SUMMARY
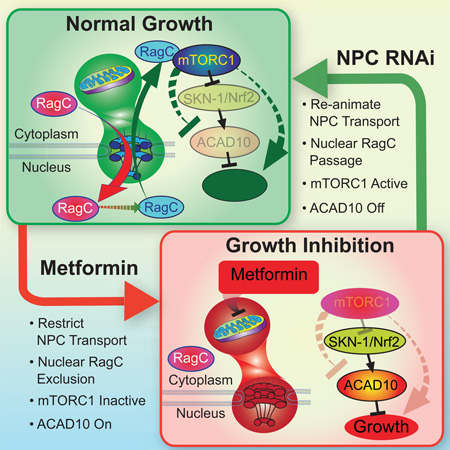
Metformin has utility in cancer prevention and treatment, though the mechanisms for these effects remain elusive. Through genetic screening in C. elegans, we uncover two metformin response elements: the nuclear pore complex (NPC) and acyl-CoA dehydrogenase family member-10 (ACAD10). We demonstrate that biguanides inhibit growth by inhibiting mitochondrial respiratory capacity, which restrains transit of the RagA-RagC GTPase heterodimer through the NPC. Nuclear exclusion renders RagC incapable of gaining the GDP-bound state necessary to stimulate mTORC1. Biguanide-induced inactivation of mTORC1 subsequently inhibits growth through transcriptional induction of ACAD10. This ancient metformin response pathway is conserved from worms to humans. Both restricted nuclear pore transit and upregulation of ACAD10 are required for biguanides to reduce viability in melanoma and pancreatic cancer cells, and to extend C. elegans lifespan. This pathway provides a unified mechanism by which metformin kills cancer cells and extends lifespan, and illuminates potential cancer targets.
Graphical Abstract
INTRODUCTION
Metformin has been used to treat type 2 diabetes (T2D) for nearly 60 years. It also has potential benefit in cancer prevention and treatment (Evans et al., 2005; Yuan et al., 2013). The class of drugs to which metformin belongs, the biguanides, inhibit cellular growth in a variety of cancer cell lines, particularly in melanoma (Yuan et al., 2013) and pancreatic cancer cells (Kordes et al., 2015). While it is widely accepted that the mitochondrion is a primary target of metformin (Griss et al., 2015; Owen et al., 2000; Wheaton et al., 2014), exactly how mitochondrial inhibition by metformin is transduced to the drug’s other health-promoting effects, including its anticancer properties, remains unclear.
Mitochondrial inhibition by metformin causes energetic stress, which results in activation of the energy sensor adenosine monophosphate-activated protein kinase (AMPK) (Zhou et al., 2001). However, multiple lines of evidence indicate that AMPK is dispensable for metformin’s beneficial effects (Foretz et al., 2010; Griss et al., 2015; Kalender et al., 2010), invoking other major metformin effectors downstream of mitochondria.
The protein kinase mechanistic target of rapamycin complex 1 (mTORC1), which also serves as an energy and nutrient sensor, plays a central role in regulating cell growth, proliferation and survival (Schmelzle and Hall, 2000). Inhibition of mTORC1 activity has been reported in cells in culture treated with metformin, suggesting that reduced TOR activity may be important for the metabolic effects of biguanides (Kalender et al., 2010). In support of this idea, both metformin and canonical mTOR inhibitors have highly similar effects on the transcriptome, selectively decreasing mRNA levels of cell cycle and growth regulators (Larsson et al., 2012). Several, distinct pathways are known to regulate mTORC1 signaling, including TSC-Rheb and Ras-related GTP-binding protein (Rag) GTPase-mediated signaling (Sancak et al., 2008). Metformin may inhibit mTORC1 via modulation of Rag GTPases (Kalender et al., 2010), but the mechanism by which this occurs is uncharacterized. It has been suggested that the pathway that leads to metformin-mediated inhibition of mTORC1 could represent a distinct mechanism of mTORC1 regulation, since no signaling pathway has been identified that connects the mitochondrion to mTORC1 without involvement of AMPK (Sengupta et al., 2010). Whether a mitochondrial-mTORC1 signaling relay plays a role in the action of metformin is still unknown.
As in mammals, metformin promotes health and extends lifespan in C. elegans (Cabreiro et al., 2013; De Haes et al., 2014; Onken and Driscoll, 2010), raising the possibility of conservation of genetic pathways responsible for metformin’s beneficial effects. Using unbiased, iterative genetic screens in C. elegans, we identified a single, central genetic pathway by which metformin regulates growth. We report two elements absolutely required for the anti-growth properties of metformin: the nuclear pore complex (NPC), and acyl-CoA dehydrogenase family member 10 (ACAD10). These two metformin response elements were used to illuminate the major, biological pathway through which metformin induces its favorable effects. Remarkably, this ancient pathway unifies mitochondria, the NPC, mTORC1, and ACAD10 into a single signaling relay that mediates metformin’s anti-aging effects in C. elegans and inhibits growth in C. elegans and human cancer cells alike.
RESULTS
Metformin Induces Growth Inhibition by Increasing CeACAD10 Expression in C. elegans
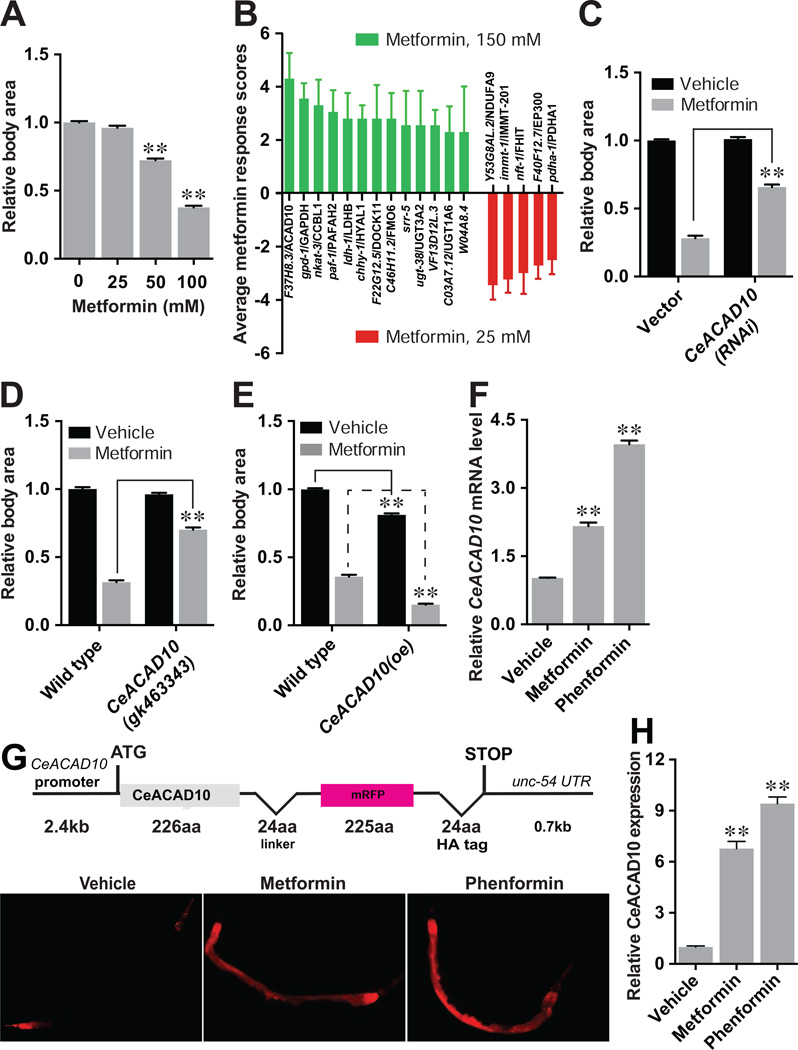
Metformin elicits a dose-dependent decrease in C. elegans growth (Figure 1A), unlike its non-dose-dependent effect on lifespan (Cabreiro et al., 2013). This result parallels metformin’s ability to inhibit growth of certain cancers (Yuan et al., 2013), leading us to hypothesize that we could use the worm to unearth mechanistic targets of biguanides, including metformin and phenformin, in neoplasia. To identify conserved targets of biguanides involved in growth inhibition, we conducted a RNA interference (RNAi) screen in C. elegans of 1,046 genes annotated to have a role in metabolism by gene-ontology term. Metformin sensitivity RNAi induce slow growth and reduce body size with 25 mM metformin, a dose that has no effect on controls, whereas metformin resistance RNAi permit animals to grow on plates with 150 mM metformin, a dose that elicits profound developmental delay and growth inhibition in controls (Figures 1B, S1A and S1B). RNAi knockdown of 13 genes leads to metformin resistance, whereas RNAi of 5 genes causes metformin sensitivity (Figures 1B, S1B, S1C, and S1D).
Figure 1. CeACAD10 Is Required for Metformin to Impede Growth in C. elegans.
(A) Metformin induces growth inhibition of C. elegans in a dose-dependent manner. n = 3 independent tests. **p < 0.01 by one-way ANOVA.
(B) Positive hits from the RNAi screen lead to resistance to 150 mM metformin (green, left) or sensitivity to 25 mM metformin (red, right). n = 4 biological replicates. Scoring system as in Figure S1B.
(C) RNAi to CeACAD10 makes C. elegans resistant to 100 mM metformin. **p < 0.01, by two-way ANOVA.
(D) Nonsense mutation of CeACAD10 suppresses the growth inhibition mediated by metformin (100 mM). **p < 0.01, by two-way ANOVA.
(E) Over-expression (oe) of CeACAD10 makes animals hypersensitive to 100 mM metformin. **p < 0.01, by two-way ANOVA.
(F)–(H), Biguanides (50 mM metformin or 5 mM phenformin) elevate CeACAD10 expression at the mRNA (F), and protein (G and H) levels using the CeACAD10 reporter. n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
For (A), (D–F), and (H), n = 33–108 animals were analyzed. All bars indicate means and SEM.
Consistent with the idea that mitochondria are the primary target of metformin (Madiraju et al., 2014; Owen et al., 2000), three metformin sensitive RNAi encode mitochondrial proteins, including a complex I subunit (Y53G8AL.2), a subunit of pyruvate dehydrogenase (pdha-1) and a component of the proton-transporting ATP synthase complex (immt-1) (Figures 1B, S1B and S1D). We also find that genetic loss-of-function mutants of mitochondrial respiration components gas-1 and mev-1 are hypersensitive to 25 mM metformin (Figure S1E).
Knockdown of the C. elegans ortholog of acyl-CoA dehydrogenase family member 10 (CeACAD10, F37H8.3) by RNAi led to the greatest degree of metformin resistance (Figures 1B, S1B and S1C). CeACAD10 RNAi and a genetic loss-of-function mutation in CeACAD10 render animals 3-fold more resistant to 100 mM metformin relative to controls (Figures 1C and 1D). In order to study CeACAD10 protein expression, we generated transgenic C. elegans CeACAD10 translational fluorescent reporter (Figure 1G). Consistent with CeACAD10 being both necessary and sufficient for biguanide-induced growth inhibition, CeACAD10 overexpression itself leads to slow growth of C. elegans, and makes the animals hypersensitive to metformin (Figure 1E).
CeACAD10 mRNA expression is elevated over 2-fold by 50 mM metformin, and ~4-fold by 5 mM phenformin (Figure 1F). The intensity of the CeACAD10 reporter is induced ~7 and ~9 fold by metformin and phenformin, respectively, indicating that a more pronounced increase in CeACAD10 protein accompanies the mRNA increase (Figures 1G and 1H). Microscopy of C. elegans CeACAD10 reporter animals indicates predominantly cytoplasmic localization (Figures 1G and S1F). Unbiased co-immunoprecipitation (co-IP) and mass spectrometry of CeACAD10::mRFP revealed enrichment of interacting proteins involved in growth and post-embryonic development, as well as proteins harboring ribosomal and mitochondrial cellular component gene ontology terms (Table S1). These data are supportive of the idea that CeACAD10 may play a role in growth through regulation of protein translation and fundamental cellular energetics.
