Abstract
Recombinant adeno-associated virus (rAAV) vectors can mediate long-term stable transduction in various target tissues. However, with rAAV serotype 2 (rAAV2) vectors, liver transduction is confined to only a small portion of hepatocytes even after administration of extremely high vector doses. In order to investigate whether rAAV vectors of other serotypes exhibit similar restricted liver transduction, we performed a dose-response study by injecting mice with β-galactosidase-expressing rAAV1 and rAAV8 vectors via the portal vein. The rAAV1 vector showed a blunted dose-response similar to that of rAAV2 at high doses, while the rAAV8 vector dose-response remained unchanged at any dose and ultimately could transduce all the hepatocytes at a dose of 7.2 × 1012 vector genomes/mouse without toxicity. This indicates that all hepatocytes have the ability to process incoming single-stranded vector genomes into duplex DNA. A single tail vein injection of the rAAV8 vector was as efficient as portal vein injection at any dose. In addition, intravascular administration of the rAAV8 vector at a high dose transduced all the skeletal muscles throughout the body, including the diaphragm, the entire cardiac muscle, and substantial numbers of cells in the pancreas, smooth muscles, and brain. Thus, rAAV8 is a robust vector for gene transfer to the liver and provides a promising research tool for delivering genes to various target organs. In addition, the rAAV8 vector may offer a potential therapeutic agent for various diseases affecting nonhepatic tissues, but great caution is required for vector spillover and tight control of tissue-specific gene expression.
Liver-directed gene transfer with viral and nonviral vectors has been explored for the treatment of a variety of inherited and acquired diseases, including hemophilia (51), various metabolic diseases such as mucopolysaccharidosis (42), hyperlipidemia (24), tyrosinemia (41), and diabetes mellitus (25), and chronic viral hepatitis (29). Among the vectors used to deliver genes to hepatocytes in vivo, recombinant adeno-associated virus (rAAV) vectors are one of the most promising vehicles because they are based on nonpathogenic viruses, transduce both dividing and nondividing cells, and achieve long-term stable transgene expression with minimal toxicity and cellular immune response in animals.
AAV is a small, nonpathogenic, replication-defective parvovirus with a single-stranded DNA genome. Among the various serotypes of AAV, rAAV vectors based on AAV serotype 2 (rAAV2) have been most extensively investigated as gene delivery vectors in vivo, demonstrating efficacy and safety. Based on successful results in a series of preclinical studies for rAAV2-mediated gene therapy, several clinical trials were initiated for the treatment of inherited diseases, including hemophilia B (22).
Despite such recent advances, rAAV2-mediated hepatic gene transfer has still been suboptimal and transduction efficiency in the liver remains unsatisfactory, particularly in cases that require higher transduction efficiency. One major drawback in this system is that only ≈10% of hepatocytes are stably transducible with rAAV2 vectors (5, 39, 59). In other words, it is not possible to increase transduction efficiency in mice (either number of transduced hepatocytes or expression of transgene products) in proportion to given doses when vector doses of 3.0 × 1011 vector genomes (vg) or more are administered per mouse. Liver transduction becomes saturated at higher doses, with transduction efficiencies of around 10% of total hepatocytes (39). The mechanism of this restricted liver transduction has not been elucidated but is not related to impaired uptake of vector particles by hepatocytes because rAAV2 vector genomes are found in a majority of hepatocytes within 24 h after vector infusion (32). We have reasoned from our observations that, in the liver, there are two distinct hepatocyte subpopulations with different metabolic states. That is, only a small subset of hepatocytes have all the machinery required for establishing stable rAAV2 transduction, while the other subset is devoid of some machinery for the process or has some inhibitory machinery that prevents the process (32). Recently, Thomas et al. proposed a model in which the rate of capsid uncoating determines the transduction efficiency with rAAV vectors in the liver (54).
In the past 5 years, there have been several major breakthroughs in rAAV vector technologies that include productions of rAAV vectors derived from alternative serotypes (6, 7, 15, 17, 46, 47, 60) and the development of self-complementary (or double-stranded) rAAV vectors (13, 30, 31, 57). Over a hundred different AAV sequences have been isolated thus far from human and nonhuman primates (14). Their recombinants have been investigated extensively for tissue tropism and transduction efficiency, enabling a dramatic increase in transduction efficiency (4, 14, 15) and a change of tissue or cell type tropism or vector distribution patterns in a given tissue (7, 53, 56). Now, finding the optimal AAV serotypes for efficient and tissue-specific transduction has become imperative for successful gene therapy. The other breakthrough, packaging of double-stranded vector genomes into virions, i.e., self-complementary rAAV, has greatly enhanced transduction efficiency, although the vectors can only hold half of the genome.
We and others have established a method by which rAAV2 vector genomes can be cross-packaged into heterologous capsid proteins derived from alternative serotypes, making chimeric virions, so-called pseudo-serotyped rAAV vectors (17, 46). This allowed us to conduct a thorough side-by-side study to compare liver transduction efficiency among different pseudo-serotyped rAAV vectors, types 1 to 6, in mice. Although the rAAV1, -2, and -6 vectors achieved similar expression levels and none of the other serotypes resulted in a dramatic increase in stable liver transduction efficiency (17), the rAAV1, -2, and -6 vectors each exhibited distinct dose-response profiles (17). We have established in two independent studies that rAAV2-mediated stable liver transduction is proportional to given vector doses ranging from 2 to 4 × 109 to 3 to 4 × 1011 vg/mouse (17, 39). Interestingly, and unlike the rAAV2 vector, rAAV1 vector administration into the liver resulted in a disproportionately greater increase in stable liver transduction, as vector doses increased within the same dose range (17), although this dose-response profile for rAAV1 was blunted at doses higher than 4 × 1011 vg/mouse. Since we quantified the transgene product and not the number of transduced hepatocytes in the previous study, it is not known if rAAV1-mediated liver transduction is also confined to a small population of hepatocytes.
In the present study, we investigated whether restricted liver transduction is also the case for pseudo-serotyped rAAV1 and rAAV8 vectors. The rAAV1 vector was selected because it exhibited a distinct dose-response profile, while the rAAV8 vector was chosen because it has been shown to transduce mouse hepatocytes better than rAAV2 (15, 49, 54). As a result, in contrast to rAAV2 vectors, we find that all the hepatocytes are permissive to stable rAAV8 transduction and ≈100% hepatocyte transduction is achievable by portal vein injection at a dose of 7.2 × 1012 vg/mouse. In addition, and unlike the situation with rAAV2 vectors, we find that such high transduction efficiency is achievable by a single tail vein injection of rAAV8 vectors. Finally, we elucidate that the rAAV8 vector can transduce skeletal muscle throughout the body, the entire cardiac muscle, and substantial numbers of pancreatic cells, smooth muscle cells, and brain cells after intravenous injection. These observations not only help us understand the mechanisms of rAAV vector transduction but also emphasize both the utility and promiscuity of rAAV8 vectors.
MATERIALS AND METHODS
Construction of rAAV vectors.
The construction and production of the rAAV2 vectors AAV2-EF1α-nlslacZ, AAV2-hF.IX16, and AAV2-CMV-lacZ were described elsewhere (23, 37, 39, 40), although we did not clearly denote the serotype in the vector names in our previous publications. Briefly, AAV2-EF1α-nlslacZ is a bacterial β-galactosidase-expressing rAAV2 vector harboring the human elongation factor 1α (EF1α) enhancer-promoter, the Escherichia coli lacZ gene with a nuclear localization signal (nls), and the simian virus 40 poly(A) signal. AAV2-hF.IX16 is a human coagulation factor IX (hF.IX)-expressing rAAV2 vector comprising a liver-specific promoter (the apolipoprotein E hepatic locus control region-human α1-antitrypsin gene promoter) (32), hF.IX minigene (containing a 1.4-kb DNA fragment of the first intron from the hF.IX gene), and the bovine growth hormone poly(A) signal. AAV2-CMV-lacZ is a β-galactosidase-expressing rAAV2 vector harboring the human cytomegalovirus enhancer-promoter with an intron from the human growth hormone gene, the cytosolic lacZ gene, and the simian virus 40 poly(A) signal.
For AAV1-EF1α-nlslacZ, AAV8-EF1α-nlslacZ, and AAV8-CMV-lacZ, rAAV2 vector genomes were cross-packaged into capsids derived from AAV serotype 1 or 8 with the corresponding AAV helper plasmids (15, 17) (the AAV8 helper plasmid was kindly provided by James M. Wilson). All the vectors were produced by the triple transfection method, purified by two cycles of cesium chloride gradient centrifugation, and concentrated as outlined elsewhere (3, 17). The final viral preparations were kept in phosphate-buffered saline (PBS) containing 5% sorbitol. The physical particle titers were determined by a quantitative dot blot assay.
