ABSTRACT
The impact of quorum sensing on polymyxin and azithromycin pharmacodynamics was assessed in Pseudomonas aeruginosa PAO1 and an isogenic rhlR/lasR double knockout. For polymyxin B, greater killing against the rhlR/lasR knockout than against PAO1 was observed at 108 CFU/ml (polymyxin B half-maximal effective concentration [EC50], 5.61 versus 12.5 mg/liter, respectively; P < 0.005). Polymyxin B combined with azithromycin (256 mg/liter) was synergistic against each strain, significantly reducing the respective polymyxin B EC50 compared to those with monotherapy (P < 0.005), and is a promising strategy by which to combat P. aeruginosa.
KEYWORDS: azithromycin, P. aeruginosa, polymyxin B, colistin, quorum sensing, pharmacodynamics, mechanism-based modeling, Pseudomonas aeruginosa, polymyxins
TEXT
Pseudomonas aeruginosa is a leading nosocomial pathogen with infections that are associated with unacceptably high rates of treatment failure, up to 40% (1, 2). The polymyxin antibiotics (colistin [polymyxin E] and polymyxin B) have become important last-line agents against multidrug-resistant P. aeruginosa infections and are being increasingly utilized as salvage therapy (3). However, current dosage regimens for colistin and polymyxin B result in plasma concentrations that are suboptimal in a number of critically ill patients (4–7). Despite the initial susceptibility of many strains, mortality rates for polymyxin monotherapy remain high; however, increasing the dose may promote further resistance amplification against a high bacterial density (8–12).
Azithromycin, a macrolide antibiotic, is often used to treat community-associated respiratory tract infections but has no intrinsic activity against P. aeruginosa. Earlier in vitro time-kill and checkerboard studies suggested that azithromycin may enhance killing in combination with the polymyxins (13, 14). However, the precise time course of bacterial response and the mechanistic basis for this combination remain unknown. Furthermore, the pharmacokinetic-pharmacodynamic relationship of the polymyxin-azithromycin combination has yet to be studied at a range of clinically relevant concentrations, including those in serum (∼0.5 mg/liter) and in neutrophils (>500 mg/liter), where azithromycin is concentrated (15, 16).
Recent studies have also concluded that there is a clinical benefit to using azithromycin in patients with cystic fibrosis or diffuse panbronchiolitis who are chronically infected with P. aeruginosa (17, 18). It has been hypothesized that one mechanism for the salutary effect of azithromycin against P. aeruginosa is through inhibition of quorum sensing, which is a mechanism of bacterial communication that coordinates a multitude of cellular behaviors, such as formation of virulence factors and biofilms (19, 20). Azithromycin interferes with quorum sensing by inhibiting the synthesis of signaling molecules employed through the las and rhl systems (20), preventing intercellular coordination among P. aeruginosa cells. Since quorum sensing functions at a high bacterial density of P. aeruginosa, which has been shown to reduce the activity of polymyxins (21), understanding the interrelationships among quorum sensing, polymyxin-azithromycin pharmacodynamics, and inoculum size is of scientific relevance. Our objectives were to (i) profile the pharmacodynamic activity of colistin and polymyxin B against P. aeruginosa PAO1 and an rhlR/lasR double-knockout strain to determine the impact of quorum sensing on the rate and extent of bacterial killing by polymyxins, and (ii) compare the pharmacodynamics of polymyxin B and azithromycin combinations against quorum sensing-proficient (PAO1) and -deficient (rhlR/lasR knockout) strains.
