Abstract
Histone ubiquitination plays a non-degradative role in regulating transcription and the DNA damage response. A mechanistic understanding of this chromatin modification has lagged that of small histone modifications because of the technical challenges in preparing ubiquitinated nucleosomes. The recent structure of the DUB module of the SAGA coactivator complex bound to a nucleosome containing monoubiquitinated H2B has provided the first view of how specialized subunits target this enzyme to its substrate. Single particle electron microscopy of the intact SAGA coactivator suggests how the DUB module and histone acetyltransferase module engage a nucleosomal substrate. A cryo EM study of 53BP1 bound to nucleosomes containing ubiquitinated H2A and H4 methylated at K20 extends our understanding of recognition of biologically distinct combinations of chromatin marks through multivalent interactions.
Introduction
The information in the eukaryotic genome is accessed in the context of chromatin, a nucleoprotein complex consisting of DNA and histone proteins. The fundamental unit of chromatin, the nucleosome core particle, comprises about 147 base pairs of DNA wrapped around an octamer core containing two copies each of histones H2A, H2B, H3, and H4 [1]. Post-translational modifications of specific histone residues serve as an additional layer of “metadata” that regulates all processes requiring access to genomic DNA. Histone modifications play a central role in regulating transcription, with specific combinations of histone modifications associated with activation or repression of gene expression. These distinct patterns of modifications, known as the “histone code” [2], provide docking sites for the transcriptional machinery as well as additional chromatin-modifying enzymes, modulate nucleosome dynamics and likely play additional roles that are yet to be understood. Histone modifications also play an important role in mediating the response to DNA damage [3] and chromosome condensation during mitosis [4]. Proteins that govern the histone code can be categorized as histone “readers,” which bind to specific histone marks, “writers,” the enzymes that covalently attach the modifications, and “erasers,” the enzymes that remove the modifications.
Whereas most histone modifications consist of small chemical groups, histones can also be modified by the covalent attachment of the 76-amino acid protein, ubiquitin, to the ε-amino group of lysine [5,6]. Histone ubiquitination serves a signaling, rather than a degradative role, and its relatively large size and complexity distinguishes it from small side chain modifications such as acetylation, methylation, or phosphorylation. Ubiquitin is attached to specific lysine residues by a cascade of E1/E2/E3 conjugation machinery, with the E2/E3 pair conferring substrate specificity [7]. Ubiquitination of histone H2B, Lys120 (H2BK120ub) is enriched at actively transcribed genomic loci [8], whereas ubiquitin attached to histone H2A, Lys 119 (H2AK119ub) is a mark found in heterochromatic regions [9]. In response to DNA double strand breaks (DSBs), histone H2A is also ubiquitinated at K13/K15 (forming H2AK13ub or H2AK15ub, respectively), where this mark is required for recruiting the DSB repair machinery [10]. Histones H1, H3 and H4 have also been shown to be ubiquitinated in response to UV irradiation [11,12]. In many cases, there is “cross-talk” between histone ubiquitination and other types of histone modifications, with the ubiquitin mark either recognized in conjunction with specific marks such as methyl-lysine, or examples where histone ubiquitination is a prerequisite for deposition of other histone modifications such as methylation or acetylation [13–15].
Until recently, our structural understanding of how histone ubiquitination marks are attached, recognized and removed has been restricted to studies of the readers, writers and erasers in the absence of their chromatin targets. This changed with the 2014 report of the structure of the PRC1 E3/E2 enzyme pair bound to a nucleosome [16], which provided the first view of how a “writer” complex specifically targets histone residue H2AK119 in a chromatin context [17]. In this review, we describe recent advances that have shed light on how a “reader” recognizes ubiquitinated histones in conjunction with methyl marks, how an “eraser” complex specifically removes ubiquitin from histone H2B, as well as how the large, multifunctional chromatin-binding enzymatic complexes coordinate the structural dynamics of multiple subcomplexes to modify nucleosomes.
