Abstract
Epigenetic, transcriptional and signaling processes in the nucleolus regulate rRNA transcription and cell growth. We report here that the tumor suppressor ING1b binds rDNA, regulates rDNA chromatin modifications and affects nucleolar localization of mTOR to modulate rRNA levels. ING1 represses rDNA transcription by recruiting HDAC1 to rDNA loci, increasing its association with the NoRC complex and deacetylating the histone H3K9 and H3K27 marks of active transcription. Loss of ING1 enhances nucleolar localization of phospho-mTOR and its association with Raptor and GβL, even during rapamycin treatment. ING1 inhibits rDNA transcription by inhibiting UBF activity and its interaction with mTOR. Regulation of rDNA heterochromatin and rRNA synthesis by ING1 is also apparent during normal cell growth and during cell stress. Moreover, this function was also important during PMA induced differentiation of THP1 cells, since knocking down ING1 affected the process by inhibiting rRNA transcriptional repression. These observations show that ING1 regulates the nucleolar epigenome and rDNA transcription suggesting that regulation of protein synthesis might serve as the basis for ING1 function as a type II tumor suppressor.
INTRODUCTION
The human nucleolar organizing region (NOR) contains ∼400 copies of rDNA repeats distributed in five chromosomes in tandem arrays. RNA polymerase I transcribes each rDNA repeat into a single 47S precursor rRNA, which is subsequently processed into mature 18S, 5.8S and 28S rRNA molecules (1,2). Synthesis of rRNA, which can account for up to 60% of total cellular transcription (3), determines the number of ribosomes in cells. The rate of rRNA transcription is tightly coupled to nutrient availability, and many positive (ERK, mTOR, CBP, c-Myc) and negative (p53, Rb, PTEN, ARF, AMPK, GSK3β) growth regulatory pathways converge on RNA Pol I to couple rRNA synthesis to the metabolic state of the cell (4,5). The upstream binding factor UBF1 is the major Pol I transcription factor (6,7). Binding of UBF1 to the rRNA promoter, regulated by posttranslational modifications, is important for pre-initiation complex assembly and efficient Pol I transcription (8,9). UBF1 binds throughout the rDNA loci, determining the number of transcriptionally active repeats (10,11). In a normal diploid cell about 40–50% of these tandem units are actively transcribed while the rest are maintained inactive as heterochromatin (3). Active rDNA repeats typically contain histone marks of active transcription while inactive repeats harbor repressive marks associated with constitutive heterochromatin. Heterochromatinization of rDNA repeats is regulated by the spacer promoter that is located within the intergenic spacer (IGS) region separating consecutive rDNA coding regions. Intergenic transcripts generated from the IGS recruit the nucleolar chromatin remodelling complex (NoRC) to silence rDNA in response to environmental cues and also during terminal differentiation (12,13). Chromatin regulators like HDAC1 (14), DNMT1 (13), SMARCAD1 (15), PARP1 (16), SIRT1 and SUV39H1 (17) are mediators of NoRC-mediated transcriptional silencing of rDNA. The nucleolar proteome consists of core structural components and proteins that shuttle between the nucleus and nucleolus. Most nucleolar epigenetic and transcription regulators are non-structural proteins that are differentially targeted to the nucleolus depending on the state of cell growth or phase of the cell cycle (2,18). The mechanisms by which chromatin factors are recruited into the nucleolus to modulate rDNA epigenetic state and cell growth are only beginning to be characterized.
The mammalian target of rapamycin (mTOR), a critical regulator of cell growth, affects several processes during protein synthesis including rDNA transcription, rRNA processing, expression of ribosomal proteins, ribosomal assembly and activation of translation factors (19). mTOR regulates rDNA transcription by activating TIF-IA, while its inhibition reduces rRNA transcription (20). It also alters nucleolar chromatin structure during stress responses, establishing a Pol I-permissive environment (21). Effector proteins of the mTOR pathway, such as p70 S6K, are thought to mediate its nucleolar effects (22). Recent reports suggest a more direct role where mTOR translocates to nucleoli to bind rDNA chromatin (23,24), but the mechanisms by which it alters rDNA transcription is not known.
The inhibitor of growth (ING) family of proteins are evolutionarily conserved members of HAT and HDAC complexes that contain a plant homeodomain (PHD) that specifically binds trimethylated H3K4, a mark of active promoters (25–29). The five human ING genes encode type II tumor suppressors that are frequently downregulated or mislocalized in cancer cells (30–32). Knockout of ING1 results in development of spontaneous B-cell lymphomas, while its ectopic expression induces cell cycle arrest and apoptosis (31). ING proteins serve as histone code readers, regulating local chromatin architecture and gene expression, by targeting HAT and HDAC complexes to specific loci (29,32–36). ING1 also bridges the Sin3 co-repressor complex to DNMT1 to maintain histone hypoacetylation at pericentric regions (37) and to regulate GADD45-mediated DNA demethylation (38). ING proteins localize mainly to cell nuclei, but increased nucleolar translocation of ING1 has been described during UV induced apoptosis (39,40). This translocation was dependent on two nucleolar targeting signals (NTS) on ING1 (39), but the functions of ING1 in the nucleolus were unclear.
In this study, we characterize the effect of ING1 on rDNA transcription in the nucleolus. We show that ING1 physically associates with rDNA repeats. ING1 is required for efficient recruitment of HDAC1 to nucleoli and for its co-operation with the nucleolar remodeling complex (NoRC) to enforce heterochromatinization and silencing of rDNA repeats. Furthermore, knockdown of ING1 increases nucleolar localization of mTOR and its association with the major Pol I transcription factor UBF1. ING1 affects acetylation of histones on rDNA chromatin, and regulates the number of active rDNA repeats available for Pol I transcription. Our results, therefore, identify rDNA chromatin modification and rRNA transcription as novel epigenetic regulatory targets of ING1.
MATERIALS AND METHODS
Cell culture & transfections
Hs68 fibroblasts, HEK 293, A431 and HeLa cells were obtained from ATCC, cultured and maintained in DMEM (Lonza) supplemented with 10% fetal bovine serum (Gibco, Invitrogen) at 37°C under 5% CO2. The knock down stable cell lines were generated using the pINDUCER lentiviral tool kit (a gift from Dr Steven Elledge, Harvard Medical School) and were maintained in medium containing puromycin (41). Induction of the knock down was carried out using 100ng/ml of doxycycline for 72 h. THP-1 (ATCC: TIB-202) cells were maintained in RPMI-1640, supplemented with 10% fetal bovine serum. Differentiation of these cells to macrophages was achieved by treating them with 10 nM PMA for 3 days. The differentiation process was analyzed by monitoring the expression of CD14 protein. Electroporation of THP-1 cells were carried out with 0.3 kV and capacitance of 1.07 × 1000 μF using a BioRad electroporator. 3 × 107 cells were resuspended in 300 μl of RPMI 1640 growth medium supplemented with 20 mM HEPES. The cells were electroporated with 20 μg of plasmid DNA or 80 nM of siRNA and placed on ice for 5 min. They were then placed in excess growth medium and analyzed for knockdown efficiency after 72 h.
RNA isolation and quantitative real-time PCR
Total RNA from cells were isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. After treating with DNase I (Qiagen) to remove any genomic DNA contamination, the RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription kit (Invitrogen). Real-time PCR was carried out in triplicate using Maxima SYBR Green qPCR Mastermix (Fermentas) on an Applied Biosystems 7900HT Fast Real-time PCR system using standard protocol. β-Actin or GAPDH were used as endogenous normalization controls. Relative fold changes were determined using the comparative threshold (ΔCT) method (42).
ChIP
Chromatin enrichment of proteins was analysed using a previously described protocol (43). Antibodies used for ChIP experiments were purchased as indicated: anti-Flag (M2; Sigma), UBF (F-9; Santacruz), HDAC1 (10E2; Cell Signaling), Tip5 (Diagenode), acetyl-H3K9 (C5B11; Cell Signaling), acetyl-H3K27 (D5E4; Cell Signaling). Monoclonal antibodies to ING1 was generated in a local hybridoma facility (44). The rDNA locus primers used were those described previously (45).
Protein synthesis assay
Global protein synthesis was measured using a recently developed non-radioactive, SUnSET protocol (46). Briefly, the A431 cells stably expressing ING1 or scrambled shRNA pulsed with 1μg/ml puromycin and chased for 50 min at 37°C with 5% CO2. The lysates were resolved on a gradient gel and protein synthesis was monitored by western blotting with anti-puromycin antibody (1:20 000) and light chain specific IgG-HRP.
Stress conditions
The cellular stress conditions were imposed by either subjecting cells to growth factor deprivation by serum starvation for 48 h or by glucose deprivation, by growing them in medium containing no glucose for 24 h. Translational stress using rapamycin was by maintaining cells in serum free medium overnight and releasing them in serum containing growth medium after pre-treatment with or without 20 nM rapamycin. Cells were analyzed 24 h after release.