Animal procedure.
Six- to 8-week-old male C57BL/6 and C57BL/6 rag-1 mice were purchased from Jackson Laboratory (Bar Harbor, Maine). The portal vein and tail vein injections of rAAV vector preparations were performed as previously described (34, 35). Controls were naïve uninjected mice or mice injected with the excipient (PBS-5% sorbitol) only. Blood samples were periodically collected from the retroorbital plexus. All the animal experiments were performed according to the guidelines for animal care at Stanford University.
Protein analysis.
We extracted total liver proteins and determined expression levels of β-galactosidase in rAAV vector-transduced mouse livers with a β-galactosidase enzyme-linked immunosorbent assay kit (Roche Molecular Biochemicals, Indianapolis, Ind.) as previously described (37). We normalized β-galactosidase levels with the total protein concentration determined by the Lowry assay, using a DC protein assay kit (Bio-Rad, Hercules, Calif.), and described the values as picograms of β-galactosidase per milligram of protein. We measured human coagulation factor IX levels in mouse plasma by an enzyme-linked immunosorbent assay specific for human coagulation factor IX. We measured levels of serum alanine aminotransferase (ALT) with the ALT reagent set (Teco Diagnostics, Anaheim, Calif.).
Histological analysis.
Pieces of mouse liver lobes were embedded in Tissue-Tek Optimal Cutting Temperature compound (Sakura Finetek USA, Inc., Torrance, Calif.) and frozen on dry ice. In some instances, various mouse tissues other than the liver, i.e., brain, lung, heart, spleen, kidney, intestine, testis, pancreas, and skeletal muscle (quadriceps, tibialis anterior, or tongue), were also collected and processed in the same way. For histochemical detection of β-galactosidase expression, 10-μm sections were cut, fixed with 1.25% glutaraldehyde, stained with 5-bromo-4-chloro-3-indolylphosphate (X-Gal) as described (21), and counterstained with light hematoxylin or nuclear fast red. To determine transduction efficiency in the liver, at least 2,000 nuclei per section were examined for β-galactosidase expression from each animal.
In order to determine cell types in the brain transduced with the rAAV8 vector, we performed immunohistochemical analysis of brain sections. The blocks were cut into coronal sections 12-μm thick with a cryostat. The sections were placed on gelatin-coated slides and dried at room temperature for 30 min. The sections were fixed in 4% paraformaldehyde for 15 min and washed three times in PBS for 5 min. The sections were blocked with PBS containing 5% goat serum at room temperature for 30 min. The sections were incubated with primary antibodies against β-galactosidase (1:200; rabbit immunoglobulin G fraction; Invitrogen, Carlsbad, Calif.), with a mixture of mouse monoclonal anti-NeuN antibody (1:200; Chemicon International Inc.), or mouse monoclonal anti-glial fibrillary acidic protein (GFAP) antibody (1:200; Chemicon International Inc.) in PBS containing 5% goat serum at room temperature for 1 h. NeuN and GFAP served as markers for neurons and astrocytes, respectively. The sections were washed three times in PBS for 5 min. Then they were incubated with Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (2 μg/ml; Invitrogen) and Alexa Fluor 594-conjugated goat anti-mouse immunoglobulin G (2 μg/ml; Invitrogen) in PBS at room temperature for 30 min. The sections were washed three times with PBS for 5 min. Finally, the sections were mounted in Vectashield (Vector Laboratories, Burlingame, Calif.). Immunoreactivity was assessed and viewed under a confocal laser-scanning microscope (TCS NT; Leica).
For detection of pancreatic islet cells, we performed X-Gal and insulin double staining as previously described (55). For detection of Kupffer cells in the liver, we performed X-Gal and F8/40 double staining. Briefly, frozen sections in Optimal Cutting Temperature compound were cut 10-μm thick, air dried for 15 min, and stored at −20°C. Samples were thawed for 1.5 h and then fixed in chilled acetone at −20°C for 20 min. Samples were dried for 6 min and rehydrated in PBS for 10 min. Slides were stained for X-Gal overnight and washed three times in PBS for 5 min. Samples were incubated in 0.3% hydrogen peroxide in methanol for 10 min, rinsed in water, and washed again in PBS. Sections were blocked in 10% normal rabbit serum (Vector Laboratories) with avidin (avidin/biotin blocking kit, Vector Laboratories) in PBS-1% bovine serum albumin for 45 min. Slides were blotted and incubated with F4/80 (1:50 dilution; Serotec, Oxford, United Kingdom) with biotin in PBS-1% bovine serum albumin at room temperature for 1 h. Samples were washed in PBS and incubated for 30 min in anti-rat immunoglobulin G (Vector Laboratories) at a 1:500 dilution in PBS-1% bovine serum albumin. Slides were washed in PBS and incubated for 30 min in the Vectastain Elite ABC kit (Vector Laboratories) according to the manufacturer's instructions. Sections were washed in PBS, rinsed in water, and developed with diaminobenzidine with a diaminobenzidine substrate kit (Vector Laboratories) for 2 to 5 min. Slides were then counterstained with hematoxylin, dehydrated, and coversliped.
DNA analysis.
We extracted total genomic DNA from each tissue by a standard phenol chloroform method. We performed Southern blot analysis to determine double-stranded vector genome copy number per diploid genomic equivalent (ds-vg/dge) and to analyze vector forms in transduced tissues as previously described (35, 39). Briefly, we digested 10 μg of total genomic DNA with BglI, which cuts the vector genome seven times, and separated the digests on 0.8% agarose gels, transferred the DNA onto nylon membranes, and hybridized the blotted membrane with a 2.1-kb radioactive vector sequence-specific lacZ probe (2.1-kb BglI/BglI fragment, nucleotide positions 1518 to 3639 of the 4,828-base AAV2-EF1α-nlslacZ vector genome). We detected and quantified the signals with a Phosphorimager and Quantity One software (Bio-Rad). For analysis of the molecular forms of the vector genomes, we digested sample DNA with BamHI, a single cutter that asymmetrically cleaves the 4,828-base vector genome at nucleotide position 1362, or KpnI, which does not cut the vector genome, and probed with the same lacZ probe. The double-stranded vector genome copy number standards were prepared by adding an equivalent number of pAAV-EF1α-nlslacZ (37) plasmid molecules to 10 μg of total genomic DNA extracted from naive mouse liver. The net vector copy number per hepatocyte represents the number of double-stranded vector genomes per transduced hepatocyte and is calculated based on a presumption that double-stranded vector genomes in the liver are carried only by transduced hepatocytes (33, 52).
RESULTS
All the hepatocytes were permissive to stable rAAV8 vector transduction.
We previously demonstrated that only a small portion of hepatocytes are permissive to stable rAAV2 vector transduction, which restricts a linear vector dose-response at doses higher than 3.0 × 1011 vg/mouse (39). The maximum number of stably transducible hepatocytes may vary depending on the experimental settings, but it normally plateaus at ≈10% of total hepatocytes. In order to investigate the correlation between vector dose and transduction efficiency with other pseudo-serotyped rAAV vectors, we injected mice with AAV1-EF1α-nlslacZ or AAV8-EF1α-nlslacZ via the portal vein at four different doses ranging from 5.0 × 1010 to 7.2 × 1012 vg/mouse, i.e., 5.0 × 1010, 3.0 × 1011, 1.8 × 1012 and 7.2 × 1012 vg/mouse (n = 3 to 6 per group). As a reference, we also injected six mice with 3.0 × 1011 vg of AAV2-EF1α-nlslacZ. Control mice were injected with the excipient (PBS-5% sorbitol) only (n = 3). Six weeks after vector injection, we harvested the liver samples and determined the transduction efficiency by counting β-galactosidase-expressing hepatocytes, measuring β-galactosidase antigen levels by enzyme-linked immunosorbent assay and quantifying the double-stranded vector genome copy numbers in the liver by Southern blot. The results are summarized in Table 1 and Fig. 1, and representative microscopic pictures are shown in Fig. 2.
TABLE 1.