Docking of the SAGA DUB module on ubiquitinated nucleosomes
The SAGA (Spt-Ada-Gcn5 acetyltransferase) transcriptional coactivator complex integrates histone code reader, writer and eraser functions and is conserved across all eukaryotes [18]. The 19-protein SAGA complex acetylates histone H3 [19], removes monoubiquitin from histone H2B-K123(yeast)/K120(humans) [20], and recognizes a variety of histone modifications including methyl lysine [21] and acetyl lysine [22] via distinct reader domains. SAGA also promotes formation of the transcription pre-initiation complex and binds directly to the TATA-binding protein (TBP) subunit of the general transcription factor, TFIID [23]. Mass spectrometry and deletion studies [24,25] have shown that the proteins in the 1.8 MDa yeast SAGA complex are organized into four distinct subcomplexes: the deubiquitinating (DUB) module; the histone acetyltransferase (HAT) module; the Spt module, which interacts with transcription factors, including TBP; and the TAF module, which is thought to maintain the architecture of the overall complex. These activities are coordinated to transition chromatin to an “open” state favoring recruitment of RNAPII and its cofactors.
While there is no high-resolution structure of the intact SAGA complex, crystal structures of small pieces of the SAGA complex have provided snapshots of some of its functional groups. These include structures of the Gcn5 acetyltransferase domain in the presence [26] and absence [27] of its cofactor, acetyl-CoA, and H3 peptide, the acetyllysine-bound bromodomain of Gcn5 [28], and the Sgf29 tandem Tudor domain bound to methylated histone peptides [21]. The largest portion of SAGA for which there is atomic resolution information is the four-protein yeast DUB module, which consists of the catalytic subunit, Ubp8, complexed with Sgf11, Sus1 and Sgf73 [20,29–31]. Crystal structures of the DUB module with and without bound ubiquitin [32,33] revealed how the four subunits adopt an interwoven structure containing two lobes: a catalytic lobe containing the Ubp8 cysteine protease USP domain and the C-terminal zinc finger of Sgf11, and an assembly lobe containing the Ubp8 ZnF-UBP domain, the Sgf11 N-terminus, Sus1 and Sgf73.
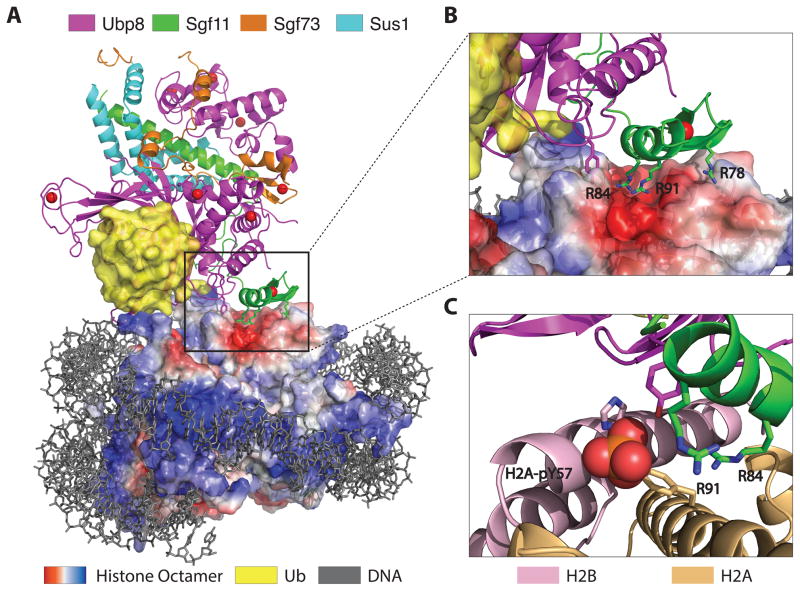
The way in which the subunits of the SAGA DUB module target this complex to its chromatin target was revealed by the recent crystal structure of the yeast DUB module bound to nucleosomes containing ubiquitinated H2B (Figure 1A) [34]. Ubiquitin was conjugated to H2B K120 via a non-hydrolyzable linkage, generating the necessary material for the 3.9 Å resolution structure of this ~390 kDa complex. The overall conformation of the DUB module is unchanged from the apo enzyme [33] or the structure with ubiquitin alone [32], indicating that the complex provides a rigid scaffold for recognizing its ubiquitinated substrate. The arginine-rich Sgf11 zinc finger, which is located adjacent to the Ubp8 active site, plays a pivotal role in docking the DUB module on the conserved nucleosome acidic patch, which is located in a cleft between histones H2A and H2B (Figure 2B). This mode of interaction, in which an “arginine anchor” [17] binds to the acidic patch, is shared by nearly all structures determined to date of proteins or peptides bound to nucleosomes: the LANA peptide [35], RCC1 [36], the Sir3-BAH domain [37], and the PRC1 complex composed of Ring1B and Bmi1 [16]. Ubp8 mediates additional contacts with the C-terminal helix of H2B, as well as with the conjugated ubiquitin. Interestingly, there are no contacts with the nucleosomal DNA other than a single predicated contact mediated by Ubp8 residue, Arg 374.
