Abstract
The family of the p53 tumor suppressive transcription factors includes p73 and p63 in addition to p53 itself. Given the high degree of amino-acid-sequence homology and structural organization shared by the p53 family members, they display some common features (i.e., induction of cell death, cell-cycle arrest, senescence, and metabolic regulation in response to cellular stress) as well as several distinct properties. Here, we describe the structural evolution of the family members with recent advances on the molecular dynamic studies of p53 itself. A crucial role of the carboxy-terminal domain in regulating the properties of the DNA-binding domain (DBD) supports an induced-fit mechanism, in which the binding of p53 on individual promoters is preferentially regulated by the KOFF over KON.
The transcription factors p63 and p73 have domain structures that are similar to those of p53. Recent studies on p53 have emphasized the critical role of the carboxy-terminal domain in regulating the DNA-binding domain.
STRUCTURE OF INDIVIDUAL p53 DOMAINS
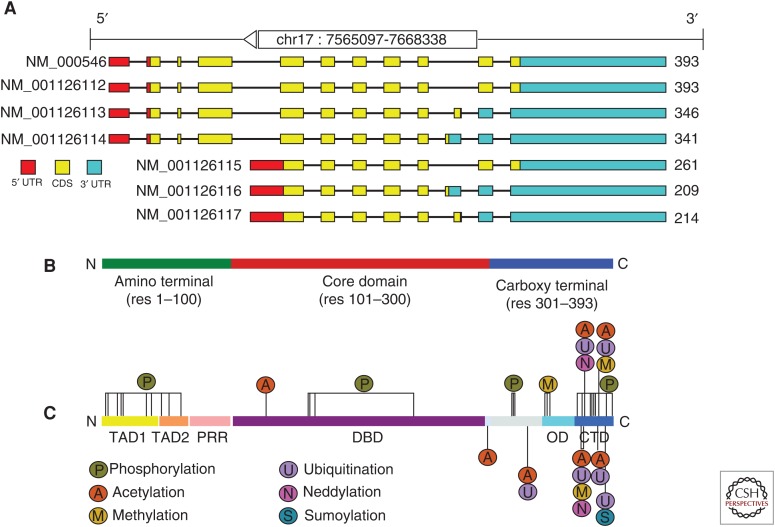
The p53 tumor-suppressor gene encodes a DNA-sequence-dependent transcription factor that preserves genome integrity through the regulation of relevant cellular pathways, including cell cycle, apoptosis, and cellular senescence (Dötsch et al. 2010; Joerger and Fersht 2010). The TP53 gene encodes nine different protein isoforms as a result of alternative splicing, alternative promoter usage, and alternative initiation sites of translation (Fig. 1A) (Bourdon et al. 2005; for more details, see Joruiz and Bourdon 2016). p53 is a nuclear phosphoprotein that is capable of either transactivation through the binding to specific DNA responsive elements (REs) (el-Deiry et al. 1992), or repressing transcription of promoters that do not contain binding sequences (see Fig. 1). Specificity, affinity, and cooperativity of p53 binding to DNA are all tightly related to its structural features.
Figure 1.
Structural organization of human p53. (A) Alternative splicing forms of the Trp53 gene. (B) Organization of the 393 residues of p53 protein into three major regions. (C) The sites of the most frequent posttranslational modifications are distributed all along the sequence, with the greatest concentrations at the transactivation domains (TADs), in the amino-terminal and carboxy-terminal domain (CTD) in the carboxy terminal. They include phosphorylation, acetylation, ubiquitination, methylation, neddylation, and sumoylation. PRR, Proline-rich region; DBD, DNA-binding domain; OD, oligomerization domain.
In its active conformation, the p53 protein is a tetramer formed by four identical chains of 393 residues each (Joerger and Fersht 2008). The p53 polypeptide displays a modular domain structure (Fig. 1B), comprising an amino-terminal transactivation domain (TAD, residues 1–61), a proline-rich region (PRR, residues 61–92), a central DBD (residues 94–292), a short tetramerization region (OD, residues 326–353), and a carboxy-terminal regulatory domain (CTD, residues 353–390) (Joerger and Fersht 2008).
The structure of the full-length p53 tetramer has been elucidated by a combination of experimental approaches, including NMR, electron microscopy, small-angle X-ray scattering, and FRET (Okorokov et al. 2006; Huang et al. 2009; Melero et al. 2011). In the absence of DNA, p53 forms an open cross-shaped structure with loosely coupled dimers interacting via the core domain. On DNA binding, the structure of p53 rigidifies, becoming more compacted. Because the high proportion of intrinsically disordered regions hampers crystallization, the majority of the structural studies have addressed only single domains or fractions of them.
The TAD, which is relevant for interaction with transcriptional coactivators and corepressors (Raj and Attardi 2016), is natively unfolded (Dawson et al. 2003; Chillemi et al. 2013). This region can be further divided into two subdomains: TAD1 (residues 1–40) and TAD2 (residues 41–60), which can independently activate transcription (Candau et al. 1997). The presence of intrinsically disordered segments in the TAD allows this domain to bind to a range of interacting proteins with high specificity. Binding partners of the p53 TAD include the transcription factors TFIID and TFIIHA, TATA box-binding, protein-associated factors (TAFs), and several protein modifiers, such as MDM2, CBP/p300, and PCAF (Chang et al. 1995; Di Lello et al. 2006; Ferreon et al. 2009). Residual secondary structure can be observed in regions containing functionally relevant hydrophobic residues. In response to DNA damage, the TAD can be phosphorylated at multiple sites by distinct kinases (Fig. 1C), which regulate p53 protein stability, subcellular localization, and function (Meek and Anderson 2009).
The PRR that links the TAD to the DBD in human p53 contains 12 proline residues, including four copies of the sequence PXXP (Walker and Levine 1996). These motifs create a binding site for Src-homology-3 (SH3) domains that mediates protein–protein interactions in signal transduction (Yu et al. 1994). Unlike other domains of p53, the PRD is relatively unconserved. Functionally, it has been shown that the PRD is necessary for apoptosis and efficient growth suppression elicited by p53 (Venot et al. 1998).
