Abstract
Background
Placental defects in somatic cell nuclear transfer (SCNT) are a major cause of complications during pregnancy. One of the most critical factors for the success of SCNT is the successful epigenetic reprogramming of donor cells. Recently, it was reported that the placental weight in mice cloned with the aggregated SCNT method was significantly reduced. Here, we examine the profile of abnormal gene expression using microarray technology in both regular SCNT and aggregated SCNT placentas as well as in vivo fertilization placentas. One SCNT embryo was aggregated with two 2 to 4 -cell stage tetraploid embryos from B6D2F1 mice (C57BL/6 × DBA/2).
Results
In SCNT placentas, 206 (1.6%) of the 12,816 genes probed were either up-regulated or down-regulated by more than two-fold. However, 52 genes (0.4%) showed differential expression in aggregated SCNT placentas compared to that in controls. In comparison of both types of SCNT placentas with the controls, 33 (92%) out of 36 genes were found to be up-regulated (>2-fold) in SCNT placentas. Among 36 genes, 13 (36%) genes were up-regulated in the aggregated SCNT placentas. Eighty-five genes were down-regulated in SCNT placentas compared with the controls. However, only 9 (about 10.5%) genes were down-regulated in the aggregated SCNT placentas. Of the 34 genes known as imprinted genes, expression was lower in SCNT placentas than that in the controls. Thus, these genes may be the cause of placentomegaly in mice produced post SCNT.
Conclusions
These results suggest that placentomegaly in the SCNT placentas was probably caused by abnormal expression of multiple genes. Taken together, these results suggest that abnormal gene expression in cloned placentas was reduced in a genome-wide manner using the aggregation method with tetraploid embryos.
Electronic supplementary material
The online version of this article (doi:10.1186/s12896-017-0355-4) contains supplementary material, which is available to authorized users.
Keywords: Aggregated SCNT, Placenta, Abnormal gene expression
Background
Somatic cell nuclear transfer (SCNT) in animals has the potential to be used in a wide range of applications such as species preservation, livestock propagation, and gene targeting [1]. However, this technology is inefficient and results in various abnormalities, leading to high pregnancy losses and neonatal deaths [2]. Although the cloned fetuses attain full term, placentomegaly is a common phenotype observed in cloned animals, irrespective of donor cell type and strain [3–5]. Placentomegaly mainly seems to arise from an abnormally expanded spongiotrophoblast layer with an increased number of glycogen cells, and irregular borderlines between the labyrinthine and spongiotrophoblast layers [6]. Interestingly, the same pattern of placentomegaly was also observed in interspecies hybridization [7, 8], during sperm injection following introduction of somatic cell cytoplasm into an oocyte [9], and in knockout mice with imprinting genes such as EsxI [10], Ipl [11], and H19 [12]. Thus, reduction in placental weight is necessary to obtain live and normal fetuses in SCNT.
Several global gene expression analyses using microarrays of more than 10,000 genes were conducted on samples from neonatal placenta to reveal a cluster of abnormally expressed genes [13–15] in the placentas of cloned mice. Of those SCNT-derived embryos that develop to full term, up to 40% have large offspring syndrome (LOS), characterized by hydrops of the fetus, lethargy, and respiratory distress [15–17]. Aggregation of embryonic stem (ES) cells with tetraploid blastocysts has been successfully conducted in mice [18, 19], and chimeric monkeys were produced by the aggregation of 4-cell embryos [20]. We also reported that aggregated SCNT significantly reduced placental weight of cloned mice and improved SCNT efficiency [5]. However, the differences in the genetic pattern of aggregated SCNT embryos and SCNT embryos are not clearly identified. It is therefore very important to analyze the differences in gene expression between the two types of SCNT embryos. In addition, these results will offer important information in solving the problem of lethality in cloned mice production.
In this study, the mRNA expression profiles of SCNT and aggregated SCNT placentas were analyzed using microarray technology. Many genes were found to be differentially expressed between the SCNT and aggregated SCNT placenta. These results further provide evidence supporting the importance of placental abnormalities in cloned animal production.
Methods
Placental samples
B6D2F1 mice (C57BL6 × DBA/2) were used to prepare oocytes and cumulus cells. Two-celled embryos were electrofused to produce one-cell tetraploid embryos. Tetraploid embryos were then aggregated with SCNT embryos. One embryo was aggregated with two 2 to 4 cell tetraploid embryos. Detailed methods are described in a previous report [5]. MII oocytes were collected from 6 to 12-week-old females (69 mice). Embryos electrofused were transferred to the foster mothers (47 mice). All recipient females were euthanized at 19.5 dpc and placentas were obtained. Finally, we produced a total of 36 clone mice and placental samples. The protocol was approved by the Committee on Ethics of Animal Experiments at the Hankyong National University (Permit Number: 2014–4).
Microarray analysis
Total RNA was extracted from five SCNT placentas, six aggregated SCNT placentas, and four controls by using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, USA) and purified using RNeasy columns (Qiagen, Valencia, USA), according to the manufacturers’ protocols.
Labeling and purification
Total RNA was amplified and purified using the Ambion Illumina RNA amplification kit (Ambion, Austin, USA) to yield biotinylated cRNA, according to the manufacturer’s instructions. Briefly, 550 ng of total RNA was reverse-transcribed to cDNA using a T7 oligo(dT) primer. Second-strand cDNA was synthesized, transcribed in vitro, and labeled with biotin-NTP.
Hybridization and data export
The labeled cRNA samples (0.75 μg) were hybridized to the Illumina MouseRef-8 v2 expression BeadChip (Illumina, Inc., San Diego, USA) for 16–18 h at 58 °C, according to the manufacturer’s instructions. Detection of the array signals was carried out using Amersham Fluorolink Streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, UK), following the bead array manual. Arrays were scanned with an Illumina bead array reader confocal scanner. Array data analysis was performed using Illumina Genome Studio v.2009.2 (Gene Expression Module v.1.5.4).
Raw data preparation and statistical analysis
The raw data were extracted using the software provided by the manufacturer (Illumina Genome Studio v.2009.2). The array data were filtered using a detection p-value < 0.05 (a signal value higher than that of the background was required to obtain a detection p-value < 0.05). The selected gene signal value was logarithmically transformed and normalized. Comparative analysis between 2 groups was carried out by p-value evaluation, using the local-pooled-error test (adjusted Benjamini-Hochberg false discovery rate controlled by 5%) and fold-change. Biological ontology-based analysis was performed using the Panther database (http://www.pantherdb.org). In addition to these statistical criteria, genes whose expression differed by more than two-fold were considered differentially expressed between the two groups.
Quantitative real-time PCR (qRT-PCR)
To validate the microarray data, 12 genes (viz., Plac1, Slc38a4, Rprml, Pla2g4f, Pla2g4d, Hsd17β7, Hmox1, Chac1, Car2, Slpi, Nrn1l, and H19) from different categories were chosen for qRT-PCR analyses. The expression of these genes was either up- and down-regulated by more than two-fold. qRT-PCR was performed with the same placenta used in the microarray analyses.