The bacterial strains utilized in this study were wild-type P. aeruginosa PAO1 and an isogenic rhlR/lasR knockout (ΔrhlR::GmR and ΔlasR::TcR cassettes) (22). Colistin, polymyxin B, and azithromycin MICs were determined using broth microdilution in duplicate according to CLSI (23). The colistin, polymyxin B, and azithromycin MICs for PAO1 were 1, 1, and 512 mg/liter, respectively, and the MICs for the rhlR/lasR knockout were 2, 2, and 512 mg/liter, respectively. Colistin (sulfate) and polymyxin B (sulfate) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and azithromycin was purchased from AK Scientific, Inc. (Union City, CA, USA). Time-kill experiments were carried out as previously described (24). LB broth supplemented with magnesium chloride (12.5 Mg2+/liter final concentration) and calcium chloride (25 Ca2+/liter final concentration) acted as growing media in all experiments. Serial samples throughout the 48-h experiment were withdrawn to quantify viable cell density after vortexing and visual inspection, which verified that the system was homogenous and planktonic. Colistin or polymyxin B killing was evaluated at different bacterial densities (CFU0 h) of ∼106, 108, or 109 CFU/ml. Polymyxin B and azithromycin combination experiments were performed at a CFU0 h of ∼108 CFU/ml. Polymyxin concentrations ranging from 0 to 128 mg/liter (4, 25) and azithromycin concentrations of 0, 0.5, 2, 128, and 256 mg/liter (15, 16) were used.
Synergy was defined as a ≥2 log10-CFU/ml reduction compared to the more active agent as monotherapy at 24 h. To characterize pharmacodynamic activity over time (0 to 48 h), the area under the CFU (AUCFU) curve was calculated using the linear-up, log-down trapezoidal rule. The log ratio change was calculated to compare killing at individual time points, whereas the log ratio area was used to assess activity throughout the time-kill experiments, as described previously (26). To characterize the interplay between inoculum effect and quorum sensing, previously developed mechanism-based models for colistin were used (21). Three preexisting subpopulations with different susceptibilities to colistin or polymyxin B were considered with the modification of the growth model. Data were modeled using a population approach in S-ADAPT software (version 1.57) with the SADAPT-TRAN pre- and postprocessing tool to fit all data simultaneously (see mathematical model development in the supplemental material).
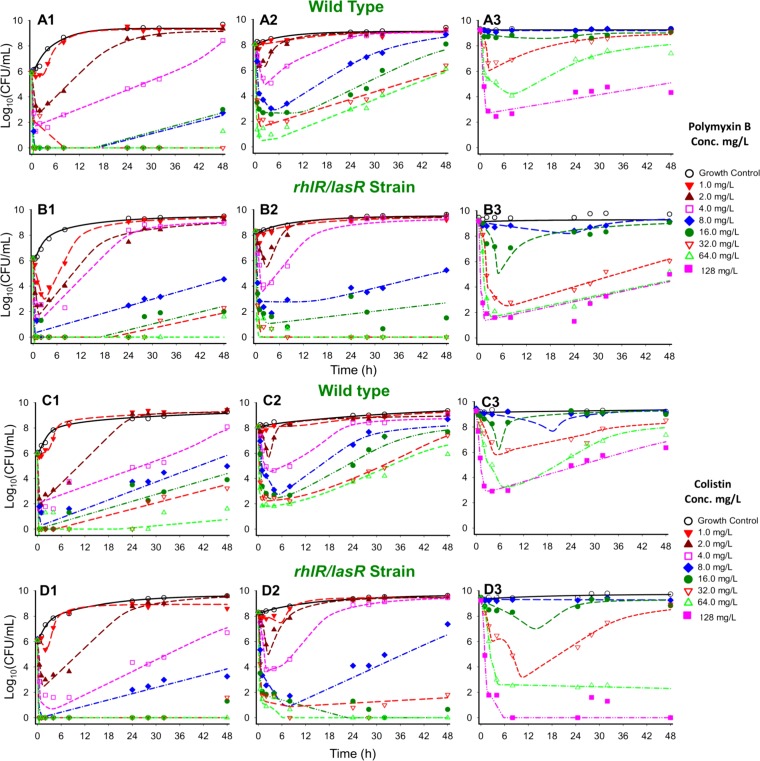
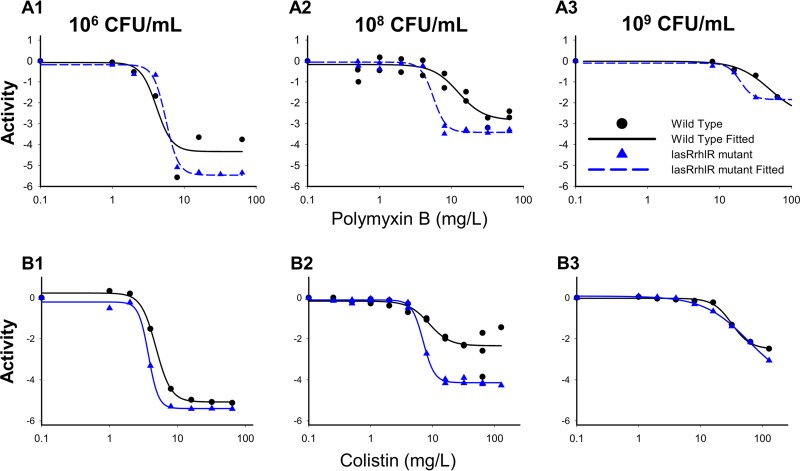