Statistical analyses
All data are expressed as mean ± standard deviation from at least three independent experiments. The statistical analyses were done using Student's t tests for two samples and one-way analysis of variance (ANOVA) for differences among groups, using GraphPad Prism software. A probability of P < 0.05 was taken to be statistically significant.
RESULTS
Regulation of 47S pre-rRNA levels by ING1
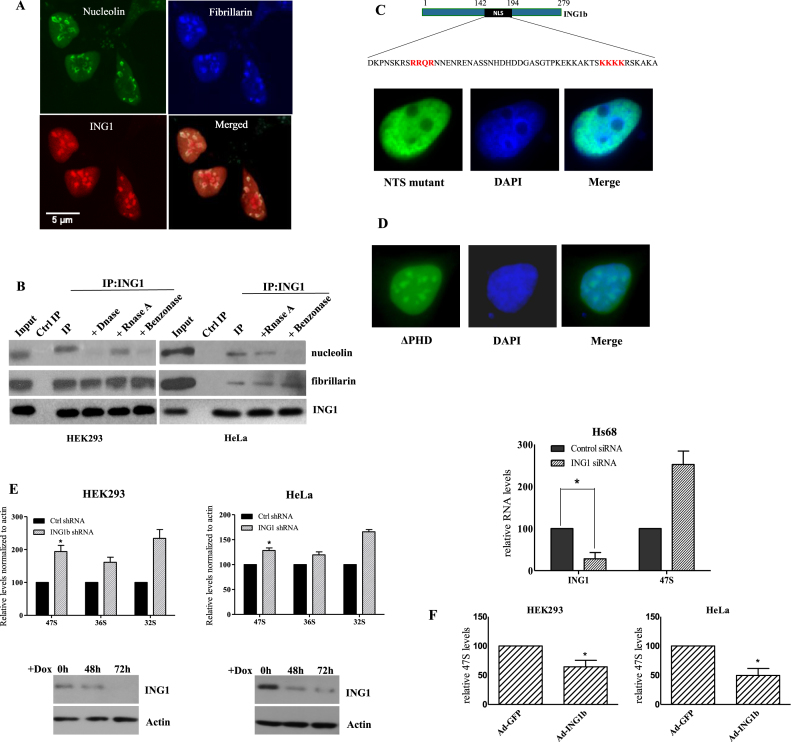
We initially confirmed the nucleolar localization of ING1 (39), and analyzed whether it affects rRNA levels. A significant fraction of both mCherry-ING1 and ING1-GFP fusion proteins co-localized with the nucleolar proteins nucleolin and fibrillarin in HeLa cells (Figure 1A and Supplementary Figure S1). We also observed that endogenous ING1 protein in both HeLa and HEK293 cells coimmunoprecipitated with nucleolar proteins nucleolin and fibrillarin (Figure 1B). This co-precipitation was independent of RNA since RNase A treatment of lysates did not affect the interaction. Association of ING1 with nucleolin was DNA-dependent as DNase I reduced the co-precipitation, but its interaction with fibrillarin was unaffected by either nuclease. Mutating the two ING1 NTS motifs (39) blocked nucleolar, but not nuclear, localization (Figure 1C). Deletion of the PHD motif of ING1, implicated in chromatin binding, did not affect its nucleolar localization and its cellular distribution resembled overexpressed wild type ING1 (Figure 1D).
Figure 1.
ING1 localization to the nucleolus requires the NTS. (A) HeLa cells transfected with mCherry-ING1b were immunostained for nucleolin and fibrillarin. (B) HEK293 and HeLa cells were lysed using RIPA buffer or RIPA buffer containing DNAse, RNAse A or benzonase. Lysates were immunoprecipitated with ING1 antibody (44) and precipitates were blotted for nucleolin and fibrillarin. (C) NTS motifs (red) found in the NLS of ING1. Residues were mutated to asparagine in the NTS mutant. The GFP-ING1-NTS mutant (panel C) and GFP-ING1-PHD deletion mutant (D) were analyzed by immunofluorescence. DAPI stained nuclear DNA. (E) HEK293 or HeLa cells stably expressing shControl or shING1 were induced with doxycycline for 3 days and checked for 47S, 36S and 32S pre-rRNA levels by qRT-PCR (P < 0.05). Western blots show efficiency of ING1 knockdown. Hs68 fibroblasts transfected with control or siING1 were analyzed for 47S levels by qRT-PCR (P < 0.04). The level of ING1 mRNA shows knockdown efficiency. (F) HEK293 or HeLa cells infected with control-GFP or GFP-ING1 adenoviral constructs were analyzed for 47S levels after 48 h (P < 0.05).
The primary function of nucleoli, which contain rDNA repeats, is rRNA synthesis. Hence, we analysed the effect of ING1 levels on rRNA synthesis using qPCR. HEK293 and HeLa cells stably transfected with doxycycline-inducible shING1 constructs, and Hs68 primary fibroblasts transfected with ING1-targeting siRNA, showed consistent increase in 47S pre-rRNA levels (Figure 1E), while overexpression of ING1 reduced pre-rRNA transcripts in the cells (Figure 1F). This suggested that ING1 regulated levels of 47S rRNA. ING1 knockdown maintained the normal fibrillarin distribution in nucleoli of HeLa cells suggesting an intact nucleolar structure (Supplementary Figure S2). Since we have observed UV-dependent translocation of ING1 to the nucleolus, we also asked if UV would counter the effects of ING1 knockdown by directing residual ING1 to the nucleolus. However, UV treatment did not significantly reduce the ability of ING1 knockdown to increase rRNA levels in different cell lines (Supplementary Figure S3).
ING1 binds rDNA repeats and targets HDAC1 to the nucleolar remodelling complex (NoRC)
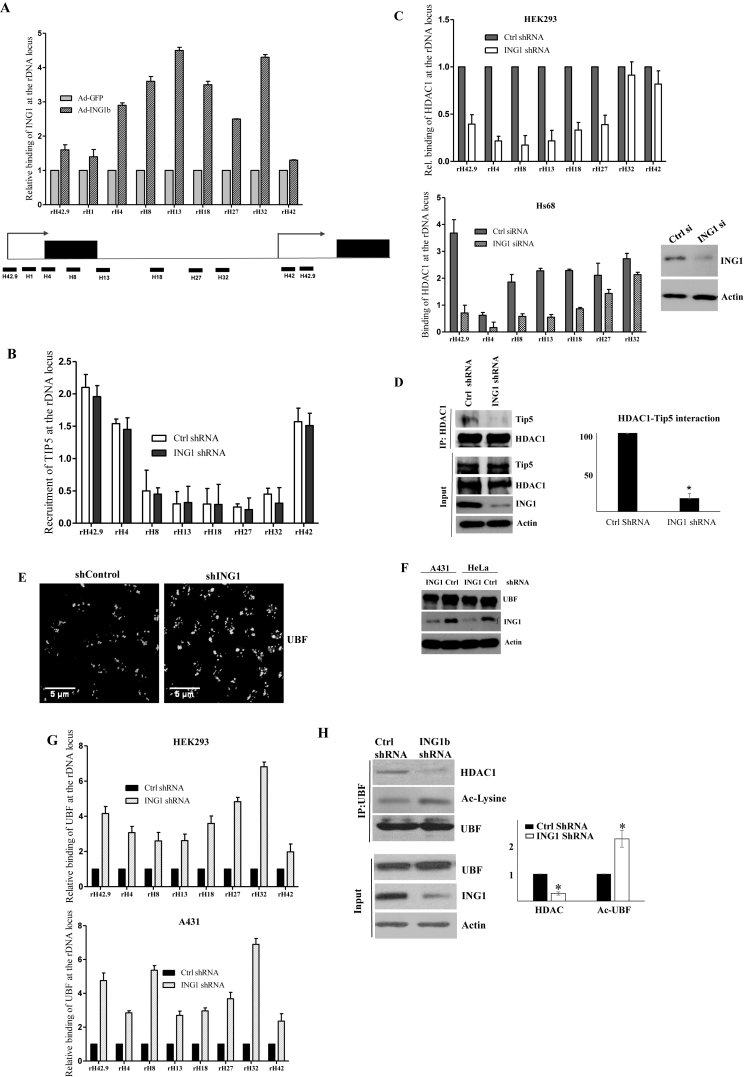
We next asked whether nucleolar ING1 functioned by binding the rDNA promoter. Chromatin immunoprecipitation and quantitative PCR (qChIP) analysis of Hs68 diploid fibroblast strains showed that ING1 associated with rDNA across the length of the repeats, although it was more abundant in the coding and IGS region than the upstream control element (UCE) or the transcriptional start site (TSS) (Figure 2A). Previous studies have suggested that intergenic spacer repeat transcripts recruit the NoRC complex to regulate the epigenetic state of rDNA (12,47). As NoRC is a major regulator of rDNA chromatin, we analyzed whether ING1 affected recruitment of the NoRC to the repeats. qChIP analysis showed no significant difference in the binding of the NoRC subunit Tip5 to rDNA when ING1 was knocked down (Figure 2B).