Primer sequences are outlined in Additional file 1: Table S1 along with the primer annealing temperatures. The primers were designed with the help of Primer 3 software (www.bioneer.co.kr/products/Oligo/CustomOligonucleotides-overview.aspx). Gene expression was analyzed from 5 SCNT placentas and 3 control placentas. The β-actin (ACTB) gene was used as the endogenous control, and the results of the analysis were calculated by using the 2-ΔΔCT method for quantitative relationships.
Results
Abnormal gene expression profiles between SCNT, aggregated SCNT, and control placentas
The mouse placentas derived from SCNT, aggregated SCNT, and in vivo fertilized controls were analyzed for their global gene expression patterns using the microarray method. The placental weight of the control, aggregated SCNT, and SCNT was 0.147 g (n = 8), 0.215 g (n = 9), and 0.287 (n = 27), respectively, as previously described [5].
Gene transcription levels were detected in the microarray analysis using 12,816 gene probes. Genes showing >2-fold difference in expression were identified for 15 placentas (SCNT, 5; aggregation SCNT, 6; control 4). Figure 1a shows the gene expression differences between control and SCNT placentas. The expression of 206 (1.6%) of 12,816 genes was found to differ by at least two-fold between the SCNT placentas and the controls. Similarly, 159 genes showed different expression between the SCNT placentas and the aggregated SCNT placentas (Fig. 1b). However, 52 (0.4%) genes showed difference (>2-fold) in expression between the aggregated SCNT placentas and the controls (Fig. 1c).
Fig. 1.
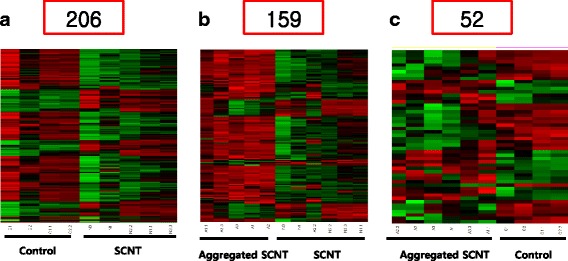
Hierarchical cluster of expression profiles in placentas of both types of cloned mice and the in vivo fertilization placenta (control). a A cluster of 206 genes between the control and the SCNT placentas, (b) a cluster of 159 genes between aggregated SCNT and SCNT placentas, and (c) a cluster of 52 genes between aggregated SCNT and control placentas. Expression of more than 2-fold difference is indicated by increasing red intensity and green indicates reduced expression. SCNT, con = control
Co-up- and down-regulated genes in SCNT and aggregated SCNT placentas compared with controls
Many of the differentially expressed gene probes were common to the two types of clones, whereas some were deregulated either in SCNT placentas or in the aggregated SCNT placentas alone. Of the 36 genes up-regulated in SCNT placentas, 10 (27.7%) were commonly up-regulated in the aggregated SCNT placenta group. Ten genes [Pla2g4f, Car2, Tekt1 (probe1), Tekt1 (probe2), Pla2g4d, Rprm1, Hsd17b7, Hic1, Hmox1, and Coll15a1] were up-regulated (>2-fold) in both SCNT and aggregated SCNT placentas (Table 1). However, three genes (Abcc10, Prss22, and Slc22a18) were only up-regulated in the aggregated SCNT placentas. The number of genes showing >2-fold up-regulation in the SCNT placentas was decreased by 33% (from 39 to 13) in the placentas obtained by aggregated SCNT.
Table 1.
Genes that were up-regulated (fold change) in SCNT placentas versus in control placentas
| Gene symbol | Accession No. | Folder Δ SCNT/Con | Folder Δ Agg/Con |
|---|---|---|---|
| Significantly elevated in NT alone | |||
| Tiam1 | NM_009384.2 | 2.32* | 1.91 |
| Dao1 | NM_010018.2 | 2.36* | 1.88 |
| Mmp15 | NM_008609.3 | 2.27* | 1.78 |
| Cxcl1 | NM_008176.1 | 2.42* | 1.84 |
| Aldh1a3 | NM_053080.2 | 2.08* | 1.54 |
| E130203B14Rik | NM_178791.4 | 2.05** | 1.50 |
| Cdx2 | NM_007673.3 | 2.74* | 1.98 |
| Plac1 | NM_019538.3 | 2.73** | 1.86 |
| Slc38a4 | NM_027052.3 | 2.32** | 1.55 |
| Irs3 | NM_010571.3 | 2.23* | 1.46 |
| Ldoc1 | NM_001018087.1 | 2.78* | 1.78 |
| Gna14 | NM_008137.3 | 3.05** | 1.87 |
| Galk1 | NM_016905.2 | 2.32* | 1.40 |
| Serpinb9d | NM_011460.1 | 2.05* | 1.23 |
| Prl7c1 | NM_026206.2 | 3.22** | 1.89 |
| Prl2c1 | NM_001045532.1 | 2.40** | 1.36 |
| Ms4a10 | NM_023529.2 | 2.64* | 1.49 |
| Sbsn | NM_172205.3 | 3.07** | 1.64 |
| Prl2c5 | NM_181852.1 | 2.51* | 1.33 |
| Sbsn | NM_172205.3 | 3.32* | 1.63 |
| Serpinb9d | NM_011460.1 | 3.03** | 1.46 |
| Prl4a1 | NM_011165.3 | 2.69** | 1.26 |
| Ghrh | NM_010285.2 | 2.15* | −1.06 |
| Ada (probe 1) | NM_007398.3 | 2.86** | 1.54 |
| Ada (probe 2) | NM_007398.3 | 3.11** | 1.92 |
| Ermap | NM_013848.1 | 2.84* | 1.12 |
| Col15a1 | NM_009928.3 | 2.70* | 2.59 |
| Hmox1 | NM_010442.1 | 2.73** | 2.35 |
| Rprml | NM_001033212.1 | 3.25** | 2.69 |
| Hsd17b7 | NM_010476.3 | 3.24** | 2.45 |
| Hic1 | NM_010430.2 | 3.02* | 2.25 |
| Pla2g4f | NM_001024145.1 | 4.10** | 2.69 |
| Pla2g4d | NM_001024137.1 | 3.70** | 2.32 |
| Tekt1 (probe 1) | NM_011569.2 | 3.40* | 2.05 |
| Tekt1 (probe 2) | NM_011569.2 | 3.90* | 2.26 |
| Car2 | NM_009801.3 | 3.95* | 2.09 |
| Significantly elevated in aggregation | |||
| Abcc10 | NM_170680.2 | 1.01 | 2.96* |
| Prss22 | NM_133731.1 | −1.11 | 2.44* |
| Slc22a18 | NM_008767.2 | 1.32 | 2.95* |
SCNT somatic cell nuclear transfer
*Significantly elevated in NT alone
*p < 0.05; **p < 0.01
In Table 2, we have identified the genes showing significant downregulation (>2-fold) in the SCNT, aggregated SCNT, and control placentas. Eighty-five genes were found to be downregulated in SCNT placentas in comparison with their expression in the controls. In contrast, 16 of these genes (18%) (Sftpd, Tph, 2010109103Rik, Cbx7, Osta, Serpina10, Macb, Fcgrt, Bex2, 84304C8G22Rik, Acox2, Vdr, Dab2, Cfi, Ltf, and Dab2) were upregulated in aggregated SCNT placentas in comparison with their expression in the controls. Five of the 85 genes (6%) (Inhba, Chac1, Nrn11, Tnfrsf11b, and Slpi) were down-regulated (>2-fold) in both types of SCNT placentas compared to their expression in the controls (Table 2). Four genes (Nppb, 1200015F23Rik, Uap1, and Ctsm) were only downregulated (>2-fold) in the aggregated SCNT placentas. These abnormalities in gene expression were significantly reduced with the use of the aggregated SCNT method.