Hepatocyte transduction with AAV1-, AAV2-, or AAV8-EF1α-nlslacZ 6 weeks postinjection
| Vector | Dose (vg/mouse) | No. of mice | Mean transduction efficiency (%)a ± SD | Mean β-galactosidase expression (pg/mg of protein)b ± SD | Mean vector copy no. (ds-vg/dge) ± SD | Mean net vector copy no. per transduced cell (ds-vg/dge)c ± SD | Mean β-galactosidase expression rated ± SD | Mean vector genome activitye ± SD |
|---|---|---|---|---|---|---|---|---|
| AAV1 | 5.0 × 1010 | 4 | 4.2 ± 1.0 | 426 ± 110 | 1.1 ± 0.2 | 27.0 ± 3.1 | 100.5 ± 9.5 | 376.7 ± 67.4 |
| 3.0 × 1011 | 6 | 7.7 ± 0.7 | 1215 ± 263 | 9.3 ± 0.9 | 121.6 ± 14.2 | 160.1 ± 40.5 | 133.3 ± 38.4 | |
| 1.8 × 1012 | 4 | 13.2 ± 2.5 | 2078 ± 389 | 54.9 ± 18.1 | 413.5 ± 83.1 | 159.7 ± 27.8 | 40.1 ± 12.1 | |
| 7.2 × 1012 | 3 | 23.5 ± 1.0 | 2057 ± 99 | 92.3 ± 12.2 | 412.1 ± 69.6 | 88.9 ± 8.0 | 21.5 ± 1.6 | |
| AAV2 | 3.0 × 1011 | 6 | 3.9 ± 0.5 | 888 ± 228 | 5.1 ± 2.0 | 132.2 ± 47.5 | 235.8 ± 72.4 | 197.0 ± 92.5 |
| AAV8 | 5.0 × 1010 | 4 | 8.1 ± 1.8 | 1669 ± 201 | 7.2 ± 1.5 | 90.6 ± 5.3 | 211.7 ± 31.5 | 223.5 ± 23.5 |
| 3.0 × 1011 | 6 | 14.9 ± 3.4 | 2963 ± 517 | 58.2 ± 10.9 | 413.1 ± 134.8 | 208.2 ± 63.5 | 53.0 ± 14.6 | |
| 1.8 × 1012 | 3 | 65.8 ± 9.0 | 9052 ± 1541 | 207.5 ± 56.0 | 318.4 ± 87.5 | 140.8 ± 40.2 | 44.8 ± 7.4 | |
| 7.2 × 1012 | 4 | 97.4 ± 0.3 | 21686 ± 3051 | 620.6 ± 42.8 | 637.2 ± 42.6 | 222.7 ± 31.4 | 35.3 ± 7.2 | |
| None (excipient) | 3 | 0.0 | <4 | 0.0 | 0.0 | NA | NA |
X-Gal-positive nuclei/total hepatocyte nuclei counted (at least 2,000 hepatocyte nuclei were counted).
β-Galactosidase antigen levels in liver extracts were normalized to the amount of total protein in samples.
Net vector copy number is defined as the number of double-stranded vector genomes per transduced hepatocyte; i.e., (vector copy number per cell [vg/dge]/transduction efficiency [%]) × 100.
β-Galactosidase expression rate is defined as picograms of β-galactosidase protein per milligram of cellular protein produced from transduced hepatocytes corresponding to 1% of total hepatocytes; i.e., β-galactosidase expression divided by transduction efficiency. The values reflect β-galactosidase production per transduced hepatocyte. NA, not applicable.
Vector genome activity is defined as β-galactosidase production per double-stranded vector genome; i.e., β-galactosidase expression divided by vector copy number (ds-vg/dge).
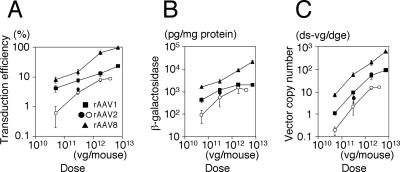
FIG. 1.
Vector dose-response profiles in AAV1-, AAV2-, and AAV8-EF1α-nlslacZ-transduced mouse livers. The percentage of transduced hepatocytes in the livers (A), total β-galactosidase antigen levels (B), and number of double-stranded vector genomes per diploid genomic equivalent (ds-vg/dge) (C) are shown as a function of injected vector doses. Solid markers represent the values obtained from the present study. The dose-response profiles in AAV2-EF1α-nlslacZ-mediated liver transduction were obtained from our previous study (39) for comparison and are depicted with open circles. Values are means ± standard deviation.
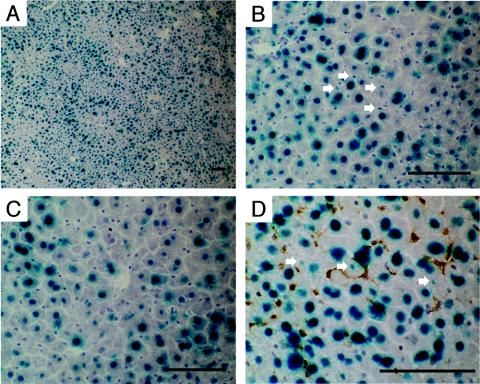
FIG. 2.
Liver transduction with 7.2 × 1012 vg of AAV8-EF1α-nlslacZ delivered via the portal vein. The liver was harvested 6 weeks postinjection and stained with X-Gal and light hematoxylin. A representative result is shown (A and B). Virtually all hepatocytes were transduced with rAAV8 throughout the liver. The liver was stained heterogeneously with X-Gal, with central vein areas being less intense. (C) X-Gal-stained hepatocytes around a central vein area. Although gene expression near central veins was not as strong as in portal areas, most of the hepatocytes express the transgene. (D) X-Gal and F4/80 double staining. None of the Kupffer cells (brown) were transduced. Small nuclei positive for β-galactosidase are indicated with arrows in panels B and D. These might be portions of hepatocyte nuclei or represent rAAV8-transduced nonparenchymal cells besides Kupffer cells. Scale bars, 100 μm.
As demonstrated, all the hepatocytes were permissive to stable transduction with the rAAV8 vector, reaching ≈100% hepatocyte transduction at 7.2 × 1012 vg/mouse. β-Galactosidase-positive cells with a small nucleus were occasionally found in the rAAV8-transduced liver (Fig. 2B). X-Gal/Kupffer cell double staining revealed no transduction in Kupffer cells (Fig. 2D). The origin of β-galactosidase-positive small nuclei could not be determined conclusively. These might be portions of hepatocyte nuclei or represent rAAV8-transduced nonparenchymal cells besides Kupffer cells. The number of hepatocytes transduced with the rAAV1 vector was 24% at a dose of 7.2 × 1012 vg/mouse. Since injection of a vector dose higher than 1013 vg/mouse was not feasible, it was not possible to determine the maximum number of hepatocytes permissive to stable rAAV1 transduction. Nonetheless, the results suggest that all the hepatocytes can process incoming single-stranded rAAV vector genomes into transgene-expressible double-stranded genomes without extrinsic assistance to augment transduction, such as providing adenovirus helper functions (11, 12), genotoxic treatment (1, 2), or forced biochemical modification of single-stranded DNA binding proteins (43-45, 61).
Dose-response profile with the rAAV8 vector did not change until all the hepatocytes were transduced.
In rAAV2-mediated liver transduction, a proportional dose-response profile does not change at doses of less than 3.0 × 1011 vg/mouse, but the vector dose-response reaches saturation at higher doses (39). Likewise, our previous and present studies showed that the dose-response profile in rAAV1-mediated liver transduction changes between a low dose range (showing a disproportionately greater dose-response) and a high dose range (showing a blunted dose-response) (17) (Fig. 1B).
In the rAAV8-mediated liver transduction, log/log plots of the vector doses and the number of transduced hepatocytes (Fig. 1A) or the level of transgene expression (Fig. 1B) exhibited linearity throughout the given vector doses, with regression coefficients (r) of 0.945 (Fig. 1A) and 0.995 (Fig. 1B). The log y/log x slopes of the dose-response curves were 0.450 (Fig. 1A) and 0.585 (Fig. 1B). The log y/log x slope for a proportional dose-response should be ≈1.0. Therefore, neither the dose-response determined by measuring the number of transduced hepatocytes nor that determined by measuring the transgene protein product was directly proportional to various vector doses. The similar values of both slopes (both are around 0.5) imply that the total amount of transgene product and the number of transduced hepatocytes correlate with each other. Although the dose-response with rAAV8 was not proportional, both the number of transduced hepatocytes and transgene expression levels predictably increased with a constant factor of exp(≈0.5). In other words, when the AAV8-EF1α-nlslacZ vector is injected into mice at doses of 5.0 × 1010 vg/mouse or higher, an x-fold increase in the vector dose results in x≈0.5-fold increase in the number of transduced hepatocytes and transgene expression until all the hepatocytes are transduced.