Mitochondrial Oxidative Phosphorylation, CeTORC1 and CeACAD10 Act in the Same Pathway Mediating the Response to Metformin
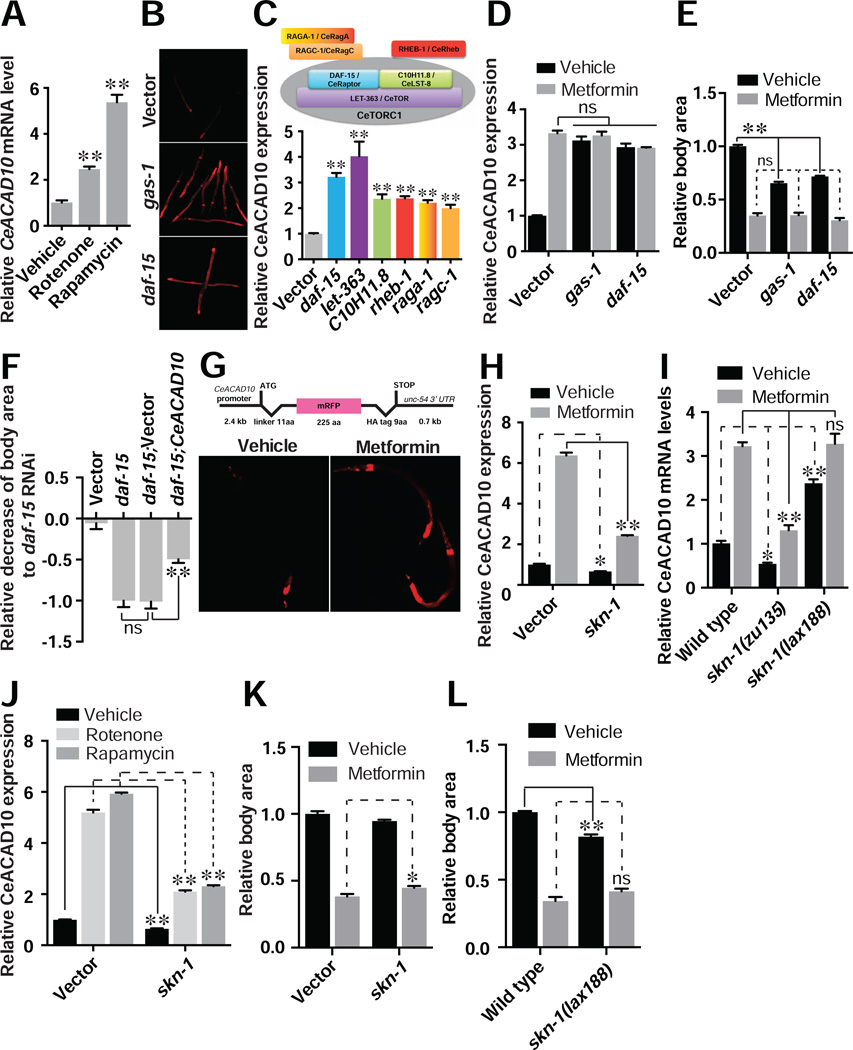
To determine whether CeACAD10 lies in the same pathway with previously reported metformin targets, we examined the effects of targeting AMPK, mitochondria, and CeTORC1 on CeACAD10 expression. Inhibition of mitochondrial complex I with rotenone or gas-1 RNAi in C. elegans elevates CeACAD10 mRNA and CeACAD10 reporter expression (Figures 2A and 2B). Mitochondrial inhibitors antimycin A and oligomycin A also induce a robust increase in CeACAD10 mRNA (Figure S2A). Similarly, reduced function of CeTORC1 by RNAi or chemical inhibition using rapamycin leads to induction of CeACAD10 mRNA and the CeACAD10 reporter (Figures 2A–2C).
Figure 2. CeACAD10 Is Induced by Inhibition of Mitochondria or CeTORC1.
(A) CeACAD10 mRNA was measured following chemical inactivation of mitochondrial complex I and CeTORC1 with rotenone (5 µM) and rapamycin (100 µM), respectively. n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(B) Genetic deficiencies of mitochondria (gas-1) and CeTORC1 (daf-15) by RNAi lead to CeACAD10 reporter induction.
(C) RNAi to the indicated CeTORC1 components and its regulators upregulates CeACAD10 reporter expression. The bar graph is color-matched with the components indicated in the illustration. n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(D) Metformin treatment (50 mM) and RNAi to gas-1 or daf-15 have no additive effect on CeACAD10 reporter induction. n = 3 biological replicates; **p < 0.01, by two-way ANOVA.
(E) RNAi of gas-1 and daf-15 decreases body size of C. elegans and has no additive effect with metformin co-treatment (100 mM). n = 37–64; **p < 0.01 and non significant (ns), by two-way ANOVA.
(F) CeACAD10 RNAi suppresses the effect of CeTORC1 deficiency on body size reduction by daf-15 RNAi (see Figure S2B for knockdown efficiencies). Data are presented as a fraction mean of the body size reduction by daf-15 RNAi ± SEM. n = 27–44; **p < 0.01 and no significance (ns), by two-way ANOVA.
(G) Sequences of the CeACAD10 promoter reporter (top) are sufficient to confer induction by metformin (bottom). The promoter reporter is only used in this figure.
(H) RNAi of skn-1 partially suppresses CeACAD10 expression and the effects of metformin on CeACAD10 induction. n = 3 biological replicates; *p < 0.05 and **p < 0.01, by two-way ANOVA.
(I) Null mutants in skn-1(zu135) partially suppress CeACAD10 baseline and metformin-induced mRNA expression. Basal CeACAD10 mRNA is increased in gain-of-function skn-1(lax188) mutants. n = 3 biological replicates; *p < 0.05, **p < 0.01 and no significance (ns), by two-way ANOVA.
(J) RNAi of skn-1 partially suppresses the effects of 50 mM metformin, 5 µM rotenone and 100 µM rapamycin on CeACAD10 expression. n = 3 biological replicates; **p < 0.01, by two-way ANOVA.
(K) Effect of skn-1 RNAi on 100 mM metformin-induced growth suppression. n = 3 biological replicates; *p < 0.05, by two-way ANOVA.
(L) skn-1 gain-of-function mutants show decreased body size and no change in metformin (100 mM)-induced body size reduction. n = 3 biological replicates; **p < 0.01 and no significance (ns), by two-way ANOVA.
For (E and F) and (K and L), n= 29–36 animals. All bars indicate means and SEM.
We next conducted combinatorial analysis to determine whether mitochondria, CeTORC1, and CeACAD10 lie in a single metformin response pathway. RNAi of gas-1 or daf-15 does not have an additive effect with metformin on CeACAD10 induction (Figure 2D). Additionally, animals treated with gas-1 RNAi do not further increase CeACAD10 expression or further reduce body size when exposed to rapamycin, supporting the idea that the mitochondria, CeTORC1 and CeACAD10 lie in a single genetic pathway (Figures S2B and S2C). RNAi of gas-1 or daf-15 significantly decreases the body size of C. elegans, but has no additive reduction when co-administered with metformin (Figure 2E). While it is not clear whether the non-additive effect of metformin with gas-1 and daf-15 RNAi is because growth inhibition is near maximal with metformin or because mitochondria and CeTORC1 truly lie in a pathway with metformin, stronger evidence is provided by the fact that RNAi to CeACAD10 suppresses 50% of the slow growth evident in animals deficient in daf-15 (Figures 2F and S2D).
In keeping with reports indicating that metformin exerts its effects independently of AMPK (Foretz et al., 2010; Kalender et al., 2010), metformin is still able to induce CeACAD10 expression in mutants lacking the AMPKα catalytic subunit aak-2 (Figure S2E). Further, the AMPK agonist AICAR does not induce CeACAD10 mRNA or protein expression (Figures S2F and S2G). Finally, mutants lacking either one or both of the AMPKα subunits aak-1 and aak-2 show growth inhibition comparable to wild type animals when treated with metformin (Figure S2H).
Metformin has been reported to extend lifespan indirectly through effects on the bacterial E. coli diet of C. elegans (Cabreiro et al., 2013). In contrast, metformin induces the CeACAD10 reporter equally well when added to cultures of live or UV-killed bacteria (Figure S2I). Direct injection of metformin into the intestine of C. elegans also induces the CeACAD10 reporter (Figure S2J). Finally, metformin blocks growth in C. elegans whether animals are fed cultures of live or UV-killed bacteria (Figure S2K). These data are consistent with a direct effect of metformin on CeACAD10 induction and growth inhibition in C. elegans.
The Stress-Responsive Transcription Factor SKN-1 Partially Mediates CeACAD10 Induction by Metformin
Analysis of a CeACAD10 promoter reporter indicates that metformin activates transcription of the CeACAD10 gene (Figure 2G). An RNAi screen of 708 transcription factors identified skn-1 as the only knockdown that blocks the effect of metformin on CeACAD10 induction (Figure 2H). A null mutation in skn-1 also partially suppresses metformin effects on CeACAD10 mRNA induction (Figure 2I). A gain-of-function mutation in skn-1 (Paek et al., 2012) induced CeACAD10 mRNA ~2.5 fold and was non-additive with metformin (Figure 2I). While skn-1 has been previously invoked in the lifespan extension attributable to metformin and inhibition of CeTORC1 (Cabreiro et al., 2013; De Haes et al., 2014; Onken and Driscoll, 2010; Robida-Stubbs et al., 2012), skn-1 is only partially responsible for CeACAD10 induction, as metformin, rotenone, and rapamycin can still induce CeACAD10 2–3 fold following RNAi to skn-1 (Figures 2H–J). RNAi of skn-1 reduces metformin-mediated growth inhibition significantly, but to a much lesser extent than CeACAD10 RNAi (Figure 2K). As expected, based upon higher CeACAD10 expression, skn-1 gain-of-function mutants are smaller than controls, but do not show additive growth inhibition with metformin treatment (Figure 2L).
In order to test whether skn-1 directly mediates the increase in CeACAD10 expression, we conducted chromatin IP of SKN-1. SKN-1 associated with two regions of the CeACAD10 promoter was non-significantly reduced in abundance following treatment with metformin, with no change evident in the 3’UTR region (Figure S2L). These data agree with the absence of a SKN-1::GFP binding site in CeACAD10 gene ModENCODE data and in prior SKN-1 ChIP studies (Steinbaugh et al., 2015). We did not see nuclear translocation of a SKN-1::GFP reporter post metformin treatment, while clear translocation was evident with hydrogen peroxide (Figure S2M). In summary these data indicate that factors other than skn-1 are primarily responsible for CeACAD10 induction and the action of biguanides on growth.