Colistin and polymyxin B alone displayed concentration-dependent killing for all three initial inocula with regrowth toward the growth control (Fig. 1 and 2). Both polymyxins caused rapid initial killing of up to 6.00 (PAO1) and 6.16 (rhlR/lasR knockout) log10 CFU/ml at the 106 CFU0 h inoculum. However, at higher starting inocula (108 and 109 CFU0 h), there was attenuation of bacterial killing, which resulted in a stepwise half-maximal effective concentration (EC50) increase for each strain (Fig. 2). Colistin and polymyxin B achieved better killing against the rhlR/lasR knockout than against PAO1, especially at higher concentrations. The discordance in killing was most prominent at the 108 CFU0 h starting inoculum, where the Emax (maximal effect) increased from 2.18 to 4.03 (P < 0.001) for colistin and from 2.67 to 3.36 (P = 0.09) for polymyxin B against the PAO1 and rhlR/lasR knockout strains, respectively (Fig. 2A2 and B2). Colistin and polymyxin B also displayed a higher EC50 for PAO1 than for the rhlR/lasR knockout strain (8.72 mg/liter [24.6% standard error (SE)] versus 7.04 mg/liter [2.64% SE] for colistin, P = 0.47; 12.45 mg/liter [16.0% SE] versus 5.61 mg/liter [5.71% SE] for polymyxin B, P < 0.005) at this inoculum. At the 106 and 109 CFU0 h inocula, the strains displayed similar concentration-response curves (Fig. 2A3 and B3); however, the bacterial killing profiles against the rhlR/lasR knockout appeared greater (Fig. 1A to D3). Among the polymyxins, colistin performed marginally better than polymyxin B against the rhlR/lasR knockout, most markedly at concentrations of ≥4 mg/liter for 106 and 108 CFU0 h and ≥32 mg/liter for 109 CFU0 h. Against PAO1, the polymyxins achieved comparable killing at each inoculum, with the only perceptible difference occurring in favor of polymyxin B at 106 CFU0 h (≥8 mg/liter). The final mechanism-based pharmacodynamic model, which consists of three preexisting subpopulations and uses the target site binding of polymyxin to LPS (see Fig. S1 in the supplemental material), excellently characterized (see Table S1 in the supplemental material, correlation coefficient individual fit >0.994 and population fit >0.905) the time course of bacterial killing, inoculum effect, and regrowth (Fig. 1, dashed lines).
FIG 1.
Time course of P. aeruginosa strains PAO1 (rows A and C) and the rhlR/lasR knockout strain (rows B and D) against polymyxin B (A1 to B3) or colistin (C1 to D3) at three different initial inocula: 106 CFU0 h (column 1), 108 CFU0 h (column 2), and 109 CFU0 h (column 3). Polymyxin B and colistin concentrations range from 0 to 128 mg/liter. Individual points represent observed viable colony counts (CFU/ml) from time-kill experiments, whereas lines represent expected bacterial killing as predicted by the previously validated mechanism-based model.