Figure 2.
ING1 co-operates with the NoRC to repress rRNA transcription. (A) Chromatin from Hs68 cells infected with Ad-GFP or Ad-ING1b were analyzed for ING1 binding at rDNA loci by ChIP (P < 0.04). Location of primer sequences in the rDNA locus used in all ChIP assays is shown. (B) HEK293 cells expressing control or shING1 were analyzed for Tip5 binding at the rDNA locus by ChIP. Binding was quantified by densitometry and is shown in the graph (P < 0.05). (C) HDAC1 binding to rDNA was analyzed in HEK293 cells expressing control or shING1 (P < 0.05). Hs68 cells transfected with control or siING1 were assayed by ChIP to check relative enrichment of HDAC1 at the rDNA locus. The blot shows efficiency of ING1 knockdown. (D) HeLa cells expressing shControl or shING1 were immunoprecipitated for HDAC1 and precipitates were blotted for Tip5 (270KDa) and HDAC1 as a control. (E) Control and ING1 knockdown HeLa cells were immunostained in parallel with UBF antibody and photographed using identical settings. (F) Protein levels of UBF, ING1 and actin in A431 and HeLa cells expressing control or shING1. (G) ChIP analysis in HEK293 and A431 cells to check UBF binding in the rDNA locus, with or without ING1 knockdown (P < 0.05). (H) HEK293 cells expressing control shRNA or shING1 were immunoprecipitated with UBF antibody to analyze association with HDAC1. The IP lysates were also blotted with anti-acetyl lysine (Ac-lysine) to check for levels of acetylated UBF. The blots were quantified by densitometry (P < 0.04).
As a component of the Sin3/HDAC complex, ING1 modulates local histone acetylation at Pol II promoters by recruiting HDAC (28,29). Since NoRC-mediated silencing of rDNA repeats requires HDAC1 (14), we tested the effect of altering ING1 levels on recruitment of HDAC1 to rDNA loci, and the interaction of HDAC1 with Tip5. As shown in Figure 2C, ING1 knockdown reduced the association of HDAC1 with rDNA. Furthermore, ING1 knockdown also reduced the association of HDAC1 with the Tip5 protein (Figure 2D). This suggested that ING1 acts as a scaffold to affect the state of nucleolar DNA by recruiting chromatin modifiers.
ING1 regulates UBF function in nucleoli
UBF is a transcription factor that is required for Pol I-mediated rRNA synthesis. Since knockdown of ING1 affected rRNA levels and HDAC recruitment to rDNA, we analyzed its effect on UBF binding to rDNA repeats. Knockdown of ING1 increased the nucleolar UBF signal without affecting its total protein levels, suggesting an increase in binding of UBF to rDNA promoters (Figure 2E and F). ChIP assays with nuclear extracts of control and ING1 knockdown cells also showed 2- to 7-fold increase in UBF binding at the rDNA repeats (Figure 2G). Deacetylation of UBF by HDAC1 is known to inactivate it, resulting in inhibition of rRNA transcription (6). Since we observed reduced HDAC1 association when ING1 was knocked down, we analysed the effect of ING1 levels on UBF–HDAC1 association. Co-immunoprecipitation analysis showed that knockdown of ING1 reduced the UBF–HDAC1 association (Figure 2H) and western blotting of the immunoprecipitated UBF using anti-acetyl lysine antibody showed that it was maintained in a hyperacetylated active state in these cells.
ING1 affects the epigenetic state of rDNA
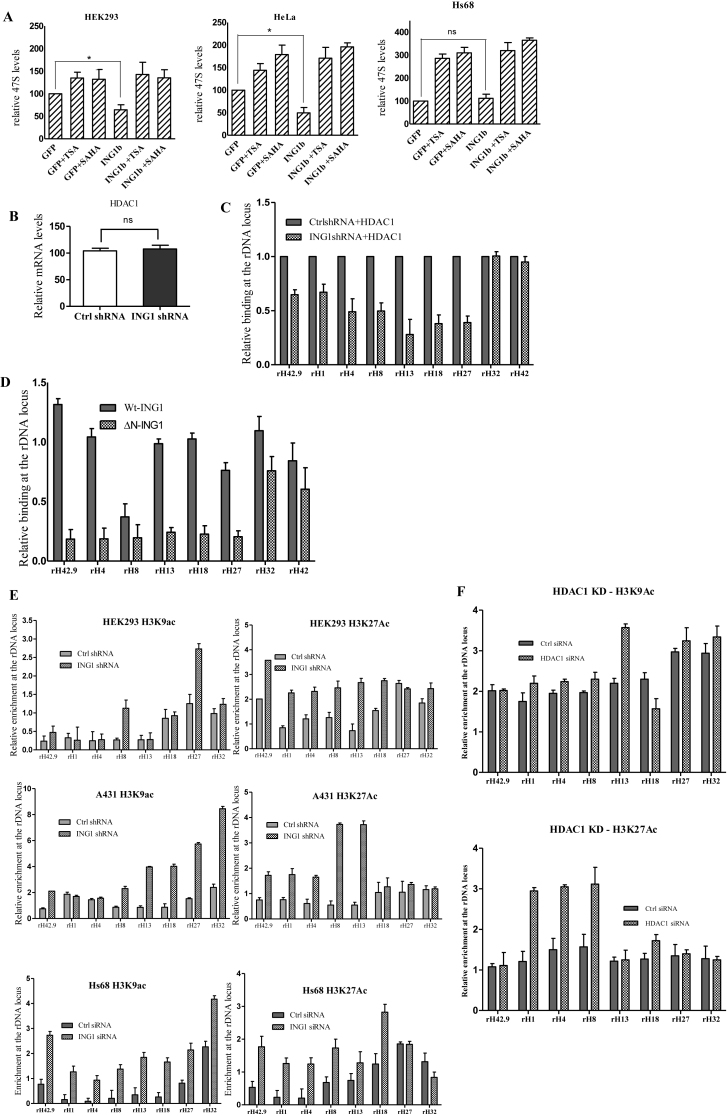
To test whether HDAC1 is required for ING1 to regulate rRNA levels we treated GFP and ING1 expressing cells with the HDAC inhibitors trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA). As shown in Figure 3A, while ING1 expression repressed rRNA transcript levels in HEK293 and in HeLa cells, HDACi treatments abrogated this effect and maintained rRNA levels similar to that of GFP expressing cells undergoing the same treatment, confirming that HDAC1 is required for this regulation. HDAC1 mRNA levels during ING1 knockdown was comparable to control conditions (Figure 3B). Furthermore, we found that overexpression of HDAC1 during ING1 knockdown did not increase HDAC1 binding to rDNA promoters (Figure 3C), showing that ING1 was required for HDAC1 to bind rDNA and to associate with UBF and NoRC.
Figure 3.