The Nuclear Pore Complex Is Required for Metformin-Induced CeACAD10 Expression and Growth Inhibition in C. elegans
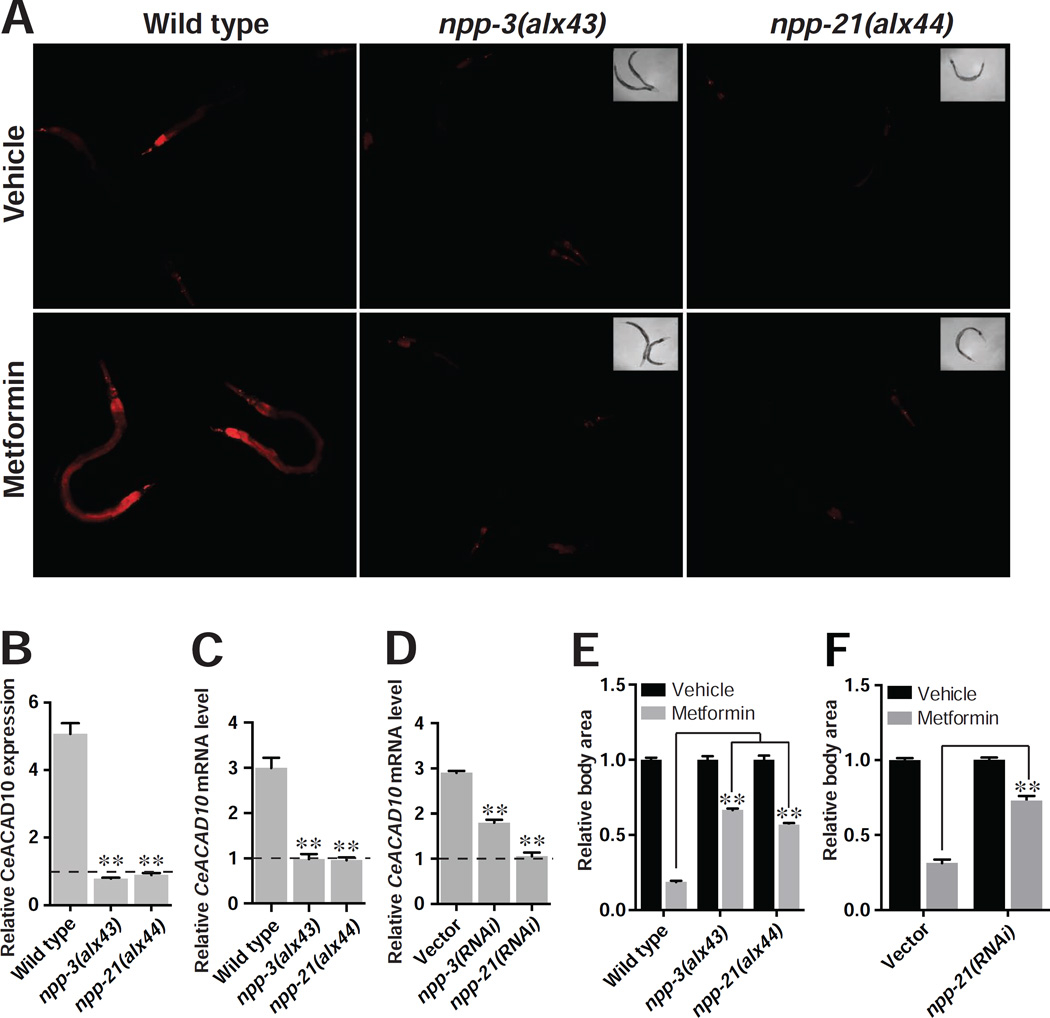
To identify the genetic machinery responsible for CeACAD10 induction by metformin, we carried out an unbiased, forward genetic screen for suppressors of metformin’s effect on the CeACAD10 reporter. Using whole-genome sequencing, we determined that the two most potent suppressive mutants carry missense mutations in genes encoding two distinct components of the NPC, npp-3/NUP205 (Nucleoporin 205 kDa) and npp-21/TPR (translocated promoter region). Both mutations, NPP-3(E1644K) and NPP-21(P1501L), as well as RNAi to npp-3 and npp-21, entirely blunt the effect of metformin on induction of CeACAD10 expression at both the mRNA and protein level (Figures 3A–3D). RNAi of npp-21 does not universally suppress transgene expression (Figures S3A–S3C).
Figure 3. The NPC Is Required for Metformin to Induce CeACAD10 Expression and Growth Inhibition in C. elegans.
(A)–(D) Missense mutations of and RNAi to npp-3 and npp-21 suppress the effect of metformin (50 mM) on CeACAD10 protein (A and B) and mRNA (C and D) induction. n = 3 biological replicates; **p < 0.01, by two-way ANOVA.
(E) npp-3 and npp-21 mutants are resistant to growth inhibition by 100 mM metformin. n = 32–74 worms analyzed per group; **p < 0.01, by two-way ANOVA.
(F) RNAi of npp-21 blunts the growth-inhibitory effects of 100 mM metformin. n = 52–62 worms analyzed per group; **p < 0.01, by two-way ANOVA.
All bars indicate means and SEM.
Strikingly, mutation of npp-3 or npp-21 also renders animals resistant to metformin’s growth-inhibiting effects (Figure 3E), comparable to CeACAD10 loss of function. RNAi of npp-21 abolished the anti-growth effect of metformin in C. elegans, indicating that the mutations obtained in npp-3 and npp-21 are loss of function alleles (Figure 3F).
An Ancient and Unified Mitochondria-NPC-mTORC1 Signal Relay Mediates the Action of Metformin
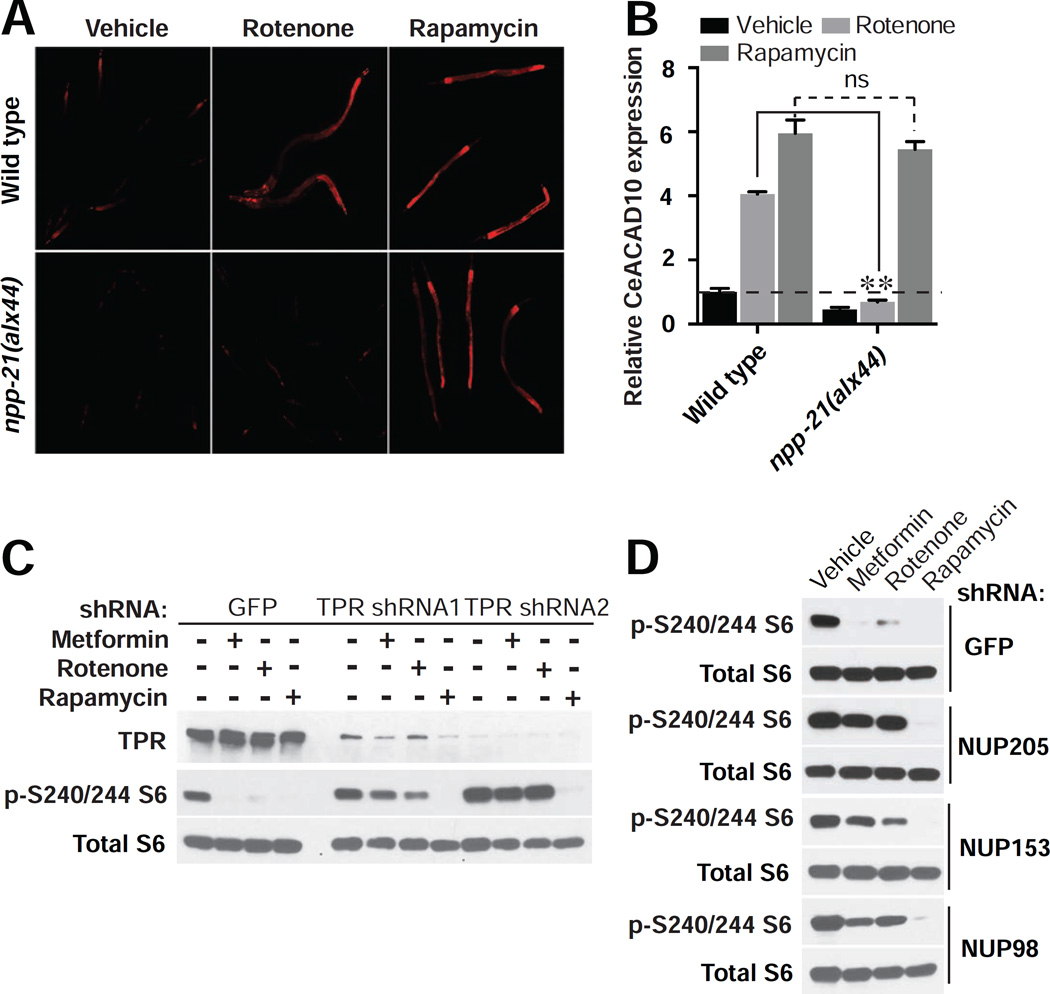
To determine the genetic relationships of mitochondria, NPC, CeTORC1, and CeACAD10 in the metformin response pathway, we examined CeACAD10 expression following treatment with rotenone or rapamycin. Both rotenone and rapamycin induce CeACAD10 in a wild type background (Figures 4A and 4B), consistent with genomic inhibition of mitochondria and CeTORC1 (Figure 2B). However, the effect of rotenone on CeACAD10 induction is abolished in the npp-21 mutant background, whereas rapamycin still induces the same increase as it does in wild type (Figures 4A and 4B). These results place the mitochondria upstream of the NPC in metformin action, and establish CeTORC1 as acting downstream of the NPC.
Figure 4. The NPC Connects Mitochondria to Regulation of mTORC1 in Metformin Action.
(A and B) NPC deficiency antagonizes the effect of rotenone (5 µM), but not rapamycin (100 µM), on CeACAD10 induction. n = 3 biological replicates; **p < 0.01, by two-way ANOVA.
(C and D) shRNA knockdown of NPC components in HEK293E cells prevents metformin (8 mM) and rotenone (40 µM) from reducing S6 protein phosphorylation, but not rapamycin (40 nM). Blots shown are representative of two biological replicates.
To determine whether this metformin growth inhibitory pathway is conserved, we examined mTORC1 activity in human cells after RNAi to NPC components. Treatment with metformin, rotenone, and rapamycin leads to potent inhibition of ribosomal S6 protein phosphorylation (Figure 4C). Upon depletion of NPC component TPR by RNAi, neither metformin nor rotenone is able to inhibit S6 phosphorylation (Figure 4C). These results confirm that the NPC is a critical effector of the metformin response pathway, connecting decreased mitochondrial activity to inhibition of mTORC1. Consistent with mTORC1 lying downstream of the NPC in the response to biguanides, RNAi knockdown of TPR does not block the inhibitory effect of rapamycin (Figure 4C). RNAi knockdown of NPC components NUP205, NUPL1, NUP153, NUP214, and NUP98 also fully suppressed the ability of metformin and rotenone to inhibit mTORC1 activity, but not that of rapamycin (Figures 4D and S4A; see knockdown efficiencies in Figure S4B).
Phenformin Restrains Passage of Macromolecules through the NPC
As certain cancer cell lines lack the metformin transporter protein OCT2 (Yuan et al., 2013) and as phenformin can induce the same or even more potent effects on CeACAD10 induction, we used phenformin to study how biguanides affect the NPC. Phenformin did not alter localization or apparent density of NPCs in HeLa cells, as assessed by immunofluorescence (Figure S5A). Phenformin does not alter the expression of NPC components at the mRNA level in C. elegans and two different cell lines, HEK293E and melanoma C8161 cells (Figures S5B–S5D) or protein level in HEK293E cells (Figures 4C and S5E).
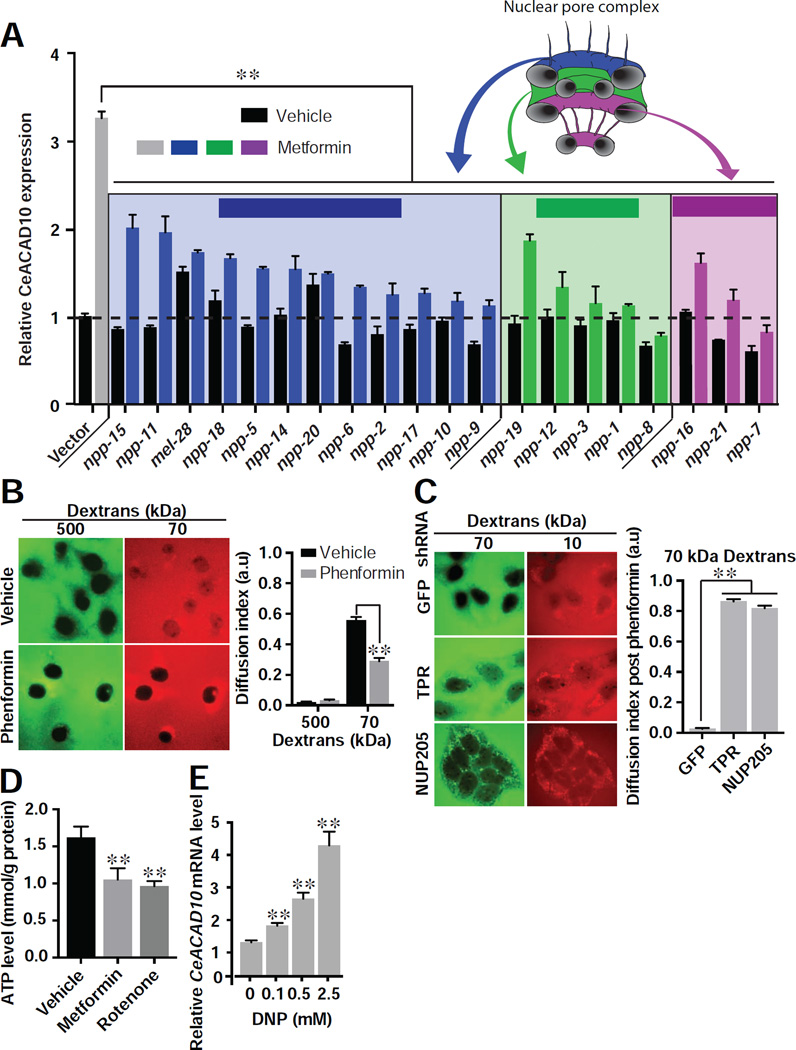
Certain components of the NPC have been reported to act in roles outside of the NPC (Simon and Rout, 2014). To determine whether it is their role in the NPC by which identified components mediate metformin-induced growth inhibition, we conducted RNAi to 18 additional functionally and structurally distinct NPC genes in C. elegans. All of these RNAi significantly suppress metformin’s effect on CeACAD10 induction (Figure 5A). Thus, reduced NPC component expression produces metformin resistance, and npp-3 and npp-21 operate in their role specific to the NPC in order to transduce metformin’s effects.
