Abstract
We report the fabrication and biological evaluation of non-woven polymer nanofiber coatings that inhibit quorum sensing (QS) and virulence in the human pathogen Staphylococcus aureus. Our results demonstrate that macrocyclic peptide 1, a potent synthetic non-bactericidal quorum sensing inhibitor (QSI) in S. aureus, can be loaded into degradable polymer nanofibers by electrospinning, and that this approach can deposit QSI-loaded nanofiber coatings onto model non-woven mesh substrates. QSI was released over ∼3 weeks when these materials were incubated in physiological buffer, and retained its biological activity and strongly inhibited agr-based QS in a GFP reporter strain of S. aureus for at least 14 days without promoting cell death. These materials also inhibited production of hemolysins, a QS-controlled virulence phenotype, and reduced the lysis of erythrocytes when placed in contact with wild-type S. aureus growing on surfaces. This approach is modular, and can be used with many different polymers, active agents, and processing parameters to fabricate nanofiber coatings on surfaces important in healthcare contexts. S. aureus is one of the most common causative agents of bacterial infections in humans, and strains of this pathogen have developed significant resistance to conventional antibiotics. The QSI-based strategies reported here thus provide springboards for the development of new anti-infective materials and novel treatment strategies that target virulence as opposed to growth in S. aureus. This approach also provides porous scaffolds for cell culture that could prove useful in future studies on the influence of QS modulation on the development and structure of bacterial communities.
Keywords: anti-virulence, coatings, controlled release, electrospinning, nanofibers, polymers, quorum sensing
Graphical Abstract
Introduction
Bacterial fouling and infection remain a persistent challenge confronting the use of interventional devices, implanted objects, dressings, and a wide range of other materials used in healthcare-related applications.1,2 One useful strategy to prevent or treat these infections is to design materials and surface coatings that release agents that can prohibit or attenuate bacterial cell proliferation or the upregulation of virulent behaviors,3–5 such as biofilm formation.2 These ‘surface-mediated’ approaches to release and delivery have several practical advantages relative to the systemic administration of antimicrobial agents, including reducing the likelihood of host cell toxicity and other side effects by enabling the local delivery of reduced concentrations of antimicrobial agents directly to areas that are more prone to bacterial infection (e.g., near the surface of an indwelling device or topical dressing).3,4
Regardless of the approach used for delivery, the use (and over-use) of conventional antimicrobial agents has lead to a sharp increase in the prevalence of antibiotic-resistant strains of many common human pathogens—a problem that is the result, at least in part, of the bactericidal nature of those agents, which can contribute to evolved resistance.6,7 A frightening statistic in this regard is that methicillin-resistant strains of the Gram-positive pathogen Staphylococcus aureus (i.e., MRSA) kill more people in the U. S. annually than Parkinson’s disease, HIV/AIDS, emphysema, and homicide combined.8 In view of this rapidly emerging threat, new methods for treating or preventing bacterial infections that move beyond conventional antibiotics and other biocidal strategies are urgently needed.9,10 One potential alternative to these biocidal strategies is the use of agents that can interfere with or inhibit bacterial ‘quorum sensing’ (QS) circuits that govern many infection phenotypes in common pathogens,11 including virulence factor production and biofilm formation, but do not promote bacterial cell death.12–14 This new ‘anti-virulence’ approach has been demonstrated to be promising in many clinically relevant bacteria, including S. aureus15–17 and the Gram-negative pathogens Pseudomonas aeruginosa18,19 and Acinetobacter baumannii.20
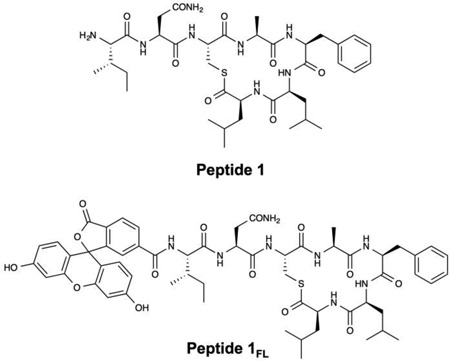
Recent studies by our group21–25 and others26–36 have demonstrated that synthetic inhibitors of bacterial QS (QSIs) can be imbedded in or immobilized onto a variety of materials, and that this approach can be used to design surfaces and coatings that inhibit QS and reduce virulent behaviors in human pathogens. Of particular relevance to the work reported here, we recently reported materials-based approaches to the release of a highly potent synthetic mimic of the auto-inducing peptide (AIP) signal that regulates the accessory gene regulator (agr) QS system in S. aureus (peptide 1).23,24 Peptide 1 inhibits QS at sub-nanomolar concentrations in all four specificity groups of S. aureus (including group-I and group-III S. aureus, which are the groups commonly associated with methicillin resistance and toxic shock syndrome, respectively).15 This peptide acts via competitive inhibition of the native AIP signal by binding its cognate transmembrane receptor, AgrC. Inhibition of S. aureus AgrC activity blocks the downstream transcription of an arsenal of virulence factors that are critical for infection, including proteases, hemolysins, enterotoxins, immunomodulatory peptides, regulatory RNAs, and surface factors.37 Modulation of AgrC activity also alters the production and maturation of S. aureus biofilms and, interestingly, inhibition of AgrC has been shown to activate biofilm formation under certain in vitro conditions.38 This inverse relationship between QS-controlled toxin production and biofilm formation (at least under select conditions), has provoked many questions about optimal strategies to attenuate agr-QS in both acute and chronic infections,38 but past in vivo studies demonstrate that the inhibition of AgrC can significantly block infection using a range of animal models,39,40 revealing AgrC as a central and attractive target for infection control in S. aureus.41 The work reported here sought to advance the design of surface coatings containing peptide 1 as a step toward new tools that could help shed light on the complex relationship between biofilm formation, toxin production, and agr-type QS modulation in in vitro and in vivo settings. In the long term, such materials and insights could contribute to the development of new approaches to the treatment of bacterial infection, including the treatment of skin-related infections caused by S. aureus.17,39,40
In our past work, we developed peptide 1-loaded coatings using either conventional solvent-casting approaches23 or new methods for the fabrication of porous superhydrophobic materials24 to promote the release of biologically-active peptidic QSIs rapidly (e.g., within ∼5 min) or over prolonged periods (e.g., over ∼8 months), respectively, when exposed to aqueous environments. However, many potential applications are likely to require (or could benefit considerably from) materials that can promote the local release of QSIs, or combinations of QSIs and other anti-microbial agents, over intermediate time frames (e.g., over several days or several weeks) or with form factors and physicochemical properties that are difficult to achieve using those past methods (e.g., to allow the facile coating of woven gauze, dressings, and other objects or devices where bacterial infection is endemic. Here, we report new materials design strategies that can (i) permit control over the release of peptidic QSIs at such intermediate time scales (i.e., over ∼14 days), and (ii) provide robust QSI-eluting materials using methods of fabrication that are scalable and more amenable to deposition on complex substrates than methods used in our past studies (i.e., dip-coating23 or layer-by-layer assembly methods24). Our approach is based on the electrospinning of porous, non-woven mesh coatings (or ‘mats’) of degradable QSI-loaded polymer nanofibers.