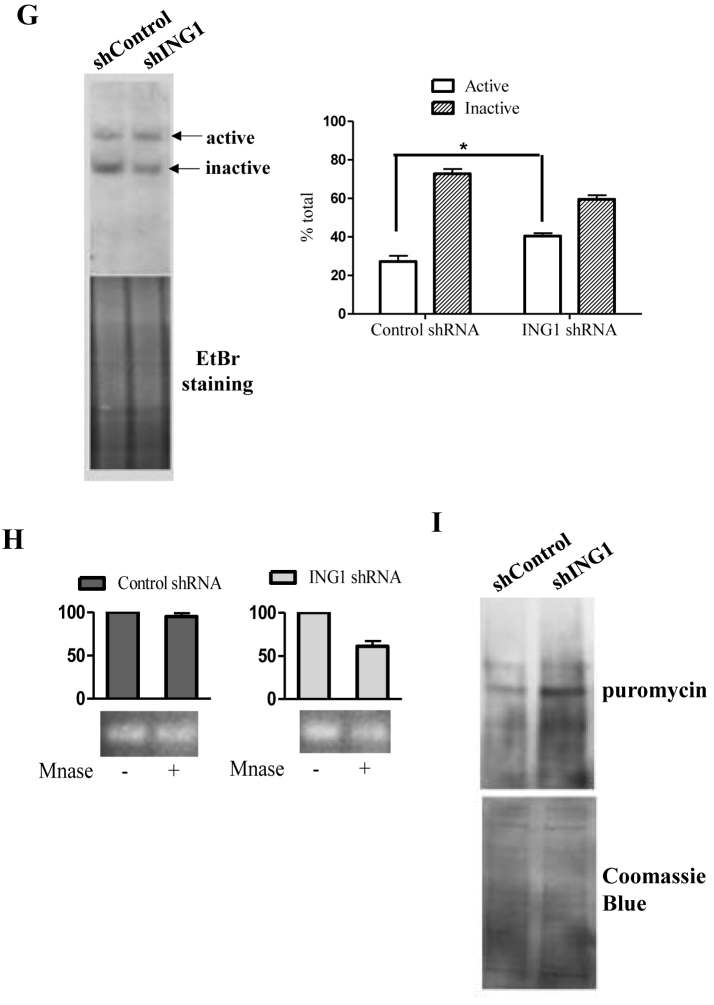
ING1 affects the epigenetic state of rDNA. (A) HEK293 and HeLa cells transfected with control GFP or ING1 plasmid were treated with DMSO, TSA or SAHA for 24 h and checked for 47S rRNA levels by qRT-PCR (P < 0.05). Lower panel: Hs68 cells infected with Ad-GFP or Ad-ING1 for 24 h were treated with DMSO, SAHA or TSA. 47S rRNA levels were assayed using qRT-PCR (P < 0.05). (B) mRNA levels of HDAC1 in control and ING1 knockdown cells by qRT-PCR. (C) HEK293 cells treated with control or shING1 for 48 h were transfected with pcDNA-HDAC1-flag. Relative enrichment of HDAC1 was analyzed at the rDNA locus by ChIP (P < 0.05). (D) HEK293 cells transfected with pCI-Wt-ING1-flag or pCI-ING1ΔN-flag (lacks aa 1–125 constituting the SAID domain) were assayed for HDAC1 binding at rDNA by ChIP using HDAC1 antibody (P < 0.04). (E) HEK293 and A431 cells expressing shING1 and Hs68 cells transfected with control or siING1 were assayed by ChIP for relative enrichment of acetyl H3K9 and acetyl H3K27 at the rDNA locus. (F) Hs68 cells transfected with either control siRNA or HDAC1 siRNA were analysed for acetylation at H3K9 and K27 residues by ChIP assay (P < 0.05). (G) Nuclei of A431 cells stably expressing control or shING1 were crosslinked with psoralen and analyzed by biotin-labelled probe to the core rDNA promoter. The agarose gel stained with ethidium bromide serves as a loading control. The graph shows the means for quantification of active versus inactive rDNA repeats from three experiments (P < 0.05). (H) Nuclei from A431 control or shING1 cells were digested with MNase to estimate relative amounts of accessible rDNA repeats. Amplicons of the MNase digested samples were normalized with respect to undigested controls (P < 0.05). (I) A431 cells expressing control or shING1 were checked for puromycin incorporation as a measure of protein synthesis. The membrane stained with Coommassie Brilliant Blue serves as a loading control.
We next analysed the role of the Sap30 interacting domain (SAID) of ING1, which associates with the mSIN3A/HDAC1 complex (34) in regulating rDNA expression. We observed that cells transfected with ΔN-ING1 (lacking the SAID) showed reduced association of HDAC1 with rDNA compared to cells expressing WT-ING1 (Figure 3D). Association of HDAC1 in cells expressing ΔN-ING1 was comparable to that observed in ING1 knockdown cells, demonstrating that ING1 mediated its effects on rDNA through HDAC1 (Figure 3D). HDAC1 regulates heterochromatin formation and promoter activity through altering histone acetylation at target sites. Hence we analysed the acetylation status of H3K9 and H3K27 residues, which are associated with increased transcription, at rDNA loci. Knockdown of ING1 increased acetylation of both these residues along most of the rDNA repeats in all three cell lines tested (Figure 3E). Although the acetylation status at H3 was affected by knockdown of ING1, the density of total histone H3 at the rDNA loci did not differ significantly between the control and the ING1 knockdown lines (Supplementary Figure S4). This suggested that reduced levels of ING1 increased the signatures associated with open chromatin and efficient Pol I transcription on rDNA. We further compared the histone acetylation pattern at the rDNA loci induced by knockdown of ING1 to the pattern induced by siRNA-mediated knockdown of HDAC1 itself. As shown in Figure 3F, HDAC1 knockdown also resulted in increased acetylation of H3K9 and K27 residues, but the patterns observed were distinct from that of ING1 knockdown. This difference might be due to the presence of HDAC2, which is functionally redundant with HDAC1 in many conditions, while ING1 targets both HDAC1 and 2 to chromatin as the part of mSIN3A/HDAC1 complex.
We analyzed whether these modifications altered the organization of rDNA chromatin and increased the number of active rDNA repeats available for Pol-1 transcription. We used two assays, psoralen crosslinking and micrococcal nuclease (MNase) digestion, which are based on susceptibility of open chromatin to the intercalating agent psoralen and MNase activity, respectively. A431 cells showed an increased proportion of slower migrating, psoralen-crosslinked active rDNA units (from 28% to 43% of the total, ∼1.7-fold increase), suggesting decondensation or reduced heterochromatinization of rDNA when ING1 was knocked down (Figure 3G). MNase digestion of rDNA chromatin confirmed the increase in susceptibility of rDNA in ING1 knockdown cells (Figure 3H). Hence, ING1 promotes establishing the heterochromatinization of rDNA repeats by facilitating NoRC-HDAC1 association.
Since rRNA levels are a limiting factor in ribosome biogenesis and protein synthesis (48), we tested if increased rRNA levels seen during ING1 knockdown affected protein synthesis. Using the recently developed SUnSET protocol which measures incorporation of puromycin into nascent peptides, we observed an almost 2-fold increase in global protein synthesis in A431 cells expressing ING1 shRNA (Figure 3I). This suggested that regulation of rRNA expression by ING1 affects cell growth through regulating total protein synthesis.
ING1 regulates rRNA expression during glucose deprivation and rapamycin treatment
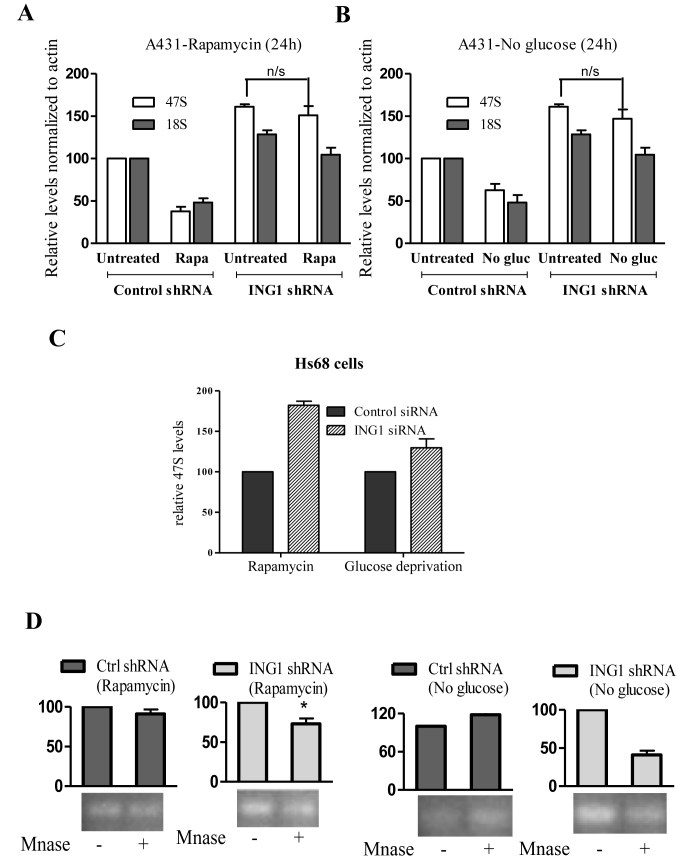
Cells respond to several stresses by reducing rRNA synthesis either by inhibiting Pol I machinery through post-translational modifications or by increasing heterochromatin content of rDNA repeats. Stress conditions such as glucose deprivation and rapamycin treatment lead to epigenetic modifications that increase rDNA heterochromatin formation and reduce rRNA synthesis (17,18,21,49). We used these conditions to test the role of ING1 in rDNA heterochromatinization during stress. Both rapamycin and glucose deprivation reduced rRNA levels in A431 cells and Hs68 primary diploid fibroblasts, and knockdown of ING1 during stress inhibited this effect (Figure 4A–C). This failure of transcriptional repression was mainly due to lack of decondensation of rDNA chromatin, as evident from the susceptibility of these cells to MNase digestion (Figure 4D). Interestingly, we noted marginal increases in ING1 mRNA levels during these stress conditions, consistent with ING1 functioning as part of a cellular stress response program (50) (Supplementary Figure S5) that can be altered by changing subcellular localization (32).
Figure 4.
ING1 regulates rDNA transcription during stress. (A and B) A431 cells expressing control or shING1 constructs were treated with 20 nM rapamycin (24 h) or cultured in medium lacking glucose (24 h). Cells were assayed for relative levels of 47S and 18S rRNA by qRT-PCR. Values were normalized to actin (P < 0.05). (C) Levels of 47S rRNA were assayed in HS68 cells transfected with siING1 or siControl that were subjected to rapamycin treatment or glucose deprivation (P < 0.04). (D) Nuclei from cells treated as above were digested with MNase and subjected to PCR using primers for the 47S core promoter. PCR amplicons of MNase digested samples were normalized to undigested controls (P < 0.04).
