Abstract
Protein–protein interactions govern biological functions in cells, in the extracellular milieu, and at the border between cells and extracellular space. Viruses are small intracellular parasites and thus rely on protein interactions to produce progeny inside host cells and to spread from cell to cell. Usage of host proteins by viruses can have severe consequences e.g. apoptosis, metabolic disequilibria, or altered cell proliferation and mobility. Understanding protein interactions during virus infection can thus educate us on viral infection and pathogenesis mechanisms. Moreover, it has led to important clinical translations, including the development of new therapeutic and vaccination strategies. Here, we will discuss protein interactions of members of the Flaviviridae family, which are small enveloped RNA viruses. Dengue virus, Zika virus and hepatitis C virus belong to the most prominent human pathogenic Flaviviridae. With a genome of roughly ten kilobases encoding only ten viral proteins, Flaviviridae display intricate mechanisms to engage the host cell machinery for their purpose. In this review, we will highlight how dengue virus, hepatitis C virus, Japanese encephalitis virus, tick-borne encephalitis virus, West Nile virus, yellow fever virus, and Zika virus proteins engage host proteins and how this knowledge helps elucidate Flaviviridae infection. We will specifically address the protein composition of the virus particle as well as the protein interactions during virus entry, replication, particle assembly, and release from the host cell. Finally, we will give a perspective on future challenges in Flaviviridae interaction proteomics and why we believe these challenges should be met.
The Flaviviridae family of viruses comprises at least 40 human-tropic virus species. These human infectious viruses belong to the genera flavivirus (40 human pathogenic species) and hepacivirus (one human pathogenic species) and include e.g. Zika virus (ZIKV), which caused the 2015 pandemic in Brazil and the Americas (1). Dengue virus (DENV), hepatitis C virus (HCV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV), West Nile virus (WNV), and yellow fever virus (YFV) are further prominent members of the Flaviviridae family and cause millions of infections annually with symptoms ranging from febrile illness to fatal hemorrhagic, neurologic and gastrointestinal disease (Box 1) (2–7). Historically, all six flaviviruses discussed here were identified and first isolated before 1950, while it was not until 1989 that the hepacivirus HCV was discovered (8–10) (Fig. 1). Human protective vaccines exist for DENV, JEV, TBEV, and YFV (11). However, with the exception of HCV, no specific therapeutic drugs are available to treat infections with Flaviviridae species (Table 1) (12). In the case of HCV, the successful development of direct-acting antivirals has been driven by the elucidation of HCV polyprotein processing and the development of an infectious cell culture system (13–17).
Table II. Box 1. Flaviviridae species discussed in this review. The acronym for each viral species is listed in the left column and the reservoir hosts, the transmission vectors, and the pathology caused in humans is briefly described in the right column.
| DENV | DENV is an emerging pathogen that naturally infects primates and affects humans in urban (sub-) tropical regions. Upon transmission by mosquitoes (predominantly Aedes aegypti and A. albopictus), DENV targets primarily myeloid cells. Most DENV infections are asymptomatic but can lead to flu-like dengue fever and, in some cases, life-threatening severe dengue (also called dengue hemorrhagic fever or dengue shock syndrome). Severe dengue cases are often associated with secondary infections due to antibody-dependent enhancement of infection. There is no antiviral therapy against DENV available. A first vaccine has been licensed in late 2015 in some Latin American and Asian countries (3, 188). |