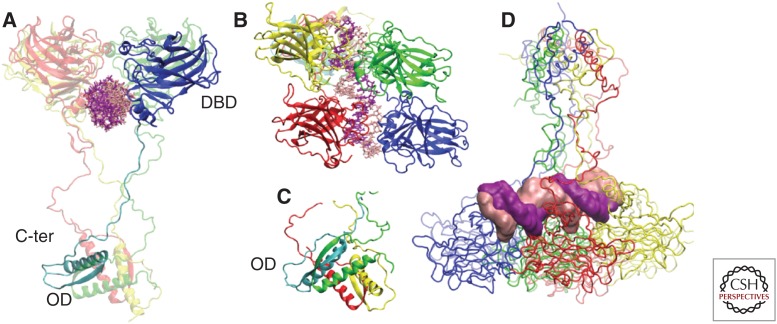
p53 binds DNA as a homotetramer, whereas its oligomerization is mediated by the OD. The DBD and the OD domain are folded regions that are connected through a flexible linker (Fig. 2A,C). The DBD core domain adopts an immunoglobulin-like β sandwich architecture that provides a scaffold for a DNA-binding surface, consisting of a loop–sheet–helix motif and two loops stabilized by a zinc ion (Cho et al. 1994; Joerger and Fersht 2010). Several crystal structures of a p53 core tetramer bound to DNA have been resolved (Ho et al. 2006; Kitayner et al. 2006; Malecka et al. 2009). The p53 RE contains two decameric half-site palindromes of the general sequence 5′-RRRCWWGYYY-3′ (R = A, G; W = A, T; Y = C, T), separated by 0–13 base pairs (el-Deiry et al. 1992). Two core domains associate with a half-site DNA motif, forming a symmetrical dimer (Fig. 2A,B). The dimers assemble to form a tetramer, which is stabilized by protein–protein and base-stacking interactions. The four p53 DBDs bind to DNA in a highly cooperative manner (Nagaich et al. 1999). With this stoichiometry, the affinity of the tetramer for DNA is increased up to 100-fold as compared with the single monomer.
Figure 2.
3D structure of the p53–DNA complex in tetrameric form. (A) Monomer 1 is divided in the DNA-binding domain (DBD) (blue), in interaction with DNA, and carboxy terminal with the OD (oligomerization domain) (cyan). The other three monomers are depicted in transparent mode. (B) Immunoglobulin-like β sandwich architecture of DBD adopted by the four monomers. (C) Tetramerization helices in the carboxy terminal. (D) Projection of the tetramer MD trajectory along eigenvector 1. Specific conformations visited by the tetramer correlate with the DNA deformation measured by the roll and twist parameters (D’Abramo et al. 2015). Therefore, p53 is capable of creating moderate deformities of the DNA, even in the absence of additional molecular partners. C-ter, Carboxy terminal.
The most frequently mutated region of p53 in human cancers is the DBD, with ∼90% of the oncogenic mutations lying in this domain (see Hainaut and Pfeifer 2016). This observation highlights the importance of sequence-specific DNA binding by the DBD for p53 to display its tumor-suppressor activities.
The carboxy-terminal region of p53 crucially controls the structure and function of the entire molecule (Fig. 2A). Tetramerization is a function of the OD that can by itself form tetramers in solution (Jeffrey et al. 1995). The monomeric OD domain comprises a short β-strand (Glu326-Arg333), and an α-helix (Arg335-Gly356) linked by a tight turn (Gly-334) and two monomers form a dimer through antiparallel interaction of β-strands. Two dimers associate through their helices to form a tightly packed tetramer, which can bind efficiently to DNA (Jeffrey et al. 1995). The oligomeric structure of p53 is thought to position in close proximity of the DBD and the CTD and to promote DNA bending when the four core domains bind the four REs (Fig. 2C) (Chen et al. 2012; D’Abramo et al. 2015). In addition, the OD of p53 contains a nuclear export signal (NES) sequence (residues 340–351), which is masked on tetramer assembly (Stommel et al. 1999). Hence, p53 tetramerization and nuclear export are functionally connected (Fig. 2D).
The CTD is highly basic, is intrinsically disordered (Bell et al. 2002; Chillemi et al. 2013), and provides a platform for the relevant posttranslational modifications and protein–protein interactions, which regulate p53 activity. The CTD can undergo disorder-to-order transitions as a result of binding to protein interactors or nonspecific DNA (Rustandi et al. 2000; Weinberg et al. 2004). In vitro nonspecific binding of the p53 CTD to DNA relies on the low-affinity electrostatic interactions between several carboxy-terminal lysine residues and DNA. Interaction of the p53 CTD with no specific DNA sequences regulates binding of the core domain to specific recognition sequences in promoters (Weinberg et al. 2004). Evidence supportive for both negative and positive regulation of sequence-specific DNA binding by the CTD has been reported in the last few decades (Hupp et al. 1992; McKinney et al. 2004; Kitayner et al. 2006; Laptenko et al. 2015). According to the model proposed by Kitayner et al. (2006), the sequence-specific complex of p53 with DNA is stabilized by multiple nonspecific electrostatic interactions between the positively charged proximal CTD and the DNA backbone. A recent report by Laptenko et al. (2015) has shown that the unmodified CTD is required in vivo for the core domain of p53 to recognize the full repertoire of its REs, comprising those that significantly diverge from its consensus sequence. An additional mechanism by which the CTD stabilizes p53 association to its binding sites is the ability of the CTD to induce conformational changes within the DBD following association with DNA (Laptenko et al. 2015). These structural changes would stabilize cooperative contacts between single-core domains within the tetramer, which are required for the stability of the p53–DNA complexes. In line with these observations, molecular dynamic simulations have revealed that the CTD of p53 can create an induced-fit mechanism on the DBD (D’Abramo et al. 2015). Further structural details of the CTD and of the intramolecular interactions among individual domains of p53 will be discussed in the next sections.