The electrospinning of polymer solutions is widely used to produce non-woven meshes or mats of randomly aligned polymer nanofibers.42–44 These materials are fabricated by using a strong electrical potential to extrude a concentrated solution of polymer from a needle to form extremely thin (nanometer-scale) fibers that can be deposited onto electrically grounded surfaces or collected onto other objects. This electrospinning approach is compatible with the use of many different types of natural and synthetic polymers, and it provides multiple means to tune the physical properties of the resulting fibers (e.g., by control over solution concentrations, flow rates, and other process variables).42–44 In the specific context of drug delivery, this approach also enables the fabrication of nanofibers loaded with active agents simply by adding those agents directly to the polymer solutions prior to electrospinning.45,46 This combination of features, combined with the ability to deposit materials over large areas and on topologically complex objects, renders electrospinning attractive for the design of fiber-based coatings of potential utility in many biomedical contexts, including the controlled release of small-molecule and macromolecular agents45–47 and the design of new functionalized or drug-eluting porous polymer platforms for cell culture and tissue engineering.46,48 Depending on the type of polymer, agent, and processing conditions used, this approach can be used to produce drug-loaded nanofiber mats and coatings that release embedded agents by diffusion, by polymer degradation, or by a combination of the two, over a broad range of time scales.46,47 Such mats and coatings have broad potential in the context of combating acute and chronic infections.49–51
Here, we report the design and fabrication of non-woven and hydrolytically degradable nanofiber mesh coatings that release peptidic QSIs active in S. aureus when exposed to physiologically relevant environments. We demonstrate that solutions of peptide 1 and poly(lactide-co-glycolide) (PLGA) can be electrospun onto planar surfaces and other non-woven supports to produce coatings that release peptide 1 for approximately three weeks, and that objects coated with these peptide-loaded mats can modulate agr-type QS signaling in S. aureus using a GFP reporter. We demonstrate further that these materials can be used to strongly inhibit the production of bacterial toxins (i.e., hemolysins) that are under the control of QS in S. aureus, both in suspensions of wild-type planktonic bacteria and when fiber-coated materials are placed into contact with wild-type bacteria growing on surfaces. Our results provide the basis of a new and highly adaptable approach that could be used to tune the release of QSIs from the surfaces of topologically complex objects, including fabric-based dressings or gauzes, interventional devices, and implantable objects. With further development, we anticipate that this electrospinning approach could contribute to the development of new and non-bactericidal anti-virulence approaches that provide novel strategies for combatting bacterial infections, with the potential to side-step, or at least substantially delay, issues associated with the development of evolved resistance that currently plague approaches based on bactericidal agents.
Materials and Methods
Materials
Poly(D,L-lactide-co-glycolide) (PLGA; 50:50, MW = 30,000–60,000), N,N-dimethylformamide (DMF; ACS reagent grade), and tetrahydrofuran (THF; HPLC grade) were purchased from Sigma-Aldrich (Milwaukee, WI) and used as received. Peptide 1 and fluorescently labeled peptide 1FL were synthesized and purified as described previously.15,24 Blood agar plates (tryptic soy agar, with 5% sheep blood) were purchased from Fisher Scientific (Hanover Park, IL). Pooled rabbit blood cells were purchased from Lampire Biological Labs (Piperville, PA). Non-woven bonded carded web mesh composed of fibers of poly(ethylene terephthalate) and polyethylene (Vliesstoffwerk Sandler 12 gsm Sawabond NW Cover – Velvet; calendared) was obtained from Sandler AG (Schwarzenbach/Saale, Germany).
Instrumentation and Related Considerations
Fluorescence microscopy images were acquired using an Olympus IX70 microscope and analyzed using the Metavue version V7.7.8.0 software package (Molecular Devices). Top-down scanning electron micrographs were acquired using a LEO-1550 VP field-emission SEM operating with an accelerating voltage of 2.00 kV. Samples were coated with a thin layer of gold using a Hummer Junior Sputtering (Technics) system operating at 10 mA under a vacuum pressure of 70 mTorr for 60 s prior to imaging. Fiber diameters were measured using the ImageJ version 1.49r software package. Solution fluorescence was measured using a Jobin Yvon FluoroMax-3 fluorometer with DataMax version 2.2 software. A Biotek Synergy 2 microplate reader with Gen5 software was used to measure the absorbance and fluorescence of bacterial cultures.
Biological Reagents and Strain Information
All biological reagents, with the exception of Brain-Heart Infusion (BHI) medium, were purchased from Sigma-Aldrich and used according to enclosed instructions. S. aureus strains AH1677 (methicillin-resistant group-I with a P3-gfp reporter plasmid) and RN6390B (wild-type group-I) were grown in BHI medium purchased from Teknova (Hollister, CA). Bacterial cultures were grown in a standard laboratory incubator at 37 °C with shaking (200 rpm) unless otherwise noted.
Fabrication of Non-Woven Nanofiber Mats
A 300 mg/mL solution of PLGA was prepared using a THF:DMF solvent mixture (3:1, v/v) and stirred overnight. For fabrication of nanofibrous mats containing peptide 1 or peptide 1FL, these solutions were prepared using DMF having a 1 mM concentration of the peptide instead of neat DMF. Electrospinning was conducted using a custom-built electrospinning device with a digital syringe pump (Harvard Bioscience Company) at a flow rate of 0.2 mL/h. A 15 cm working distance separated the blunt 20G needle and the 10 × 10 cm grounded collector covered with the non-woven mesh substrate. A 20 kV potential was applied between the needle tip and collector. All materials used in release experiments were fabricated by electrospinning 0.25 mL of solution. The resulting electrospun mats were stored in the dark at room temperature until further use.
Characterization of Peptide Release Kinetics
Experiments used to characterize the release of imbedded peptides were conducted using PLGA mats containing peptide 1FL. Strips of non-woven mesh coated with electrospun nanofibers (0.8 × 2.0 cm; 1.6 cm2) were incubated in 1 mL of PBS buffer (pH = 7.4) at 37 °C. At designated time points, the buffer was removed for analysis and replaced with fresh buffer. Removed buffer was briefly centrifuged to pellet and remove any solid polymer debris that may have dislodged from the mats and could complicate analysis of the peptide release. Concentrations of released peptide 1FL were measured using a fluorometer. All release experiments were conducted with n = 4.
Release of Peptides for Liquid Culture Assays
Characterization of the biological activities of released peptides was conducted using PLGA mats containing peptide 1. Small strips of non-woven mesh coated with electrospun nanofibers (0.6 × 1.5 cm; 0.9 cm2) were cut and UV sterilized by exposure to UV light for 15 min in a biological safety cabinet. In the 24 h immediately prior to a designated time point in the biological assay, the strips were incubated statically in 500 µL of either BHI media (for GFP reporter assays) or Tryptic Soy Broth media (TSB; for hemolysis assays) at 37 °C. At all other times prior to the 24 h period before a measured time point, the strips were maintained statically in 500 µL of 7.4 pH 1x PBS buffer at 37 °C. PLGA mats fabricated without peptide 1 were used as controls for the biological assays. Each release experiment was conducted with n = 4, and three release experiments were performed for each biological assay to generate three biological replicates. All manipulations of sterilized samples were performed using flame-sterilized forceps.
GFP Reporter Assays
GFP reporter assays in S. aureus were performed using our previously reported protocol.15 In brief, an overnight culture of the GFP-reporter strain S. aureus AH167752 was diluted 1:50 to a total volume of 200 µL with BHI medium used in peptide release experiments and placed into the wells of a black 96-well polystyrene microtiter plate (Costar). Dilution with fresh BHI medium was used as a control. The plates were incubated at 37 °C for 24 h with shaking. The fluorescence and OD600 of each well were then measured using a plate reader, with fluorescence values normalized to the fresh BHI control.
Hemolysis Assays
For liquid culture hemolysis assays, an overnight culture of the wild-type strain S. aureus RN6390B was diluted 1:100 to a total volume of 200 µL with TSB medium used in peptide release experiments and placed into the wells of a clear 96-well polystyrene microtiter plate (Costar). Dilution with fresh TSB medium was used as a control. The plates were incubated statically at 37 °C for 6 h, and OD600 values were then measured using a plate reader. A 13 µL aliquot of rabbit red blood cells (10%, suspended in PBS) was then added to each well, and the plate was incubated statically at 37 °C for 15 min. The plate was centrifuged at 2000 rpm at room temperature for 4 min, and a 150 µL aliquot of the supernatant from each well was transferred to wells of a clean, clear 96-well plate to measure absorbance at 420 nm using a plate reader. For blood agar plate assays, an overnight culture of S. aureus RN6390B was diluted 1:2×105 with fresh BHI. A 500 µL aliquot of the diluted culture was added to the blood agar plate and spread evenly over the surface using a sterilized Pasteur pipette that was bent at a 90° angle using a flame. Segments of non-woven mesh coated with electrospun nanofibers, fabricated both with and without peptide 1, were UV sterilized for 15 min and placed on top of the agar plate such that the nanofiber-coated side was facing the agar surface. The agar plate was then incubated at 37 °C for 24 h, after which digital images of the plate were acquired.