| HCV | Approximately 2% of the world's population is chronically infected with HCV. The virus, which is naturally only infecting humans, is transmitted parenterally. Upon transmission, HCV infects the liver, in particular hepatocytes. Acute infection is asymptomatic in most cases, but chronicity is established in about 80% of the cases, leading to liver fibrosis, cirrhosis and hepatocellular carcinoma. Thus HCV is one of the leading causes for liver transplantations worldwide. Effective antiviral treatment licensed since 2014, cures over 90% of patients treated. However, treatment costs are high and there is no vaccine available yet (189, 190). |
| JEV | JEV is maintained in an enzootic cycle between mosquitoes and pigs or birds. It is transmitted to humans through mosquitoes (mostly of the Culex type), mainly in rural areas of Southeast and East Asia. Normally, infection with the virus is mild. However, JEV is the leading cause of viral encephalitis in Asia. There is no specific therapy for Japanese encephalitis, but safe and effective vaccines are available (191). |
| TBEV | TBEV is endemic in forested areas of Central and Eastern Europe as well as in Northern Asia. TBEV infects small mammals, birds, and ticks (Ixodidae familiy), which are the natural reservoir of the virus and also predominantly transmit it. Humans are only accidental hosts; however, infection rates are increasing and tick-borne encephalitis is the most important viral tick-borne disease in Europe. Upon transmission, TBEV first replicates at the site of inoculation and the draining lymph nodes, from where it spreads to many extraneural parts of the body, which causes unspecific flu-like symptoms. Neuroinvasion occurs in approximately one-third of the patients, leading to the development of neurological diseases like encephalitis, meningitis, and meningoencephalitis. The severity of the disease and long-term consequences depend on the virus strain a patient is infected with. There is a potent vaccine available (192, 193). |
| WNV | WNV is commonly found in Africa, Europe, the Middle East, North America, and Asia. It circulates in an enzootic cycle between birds and mosquitoes but can also infect humans and other vertebrates. Transmission occurs predominantly by mosquitoes (Aedes and Culex species). In the host, WNV enters various cell types like neurons, keratinocytes, and skin-resident dendritic cell. Infections are mainly asymptomatic but can lead to the mostly self-limiting West Nile fever, which in some cases progresses to severe neurological illnesses, including meningitis and encephalitis. A specific antiviral treatment or vaccine against WNV infections is not available yet (194, 195). |
| YFV | YFV is endemic in (sub-) tropical areas of Africa and Central and South America. The only known natural hosts are primates, from which the virus spreads through mosquitoes (mainly A. aegypti), also infecting humans. In humans the virus first infects lymph nodes, in particular dendritic cells, and then the liver. Infection is mostly asymptomatic or causes mild flu-like symptoms. However, in a small percentage of patients, infection causes acute yellow fever leading to jaundice (hence the name) and hemorrhagic fever, which is lethal in half of the cases. A specific treatment for yellow fever is not available, but there is a safe and highly effective vaccine (196, 197). |
| ZIKV | ZIKV was first discovered in Africa and then spread to Asia and Latin and South America. The virus has emerged very fast in the last ten years, causing, for instance, the 2015 epidemic in Brazil. Notably, it spreads through the same mosquito species as DENV and YFV (Aedes sp.). ZIKV can also be transmitted sexually and from mother to child during pregnancy, which is linked to birth defects, in particular with microcephaly. Infections lead to relatively mild dengue-like disease but have also been associated to neurological complications as the Guillian-Barré syndrome. There is no specific treatment or vaccine available yet (198, 199). |
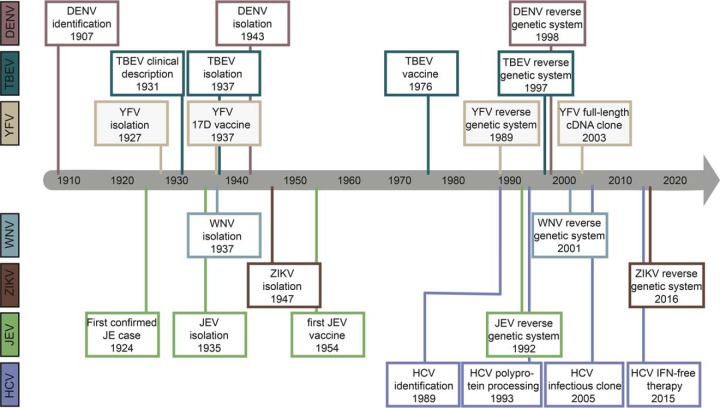
Fig. 1.
Milestones in flavivirus and hepacivirus research. The identification, development of reverse genetic systems to study the protein biochemistry and cell biology of infection as well as the first vaccines are shown for the seven human-pathogenic Flaviviridae family members discussed in this review. See text for references.
Table I. Human pathogenic flaviviruses included in the study. The date of discovery, transmission mode, and therapy and vaccination options are listed.