ING1 regulates nucleolar translocation and activity of mTOR
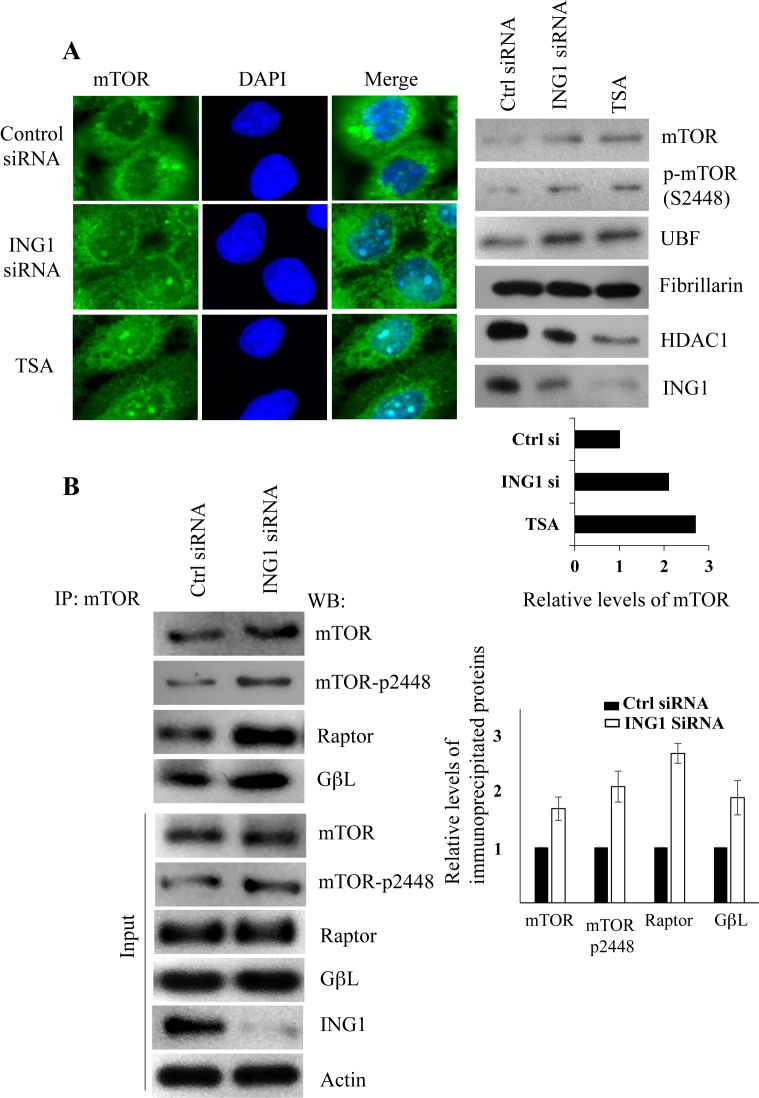
mTOR is a critical node in a cell signalling network that links cellular metabolic state to rDNA transcription and protein synthesis. Since we observed that ING1 was required for efficient rRNA repression during rapamycin treatment and glucose deprivation, two conditions that inhibit mTOR function, we tested whether knockdown of ING1 affected mTOR-mediated regulation of rRNA levels. Immunofluorescence analysis showed that ING1 knockdown and TSA treatment increased mTOR staining in the nucleoli of A431 cells (Figure 5A). Western blotting of purified nucleoli of A431 cells transfected with control or ING1-specific siRNA, or treated with TSA, showed that ING1 knockdown induced a 2-fold increase in the nucleolar levels of mTOR (Figure 5A, right panels). Both ING1 knockdown and TSA treatment increased the levels of mTOR phosphorylated on the S2448 residue in nucleoli. Consistent with our previous observations, increased UBF levels and reduced HDAC1 levels were also evident in these nucleolar lysates (Figure 5A).
Figure 5.
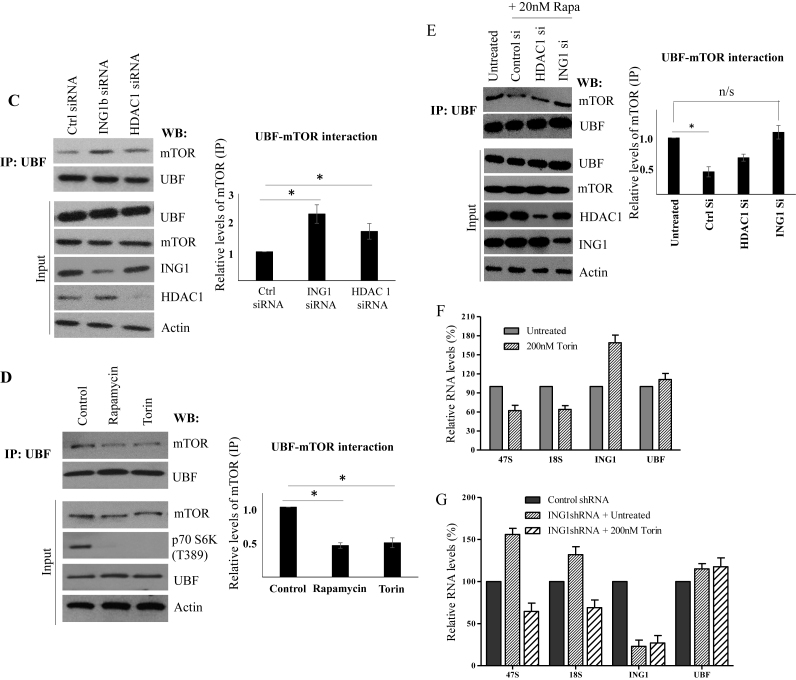
ING1 regulates mTOR localization and function. (A) A431 cells treated with TSA, transfected with siControl or siING1 were fixed and stained using mTOR antibody. Nucleolar fractions isolated from ING1 or control siRNA-transfected, or TSA treated A431 cells were analyzed in parallel for levels of the indicated proteins by western blotting. The graph shows relative levels of mTOR as estimated by scanning densitometry. (B) Control and ING1b knockdown cells were treated with rapamycin and immunoprecipitation was carried out using anti-mTOR antibodies. The proteins indicated were detected in the precipitates by western blotting. Blots were quantified by densitometry and are presented as graphs. Total cell lysate was used to determine the levels of these proteins in the cells and as loading controls. (C) Lysates from A431 cells transfected with siControl, siING1 or siHDAC1 were immunoprecipitated with UBF antibody and blotted for mTOR. Blots of whole cell lysates serve as controls. The quantification of UBF-mTOR interaction under each condition are presented in the graph. (D) A431 cells treated with rapamycin or Torin were lysed, immunoprecipitated with UBF antibody and precipitates were blotted for the indicated proteins and quantified by densitometry (P < 0.05). Whole cell lysates show expression of proteins and p70 S6K T389 phosphorylation indicates mTOR activity. (E) A431 cells transfected with siControl, siHDAC1 or siING1 were treated with rapamycin, lysed, immunoprecipitated with UBF1 antibody and blotted for mTOR and UBF1. Untreated cells were used as a reference. Blotting of lysates was used to confirm ING1 and HDAC1 knockdown. (F) A431 cells were treated with 200 nM Torin for 4 h and qRT-PCR was used to estimate relative abundance of 47 and 18S rRNA, ING1 and UBF in total RNA using actin as a loading control. (G) Relative levels of 47 and 18S rRNA, ING1 and UBF were determined by quantitative real time PCR using actin as a control, using RNA from A431 cells expressing shControl or shING1 ± Torin treatment. The graphs represent the average of three independent experiments.
We also observed that knockdown of ING1 increased association of mTOR with the TORC1 components, Raptor and GβL in the presence of rapamycin (Figure 5B), consistent with ING1 being required for efficient inhibition of mTOR by rapamycin. Recent reports have suggested that, in addition to regulating downstream effector kinases, mTOR can directly interact with rDNA chromatin (23,24,51). Activation of UBF is one of the mechanisms by which mTOR increases rRNA synthesis, and since knockdown of ING1 increased nucleolar localization and association of UBF with rDNA chromatin, we tested for UBF–mTOR association by co-immunoprecipitation. As shown in Figure 5C, knockdown of ING1 increased association of UBF with mTOR. Consistent with the immunofluorescence data on mTOR localization (Figure 5A), knockdown of HDAC1 also increased mTOR–UBF association, but mTOR inhibitors Torin and rapamycin reduced this association (Figure 5D). In order to analyze whether ING1 inhibited the mTOR–UBF interaction during rapamycin treatment, we immunoprecipitated UBF from rapamycin-treated cells after ING1 or HDAC1 knockdown. Blotting these immunoprecipitates showed that rapamycin-mediated inhibition of mTOR–UBF association was reduced when either ING1 or HDAC1 was knocked down (Figure 5E). This suggests that the ING1–HDAC1 complex and mTOR have opposing roles in regulating rDNA transcription and that rapamycin-mediated rDNA transcriptional repression requires ING1/HDAC1 activity.