Results and Discussion
Fabrication of Nanofiber Mats Containing Synthetic Peptidic QSIs 1 and 1FL
Our approach to the design of non-woven nanofiber mats that can inhibit bacterial QS is based upon the electrospinning of polymer solutions containing peptide 1. As outlined above, peptide 1 is a potent synthetic inhibitor of agr-type QS in the four groups of S. aureus, with an IC50 of 0.485 nM against MRSA-associated group-I strains and an IC50 of 0.0506 nM against toxic shock syndrome-associated group-III strains,15 strains that are of importance in the context of bacterial infections. To explore the feasibility of this new approach and demonstrate proof of concept, we selected the commercially available polyester PLGA (with a 50:50 ratio of lactide and glycolide repeat units) as a model degradable polymer for several reasons: (i) this polymer is FDA-approved and has a long history of use for drug delivery and other biomedical applications,53–55 (ii) the structure and composition of this polymer can be easily manipulated to exert useful control over the release of encapsulated agents,56,57 and (iii) the use of this polymer as a basis for the electrospinning of degradable polymer nanofibers using a range of solvents and processing parameters is well established.46,48,58–60
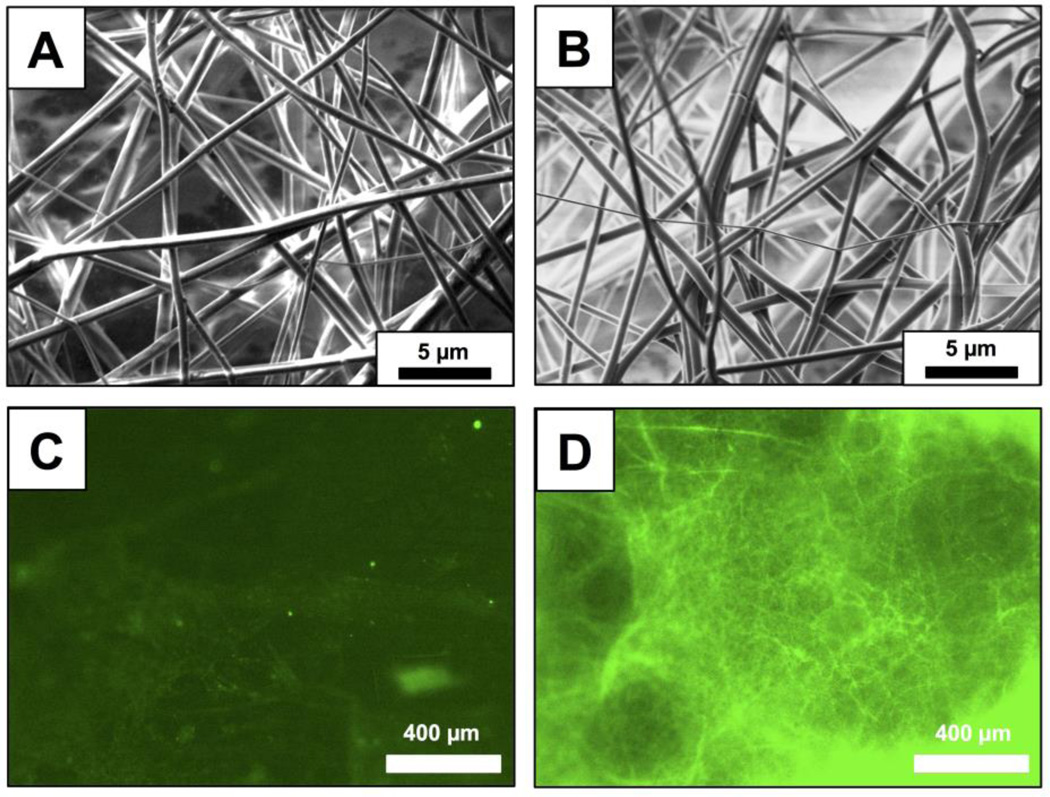
For all experiments described below, PLGA nanofibers were fabricated by electrospinning solutions of PLGA containing 0.25 mM of peptide 1 onto model non-woven mesh substrates composed of poly(ethylene terephthalate) and polyethylene fibers (see Materials and Methods for additional details of electrospinning process parameters used in these experiments). For experiments designed to characterize the loading and release of the peptide from the nanofibers, we used PLGA solutions containing peptide 1FL, a fluorescein-labeled derivative of peptide 1,24 to facilitate characterization by both fluorescence microscopy and solution fluorometry. Figure 1A–B shows SEM images of nanofiber mats electrospun from PLGA solutions containing peptide 1 (A) and peptide 1FL (B). These images reveal a network of smooth and randomly aligned nanofibers with median diameters of approximately 780 nm (± 410 nm) and 770 nm (± 330 nm), respectively (see also Table 1; results for PLGA nanofibers alone (no QSI) are shown for comparison; additional characterization of nanofiber distributions is shown in Figure S1 of the Supporting Information). We did not observe substantial non-uniformities or the presence of beaded fiber morphologies43,46,58 in PLGA/QSI fibers fabricated under the conditions used in this study. Figure 1D shows a representative fluorescence microscopy image of a fiber mat fabricated using a PLGA solution containing peptide 1FL. The green fluorescent network of filamentous structures in this image is consistent with the encapsulation of this fluorescently-labeled QSI in the electrospun fibers (Figure 1C shows an image of fibers electrospun using polymer solutions containing non-fluorescent peptide 1, and is dark by comparison).
Figure 1.
(A,B) Representative SEM images of electrospun PLGA nanofibers loaded with peptide 1 (A) or peptide 1FL (B). (C,D) Representative fluorescence microscopy images of electrospun PLGA nanofibers loaded with non-fluorescent peptide 1 (C) or fluorescently-labeled peptide 1FL (D).
Table 1.
Measurements of Fiber Diameters
| Material | Average (nm) |
Median (nm) |
St. Dev (nm) |
|---|---|---|---|
| PLGA | 930 | 830 | 380 |
| PLGA+Peptide 1 | 860 | 780 | 410 |
| PLGA+Peptide 1FL | 830 | 770 | 330 |
Characterization of QSI Release Profiles
We characterized the release of peptide 1FL from the PLGA nanofiber mats described above by incubating small swatches of fiber-coated mesh substrates in PBS at 37 °C. Figure 2 reveals peptide release to occur over a period of approximately three weeks (∼210 pmol/cm2 of peptide 1FL was released over the first 24 h, with ∼350 pmol/cm2 released over the course of the first 24 days). This period of release was followed by a plateau, after which very little of the peptide (∼0.56 pmol/cm2 per day) was released. This general behavior is consistent with the release profiles of other small molecules from PLGA-based polymer/drug matrices,57 including similar PLGA electrospun nanofiber-based materials.61,62
Figure 2.