Figure 1.
Docking of the SAGA DUB module on H2B-ubiquitinated nucleosomes. (A) Structure of the yeast DUB module (Ubp8, Sgf11, Sgf73, Sus1) bound to a nucleosome core particle containing monoubiquitinated H2B. Surface representation of the histone octamer shows negative (red) and positive (blue) electrostatic potential. (B) Docking of the arginine-rich Sgf11 zinc finger (green) on the acidic cleft (red) between H2A and H2B. (C) Modeling showing position of phosphorylated H2A-Y57 at the interface between the DUB module and histones H2A and H2B.
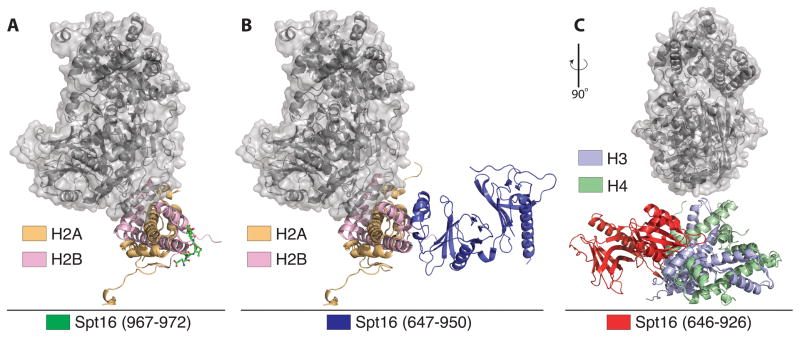
Figure 2.
Structural insights into relation of FACT and DUB module binding to histones. All panels show relative position of DUB module (gray surface) on either H2A/H2B or H3/H4. (A) Docking of DUB module relative to a peptide derived from the C-terminus of Spt16 solved to 1.80 Å resolution (PDB ID: 4WNN). (B) The Spt16M domain from Chaetomium thermophilum fused to H2B solved to 2.35 Å resolution (PDB ID: 4KHA). (C) Structure of the human FACT mid-AID domain bound to an H3/H4 tetramer solved to 2.98 Å resolution (PDB ID: 4Z2M).
The observed docking of the DUB module bound to a nucleosome explains the recent finding that phosphorylation of H2A-Y57 (H2A-Y57p) by casein kinase 2 (CK2) inhibits DUB module-dependent deubiquitination of H2B-Ub in both yeast and human cells [38]. Modeling of H2A-Y57p in the structure of the DUB module bound to a ubiquitinated nucleosome [34] indicated that this newly discovered histone PTM has the potential to alter interactions between the arginine anchor and the acidic patch (Figure 1C). This hypothesis was confirmed through use of a fully synthetic strategy to incorporate phosphotyrosine into histone H2A at Y57 [39]. Nucleosomes containing H2A-Y57p and H2B-Ub made by native chemical ligation [40] were used to confirm that this phosphorylation indeed inhibits DUB module activity in vitro [39].