To confirm that mTOR activity increased rRNA transcription during ING1/HDAC1 knockdown, we analyzed the 47S rRNA transcript levels in ING1 knockdown cells in the presence or absence of the mTOR kinase inhibitor, Torin. The increase in 47S transcript levels that we previously observed upon ING1 knockdown was inhibited by Torin (Figure 5F and G), confirming that loss of ING1 and HDAC1 increased the localization and activation of mTOR in the nucleolus, where it associated with UBF to increase rDNA transcription.
Epigenetic silencing during monocyte differentiation is mediated by ING1
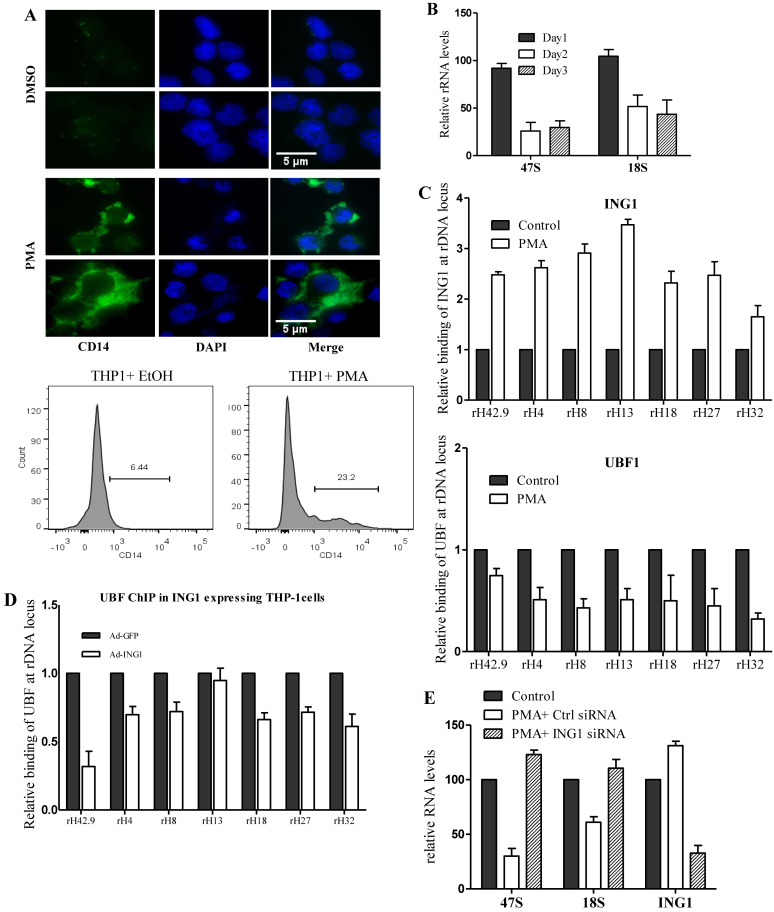
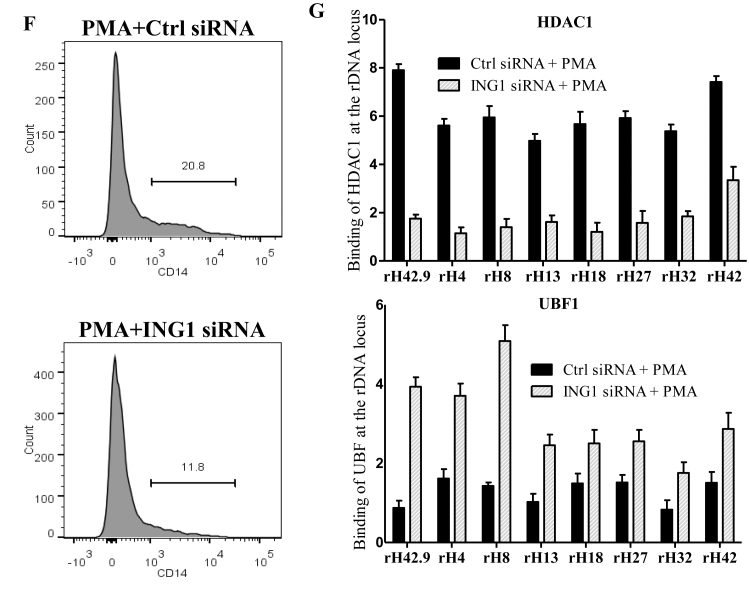
Cells exiting the cell cycle during terminal differentiation often have reduced requirements for protein synthesis, and they turn off a fraction of their rRNA genes through epigenetic modifications (52–55). We used phorbol 12-myristate 13-acetate (PMA)-induced differentiation of THP-1 monocyte cells to macrophages as a model to analyze the role of ING1 in this process. The PMA-induced differentiation, confirmed by the expression of macrophage-surface marker CD14 (Figure 6A) using immunofluorescence and flow cytometry, resulted in reduced rRNA levels in THP-1 cells by day 3 (Figure 6B). ChIP analyses showed that association of ING1 with rDNA increased several fold on the second day of PMA treatment, with a concomitant decrease in UBF binding (Figure 6C). This difference corroborated the observation that ING1 and UBF showed reciprocal association and function on rDNA loci. Moreover, adenoviral overexpression of ING1 in undifferentiated THP-1 cells reduced UBF binding to rDNA (Figure 6D). Conversely, PMA treatment of siING1-transfected THP-1 cells failed to repress rDNA transcription, with cells showing higher levels of 47S and 18S rRNA (Figure 6E), resulting in reduced differentiation of THP-1 cells as shown by flow cytometric analysis of CD14 surface marker (Figure 6F) and increased THP1 cell proliferation (Supplementary Figure S6). This alleviation of rRNA repression in cells with reduced ING1 levels, concomitant with defective HDAC1 recruitment and increased UBF binding to rDNA, was evident in qChIP analysis (Figure 6G). These observations show that the nucleolar ING1–HDAC axis is a significant regulator of rDNA epigenetics and transcription.
Figure 6.
ING1 is required for rDNA silencing during monocyte differentiation. (A) Immunostaining of THP-1 cells for expression of the CD14 macrophage marker. Two representative fields are shown. Nuclei were co-stained with DAPI. Differentiation of THP1 cells was also tested by flow cytometric analysis using a CD14-PEcy7 antibody (lower panel). (B) 47S and 18S rRNA levels were assayed using qRT-PCR during the 3 days of PMA treatment. (C) ChIP of day 2 PMA treated cells using ING1 or UBF antibodies to check relative rDNA occupancy (P < 0.05). (D) Undifferentiated THP-1 cells were infected with Ad-GFP or Ad-ING1 and levels of UBF at the rDNA loci shown were determined by ChIP. (E) THP-1 cells transfected with siControl or siING1 were induced to differentiate with PMA and levels of 47S and 18S transcripts were quantified on day 3 by qRT-PCR. The levels of ING1 mRNA are included to show knockdown efficiency (P < 0.05). (F) THP-1 cells transfected with siControl or siING1 were induced to differentiate in the presence of PMA for 3 days and were tested for differentiation by flow cytometric analysis for CD14 expression. (G) ING1 or control knockdown cells were assayed for HDAC1 (top panel) and UBF (lower panel) binding at the rDNA locus by ChIP (P < 0.05). Error bars are from three independent experiments.
DISCUSSION
In this study, we describe a novel role for ING1 in regulating rRNA transcription by modulating nucleolar localization and function of HDAC1 and mTOR. ING1 bound throughout the rDNA loci, but was enriched at intergenic spacer regions. Knockdown of ING1 reduced binding of HDAC1 to rDNA, resulting in attenuated NoRC-mediated transcriptional silencing, and increased H3K9 and H3K27 acetylation, rDNA decondensation, and the number of active rDNA copies. Increase in the acetylation status of rDNA chromatin that we observed correlates with active transcription (56) while silent NORs have repressive methyl H3K9 (57). ING1 knockdown altered chromatin structure and alleviated HDAC1-mediated deacetylation, increasing UBF binding across the rDNA locus. Knockdown of HDAC1 also resulted in hyperacetylation of histones at the rDNA loci, although in a different pattern compared to ING1 knockdown, most likely due to the activity of the closely related HDAC2 in these cells. We also observed increased nuclear localization of mTOR and its interaction with UBF upon ING1 knockdown. Furthermore, ING1 knockdown partially alleviated the inhibitory effect of rapamycin on mTOR in the nucleolus, suggesting functional antagonism between nucleolar ING1 and mTOR. The effect of ING1 on rRNA levels during stress and differentiation, and increase in protein synthesis observed during ING1 knockdown, suggest that nucleolar ING1 is an important regulator of cell growth.