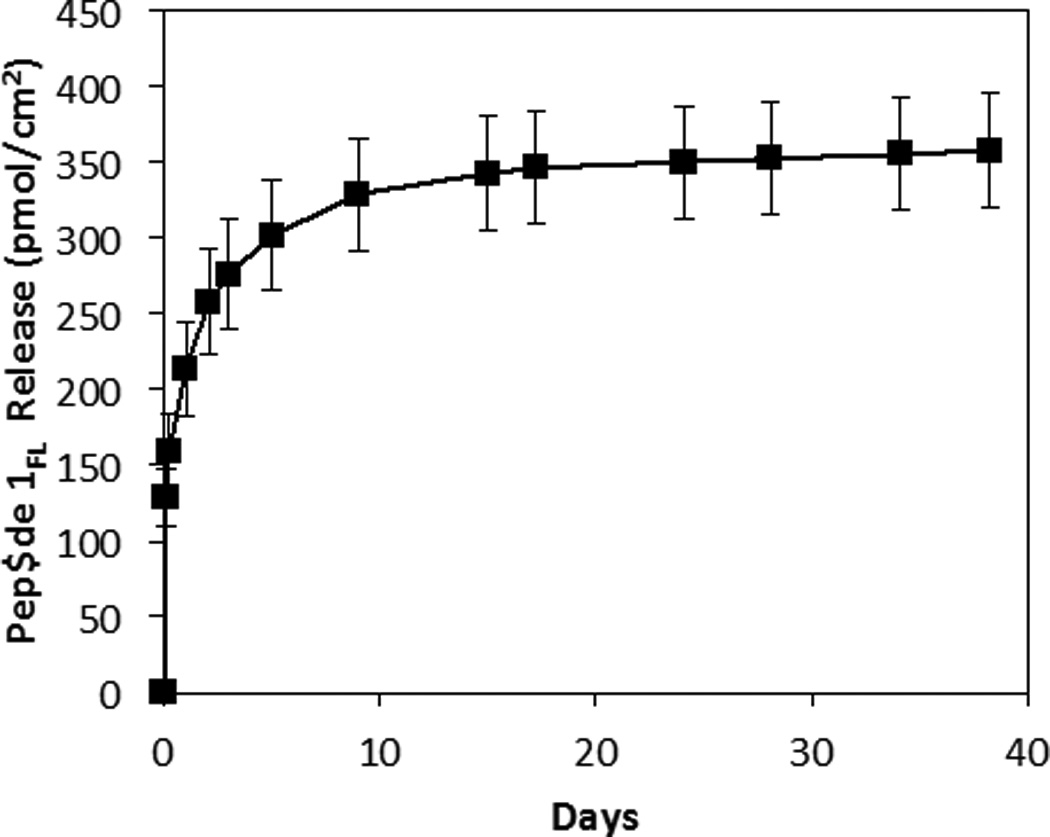
Plot showing the release of peptide 1FL from PLGA nanofiber mats versus time upon incubation in PBS at 37 °C. Each point represents the mean values obtained from four different replicates with error bars representing standard error. Total release is normalized to the surface area of the fiber-coated substrates used in these experiments.
Upon more extended incubation, we visually observed segments of the nanofibers to detach from the mats (we note that at least some portion of this mass loss was likely promoted by repeated stresses associated with the physical manipulation and transfer of these fiber-coated substrates during experiments designed to characterize release profiles). In view of these observations, we restricted proof-of-concept characterization of the biological activities of QSI-loaded coatings in the studies described below to periods of no more than 14 days, over which no potentially confounding mass loss was observed. Notably, this shorter time frame is relevant in the context of infection control, and the amounts of peptide released during this 14-day period (Figure 2) were substantially higher than those needed to inhibit QS in S. aureus using peptide 1 (IC50 of 0.485 nM in group I S. aureus;15 as described below).
QSI-Loaded Nanofiber Mats Inhibit QS in S. aureus
One potential limitation of peptide 1 in the context of integration into materials-based strategies for extended release in aqueous environments is that this QSI contains a water-labile thioester bond that maintains the peptide in a macrocyclic conformation that is critical to its biological activity.16,37,63 Hydrolysis of this thioester bond renders peptide 1 completely inactive as a QSI and occurs with a half-life of ∼72 h at neutral pH64 (and with a half-life of ∼4 h in alkaline media64). In past studies, we demonstrated that embedding this peptide in extremely non-wetting ‘superhydrophobic’ coatings that limit contact between the peptide and bulk water can preserve the biological activities of peptide 1 (e.g., the ability to inhibit QS and regulate biofilm formation in S. aureus) for at least 40 days.24 Past studies establish, however, that the transport of bulk water into PLGA matrices occurs rapidly.57,65 Thus, although the total amount of fluorescent peptide released in the above experiments was high relative to the IC50 of peptide 1, it was not clear at the outset of these studies (because no distinction between intact and hydrolyzed peptide is made in the fluorescence detection method) that this PLGA-based approach could be used to sustain the release of biologically-active peptide in quantities sufficient to inhibit QS.
To determine whether peptide was released from QSI-loaded PLGA fiber mats in a bioactive form following electrospinning and incubation in aqueous environments, we conducted a series of experiments in which aliquots of released peptide were collected at pre-determined time points and incubated with cultures of group-I S. aureus. In initial experiments, we used a reporter strain of S. aureus that produces GFP when QS is activated,52 thus permitting quantification of QSI-induced reductions in QS by fluorometry. These and all other biological studies reported here were performed using PLGA nanofiber mats loaded with peptide 1; fibers loaded with peptide 1FL were not used in these experiments because this fluorescently-labeled derivative has a lower activity than peptide 1 and the presence of the fluorescein tag would complicate the characterization and quantification of GFP production. We used an experimental design in which strips of nanofiber-coated substrates were incubated initially in PBS for a defined period, and then incubated in assay media during the 24 h immediately prior to characterization of biological activity (see Materials and Methods for additional details related to these biological assays). This overall experimental design permitted characterization of the biological activity of only that peptide released during the most recent preceding 24-hour period.
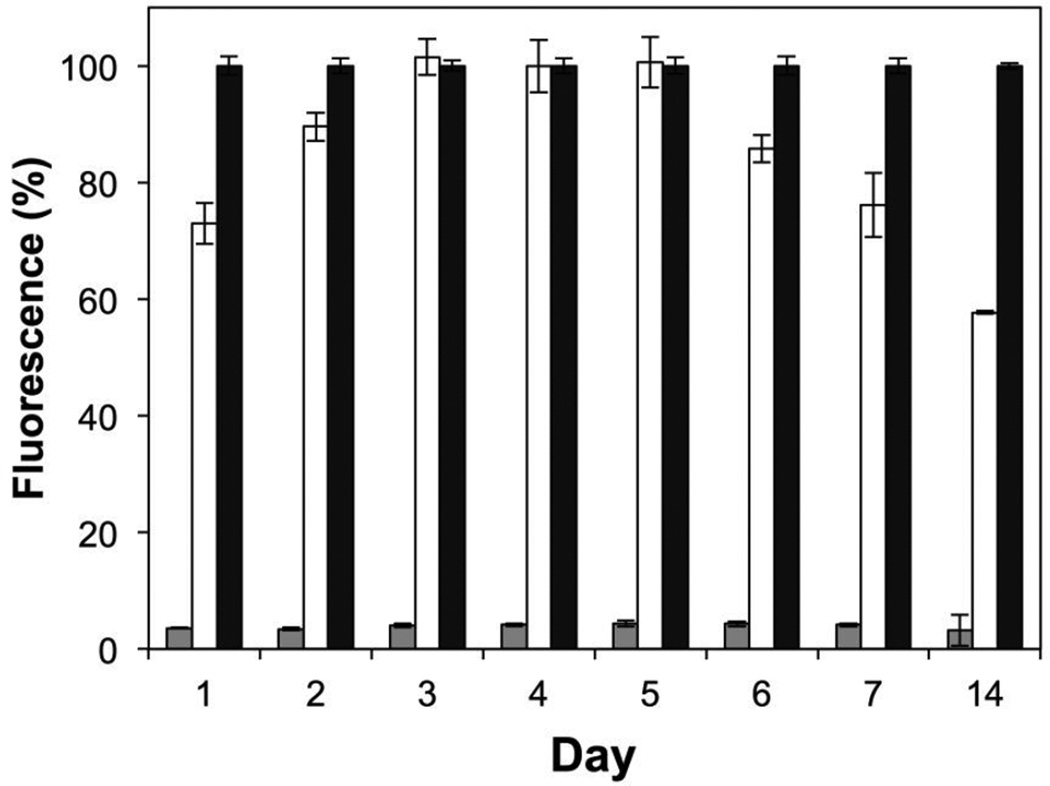
Figure 3 shows a plot of percent fluorescence, relative to results obtained using media alone (black bars), in cultures of S. aureus incubated with aliquots collected from nanofiber-coated substrates for up to 14 days of incubation. Inspection of these results reveals samples obtained from peptide 1-loaded mats (gray bars) to inhibit the production of GFP by >95% at all time points [in contrast, levels of GFP production were >70% for at least 7 days in experiments conducted using otherwise identical PLGA-only nanofiber mat control that were free of peptide 1 (white bars)]. Further, bacterial cultures incubated with samples obtained from the incubation of peptide 1-loaded mats exhibited no decrease in optical density (OD600) over these time periods (see Figure S2 of the Supporting Information), suggesting that, as expected, neither the peptidic QSI nor the PLGA have biocidal effects. These results, when combined, demonstrate that this degradable nanofiber approach can be used to sustain the release of peptide 1 in a form that remains biologically active, and at concentrations that are able to strongly inhibit QS in a model S. aureus strain, after incubation in bulk aqueous media for at least 14 days. These results also hint that the hydrolysis of the thioester bond in peptide 1, which, as noted above, is critical to its biological activity, may occur more slowly upon exposure to water than it does when dissolved in pH neutral media. Although additional analyses will be required to understand processes that could lead to the inactivation of peptide 1 upon incorporation or long-term encapsulation in these materials, it is possible that the preservation of biological activity observed here could be aided, at least in part, by the presence of an acidic microclimate in the PLGA nanofiber matrix (which has been demonstrated in past studies to stabilize or prolong the activities of other hydrolytically-labile agents under certain conditions).66–68
Figure 3.