Total number of stably transduced double-stranded vector genomes in the liver increased in proportion to given vector doses irrespective of serotype.
Next we determined double-stranded vector genome copy numbers per diploid genomic equivalent (ds-vg/dge) in the livers by Southern blot analysis. The results are summarized in Table 1 and Fig. 1C. In Fig. 1C, all vectors showed similar log y/log x slopes close to 1.0 (i.e., 0.775, 1.005, and 1.119 for rAAV8, rAAV1 and rAAV2, respectively) with a regression coefficient (r) of 0.999 in each case until the slopes of the dose-response curves started decreasing. This demonstrates that the total number of stably transduced double-stranded vector genomes in the liver increased proportionally irrespective of serotype until the saturation dose was reached. Considering that each of the three serotypes display distinct dose-response profiles, it is conceivable that the quality or state but not the absolute quantity of the double-stranded vector genomes in hepatocytes determines the dose-response profiles.
Decreased specific activity of vector genomes in the liver was concordant with the emergence of vector genome concatemers.
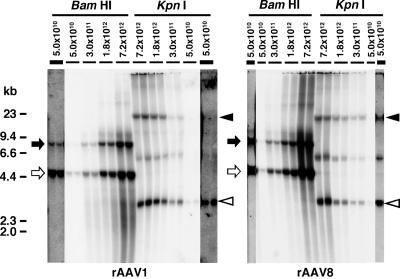
It is possible to deduce the specific activity of the double-stranded vector genomes by dividing β-galactosidase expression levels by vector genome copy numbers (Table 1). In rAAV1 or rAAV8-mediated liver transduction, vector genome specific activity decreased as the vector dose increased. This was also the case for rAAV2 vectors (39). The mechanism of this decreased genome specific activity is not yet clear but is likely related to the formation of less active double-stranded vector molecules, presumably concatemers (39). The present study, combined with the results from our previous study (39), showed that decreased vector genome specific activity is concordant with the emergence of concatemers. At the lowest dose, 5.0 × 1010 vg/mouse, the rAAV8 vector genomes formed substantial concatemers, while rAAV1 and rAAV2 formed exclusively circular monomers (Fig. 3) (39). The vector genome specific activities of rAAV1, rAAV2, and rAAV8 at this dose were 377, 489, and 223 (pg/mg of protein)/(ds-vg/dge), respectively, which is consistent with our presumption that concatemer formation decreases vector genome specific activity.
FIG. 3.
Southern blot analysis of rAAV vector genomes in liver transduced with AAV1- or AAV8-EF1α-nlslacZ at various doses. The left and right panels show the results obtained with AAV1-EF1α-nlslacZ- and AAV8-EF1α-nlslacZ-injected mice. Total genomic DNA was extracted from the livers harvested 6 weeks postinjection and separated on 0.8% agarose gels following BamHI or KpnI digestion. BamHI cleaves the vector genome only once at nucleotide position 1362, while KpnI does not cut the 4,828-base genome. The vector genomes were detected with a 2.1-kb lacZ probe (nucleotide positions 1518 to 3639). Each lane represents an individual mouse. Injected vector doses (vg per mouse) are indicated above each lane. For the results obtained from the mice injected with 5.0 × 1010 vg/mouse, strips from overexposed blots are also shown to demonstrate the presence or absence of concatemers. They are indicated with thicker lines above the lanes. Open and solid arrows indicate head-to-tail and tail-to-tail molecules, respectively. Open and solid arrowheads indicate supercoiled double-stranded circular monomer vector genomes and concatemers, respectively. Head-to-tail molecules include both circular monomer genomes and concatemers, while tail-to-tail molecules represent concatemers exclusively. Therefore, the intensity of tail-to-tail molecules well correlates with the abundance of concatemers.
Comparison of tail (peripheral) vein and portal vein injection of rAAV8 vectors.
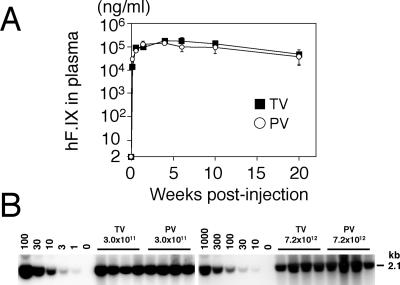
Recently, Sarkar et al. reported that tail vein injection was as efficient as portal vein injection in the context of the canine coagulation factor VIII-expressing rAAV8 vector at a dose of 1.0 × 1011 vg/mouse (49). In our previous studies, we found that, with multiple serotypes, rAAV transduction was more efficient when delivered by the portal vein compared to the tail vein (17, 35). To further address this issue, we injected male C57BL/6 mice with 3.0 × 1011 vg of the rAAV8-hF.IX16 vector via the portal vein or the tail vein and monitored plasma human coagulation factor IX levels. Figure 4A summarizes the results. Both routes of injection resulted in rapid and robust expression of human coagulation factor IX in mouse plasma with no significant difference in transgene expression (Student's t test, P = 0.28, with 4-week time point values). Human coagulation factor IX expression peaked 4 weeks postinjection at levels of around 200 μg/ml, one log higher than that obtainable with the corresponding rAAV2 vector, AAV2-hF.IX16 (17, 39). However, transgene expression declined thereafter to levels of 40 to 50 μg/ml. Such a decline in human coagulation factor IX expression has not been observed with AAV2-hF.IX16. The mechanism of the decline is currently unknown.
FIG. 4.
Comparison of efficiency of rAAV8-mediated liver transduction between tail vein and portal vein injections. (A) Plasma human coagulation factor IX (hF.IX) levels after tail vein (TV) or portal vein (PV) injection of AAV8-hF.IX16 into male C57BL/6 mice. Robust human coagulation factor IX expression with no lag phase was observed with both routes. Expression peaked 4 weeks after injection, followed by a substantial (≈75%) decline. Vertical bars indicate standard deviations. (B) Vector genome copy numbers (ds-vg/dge) in livers transduced with AAV8-EF1α-nlslacZ via tail vein or portal vein injection at 3.0 × 1011 or 7.2 × 1012 vg/mouse. Total liver DNA was extracted 6 weeks postinjection, and 10 μg of DNA was analyzed by Southern blot with BglI digestion and a 2.1-kb lacZ probe (BglI-BglI fragment). The left and right blots were analyzed separately with a different series of vector copy number standards. The double-stranded vector copy number standards (0 to 100 and 0 to 1,000 ds-vg/dge) were prepared by adding the corresponding amount of plasmid, pAAV-EF1α-nlslacZ, to 10 μg of liver DNA extracted from a naïve mouse. Each lane represents an individual mouse. Routes of administration and vector doses are indicated above the lanes.
Next, in order to determine the transduction efficiency of rAAV8 in the liver, we injected male C57BL/6 rag-1 mice with two different doses of AAV8-EF1α-nlslacZ (3.0 × 1011 and 7.2 × 1012 vg/mouse) via two different routes (tail vein and portal vein, n = 4 each), and determined liver transduction efficiency 6 weeks postinjection through histochemical and molecular analyses. Control mice received excipient only. Serum samples were collected for measurement of ALT levels at days 1, 3, and 10 after injection. As summarized in Table 2, liver transduction efficiency was comparable between the two routes of injection at any vector dose (Student's t test, P = 0.27 and 0.47 for the doses of 3.0 × 1011 and 7.2 × 1012 vg/mouse, respectively), achieving ≈90% transduction efficiency with 7.2 × 1012 vg/mouse. These results were similar but not identical to the transduction efficiency found in the dose-response study (Table 1). This was presumed to result from a lot-to-lot variation of the two vector preparations and/or the use of a different batch of animals.
TABLE 2.
Comparison of portal vein and tail vein injection of AAV8-EF1α-nlslacZ 6 weeks postinjection
| Vector dose (vg/mouse) | Mean transduction efficiency (%) ± SD (n)
|
|
|---|---|---|
| Portal vein | Tail vein | |
| 3.0 × 1011 | 24.2 ± 13.1 (4) | 15.5 ± 6.2 (4) |
| 7.2 × 1012 | 85.4 ± 7.3 (4) | 89.1 ± 6.3 (4) |
| 0 (excipient) | 0.0 (2) | 0.0 (2) |
Southern blot analysis of liver DNA to assess vector genome copy numbers in the liver also supported the histological observations (Fig. 4B). The average vector copy numbers determined by densitometric analysis were 37.6 ± 8.8 (tail vein, 3.0 × 1011 vg/mouse), 47.1 ± 6.0 (portal vein, 3.0 × 1011 vg/mouse), 815.6 ± 203.7 (tail vein, 7.2 × 1012 vg/mouse), and 1,044.3 ± 410.2 (portal vein, 7.2 × 1012 vg/mouse) (mean ± standard deviation). There was no statistical difference in vector genome copy numbers between tail vein and portal vein injections (Student's t test, P = 0.12 and 0.36 for 3.0 × 1011 and 7.2 × 1012 vg/mouse, respectively). Importantly, we did not observe any significant increase in the ALT levels at any time point (data not shown), suggesting that these vector doses did not cause liver damage. In addition, all the mice tolerated such a high dose well, and we did not find any histological evidence of cell damage or inflammation in the liver.