EVOLUTION OF THE p53 PROTEIN FAMILY
Although greater emphasis has probably been placed on the structural and functional characteristics of p53, largely as a result of its relevance to human cancer, p63 and p73 are evolutionarily older homologs of p53. TP63 and TP73 genes are expressed as multiple protein isoforms generated by the presence of alternative promoters and splicing sites (Murray-Zmijewski et al. 2006). Both p63 and p73 proteins display domain structures similar to that of p53, and significant amino-acid sequence homology in the TAD, DBD, and OD domains, with the DBD showing the highest degree of conservation (Tomasini et al. 2008a; Dötsch et al. 2010; Levine et al. 2011; Melino 2011).
Knockout mouse studies of all three members of the p53 protein family have shown that p63, p73, and p53 serve different functions in mammals (Mills et al. 1999; Yang et al. 1999, 2000; Tomasini et al. 2008b; Wilhelm et al. 2010; Rufini et al. 2012). The bewildering complexity of these different functions in light of the very high-sequence identity of the DBD—leading to similar transcriptional targets—has sparked speculation about the evolutionary origin of this protein family. The discovery that p63 is not only expressed in the basal compartment of stratified epithelial tissues but also in oocytes (Suh et al. 2006) has suggested that the original ancestor of the mammalian family members was a quality-control factor of germ cells (Fig. 3). Indeed, p53-like proteins have been identified in short-lived animals that are not threatened by the development of cancer and therefore do not need a tumor suppressor (Brodsky et al. 2000; Ollmann et al. 2000; Derry et al. 2001; Nedelcu and Tan 2007; Joerger et al. 2014). In the nematode Caenorhabditis elegans, the p53-like protein Cep-1 is expressed in its germ cells in which it is required for DNA damage–induced apoptosis in the late-stage pachytene phase of meiotic cells (Greiss et al. 2008). Structural investigations of Cep-1 have revealed that it contains a SAM domain, similar to mammalian p63 and p73, suggesting that Cep-1 is more closely related to p63/p73 than to p53 (Ou et al. 2007). In agreement with the hypothesis that p63-like proteins developed first as quality-control factors of germ cells, the eggs of the tunicate species, Ciona intestinalis, express two different p53-like isoforms (Noda 2011; Heering et al. 2015). Because germ cells are transferred from generation to generation, they must be kept under very tight quality control to ensure the survival even of short-lived species.
Figure 3.
Functional evolution and regulation within the p53 family highlighting the unique regulatory mechanism of the most ancient family member TAp63α. (A) All family members and isoforms have acquired distinct regulatory roles in the functional evolution from germ-line quality control via embryonic development and stem-cell maintenance to tumor suppression. (B) Although activation leads to a very similar transcriptional program inducing cell-cycle arrest and apoptosis, the transactivation (TA) isoforms display fundamental differences in the regulation of their transactivation potential. (C) In TAp63α, the TA and transactivation inhibitory (TI) domains of a dimer interact on top of the tetramerization interface establishing a closed, dimeric autoinhibitory conformation. TAp63α monomers are indicated in different colors for clarity. DNA-damage-induced phosphorylation disrupts the interdomain interaction network allowing formation of active tetrameric protein that subsequently induces oocyte death. Protein phosphatases (PPases) may revert activating phosphorylations, preventing the disruption of the autoinhibitory conformation. DN, Dominant negative; PTM, posttranslational modification.
In mammals, the expression of the TAp63α isoform starts around day E18.5 and remains at a high level during the dictyate arrest stage (Suh et al. 2006). The expression of TAp63α is correlated with a dramatic increase in the sensitivity of the developing oocytes toward DNA double-strand breaks. Although prenatal oocytes tolerate hundreds of DNA double-strand breaks as part of the process of homologous recombination, postnatal oocytes expressing TAp63α are killed by fewer than 10 DNA lesions (Suh et al. 2006). The high expression level of p63 makes oocytes far more vulnerable than the surrounding follicular cells. Because oocytes are limited in number and are no longer generated after birth, several control mechanisms ensure that unintended cell death of oocytes is prevented. TAp63α is expressed in a closed and inhibited conformation in which the protein is transcriptionally inactive (Serber et al. 2002). Although all transcriptionally active forms of the mammalian p53 protein family are tetrameric (Luh et al. 2013), TAp63α in oocytes forms inactive dimers (Fig. 3B) (Deutsch et al. 2011). These dimers are stabilized by the interaction of the carboxy-terminal transactivation inhibitory (TI) domain and the amino-terminal TAD, which together block the tetramerization interface of the central tetramerization domain (Fig. 3C). This inhibition allows oocytes to survive for decades in humans despite the high concentration of the proapoptotic factor TAp63α. Detection of DNA double-strand breaks, however, activates p63 through phosphorylation (Fig. 3B,C). Several kinases, including ATM, c-Abl, and Chk2, have been reported to be involved in this process (Fig. 3C) (Suh et al. 2006; Gonfloni et al. 2009; Bolcun-Filas et al. 2014). Activation results in the formation of an open and tetrameric form of TAp63α with a 20-fold higher DNA-binding affinity that leads to the induction of apoptosis mediated by the two BH3-only proteins PUMA and NOXA, which are direct transcriptional targets of activated TAp63α (Fig. 3B,C) (Kerr et al. 2012). Presumably, the p63 quality-control system developed originally to ensure that oocytes, which still have DNA double-strand breaks after chromosomal repair following homologous recombination, undergo removal. The end-point of homologous recombination, however, overlaps with the beginning of the expression of TAp63α in oocytes. Phosphatases (PPases) ensure that p63 stays in the inhibited and closed conformation during that stage (Kim and Suh 2014) to prevent the premature induction of cell death in oocytes that are still actively repairing their chromosomes (Fig. 3C).
Following maturation into pre-antral follicles, TAp63α expression is lost, and instead p73 starts to play an important quality-control function. In vitro fertilization experiments with oocytes obtained from knockout mice lacking only the TA isoform of p73 have shown that the development of the blastocyst is severely impaired (Tomasini et al. 2008b, 2009), often resulting in the formation of multinucleated blastomeres (Fig. 3A). Detailed analysis suggested that this effect is because of problems in proper formation of the spindle assembly complex resulting in genomic instability associated with enhanced aneuploidy. The molecular basis of this effect seems to be the direct interaction of TAp73α with several components of the spindle assembly checkpoint (SAC) complex (Tomasini et al. 2009). Interestingly, and consistent with the mouse studies, morpholino knockdown experiments with the two p53-like proteins from the tunicate C. intestinalis showed that both proteins are involved in the development of the blastopore by inhibiting gastrulation movement, important for closing of the blastopore (Noda 2011).