| Virus species | Acronym | Genus | Discovery | Transmission | Antiviral therapy | Vaccine |
|---|---|---|---|---|---|---|
| Dengue virus | DENV | Flavivirus | 1907 | Mosquitoes | - | Yes |
| Hepatitis C virus | HCV | Hepacivirus | 1989 | parenteral | Direct acting antivirals | Clinical trial |
| Japanese encephalitis virus | JEV | Flavivirus | 1935 | Mosquitoes | - | Yes |
| Tick-borne encephalitis virus | TBEV | Flavivirus | 1937 | Ticks | - | Yes |
| West Nile virus | WNV | Flavivirus | 1937 | Mosquitoes | - | Veterinary vaccine, human vaccine in clinical trail |
| Yellow fever virus | YFV | Flavivirus | 1927 | Mosquitoes | - | Yes |
| Zika virus | ZIKV | Flavivirus | 1947 | Mosquitoes | - | Clinical trial |
Flaviviridae are small enveloped positive strand RNA viruses of an approximate diameter of 50 nm, which replicate in the cytoplasm of their respective host cell. The development of reverse genetic systems (18) spurred the elucidation of the viral life cycle, including protein functions and protein–protein interactions (PPI). Flaviviruses and hepaciviruses share common mechanisms of propagation in their host cells. Initially, they attach with their surface glycoproteins to the cell surface, then they interact with one or several receptors, which trigger endocytosis. After endocytic uptake and acidification of the endosomal lumen, the viral surface glycoproteins undergo a conformational change and induce fusion of the limiting endosomal membrane and the viral envelope. Disassembly of the viral capsid (uncoating) delivers the RNA genome to the cytoplasm, which completes the entry process. The positive polarity genomes subsequently serve as translation and replication templates. Flaviviridae genomes are 10 to 11 kb in size and encode a single polyprotein. After translation at the endoplasmatic reticulum (ER), viral and host proteases cleave the polyprotein into three structural and seven nonstructural (NS) proteins. Like all Baltimore class IV viruses, flaviviruses and hepaciviruses encode their own RNA-dependent RNA polymerase, which copies the viral genome through a negative strand RNA intermediate. Replication takes place at ER-derived, specialized membrane compartments, which the virus itself induces. Viral genomes are then packaged into a capsid composed of the core (HCV) or capsid (flaviviruses) protein and bud into the ER, thereby acquiring a lipid envelope embedding multiple copies of the two structural glycoproteins. Viral particles are finally released from the cell in a nonlytic fashion through the secretory pathway (Fig. 2) (19–21). Individual life cycle steps and peculiarities of specific viruses are detailed below.
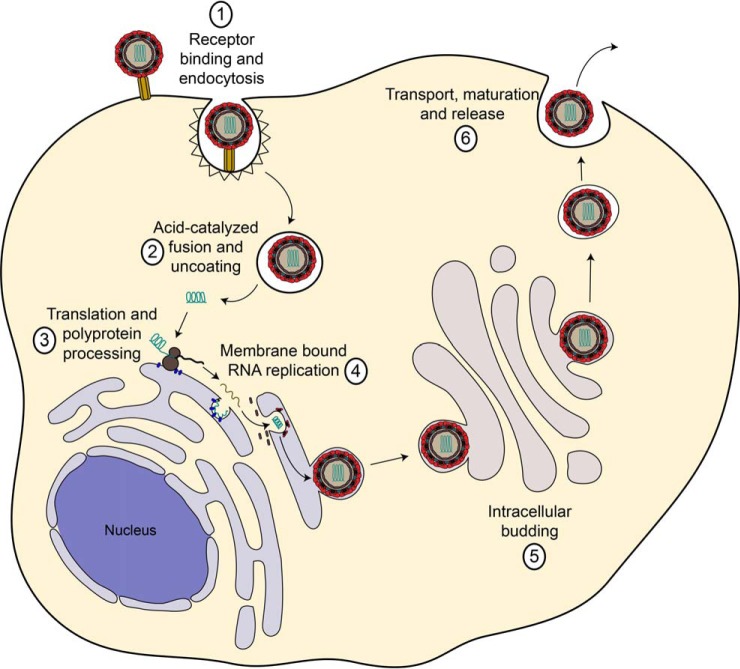
Fig. 2.
The Flaviviridae life cycle. The viruses attach to receptors on the host cell and internalize by endocytosis. In endosomes the viral envelope fuses with the host membrane and the viral capsid disassembles, releasing the viral genome into the cytoplasm. At the ER, viral proteins are translated by host ribosomes. The viral RNA-dependent RNA polymerase replicates the viral genome in specialized ER-derived membrane compartments. Assembled viral nucleocapsids bud into the ER lumen and get released from the cell through the secretory pathway. During GOLGI transit of flaviviruses, the host protease furin processes the viral envelope protein prM. In contrast, HCV does not require proteolytic processing but instead tightly associates with the host lipoprotein release pathway. See text for details.