FIG 2.
Comparative pharmacodynamic responses between PAO1 wild-type (black) and rhlR/lasR knockout (blue) strains at each inoculum to either polymyxin B (row A) or colistin (row B). Activity is defined as the log10 ratio of the AUCFU of treatment to the AUCFU of growth control (log ratio area) based on the observed response throughout the 48-h time-kill experiments. Parameter estimates for the Hill-type models are found in Table S2 in the supplemental material.
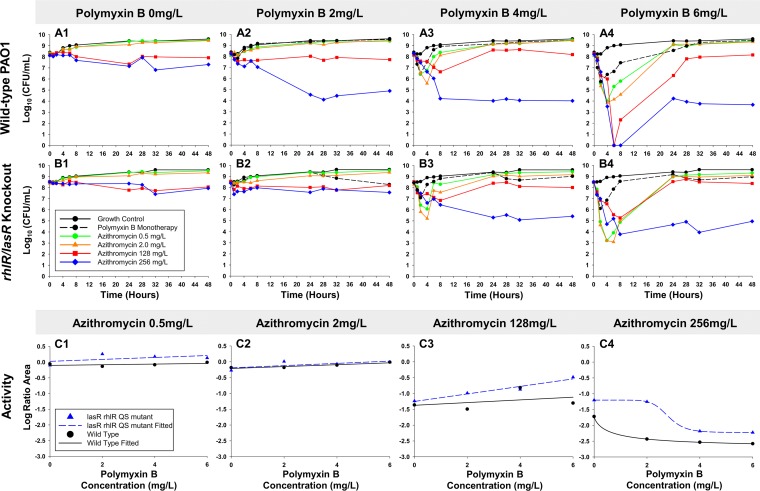
Our time-kill data and pharmacodynamic modeling of colistin and polymyxin B monotherapies show that the rapid bactericidal activity achieved was not sustained, suggesting potential utility for combinations. Polymyxin B was investigated in addition to azithromycin at 108 CFU0 h due to the favorable pharmacokinetics of polymyxin B (not administered as a prodrug) (27) and the similar performance of both polymyxins at clinically achievable concentrations at this inoculum (Fig. 2A2 and B2). In contrast to the regrowth seen against polymyxins alone, azithromycin (256 mg/liter) and polymyxin B achieved synergy at 24 h against PAO1 regardless of the polymyxin B concentration (Fig. 3A1 to 4). Remarkably, after the rapid initial bacterial killing within 8 h by polymyxin B and azithromycin (128 or 256 mg/liter) combinations, apparent bacteriostasis was achieved and persisted through 48 h. Against the rhlR/lasR knockout, the combination with azithromycin 256 mg/liter was synergistic at polymyxin B concentrations of 4 and 6 mg/liter. Comparison of bacterial killing seen in PAO1 and the rhlR/lasR knockout using the log ratio area, showed minimal differentiation (<0.82) at azithromycin 0.5, 2, and 128 mg/liter (Fig. 3C1 to 3). The progressive decreases in log ratio area between these azithromycin concentrations were well fit to linear functions (standard error of estimate, <0.2). The highest azithromycin concentration (256 mg/liter) displayed less total killing against the rhlR/lasR knockout than against PAO1, and both were excellently fit to a Hill-type model (R2, >0.99). At an azithromycin concentration of 256 mg/liter, the largest difference in killing was seen at 2 mg/liter of polymyxin B, in which combination treatment achieved 2.86 and 2.51 log10 CFU/ml less killing against the rhlR/lasR knockout strain at 24 and 48 h, respectively. For the polymyxin combinations with 256 mg/liter azithromycin, the polymyxin B EC50s against the rhlR/lasR knockout and PAO1 increased from 0.66 to 2.80 mg/liter (P < 0.001).