Figure 5. Phenformin Leads to CeACAD10 Induction by Restraining Transport through the NPC.
(A) RNAi knockdowns of NPC components suppress the effect of metformin (50 mM) on CeACAD10 reporter induction (color-coded based upon location and role). n = 3 biological replicates; **p < 0.01, by two-way ANOVA.
(B) Phenformin (1 mM) restricts passive transport of 70 kDa cargo through the NPC. n = 42 cells for vehicle, 46 for phenformin from 3 biological replicates. **p < 0.01, by one-way ANOVA. Scale bar, 10 µm.
(C) Lentiviral shRNA to TPR and NUP205 in HeLa cells permits passive diffusion of 70 kDa dextran into the nucleus, in spite of 1 mM phenformin treatment. n = 26 cells for GFP shRNA, 29 for TPR and 29 for NUP205, from 3 biological replicates. **p < 0.01 by one-way ANOVA. Scale bar, 10 µm.
(D) C. elegans ATP levels are significantly reduced by 50 mM metformin and 5 µM rotenone. n = 3 biological replicates. **p < 0.01, by one-way ANOVA.
(E) CeACAD10 mRNA is induced in a dose-dependent manner by DNP. n = 3 biological replicates. **p < 0.01, by one-way ANOVA.
All bars indicate means and SEM.
We next asked whether biguanides exert their antigrowth activities by altering NPC activity. Loss of function of NPC components such as NUP205 is known to increase passage of macromolecules through the nuclear pore (Galy et al., 2003). Indeed, biguanides have the opposite effect, restricting passive transport of macromolecules through the NPC in melanoma cells (Figure 5B). In contrast, biguanides had no effect on active nuclear import of a constitutive NLS::GFP (Figure S5F). Consistent with our earlier results, protein levels of the NPC subunit NUP98, which, along with other FG motif proteins, is critical for maintaining the permeability barrier of the NPC (Labokha et al., 2013), are unchanged by phenformin (Figure S5E).
As biguanides restrict passive transport through the NPC and loss of function of NPC components suppresses biguanide effects, loss of function of various nuclear pore proteins should reverse the ability of biguanides to restrict passive NPC transport. Indeed, RNAi to TPR, NUP205, NUP214, NUP98, and NUP85 in HeLa cells prevented phenformin from restricting nuclear entry of fluorescent dextrans of 70 kDa (Figures 5C and S5G). We found this surprising given that many of these NPC components have not been reported to play a role in nuclear passive transport (Timney et al., 2016). RNAi to these same components did not affect active nuclear transport of a constitutive NLS::GFP (Figure S5H).
As metformin has well reported effects on mitochondrial function (Madiraju et al., 2014; Wheaton et al., 2014), we next sought to mechanistically tie biguanide action at the mitochondrion to changes in NPC activity. We reasoned that metformin might affect NPC activity through reduction in cellular energy stores or, alternatively, through altered reactive oxygen species (ROS) generation. Metformin reduced ATP levels in C. elegans comparably to rotenone, which also potently induces CeACAD10 expression (Figures 5D, 2A, and S2A). The ROS generator paraquat and ROS scavengers N-acetyl cysteine (NAC) and butylated hydroxyanisole (BHA) failed to affect CeACAD10 expression (Figures S5I and S5J). In contrast, treatment of worms with the uncoupling agent 2,4-dinitrophenol (DNP) which lowers both ATP and ROS increases CeACAD10 mRNA in a dose-dependent manner (Figure 5E). These data suggest that biguanides alter NPC transport by reducing cellular ATP levels.
Phenformin Inhibits Rag GTPase-Mediated mTORC1 Signaling by Restricting RagA/RagC Nuclear Access
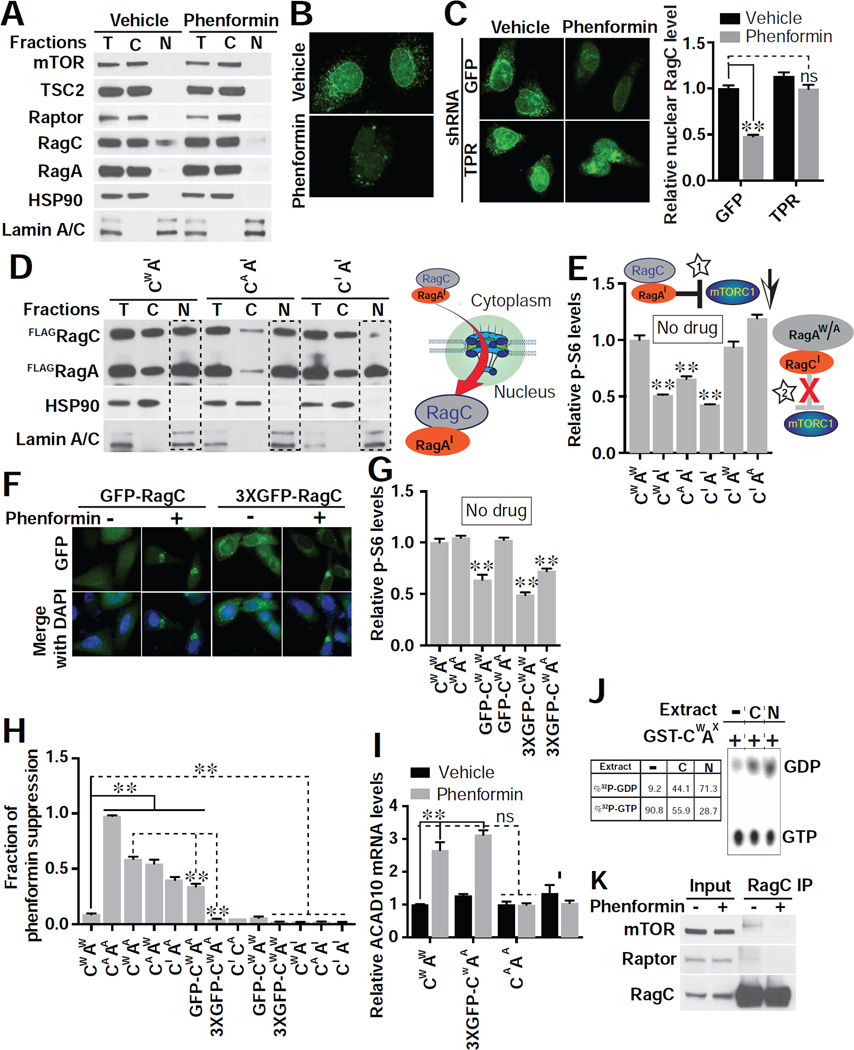
Inhibition of nuclear export with leptomycin is known to inhibit mTORC1 (Kim and Chen, 2000). We hypothesized that biguanides inhibit mTORC1 activity in part by altering nucleocytoplasmic partitioning of essential mTORC1 pathway proteins. Of mTORC1 pathway proteins analyzed, only RagC showed a significant nuclear proportion under basal conditions (Figure 6A). Following treatment with phenformin, the majority of RagC was excluded from the nucleus, while the overall level of RagC was not altered (Figures 6A, 6B and S6A).
Figure 6. Phenformin Inhibits mTORC1 through Nuclear Exclusion of RagC GTPase.
(A) Subcellular localization of endogenous mTORC1 pathway components post phenformin (1 mM) treatment in HEK293E cells. Equal amounts of protein from each fraction were loaded (T, total lysate; C, cytosol fraction; N, nuclear fraction). Hsp90 specifically marks the cytosolic fraction, and Lamin A/C the nuclear fraction. Blots shown are representative of two biological replicates.
(B) Nuclear RagC is excluded by phenformin (1mM), assessed by immunofluorescence in HeLa cells. Images are representative of 3 biological replicates. Scale bar, 10 µm.
(C) RNAi to TPR allows RagC to enter the nucleus with or without phenformin treatment (1 mM). Nuclear RagC was quantified with ImageJ (right). Images are representative of 2 biological replicates. Scale bar, 10 µm. n = 17 cells quantified for GFP/vehicle, 14 for GFP/phenformin, 16 for TPR/vehicle, and 12 for TPR/phenformin; **p < 0.01 and no significance (ns), by two-way ANOVA.
(D) Overexpression of inactive RagA (AI) drives RagC in wild type (CW), activated (CA), or inactive (CI) form to the nucleus in HEK293E cells (see Methods for molecular identity of mutants). A schematic model indicating that RagAI is the stimulus for RagC nuclear entry is shown. Blots shown are representative of three biological replicates.
(E) Overexpression of RagAI acts as a dominant negative inhibitor of mTORC1. HEK293E cells were transfected with the indicated combination of RagA (AW, wild type; AI, inactive; AA, active) and RagC (CW, wild type; CI, inactive; CA, active), and mTORC1 activity assessed (p-S6/total S6). A cartoon is shown of RagAI-RagC inhibiting mTORC1 (star 1) but not RagA-RagCI (star 2). n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(F) Expression of RagC fused to one (GFP-RagC) or three (3xGFP-RagC) GFP molecules leads to pronounced nuclear exclusion upon treatment with phenformin (1 mM). Images (GFP in green and DAPI in blue) are representative cells from 3 biological replicates.
(G) mTORC1 activity (p-S6/total S6) is reduced by adding bulky GFP modifications to RagC as in (F). No biguanide treatment was used. n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(H) Suppression of phenformin effect on mTORC1 activity (p-S6/total S6) by different combinations of RagA and RagC was assessed after treating transfected HEK293E cells with 1 mM phenformin as in (F). n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(I) Overexpression of activated RagAA/RagCA but not RagAA/3xGFP-RagC fully suppresses the induction of ACAD10 mRNA evident with 1 mM phenformin in HEK293E cells. **p < 0.01, by one-way ANOVA.
(J) Nuclear GTPase-activating protein (GAP) activity is present towards RagC. GST-RagCW/RagAX was incubated with buffer (lane 1), total cytoplasmic (C, lane 2) or nuclear (N, lane 3) lysates.
(K) RagC from phenformin (1 mM) treated HEK293E cells fails to associate with mTORC1 indicated by co-IP analysis. Blots shown are representative of two biological replicates.
Given that RNAi to NPC components suppresses biguanide effects on ACAD10 expression and increases nuclear entry of 70 kDa dextrans, we reasoned that RNAi to TPR should allow RagC to enter the nucleus even upon phenformin treatment. RNAi to TPR fully reversed reduced nuclear RagC levels evident with phenformin (Figure 6C) and moreover re-animated mTORC1 in spite of phenformin treatment (Figure S6B).
Analysis of the Rag protein sequences revealed that only RagC harbors a predicated nuclear export signal (NES). Mutation of its predicted NES led to a significant rise in nuclear RagC, whereas wild type RagC is largely cytoplasmic (Cw versus CΔNES, Figure S6C). Demonstrating that phenformin prevents RagC nuclear localization, even RagCΔNES fails to localize to the nucleus with phenformin pretreatment.