Plot showing the fluorescence of a GFP reporter strain of S. aureus, normalized to positive media controls, versus time for the incubation of substrates coated with nanofiber mats (see main text for additional details). Gray bars show the average normalized fluorescence for peptide 1-loaded PLGA nanofiber mats; white bars show average normalized fluorescence for control PLGA nanofiber mats (no peptide); black bars show average normalized fluorescence for positive media controls. All experiments were performed in three sets of four replicates; error bars represent standard error. All results obtained for peptide-loaded nanofiber mats were significant at the 95% confidence interval (p < 0.05) versus control PLGA mats and positive media controls.
QSI-Loaded Nanofiber Mats Inhibit QS-Controlled Hemolysis in Solution and at Surfaces
Further studies demonstrated that, in addition to being able to inhibit the artificial QS reporter system described above, these QSI-loaded nanofiber mats are also capable of modulating QS phenotypes relevant to S. aureus virulence in wild-type cells. As noted above, inhibition of AgrC receptor activity has been demonstrated to inhibit the production of many toxins that play important roles in infection. One particularly salient AgrC-controlled phenotype of S. aureus is the production of hemolysins (e.g., alpha-toxin), which promote extensive lysis of mammalian host red blood cells.69–71 Hemolysin production can be readily monitored using straightforward and robust assays in the laboratory, and we have shown that peptide 1 can strongly inhibit hemolysin production in prior studies.15 We thus chose to characterize the ability of our peptide 1-loaded nanofiber mats to prevent or reduce hemolysis of red blood cells as an initial proof-of-concept experiment to demonstrate the ability of these materials to block an AgrC-controlled QS phenotype.
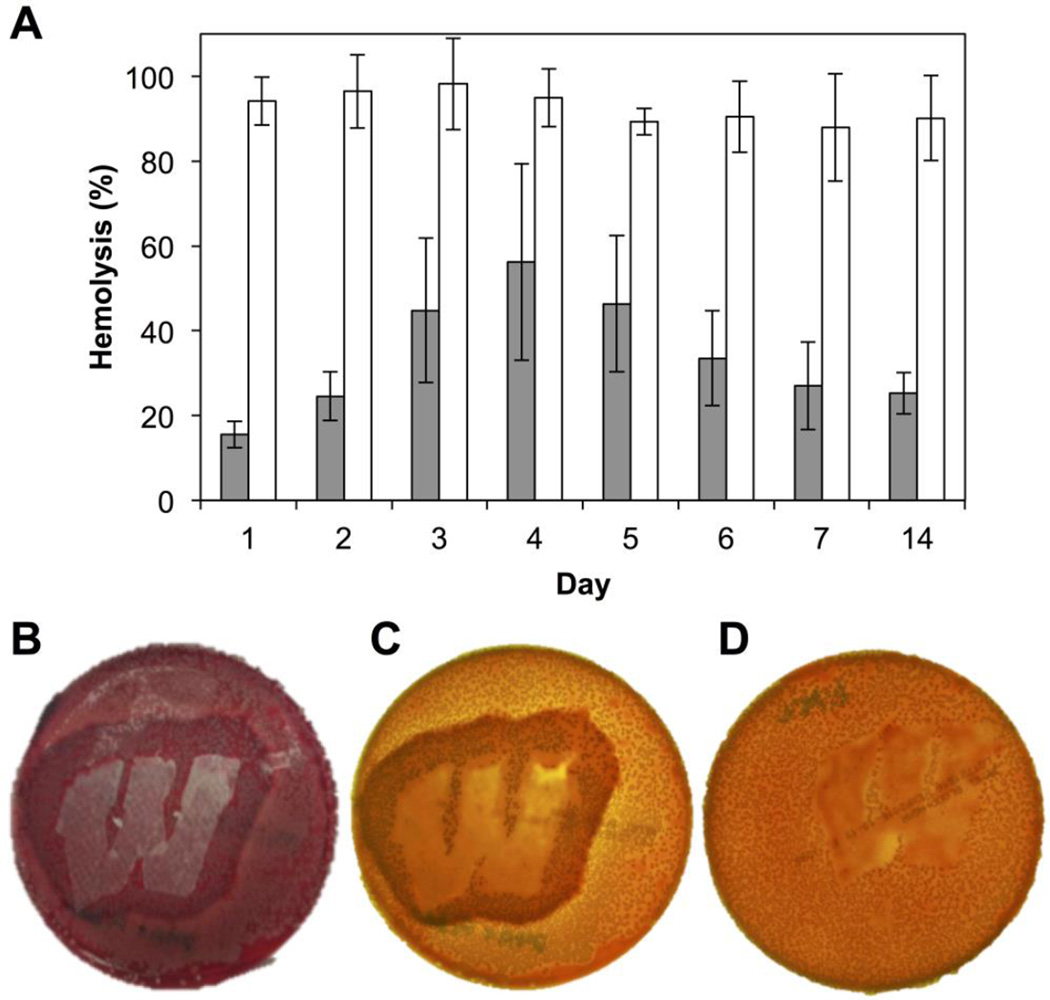
We first investigated the potential of our peptide 1-loaded mats to inhibit hemolysis using a liquid culture assay. Using an experimental design similar to that used in the GFP-reporter assays described above, we incubated wild-type S. aureus in media obtained during peptide 1 release experiments for 6 hr before using those cells in quantitative solution-based red blood cell hemolysis assays. As shown in Figure 4A, hemolysis was inhibited, relative to media controls, by an average of 65% over a period of 14 days (gray bars). In comparison, PLGA-only (no peptide) control samples promoted an average inhibition of ∼7% under otherwise identical conditions (white bars). The differences in the levels of inhibition exhibited by the peptide 1- loaded mats relative to these PLGA controls was statistically significant (p < 0.05; 95% confidence interval) at all time points. It is likely that these levels of inhibition could be improved further by manipulation of peptide loading levels or through changes to polymer structure or electrospinning parameters to tune release profiles; further optimization of this solution-based analytical assay was not pursued as part of the proof-of-concept studies reported here. In addition, we note that, as observed during the GFP assays above, bacterial densities (OD600 values) did not change significantly during these experiments (see Figure S3 of the Supporting Information). On the basis of these results, we again conclude that the decreases in hemolytic activity observed in Figure 4A are the result of the inhibition of agr-type QS by the released peptide, and not a result of cell death.
Figure 4.