High-dose rAAV8 injection transduced multiple organs with considerable efficiency.
The experimental results described above suggested that peripheral vein injection of the rAAV8 vector is a promising strategy that can safely yield ≈100% hepatocyte transduction. Although the results are pertinent to future gene therapy trials, we need to take into consideration that this strategy may increase the chance of vector spillover into nonhepatic tissues at these higher doses. To address this issue, we performed a tissue distribution study with mice injected via the tail vein or portal vein with two different doses (3.0 × 1011 and 7.2 × 1012 vg/mouse) of AAV8-EF1α-nlslacZ (four different combinations, as shown in Table 2). Six weeks postinjection, the brain, lung, heart, spleen, kidney, pancreas, testis, intestines, and skeletal muscle were examined in addition to the liver. Tissue distribution was assessed by both histological X-Gal staining of tissue sections and Southern blot analysis of vector genomes in the tissue DNA. We initially used the human EF1α enhancer-promoter-driven marker gene because it has been shown to be ubiquitously expressed in transgenic animals in a wide range of mouse cell types (18). Representative results of X-Gal staining of each tissue harvested from the mice injected with excipient only (control) and 7.2 × 1012 vg/mouse via the tail vein are shown in Fig. 5A and B.
FIG. 5.
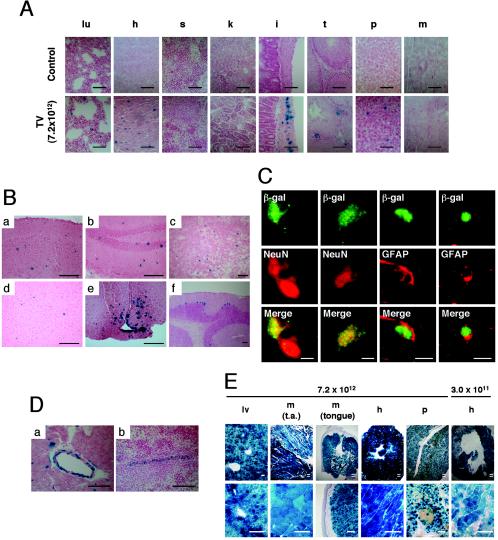
Representative photomicrographs of sections of various mouse tissues 6 weeks after tail vein injection of AAV8-EF1α-nlslacZ at a dose of 7.2 × 1012 vg/mouse (A to D) or 3 weeks after tail vein injection of AAV8-CMV-lacZ at a dose of 3.0 ×1011 or 7.2 × 1012 vg/mouse (E). The sections were either X-Gal stained (A, B, D, and E) or stained with designated antibodies (C). (A) Tissue distribution of β-galactosidase-positive cells: lu, lung; h, heart; s, spleen; k, kidney; i, intestine; t, testis; p, pancreas; and m, skeletal muscle (quadriceps). The top row represents tissues from a mouse injected with excipient only, while the bottom row shows samples from vector-injected mice. (B) Brain transduction with rAAV8. (a) Cerebral cortex. Positive cells are scattered throughout the region. (b) Hippocampus. Positive cells are observed in both granule and pyramidal cell layers. (c) Striatum. (d) Amygdala. (e) Hypothalamus. β-Galactosidase-positive neurons and glial cells are clustered in the arcuate nucleus and median eminence. Some ependymal cells of the third ventricle are also positive. (f) Cerebellum. Purkinje cells are regionally well transduced. (C) Confocal microscopy to assess colocalization of β-galactosidase and either NeuN (a marker for neurons) or GFAP (a marker for astrocytes) to determine rAAV8-transduced cell types in the cerebral cortex of the brain. Both neurons and glial cells were transduced with rAAV8. Scale bars, 5 μm. (D) Transduction of vascular smooth muscle cells in the walls of a branch of the coronary artery (a) and a branch of the splenic artery (b). (E) Tissue distribution of β-galactosidase-positive cells in mice injected with AAV8-CMV-lacZ via the tail vein. The vector doses (vg/mouse) are indicated above the pictures. The section of the pancreas was also stained with anti-insulin antibody (brown cells). Scale bars (duplicated lines), 250 μm. lv, liver; t.a., tibialis anterior limb muscle. The tissues in panels A, B, D, and E were counterstained with nuclear fast red or light hematoxylin. Scale bars represent 100 μm unless otherwise noted.
The study revealed that, at 3.0 × 1011 vg/mouse, very few β-galactosidase-expressing transduced cells were observed outside the liver. However, at 7.2 × 1012 vg/mouse, multiple organs contained many β-galactosidase-expressing cells (Fig. 5A and B). In particular, the brain, heart, and smooth muscles of the intestinal wall were relatively well transduced. The lung and pancreas also contained a considerable number of positive cells. In addition, vascular smooth muscle cells in a variety of tissues were often found to be strongly positive (Fig. 5D). In the pancreas, most of the positive cells were acinar cells and found outside the Langerhans islets. There was no difference in the distribution between tail vein and portal vein injections (data not shown).
In the brain, immunohistochemical analyses demonstrated that both neurons and glial cells were transduced (Fig. 5C). The transduced cells were distributed throughout the brain, including the cerebral cortex, striatum, hippocampus, thalamus, and cerebellum. There were several small foci where transduced cells were clustered (Fig. 5Be). Such foci include the median eminence and arcuate nucleus of the hypothalamus and basolateral nucleus of the amygdala. Purkinje cells in the cerebellum were regionally well transduced (Fig. 5Bf), as previously reported by others using rAAV2 vectors (13, 20).
In the testis, β-galactosidase-positive cells were occasionally observed but restricted to cells residing in the interstitial space. None of the cells in the seminiferous tubules were positive for β-galactosidase activity.
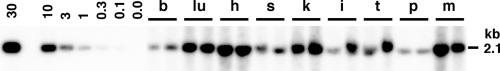
Southern blot analysis of DNA extracted from these tissues revealed that double-stranded vector genomes were detected in all tissues analyzed at relatively high levels (Fig. 6). The average double-stranded vector copy numbers in each tissue were 19.6 ds-vg/dge in the heart, 19.0 ds-vg/dge in the skeletal muscle, 14.4 ds-vg/dge in the lung, 13.7 ds-vg/dge in the kidney, 7.6 ds-vg/dge in the testis, 5.1 ds-vg/dge in the intestine, 4.2 ds-vg/dge in the spleen, 2.7 ds-vg/dge in the brain, and 2.1 ds-vg/dge in the pancreas (from the highest to the lowest). It should be noted that even in the tissues with few positive cells, such as the spleen, testis, and skeletal muscle, double-stranded rAAV8 vector genomes were detected at levels comparable to those in the tissues with many β-galactosidase-expressing cells, suggesting that the extent of β-galactosidase expression does not necessarily correlate with the level of vector genome dissemination.
FIG. 6.
Tissue distribution analysis by Southern blot. Various tissues were harvested from mice injected with 7.2 × 1012 vg of AAV8-EF1α-nlslacZ via the tail vein or the portal vein (one mouse each). Total genomic DNA was extracted from tissues, and 10 μg of each DNA was digested with BglI and separated on a 0.8% agarose gel. The vector genomes were detected with a 2.1-kb BglI-BglI lacZ probe. The double-stranded vector genome copy number standards (0 to 30 ds-vg/dge) were prepared as described in the legend to Fig. 4. Abbreviations: b, brain; lu, lung; h, heart; s, spleen; k, kidney; i, intestine; t, testis; p, pancreas; and m, skeletal muscle. In each set of tissues, the left and right lanes represent samples from mice injected via the tail vein and portal vein, respectively. For densitometric analysis, see the Results section.
Peripheral injection of a rAAV8 vector transduced all the skeletal and heart muscles and a majority of pancreatic cells in the context of the cytomegalovirus promoter.