RagC does not, alternatively, have a predicted, canonical nuclear localization signal (NLS). Curiously, even the addition of an NLS to RagC failed to drive it to the nucleus when co-expressed with wild-type RagA (Figure S6D). The majority of Rag proteins exist in the cell in heterodimeric form, with RagA and RagB associating with either RagC or RagD (Sancak et al., 2008). This raised the possibility that the Rag binding partner of RagC rather than RagC itself is the trigger for nuclear entry.
Activation state and conformation of Rag family proteins is driven by guanine nucleotide-bound state. Previous studies have made use of mutations in Rag proteins that mimic the activated and inactive conformations (Oshiro et al., 2014; Sancak et al., 2008). When RagA is in the GTP bound state and RagC the GDP bound state, the RagA/RagC heterodimer is fully capable of activating mTORC1 (Sancak et al., 2008). Alternatively, RagA (or RagB) carrying mutations mimicking the GDP bound state act as dominant negative suppressors of mTORC1 activation (Figures 6E and S6E) (Sancak et al., 2008). Here we also made use of RagA and RagC mutations that mimic the activated and inactive conformations (RagAA: RagA(Q66L) mimics active RagAGTP; RagAI: RagA(T21L) mimics inactive RagAGDP; RagCA: RagC(S75L) mimics active RagCGDP; RagCI: RagC(Q120L) mimics inactive RagCGTP; RagAW and RagCW: wild type proteins).
We hypothesized that inactive RagAI might act as an inhibitor of mTORC1 because RagAGDP serves as the normal trigger for nuclear entry of the RagA/RagC heterodimer. Thus, mutant RagAI could provide an unyielding stimulus to localize RagA/C in the nucleus, preventing its exit to activate mTORC1. Indeed, RagAI stimulated a massive, 51% shift in RagCW nuclear localization, consistent with this form of RagA being the signal for RagC nuclear entry (Figures 6D and S6F). RagC is driven to the nucleus by RagAI regardless of its activation state, although the RagCI form showed less propensity to go to the nucleus (Figure 6D). Virtually no RagC was detected in the nucleus when co-expressed with RagAW or RagAA (Figure S6G). Pre-treating cells with phenformin prevents RagAI from driving RagC to the nucleus, supporting the conclusion that phenformin-mediated NPC changes prevent the RagA/RagC complex from entering the nucleus (Figure S6F). We further confirmed that RagAI acts as a dominant negative inhibitor of mTORC1 signaling, no matter what form of RagC it is co-expressed with (Figure 6E).
We next attempted rescue of phenformin-mediated mTORC1 suppression by expressing variants in RagA and RagC. Expression of any single isoform of RagA or RagC is not capable of suppressing the phenformin effect (Figure S6E). Full suppression of the phenformin effect requires both RagAA and RagCA (Figures 6H and S6E). RagAA is only partially able to suppress phenformin effect when co-expressed with RagCW, similar to the result with RagCA co-expression with RagAW (Figure 6H).
Our data support that RagC requires nuclear passage in order to be fully capacitated to activate mTORC1. If this is correct, then bulky modification of RagC that further restricts its nuclear passage should not only prevent RagAA-mediated activation of mTORC1 in phenformin treated cells, but we predict it might also act as a dominant-negative inhibitor of mTORC1 action. Indeed, increasing RagC molecular weight by ~27 kDa and 78 kDa by coupling one or three GFP molecules to RagC, largely and nearly completely restricts RagC from the nucleus of phenformin treated cells, respectively (Figure 6F). In support of the idea that nuclear exclusion of RagC prevents its activation, GFP-RagCW and 3xGFP-RagCW inhibit mTORC1 in the basal state, even when co-expressed with RagAA (Figure 6G). 3xGFP-RagCW renders RagAA completely incapable of suppressing phenformin’s effects on mTORC1 activity (Figure 6H and S6H). Thus, when RagA cannot partner with any nuclear-potentiated RagC, it cannot block biguanide-mediated suppression of mTORC1. As predicted, the induction of ACAD10 mRNA by phenformin, which is conserved in mammalian cells, is fully suppressed by the RagAA/RagCA complex but not the 3XGFP-RagCW/RagAA (Figure 6I).
From these data it logically flows that RagC acquires activation in the nucleus that potentiates is ability to activate mTORC1. Examination of immunopurified Flag-RagC by mass spectrometry did not identify any phosphorylation, acetylation, oxidation, or mono-, di- and tri-methylation that distinguished control versus phenformin treatment, or cytoplasmic versus nuclear fractions (data not shown). Our next hypothesis was that RagC transitions from the GTP-bound inactive form to the GDP-bound active form during nuclear transit. To this end, we examined nuclear extracts for GTPase activating protein (GAP) activity for RagC. Indeed, the majority of RagC GAP activity was seen in nuclear extract (Figure 6J), and the reported GAP for RagC, folliculin (FLCN) (Tsun et al., 2013), was detected in the nucleus (Figure S6I). These data indicate that RagC is potentiated by nuclear GAP activity that facilitates conversion of RagCGTP to activated RagCGDP. In support of this, treatment of cells with phenformin renders RagC incapable of binding mTORC1 by co-IP (Figure 6K).
An Ancient Pathway Defined by ACAD10 and the NPC Is Required for Metformin-Mediated Lifespan Extension and Growth Inhibition in Cancer
Metformin promotes longevity in organisms as diverse as C. elegans and mice (Barzilai et al., 2016). Accordingly, we next analyzed whether the ACAD10-NPC signaling pathway is involved in lifespan extension in C. elegans. Either RNAi or genetic loss of function mutation in CeACAD10 or npp-21 completely abrogates lifespan extension mediated by metformin (Figures 7A–7D and Table S2). These data suggest that this signaling axis is involved in whole organism health, and so we next moved to study the role of this pathway in human cancer cells.
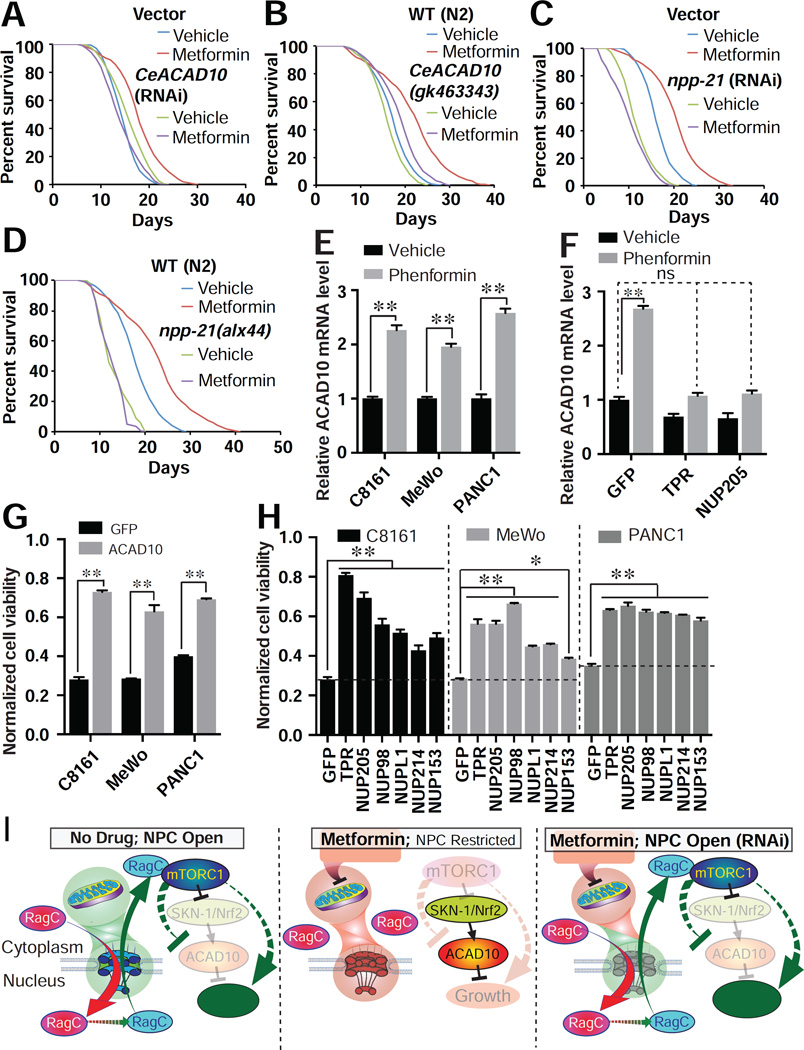
Figure 7. Phenformin Promotes Lifespan and Retards Cancer Growth through an Ancient and Unified Mechanism.
(A – D) Deficiency of CeACAD10 by RNAi (A) or nonsense mutation (B) or npp-21 by RNAi (C) or missense mutation (D) in C. elegans suppresses metformin’s effect on lifespan extension. Results are representative of 2 – 3 biological replicates (see also table S2).
(E) ACAD10 mRNA increases in melanoma (C8161 and MeWo) and pancreatic cancer (PANC1) cell lines treated with 1 mM phenformin relative to the level of untreated cells. n = 3 biological replicates; **p < 0.01, by one-way ANOVA.
(F) shRNA knockdown of TPR and NUP205 suppresses phenformin’s effect (1 mM) on ACAD10 induction in C8161 cancer cells (relative to the GFP shRNA group). n = 3 biological replicates; **p < 0.01 and no significance (ns), by two-way ANOVA.
(G) ACAD10 shRNA knockdown renders C8161, MeWo, and PANC1 tumor cell lines resistant to 1 mM phenformin n = 4 biological replicates; **p < 0.01, by two-way ANOVA.
(H) Knockdown of NPC components by lentiviral shRNA mitigates the antigrowth activity of phenformin in cancer cells. n = 4 biological replicates; **p < 0.01, by two-way ANOVA.
(I) Model of the ancient pathway by which biguanides promote lifespan and inhibit growth.
We first confirmed that ACAD10 expression is induced by biguanides in human cells, including HEK293E (Figure 6I), two melanoma cell lines C8161 and MeWo, and one pancreatic cancer cell line PANC1 (Figure 7E). Remarkably, RNAi to NPC components TPR and NUP205 abolished the increase of ACAD10 mRNA by phenformin in melanoma cells (Figure 7F), indicating conservation of the NPC-ACAD10 signaling axis.
Working from our prior observation that phenformin inhibits the growth of melanoma cells (Yuan et al., 2013), we next asked whether the ancient mitochondria-NPC-mTORC1-ACAD10 genetic pathway is a conserved mediator of the growth-inhibitory properties of biguanides. Indeed, loss of function of the NPC or ACAD10 leads biguanide-sensitive cancer cells to become resistant to phenformin-mediated killing (Figures 7G and 7H; knockdown efficiency in Figure S7).
DISCUSSION
Ancient and Unified Mechanism for Metformin’s Lifespan-Extending and Anticancer Activities Uncovered by C. elegans Genomics
Using iterative genomic screening in C. elegans, we identify a genetic pathway linking metformin to inhibition of cancer growth and lifespan extension (Figure 7I). We put forward an ancient, conserved pathway that unifies metformin response elements including the mitochondrion and mTORC1, adding mechanistic understanding of how metformin inhibits mTORC1 by restraining NPC transport. The reduction in mitochondrial function signals to the NPC, which connects metformin to nuclear exclusion of RagC, ultimately preventing activation of mTORC1. Finally, we identify that inhibition of mTORC1 by biguanides leads to transcriptional upregulation of the previously uncharacterized gene ACAD10, a step absolutely required for the lifespan-extending and anticancer activities of metformin (Figure 7I).
The Role of ACAD10 in Metformin Action and the TORC1 Pathway
From data presented here, we conclude that CeACAD10 expression levels are central to the effects of metformin in C. elegans. Although function of human ACAD10 and its nematode ortholog CeACAD10 are not fully characterized, our early results indicate that proteins interacting with CeACAD10 are implicated in growth, development, and in fundamental regulation of protein translation and cellular energetics. Importantly, we show that ACAD10 is a critical effector of TORC1-regulated organismal growth. Further investigation will be needed to determine how ACAD10 affects anabolic and catabolic processes downstream of TORC1.