In mammals, at an even later phase, during the implantation stage of the embryo, p53 also gets involved by controlling the expression of leukemia inhibitory factor (LIF), a cytokine important for implantation (Fig. 3A). Indeed, in p53 null mice, the number of pregnancies and the litter size are reduced because of impaired implantation, demonstrating that all three family members are involved in controlling different stages of maternal reproduction (Hu et al. 2007). In addition, p73 is also involved in the process of spermatogenesis. Specific knockout of the TA isoforms result in male infertility because of severe impairment of spermatogenesis (Inoue et al. 2014). Detailed analysis has revealed that TAp73α functions as a critical factor for adhesion and maturation of the seminiferous epithelium (Holembowski et al. 2014). In Hominidae, a special isoform of p63 with an amino-terminal elongation is also involved in quality control of male germ cells (Mattia et al. 2007; Beyer et al. 2011). This special variant, GTAp63α, was created relatively recently during evolution by fusing the 5′ end of the p63 gene with the long terminal repeat (LTR) region of the human endogenous retrovirus 9 (ERV9). GTAp63α is strongly expressed in spermatogenic precursors but not in mature spermatozoa, indicating that this isoform serves a similar quality-control function as TAp63α in the female germ line.
Although germ-cell quality control is likely to be the original function of the p53 family, it is not the only developmental process in which it is involved. The p63 and p73 knockout mouse studies have revealed that both proteins are master regulators of the development of epithelial tissues and neuronal cells, respectively (Fig. 3A). As the average lifetime of organisms increased and started to exceed the average lifetime of individual cells, evolution developed renewable tissue, which required the establishment of stem cells. Because germ cells are the prototype of a stem cell and p63 (and p73) was already involved in the maintenance of the genetic quality of germ cells, it probably got reused for the new tasks of controlling the proliferative potential of stem cells. In the basal layer of mammalian epithelial tissue, the ΔNp63α isoform plays a central role by orchestrating a transcriptional program that is essential for the maintenance of stratified epithelial tissues (Mills et al. 1999; Yang et al. 1999). Likewise, p73 has developed into an essential factor for neuronal stem-cell maintenance (Fujitani et al. 2010). With the appearance of renewable tissue, however, tumorigenesis became an increasing problem with ∼80% of human cancers originating from epithelial tissues. In the most recent stage of the evolution of the p53 protein family, the tumor-suppressor function was added to its repertoire with p73 (Tomasini et al. 2008) and, in particular, p53 (Levine 1997; Vousden and Lane 2007) being assigned to the surveillance of the genetic and cellular quality. Although p63 seems to play a minor role as a tumor suppressor, the ΔNp63α isoform has been shown to suppress metastasis. In addition, recent studies have also assigned stem-cell maintenance functions to p53 (Cicalese et al. 2009; Levine et al. 2016), demonstrating that all three family members have similar functions in quality control in reproduction, stem-cell maintenance, and tumor suppression, albeit with different specializations acquired during evolution.
STRUCTURE AND MOLECULAR DYNAMICS OF p53
Molecular dynamics (MD) has been extensively used in p53 research, with the great majority of in silico studies dealing with the DNA-binding region (Lu et al. 2007; Madhumalar et al. 2008) and its cancer-related mutations (Rohani et al. 2015; Thayer and Quinn 2015).
The structural and dynamic characterizations of p53 mutants is a first step toward the rational drug design of anticancer molecules that stabilize the mutants, thus recovering wild-type activity (Boeckler et al. 2008; Basse et al. 2010), but MD also has the potential to find new drug targets, sampling transient protein states not detected by X-ray crystallography (Joerger et al. 2015).
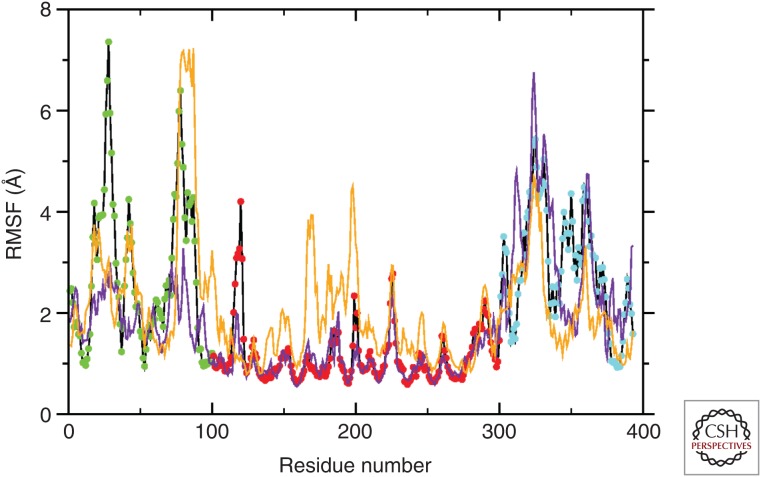
Mutation R175H, located in the helix H1 in the DBD, is close to the Zn-binding residue H179. Therefore, we modeled two possible scenarios, the first in which the binding site for the ion is conserved (purple line in Fig. 4) and the other in which it is coordinated to H175 (orange line in Fig. 4). These two full-length models were compared with the native proteins in the monomeric state.
Figure 4.