To further address the discrepancy of the histological and Southern blot analyses in the context of the AAV8-EF1α-nlslacZ vector, we repeated the tissue distribution study with the AAV8-CMV-lacZ vector. We injected C57BL/6 rag-1 male mice via the tail vein with the AAV8-CMV-lacZ vector at a dose of 3.0 × 1011 or 7.2 × 1012 vg/mouse (n = 2 each). Three weeks after vector injection, we analyzed various tissues by X-Gal staining. At a dose of 3.0 × 1011 vg/mouse, hepatocytes were transduced at levels comparable to that with AAV8-EF1α-nlslacZ. However, interestingly, the best-transduced organ was not the liver but the heart. A majority of cardiomyocytes were transduced with AAV8-CMV-lacZ at a dose of 3.0 × 1011 vg/mouse (Fig. 5E), although not many β-galactosidase-expressing positive cells were observed in other nonhepatic tissues, including skeletal muscle.
At a dose of 7.2 × 1012 vg/mouse, the whole liver was transduced (Fig. 5E). Amazingly, AAV8-CMV-lacZ transduced the heart and skeletal muscles with an extremely high efficiency at this dose. The entire heart muscle was transduced, and virtually all the myofibers in skeletal muscles were transduced (Fig. 5E). The whole-body X-Gal staining of a mouse injected intravenously with AAV8-CMV-lacZ revealed that all the skeletal muscles throughout the body, including the diaphragm, were completely transduced (data not shown). In addition, we observed substantial transduction of pancreatic acinar cells and islet cells (Fig. 5E).
Thus, the rAAV8 vector has strong tropism to the liver, but when high vector doses are systemically administered, the tropism becomes promiscuous, leading to undesirable transduction in nontarget tissues, particularly the heart, skeletal muscles, smooth muscles, pancreas, and brain. However, this opens up a new possibility that systemic administration of the rAAV8 vector can yield widespread transduction, with considerable efficiency, of a given target tissue.
DISCUSSION
The present study was conducted to investigate whether rAAV vectors of alternative serotypes can stably transduce all hepatocytes by administration of a high vector dose. To address this issue, we have chosen the rAAV1 and rAAV8 vectors and performed a dose-response study with a nuclear localizing LacZ-expressing vector. Both vectors have been shown to have liver transduction kinetics distinct from that of rAAV2 vectors (15, 17, 54). Although the rAAV1 vector failed to transduce all the hepatocytes due to a blunted dose-response at high vector doses, the present study clearly demonstrated that the dose-response profile of rAAV8 remained unchanged throughout a wide range of vector doses until all the hepatocytes were stably transduced at 7.2 × 1012 vg/mouse. Excluding the self-complementary vectors discussed below, this is the first report that a rAAV vector can yield ≈100% hepatocyte transduction with no obvious toxicity after a simple, noninvasive peripheral vein injection. In addition, we demonstrated that intravenous injection of the rAAV8 vector into mice could transduce entire skeletal muscles throughout the body, the entire heart muscle, and substantial numbers of pancreatic cells, smooth muscle cells, and brain cells.
Our knowledge about the mechanisms of rAAV transduction is still very limited. We have been investigating why rAAV2 vectors can stably transduce only a subset of hepatocytes. Impaired vector uptake in a subset of hepatocytes cannot explain this observation because vector genomes were found in most of the hepatocytes a day following vector administration (32). Therefore, blocks at the level of post-vector entry processing should contribute to the restricted liver transduction with rAAV2. Such barriers include endosomal processing and viral coat modification, which involves the ubiquitin/proteasome system (9, 19), cytoplasmic trafficking (48, 50), nuclear entry, uncoating (54), conversion from single-stranded to double-stranded vector genomes (11, 12, 38), processing of double-stranded vector genomes into transcriptionally active molecules via vector genome recombination (8, 36), and stable residence in the nuclear environment, which allows transgene expression. It is not easy to reconcile various observations with some conflicts from different laboratories at this time, but several recent observations or innovations have provided several important clues to this issue.
Recently, self-complementary or double-stranded rAAV vectors have been developed (13, 30, 31, 57). Self-complementary rAAV vectors possess half-sized hairpin-like double-stranded vector genomes. The important feature of this vector is that it skips the requirement for duplex DNA formation from single-stranded vector genomes, which is one of the fundamental limiting steps for rAAV vector transduction (11, 12). It has recently been shown that ≈90% of hepatocytes could be stably transduced when the self-complementary rAAV serotype 2 vector carrying a marker gene was injected into mouse livers (57). This suggests that release of vector genomes from viral virions occurs in most of the hepatocytes, and therefore the mechanisms for restricted liver transduction should be related to the inefficiency of duplex DNA formation from single-stranded vector genomes. However, and importantly, this does not necessarily exclude the possibility that factors upstream of duplex DNA formation may contribute to the inefficiency of liver transduction with rAAV2.
At least two models have been proposed to address the inefficient duplex DNA formation in rAAV2-mediated liver transduction. The first model involves cellular machinery that directly regulates duplex vector genome formation. Recently, Zhong et al. demonstrated that, in T-cell protein tyrosine phosphate transgenic mice and FKBP52-knockout mice, the rAAV2 vector transduced 12 to 16 times more hepatocytes than the wild-type counterparts (61). In these mouse models, phosphorylated forms of FKBP52, known to bind to the AAV inverted terminal repeat and block second-strand synthesis (43), are downregulated or deficient. From their observations, they claimed that impaired duplex DNA formation by second-strand synthesis (11, 12) precludes efficient transduction. The second model involves an upstream factor, i.e., the rate of capsid uncoating indirectly determines the efficiency of duplex DNA formation. In this model proposed by Thomas et al. (54), slower capsid uncoating of rAAV2 than of rAAV8 limits the formation of duplex DNA in hepatocytes through annealing of complementary plus and minus single-stranded rAAV2 genomes (38), resulting in inefficient liver transduction with rAAV2 vectors. At present, however, we do not have a clear answer that explains why stable liver transduction with rAAV2, but not rAAV8, is restricted to a fraction of hepatocytes.
Nonetheless, the present study demonstrated that all hepatocytes are capable of processing single-stranded rAAV2 genomes (delivered with AAV8 capsids) into duplex DNA. This implies that viral capsid proteins, and not cellular factors by themselves, substantially influence the efficiency of duplex DNA formation in each hepatocyte, at the level of post-vector entry processing of rAAV vectors. In other words, in rAAV2-nonpermissive hepatocytes, which account for ≈90% of total hepatocytes and can take up rAAV2 vectors but not express transgene product, duplex rAAV2 vector genome formation is impaired at the level of post-vector entry when vector genomes are delivered with AAV2 capsids but not impaired when they are delivered with AAV8 capsids.
The tissue biodistibution profile after systemic administration of the rAAV8 vector has been reported in a study with a hemophilia A mouse model (49). They injected mice with 1010 to 1011 vg of canine coagulation factor VIII-expressing vector and quantified vector genome copy numbers by TaqMan PCR. They concluded that rAAV8 has strong tropism to the liver, but no apparent vector genome dissemination was observed, although they found 0.26 to 0.70 copy/cell signals in the hearts of 2 out of 21 mice examined and 0.41 to 0.77 copy/cell signals in the lungs of 3 out of 21 animals examined, the significance of which was not discussed in their report. It should be noted that these two organs are among the four organs that had vector genomes at levels over 10 ds-vg/dge in our study.
In our study, we have clearly demonstrated that, in the context of the cytomegalovirus promoter, rAAV8 transduced the heart with an extremely high efficiency even at a dose of 3.0 × 1011 vg/mouse, and the heart was the best-transduced tissue among all the tissues analyzed including the liver at 3.0 × 1011 vg/mouse. At a dose of 7.2 × 1012 vg/mouse, 100% of cardiomyocytes were transduced. Although we could not determine the minimum rAAV8 vector dose required for 100% cardiomyocyte transduction, it is presumed to be much less than 7.2 × 1012 vg/mouse, given that 3.0 × 1011 vg/mouse was sufficient to transduce a majority of cardiomyocytes. Skeletal muscles were also well transduced at a high vector dose. At a dose of 7.2 × 1012 vg/mouse, virtually all the myofibers in the entire skeletal muscle system throughout the body were transduced, although they were less susceptible to rAAV8 than cardiac muscle, given that not many myofibers were transduced at a dose of 3.0 × 1011 vg/mouse. Recently, Gregorevic et al. have shown that intravascular administration of rAAV6 vectors resulted in widespread skeletal muscle transduction and entire cardiac muscle transduction, as we have observed with rAAV8 in our present study, and they also have established proof of principle that systemic administration of a rAAV6 vector can be used to treat Duchenne muscular dystrophy (16). It should be noted that, in order to increase the permeability of the peripheral microvasculature, their method required simultaneous injection of vascular endothelium growth factor, which was not needed in the context of the rAAV8 vectors.