During each step of the virus life cycle, PPIs play a critical role. Viral structural proteins interact homotypically and heterotypically to form the virus particle. Viral glycoproteins bind to cell surface attachment factors and receptors to trigger cell penetration. Translation takes place at host ribosomes, and replication requires not only viral non-structural (NS) proteins but also several host proteins. During assembly and release of particles, host proteins coordinate intracellular trafficking and viral glycoprotein processing. While for HCV the molecular understanding of the function of viral proteins, e.g. the NS3/4A protease, has led to the development of efficient drugs (20), for newly emerging flaviviruses like ZIKV, reverse genetic systems have just been developed (22, 23). It is conceivable that experimental approaches developed for HCV and DENV are applicable to less-well-studied flaviviruses like ZIKV. Here, we will review, how mass spectrometry (MS)-based proteomics contributed to the understanding of the molecular biology of Flaviviridae infections and will discuss chances and caveats of the application of interaction proteomics to the field of virology.
FLAVIVIRUS INFECTION AND INTERACTIONS WITH HOST PROTEINS
Composition of the Virus Particle
Flaviviruses and hepaciviruses egress from infected cells through the secretory pathway, which is repurposed by the structural components of the virus particle. During egress, virus particles can undergo diverse maturation steps, including modifications of their envelope proteins by glycosylation or proteolysis. Furthermore, host proteins may associate with the virus particle. Thus, extracellular infectious flavivirus and hepacivirus particles comprise of viral structural components, i.e. multiple copies of a core or capsid protein and of two envelope proteins plus a virus-specific set of host proteins (Fig. 3A) (24, 25).
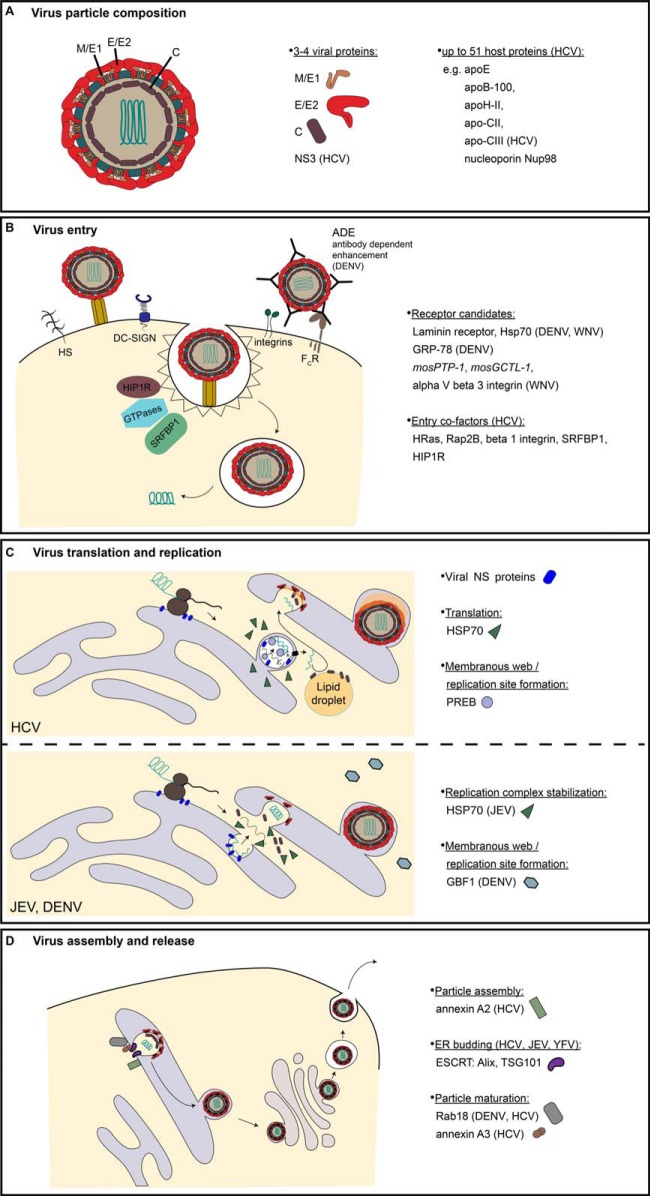
Fig. 3.