Table 2.
Genes that were down-regulated (fold change) in SCNT placentas versus in control placentas
| Gene symbol | Accession No. | Folder Δ SCNT/Con | Folder Δ Agg/Con | Gene symbol | Accession No. | Folder Δ SCNT/Con | Folder Δ Agg/Con |
| 1300017J02Rik | NM_027918.1 | −2.67** | −1.34 | Cbx7 | NM_144811.3 | −3.10** | 1.12 |
| Ly6g6c | NM_023463.3 | −3.82** | −1.92 | Osta | NM_145932.3 | −2.88* | 1.18 |
| Pcyox1 | NM_025823 | −2.16** | −1.10 | Entpd2 | NM_009849.1 | −3.64** | −1.10 |
| Lamb3 | NM_008484.2 | −2.94* | −1.53 | Bex4 | NM_212457.1 | −3.50** | −1.07 |
| Ang | NM_007447.2 | −2.26* | −1.20 | Serpina10 | NM_144834.3 | −2.39* | 1.36 |
| Bmp4 | NM_007554.2 | −2.59** | −1.42 | Fga | NM_010196.1 | −3.51** | −1.10 |
| LOC100046120 | XM_001475611.1 | −2.62** | −1.51 | 1700045I19Rik | NM_028842.1 | −3.47** | −1.11 |
| Aqp8 | NM_007474.1 | −2.32** | −1.36 | Fga | NM_010196.2 | −3.47** | −1.12 |
| Heph | NM_010417.1 | −2.52* | −1.51 | BC040758 | NM_001033364.1 | −3.34** | −1.12 |
| Serping1 | NM_009776.1 | −2.35** | −1.43 | Maob | NM_172778.1 | −2.95* | 1.01 |
| Doxl2 | NM_001029987.1 | −2.18** | −1.33 | BC040758 | NM_001033364.1 | −3.34** | −1.12 |
| Saa3 | NM_011315.3 | −2.95** | −1.79 | Fcgrt | NM_010189.1 | −2.75** | 1.07 |
| Aqp8 | NM_007474.1 | −2.19** | −1.34 | Gdpd3 | NM_024228.2 | −3.60** | −1.23 |
| Klk4 | NM_019928.1 | −2.79** | −1.72 | Cldn2 | NM_016675.3 | −2.93** | −1.01 |
| Serpind1 | NM_008223.2 | −2.67* | −1.67 | Psca | NM_028216.1 | −4.43** | −1.53 |
| Abp1 | NM_029638.1 | −2.06** | −1.33 | 1600015I10Rik | NM_001081273.1 | −4.64** | −1.61 |
| Krt14 | NM_016958.1 | −2.11* | −1.42 | Bex2 | XM_977338.1 | −2.52* | 1.14 |
| Scg5 | NM_009162.3 | −2.77* | −1.95 | Apom | NM_018816.1 | −3.27** | −1.16 |
| Mustn1 | NM_181390.2 | −2.05* | −1.65 | 8430408G22Rik | NM_145980.1 | −2.12* | 1.31 |
| Tacstd2 | NM_020047.3 | −2.07** | −1.71 | Acox2 | NM_053115.1 | −2.53** | 1.08 |
| Sfrp5 | NM_018780.2 | −2.02** | −1.88 | Spp2 | NM_029269.1 | −2.91** | −1.06 |
| Sftpd | NM_009160.1 | −4.14** | 1.13 | Vdr | NM_009504.3 | −2.19* | 1.21 |
| Tph1 | NM_009414.2 | −2.65* | 1.59 | Itih3 | NM_008407.1 | −2.64* | −1.00 |
| 2010109I03Rik | NM_025929.2 | −2.75** | 1.47 | Spink3 | NM_009258.2 | −2.81* | −1.08 |
| Amn | NM_033603.2 | −4.03** | −1.14 | Apoa2 | NM_013474.1 | −2.77* | −1.07 |
| Gene symbol | Accession No. | Folder Δ SCNT/Con | Folder Δ Agg/Con | Gene symbol | Accession No. | Folder Δ SCNT/Con* | Folder Δ Agg/Con |
| Significantly reduced in NT | Significantly reduced in NT | ||||||
| Tfrc | NM_011638.3 | −3.08** | −1.20 | C3 | NM_009778.1 | −2.23* | −1.06 |
| Kng1 | NM_023125.2 | −3.07* | −1.20 | Igfbp6 | NM_008344.2 | −3.64** | −1.76 |
| Fcgr3 | NM_010188.4 | −2.56* | −1.02 | Popdc3 | NM_024286.1 | −3.68** | −1.79 |
| Dab2 | NM_001008702.1 | −2.34** | 1.06 | Muc13 | NM_010739.1 | −2.09** | −1.03 |
| Gldc | NM_138595.1 | −2.66** | −1.08 | Lcn2 | NM_008491.1 | −2.42* | −1.20 |
| Serpina1b | NM_009244.4 | −2.80** | −1.14 | Inhba | NM_008380.1 | −2.81** | −2.25 |
| Tfrc | NM_011638.3 | −2.87** | −1.18 | Chac1 | NM_026929.3 | −2.27* | −2.09 |
| Cfi | NM_007686.2 | −2.20* | 1.10 | Nrn1l | NM_175024.3 | −2.49* | −2.49* |
| Ltf | NM_008522.3 | −2.30** | 1.05 | Tnfrsf11b | NM_008764.3 | −2.15* | −2.30 |
| Gpc3 | NM_016697.2 | −2.50* | −1.05 | Slpi | NM_011414.2 | −4.56** | −2.09 |
| Mgst1 | NM_019946.3 | −2.51* | −1.06 | ||||
| Fgg | NM_133862.1 | −2.95** | −1.25 | Significantly reduced in aggregation | |||
| Nr1h4 | NM_009108.1 | −3.16* | −1.35 | Nppb | NM_008726.3 | −1.46 | −3.14** |
| Dab2 | NM_023118.1 | −2.31 | 1.01 | 1200015F23Rik | NM_001033136.2 | −1.00 | −2.71* |
| Kng1 | NM_023125.2 | −2.87 | −1.24 | Uap1 | NM_133806.4 | 1.27 | −2.72** |
| Slc7a9(prob1) | NM_021291.1 | −3.04 | −1.34 | Ctsm | NM_022326.3 | 1.12 | −4.82** |
| Slc7a9(prob2) | NM_021291.1 | −2.82 | −1.24 | ||||
| Irf6 | NM_016851.2 | −2.43 | −1.08 | ||||
| Trf | NM_133977.2 | −2.37 | −1.08 | ||||
| Serpina1d | NM_009246.3 | −2.59 | −1.19 | ||||
| Sema4a | NM_013658.2 | −2.60 | −1.20 | ||||
| Serpina1b | NM_009244.4 | −2.62 | −1.21 | ||||
| Gipc2 | NM_016867.1 | −2.56 | −1.19 | ||||
| Kng2 | NM_201375.1 | −2.59 | −1.20 | ||||
| Slc27a2 | NM_011978.2 | −2.96 | −1.40 | ||||
*Significantly reduced in NT alone, *p < 0.05; **p < 0.01
Up-and down-regulated genes in SCNT and aggregated SCNT placentas
We next compared the expression patterns of the up-regulated (>2-fold) genes between SCNT and aggregated SCNT placentas. Twenty-one genes showed higher expression in SCNT placentas than in aggregated SCNT placentas. Four of these genes (19%; Ermap, Prl4a1, Sbsn, and Serpinb9d) also showed >2-fold higher expression in the SCNT placentas than in the controls. Additional seven genes (30%; Atf4, Atp6v1d, Fmr1nb, Gnaq, Riok1, Tomm22, and Zfp330) showed >2-fold lower expression in the aggregated SCNT placentas than in the controls (Table 3). We also analyzed the expression patterns of the down-regulated (p<0.05 and >2-fold) genes between both SCNT groups. Of the 102 genes that showed lower expression in the SCNT placentas than in the aggregated SCNT group, 53 (51%) had even lower expression than that observed in the controls. However, four genes were up-regulated and one gene (Slpi) was down-regulated in the aggregated SCNT placentas compared with that in the controls (Table 4).