Genetic analysis of CeACAD10 enabled us to demonstrate that mitochondria, the NPC, TORC1, and ACAD10 all participate in a single metformin response pathway. Importantly, SKN-1, a proven regulator of metformin action in worms (Cabreiro et al., 2013; De Haes et al., 2014; Onken and Driscoll, 2010), is partially responsible for metformin-induced CeACAD10 upregulation, but based upon our analysis, regulation of CeACAD10 by SKN-1 is likely to be indirect. As reduction of skn-1 only modestly inhibits the growth inhibitory properties of metformin, we conclude that residual CeACAD10 induction is sufficient to mediate metformin effects, and additional transcription factors are responsible for the full spectrum of biguanide-mediated growth inhibition.
NPC Conformational Changes Are Linked to Metformin and Altered Cellular Energetics
Substantial evidence supports the conclusion that the anticancer activity of metformin is attributable to its inhibition of mitochondrial complex I (Wheaton et al., 2014). Cancer cells become markedly resistant to metformin when expressing high levels of the metformin-resistant S. cerevisiae mitochondrial complex I component, NADH reductase (NDI1). Mitochondria-directed metformin analogs are 1,000 times more potent than metformin at killing cancer cells (Cheng et al., 2016). It has been previously shown that metformin can decrease cellular ATP levels by inhibiting mitochondrial oxidative phosphorylation (OXPHOS) in a dose-dependent manner (Foretz et al., 2010), consistent with its dose-dependent antigrowth property in C. elegans and cancer.
In a surprising twist, we show that it is metformin’s effects on the nuclear pore that are the essential determinant of cell growth, not the drop in ATP levels and OXPHOS activity themselves. This conclusion is in line with prior observations in the literature relating mitochondrial function and ATP levels to nucleocytoplasmic transport. Mitochondrial OXPHOS produces the majority of cellular ATP that is required for NPC conformational changes that dictate nuclear pore opening or closure (Rakowska et al., 1998). Direct visualization of NPC by atomic force microscopy has been used to show that ATP depletion alters NPC conformation (Rakowska et al., 1998). Mitochondria are frequently observed in close contact with the nuclear envelope, and this juxtaposition may serve to as a conduit by which mitochondria provide energy for nucleocytoplasmic transport through the NPC (Prachar, 2003). Thus, alterations in the NPC in upon ATP deprivation are well poised to modulate NPC-regulated signaling.
Nucleocytoplasmic Shuttling Regulates mTORC1 Activity via RagC
Nucleus-localized components of the mTORC1 signaling pathway have been widely reported (Betz and Hall, 2013), including mTOR, Raptor, and TSC. Nuclear distribution of these proteins has substantial effects on mTOR signaling (Back and Kim, 2011), especially mTOR itself (Kim and Chen, 2000). Consistent with our observations, the yeast ortholog RagC/Gtr2 has been reported in the nucleus in previous studies (Hirose et al., 1998).
Based upon our results, we conclude that biguanides exclude RagC from the nucleus by restricting passive transport through nuclear pores. It has been previously established that RNAi to nuclear pore components generally enhances rather than restricts passive nucleocytoplasmic transport (Galy et al., 2003). Indeed our work suggests that loss of function of a larger group of NPC components than previously appreciated (Timney et al., 2016) affects passive diffusion through the NPC, re-animating nuclear RagC and mTORC1 activity. We cannot conclusively rule out the possibility that other aspects of NPC transport are modulated by biguanides or that these might play a role in nuclear transport of the Rag proteins. Future work will be needed to determine the full spectrum of biguanide-mediated effects on the NPC.
We propose that RagC enters the nucleus with RagA in its inactive conformation, RagC acquires the activated, GDP bound form and, that RagA/RagC exits the nucleus to activate mTORC1 signaling. As would be expected, modifications of RagC that prevent nuclear entry (bulky 3xGFP tagging) or that prevent subsequent exit of activated RagC (co-expression of inactive RagAI) lead to dominant-negative like inhibition of mTORC1 activity. The importance of subsequent nuclear export is underscored by similar mTORC1 inhibition when we mutate the putative NES on RagC. In support of our model, others’ work corroborates our observation that RagAI is found in the nucleus (Hirose et al., 1998). Demonstrable nuclear RagC GAP activity and the nuclear existence of the RagC GAP FLCN support the conclusion that RagC nuclear transit is critical for activation of mTORC1. Other studies have reported the presence of nuclear FLCN by immunofluorescence (Tsun et al., 2013). While our data do not rule out that cytoplasmic GAP activity also activates RagC, under the circumstances tested, our data suggest the nuclear context of FLCN rather than its bulk amount is important for its GAP activity towards RagC. Finally, our data neither confirm nor deny the exciting possibility of other modes of nuclear potentiation of RagC.
The underlying mechanisms how Rag GTPases mediate mTORC1 signaling are still unclear. The Rag heterodimer is rendered fully capable of activating mTORC1 when RagA/B is bound to GTP and RagC/D to GDP (Sancak et al., 2008). Prior work suggests that activating mutations in RagC strengthen its affinity for Raptor, an important step in mTORC1 activation (Oshiro et al., 2014). We suggest that activating mutations in RagA and RagC bypass the need for nuclear passage of RagC by mimicking the activation step normally acquired in the nucleus, fully animating mTORC1 even in the face of biguanide treatment.
An overarching implication of our findings is that the RagC-NPC signaling axis is a major means by which TORC1 activity is intimately linked to levels of cellular ATP. We suggest that disruption of RagC-mediated mTORC1 activation might represent the elusive link between kinase activity and cellular energy (ATP) levels.
Nuclear Transport in Aging, Cancer, and Aging-Related Metabolic Disease
The NPC serves as the only conduit through which material exchanges between the cytoplasm and nucleus in eukaryotic cells. It has been postulated that modifying nuclear transport could be an effective way to block tumor formation and growth, given that increased nucleocytoplasmic transport has been invoked in cancer progression and antineoplastic agent resistance (Kinoshita et al., 2012; Vargas et al., 2012). Here, we demonstrate that metformin induces growth inhibition in cancer by restricting NPC transport, and that this effect can be reversed by RNAi to NPC components. We conclude that modulating nuclear transport machinery could be a general strategy for halting progression of melanoma and other tumor types.
Nuclear leakiness has been previously implicated in age-dependent deterioration in mammalian cells (D'Angelo et al., 2009), and altering nuclear transport impacts yeast longevity (Lord et al., 2015). In the current study, we prove that the NPC is absolutely required for metformin to extend lifespan in C. elegans. Thus, it is plausible that restraining nuclear transport either by treatment with metformin or through other means might also be a general strategy for promoting healthy aging.
We speculate that the NPC and ACAD10 may be involved in insulin action and regulation of blood glucose levels. Though little is known about ACAD10, it lies within a susceptibility locus for T2D in Pima Indians (Bian et al., 2010). The NPC has also been suggested to act in the insulin signaling pathway (Pinkston-Gosse and Kenyon, 2007). It remains an intriguing possibility that the ancient pathway defined by the NPC and ACAD10 underlies metformin’s antidiabetic actions and its ability to improve insulin sensitivity.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents will be addressed by the corresponding author Alexander A. Soukas (asoukasmgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Nematode Strains
All nematode strains were grown at 20°C, and mainta ined following standard procedures (Brenner, 1974). N2 Bristol was used as the wild-type strain. The listed mutant strains were used: MGH325 CeACAD10(gk463343) II 4x, MGH275 aak-1(tm1944) III 4X, MGH274 aak-2(ok524) X 4X, MGH276 aak-1(tm1944) III;aak-2(ok524) X 4X, MGH326 npp-3(alx43) II 3X, and npp-21(alx44) II 4X. The following transgenic lines were used: MGH249 alxIs19[CeACAD10p::CeACAD10::mRFP3-HA myo-2p::GFP] 8X, MGH328 alxEx90[CeACAD10p::CeACAD10::mRFP3-HA myo-2p::GFP], MGH322 npp-3(alx43);alxIs19 3X, MGH323 npp-21(alx44);alxIs194X, MGH277 aak-2(ok524);alxIs194X, CF1553 muIs84 [(pAD76) sod-3p::GFP + rol-6] 3X and MGH50 mgIs48[ges-1p::MITO::GFP] 0X, LG326 skn-1(zu169) IV; geIs7[skn-1b::GFP].
Human Cell Lines and Culture Conditions
C8161 cells (ATCC) were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1% penicillin-streptomycin (PS, Life Technologies) in a 5% CO2 atmosphere at 37°C. All other cell lines used in this study were grown in high glucose Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% FBS and 1% PS in a 5% CO2 atmosphere at 37°C. For western blotting and cell fractions, HEK293E cells (a gift from Dr. Ramnik Xavier) were used. For cell viability measurements, C8161 (ATCC), MeWo (ATCC) and PANC1 (a gift from Dr. Nabeel Bardeesy) cells were used. For nuclear permeability testing, C8161 and HeLa cells were used. For GST-RagCW/FLAG-RagAX expression, HEK293T cells (ATCC) were used. For ACAD10 mRNA level determination, HEK293E and C8161 cells were used. For immunofluorescence assays, HeLa cells (ATCC) were used.
Chemicals and Antibodies
Metformin and phenformin (Sigma) were dissolved in water with the stock solutions as 1 M and 0.2 M respectively. AICAR (LC Laboratories) was dissolved in water at 50 mM. Rotenone (Sigma) and rapamycin (LC laboratories) were dissolved in ethanol to make 2.5 mM and 10 mM stock solutions, respectively. For transfection, Lipofectamine 3000 was used according to the instructions of manufacturer (Invitrogen). For western blotting, we used the anti-TPR antibody (NB100-2866, Novus Biologicals), the self-generated anti-RagC antibody (Oshiro et al., 2014), and antibodies against p-S244/240 S6 (#2215), total S6 (#2217), mTOR, TSC2, Raptor as well as RagA from Cell Signaling Technology, and FLAG-Tag from Sigma. For immunostaining of RagC, the same self-made Rabbit RagC mAb and the corresponding secondary Anti-Rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate, Cell Signaling Technology) was used, while Alexa Fluor® 594 anti-nuclear pore complex proteins antibody, mAb414, was used to stain NPCs (BioLegend).
METHOD DETAILS
Feeding RNAi in C. elegans
The RNAi clones used in this study were all isolated from a genome-wide E. coli feeding RNAi library and fed to C. elegans as described previously (Pino et al., 2013). Synchronous C. elegans were obtained by bleach treatment of gravid adults, and after 72 hours of RNAi feeding, animals were harvested and examined as day-1 adults unless noted otherwise. For the RNAi screen of metformin response genes, the RNAi bacteria were seeded onto 96-well plates using NGM agarose supplemented with 5 mM IPTG (US Biologicals) and 100 µg/mL carbenicillin (Sigma), free liquid was driven off in a laminar flow hood, and RNAi plates were incubated at room temperature overnight. The following day, the indicated dose of metformin was added into 96-well RNAi plates and free liquid was driven off in a laminar flow hood. Synchronous populations of L1 stage, wild type worms were dropped onto 96-well RNAi plates with or without metformin, and imaged directly from the RNAi plates after 3 days exposure to drugs using a Leica DM6000 microscope at a magnification of 40×. Imaged worms were scored based on their stages and sizes compared to controls.