It is also intriguing that we observed extensive transduction in the pancreas with rAAV8 without any histological evidence suggestive of cell damage or inflammation. In agreement with the previous report on rAAV8 (55), pancreatic acinar cells were the major target, but insulin-producing pancreatic islet cells were also transduced to a certain extent. Our study has demonstrated that systemic administration of rAAV8 vectors could achieve pancreatic transduction at levels equivalent to or even higher than that achievable with adenovirus vectors (55).
It was surprising to us that tail vein or portal vein injection of the rAAV8 vector could transduce broad regions of the brain, since in general it is not possible to transduce this organ by systemic intravenous administration of viral vectors due to the presence of the blood-brain barrier. rAAV vector shedding with negligible levels in the brain has occasionally been reported in tissue distribution preclinical studies (10, 17, 32, 49, 58). However, none of these studies have investigated the origins of the PCR-positive signals. Whether rAAV traversed the blood-brain barrier and transduced neurons and glial cells or remained in the connective tissues including blood vessels has not been addressed. It is intriguing that the median eminence and arcuate nucleus of the hypothalamus and basolateral nucleus of the amygdala were focally transduced with high efficiency. The mechanism(s) underlying focal high transduction is not clear but may be related to a rich blood supply. Interestingly, the hypothalamus is known to have fenestrated capillaries that have numerous small pores increasing vascular permeability.
Dissemination of the viral vectors to the brain is a serious concern in terms of liver gene therapy, but gene delivery to neurons and glial cells by viral vectors holds great promise for gene therapy for central nervous system diseases. Direct intracranial injection of vectors allows efficient transduction of brain tissue, but the transduction is normally limited to the vicinity of the injection site. Many central nervous system diseases broadly affect brain tissue, and therefore global brain transduction by alternative approaches is often preferred. However, the presence of the blood-brain barrier has precluded widespread transduction of the brain. Recently, two strategies, in utero gene transfer (26, 27) and systemic or regional viral administration after mannitol infusion (13, 28), have been shown to successfully overcome this hurdle. The former approach takes advantage of the immaturity of the fetus's blood-brain barrier with increased permeability, and the latter transiently disrupts the blood-brain barrier by making a hyperosmotic environment in the brain capillaries. The mechanisms by which rAAV8 could efficiently traverse the intact blood-brain barrier have yet to be elucidated, and whether rAAV8 particles were actively escorted by a not-yet-defined system or the high dose of rAAV8 vector infusion itself damaged the blood-brain barrier needs to be addressed.
It should be noted that the viral preparation we used for this study contained 5% sorbitol in PBS. Sorbitol is a carbohydrate with the same molecular weight as mannitol and is used clinically to introduce a hyperosmotic environment. Fu et al. reported that, in order to open the blood-brain barrier and transduce mouse brain tissue with intravenously administered rAAV2, preinfusion of 200 μl of 25% mannitol (corresponding to 50 mg of mannitol) was required, and simultaneous infusion of the same amount of 12.5% mannitol (corresponding to 25 mg) had no effect. In our study, we injected 300 μl of vector preparations (equivalent to 15 mg of mannitol), and therefore it is unlikely that our excipient contributed to the transient disruption of the blood-brain barrier. Nonetheless, our study clearly demonstrated that intravenous administration of rAAV8 vectors can transduce neurons and glial cells in broad regions of the adult mouse brain without any treatment that disrupts the blood-brain barrier. Although the mechanism is not clear, rAAV8 will offer an alternative approach to global central nervous system gene delivery in combination with currently available strategies.
Except for the liver, direct injection into the target tissue is a standard approach for transduction with rAAV vectors. This approach is desirable because it can minimize the possibility of vector dissemination to remote organs. However, it has often suffered from the confinement of vectors to the injection site, precluding widespread transduction in a target organ. In this regard, rAAV8 may be applied for global transduction in a given nonhepatic target organ. All the tissues analyzed had double-stranded rAAV8 vector genomes at levels of at least 2 ds-vg/dge. It is important to emphasize that vector dissemination was determined by genomic DNA Southern blot analysis and not a PCR-based assay. This method is superior in detecting double-stranded vector genomes formed within cells. It should be noted that transduction efficiency determined by transgene expression and vector genome copy numbers were not correlated. Presumably the promoter activities vary among the tissues, and it is possible that the tissues with a limited number of β-galactosidase-positive cells in the context of the EF1α or cytomegalovirus enhancer-promoter would have substantial transduction if a different enhancer-promoter is used. Further investigation will be needed to address these discrepancies.
In summary, we demonstrate that all hepatocytes are able to convert incoming single-stranded vector genomes to duplex DNA and are permissive to stable transduction with rAAV8 vectors. In contrast to rAAV2 vectors, ≈100% hepatocyte transduction with the rAAV8 vector could be achieved, and multiple organs could be transduced with extremely high efficiencies following a peripheral vein injection simply by increasing the vector dose. These results not only provide new insights into the mechanisms of liver transduction with rAAV vectors but also open up new applications for rAAV8 vectors in gene therapy, functional genomics, and generating various disease animal models, although, from a safety point of view, a high-dose systemic rAAV8 vector injection strategy will need to take into account the promiscuous tropism of this vector.
Acknowledgments
We thank Makoto Yamakado for histological analysis.
This work was supported by a National Hemophilia Foundation Career Development Award to H.N. and an NIH grant HL66948 to M.A.K.
REFERENCES
- 1.Alexander, I. E., D. W. Russell, and A. D. Miller. 1994. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 68:8282-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, I. E., D. W. Russell, A. M. Spence, and A. D. Miller. 1996. Effects of gamma irradiation on the transduction of dividing and nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum. Gene Ther. 7:841-850. [DOI] [PubMed] [Google Scholar]
- 3.Burton, M., H. Nakai, P. Colosi, J. Cunningham, R. Mitchell, and L. Couto. 1999. Coexpression of factor VIII heavy and light chain adeno-associated viral vectors produces biologically active protein. Proc. Natl. Acad. Sci. USA 96:12725-12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S. J., J. Tazelaar, and J. M. Wilson. 2001. Selective repopulation of normal mouse liver by hepatocytes transduced in vivo with recombinant adeno-associated virus. Hum. Gene Ther. 12:45-50. [DOI] [PubMed] [Google Scholar]
- 6.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, B. L., C. S. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan, D., Y. Yue, Z. Yan, J. Yang, and J. F. Engelhardt. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Investig. 105:1573-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favre, D., N. Provost, V. Blouin, G. Blancho, Y. Cherel, A. Salvetti, and P. Moullier. 2001. Immediate and long-term safety of recombinant adeno-associated virus injection into the nonhuman primate muscle. Mol. Ther. 4:559-566. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, H., J. Muenzer, R. J. Samulski, G. Breese, J. Sifford, X. Zeng, and D. M. McCarty. 2003. Self-complementary adeno-associated virus serotype 2 vector: global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol. Ther. 8:911-917. [DOI] [PubMed] [Google Scholar]
- 14.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregorevic, P., M. J. Blankinship, J. M. Allen, R. W. Crawford, L. Meuse, D. G. Miller, D. W. Russell, and J. S. Chamberlain. 2004. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 10:828-834. [DOI] [PMC free article] [PubMed]
- 17.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102:2412-2419. [DOI] [PubMed] [Google Scholar]