Inhibition of hemolysis by group-I S. aureus promoted by QSI released from substrates coated with electrospun PLGA nanofiber mats. (A) Inhibition of hemolysis by S. aureus in liquid-culture assays over 14 days. Gray bars: peptide 1-loaded mats; white bars: control PLGA nanofiber mats (no peptide). Data are presented as averages of three biological replicates; error bars represent standard error. All results were significant at the 95% confidence interval (p < 0.05). (B–D) Representative images of hemolysis inhibition as observed on a solid blood agar plate inoculated with S. aureus. (B) Zone of hemolysis inhibition (dark red) observed around a non-woven substrate coated with a peptide 1-loaded mat; photographed from above under ambient lighting; the lighter red background indicates active hemolysis by S. aureus colonies in areas not adjacent to the substrate. (C) The same plate as in (B), but photographed when backlit to enhance contrast (see text); the yellow-orange background indicates active hemolysis by S. aureus colonies in areas not adjacent to the substrate. (D) No inhibition of hemolysis was observed in experiments using an otherwise identical uncoated mesh substrate; the plate was also photographed when backlit for clarity.
To supplement the liquid culture hemolysis assay, we also characterized the ability of our QSI-loaded nanofiber coatings to inhibit hemolysis by wild-type S. aureus when placed in direct contact with each other on a surface, thus better mimicking interactions that would occur between bacteria and a nanofiber coating at an infection site. To test for this function, we characterized the ability of peptide 1-loaded mats to inhibit hemolysis in S. aureus grown on blood agar plates. These experiments were performed by manually placing samples of nanofiber-coated mesh substrates on top of a blood agar plate inoculated with S. aureus and incubating these two contacting surfaces for 24 h (see Materials and Methods for additional details). Over this time frame, S. aureus grows to cover the entire plate surface, reaches a (presumably) quorate population, and produces hemolysins to promote a change in the color of the underlying blood agar plate from dark red (a color indicative of intact red blood cells) to light red/orange (a color indicative of lysed red blood cells).72 This change in color can thus be used to visualize and characterize qualitative changes in the production of hemolysins by S. aureus growing on a solid surface.72
Figure 4B–D shows representative results of this assay 24 h after the incubation of inoculated blood agar plates with non-woven mesh substrates coated with peptide 1-loaded nanofibers (B–C) or control PLGA nanofibers electrospun in the absence of peptide 1 (D). The image in panel B shows a plate imaged under normal light prior to removal of the coated substrate (approximately 2.5 × 3.8 cm, shown in white); the images in panels C and D show plates imaged using backlighting and after the removal of the coated substrate for clarity. As anticipated, we observed a zone of inhibition of hemolysis surrounding substrates coated with peptide 1-loaded nanofibers (darker red color in Figure 4B and darker orange color in Figure 4C, corresponding to regions with intact red blood cells; the apparent differences in color in these images arise from differences in the lighting used to acquire these two images, as noted above). Beyond this zone of inhibition, the remainder of the plate was lighter in color (e.g., lighter red in Figure 4B and light orange/yellow color in Figure 4C), consistent with extensive hemolysis of the red blood cells in areas remote from the coated substrates.72 The spatial extent of the zone of inhibition observed in these studies reflects, in part, the rate and extent of the diffusion of peptide 1 within these agar plates after it is released from the nanofibers. In contrast, when similar experiments were performed using substrates coated with PLGA-only (no peptide) nanofiber controls, the entire plate appeared light orange/yellow (Figure 4D), indicative of uniform red blood cell hemolysis in all areas surrounding the substrate. Bacterial colonies were visible on the plates, both within and around the zones of inhibition, suggesting no decreases in cell viability as a result of presence of peptide 1 (bacterial colonies were not observed in the areas directly beneath the film-coated substrates in either the peptide-loaded or the PLGA control cases under the conditions used in these assays; bacterial colonies were, however, observed to be attached to the plate-facing sides of the removed substrates, suggesting preferential growth on the nanofiber mats in those regions). When combined, these results demonstrate that peptide 1-loaded PLGA nanofibers can be used to inhibit the production of a critical, QS-controlled virulence factor in wild-type S. aureus, both in liquid-culture and when placed directly in contact with bacteria growing on a soft surface. Because AgrC controls the production of numerous other virulence factors and biofilm growth in S. aureus, we anticipate that these peptide 1-loaded coatings should also modulate the production of these other phenotypes, thereby expanding the potential future utility of these materials beyond the proof-of-concept results reported here.
Summary and Conclusions
We have reported the fabrication, characterization, and biological evaluation of degradable polymer nanofiber mats that release potent peptide-based inhibitors of bacterial QS and promote the localized, surface-mediated inhibition of a salient, QS-controlled virulence phenotype in the pervasive human pathogen S. aureus. This study is, to our knowledge, the first to evaluate the use of electrospun polymer nanofiber mats for the distribution and dissemination of small-molecule or peptidic QSIs, and has implications for the development of new anti-infective materials, coatings, and treatment strategies that target bacterial virulence rather than bacterial growth. Our results demonstrate that peptide 1, a synthetic macrocyclic QSI that is active against S. aureus, can be readily loaded into PLGA nanofiber mats using conventional polymer electrospinning techniques, and that this approach can be used to readily deposit nanofiber-based coatings on the surfaces of model non-woven mesh substrates. Peptide 1 was released over a period of approximately three weeks when nanofiber mats were incubated in physiologically relevant media. Liquid-culture cell-based reporter assays revealed released peptide to retain its biological activity and strongly inhibit agr-based QS in an S. aureus strain containing a GFP reporter for periods of at least 14 days. We also demonstrated that these QSI-loaded nanofiber mats can inhibit the production of hemolysins, an important QS-controlled virulence phenotype in S. aureus.73 Our results reveal that the QSIs released from these materials not only reduce levels of hemolysis in red blood cells in liquid cultures of wild-type S. aureus, but also strongly block hemolysis when placed in direct contact with wild-type S. aureus growing on the surface of a blood agar plate, an experimental geometry designed to more closely mimic interactions that would occur between bacteria and a nanofiber mat at an infection site. In view of the central role of the agr-QS system in the production of virulence factors and biofilm formation in S. aureus, it is likely that these materials could also modulate other associated virulence phenotypes pertinent in both acute and chronic infections.
To close, S. aureus is a causative agent of life-threatening infections in burn wounds, diabetic ulcers, and other skin lesions, and strains of this pathogen have developed substantial resistance to conventional antibiotic treatments (e.g., MRSA).74 Our findings, and this general non-bactericidal approach to the design of anti-infective materials, are thus timely and provide a useful springboard for the continued development of new QSI-based strategies to assist in the prevention or treatment of infectious diseases and test hypotheses and dissect relationships between the agr system, toxin production and biofilm formation, and infection in S. aureus. In this specific context, we note that the electrospinning approach reported here is modular, and can be used with many different types of polymers, active agents (or combinations of agents), and processing parameters to deposit nanofiber coatings with tunable physical or chemical properties on a broad range of topologically complex objects important in healthcare-related applications (including indwelling interventional devices, implants, wound dressings, and even the surfaces of open wounds).46,49–51 For example, it should be straightforward using this approach to fabricate non-woven mats containing combinations of QSIs and one or more conventional antibiotic agents to identify potential synergies and design new generations of materials that combat infection more effectively. More broadly, the overall approaches and materials reported here could also provide novel porous scaffolds for bacterial cell culture useful for fundamental studies of the influence of agr-type QS modulation (either inhibition or activation) on the development of bacterial communities.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Office of Naval Research (N00014-16-1-2185), the NSF (through a grant to the UW-Madison Materials Research Science and Engineering Center; MRSEC; DMR-1121288), and the Burroughs Wellcome Fund, and made use of NSF-supported facilities (DMR-1121288). M. J. K. acknowledges the UW-Madison Biotechnology Center for a Morgridge Biotechnology Fellowship. T. Y. acknowledges support from the UW-Madison NIH Biotechnology Training Program (T32 GM08349). We thank Prof. Tom Turng for providing access to electrospinning instrumentation, and acknowledge support for those facilities provided by the Wisconsin Institutes of Discovery and the Vice Chancellor for Research and Graduate Education at UW-Madison. We also thank Dr. Matthew C. D. Carter for assistance with the acquisition of SEM images, and Eric Codner for assistance with the laser cutting of substrates used in hemolysis blood agar assays.