H2B ubiquitination has been shown to promote recruitment of the histone chaperone, FACT [41], which promotes H2A/H2B eviction in advance of the transcribing RNA polymerase II and nucleosome reassembly in its wake [42]. It is not known whether SAGA deubiquitinates H2B before or after the nucleosome is disassembled, or whether an ejected H2A/H2B-Ub dimer bound to FACT could also be a substrate. Since interactions between the DUB module and the nucleosome are limited to the surface of the H2A/H2B heterodimer, ubiquitinated dimers could, in theory, also be targeted during transcription. While there is, as yet, no structural information for the full-length FACT Spt16/Pob3 heterodimer bound to H2A/H2B, several recent structures of FACT have mapped interactions between FACT domains and the histone octamer [43–45]. None of these structures indicate overlap between FACT and the DUB module docking site (Figure 2), leaving open the possibility that SAGA could remove ubiquitin from nucleosomes while FACT is still bound. FACT subunits have been shown to physically associate with SAGA in mass spectrometry experiments [24], consistent with a scenario in which FACT and SAGA may act on a nucleosome simultaneously. In vitro, the SAGA DUB module can deubiquitinate H2B in the presence of FACT when assayed on either H2A/H2B-Ub heterodimers or intact ubiquitinated nucleosomes [34]. Further studies will be needed to sort out the sequence of biochemical events during transcription in the presence of intact SAGA and the transcribing polymerase.
A broader role for SAGA subunits in directing H2B deubiquitination in humans
The pivotal role played by the Sgf11 subunit in both directing the catalytic Ubp8 subunit to its substrate [34] and maintaining Ubp8 in its active conformation [33,46] explains a recent report that multiple DUBs regulate H2B ubiquitination levels in human cells [47]. The human SAGA DUB module is composed of USP22, ATXN7L3, ENY2 and ATXN7, homologues of yeast Ubp8, Sgf11, Sus1 and Sgf73, respectively. A recent study showed that ATXN7L3 and ENY2 can also form complexes with two additional USP DUBs, USP27x and USP51, and target them to H2B-Ub [47]. The requirement of ATXN7L3 for H2B deubiquitination by USP22, USP27x, and USP51 suggests that all three use the ATXN7L3 zinc finger to dock the H2A/H2B acidic patch in a manner similar to that shown in the structure of the yeast DUB module bound to ubiquitinated nucleosomes.
Locating the DUB module in intact SAGA
A clear picture of the three-dimensional organization of the entire 19-protein SAGA complex will ultimately be needed to determine how histone deubiquitination is coordinates with SAGA’s other activities, including histone acetylation and preinitiation complex assembly [48,49]. A recent negative stain electron microscopy (EM) structure determined to 30.3 Å (EMDB ID: 2693) was used to locate the DUB module within intact yeast SAGA as well as determine how SAGA docks on an unmodified nucleosome core particle [50]. By determining structures of intact SAGA as well as deletions of Sgf73 solved to 25.8 Å (EMDB ID: 2694), which anchors the DUB module to the rest of the coactivator complex, the authors used observed differences in density to locate the DUB module proximal to the HAT module. This could explain why deletions of components of either module lead to reduced activity in the other subcomplex, even though they appear to fold independently [50]. In the negative stain structure, unmodified recombinant nucleosomes associate with the region identified as the DUB module, suggesting that DUB module can associate with nucleosomes even in the absence of conjugated ubiquitin. The position of the nucleosome relative to the DUB module does not, however, agree with the crystal structure of the isolated DUB module bound to ubiquitinated nucleosome [34]. This discrepancy may arise from the absence of ubiquitin, which forms extensive contacts with Ubp8, non-specific binding by SAGA, or from the difficulty in docking the crystal structure of the apo DUB module in a low resolution map. High resolution studies with nucleosomes containing H2B-Ub, and possibly additional histone modifications, will ultimately be needed to resolve this issue.