Per-residue root-mean-square fluctuation (RMSF) for wild-type p53 and the R175H mutant. The average fluctuation observed by molecular dynamics (MD) (200 nsec) shows different degrees of movement depending on the distinct region of p53 in the wild-type form (black line); point color code as in Fig. 1 (amino terminal, green; DBD [DNA-binding domain], red; carboxyl terminal, blue). The two TADs (transactivation domains), PRR (proline-rich domain), OD (oligomerization domain), and CTD (carboxy-terminal domain) are clearly distinguishable. The DBD is highly stable. Mutant p53 isoforms shows a drastic change in flexibility. The p53 R175H mutant that maintains the native Zn binding (yellow line) has increased the flexibility of DBD, indicating a potential abnormal binding to the selected promoter. The p53 R175H mutant that has an alternative Zn-binding site (purple line) has reduced flexibility of the carboxyl terminus, suggesting abnormal transactivation properties. (From D’Abramo et al. 2015; modified, with permission.)
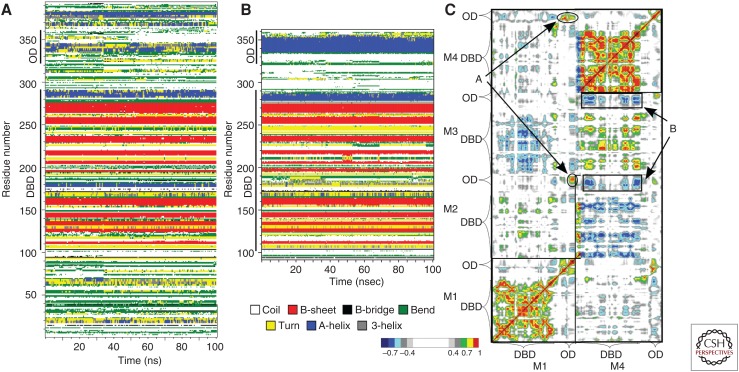
Computational studies have also been performed on the p53 amino-terminal (Mavinahalli et al. 2010) and carboxy-terminal fragments (Gordo et al. 2008; Allen et al. 2010). The complexity of posttranslational regulations targeting p53 (Fig. 1C) makes it the ideal system to be studied by MD, especially when structural information is not obtainable by any experimental technique. Comparison of the full-length p53 in monomeric form with the tetramer–DNA complex shows, for example, that the OD regions in the carboxy-terminal domain are fully structured only in the tetrameric form, whereas the DBD secondary structure is already formed in the isolated monomeric form (OD and DBD in Fig. 5; compare panels A and B).
Figure 5.
Secondary structure content as a function of simulation time. (A) Molecular dynamics (MDs) of the full-length p53 in isolated monomeric form (Chillemi et al. 2013). (B) MD of monomer 1 in the p53–DNA tetrameric complex, taken as representative of the secondary structure in the tetramer (D’Abramo et al. 2015). Comparison of the two systems shows that the oligomerization domain (OD) region is fully structured only in the tetrameric form, whereas nascent helix structure is observed in the monomeric form both in transactivation domains (TADs) and OD. (C) Dynamic cross correlation map of monomers 1 and 4 showing the intermonomeric motions M4-M1, M2-M1 (A), and the anticorrelated motions of the TET-DBD between M3-M4 and M2-M4 (B) (see D’Abramo et al. 2015).
LONG-RANGE COMMUNICATION BETWEEN DIFFERENT DOMAINS: OD AND DBD
We have recently performed an MD simulation of the p53 protein as a tetramer bound to DNA, in which we highlighted the existence of long-range interactions between different domains of p53 (Fig. 5). In particular, the p53 carboxyl terminus, including the OD, shows clear intramonomer-3 (M3) interactions with the DBD as well as intermonomer level interactions between M2-OD and M3-DBD and also between M3-OD and M4-DBD (Fig. 5C). As previously described (D’Abramo et al. 2015), the protein shows a double interaction with DNA. First, distinct interaction is formed by nonspecific noncovalent interaction with DNA; this seems to occur via the β strands S9 in the β sandwich and the carboxyl terminus of the β strand S10 in the loop–sheet–helix motif (Wassman et al. 2013). Second, specific sequence recognition occurs at the level of the helix H2 and loop L1, which are within the loop–sheet–helix motif. When we analyzed the molecular dynamics of this interaction, the loop L1 indicates nonsymmetrical dynamics in the four p53 monomers; this is shown by the essential dynamics of the concatenated DBD trajectory along the second eigenvector. MD indicates a switch from a conformation where L1 tucks into the major groove with an outward projection away from the major groove, whereas the other monomers move in the opposite direction. These data provides a molecular mechanism for the regulation of p53 functions through posttranslational modifications of the carboxyl terminus over the DBD.
As suggested above by the MD analysis (D’Abramo et al. 2015) and by very recent evidence (Hamard et al. 2013; Laptenko et al. 2015), p53 carboxyl terminus has a significant effect on the transcriptional activity of p53 by regulating the function of the DBD, p53 stability and p53 subcellular localization, as well as on the recruitment of additional co-factors. The investigators attribute this effect to changes in the affinity of electrostatic interactions between the lysines at the carboxyl terminus during specific binding with other p53 regions (Laptenko et al. 2015). In agreement, the MD changes indicate the biological value of the lysine interaction on the stability of the p53–DNA complex through the facilitation of cooperative contacts with the DBD (D’Abramo et al. 2015). Both papers are in line with the hypothesis of the “induced-fit mechanism.” Indeed, Petty et al. (2011) showed a conformational change within the DBD involving the L1 loop, which adopted either an extended or a recessive conformation. The dynamic flexible movement of the four monomers shows how the carboxyl terminus can modulate the electrostatic surface regardless of the presence of the lysines (Lys370, 372, 373, 381, 382, and 386 are absent in the MD model) according to the two different conformations at the extreme of the first essential eigenvector. This is the biophysical rationale for the understanding of the carboxyl terminus interaction with the DBD. The in vivo role for the carboxyl terminus of p53 is finally highlighted by the recent knockout mouse lacking the carboxyl terminal 24 amino acids (Hamard et al. 2013). These mice show premature death at 2 weeks with hematopoietic failure and impaired cerebellar development related to enhanced expression of the proapoptotic proteins Puma and Noxa in the bone marrow, while in the liver and in the spleen the phenotype is related to alternative gene expression mechanisms (Hamard et al. 2013).