Footnotes
Supporting Information. Results of additional characterization of peptide-loaded nanofibers and biological assays. This material is available free of charge via the Internet at: DOI:
References
- 1.Percival SL, Suleman L, Vuotto C, Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Wu P, Grainger DW. Drug/device combinations for local drug therapies and infection prophylaxis. Biomaterials. 2006;27:2450–2467. doi: 10.1016/j.biomaterials.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 4.Hetrick EM, Schoenfisch MH. Reducing implant-related infections: active release strategies. Chem. Soc. Rev. 2006;35:780–789. doi: 10.1039/b515219b. [DOI] [PubMed] [Google Scholar]
- 5.Zilberman M, Elsner JJ. Antibiotic-eluting medical devices for various applications. J. Control. Release. 2008;130:202–215. doi: 10.1016/j.jconrel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Wright GD. The origins of antibiotic resistance. Handbook of experimental pharmacology. 2012:13–30. doi: 10.1007/978-3-642-28951-4_2. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT, Wright GD. Antimicrobials. Curr. Opin. Microbiol. 2009;12:473–475. doi: 10.1016/j.mib.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P. T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 9.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Disc. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 10.Curtis MM, Russell R, Moreira CG, Adebesin AM, Wang C, Williams NS, Taussig R, Stewart D, Zimmern P, Lu B, Prasad RN, Zhu C, Rasko DA, Huntley JF, Falck JR, Sperandio V. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio. 2014;5:e02165. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor perspectives in medicine. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njoroge J, Sperandio V. Jamming bacterial communication: New approaches for the treatment of infectious diseases. EMBO Mol. Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 15.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed Through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J. Am. Chem. Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Muir TW. Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles. Cell Chem. Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sully EK, Malachowa N, Elmore BO, Alexander SM, Femling JK, Gray BM, DeLeo FR, Otto M, Cheung AL, Edwards BS, Sklar LA, Horswill AR, Hall PR, Gresham HD. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLOS Pathog. 2014;10:e1004174. doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welsh MA, Eibergen NR, Moore JD, Blackwell HE. Small Molecule Disruption of Quorum Sensing Cross-Regulation in Pseudomonas aeruginosa Causes Major and Unexpected Alterations to Virulence Phenotypes. J. Am. Chem. Soc. 2015;137:1510–1519. doi: 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stacy DM, Welsh MA, Rather PN, Blackwell HE. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem. Biol. 2012;7:1719–1728. doi: 10.1021/cb300351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitbach AS, Broderick AH, Jewell CM, Gunasekaran S, Lin Q, Lynn DM, Blackwell HE. Surface-mediated release of a synthetic small-molecule modulator of bacterial quorum sensing: gradual release enhances activity. Chem. Commun. 2011;47:370–372. doi: 10.1039/c0cc02316g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick AH, Breitbach AS, Frei R, Blackwell HE, Lynn DM. Surface-mediated release of a small-molecule modulator of bacterial biofilm formation: a non-bactericidal approach to inhibiting biofilm formation in Pseudomonas aeruginosa . Adv. Healthcare Mater. 2013;2:993–1000. doi: 10.1002/adhm.201200334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broderick AH, Stacy DM, Tal-Gan Y, Kratochvil MJ, Blackwell HE, Lynn DM. Surface Coatings that Promote Rapid Release of Peptide-Based AgrC Inhibitors for Attenuation of Quorum Sensing in Staphylococcus aureus . Adv. Healthcare Mater. 2014;3:97–105. doi: 10.1002/adhm.201300119. [DOI] [PubMed] [Google Scholar]
- 24.Kratochvil MJ, Tal-Gan Y, Yang T, Blackwell HE, Lynn DM. Nanoporous Superhydrophobic Coatings that Promote the Extended Release of Water-Labile Quorum Sensing Inhibitors and Enable Long Term Modulation of Quorum Sensing in Staphylococcus aureus . ACS Biomat. Sci. Eng. 2015;1:1039–1049. doi: 10.1021/acsbiomaterials.5b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kratochvil MJ, Welsh MA, Manna U, Ortiz BJ, Blackwell HE, Lynn DM. Slippery Liquid-Infused Porous Surfaces that Prevent Bacterial Surface Fouling and Inhibit Virulence Phenotypes in Surrounding Planktonic Cells. ACS Infect. Dis. 2016;2:509–517. doi: 10.1021/acsinfecdis.6b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hume EBH, Baveja J, Muir BW, Schubert TL, Kumar N, Kjelleberg S, Griesser HJ, Thissen H, Read R, Poole-Warren LA, Schindhelm K, Willcox MDP. The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones. Biomaterials. 2004;25:5023–5030. doi: 10.1016/j.biomaterials.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 27.Baveja JK, Wilcox MDP, Hume EBH, Kumar N, Odell R, Poole-Warren LA. Furanones as potential anti-bacterial coatings on biomaterials. Biomaterials. 2004;25:5003–5012. doi: 10.1016/j.biomaterials.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 28.Melander C, Moeller PDR, Ballard TE, Richards JJ, Huigens RW, III, Cavanagh J. Evaluation of dihydrooroidin as an antifouling additive in marine paint. Int. Biodeterior. Biodegradation. 2009;63:529–532. doi: 10.1016/j.ibiod.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho KKK, Cole N, Chen R, Willcox MDP, Rice SA, Kumar N. Characterisation and in vitro activities of surface attached dihydropyrrol-2-ones against Gram-negative and Gram-positive bacteria. Biofouling. 2010;26:913–921. doi: 10.1080/08927014.2010.531463. [DOI] [PubMed] [Google Scholar]
- 30.Ho KKK, Chen R, Willcox MDP, Rice SA, Cole N, Iskander G, Kumar N. Quorum sensing inhibitory activities of surface immobilized antibacterial dihydropyrrolones via click chemistry. Biomaterials. 2014;35:2336–2345. doi: 10.1016/j.biomaterials.2013.11.072. [DOI] [PubMed] [Google Scholar]
- 31.Nowatzki PJ, Koepsel RR, Stoodley P, Min K, Harper A, Murata H, Donfack J, Hortelano ER, Ehrlich GD, Russell AJ. Salicylic acid-releasing polyurethane acrylate polymers as anti-biofilm urological catheter coatings. Acta Biomater. 2012;8:1869–1880. doi: 10.1016/j.actbio.2012.01.032. [DOI] [PubMed] [Google Scholar]
- 32.Gomes J, Grunau A, Lawrence AK, Eberl L, Gademann K. Bioinspired, releasable quorum sensing modulators. Chem. Commun. 2013;49:155–157. doi: 10.1039/c2cc37287h. [DOI] [PubMed] [Google Scholar]
- 33.Nafee N, Husari A, Maurer CK, Lu C, de Rossi C, Steinbach A, Hartmann RW, Lehr C-M, Schneider M. Antibiotic-free nanotherapeutics: Ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J. Control. Release. 2014;192:131–140. doi: 10.1016/j.jconrel.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 34.Shenderovich J, Feldman M, Kirmayer D, Al-Quntar A, Steinberg D, Lavy E, Friedman M. Local sustained-release delivery systems of the antibiofilm agent thiazolidinedione-8 for prevention of catheter-associated urinary tract infections. Int. J. Pharm. 2015;485:164–170. doi: 10.1016/j.ijpharm.2015.02.067. [DOI] [PubMed] [Google Scholar]
- 35.Lu HD, Spiegel AC, Hurley A, Perez LJ, Maisel K, Ensign LM, Hanes J, Bassler BL, Semmelhack MF, Prud’homme RK. Modulating Vibrio cholerae Quorum-Sensing-Controlled Communication Using Autoinducer-Loaded Nanoparticles. Nano Lett. 2015;15:2235–2241. doi: 10.1021/acs.nanolett.5b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YC, Quan XC, Si XR. Incorporation of brominated furanone into Nafion polymer enhanced anti-biofilm efficacy. Int. Biodeterior. Biodegradation. 2015;99:39–44. [Google Scholar]