A separate EM study of the intact SAGA complex [51] pinpointed a role for the DUB module subunit, Sgf73, in modulating conformational flexibility of the entire complex. The first ~100 residues of Sgf73 form an integral part of the DUB module [31–33], while the remaining C-terminal portion of Sgf73 anchors this protein to the remainder of SAGA, contacting multiple subunits as judged by mass spectrometry and cross-linking studies [25]. Setiaputra and colleagues [51] utilized the GraFix cross-linking technique [52] to limit conformational exchange of SAGA particles. The authors thereby captured three discrete forms of SAGA, which they call the “arched” (EMDB ID: 6299), “curved” (EMDB ID: 6300), and “donut” (EMDB ID: 6301) conformations, determined at 45.3 Å, 41.7 Å, and 38.9 Å resolution, respectively. These conformations result from mobility of three distinct regions of the complex. The “head” appears largely globular; the elongated “tail” region, which was largely absent in the particles analyzed in previous studies; and the “torso” region, which connects the head and tail, composed of a “hinge” that associates with the tail and “shoulder” which does not. The tail travels over 50 Å throughout the dynamic range of conformations, with intermediates of each observed in class averaging, suggesting that the motions of SAGA are coordinated. Loss of the DUB module by deletion of Sgf73 significantly alters the distribution of conformations, making the donut form less frequent and reducing the number of particles in which the HAT module is present. The authors suggest that the DUB module may occupy a central region of SAGA toward which the tail curls, which contains the HAT complex. This further suggests that the DUB and HAT modules may contact one another in the donut conformation, enabling both enzymatic subcomplexes to act on the same nucleosome substrate.
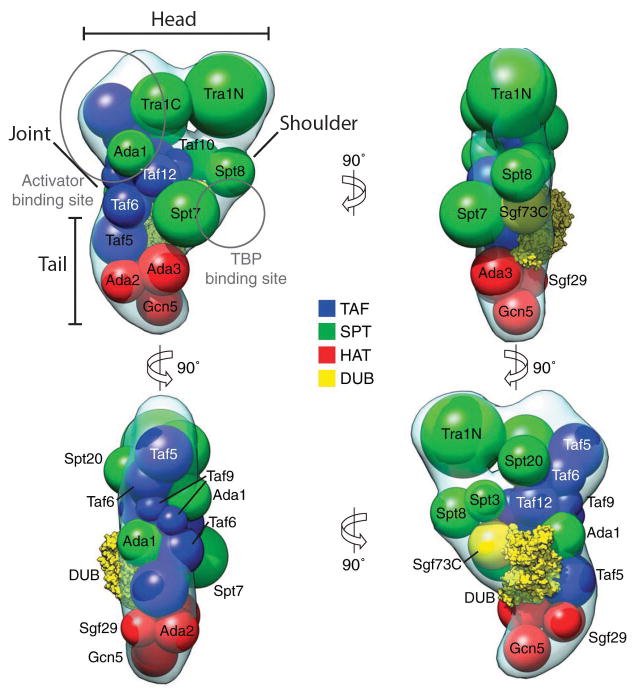
Setiaputra and colleagues also determined where each subunit is located within the complex by GFP-tagging proteins within each module and searching for additional density [51]. This approach, coupled with cross-linking mass spectrometry allowed the authors to construct a detailed map of SAGA in which the TAF module and Spt7, Spt20, and Tra1 of the SPT module form a highly interconnected core around which the rest of the subunits are peripherally arranged (Figure 3). Taken together, these results demonstrate that SAGA exists in a dynamic state, adopting discrete conformational intermediates that are coordinated by its subunits. The observed conformational exchange is likely an important mechanism whereby SAGA coordinates its multiple functionalities to properly regulate transcription.
Figure 3.
Model of the arrangement of SAGA subunits as determined by crosslinking mass spectrometry and electron microscopy subunit localization experiments. Sphere size is proportional to molecular weight. Binding sites of transcription factors are indicated, as determined by pull-down experiment and electron microscopy. Adapted from D. Setiaputra et al., 2015, Journal of Biological Chemistry, 290, p. 10057. © 2015 The American Society for Biochemistry and Molecular Biology with permission.