FIG 3.
Time-kill experiments evaluating bacterial killing activity of polymyxin B and azithromycin combinations versus the P. aeruginosa wild-type PAO1 (row A) and the rhlR/lasR knockout (row B) strains. Comparative pharmacodynamic responses between PAO1 (black) and the rhlR/lasR knockout (blue) at increasing azithromycin concentrations in combination with polymyxin B and data fit with linear (C1 to 3) or Hill-type (C4) functions. Data for polymyxin B monotherapy are not shown for C1 to C4. Activity is defined as the log10 ratio of the AUCFU of treatment to the AUCFU of growth control (log ratio area) based on observed response throughout the 48-h time-kill experiments.
In the current work, we explored the interrelationship between quorum sensing and polymyxin pharmacodynamics at various bacterial densities. Interestingly, we determined that rhlR/lasR deficiency enhanced the pharmacodynamic activity of colistin and polymyxin B monotherapies, especially with the 108 CFU0 h inoculum. The hypothesized quorum sensing-driven alterations to killing were well characterized by our mathematical model, which was based on the known mechanisms of action of the polymyxin antibiotics. Furthermore, the model used signaling compartments that altered killing and growth to account for the observed inoculum effect. Consistent with these results, quorum sensing has been shown to influence susceptibility to other antimicrobials. Kayama et al. (28) demonstrated that quorum sensing-deficient P. aeruginosa strains (lasR/lasI but not rhlR/rhlI) exposed to ofloxacin had >40 times lower survival rates than the quorum sensing-proficient strain. Modulation of gene expression is thought to be important in adaptation to polymyxin exposure and has been proposed to be regulated by the two-component regulatory systems phoP-phoQ (29), pmrA-pmrB (30), and ParR-ParS (31). The role of such systems may be inoculum dependent, as we previously found that colistin exposed to pmrA and phoP mutant strains achieved greater killing than the wild-type strain at low initial inocula (32). Unlike our discovery in the current study, Ly et al found that there was no disparity of bacterial killing between the mutant and wild-type strains at the 108 CFU0 h inoculum. Collectively, these observations suggest that the adaptive response to polymyxin exposure may be regulated by rhl- and las-mediated quorum sensing directly or by downstream oversight of other two-component regulatory systems associated with polymyxin resistance.
As an adjuvant to polymyxin B, high concentrations (128 or 256 mg/liter) of the known quorum sensing inhibitor azithromycin caused bacterial killing of P. aeruginosa that persisted below the growth control level. Thus, early bactericidal activity by polymyxin B monotherapy against a high-density infection may be sustained longer with polymyxin-azithromycin combinations. Perhaps the static concentrations of P. aeruginosa below growth control levels indicate a persister phenotype. Persister cells become tolerant to antibiotic therapy but are unable to replicate (33). By lowering the bacterial burden and forcing P. aeruginosa into a static phenotype, the polymyxin B-azithromycin combination may improve the immune system's likelihood of eradicating the infection.
In contrast to our results with polymyxin B monotherapy, we determined that combinations with high azithromycin concentrations were more active against the wild-type strain versus the rhlR/lasR knockout. This finding supports previous data that showed that increasing azithromycin concentrations inhibited growth of a P. aeruginosa wild-type strain more than quorum sensing-deficient mutants (lasR) (19). Similar bacterial killing may have been anticipated between the wild-type and rhlR/lasR knockout strains in the presence of complete quorum sensing inhibition by azithromycin. Therefore, an enhanced susceptibility of the wild-ype strain to the polymyxin-azithromycin combination may be explained in part by the production of rhamnolipids, an exoproduct that is controlled by quorum sensing (rhl and las systems) and thus only expressed in the wild-type strain before inhibition by azithromycin. Rhamnolipids may increase the uptake of azithromycin into P. aeruginosa, making the wild-type strain more susceptible to the polymyxin-azithromycin combination, especially at higher concentrations (34). Furthermore, evidence supports the benefit of azithromycin in prevention of P. aeruginosa ventilator-associated pneumonia in patients colonized with P. aeruginosa producing high levels of rhamnolipids (35). A potential limitation of this study is that we did not directly quantify quorum sensing activity or rhamnolipid expression in our in vitro system. Additional studies are warranted to fully elucidate the mechanism(s) of azithromycin attack on P. aeruginosa in the presence of polymyxin B.