Table 3.
Comparison of the genes that were up-regulated (fold change) in SCNT placentas versus in aggregated SCNT placentas
| Gene symbol | Accession No. | Folder Δ SCNT/Agg | Folder Δ SCNT/Con | Folder Δ Agg/Con |
|---|---|---|---|---|
| Cmas | NM_009908.1 | 3.15** | 1.85** | −1.70 |
| Ermap | NM_013848.1 | 2.53** | 2.84* | 1.12 |
| Prl2a1 | NM_019991.1 | 2.22** | 1.30* | −1.70 |
| Prl4a1 | NM_011165.3 | 2.13** | 2.69** | 1.26 |
| Sbsn | NM_172205.3 | 2.03* | 3.32** | 1.63 |
| Serpinb9d | NM_011460.1 | 2.08** | 3.03** | 1.46 |
| 2310039H08Rik | NM_025966.3 | 2.11* | 1.28 | −1.65 |
| Gpn2 | NM_133884.1 | 2.14* | 1.14 | −1.88 |
| H2-Q5 | NM_010393.1 | 2.10** | 1.45 | −1.45 |
| Matn1 | NM_010769.1 | 2.30* | 1.81 | −1.27 |
| Mlycd | NM_019966.2 | 2.08** | 1.19 | −1.74 |
| Pacsin1 | NM_011861.2 | 2.20* | 1.28 | −1.71 |
| Prcp | NM_028243.2 | 2.12** | 1.37 | −1.55 |
| Stab2 | NM_138673.2 | 2.03* | 1.32 | −1.54 |
| Atf4 | NM_009716.2 | 2.42* | 1.02 | −2.36 |
| Atp6v1d | NM_023721.2 | 2.43** | −1.01 | −2.46 |
| Fmr1nb | NM_174993.1 | 2.35** | 1.14 | −2.07 |
| Gnaq | NM_008139.5 | 2.75** | 1.23 | −2.25 |
| Riok1 | NM_024242.2 | 2.37** | 1.08 | −2.19 |
| Tomm22 | NM_172609.3 | 2.35** | 1.04 | −2.26 |
| Zfp330 | NM_145600.1 | 2.58** | 1.14 | −2.26 |
Significantly elevated in SCNT
*p < 0.05; **p < 0.01
Table 4.
Comparison of the genes that were down-regulated (fold change) in SCNT placentas versus in aggregated SCNT placentas
| Gene symbol | Accession No. | Folder Δ SCNT/Agg | Folder Δ SCNT/Con | Folder Δ Agg/Con |
|---|---|---|---|---|
| C6 | NM_016704.1 | −2.05** | −1.75* | 1.17 |
| 5033414D02Rik | NM_026362.1 | −2.04** | −1.86* | 1.10 |
| Sftpd | NM_009160.1 | −4.68** | −4.14** | 1.13 |
| Tph1 | NM_009414.2 | −4.20** | −2.65* | 1.59 |
| 2010109I03Rik | NM_025929.2 | −4.04** | −2.75** | 1.47 |
| Amn | NM_033603.2 | −3.55** | −4.03** | −1.14 |
| Cbx7 | NM_144811.3 | −3.46** | −3.10** | 1.12 |
| Osta | NM_145932.3 | −3.40** | −2.88* | 1.18 |
| Entpd2 | NM_009849.1 | −3.30** | −3.64** | −1.10 |
| Bex4 | NM_212457.1 | −3.26** | −3.50** | −1.07 |
| Serpina10 | NM_144834.3 | −3.25** | −2.39* | 1.36 |
| Fga | NM_010196.1 | −3.18** | −3.51** | −1.10 |
| 1700045I19Rik | NM_028842.1 | −3.13** | −3.47** | −1.11 |
| Fga | NM_010196.2 | −3.10** | −3.47** | −1.12 |
| BC040758 | NM_001033364.1 | −2.99** | −3.34** | −1.12 |
| Maob | NM_172778.1 | −2.99* | −2.95* | 1.01 |
| BC040758 | NM_001033364.1 | −2.97** | −3.34* | −1.12 |
| Fcgrt | NM_010189.1 | −2.94** | −2.75** | 1.07 |
| Cldn2 | NM_016675.3 | −2.91** | −2.93** | −1.01 |
| Psca | NM_028216.1 | −2.90** | −4.43** | −1.53 |
| 1600015I10Rik | NM_001081273.1 | −2.89** | −4.64** | −1.61 |
| Bex2 | XM_977338.1 | −2.88** | −2.52* | 1.14 |
| Apom | NM_018816.1 | −2.81** | −3.27** | −1.16 |
| 8430408G22Rik | NM_145980.1 | −2.77** | −2.12* | 1.31 |
| Acox2 | NM_053115.1 | −2.74** | −2.53** | 1.08 |
| Spp2 | NM_029269.1 | −2.74** | −2.91** | −1.06 |
| Muc13 | NM_010739.1 | −2.03* | −2.09** | −1.03 |
| Lcn2 | NM_008491.1 | −2.01* | −2.42** | −1.20 |
| Slpi | NM_011414.2 | −2.18** | −4.56** | −2.09 |
| Aig1 | NM_025446.1 | −2.81** | −1.09 | 2.57 |
| Wfdc2 | NM_026323.2 | −2.02* | −1.01 | 2.00 |
| Snca | NM_009221.2 | −2.86** | −1.56 | 1.84 |
| Eraf | NM_133245.1 | −2.84** | −1.61 | 1.77 |
| Snca | NM_009221.2 | −2.66** | −1.42 | 1.87 |
| Slco2b1 | NM_175316.3 | −2.64** | −1.88 | 1.41 |
| Hpx | NM_017371.1 | −2.52* | −1.70 | 1.49 |
| 1810007E14Rik | NM_025308.1 | −2.52** | −1.66 | 1.52 |
| Slc4a1 | NM_011403.1 | −2.49** | −1.48 | 1.68 |
| Alas2 | NM_009653.1 | −2.46** | −1.41 | 1.75 |
| Ctsh | NM_007801.1 | −2.43** | −1.97 | 1.24 |
| Ttr | NM_013697.3 | −2.34* | −1.65 | 1.42 |
| Dnmt3l | NM_019448.2 | −2.31** | −1.36 | 1.69 |
| Abhd14b | NM_029631.2 | −2.25* | −1.77 | 1.27 |
| Cish | NM_009895.3 | −2.22* | −1.99 | 1.12 |
| Vdr | NM_009504.2 | −2.22** | −1.89 | 1.17 |
| Dnmt3l | NM_001081695.1 | −2.21** | −1.40 | 1.58 |
| Gstm1 | NM_010358.4 | −2.19* | −1.67 | 1.31 |
| Magi1 | NM_001029850.2 | −2.18** | −1.21 | 1.80 |
| Upk3b | NM_175309.3 | −2.16* | −1.70 | 1.27 |
| Clic6 | NM_172469.3 | −2.16* | −1.73 | 1.24 |
| LOC100044204 | XM_001471696.1 | −2.14** | −1.88 | 1.14 |
| Igfbp2 | NM_008342.2 | −2.12** | −1.65 | 1.29 |
| Vdr | NM_009504.3 | −2.65** | −2.19* | 1.21 |
| Itih3 | NM_008407.1 | −2.63** | −2.64* | −1.00 |
| Apoa2 | NM_013474.1 | −2.59** | −2.77* | −1.07 |
| Tfrc | NM_011638.3 | −2.56** | −3.08** | −1.20 |
| Kng1 | NM_023125.2 | −2.56** | −3.07* | −1.20 |
| Fcgr3 | NM_010188.4 | −2.52** | −2.56* | −1.02 |