Recognition of histone ubiquitination and methylation in the DNA damage response
In addition to its role in transcription regulation, histone ubiquitination plays an important role in signaling the presence of DNA double strand breaks (DSBs) and in recruiting the necessary repair enzymes [3,53]. The ubiquitin E3 ligase, RNF168, ubiquitinates histone H2A at Lys 13/15 (forming H2AK13ub or H2AK15ub) near DNA double strand breaks (DSBs) [10]. This event triggers the recruitment of 53BP1 [54], a large protein that serves to determine the choice between non-homologous end joining and homologous recombination in response to a DSB [55]. 53BP1 is also essential to the accumulation of DNA damage response proteins, including EXPAND1 [56] and RIF1 [57]. 53BP1 uses its ubiquitination-dependent recruitment (UDR) domain to engage the H2AK13/15ub marks, while the proximal tandem Tudor domain of 53BP1 binds to H4 dimethylated at Lys 20 (forming H4K20me2) [54]. H4K20me2 is an abundant mark in the genome, found in 80% of nucleosomes in asynchronous, undamaged cells [58]. The 53BP1 protein associates weakly with H4K20me2 alone. However, binding studies indicate that 53BP1 affinity for chromatin is increased when ubiquitin is conjugated to H2AK13/15, triggering the accumulation of 53BP1 at damage foci.
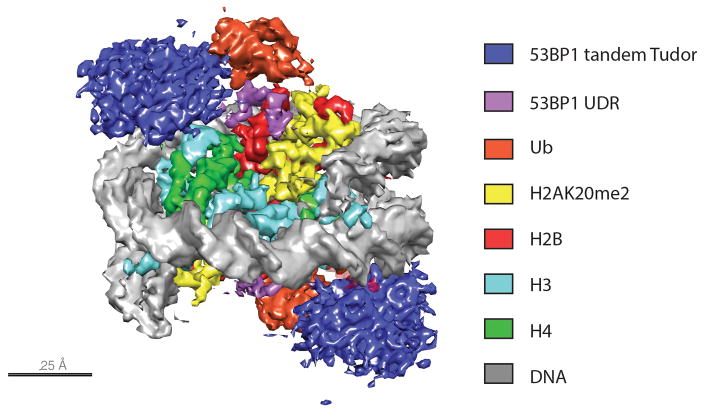
A recent cryo electron microscopy (cryo-EM) structure has provided insights into how 53BP1 recognizes tandem ubiquitin and methyl marks. Using nucleosomes ubiquitinated at H2AK15 and containing a methyl lysine mimic at residue 20 of histone H4, Wilson et al. [59] generated complexes with the minimal required fragment of 53BP1 (residues 1484–1631), which contains both the UDR and tandem Tudor domain, fused to glutathione-S-transferase (GST). The single particle reconstruction of the 53BP1-bound nucleosome was determined at 4.5 Å resolution (Figure 4) and the structure of a ubiquitinated nucleosome alone was determined at a resolution of 7.7 Å. A comparison of the structures revealed that ubiquitin is dynamic in the absence of 53BP1, whereas its position is constrained when 53BP1 is bound. Density corresponding to the UDR domain of 53BP1 was threaded between the ubiquitin and the H2B C-terminal helix, where its position was consistent with predicted electrostatic complementarity between the UDR, ubiquitin and the H2A/H2B acidic patch. The concerted engagement of the 53BP1 UDR with H2AK15ub and its tandem Tudor domain with H4K20me2 is required for high-affinity binding, in addition to contacts made with the H2A/H2B acidic patch and the H2B/H4 cleft. While electron density for the tandem Tudor domain was insufficient to unambiguously position it in the model, its corresponding density was centered upon H4K20me2. The authors tested whether H2BK120 ubiquitination might interfere with 53BP1 binding, due to the close approach of the UDR to the H2B C-terminal helix; however 53BP1 recruitment is unaffected by H2BK120ub, suggesting that these marks are not mutually exclusive. This recent structure demonstrates how chromatin-binding proteins attain specificity through coordinated binding of multiple histone PTMs, and suggests the potential for binding events to couple conformational changes of the modified nucleosome with downstream processes.