In conclusion, our study demonstrated that bacterial modulation by quorum sensing may decrease P. aeruginosa susceptibility to polymyxin antibiotics. Considering the utility of azithromycin in cystic fibrosis exacerbations and its ability to inhibit quorum sensing, expanding its niche to other P. aeruginosa infections is worth consideration. Azithromycin may therefore provide a multifactorial attack on P. aeruginosa and prove useful as an adjuvant to polymyxins in a range of clinical scenarios. Further in vivo investigations with polymyxin-azithromycin combinations are needed to better understand the dual benefit of quorum sensing inhibition and potentiation of polymyxin activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Peter Greenberg, University of Washington, for kindly providing the P. aeruginosa strains. We also thank Poornima Subramanian, Jonathan S. Gall, and Alan Forrest for their important contributions to this project.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI111990.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00096-16.
REFERENCES
- 1.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Planquette B, Timsit JF, Misset BY, Schwebel C, Azoulay E, Adrie C, Vesin A, Jamali S, Zahar JR, Allaouchiche B, Souweine B, Darmon M, Dumenil AS, Goldgran-Toledano D, Mourvillier BH, Bedos JP. 2013. Pseudomonas aeruginosa ventilator-associated pneumonia: predictive factors of treatment failure. Am J Respir Crit Care Med 188:69–76. doi: 10.1164/rccm.201210-1897OC. [DOI] [PubMed] [Google Scholar]
- 3.Linden PK, Kusne S, Coley K, Fontes P, Kramer DJ, Paterson D. 2003. Use of parenteral colistin for the treatment of serious infection due to antimicrobial-resistant Pseudomonas aeruginosa. Clin Infect Dis 37:e154–e160. doi: 10.1086/379611. [DOI] [PubMed] [Google Scholar]
- 4.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 5.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Saitovitch D, Wang J, Forrest A, Nation RL, Zavascki AP, Li J. 2013. Pharmacokinetics of polymyxin B in patients on continuous venovenous haemodialysis. J Antimicrob Chemother 68:674–677. doi: 10.1093/jac/dks437. [DOI] [PubMed] [Google Scholar]
- 6.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi: 10.1128/AAC.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwa AL, Abdelraouf K, Low JG, Tam VH. 2011. Pharmacokinetics of polymyxin B in a patient with renal insufficiency: a case report. Clin Infect Dis 52:1280–1281. doi: 10.1093/cid/cir137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 10.Qureshi ZA, Hittle LE, O'Hara JA, Rivera JI, Syed A, Shields RK, Pasculle AW, Ernst RK, Doi Y. 2015. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis 60:1295–1303. doi: 10.1093/cid/civ048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barin J, Martins AF, Heineck BL, Barth AL, Zavascki AP. 2013. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann Clin Microbiol Antimicrob 12:15. doi: 10.1186/1476-0711-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuji BT, Landersdorfer CB, Lenhard JR, Cheah SE, Thamlikitkul V, Rao GG, Holden PN, Forrest A, Bulitta JB, Nation RL, Li J. 2016. Paradoxical effect of polymyxin B: high drug exposure amplifies resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 60:3913–3920. doi: 10.1128/AAC.02831-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bratu S, Quale J, Cebular S, Heddurshetti R, Landman D. 2005. Multidrug-resistant Pseudomonas aeruginosa in Brooklyn, New York: molecular epidemiology and in vitro activity of polymyxin B. Eur J Clin Microbiol Infect Dis 24:196–201. doi: 10.1007/s10096-005-1294-x. [DOI] [PubMed] [Google Scholar]