| Dab2 | NM_001008702.1 | −2.48** | −2.34* | 1.06 |
| Gldc | NM_138595.1 | −2.47* | −2.66** | −1.08 |
| Serpina1b | NM_009244.4 | −2.45** | −2.80** | −1.14 |
| Tfrc | NM_011638.3 | −2.44** | −2.87** | −1.18 |
| Cfi | NM_007686.2 | −2.43** | −2.20* | 1.10 |
| Ltf | NM_008522.3 | −2.40** | −2.30** | 1.05 |
| Gpc3 | NM_016697.2 | −2.38** | −2.50* | −1.05 |
| Mgst1 | NM_019946.3 | −2.37** | −2.51* | −1.06 |
| Fgg | NM_133862.1 | −2.36** | −2.95** | −1.25 |
| Nr1h4 | NM_009108.1 | −2.33** | −3.16* | −1.35 |
| Kng1 | NM_023125.2 | −2.32** | −2.87** | −1.24 |
| Slc7a9 | NM_021291.1 | −2.28* | −3.04* | −1.34 |
| Slc7a9 | NM_021291.1 | −2.27** | −2.82* | −1.24 |
| Irf6 | NM_016851.2 | −2.25** | −2.43* | −1.08 |
| Trf | NM_133977.2 | −2.19** | −2.37** | −1.08 |
| Sema4a | NM_013658.2 | −2.16* | −2.60* | −1.20 |
| Serpina1b | NM_009244.4 | −2.16** | −2.62** | −1.21 |
| Gipc2 | NM_016867.1 | −2.15** | −2.56* | −1.19 |
| Kng2 | NM_201375.1 | −2.15** | −2.59* | −1.20 |
| Igfbp6 | NM_008344.2 | −2.06** | −3.64* | −1.76 |
| Ostb | NM_178933.2 | −2.08** | −1.83 | 1.14 |
| Pmp22 | NM_008885.2 | −2.06* | −1.98 | 1.04 |
| Fbp2 | NM_007994.3 | −3.62** | −2.27 | 1.59 |
| Apoa4 | NM_007468.2 | −3.25** | −3.06 | 1.06 |
| Bex2 | NM_009749.1 | −3.19** | −2.71 | 1.18 |
| Eps8l3 | NM_133867.1 | −3.00** | −2.13 | 1.40 |
| Slc23a3 | NM_194333.3 | −2.92** | −2.97 | −1.02 |
| Lbp | NM_008489.2 | −2.89** | −2.39 | 1.21 |
| Pdzk1ip1 | NM_026018.2 | −2.81** | −2.62 | 1.07 |
| Upk3b | NM_175309.3 | −2.78** | −2.27 | 1.23 |
| Sepp1 | NM_009155.3 | −2.74** | −2.60 | 1.06 |
| Cyp2c70 | NM_145499.1 | −2.65** | −2.28 | 1.16 |
| Fmo1 | NM_010231.2 | −2.59** | −2.44 | 1.06 |
| Upk3b | NM_175309.3 | −2.43** | −2.05 | 1.19 |
| Rnase4 | NM_201239 | −2.32** | −2.76 | −1.19 |
| Apoc1 | NM_007469.3 | −2.31* | −2.07 | 1.11 |
| Apoc1 | NM_007469.3 | −2.30* | −2.07 | 1.11 |
| Gpc3 | NM_016697.2 | −2.25* | −2.23 | 1.01 |
| Timd2 | NM_134249.3 | −2.16* | −2.39 | −1.11 |
| Pcdh24 | NM_001033364.2 | −2.15** | −2.29 | −1.07 |
| Gldc | NM_138595.1 | −2.12** | −2.18 | −1.03 |
| Bglap-rs1 | NM_031368.3 | −2.03* | −2.13 | −1.05 |
| Abcc10 | NM_170680.2 | −2.93** | 1.01 | 2.96* |
| Prss22 | NM_133731.1 | −2.70** | −1.11 | 2.44* |
Significantly reduced in SCNT
*p < 0.05; **p < 0.01
Biological process and functional prediction of the differentially expressed genes
We further analyzed the data to obtain more insights into the biological processes and functions of the differentially expressed genes. The distribution of 206 genes that showed differential expression (of at least 2-fold) between the SCNT placentas and the controls, as well as their distribution in different gene ontology (GO) categories, is given in Additional file 2: Figure S1 and Additional file 3: Figure S2. GO-based analysis was performed using the Panther database (http://www.pantherdb.org). The GO terms under the category “biological process” that were most represented (>7%) in the SCNT placentas included “signal transduction (14%),” “immunity and defense (10%),” “transport (8%),” “protein metabolism and modification (8%),” and “developmental processes (7%).” In particular, the proportion of genes under “biological process unclassified” was 12% (Additional file 2: Figure S1). Under the category of “molecular function,” genes were classified into 27 categories by GO, the most represented ones being those for “select-regulatory molecule (9%),” “signaling molecule (9%),” “transporter (7%),” and “oxidoreductase (7%).” Sixteen percent of genes were categorized under “molecular function unclassified” (Additional file 3: Figure S2). The number of classified genes constitutes the number of categories calculated after excluding the overlapping ones.
Imprinting gene expression in the clones
Next, we focused on the expression profiles of imprinted genes in placentas. Of the 34 imprinted genes identified, none showed differences (>2-fold) in expression between the SCNT and aggregated SCNT placentas. Of these 34 genes, two Slc22a18; Slc38a4 showed higher expression (>2-fold) in the aggregated SCNT placenta than in the controls. Conversely, Igfbp6 was down-regulated (>2-fold) in the SCNT placentas than in the controls. Six genes Ppp1r9a, Tssc4, Ascl2, Cd81, Pon2, and Slc22a2 were placenta-specific imprinted genes that are expressed on the maternal allele in mice and humans. All these six genes were similarly expressed between the SCNT, aggregated SCNT, and control placentas. Most of the imprinted genes showed lower expression in SCNT placentas than in the controls (Table 5).