Figure 4.
Electron density at 4.5 Å resolution for the complex of the 53BP1 tandem Tudor-UDR construct bound to nucleosomes containing H2A ubiquitinated at K15 and H4 methylated at K20. All density is shown at threshold of 0.37 except that which corresponds to the 53BP1 tandem Tudor domain, shown at a threshold of 0.17. The 53BP1 UDR (purple) can be clearly seen threading between the disk face of the nucleosome and the tethered ubiquitin.
Conclusions
The recent findings discussed above leave the field poised for further interrogation, thanks to dual advances in chemistry and structural biology. Thanks to robust methodologies for incorporating ubiquitin at specific histone residues with either non-hydrolyzable [34,59] or native isopeptide linkages [14,40], as well other histone modifications, there are now multiple approaches to generating the needed chromatin templates. The recent advances in high-resolution cryo-electron microscopy [60] make it possible to obtain structural information on very large complexes that are typically not amenable to study by x-ray crystallography or NMR spectroscopy. While the challenges of determining atomic-resolution structures of transcriptional coactivators such as SAGA and DNA repair complexes bound to chromatin remain significant, in part due to the flexibility of the histone tails, the near future prospects for further advances are highly promising.
Highlights.
New advances have made it possible to determine structure of complexes containing ubiquitinated nucleosomes.
X-ray crystallographic and EM studies shed light on how the SAGA complex engages nucleosomes containing monoubiquitinated H2B.
The intact SAGA complex adopts multiple discrete conformations.
An EM study shows how 53BP1 recognizes tandem ubiquitin and methyl marks in the nucleosome.
Acknowledgments
Supported by grant GM-095822 from the National Institute of General Medical Sciences (C.W.) and by a Ruth L. Kirschstein National Research Service Award (M.T.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W, Neumann H. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343:77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 5.Wu RS, Kohn KW, Bonner WM. Metabolism of ubiquitinated histones. J Biol Chem. 1981;256:5916–5920. [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A, Rose IA. Identification of the active amino acid residue of the polypeptide of ATP-dependent protein breakdown. J Biol Chem. 1981;256:1525–1528. [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 8.Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 10.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 12*.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. This study showed that linker histone, H1, is modified by K63-linked polyubiquitin by the E3-E2 pair, RNF8-UBC13, during the DNA damage response. This is a critical event in the recruitment of downstream DNA repair machinery, including RNF168, which in turn ubiquitinates H2AK13/15. [DOI] [PubMed] [Google Scholar]
- 13.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 14.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. This structure of the ubiquitin E3 ligase complex, Ring1B-Bmi1, and the E2, UBCH5C, bound to a nucleosome core particle revealed revealed the multivalent interaction surface that facilitates binding and catalytic specificity of the PRC1 complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGinty RK, Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Curr Opin Struct Biol. 2016;37:54–61. doi: 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 20.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 23.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Han Y, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. This study utilized cross-linking and mass spectrometry to produce of comprehensive map of the connectivity between all of the SAGA subunits plus TATA-binding protein (TBP). Accompanying deletions studies also revealed cross-talk between the enzymatic activity of the DUB and HAT modules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 27.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 32.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. This structure of the four-protein SAGA DUB module bound to ubiquitinated nucleosomes reveals the role of one of the non-enzymatic subunits, Sgf11, in directing this complex to the ubiquitinated H2B within the nucleosome core particle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 36.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3. 0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Basnet H, Su XB, Tan Y, Meisenhelder J, Merkurjev D, Ohgi KA, Hunter T, Pillus L, Rosenfeld MG. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516:267–271. doi: 10.1038/nature13736. The authors describe a novel histone modification, phospho-H2A-Y57, which they show descreases Ubp8-dependent H2B deubiquitination and inhibits transcription in both yeast and human cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Jbara M, Maity SK, Morgan M, Wolberger C, Brik A. Chemical Synthesis of Phosphorylated Histone H2A at Tyr57 Reveals Insight into the Inhibition Mode of the SAGA Deubiquitinating Module. Angew Chem Int Ed Engl. 2016;55:4972–4976. doi: 10.1002/anie.201600638. This paper describes a method for generating fully synthetic histones, thus making it possible to incorporate a phosphotyrosine in the middle of histone H2A. This material made it possible to demonstrate that phospho-H2A-Y57 directly inhibits the ability of the SAGA DUB module to deubiquitinate H2B in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar KS, Spasser L, Ohayon S, Erlich LA, Brik A. Expeditious chemical synthesis of ubiquitinated peptides employing orthogonal protection and native chemical ligation. Bioconjug Chem. 2011;22:137–143. doi: 10.1021/bc1004735. [DOI] [PubMed] [Google Scholar]
- 41.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 42.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 43.Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, Zhang ET, Nijmeijer B, Kotthoff C, Rybin V, Amlacher S, et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–114. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 44*.Kemble DJ, McCullough LL, Whitby FG, Formosa T, Hill CP. FACT Disrupts Nucleosome Structure by Binding H2A–H2B with Conserved Peptide Motifs. Mol Cell. 2015;60:294–306. doi: 10.1016/j.molcel.2015.09.008. This paper presents a crystal structure of a complex between the peptide from the acidic C-terminus of Spt16, a subunit of the histone chaperone, FACT, bound to an H2A/H2B heterodimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016;30:673–686. doi: 10.1101/gad.274183.115. This structure of the FACT Mid-AID domain bound to an H3/H4 tetramer reveals an orientation that is mutually exclusive with the presence of H2A/H2B, suggesting that this FACT domain aids in H2A/H2B displacement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samara NL, Ringel AE, Wolberger C. A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity. Structure. 2012;20:1414–1424. doi: 10.1016/j.str.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, McIvor E, Li W, Zhang Y, Florens L, et al. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol Cell. 2016;62:558–571. doi: 10.1016/j.molcel.2016.03.030. The authors show that the human SAGA components ATXN7L3 and ENY2 (homologues of yeast Sgf11 and Sus1, respectively) can bind to mutiple USP-family deubiquitinating enzymes, targeting them to deubiquitinate histone H2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Dent SY. Functions of SAGA in development and disease. Epigenomics. 2014;6:329–339. doi: 10.2217/epi.14.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 50**.Durand A, Bonnet J, Fournier M, Chavant V, Schultz P. Mapping the deubiquitination module within the SAGA complex. Structure. 2014;22:1553–1559. doi: 10.1016/j.str.2014.07.017. This negative stain electron microscopy study locates the DUB module within the density of the full SAGA complex and showsg that unmodified nucleosomes bind near the putative DUB module site. [DOI] [PubMed] [Google Scholar]
- 51**.Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK. Conformational flexibility and subunit arrangement of the modular yeast Spt-Ada-Gcn5 acetyltransferase complex. J Biol Chem. 2015;290:10057–10070. doi: 10.1074/jbc.M114.624684. An electron microscopy study of yeast SAGA complex that has been stabilized by cross-linking reveals several distinct states, providing insight into the dynamic intermediates of this large complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 53.Smeenk G, Mailand N. Writers, Readers, and Erasers of Histone Ubiquitylation in DNA Double-Strand Break Repair. Front Genet. 2016;7:122. doi: 10.3389/fgene.2016.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huen MS, Huang J, Leung JW, Sy SM, Leung KM, Ching YP, Tsao SW, Chen J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell. 2010;37:854–864. doi: 10.1016/j.molcel.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59**.Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. The structural basis of modified nucleosome recognition by 53BP1. Nature. 2016;536:100–103. doi: 10.1038/nature18951. The authors use cryo-electron microscopy to map the binding of a fragment of 53BP1 on a nucleosome that includes histone H2A ubiquitinated at K15 and histone H4 dimethylated at K20, providing insights into the structural basis for multivalent recognition of histone marks. [DOI] [PubMed] [Google Scholar]
- 60*.Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. An excellent review of recent advances that have made it possible to use cryo-electron microscopy to determine structures at atomic resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]