Quantitative RT-PCR
To quantify changes in mRNA abundance in nematode and human cells, total RNA was extracted using TRIzol (Invitrogen) according to manufacturer instructions. RNA was treated with RNAse free DNAse prior to reverse transcription with the Quantitect reverse transcription kit (Qiagen). Quantitative PCR was conducted in triplicate using Quantitect SYBR Green PCR reagent (Qiagen) following manufacturer instructions on a Bio-Rad CFX96 Real-Time PCR system (Bio-Rad). For C. elegans, all assays were performed with 2000 animals at late L4 stage per sample. Drug treatments were started from synchronous L1 larvae stage and ended as late-L4 stage. Drug treated worms were harvested by washing off of plates and washed an additional 4 times with M9 buffer, allowing worms to settle by gravity between washes. RT-PCR assays for human cells were conducted with 2 × 105 cells post 24-hour drug treatments or 96-hour shRNA transduction. Both worms and cells were flash frozen in liquid nitrogen and kept in −80 °C until RNA preparation. The sequences of used primer sets were: for C. elegans: CeACAD10: 5’-GCCATGCTGACAAACAACAT-3’ and 5’-TCAAATCGCATGGAAGTCTG-3’, npp-3: 5’-CCTCGacTTCTCGATCTTCG-3’ and 5’-GTTGAActTTTGCATCAACACAC-3’, npp-21: 5’-ACAGCACGCCAAGAATCAG-3’ and 5’-TCCGTATtcTCGTTGCGATA-3’, act-1: 5’-TGCTGATCGTATGCAGAAGG-3’ and 5’-TAGATCCTCCGATCCAGACG-3’, daf-15: 5’-CTGAGAAggCATGTATGCATC-3’ and 5’-ATGGGCAGAATGGATTTGAA-3’; for human cells: ACAD10: 5’-CGAGTGGCAAAGCAGTTCC-3’ and 5’-CCATGCAGGACTCCACAATCA-3’, TPR: 5’-AACGCCAGCGTGAGGAATATG-3’ and 5’-ATTACGTGGTTACCCCTTGCT-3’, NUP205: 5’-GTACTGGGATGGAAAGCGATG-3’ and 5’-GCTCTGGACTGAGTTCTAGGG-3’, NUP153: 5’-CAGGGGCCAATTAAGCCTTAC-3’ and 5’-ACCTCGCTTGTGTCTGTTGAA-3’, NUP98: 5’-CTCCACCACTAATTCAGGCTTT-3’ and 5’-GAGGCTGGTAGTCTGCTGATT-3’, NUP214: 5’-TGACTCCCCTGAGGAATTGC-3’ and 5’-GCGAAGACCAGACCATATTTGTT-3’, beta-actin: 5’-CATGTACGTTGCTATCCAGGC-3’ and 5’-CTCCTTAATGTCACGCACGAT-3’.
Expression levels of tested genes were presented as normalized fold changes to the mRNA abundance of act-1 for worms and beta-actin for human cells respectively by the ΔΔCt method.
Lentiviral shRNA Infection
Lentiviral shRNA constructs based in pLKO1 were obtained from the Broad Institute of Harvard and MIT Genetic Perturbation Platform and are commercially available from Sigma. shRNA delivery was conducted as previously published (Yuan et al., 2012). shRNA knockdown cells post 24 hour-selection with 2 µg/ml puromycin were treated in medium containing 0.5 µg/ml puromycin with or without drugs for the indicated times followed by western blotting or cell viability measurement. The knockdown efficiency of targeted genes by shRNA in different cell lines was examined by RT-PCR for mRNA levels under identical experimental conditions at the identical time of experiments.
Western Blotting
Cell lysates were prepared immediately following the indicated treatments by boiling in 0.5% SDS for 10 minutes, followed by incubation on ice. Protein concentration was determined using the Pierce BCA assay (Thermo Fisher). SDS-PAGE was conducted on the same day that lysates were made, followed by electrophoretic transfer to nitrocellulose membrane at 50 volts for 16 hours at 4°C. Immunoblots were performed according to primary antibody manufacturers’ protocols. For immunodetection of primary antibodies, goat-anti-rabbit-HRP conjugate (GE Healthcare) was used at 1:5000 in 5% BSA dissolved in TBST, and HRP was detected using West-Pico chemiluminescence substrate (Thermo Pierce). The western blot results shown are representatives of at least two independent biological replicates.
Immunofluorescence
HeLa cells were plated on glass coverslips in 6 well cell culture plates grown to about 50% confluence. Cells were treated with vehicle and 1 mM phenformin for 24 hours, then washed twice in PBS and fixed with 4% paraformaldehyde in PBS for 15 minutes. Cells were rinsed twice with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 minutes. After rinsing with PBS, cells were blocked in 3% BSA/PBS for 30 minutes and then incubated in 3% BSA with anti-RagC antibody overnight. Cells were washed three times in PBS and then incubated in 3% BSA with Anti-Rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) for 1 hour. For NPC immunostaining with mAb414 antibody, the experiment was done exactly following the instructions from the manufacturer (BioLegend). Cells were then rinsed three times for 5 minutes each in PBS and mounted using ProLong® Diamond Antifade mounting medium with DAPI (Life Technologies). Images were captured using a Leica DM6000 microscope with a 63× oil immersion objective.
Body Size Determination of C. elegans
Animals, post RNAi feeding, drug treatment, or combinations of both for 72 hours, were paralyzed in M9 buffer with 1 mg/ml levamisole (Sigma), and then imaged at 5× magnification on a Leica DM6000 microscope. Maximal, longitudinal cross-sectional area of the imaged C. elegans was determined by using MetaMorph software for a minimum of 27 animals per condition in each experiment. Results of single experiments are shown. Each experiment was performed at least twice, and results were consistent between experiments.
Creation of the CeACAD10 Fusion and Promoter Transgenic Lines
For the CeACAD10 reporter (Figure 1G), the entire genomic sequence of CeACAD10 locus (1367 bp), including introns and exons, plus 2418 bp of promoter were amplified and cloned into a modified version of the commercial Fire vector pPD95.77 (the GFP region was swapped with monomeric red fluorescent protein (mRFP) with a C-terminal HA epitope tag) at the SbfI and SalI sites. The following cloning primers were used for the fusion reporter: forward: 5’-AGCCTGCAGGACCGTAGCTCCAGTGTCGATT-3’ and reverse: 5’-AGCCTGCAGGTCCACAAAAATTGCTCATTGC-3’. The same forward primer above was used for cloning the promoter only transgenic line (Figure 2G), while the reverse one was: 5’-CCGGGTACCTGCTGGGTCATAGTCATTTTG-3’. Both fragments were injected at 20 ng/µl into the gonad of wild type adult animals. The CeACAD10 reporter was integrated into the genome with 250 J m−2 UV-C using a Stratalinker 2400 UV crosslinker (Stratagene, La Jolla, CA, USA) and screened for the successfully integrated lines, while the promoter only transgenic was used directly for expression tests post metformin treatment.
Isolation of Metformin Suppressor Mutants
The reporter line was mutagenized with ethyl methanesulfonate (EMS, Sigma), and the resultant F2 generation was screened on plates supplemented with 50 mM metformin for suppressors of CeACAD10 expression. Whole genome Illumina sequencing was employed followed by analysis and variant calling in Galaxy (Kim et al., 2008) to identify causal mutations. Causal mutations were confirmed by Sanger sequencing and RNAi.
CeACAD10 Reporter Fluorescence Intensity Analysis
Animals of the reporter line in either wild type or mutant background were treated with drugs or RNAi for indicated times, paralyzed with 1 mg/ml of levamisole, and then imaged in 96-well format with a Leica DM6000 microscope outfitted with a mCherry filter set and MMAF software. Image analysis was conducted with MATLAB (MathWorks, Inc.) as described previously (Pino et al., 2013). All RFP or GFP intensity tests were carried out in biological triplicate or quadruplicate with at least 30 animals tested per replicate.
ATP Measurement
500 synchronized young adult worms were collected in M9 buffer and washed three times. Worm pellets were treated with three freeze/thaw cycles and boiled for 15 min to release ATP and destroy ATPase activity and then spun at 4° at 12,000 × g for 10 min. ATP content was mea sured with an ATP detection kit according to the manufacturer's instructions (ThermoFisher, A22066), and normalized to total protein content determined by BCA assay.
SKN-1 Chromatin Immunoprecipitation (ChIP)
Synchronous L4 animals (~6000 per plate × 8 large plates) were generated from three independent egg preps of SKN-1::GFP transgenic animals. Animals were washed in M9 with 0.005% triton X-100 three times, washed once with M9 with 0.005% triton with 1% formaldehyde, then frozen drop wise in 3 pellet volumes of the same cross-linking buffer (M9 plus 0.005% triton X-100 plus 1% formaldehyde) in liquid nitrogen and kept at −80C. Pea-size frozen pellets were crushed in a dry-ice-chilled mortar and pestle, and ground into powder until worm carcasses are disturbed. After thawing, animals were formaldehyde crosslinked at room temperature for 20 minutes, followed by quenching with 125 mM glycine (final concentration) at room temperature for 5 minutes. Pellets were washed three times in ice cold PBS, spinning each time 2 minutes 4400 rpm between washes at 4C. Pellets were then resuspended in 2 pellet volumes ice cold FA lysis buffer (50 mM HEPES: adjust pH to 7.5 with KOH, 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.2% Sodium Lauryl Sarcosine (AKA Sarkosyl; sodium lauryl sarcosinate), and 2mM PMSF added immediately before use. Sonication was conducted in a Qsonica Q800R with total sample volume not exceeding 180ul per tube with total sonication time of 25 minutes, amplitude 70%, 30 second pulses on alternating with 30 seconds off. The resulting chromatin was cleared by centrifugation at 14,000rpm (21,000 g) for 15min at 4C. The supernatant was retained and protein quantified. One mg of chromatin was used for chromatin immunoprecipitation diluted to 1 mL final volume with no detergent FA Lysis buffer (50 mM HEPES: adjust pH to 7.5 with KOH, 150 mM NaCl, 1 mM EDTA, and 2 mM PMSF added immediately before use). Triton X-100 was added from 25% stock to 1% final concentration. Each 1 mL chromatin was pre-cleared with 30 µl of magnetic protein G Sepharose at 4°C for 1hr. Fifty microliters were removed and used to assess input DNA. Pre-blocked 30uL magnetic protein G-sepharose (with 1mg/ml BSA and 0.3 mg/ml of salmon sperm DNA in FA lysis buffer + 1% Triton X-100) were added per IP (primary antibody and negative control IgG antibody) along with 5 µg of A-11120 mouse anti-GFP monoclonal antibody (Invitrogen) or IgG2a isotype control (02-6200, Invitrogen). Chromatin IPs were incubated overnight at 4°C on a Nutator. Beads were washed five times with 1 ml of no detergent FA lysis buffer + 1% Triton X-100, followed by 1 ml of no detergent FA lysis buffer +1% Triton X100 + 0.5 M NaCl (3 minute wash end over end rotator). Beads were washed with 1 ml TE and immune complexes were eluted with 100 µL ChIP elution buffer 50 mM Tris-Cl, pH 7.5, 10 mM EDTA, 1% SDS 10 minutes at 65C. An equal volume of RNAse A mix (50 mM Tris pH 8.0 99.5 parts to 0.5 part RNAse A sigma R4642 in 50% glycerol (stock 33 mg/mL)) was added to eluted ChIP and input samples and incubated for 1 hour at 37 degrees C. Thereafterer 2 µL per 100uL of 20mg/mL ChIP Proteinase K (4 µL for 200uL of total volume, chromatin plus ChIP elution buffer; PK stock in water +50% glycerol, Sigma P4850) was added to each sample followed by incubation 2 hours at 55C and then 6 hours to overnight at 65C to digest protein and reverse cross links. DNA was cleaned in a Qiagen PCR clean up column and eluted in 50 µL elution buffer. One half µL was added to each qPCR reaction. Signal minus IgG background was expressed relative to the amount of input DNA after calculating the amount of each qPCR reaction from an 8-point standard curve. Primers used for qPCR of ACAD10 genomic loci were as follows: ACAD10p −460nt F, tttttgaacattttggttttgacc; ACAD10p −460nt R, ttccctcgctggaactgaac; ACAD10p −105nt F, gaccaactcgttcagttttgatg; ACAD10p −105nt R, tttgctgggtcatagtcattttg; ACAD10 3'UTR +1.7kb F, tattatgcggcggaatgtcac; ACAD10 3'UTR +1.7kb R, tgcaagcagtgtgattacttgg.