Table 5.
Imprinted gene expression
| Gene symbol | Accession No. | Folder Δ SCNT/Con | Folder Δ Agg/Con | Gene symbol | Accession No. | Folder Δ SCNT/Con* | Folder Δ Agg/Con |
|---|---|---|---|---|---|---|---|
| Atp10a | NM_009728.1 | 1.22 | 1.25 | Slc22a4 | NM_019687.3 | −1.11 | 1.35 |
| Cdkn1c | NM_009876.3 | 1.01 | −1.02 | Ube3a | NM_011668.2 | −1.05 | −1.03 |
| Dcn | NM_007833.4 | −1.56 | −1.18 | Zim1 | NM_011769.3 | −1.32 | −1.27 |
| Gnas | NM_201617.1 | −1.10 | 1.04 | Slc38a4 | NM_027052.3 | 2.32** | 1.55 |
| Meg1/Grb10 | NM_010345 | −1.75 | −1.05 | Cdkn1c | NM_009876.3 | 1.01 | −1.02 |
| H19 | NR_001592.1 | 1.02 | 1.10 | Igfbp6 | NM_008344.2 | −3.64** | −1.76 |
| Igf2 | NM_010514.2 | −1.01 | −1.02 | Igfbp2 | NM_008342.2 | −1.65 | 1.29 |
| Igf2r | NM_010515.1 | −1.35 | −1.11 | Xist | NR_001463.2 | 1.24 | 1.03 |
| Impact | NM_008378.2 | −1.20 | 1.04 | Ppp1r9aa | NM_181595.3 | −1.44 | −1.05 |
| Ins1 | NM_008386.3 | −1.03 | −1.01 | Tssc4a | NM_020285.1 | 1.06 | −1.02 |
| Ins2 | NM_008387.3 | 1.26 | 1.10 | Ascl2a | NM_008554.2 | 1.81 | 1.52 |
| Peg1/Mest | NM_008590.1 | −1.41 | 1.00 | Cd81a | NM_133655.1 | −1.42 | −1.18 |
| Peg10 | NM_001040611.1 | 1.10 | 1.07 | Pon2a | NM_183308.2 | 1.20 | 1.16 |
| Peg3 | NM_008817.2 | −1.16 | 1.04 | Slc22a2a | NM_013667.2 | −1.02 | 1.06 |
| Rasgrf1 | NM_011245.1 | 1.17 | 1.24 | ||||
| Ndn | NM_010882.3 | −1.23 | −1.06 | ||||
| Nnat | NM_010923.2 | −1.62 | 1.13 | ||||
| Slc22a18 | NM_008767.2 | 1.32 | 2.95* |
Significantly reduced in SCNT
aPlacenta-specific imprinted genes in mice and human
*p < 0.05; **p < 0.01
Gene expression analysis by qRT- PCR
To validate our microarray analysis, we performed qRT-PCR analysis for the 12 genes identified (Fig. 2). With regard to the down-regulated genes that were identified by microarray in SCNT placentas, three Chac1, Slpi, and Nrn1l were confirmed to be down-regulated in the SCNT placentas by qRT-PCR. Eight genes Plac1, Slc38a4, Rprml, Pla2g4f, Pla2g4d, Hsd17β7, Hmox1, and Car2 were identified as up-regulated by >2-fold in the SCNT placentas. Six of these genes Slc38a4, Rprml, Pla2g4f, Pla2g4d, Hsd17b7, and Car2 were confirmed to be up-regulated by qRT-PCR (Table 6). The fold change in expression of H19, a known imprinting gene, was found to be 1.1 in the microarray analysis indicating, that its expression level was similar in the SCNT placentas and controls, and confirming that the results of qRT-PCR correlated with those of the microarray analysis. However, quantitative gene expression analysis in individual placentas showed that the expression levels varied over a wide range among the five SCNT placentas from the normal placental state.
Fig. 2.
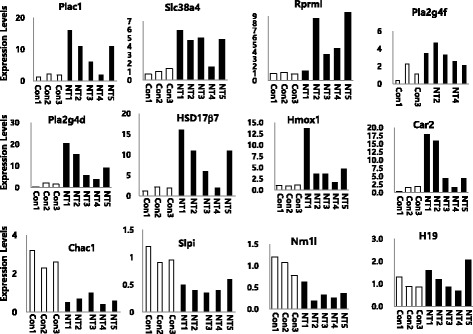
Quantitative real time PCR analysis. Twelve genes from different categories were chosen for qRT-PCR analyses. Eight genes were upregulated in the SCNT placentas. The other three genes were upregulated in the control group. H19 showed almost the same expression pattern. The β-actin gene was used as the endogenous control
Table 6.
Validation of microarray results using quantitative real-time PCR
| Gene symbol | Accession No. | Microarray folder Δ, SCNT/Con | Expression | qRT-PCR folder Δ, SCNT/Con |
|---|---|---|---|---|
| Plac1 | NM_019538.3 | 2.73** | ↑NT | 6.54 |
| Slc38a4 | NM_027052.3 | 2.32** | ↑NT | 4.04* |
| Rprml | NM_001033212.1 | 3.25** | ↑NT | 4.51 |
| Pla2g4f | NM_001024145.1 | 4.10** | ↑NT | 3.13* |
| Pla2g4d | NM_001024137.1 | 3.70** | ↑NT | 9.04 |
| Hsd17b7 | NM_010476.3 | 3.24** | ↑NT | 7.66 |
| Hmox1 | NM_010442.1 | 2.73** | ↑NT | 4.30 |
| Chac1 | NM_026929.3 | −2.27* | ↓NT | −8.41 |
| Car2 | NM_009801.3 | 3.95** | ↑NT | 6.11 |
| Slpi | NM_011414.2 | −4.56** | ↓NT | −4.06 |
| Nrn1l | NM_175024.3 | −2.49** | ↓NT | −3.05** |
| H19 | NR_001592.1 | 1.10 | 1.19 |
*Fold change values with superscripts were significantly different
*p < 0.05; **p < 0.01
Discussion
This study was undertaken to profile gene expression in the placentas of SCNT and aggregated SCNT and in vivo fertilization placentas by microarray analysis. We obtained a list of up- and down-regulated genes showing >2-fold difference between the SCNT and control placentas. The number of upregulated genes was lower by 75% in the aggregated SCNT placentas than in the SCNT placentas (206 → 52). The gene expression profiles of aggregated SCNT placentas were more similar to those of controls than to the profiles of SCNT placentas.