- 37.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem. Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le KY, Otto M. Quorum-sensing regulation in staphylococci-an overview. Frontiers in Microbiology. 2015;6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang BY, Muir TW. Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles. Cell Chem. Biol. 2016;23:214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang ZM, Zhang YZ, Kotaki M, Ramakrishna S. A review on polymer nanofibers by electrospinning and their applications in nanocomposites. Compos. Sci. Technol. 2003;63:2223–2253. [Google Scholar]
- 43.Li D, Xia YN. Electrospinning of nanofibers: Reinventing the wheel? Adv. Mater. 2004;16:1151–1170. [Google Scholar]
- 44.Greiner A, Wendorff JH. Electrospinning: A fascinating method for the preparation of ultrathin fibres. Angew. Chem. Int. Ed. 2007;46:5670–5703. doi: 10.1002/anie.200604646. [DOI] [PubMed] [Google Scholar]
- 45.Zamani M, Prabhakaran MP, Ramakrishna S. Advances in drug delivery via electrospun and electrosprayed nanomaterials. Int. J. Nanomedicine. 2013;8:2997–3017. doi: 10.2147/IJN.S43575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sill TJ, von Recum HA. Electrospinning: Applications in drug delivery and tissue engineering. Biomaterials. 2008;29:1989–2006. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Chou SF, Carson D, Woodrow KA. Current strategies for sustaining drug release from electrospun nanofibers. J. Control. Release. 2015;220:584–591. doi: 10.1016/j.jconrel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Electrospun nanofibrous structure: A novel scaffold for tissue engineering. J. Biomed. Mater. Res. 2002;60:613–621. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 49.Dong RH, Jia YX, Qin CC, Zhan L, Yan X, Cui L, Zhou Y, Jiang XY, Long YZ. In situ deposition of a personalized nanofibrous dressing via a handy electrospinning device for skin wound care. Nanoscale. 2016;8:3482–3488. doi: 10.1039/c5nr08367b. [DOI] [PubMed] [Google Scholar]
- 50.Mele E. Electrospinning of natural polymers for advanced wound care: towards responsive and adaptive dressings. J. Mater. Chem. B. 2016;4:4801–4812. doi: 10.1039/c6tb00804f. [DOI] [PubMed] [Google Scholar]
- 51.Zahedi P, Rezaeian I, Ranaei-Siadat SO, Jafari SH, Supaphol P. A review on wound dressings with an emphasis on electrospun nanofibrous polymeric bandages. Polymer. Adv. Tech. 2010;21:77–95. [Google Scholar]
- 52.Kirchdoerfer RN, Garner AL, Flack CE, Mee JM, Horswill AR, Janda KD, Kaufmann GF, Wilson IA. Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J. Biol. Chem. 2011;286:17351–17358. doi: 10.1074/jbc.M111.231258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson JM, Shive MS. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv. Drug Deliv. Rev. 1997;28:5–24. doi: 10.1016/s0169-409x(97)00048-3. [DOI] [PubMed] [Google Scholar]
- 54.Ikada Y, Tsuji H. Biodegradable polyesters for medical and ecological applications. Macromol. Rapid Commun. 2000;21:117–132. [Google Scholar]
- 55.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv. Drug Deliv. Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 56.Makadia HK, Siegel SJ. Poly Lactic-co-Glycolic Acid (PLGA) as Biodegradable Controlled Drug Delivery Carrier. Polymers. 2011;3:1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fredenberg S, Wahlgren M, Reslow M, Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems-A review. Int. J. Pharm. 2011;415:34–52. doi: 10.1016/j.ijpharm.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 58.Katti DS, Robinson KW, Ko FK, Laurencin CT. Bioresorbable nanofiber-based systems for wound healing and drug delivery: Optimization of fabrication parameters. J. Biomed. Mater. Res. Part B Appl. Biomater. 2004;70B:286–296. doi: 10.1002/jbm.b.30041. [DOI] [PubMed] [Google Scholar]
- 59.Kim K, Luu YK, Chang C, Fang DF, Hsiao BS, Chu B, Hadjiargyrou M. Incorporation and controlled release of a hydrophilic antibiotic using poly(lactide-co-glycolide)-based electrospun nanofibrous scaffolds. J. Control. Release. 2004;98:47–56. doi: 10.1016/j.jconrel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Luu YK, Kim K, Hsiao BS, Chu B, Hadjiargyrou M. Development of a nanostructured DNA delivery scaffold via electrospinning of PLGA and PLA-PEG block copolymers. J. Control. Release. 2003;89:341–353. doi: 10.1016/s0168-3659(03)00097-x. [DOI] [PubMed] [Google Scholar]
- 61.Puppi D, Piras AM, Detta N, Dinucci D, Chiellini F. Poly(lactic-co-glycolic acid) electrospun fibrous meshes for the controlled release of retinoic acid. Acta Biomater. 2010;6:1258–1268. doi: 10.1016/j.actbio.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 62.Sofokleous P, Stride E, Edirisinghe M. Preparation, Characterization, and Release of Amoxicillin from Electrospun Fibrous Wound Dressing Patches. Pharm. Res. 2013;30:1926–1938. doi: 10.1007/s11095-013-1035-2. [DOI] [PubMed] [Google Scholar]
- 63.Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. Structural Characterization of Native Autoinducing Peptides and Abiotic Analogs Reveals Key Features Essential for Activation and Inhibition of an AgrC Quorum Sensing Receptor in Staphylococcus aureus . J. Am. Chem. Soc. 2013;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Highly Stable, Amide-Bridged Autoinducing Peptide Analogues that Strongly Inhibit the AgrC Quorum Sensing Receptor in Staphylococcus aureus . Angew. Chem. Int. Ed. 2016;55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batycky RP, Hanes J, Langer R, Edwards DA. A theoretical model of erosion and macromolecular drug release from biodegrading microspheres. J. Pharm. Sci. 1997;86:1464–1477. doi: 10.1021/js9604117. [DOI] [PubMed] [Google Scholar]
- 66.Zhu GZ, Mallery SR, Schwendeman SP. Stabilization of proteins encapsulated in injectable poly (lactide-co-glycolide) Nat. Biotechnol. 2000;18:52–57. doi: 10.1038/71916. [DOI] [PubMed] [Google Scholar]
- 67.Fu K, Pack DW, Klibanov AM, Langer R. Visual evidence of acidic environment within degrading poly(lactic-co-glycolic acid) (PLGA) microspheres. Pharm. Res. 2000;17:100–106. doi: 10.1023/a:1007582911958. [DOI] [PubMed] [Google Scholar]
- 68.Shenderova A, Burke TG, Schwendeman SP. The acidic microclimate in poly(lactide-co-glycolide) microspheres stabilizes camptothecins. Pharm. Res. 1999;16:241–248. doi: 10.1023/a:1018876308346. [DOI] [PubMed] [Google Scholar]
- 69.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 70.Pragman AA, Schlievert PM. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 2004;42:147–154. doi: 10.1016/j.femsim.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 71.George EA, Muir TW. Molecular mechanisms of agr quorum sensing in virulent staphylococci . Chembiochem. 2007;8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 72.Burnside K, Lembo A, de los Reyes M, Iliuk A, BinhTran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, Tao WA, Rajagopal L. Regulation of Hemolysin Expression and Virulence of Staphylococcus aureus by a Serine/Threonine Kinase and Phosphatase. PLOS One. 2010;5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bantel H, Sinha B, Domschke W, Peters G, Schulze-Osthoff K, Janicke RU. alpha-toxin is a mediator of Staphylococcus aureus-induced cell death and activates caspases via the intrinsic death pathway independently of death receptor signaling. J. Cell Biol. 2001;155:637–647. doi: 10.1083/jcb.200105081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.