Longevity Assay
Lifespan analysis was conducted at 20°C according t o a protocol modified from our previous and other’s procedures (Cabreiro et al., 2013). Briefly, synchronized L1 animals were seeded onto the standard nematode growth media (NGM) plates until L4 stages. On day 0, 30–50 L4 worms per plate (3 plates, 90–150 worms in total per condition) were transferred onto NGM (for mutants) or RNAi plates (for RNAi) with or without metformin co-treatment. All plates were supplemented with 100 µM 5-fluorodeoxyuridine (FUdR) solution to suppress progeny production. Statistic analysis was performed with online OASIS2 resources (Han et al., 2016).
Cell Fractionation
Cell fractionation was carried out following a rapid and efficient protocol (Suzuki et al., 2010) to avoid possible loss of any nucleocytoplasmic shuttling proteins from the nucleus during the time of isolation, with the following modifications. In this study sub confluent HEK293E cells untreated or treated with 24 hours on 6-well plates were washed 3 times with ice cold PBS prior to fractionation with 0.1% NP40 in PBS. Total lysates, cytosol and nuclear fractions were collected at each step of fractionation for western blot analysis.
Cell Viability Measurement
Cell viability analysis was conducted as previously described (Yuan et al., 2013). In brief, shRNA knockdown cells were seeded onto 96-well plates at a density of 800–1000 cells per well, and drug treatment was initiated the following day. After 3 days of drug treatment, the viability measurement was carried out with the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega) according to the manufacturer’s instructions.
Nuclear Permeability Assay
C8161 cells were plated with 3 × 105 cells per well onto sterilized coverslips in the base of 6-well plates 24 hours before drug treatment. After 24 hours of drug treatment, cell membranes were permeabilized with 40 µg/ml digitonin in freshly made transport buffer (TB) containing 20 mM HEPES, pH 7.3, 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, 2 mM DTT and protease inhibitor cocktail (Sigma) at room temperature. Digitonin permeabilization times were optimized by using trypan blue (Life Technologies) staining in C8161 cells (10 minutes) to efficiently permeabilize plasma membranes while keeping the nuclear envelope intact as indicated by full exclusion of fluorescein isothiocyanate (FITC)-Dextran 500000-Conjugate (Sigma). Immediately after semi-permeabilization, cells were rinsed twice with TB and incubated with fluorescent labeled dextran mixtures at the concentration of 2 mg/ml of each dextran for 20 minutes, including FITC-Dextran 500000-Conjugate mixed with Tetramethylrhodamine isothiocyanate (TRITC)-Dextran average molecular weight 65,000–85,000 (Sigma) for C8161 cells. Then, coverslips were inverted onto a 15 µl drop of Antifade mounting medium made with 1X PBS, 90% glycerol, and 2% n-propyl gallate (Sigma) for imaging. Images were taken from randomly distributed fields of the coverslips using a Leica DM6000 microscope outfitted with a standard GFP filter set or mCherry filter set and MMAF software at a 63× oil immersion objective. Scale bars were added using ImageJ software. The fluorescence intensities of intranuclear area and extranuclear background for each nucleus were measured using ImageJ, and the intensity ratio of intranuclear and extranuclear levels was calculated to index nuclear permeability.
RagC Proteomic Analysis
Flag-tagged RagC and RagA were co-transfected into HEK293T cells in order to prepare four samples: 1) control (no drug) total RagC, 2) total RagC subjected to treatment with 1 mM phenformin, 3) control (no drug) cytoplasmic RagC, and 4) control (no drug) nuclear RagC. Flag-tagged RagC was immunopurified by large-scale immunoprecipitation with anti-Flag M2 magnetic beads (Sigma), eluted with Flag peptide and separated by SDS-PAGE. The 4 gel bands were in-gel digested with trypsin and analyzed by a Q-Exactive mass spectrometer, which provided high-resolution MS peptide mass measurement and high-resolution MS/MS peptide sequencing. We searched the 4 data files with both our in-house Sequest-based platform, as well as the commercial software PEAKS in attempt to identify modifications in RagC. Overall sequence coverage was above 60% for phenformin and control, 90% for cytoplasmic and 50% for nuclear fractions, which correlated well with gel band intensity. For our in-house platform, we used a wide mass tolerance no enzyme search and a single protein database. Identified peptides were limited to fully tryptic peptides having a ppm tolerance of less than 10 ppm and a cross-correlation score of greater than 2.5. We specifically searched for oxidation, mono-, di- and tri-methylation, acetylation and phosphorylation with our in-house software and 485 different modifications using the PEAKS software.
GAP Assay
The GAP assay was performed to detect whether there is the GAP activity present in the nucleus for the activation of RagC as described previously (Oshiro et al., 2014) with minor modifications according to other recent work (Tsun et al., 2013). Briefly, in order to specifically monitor the GAP activity on RagC GTPase, the wild type GST-RagCW construct (8 µg) was co-transfected with FLAG-RagAQ66L;D130N (RagAXTP form, which only binds xanthosine containing nucleotides, 4 µg) into HEK293T cells in 10 cm tissue culture plates using Lipofectamine 3000 (ThermoFisher). Cells were collected 48 hours post transfection for protein purification with the Glutathione Agarose. The purified heterodimer (25 µg) was first charged with 0.1 µM xanthosine triphosphate (XTP, sigma) to get RagA fully charged with XTP to avoid interruptions to the GAP assay for RagC. Then the complex was incubated with α-32P-GTP to charge RagC with labeled GTP for monitoring the GAP activity in the cell fractions. As indicated in Figure 6J, cytoplasmic and nuclear fractions were added to the pre-charged Rag GTPase complex to determine which fraction has GAP activity towards RagC GTPase.
QUANTIFICATION AND STATISTICAL ANALYSIS
All the western blotting and dextran diffusion quantifications were conducted in ImageJ. The statistical analyses were performed using Prism (GraphPad Software). All values are presented with mean ± SEM and the number (n) of samples employed is indicated in legends. The statistical differences between control and experimental groups were determined by one-way ANOVA, with p values < 0.05 considered significant. Two-way ANOVA with the Bonferroni corrected for multiple hypothesis testing was performed to determine the significant difference where genetic interferences and drug treatments were applied together. p values < 0.05 were considered statistically significant. The log rank test was used to determine significance in lifespan analyses.
Supplementary Material
Highlights.
-
➢
NPC and ACAD10 mediate biguanide-induced growth inhibition and lifespan extension
-
➢
Metformin effects on mitochondria are transduced to mTORC1 and growth by the NPC
-
➢
Biguanides restrain RagC transit through the NPC
-
➢
Nuclear transit of RagC is essential for activation of mTORC1 signaling
Metformin both suppresses cancer cell growth and promotes organismal longevity through a key transcriptional target that is induced through inhibition of mitochondrial respiration and modulation of mTOR signaling.
Acknowledgments
We thank the Broad Institute GPP for lentiviral shRNA reagents. Some worm strains were from the CGC, which is funded by NIH (P40 OD010440). This work was funded by NIH Grants R03DK098436 (A.A.S), K08DK087941 (A.A.S), R01DK072041 (to A.A.S), and R01CA166717 (B.Zheng), a Broad Institute SPARC Grant (A.A.S.), the Ellison Medical Foundation New Scholar in Aging Award (A.A.S.), by support from the NORC of Harvard (P30DK040657) and the Boston Area DERC (P30DK057521). We thank Joseph Avruch, Jose Florez, and Geoffrey Walford for critical reading and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online.
AUTHOR CONTRIBUTIONS
Conceptualization, L.W., B.Zheng, A.A.S.; Methodology, L.W., N.O-R., M.L., F.M., M.E.T., S.P.G., B.Zheng, A.A.S.; Software, M.E.T., C.E.C.; Formal Analysis, L.W., M.E.T., S.P.G., B.Zheng, A.A.S.; Investigation, L.W., B. Zhou., N.O-R., J.A.P., C.M.W., F.M., M.C.K., S.P.G, A.A.S.; Resources, M.E.T., C.E.C., S.P.G, B.Zheng, A.A.S.; Writing—Original Draft, L.W., A.A.S.; Writing—Review & Editing, L.W., B. Zhou, C.M.W., B.Zheng, A.A.S.; Visualization, L.W., M.C.K., A.A.S.; Supervision, A.A.S.; Project Administration, A.A.S.; Funding Acquisition, A.A.S., B.Zheng.
REFERENCES
- Back JH, Kim AL. The expanding relevance of nuclear mTOR in carcinogenesis. Cell Cycle. 2011;10:3849–3852. doi: 10.4161/cc.10.22.18329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA. Metformin as a Tool to Target Aging. Cell Metab. 2016;23:1060–1065. doi: 10.1016/j.cmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Hanson RL, Muller YL, Ma L, Investigators M, Kobes S, Knowler WC, Bogardus C, Baier LJ. Variants in ACAD10 are associated with type 2 diabetes, insulin resistance and lipid oxidation in Pima Indians. Diabetologia. 2010;53:1349–1353. doi: 10.1007/s00125-010-1695-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Zielonka J, Ouari O, Lopez M, McAllister DM, Boyle KA, Barrios CS, Weber J, Johnson BD, Hardy M, et al. Mitochondria-targeted analogs of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res. 2016;76:3904–3915. doi: 10.1158/0008-5472.CAN-15-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L, Temmerman L. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci U S A. 2014;111:E2501–E2509. doi: 10.1073/pnas.1321776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Hebrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, Sakamoto K, Andreelli F, Viollet B. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, Jones RG. Metformin Antagonizes Cancer Cell Proliferation by Suppressing Mitochondrial-Dependent Biosynthesis. PLoS Biol. 2015;13:e1002309. doi: 10.1371/journal.pbio.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, Kim S. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016 doi: 10.18632/oncotarget.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc Natl Acad Sci U S A. 2000;97:14340–14345. doi: 10.1073/pnas.011511898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y, Kalir T, Rahaman J, Dottino P, Kohtz DS. Alterations in nuclear pore architecture allow cancer cell entry into or exit from drug-resistant dormancy. Am J Pathol. 2012;180:375–389. doi: 10.1016/j.ajpath.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes S, Pollak MN, Zwinderman AH, Mathot RA, Weterman MJ, Beeker A, Punt CJ, Richel DJ, Wilmink JW. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16:839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A. 2012;109:8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CL, Timney BL, Rout MP, Wente SR. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J Cell Biol. 2015;208:729–744. doi: 10.1083/jcb.201412024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken B, Driscoll M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One. 2010;5:e8758. doi: 10.1371/journal.pone.0008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N, Rapley J, Avruch J. Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging. J Biol Chem. 2014;289:2658–2674. doi: 10.1074/jbc.M113.528505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, Suzuki T, Yamamoto M, Blackwell TK, Curran SP. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston-Gosse J, Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 2007;39:1403–1409. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- Pino EC, Webster CM, Carr CE, Soukas AA. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J Vis Exp. 2013;73 doi: 10.3791/50180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachar J. Intimate contacts of mitochondria with nuclear envelope as a potential energy gateway for nucleo-cytoplasmic mRNA transport. Gen Physiol Biophys. 2003;22:525–534. [PubMed] [Google Scholar]
- Rakowska A, Danker T, Schneider SW, Oberleithner H. ATP-Induced shape change of nuclear pores visualized with the atomic force microscope. J Membr Biol. 1998;163:129–136. doi: 10.1007/s002329900377. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Rout MP. Cancer and the nuclear pore complex. Adv Exp Med Biol. 2014;773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- Steinbaugh MJ, Narasimhan SD, Robida-Stubbs S, Moronetti Mazzeo LE, Dreyfuss JM, Hourihan JM, Raghavan P, Operana TN, Esmaillie R, Blackwell TK. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife. 2015;4:e07836. doi: 10.7554/eLife.07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC Res Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJ. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 2014;156:759–770. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Pino E, Wu L, Kacergis M, Soukas AA. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287:29579–29588. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Ito K, Perez-Lorenzo R, Del Guzzo C, Lee JH, Shen CH, Bosenberg MW, McMahon M, Cantley LC, Zheng B. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci U S A. 2013;110:18226–18231. doi: 10.1073/pnas.1317577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.