The implications of the above-mentioned results are evident on the induced-fit theory (see Koshland 1958; Johnson 2008). The existence of an induced-fit conformational transition alters the kinetic equilibrium from Equation 1 to Equation 2:
| (1) |
| (2) |
Multiple structural switches (p53m) induce a better conformational fit of p53 to DNA, moving from Equation 1 to Equation 2 in which K2>K1 and resulting in the stabilization of the bound conformation. Therefore, the selectivity of the promoter bound is more dependent on the KOFF, rather than on the KON. Originally, Halazonetis identified a role for L1 in this mechanism (Emamzadah et al. 2014); however, we cannot disregard that the conformationally induced fit is caused by additional partners that are included in the transcriptional complex. Nonetheless, MD shows that the carboxyl terminus can per se induce a conformational change on the DBD, even in the absence of other partners.
MD OF p53 WITH OTHER INTERACTORS
Being able to accurately model the protein interactions of p53 with its molecular partners would greatly enhance our understanding of the biological processes involved. In which direction are the structural and MD research moving? Clearly, a full structural analysis by Cryo-EM would provide a more solid base to elucidate the entire p53 complex with the DNA. This would allow a more realistic MD analysis, in which both the DNA (different promoter sequences) and p53 (phosphorylation, ubiquitylation, and other posttranslational modifications) could be studied. Already now, the interaction of p53 with other interactors is under investigation.
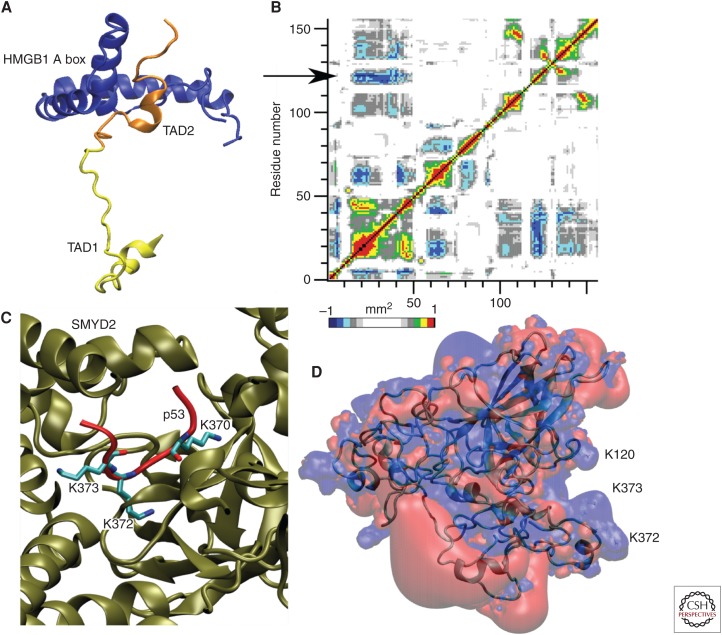
Binding of the HMGB1 A box with the TAD2 region (Fig. 6A) plays a role in DNA binding (Rowell et al. 2012). In agreement, MD results of the full-length form show an anticorrelation movement between the TADs and DBD in the helix region around Lys120 (Fig. 6B).
Figure 6.
Examples of molecular partners of p53 with different biological roles. (A) Binding of the HMGB1 A box (blue) with TAD2 influences the p53 (orange) binding with DNA (not shown). (B) Long-range communications between p53 transactivation domains (TADs) and DNA-binding domains (DBDs) have been observed in the full-length molecular dynamic (MD) simulation with an anticorrelation motion (highlighted by the arrow) that structurally connect TADs with the Lys120 region in DBD. Red, correlation motion; blue, anticorrelation motion (see color legend). (C) Among the posttranslational modifications, methylation of lysine residues in CTD (carboxy-terminal domain) plays an important role. Lys370 is specifically methylated by SMYD2. (D) Lysine residues are important also for DNA binding. The most stable conformations in the eigenvector 1–2 essential subspace present a positive charged iso-surface participated by Lys120, besides the CTD lysines.
The structural mechanism that allows the alternative methylation of Lys372 by Set9 (Chuikov et al. 2004) or Lys370 by SMYD2 (Fig. 5C) (Wang et al. 2011) is still not fully unraveled. A synergistic role for the carboxy-terminal lysine residues and Lys120 in the DBD has been highlighted by the MD simulation of the full-length monomeric form, in which conformations in the most visited essential subspace present a unique positive charged surface formed mainly by the lysines (Fig. 6D).
In keeping, the analysis of mutant isoforms of p53 (see Fig. 4) may provide additional structural information on the defective function of p53 itself in cancer.
CONCLUDING REMARKS
Although the role of the p53 core domain in DNA binding has been extensively established, the contribution of its carboxy-terminal domain remained elusive until recently. Latest data on the structural properties of p53 have shed light on the regulatory mechanisms exerted by the carboxy-terminal domain of p53 on DNA binding by the DBD. The MD simulation of the p53 tetramer in complex with DNA highlighted the presence of long-range communication between different domains. These novel data showed a dynamic flexible movement of the four monomers and showed that the carboxyl terminus can modulate the electrostatic surface between DNA and the transcription factor. The unstructured carboxy-terminal domain of p53 is indeed able to regulate the stability of site-specific DNA binding and facilitate contacts between the core DBDs of the tetramer. As a result, the carboxyl terminus of p53 has a strong biological effect on its transcriptional function by controlling the DNA-binding activity, the stability, and the subcellular localization as well as the recruitment of transcriptional cofactors.
ACKNOWLEDGMENTS
We thank Dr. R.A. Knight for his constructive criticisms, comments, and support. This work is supported by the Medical Research Council, U.K.; grants from Associazione Italiana per la Ricerca contro il Cancro (AIRC): AIRC 2014 IG15653 (to G.M.), AIRC 5xmille MCO9979 (to G.M.), Fondazione Roma malattie Non trasmissibili Cronico-Degenerative (NCD) Grant (to G.M.).