- 14.Saiman L, Chen Y, San Gabriel P, Knirsch C. 2002. Synergistic activities of macrolide antibiotics against Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. Antimicrob Agents Chemother 46:1105–1107. doi: 10.1128/AAC.46.4.1105-1107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulds G, Shepard RM, Johnson RB. 1990. The pharmacokinetics of azithromycin in human serum and tissues. J Antimicrob Chemother 25(Suppl A):73–82. [DOI] [PubMed] [Google Scholar]
- 16.Ballow CH, Amsden GW, Highet VS, Forrest A. 1998. Pharmacokinetics of oral azithromycin in serum, urine, polymorphonuclear leucocytes and inflammatory vs non-inflammatory skin blisters in healthy volunteers. Clin Drug Invest 15:159–167. doi: 10.2165/00044011-199815020-00009. [DOI] [PubMed] [Google Scholar]
- 17.Saiman L, Marshall BC, Mayer-Hamblett N, Burns JL, Quittner AL, Cibene DA, Coquillette S, Fieberg AY, Accurso FJ, Campbell PW III. 2003. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290:1749–1756. doi: 10.1001/jama.290.13.1749. [DOI] [PubMed] [Google Scholar]
- 18.Kudoh S, Azuma A, Yamamoto M, Izumi T, Ando M. 1998. Improvement of survival in patients with diffuse panbronchiolitis treated with low-dose erythromycin. Am J Respir Crit Care Med 157:1829–1832. doi: 10.1164/ajrccm.157.6.9710075. [DOI] [PubMed] [Google Scholar]
- 19.Köhler T, Perron GG, Buckling A, Van Delden C. 2010. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog 6:e1000883. doi: 10.1371/journal.ppat.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tateda K, Comte R, Pechere J-C, Köhler T, Yamaguchi K, Van Delden C. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D'Hondt RE, Jusko WJ, Forrest A, Tsuji BT. 2010. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 54:2051–2062. doi: 10.1128/AAC.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberon-Chavez G. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol 40:708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. 2009. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:46–56. doi: 10.1128/AAC.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavascki AP, Goldani LZ, Cao G, Superti SV, Lutz L, Barth AL, Ramos F, Boniatti MM, Nation RL, Li J. 2008. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 47:1298–1304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
- 26.Tsuji BT, von Eiff C, Kelchlin PA, Forrest A, Smith PF. 2008. Attenuated vancomycin bactericidal activity against Staphylococcus aureus hemB mutants expressing the small-colony-variant phenotype. Antimicrob Agents Chemother 52:1533–1537. doi: 10.1128/AAC.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nation RL, Velkov T, Li J. 2014. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 59:88–89. doi: 10.1093/cid/ciu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kayama S, Murakami K, Ono T, Ushimaru M, Yamamoto A, Hirota K, Miyake Y. 2009. The role of rpoS gene and quorum-sensing system in ofloxacin tolerance in Pseudomonas aeruginosa. FEMS Microbiol Lett 298:184–192. doi: 10.1111/j.1574-6968.2009.01717.x. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol 34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 30.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ly NS, Yang J, Bulitta JB, Tsuji BT. 2012. Impact of two-component regulatory systems PhoP-PhoQ and PmrA-PmrB on colistin pharmacodynamics in Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:3453–3456. doi: 10.1128/AAC.06380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler T, Dumas JL, Van Delden C. 2007. Ribosome protection prevents azithromycin-mediated quorum-sensing modulation and stationary-phase killing of Pseudomonas aeruginosa. Antimicrob Agents Chemother 51:4243–4248. doi: 10.1128/AAC.00613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Delden C, Kohler T, Brunner-Ferber F, Francois B, Carlet J, Pechere JC. 2012. Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: a randomized controlled trial. Intensive Care Med 38:1118–1125. doi: 10.1007/s00134-012-2559-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.