Epigenetic modifications mediated by the energy-sensing complex eNoSC is known to repress rDNA transcription during nutrient stress. The SIRT1 deacetylase, which associates with the eNoSC protein complex, deacetylates histone H3 and inhibits Pol I transcription during nutrient stress (17). Similarly, inhibition of TORC1 by rapamycin leads to association of the Sir2-containing RENT complex to rDNA loci, leading to increased deacetylation and rDNA silencing in yeast (58). Here we report that ING1 is a chromatin regulator that recruits HDAC1 to modify the epigenetic state of nucleolar DNA by histone acetylation. We analysed the role of ING1 as an epigenetic regulator under conditions of stress and THP-1 differentiation. As in normal cells, knockdown of ING1 in differentiating THP-1 cells inhibited transcriptional repression of rRNA and increased UBF binding at the rDNA locus. We observed that HDAC1 binding was reduced at rDNA promoters when ING1 was knocked down, further confirming that ING1 is required for NoRC-mediated silencing of rDNA repeats during monocyte differentiation. We also observed that PMA-induced differentiation of THP-1 cells was abrogated when ING1 was knocked down, suggesting a physiological role for this process. ING1 knockdown also resulted in increased proliferation of PMA-treated THP1 cells compared to control cells. A role for NoRC in mouse adipocyte differentiation has also been reported (59), suggesting that ING1–HDAC1–Tip5 association might function in silencing rDNA loci during terminal differentiation of multiple cell types.
This role of nucleolar ING1 on rRNA via effects of mTOR and the NoRC partly explains its role as a cell growth regulator and tumor suppressor. We have previously reported that UV increased nucleolar ING1 translocation (39), and its association with PCNA (50). This suggests that ING1 can modulate nucleolar chromatin during both transcription and repair.
Two distinct functional chromatin states of rDNA, reflecting transcriptional activity of individual repeats, are faithfully propagated epigenetically during cell division (60). Psoralen crosslinking assays suggest that at least half of the NORs in mouse cells are maintained in a transcriptionally inactive state though histone modifications and CpG methylation (13). The increase in transcriptionally active rDNA repeats during ING1 knockdown seen establishes a role for ING1 as an epigenetic regulator that can alter the ratio of active:inactive rDNA repeats by modulating nucleolar chromatin architecture.
The nucleolar remodeling complex (NoRC) is important to establish the heterochromatin state of rDNA repeats. Recruitment of Tip5, a NoRC subunit, to rDNA promoters was not affected by ING1, suggesting indirect modulation of NoRC function and epigenetic status of rDNA by ING1. This is achieved when ING1 recruits HDAC1 to rDNA, facilitating its interaction with the NoRC complex. The requirement of the SAID of ING1 in this process is reminiscent of the role of ING1 as an adaptor that recruits the Sin3/HDAC co-repressor complex to Pol II promoters (34). This was supported by TSA and SAHA-mediated alleviation of reduced rRNA levels seen in ING1 overexpressing cells. We suggest that while the PHD domain of Tip5 is sufficient to target NoRC complexs to rDNA clusters, ING1 was required for HDAC1 to bind NoRC and suppress rDNA transcription. A model of the interactions occurring between ING1 and other nucleolar proteins within the rDNA clusters is shown in Figure 7.
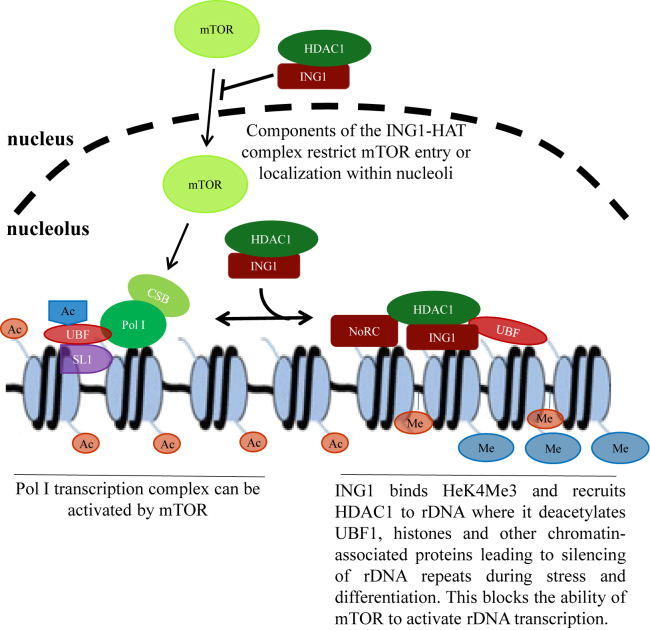
Figure 7.
Model of the role of ING1 in regulating Pol I transcription. rDNA transcription is tightly regulated by epigenetic mechanisms and growth factor/kinase signaling, partly through mTOR that integrates many growth signals. The interaction of Pol I complex subunits with the transcription machinery including UBF1 is required for the formation of enhanceosomes and promoter melting. Histone modifying enzymes acetylate proteins within the rDNA repeats to make them more accessible to the Pol I transcriptional machinery. ING1 reduces mTOR localization to nucleoli and helps recruit HDAC1 to the NoRC complex and to UBF for deacetylation and transcriptional repression. Loss of ING1 also increased H3 acetylation and increased the number of active rDNA units available for transcription. This study suggests that ING1 facilitates HDAC1 activity and antagonizes mTOR function in the nucleolus, and is required for maintaining a portion of the rDNA repeats in a silenced state in normally replicating cells and during differentiation of monocytes.
Knockdown of ING1 increased the nucleolar localization of mTOR and its interaction with UBF. Signal- and nutrient-initiated pathways converge on mTOR to regulate rRNA synthesis and processing, ribosome assembly, and protein synthesis (19). Although mTOR is reported to affect nucleolar transcription through its downstream effector p70 S6K, recent reports have also described nucleolar localization of mTOR and its association with rDNA (23,24). Consistent with this, we observed that both ING1 knockdown and TSA increased nucleolar mTOR. The increase in rRNA when ING1 is knocked down is at least partly mediated through mTOR kinase activity since Torin blocked this process. This suggests functional antagonism between the ING1 and mTOR pathways.
While we describe here that ING1 and HDAC1 regulate nucleolar localization and function of mTOR, some reports suggest an upstream role for mTOR where it controls Rpd3-Sin3 histone deacetylase to modulate rDNA heterochromatinization (51,61). Knockdown of ING1 reduced rapamycin-, but not Torin-mediated inhibition of mTOR. We did not observe physical interaction between ING1 and mTOR (data not shown) and so the mechanism by which ING1 affects mTOR activation and localization is not clear. ING1 did not affect levels of FKBP12, which modulates inhibition of mTOR by rapamycin (62). However, the effect of ING1 knockdown and HDAC1 inhibition on mTOR suggest either a chromatin-based or a posttranslational mechanism.
Nucleolar function is impacted by cancer-related proteins such as p53, which can affect rRNA transcription by interacting with SL1 (63). Other tumor suppressors that have been reported to affect rRNA levels and Pol I function include VHL (64), PTEN (65) and p14ARF (66,67). Here we show that in addition to the effect on rDNA chromatin, ING1 suppressed nucleolar mTOR activity. In summary, we report that the epigenetic regulator ING1 cooperates with the NoRC at rDNA loci, and affects mTOR localization and its interaction with UBF in nucleoli to regulate rRNA synthesis. Both HDAC1 and mTOR integrate several signals to determine cell growth and response to stimuli. Our observation that ING1 regulates rDNA through these pathways during normal cell growth, cell stress and differentiation suggests that the regulation of rDNA may constitute the major tumor suppressor function of ING1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steven Elledge for the pINDUCER lentiviral toolkit, Brent Winston for input on macrophage differentiation and Savraj Grewal for experimental suggestions.