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Allen WJ, Capelluto DGS, Finkielstein CV, Bevan DR. 2010. Modeling the relationship between the p53 C-terminal domain and its binding partners using molecular dynamics. J Phys Chem B 114: 13201–13213. [DOI] [PubMed] [Google Scholar]
- Basse N, Kaar JL, Settanni G, Joerger AC, Rutherford TJ, Fersht AR. 2010. Toward the rational design of p53-stabilizing drugs: Probing the surface of the oncogenic Y220C mutant. Chem Biol 17: 46–56. [DOI] [PubMed] [Google Scholar]
- Bell S, Klein C, Muller L, Hansen S, Buchner J. 2002. p53 contains large unstructured regions in its native state. J Mol Biol 322: 917–927. [DOI] [PubMed] [Google Scholar]
- Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M. 2011. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci 108: 3624–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci 105: 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Rinaldi VD, White ME, Schimenti JC. 2014. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 343: 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19: 2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103–113. [DOI] [PubMed] [Google Scholar]
- Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis TD, Berger SL. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15: 807–816. [DOI] [PubMed] [Google Scholar]
- Chang J, Kim DH, Lee SW, Choi KY, Sung YC. 1995. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem 270: 25014–25019. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bates DL, Dey R, Chen PH, Machado AC, Laird-Offringa IA, Rohs R, Chen L. 2012. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep 2: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillemi G, Davidovich P, D’Abramo M, Mametnabiev T, Garabadzhiu AV, Desideri A, Melino G. 2013. Molecular dynamics of the full-length p53 monomer. Cell Cycle 12: 3098–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. 1994. Crystal structure of a p53 tumor suppressor–DNA complex: Understanding tumorigenic mutations. Science 265: 346–355. [DOI] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432: 353–360. [DOI] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. 2009. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138: 1083–1095. [DOI] [PubMed] [Google Scholar]
- D’Abramo M, Bešker N, Desideri A, Levine AJ, Melino G, Chillemi G. 2015. The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain. Oncogene 10.1038/onc.2015.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R, Müller L, Dehner A, Klein C, Kessler H, Buchner J. 2003. The N-terminal domain of p53 is natively unfolded. J Mol Biol 332: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Demir O, Baronio R, Salehi F, Wassman CD, Hall L, Hatfield GW, Chamberlin R, Kaiser P, Lathrop RH, Amaro RE. 2011. Ensemble-based computational approach discriminates functional activity of p53 cancer and rescue mutants. PLoS Comput Biol 7: e1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. 2001. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- Deutsch GB, Zielonka EM, Coutandin D, Weber TA, Schafer B, Hannewald J, Luh LM, Durst FG, Ibrahim M, Hoffmann J, et al. 2011. DNA damage in oocytes induces a switch of the quality control factor TAp63α from dimer to tetramer. Cell 144: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG. 2006. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 22: 731–740. [DOI] [PubMed] [Google Scholar]
- Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. 2010. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol 2: a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49. [DOI] [PubMed] [Google Scholar]
- Emamzadah S, Tropia L, Vincenti I, Falquet B, Halazonetis TD. 2014. Reversal of the DNA-binding-induced loop L1 conformational switch in an engineered human p53 protein. J Mol Biol 426: 936–944. [DOI] [PubMed] [Google Scholar]
- Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. 2009. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci 106: 6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, Mak TW, Wojtowicz MJ, Miller FD, Kaplan DR. 2010. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol 20: 2058–2065. [DOI] [PubMed] [Google Scholar]
- Gonfloni S, Di Tella L, Caldarola S, Cannata SM, Klinger FG, Di Bartolomeo C, Mattei M, Candi E, De Felici M, Melino G, et al. 2009. Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat Med 15: 1179–1185. [DOI] [PubMed] [Google Scholar]
- Gordo S, Martos V, Santos E, Menédez M, Bo C, Giralt E, de Mendoza J. 2008. Stability and structural recovery of the tetramerization domain of p53–R337H mutant induced by a designed templating ligand. Proc Natl Acad Sci 105: 16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. 2008. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics 9: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hainaut P, Pfeifer G. 2016. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, Senturk E, Gillespie V, Aaronson SA, Merad M, et al. 2013. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev 27: 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heering J, Jonker HR, Lohr F, Schwalbe H, Dotsch V. 2015. Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain. Protein Sci 25: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WC, Fitzgerald MX, Marmorstein R. 2006. Structure of the p53 core domain dimer bound to DNA. J Biol Chem 28: 20494–20502. [DOI] [PubMed] [Google Scholar]
- Holembowski L, Kramer D, Riedel D, Sordella R, Nemajerova A, Dobbelstein M, Moll UM. 2014. TAp73 is essential for germ cell adhesion and maturation in testis. J Cell Biol 204: 1173–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Fen Z, Teresky AK, Levine AJ. 2007. p53 regulates maternal reproduction through LIF. Nature 450: 721–724. [DOI] [PubMed] [Google Scholar]
- Huang F, Rajagopalan S, Settanni G, Marsh RJ, Armoogum DA, Nicolaou N, Bain AJ, Lerner E, Haas E, Ying L, et al. 2009. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci 106: 20758–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. 1992. Regulation of the specific DNA binding function of p53. Cell 71: 875–886. [DOI] [PubMed] [Google Scholar]
- Inoue S, Tomasini R, Rufini A, Elia AJ, Agostini M, Amelio I, Cescon D, Dinsdale D, Zhou L, Harris IS, et al. 2014. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci 111: 1843–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP. 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267: 1498–1502. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. 2008. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77: 557–582. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. 2010. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb Perspect Biol 2: a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Wilcken R, Andreeva A. 2014. Tracing the evolution of the p53 tetramerization domain. Structure 22: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Bauer MR, Wilcken R, Baud MG, Harbrecht H, Exner TE, Boeckler FM, Spencer J, Fersht AR. 2015. Exploiting transient protein states for the design of small-molecule stabilizers of mutant p53. Structure 23: 2246–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. 2008. Role of induced fit in enzyme specificity: A molecular forward/reverse switch. J Biol Chem 283: 26297–26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Joruiz SM, Bourdon J-C. 2016. p53 isoforms: Key regulators of the cell fate decision. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JB, Hutt KJ, Michalak EM, Cook M, Vandenberg CJ, Liew SH, Bouillet P, Mills A, Scott CL, Findlay JK, et al. 2012. DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol Cell 48: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DA, Suh EK. 2014. Defying DNA double-strand break-induced death during prophase I meiosis by temporal TAp63α phosphorylation regulation in developing mouse oocytes. Mol Cell Biol 34: 1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. 2006. Structural basis of DNA recognition by p53 tetramers. Mol Cell 22: 741–753. [DOI] [PubMed] [Google Scholar]