The data presented here indicate that the number of differentially deregulated genes in the SCNT placentas was decreased by 94.2% (85 → 5) in the aggregated SCNT placentas. These results are consistent with those of a direct comparison between gene expression in cumulus and ES cells of SCNT placentas, where there was a similar number of deregulated genes between both cell types [13]. Inappropriate reprogramming frequently occurs in somatic cell-cloned embryos [21], resulting in various deregulated gene expression patterns and epigenetic modifications in both the placenta and fetus [13]. Thus, the differences in gene expression were remarkably reduced by the aggregated SCNT method. The majority of aberrantly expressed genes were common to placentas cloned with ES or cumulus cells [13]. This indicated that placentomegaly in cloned mice is independent of the nuclear source of donor cells [6, 22, 23]. Therefore, aggregation of the tetraploid embryos utilized in this study have the key potential to reduce aberrant gene expression during the production of cloned mice, regardless of the nucleus source.
Many abnormalities in cloned animals suggest imprinting disruptions [24]. Placentomegaly was observed upon deregulation of imprinting genes such as H19 [12], Esx1 [10], and Ipl [11]. Although H19 was shown to be one of the variable genes among cloned animals, its expression showed no variability in the presented microarray results, confirming previous results [13, 25]. It is also reported that the expression of insulin-like growth factor 2 receptor (Igf2r) was increased in placentomegaly [26]. In the present study, Igf2r expression was marginally reduced. Most of the identified imprinted genes showed a decreased expression. These results indicate that the decreased expression of those imprinted genes is caused by reduced expression of the normally active allele [25, 27]. However, only three genes Slc22a18, Slc38a4, and Igfbp6 were expressed differentially between both placentas of SCNT and control placentas.
In the “biological process” category, the largest number of deregulated genes represented signal transduction (14%) proteins. In contrast, the largest number of genes in the molecular function category remained unclassified. Thus, it was very difficult to identify the specific causative genes of placentomegaly in SCNT placenta, but the condition seems to be caused by multiple-gene dysfunction. In the present study, we assessed differences in the expression of placenta-specific genes between SCNT and controls by qRT-PCR. These differences in expression were confirmed for eight genes. Plac1, a placenta-specific gene, is known to be expressed exclusively by the cells of the trophoblastic lineage in mice [28]. The other seven genes Slc38a4, Rprml, Pla2g4f, Hsd17b7, Hmox1, Car2, and Pla2g4d were specifically expressed in SCNT placentas. In the present study, Plac1 is considered a candidate gene involved in placentomegaly in NT placentas, as reported previously [14]. Thus, these genes need to be systematically studied to resolve placentomegaly.
The altered expression of hundreds of genes in SCNT placentas may be related to the high mortality rate of cloned embryos [13]. According to Miki et al. [19], the extraembryonic lineages could be composed of tetraploid cells, the population of which was increased in full-term placental tissues. Specifically, tetraploid chimeras are considered the most outstanding result, since they enable the production of whole stem cell-derived mice offspring, whereas offspring could not be produced using the inner cell mass and pluripotent cells in chimeric rhesus monkeys [20, 29]. In the present study, our results are consistent with those of previous reports indicating that most clones show gene expression abnormalities resulting in subtle phenotype changes [30, 31], premature death [32], placental hyperplasia [19], or obesity [33].
These results are thought to be caused by aggregation of tetraploid embryos leading to the recovery of downregulated gene expression in the SCNT placentas.
Conclusions
In summary, we present list of up- and down-regulated genes in the two types of SCNT and in vivo fertilization placentas. The expression of 206 (1.6%) of the 12,816 genes was found to be different by at least 2-fold between the SCNT placentas and controls. Further, 159 genes showed differential expression between the SCNT placentas and the aggregated SCNT placentas. However, gene expression profiles of the aggregated SCNT placentas were more similar to those of the controls than to those of the SCNT placentas. These results indicate that the aggregation SCNT technique using tetraploid embryos considerably decreased the number of deregulated genes by 94.2% (85 → 5) in the SCNT placentas. Therefore, aggregation with tetraploid embryos reduced abnormal gene expression in a genome-wide manner in the cloned placentas. Further studies will be needed to outline the molecular and functional mechanisms underlying abnormal expression of placenta-specific genes derived from tetraploid and cloned embryos.
Additional files
List of primers used for real-time PCR. Genes from five SCNT placentas and three control placentas were analyzed. Twelve genes from different categories were chosen for qRT-PCR analyses. The gene for β-actin was used as the endogenous control. (PPTX 65 kb)
Gene ontology of biological process. Gene ontology (GO) pie diagram of >2-fold differentially expressed genes between control and SCNT placentas. The upregulated or downregulated genes are categorized by the GO term “biological process.” (PPTX 152 kb)
Gene ontology of molecular function. Gene ontology (GO) pie diagram of >2-fold differentially expressed genes between control and SCNT placentas. The upregulated or downregulated genes are categorized by the GO term “molecular function”. (PPTX 133 kb)
Acknowledgements
The authors thank Dr. HW Seong (Institute of Animal Science) for his helpful discussion.
Funding
This study was supported by the KRIBB Research Initiative Program (KGM4251622), Republic of Korea.
Availability of data and materials
All the data supporting the findings are contained within the manuscript and Additional files (Additional file 2: Figure S1 and Additional file 3: Figure S2, Additional file 1: Table S1).
Authors’ contributions
BWS and CWP performed the experiments and analyzed the data. MHK and KSM designed the experiments and contributed to manuscript preparation. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
The protocol was approved by the Committee on Ethics of Animal Experiments at the Hankyong National University (Permit Number: 2014–4).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ES
Embryonic stem
- GO
Gene ontology
- LOS
Large offspring syndrome
- qRT-PCR
Quantitative real-time PCR
- SCNT
Somatic cell nuclear transfer
References
- 1.Wakayama T, Rodriquez I, Perry AC, Yanagimachi R, Mombaerts P. Mice cloned from embryonic stem cells. Proc Natl Acad Sci U S A. 1999;96:14984–9. doi: 10.1073/pnas.96.26.14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3:155–63. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- 3.De Sousa PA, Dobrinsky JR, Zhu J, Archibald AL, Ainslie A. Somatic cell nuclear transfer in the pig: control of pronuclear formation and integration with improved methods for activation and maintenance of pregnancy. Biol Reprod. 2002;66:642–50. doi: 10.1095/biolreprod66.3.642. [DOI] [PubMed] [Google Scholar]
- 4.Heyman Y, Chavatte-Palmer P, LeBourhis D, Camous S, Vignon X, Renard JP. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol Reprod. 2002;66:6–13. doi: 10.1095/biolreprod66.1.6. [DOI] [PubMed] [Google Scholar]
