer. Mol Cancer. 2010;9:189. doi: 10.1186/1476-4598-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y, Zhuo Y, Fang ZF, Che LQ, Wu D. Effect of maternal dietary energy types on placenta nutrient transporter gene expressions and intrauterine fetal growth in rats. Nutrition. 2012;28:1037–43. doi: 10.1016/j.nut.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010;5:619–26. doi: 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- 39.Escrich E, Moral R, Garcia G, Costa I, Sanchez JA, Solanas M. Identification of novel differentially expressed genes by the effect of a high-fat n-6 diet in experimental breast cancer. Mol Carcinog. 2004;40:73–8. doi: 10.1002/mc.20028. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Wang X, Li M, Pan J, Jin M, Wang J, et al. Long non-coding RNA expression profile in the kidneys of male, low birth weight rats exposed to maternal protein restriction at postnatal day 1 and day 10. PLoS One. 2015;10:e0121587. doi: 10.1371/journal.pone.0121587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Li L, Xu H, Liu Y, Yang C, Cowley AW, Jr, et al. Characteristics of long non-coding RNAs in the Brown Norway rat and alterations in the Dahl salt-sensitive rat. Sci Rep. 2014;4:7146. doi: 10.1038/srep07146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Yang X, Lin Y, Suo M, Gong L, Chen J, et al. Anti-hypertensive effect of Lycium barbarum L. with down-regulated expression of renal endothelial lncRNA sONE in a rat model of salt-sensitive hypertension. Int J Clin Exp Pathol. 2015;8:6981–7. [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Sheng L, Miao H, Saunders TL, MacDougald OA, Koenig RJ, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Bio Chem. 2014;289:13000–9. doi: 10.1074/jbc.M114.564658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–52. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–29. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. The AAPS journal. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- 48.Yun J, Mullarky E, Lu C, Bosch KN, Kavalier A, Rivera K, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–6. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun A, Li C, Chen R, Huang Y, Chen Q, Cui X, et al. GSK-3beta controls autophagy by modulating LKB1-AMPK pathway in prostate cancer cells. Prostate. 2016;76:172–83. doi: 10.1002/pros.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Prisco R, Sorrentino S, Leone E, Libonati M. A ribonuclease from human seminal plasma active on double-stranded RNA. Biochim Biophys Acta. 1984;788:356–63. doi: 10.1016/0167-4838(84)90049-9. [DOI] [PubMed] [Google Scholar]
- 52.Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50:213–21. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med. 2007;232:227–34. [PMC free article] [PubMed] [Google Scholar]
- 55.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson GW, Wickramasekara S, Palomera-Sanchez Z, Black C, Maier CS, Williams DE, et al. SUV39H1/H3K9me3 attenuates sulforaphane-induced apoptotic signaling in PC3 prostate cancer cells. Oncogenesis. 2014;3:e131. doi: 10.1038/oncsis.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong CP, Hsu A, Buchanan A, Palomera-Sanchez Z, Beaver LM, Houseman EA, et al. Effects of sulforaphane and 3,3′-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PLoS One. 2014;9:e86787. doi: 10.1371/journal.pone.0086787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.