- Koshland DE. 1958. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci 44: 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Shiff I, Freed-Pastor W, Zupnick A, Mattia M, Freulich E, Shamir I, Kadouri N, Kahan T, Manfredi J, et al. 2015. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell 57: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. 1997. p53, the cellular gatekeeper for growth and division. Cell 88: 323–331. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. 2011. The p53 family: Guardians of maternal reproduction. Nat Rev Mol Cell Biol 12: 259–265. [DOI] [PubMed] [Google Scholar]
- *.Levine AJ, Puzio-Kuter AM, Chan CS, Hainaut P. 2016. The role of the p53 protein in stem-cell biology and epigenetic regulation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Tan Y, Luo R. 2007. Molecular dynamics simulations of p53 DNA-binding domain. J Phys Chem 111: 11538–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh LM, Kehrloesser S, Deutsch GB, Gebel J, Coutandin D, Schafer B, Agostini M, Melino G, Dotsch V. 2013. Analysis of the oligomeric state and transactivation potential of TAp73α. Cell Death Differ 20: 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhumalar A, Smith DJ, Verma C. 2008. Stability of the core domain of p53: Insights from computer simulations. BMC Bioinformatics 9: S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecka KA, Ho WC, Marmorstein R. 2009. Crystal structure of a p53 core tetramer bound to DNA. Oncogene 28: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C. 2007. p53-dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol 27: 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavinahalli JN, Madhumalar A, Beuerman RW, Lane DP, Verma C. 2010. Differences in the transactivation domains of p53 family members: A computational study. BMC Genomics 11: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K, Mattia M, Gottifredi V, Prives C. 2004. p53 linear diffusion along DNA requires its C terminus. Mol Cell 16: 413–424. [DOI] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. 2009. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol 1: a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero R, Rajagopalan S, Lázaro M, Joerger AC, Brandt T, Veprintsev DB, Lasso G, Gil D, Scheres SH, Carazo JM, et al. 2011. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc Natl Acad Sci 108: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G. 2011. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 18: 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. 2006. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 13: 962–972. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Durell SR, Jernigan RL, Appella E, Harrington RE. 1999. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc Natl Acad Sci 96: 1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM, Tan C. 2007. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol 217: 801–806. [DOI] [PubMed] [Google Scholar]
- Noda T. 2011. The maternal genes Ci-p53/p73-a and Ci-p53/p73-b regulate zygotic ZicL expression and notochord differentiation in Ciona intestinalis embryos. Dev Biol 360: 216–229. [DOI] [PubMed] [Google Scholar]
- Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV. 2006. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J 25: 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91–101. [DOI] [PubMed] [Google Scholar]
- Ou HD, Löhr F, Vogel V, Mäntele W, Dötsch V. 2007. Structural evolution of C-terminal domains in the p53 family. EMBO J 26: 3463–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty TJ, Emamzadah S, Costantino L, Petkova I, Stavridi ES, Saven JG, Vauthey E, Halazonetis TD. 2011. An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J 30: 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Raj N, Attardi LD. 2016. The transactivation domains of the p53 protein. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani L, Morton DJ, Wang XQ, Chaudhary J. 2015. Relative stability of wild-type and mutant p53 core domain: A molecular dynamic study. J Comput Biol 10.1089/cmb.2015.0163. [DOI] [PubMed] [Google Scholar]
- Rowell JP, Simpson KL, Stott K, Watson M, Thomas JO. 2012. HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 20: 2014–2024. [DOI] [PubMed] [Google Scholar]
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. 2012. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev 26: 2009–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustandi RR, Baldisseri DM, Weber DJ. 2000. Structure of the negative regulatory domain of p53 bound to S100B(ββ). Nat Struct Biol 7: 570–574. [DOI] [PubMed] [Google Scholar]
- Santini S, Bizzarri AR, Cannistraro S. 2011. Modelling the interaction between the p53 DNA-binding domain and the p28 peptide fragment of Azurin. J Mol Recognit 24: 1043–1055. [DOI] [PubMed] [Google Scholar]
- Serber Z, Lai HC, Yang A, Ou HD, Sigal MS, Kelly AE, Darimont BD, Duijf PH, Van Bokhoven H, McKeon F, et al. 2002. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol 22: 8601–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J 18: 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. 2006. p63 protects the female germ line during meiotic arrest. Nature 444: 624–628. [DOI] [PubMed] [Google Scholar]
- Thayer KM, Quinn TR. 2015. p53 R175H Hydrophobic patch and H-Bond reorganization observed by MD simulation. Biopolymers 10.1002/bip.22766. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G. 2008a. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 18: 244–252. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. 2008b. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G, et al. 2009. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci 106: 797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. 1998. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J 17: 4668–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. 2007. p53 in health and disease. Nat Rev 8: 275–283. [DOI] [PubMed] [Google Scholar]
- Walker KK, Levine AJ. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci 93: 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li L, Zhang H, Luo X, Dai J, Zhou S, Gu J, Zhu J, Atadja P, Lu C, et al. 2011. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J Biol Chem 286: 38725–38737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassman CD, Baronio R, Demir O, Wallentine BD, Chen CK, Hall LV, Salehi F, Lin DW, Chung BP, Hatfield GW, et al. 2013. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat Commun 4: 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. 2004. Regulation of DNA binding of p53 by its C-terminal domain. J Mol Biol 342: 801–811. [DOI] [PubMed] [Google Scholar]
- Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, et al. 2010. Isoform-specific p73 knockout mice reveal a novel role for ΔNp73 in the DNA damage response pathway. Genes Dev 24: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404: 99–103. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76: 933–945. [DOI] [PubMed] [Google Scholar]