- 5.Sim BW, Min KS. Production of cloned mice by aggregation of tetraploid embryo. Animal Cells Syst. 2014;18:324–32. doi: 10.1080/19768354.2014.948488. [DOI] [Google Scholar]
- 6.Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod. 2001;65:1813–21. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- 7.Singh U, Fohn LE, Wakayama T, Ohgane J, Steinhoff C, Lipkowitz B, Schulz R, Orth A, Ropers HH, Behringer RR, Tanaka S, Shiota K, Yanagimachi R, Nuber UA, Fundele R. Different molecular mechanisms underlie placental overgrowth phenotypes caused by interspecies hybridization, cloning, and Esx1 mutation. Dev Dyn. 2004;230:149–64. doi: 10.1002/dvdy.20024. [DOI] [PubMed] [Google Scholar]
- 8.Shinmen A, Honda A, Ohkawa M, Hirose M, Ogonuki N, Yuzuriha M, Miki H, Mochida K, Inoue K, Abe K, Ito M, Ogura A. Efficient production of intersubspecific hybrid mice and embryonic stem cells by intracytoplasmic sperm injection. Mol Reprod Dev. 2007;74:1081–8. doi: 10.1002/mrd.20612. [DOI] [PubMed] [Google Scholar]
- 9.Van Thuan N, Wakayama S, Kishigami S, Ohta H, Hikichi T, Mizutani E, Bui HIT, Wakayama T. Injection of somatic cell cytoplasm into oocytes before intracytoplasmic sperm injection impairs full-term development and increases placental weight in mice. Biol Reprod. 2006;74:865–73. doi: 10.1095/biolreprod.105.047803. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet. 1998;20:309–11. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- 11.Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B. Placental overgrowth in mice lacking the imprinted gene lpl. Proc Natl Acad Sci U S A. 2002;99:7490–5. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–42. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub T, Jaenisch R. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci U S A. 2002;99:12889–94. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suemizu H, Aiba K, Yoshikawa T, Sharov AA, Shimozawa N, Kamaoki N, Ko MS. Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev Biol. 2003;253:36–53. doi: 10.1006/dbio.2002.0870. [DOI] [PubMed] [Google Scholar]
- 15.Everts RE, Chavatte-Palmer P, Razzak A, Hue I, Green CA, Oliveira R, Vignon X, Rodriguez-Zas SL, Tian XC, Yang X, Renard JP, Lewin HA. Aberrant gene expression patterns in placentomes are associated with phenotypically normal and abnormal cattle cloned by somatic cell nuclear transfer. Physiol Genomics. 2008;33:65–77. doi: 10.1152/physiolgenomics.00223.2007. [DOI] [PubMed] [Google Scholar]
- 16.Chavatte-Palmer P, Heyman Y, Richard C, Monget P, Le Bourhis D, Kann G, Chilliard Y, Vignon X, Renard JP. Clinical, hormonal, and hematologic characteristics of bovine calves derived from nuclei from somatic cells. Biol Reprod. 2002;66:1596–603. doi: 10.1095/biolreprod66.6.1596. [DOI] [PubMed] [Google Scholar]
- 17.Constant F, Guillomot M, Heymann Y, Vignon X, Laigre P, Servely JL, Renard JP, Chavatte-Palmer P. Large offspring or large placentas syndrome? Morphometric analysis of late gestation bovine placentomes from somatic nuclear transfer pregnancies complicated by hydrallantois. Biol Reprod. 2006;75:122–30. doi: 10.1095/biolreprod.106.051581. [DOI] [PubMed] [Google Scholar]
- 18.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Tilghman SM. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1998;90:8424–8. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miki H, Wakisaka N, Inoue K, Ogonuki N, Mori M, Kim JM, Ohta A, Ogura A. Embryonic rather extraembryonic tissues have more impact on the development of placental hyperplasia in cloned mice. Placenta. 2009;30:543–6. doi: 10.1016/j.placenta.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Tachibana M, Sparman M, Ramsey C, Ma H, Lee HY, Pendedo MCT, Mitalipov S. Generation of chimeric rhesus monkeys. Cell. 2012;148:285–95. doi: 10.1016/j.cell.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y, Kono T. Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol Reprod. 2008;78:568–76. doi: 10.1095/biolreprod.107.064634. [DOI] [PubMed] [Google Scholar]
- 22.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–74. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 23.Wakayama T, Yanagimachi R. Effect of cytokinesis inhibitors, DMSO and the timing of oocyte activation on mouse cloning using cumulus cell nuclei. Reproduction. 2001;122:49–60. doi: 10.1530/rep.0.1220049. [DOI] [PubMed] [Google Scholar]
- 24.Mann MR, Chung YG, Nolen LD, Verna RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69:902–14. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- 25.Inoue K, Kohda T, Lee J, Ogonuki N, Mochida Y, Tanemura K, Kaneko-Ishino T, Ishino F, Ogura A. Faithful expression of imprinted genes in cloned mice. Science. 2002;295:297. doi: 10.1126/science.295.5553.297. [DOI] [PubMed] [Google Scholar]
- 26.Sferruzzi-Perri A, Vaughan OR, Coan PM, Suciu MC, Darbyshire R, Constancia M, Burton GJ, Fowden AL. Placental-specific Igf2 deficiency alters developmental adaptations to under nutrition in mice. Endocrinology. 2011;152:3202–12. doi: 10.1210/en.2011-0240. [DOI] [PubMed] [Google Scholar]
- 27.Thurston A, Taylor J, Gardner J, Sinclair KD, Young LE. Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage. Reproduction. 2008;135:29–40. doi: 10.1530/REP-07-0211. [DOI] [PubMed] [Google Scholar]
- 28.Fant M, Weisoly DL, Cocchia M, Huber R, Khan S, Lunt T, Schlessinger D. Plac1, a trophoblast-specific gene, is expressed throughout pregnancy in the human placenta and modulated by keratinocyte growth factor. Mol Reprod Dev. 2002;63:430–6. doi: 10.1002/mrd.10200. [DOI] [PubMed] [Google Scholar]
- 29.Polejaeva I, Mitalipov S. Stem cell potency and the ability to contribute to chimeric organisms. Reproduction. 2013;145:81–8. doi: 10.1530/REP-12-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rideout WM, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the gene. Science. 2001;293:1093–8. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 31.Tamashiro KL, Sakai RR, Yamazaki Y, Wakayama T, Yanagimachi R. Developmental, behavioral, and physiological phenotype of cloned mice. Adv Exp Med Biol. 2007;591:72–83. doi: 10.1007/978-0-387-37754-4_5. [DOI] [PubMed] [Google Scholar]
- 32.Ogonuki N, Inoue K, Yamamoto Y, Noguchi Y, Tanemura K, Suzuki O, Nakayama H, Doi K, Ohtomo Y, Satoh M, Nishida A, Ogura A. Early death of mice cloned from somatic cells. Nat Genet. 2002;30:253–4. doi: 10.1038/ng841. [DOI] [PubMed] [Google Scholar]
- 33.Tamashiro KL, Wakayama T, Akutsu H, Yamazaki Y, Lachey JL, Wortman MD, Seeley RJ, D’Alessio DA, Woods SC, Yanagimachi R, Sakai RR. Cloned mice have an obese phenotype not transmitted to their offspring. Nat Med. 2002;8:262–7. doi: 10.1038/nm0302-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for real-time PCR. Genes from five SCNT placentas and three control placentas were analyzed. Twelve genes from different categories were chosen for qRT-PCR analyses. The gene for β-actin was used as the endogenous control. (PPTX 65 kb)
Gene ontology of biological process. Gene ontology (GO) pie diagram of >2-fold differentially expressed genes between control and SCNT placentas. The upregulated or downregulated genes are categorized by the GO term “biological process.” (PPTX 152 kb)
Gene ontology of molecular function. Gene ontology (GO) pie diagram of >2-fold differentially expressed genes between control and SCNT placentas. The upregulated or downregulated genes are categorized by the GO term “molecular function”. (PPTX 133 kb)
Data Availability Statement
All the data supporting the findings are contained within the manuscript and Additional files (Additional file 2: Figure S1 and Additional file 3: Figure S2, Additional file 1: Table S1).