- 18.Hanaoka, K., M. Hayasaka, T. Uetsuki, A. Fujisawa-Sehara, and Y. Nabeshima. 1991. A stable cellular marker for the analysis of mouse chimeras: the bacterial chloramphenicol acetyltransferase gene driven by the human elongation factor 1α promoter. Differentiation. 48:183-189. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, J., K. Qing, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J. Virol. 75:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaemmerer, W. F., R. G. Reddy, C. A. Warlick, S. D. Hartung, R. S. McIvor, and W. C. Low. 2000. In vivo transduction of cerebellar Purkinje cells using adeno-associated virus vectors. Mol. Ther. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 21.Kay, M. A., Q. Li, T. J. Liu, F. Leland, C. Toman, M. Finegold, and S. L. Woo. 1992. Hepatic gene therapy: persistent expression of human α1-antitrypsin in mice after direct gene delivery in vivo. Hum. Gene Ther. 3:641-647. [DOI] [PubMed] [Google Scholar]
- 22.Kay, M. A., C. S. Manno, M. V. Ragni, P. J. Larson, L. B. Couto, A. McClelland, B. Glader, A. J. Chew, S. J. Tai, R. W. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. W. Flake, and K. A. High. 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 24:257-261. [DOI] [PubMed] [Google Scholar]
- 23.Kessler, P. D., G. M. Podsakoff, X. Chen, S. A. McQuiston, P. C. Colosi, L. A. Matelis, G. J. Kurtzman, and B. J. Byrne. 1996. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc. Natl. Acad. Sci. USA 93:14082-14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, I. H., A. Jozkowicz, P. A. Piedra, K. Oka, and L. Chan. 2001. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. USA 98:13282-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima, H., M. Fujimiya, K. Matsumura, P. Younan, H. Imaeda, M. Maeda, and L. Chan. 2003. Neuro-D-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat. Med. 9:596-603. [DOI] [PubMed] [Google Scholar]
- 26.Lai, L., B. B. Davison, R. S. Veazey, K. J. Fisher, and G. B. Baskin. 2002. A preliminary evaluation of recombinant adeno-associated virus biodistribution in rhesus monkeys after intrahepatic inoculation in utero. Hum. Gene Ther. 13:2027-2039. [DOI] [PubMed] [Google Scholar]
- 27.Lipshutz, G. S., D. Titre, M. Brindle, A. R. Bisconte, C. H. Contag, and K. M. Gaensler. 2003. Comparison of gene expression after intraperitoneal delivery of AAV2 or AAV5 in utero. Mol. Ther. 8:90-98. [DOI] [PubMed] [Google Scholar]
- 28.Mastakov, M. Y., K. Baer, R. Xu, H. Fitzsimons, and M. J. During. 2001. Combined injection of rAAV with mannitol enhances gene expression in the rat brain. Mol. Ther. 3:225-232. [DOI] [PubMed] [Google Scholar]
- 29.McCaffrey, A. P., H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion, and M. A. Kay. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21:639-644. [DOI] [PubMed] [Google Scholar]
- 30.McCarty, D. M., H. Fu, P. E. Monahan, C. E. Toulson, P. Naik, and R. J. Samulski. 2003. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 10:2112-2118. [DOI] [PubMed] [Google Scholar]
- 31.McCarty, D. M., P. E. Monahan, and R. J. Samulski. 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8:1248-1254. [DOI] [PubMed] [Google Scholar]
- 32.Miao, C. H., H. Nakai, A. R. Thompson, T. A. Storm, W. Chiu, R. O. Snyder, and M. A. Kay. 2000. Nonrandom transduction of recombinant adeno-associated virus vectors in mouse hepatocytes in vivo: cell cycling does not influence hepatocyte transduction. J. Virol. 74:3793-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao, C. H., R. O. Snyder, D. B. Schowalter, G. A. Patijn, B. Donahue, B. Winther, and M. A. Kay. 1998. The kinetics of rAAV integration in the liver. Nat. Genet. 19:13-15. [DOI] [PubMed] [Google Scholar]
- 34.Nakai, H., R. W. Herzog, J. N. Hagstrom, J. Walter, S. H. Kung, E. Y. Yang, S. J. Tai, Y. Iwaki, G. J. Kurtzman, K. J. Fisher, P. Colosi, L. B. Couto, and K. A. High. 1998. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood 91:4600-4607. [PubMed] [Google Scholar]
- 35.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 73:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai, H., T. A. Storm, S. Fuess, and M. A. Kay. 2003. Pathways of removal of free DNA vector ends in normal and DNA-PKcs-deficient SCID mouse hepatocytes transduced with rAAV vectors. Hum. Gene Ther. 14:871-881. [DOI] [PubMed] [Google Scholar]
- 37.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 18:527-532. [DOI] [PubMed] [Google Scholar]
- 38.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai, H., C. E. Thomas, T. A. Storm, S. Fuess, S. Powell, W. J. F., and M. A. Kay. 2002. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in rAAV-mediated liver transduction. J. Virol. 76:11343-11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overturf, K., M. Al-Dhalimy, R. Tanguay, M. Brantly, C. N. Ou, M. Finegold, and M. Grompe. 1996. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 42.Ponder, K. P., J. R. Melniczek, L. Xu, M. A. Weil, T. M. O'Malley, P. A. O'Donnell, V. W. Knox, G. D. Aguirre, H. Mazrier, N. M. Ellinwood, M. Sleeper, A. M. Maguire, S. W. Volk, R. L. Mango, J. Zweigle, J. H. Wolfe, and M. E. Haskins. 2002. Therapeutic neonatal hepatic gene therapy in mucopolysaccharidosis VII dogs. Proc. Natl. Acad. Sci. USA 99:13102-13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qing, K., J. Hansen, K. A. Weigel-Kelley, M. Tan, S. Zhou, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: role of cellular FKBP52 protein in transgene expression. J. Virol. 75:8968-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qing, K., W. Li, L. Zhong, M. Tan, J. Hansen, K. A. Weigel-Kelley, L. Chen, M. C. Yoder, and A. Srivastava. 2003. Adeno-associated virus type 2-mediated gene transfer: role of cellular T-cell protein tyrosine phosphatase in transgene expression in established cell lines in vitro and transgenic mice in vivo. J. Virol. 77:2741-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qing, K., X. S. Wang, D. M. Kube, S. Ponnazhagan, A. Bajpai, and A. Srivastava. 1997. Role of tyrosine phosphorylation of a cellular protein in adeno-associated virus 2-mediated transgene expression. Proc. Natl. Acad. Sci. USA 94:10879-10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar, R., R. Tetreault, G. Gao, L. Wang, P. Bell, R. Chandler, J. M. Wilson, and H. H. Kazazian, Jr. 2004. Total correction of hemophilia A mice with canine FVIII using an AAV 8 serotype. Blood 103:1253-1260. [DOI] [PubMed] [Google Scholar]
- 50.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 51.Snyder, R. O., C. Miao, L. Meuse, J. Tubb, B. A. Donahue, H. F. Lin, D. W. Stafford, S. Patel, A. R. Thompson, T. Nichols, M. S. Read, D. A. Bellinger, K. M. Brinkhous, and M. A. Kay. 1999. Correction of hemophilia B in canine and murine models using recombinant adeno-associated viral vectors. Nat. Med. 5:64-70. [DOI] [PubMed] [Google Scholar]
- 52.Snyder, R. O., C. H. Miao, G. A. Patijn, S. K. Spratt, O. Danos, D. Nagy, A. M. Gown, B. Winther, L. Meuse, L. K. Cohen, A. R. Thompson, and M. A. Kay. 1997. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat. Genet. 16:270-276. [DOI] [PubMed] [Google Scholar]
- 53.Tenenbaum, L., A. Chtarto, E. Lehtonen, T. Velu, J. Brotchi, and M. Levivier. 2004. Recombinant AAV-mediated gene delivery to the central nervous system. J. Gene Med. 6(Suppl. 1):S212-S222. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, C. E., T. A. Storm, Z. Huang, and M. A. Kay. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 78:3110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, A. Y., P. D. Peng, A. Ehrhardt, T. A. Storm, and M. A. Kay. 2004. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Hum. Gene Ther. 15:405-413. [DOI] [PubMed] [Google Scholar]
- 56.Wang, C., C. M. Wang, K. R. Clark, and T. J. Sferra. 2003. Recombinant AAV serotype 1 transduction efficiency and tropism in the murine brain. Gene Ther. 10:1528-1534. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z., H. I. Ma, J. Li, L. Sun, J. Zhang, and X. Xiao. 2003. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 10:2105-2111. [DOI] [PubMed] [Google Scholar]
- 58.White, S. J., S. A. Nicklin, H. Buning, M. J. Brosnan, K. Leike, E. D. Papadakis, M. Hallek, and A. H. Baker. 2004. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation 109:513-519. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, W., S. C. Berta, M. M. Lu, A. D. Moscioni, J. Tazelaar, and J. M. Wilson. 1998. Adeno-associated virus as a vector for liver-directed gene therapy. J. Virol. 72:10222-10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong, L., W. Li, Z. Yang, L. Chen, Y. Li, K. Qing, K. A. Weigel-Kelley, M. C. Yoder, W. Shou, and A. Srivastava. 2004. Improved transduction of primary murine hepatocytes by recombinant adeno-associated virus 2 vectors in vivo. Gene Ther. 11:1165-1169. [DOI] [PubMed] [Google Scholar]