Footnotes
Present address: Uma Karthika Rajarajacholan, Program in Cellular & Molecular Medicine, Boston Children's Hospital, Boston MA 02115, USA; Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research [#MOP-133646] and Alberta Innovates–Health Solutions (to K.R.). Alberta Cancer Foundation (to U.K.R.). Funding for open access charge: CIHR or the University of Calgary open access fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Warner J.R. Nascent ribosomes. Cell. 2001; 107:133–136. [DOI] [PubMed] [Google Scholar]
- 2.Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I.. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007; 8:574–585. [DOI] [PubMed] [Google Scholar]
- 3.Moss T., Stefanovsky V.Y.. At the center of eukaryotic life. Cell. 2002; 109:545–548. [DOI] [PubMed] [Google Scholar]
- 4.Wang L., Gout I., Proud C.G.. Cross-talk between the ERK and p70 S6 kinase (S6K) signaling pathways. MEK-dependent activation of S6K2 in cardiomyocytes. J. Biol. Chem. 2001; 276:32670–32677. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J., Yuan X., Frodin M., Grummt I.. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003; 11:405–413. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier G., Stefanovsky V.Y., Faubladier M., Hirschler-Laszkiewicz I., Savard J., Rothblum L.I., Cote J., Moss T.. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell. 2000; 6:1059–1066. [DOI] [PubMed] [Google Scholar]
- 7.Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L.I., Moss T.. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell. 2006; 21:629–639. [DOI] [PubMed] [Google Scholar]
- 8.Bazett-Jones D.P., Leblanc B., Herfort M., Moss T.. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994; 264:1134–1137. [DOI] [PubMed] [Google Scholar]
- 9.Stefanovsky V.Y., Pelletier G., Bazett-Jones D.P., Crane-Robinson C., Moss T.. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001; 29:3241–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Sullivan A.C., Sullivan G.J., McStay B.. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002; 22:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V. et al. . UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008; 183:1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R.. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006; 22:351–361. [DOI] [PubMed] [Google Scholar]
- 13.Santoro R., Grummt I.. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 2005; 25:2539–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Santoro R., Grummt I.. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002; 21:4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowbotham S.P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W.R., Webster J., Oxley D. et al. . Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell. 2011; 42:285–296. [DOI] [PubMed] [Google Scholar]
- 16.Guetg C., Scheifele F., Rosenthal F., Hottiger M.O., Santoro R.. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012; 45:790–800. [DOI] [PubMed] [Google Scholar]
- 17.Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K. et al. . Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008; 133:627–639. [DOI] [PubMed] [Google Scholar]
- 18.Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I.. The nucleolus under stress. Mol. Cell. 2010; 40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer C., Grummt I.. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006; 25:6384–6391. [DOI] [PubMed] [Google Scholar]
- 20.Mayer C., Zhao J., Yuan X., Grummt I.. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004; 18:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Fan M., Pfeffer L.M., Laribee R.N.. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 2012; 40:6534–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannan K.M., Brandenburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G.A., Pearson R.B. et al. . mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 2003; 23:8862–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Tsang C.K., Watkins M., Bertram P.G., Zheng X.F.. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006; 442:1058–1061. [DOI] [PubMed] [Google Scholar]
- 24.Tsang C.K., Liu H., Zheng X.F.. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010; 9:953–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D.G., Baetz K., Shi X., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L.. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 2006; 26:7871–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena P.V., Davrazou F., Shi X., Walter K.L., Verkhusha V.V., Gozani O., Zhao R., Kutateladze T.G.. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006; 442:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R. et al. . ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006; 442:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyon Y., Cayrou C., Ullah M., Landry A.J., Cote V., Selleck W., Lane W.S., Tan S., Yang X.J., Cote J.. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006; 21:51–64. [DOI] [PubMed] [Google Scholar]
- 29.Loewith R., Meijer M., Lees-Miller S.P., Riabowol K., Young D.. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 2000; 20:3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerillon C., Larrieu D., Pedeux R.. ING1 and ING2: multifaceted tumor suppressor genes. Cell. Mol. Life Sci.: CMLS. 2013; 70:3753–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tallen G., Riabowol K.. Keep-ING balance: tumor suppression by epigenetic regulation. FEBS Lett. 2014; 588:2728–2742. [DOI] [PubMed] [Google Scholar]
- 32.Vieyra D., Senger D.L., Toyama T., Muzik H., Brasher P.M.A., Johnston R.N., Riabowol K., Forsyth P.A.. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clin. Cancer Res. 2003; 9:5952–5961. [PubMed] [Google Scholar]
- 33.Champagne K.S., Kutateladze T.G.. Structural insight into histone recognition by the ING PHD fingers. Curr. Drug Targets. 2009; 10:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzmichev A., Zhang Y., Erdjument-Bromage H., Tempst P., Reinberg D.. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 2002; 22:835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nourani A., Doyon Y., Utley R.T., Allard S., Lane W.S., Cote J.. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 2001; 21:7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skowyra D., Zeremski M., Neznanov N., Li M., Choi Y., Uesugi M., Hauser C.A., Gu W., Gudkov A.V., Qin J.. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 2001; 276:8734–8739. [DOI] [PubMed] [Google Scholar]
- 37.Xin H., Yoon H.G., Singh P.B., Wong J., Qin J.. Components of a pathway maintaining histone modification and heterochromatin protein 1 binding at the pericentric heterochromatin in Mammalian cells. J. Biol. Chem. 2004; 279:9539–9546. [DOI] [PubMed] [Google Scholar]
- 38.Schafer A., Karaulanov E., Stapf U., Doderlein G., Niehrs C.. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev. 2013; 27:261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott M., Boisvert F.M., Vieyra D., Johnston R.N., Bazett-Jones D.P., Riabowol K.. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res. 2001; 29:2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N., Zhao G., Chen T., Xue L., Ma L., Niu J., Tong T.. Nucleolar protein CSIG is required for p33ING1 function in UV-induced apoptosis. Cell Death Dis. 2012; 3:e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T. et al. . The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 43.Rajarajacholan U.K., Thalappilly S., Riabowol K.. The ING1a tumor suppressor regulates endocytosis to induce cellular senescence via the Rb-E2F pathway. PLoS Biol. 2013; 11:e1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K., Boland D., Gong W., Riabowol K.. Domain recognition of the ING1 tumor suppressor by a panel of monoclonal antibodies. Hybridoma. 2011; 30:239–245. [DOI] [PubMed] [Google Scholar]
- 45.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J.. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005; 7:311–318. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt E.K., Clavarino G., Ceppi M., Pierre P.. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009; 6:275–277. [DOI] [PubMed] [Google Scholar]
- 47.Santoro R., Schmitz K.M., Sandoval J., Grummt I.. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010; 11:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013; 122:487–497. [DOI] [PubMed] [Google Scholar]
- 49.Zillner K., Filarsky M., Rachow K., Weinberger M., Langst G., Nemeth A.. Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res. 2013; 41:5251–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott M., Bonnefin P., Vieyra D., Boisvert F.M., Young D., Bazett-Jones D.P., Riabowol K.. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 2001; 114:3455–3462. [DOI] [PubMed] [Google Scholar]
- 51.Tsang C.K., Bertram P.G., Ai W., Drenan R., Zheng X.F.. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003; 22:6045–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayashi Y., Kuroda T., Kishimoto H., Wang C., Iwama A., Kimura K.. Downregulation of rRNA transcription triggers cell differentiation. PLoS One. 2014; 9:e98586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M., Tu X., Ferrari-Amorotti G., Calabretta B., Baserga R.. Downregulation of the upstream binding factor1 by glycogen synthase kinase3beta in myeloid cells induced to differentiate. J. Cell Biochem. 2007; 100:1154–1169. [DOI] [PubMed] [Google Scholar]
- 54.Poortinga G., Wall M., Sanij E., Siwicki K., Ellul J., Brown D., Holloway T.P., Hannan R.D., McArthur G.A.. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011; 39:3267–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali S.A., Zaidi S.K., Dacwag C.S., Salma N., Young D.W., Shakoori A.R., Montecino M.A., Lian J.B., van Wijnen A.J., Imbalzano A.N. et al. . Phenotypic transcription factors epigenetically mediate cell growth control. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:6632–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirschler-Laszkiewicz I., Cavanaugh A., Hu Q., Catania J., Avantaggiati M.L., Rothblum L.I.. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001; 29:4114–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santoro R., Li J., Grummt I.. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002; 32:393–396. [DOI] [PubMed] [Google Scholar]
- 58.Ha C.W., Huh W.K.. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011; 39:1336–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J., Langst G., Grummt I.. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006; 25:5735–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conconi A., Widmer R.M., Koller T., Sogo J.M.. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989; 57:753–761. [DOI] [PubMed] [Google Scholar]
- 61.Sun Z.W., Hampsey M.. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999; 152:921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaubitz C., Oliveira T.M., Prouteau M., Leitner A., Karuppasamy M., Konstantinidou G., Rispal D., Eltschinger S., Robinson G.C., Thore S. et al. . Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol. Cell. 2015; 58:977–988. [DOI] [PubMed] [Google Scholar]
- 63.Zhai W., Comai L.. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 2000; 20:5930–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mekhail K., Rivero-Lopez L., Khacho M., Lee S.. Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle. 2006; 5:2401–2413. [DOI] [PubMed] [Google Scholar]
- 65.Zhang C., Comai L., Johnson D.L.. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 2005; 25:6899–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayrault O., Andrique L., Larsen C.J., Seite P.. Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene. 2004; 23:8097–8104. [DOI] [PubMed] [Google Scholar]
- 67.Gonzalez L., Freije J.M.P., Cal S., Lopez-Otin C., Serrano M., Palmero I.. A functional link between the tumour suppressors ARF andp33ING1. Oncogene. 2006; 25:5173–5179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.