Protein interactions during the flavivirus life cycle elucidated by MS-based proteomics. (A) Flavivirus particle composition. Flaviviridae particles are composed of a capsid/core (C) surrounded positive strand RNA genome and enveloped by a host derived lipid bilayer in which the viral transmembrane domain glycoproteins M/E1 and E/E2 are embedded. Additionally, the virions may comprise of nonstructural proteins and host proteins. (B) Virus host protein interactions during entry. Flaviviridae members penetrate the host cell by receptor-mediated endocytosis and fusion in endosomal compartments. MS-based proteomics discovered the listed interaction partners of viral glycoproteins as receptor candidates. SILAC-approaches further revealed secondary interactors of HCV through co-IP MS of the HCV receptor CD81 (listed as entry cofactors). (C) Virus host protein interactions during translation and replication. The Flaviviridae viral genomes translate and replicate in the cytosol in close association with ER membranes, which are remodeled during infection. MS-based proteomics identified heat shock protein 70 (HSP70) as host translation factor and prolactin regulatory element binding (PREB), HSP70, and GBF1 as host replication factors for the indicated viruses. Viral NS proteins forming the replication complex are depicted by a single blue symbol. (D) Virus host protein interactions during particle assembly and release. Particles are assembled at ER-derived membranes and exit the cell through the secretory pathway. MS-based proteomics revealed annexins, ESCRT components, and the GTPase Rab18 as host factors promoting particle assembly, ER budding, and maturation of the indicated Flaviviridae members. All panels show the assumed but not necessary experimentally confirmed localization of host factors identified by MS-based proteomics.
Determining the protein composition and stoichiometry of Flaviviridae particles has proven a challenging task, as it requires the purification of virions with high specific infectivity, i.e. the fraction of particles, which gives rise to productive infection (26, 27). Release of defective particles as well as immature and partially mature mosaic particles with altered protein composition is well documented and complicates proteomic particle analysis (28–30). Additionally, the high titer preparations needed for proteomic analysis in the past were only achieved with cell-culture-produced virus, which cannot fully mimic patient isolates.
With regards to the viral protein components, their stoichiometry and number is known for most of the arthropod-borne flaviviruses. Cryo-electron microscopy and X-ray crystallography of DENV (31–34), TBEV (35, 36), and WNV (37, 38) revealed a common icosahedral array of 90 envelope glycoprotein heterodimers forming a smooth surface. The nucleocapsid composed of the core protein and the viral RNA genome is somewhat less ordered than the envelope. ZIKV shows a glycoprotein fold and particle structure similar to other flaviviruses. This fits the hypothesis that arthropod-borne flaviviruses share important structural features (39–42). HCV in contrast differs from these Flaviviridae species as its viral particles are intimately associated with serum lipoproteins (27, 43, 44). This association reflects the unique assembly process of HCV as well as the parenteral mode of transmission. HCV not only displays fewer copies of glycoproteins than flaviviruses but also has an E2 glycoprotein, which markedly differs from the typical class II fusion proteins of flaviviruses (45, 46). The heterodimerization of the two surface glycoproteins is, however, conserved throughout the Flaviviridae family, and the HCV E1E2 dimer interfaces have been mapped in detail (47–49).
Host proteins incorporated into the HCV particle have recently been described by a pioneering study by Catanese et al. (50). The authors employed two alternative affinity purification strategies and identified 46 particle-associated host proteins by MS-based proteomics. While they confirmed the association with serum lipoproteins (reviewed in (51, 52)), they surprisingly found the nucleoporin Nup98 as HCV particle component. Nup98 associated with viral core, was important for particle assembly and relocated in infected cells from the nuclear pore to virus assembly sites. This study highlights the power of unbiased proteome analysis for generating hypotheses and discovering unexpected cellular mechanisms.
For the virion composition of other Flaviviridae members, proteomic analyses are lacking to date. It is, however, conceivable that their envelope incorporates host proteins and that further host proteins might be enclosed within the envelope. Whether or not such host components are random cargo or fulfill a specific function is yet to be discovered. Examples from other enveloped viruses such as human immunodeficiency virus (HIV) show that at least a subset of virus-associated molecules plays an important role. For instance, components of the endosomal sorting complexes required for transport (ESCRT) machinery guide particle release and apolipoprotein B mRNA editing enzyme complex 3G, when incorporated into HIV-1 particles, edits viral progeny genomes, leading to inactivation of newly synthesized virions (reviewed in (53)).
Importantly, proteomics methods are only as good as the samples that are analyzed. For instance, flavivirus particles can copurify with exosomes, which are host-derived extracellular vesicles of similar size as the virions. A recent study by Cullens and coworkers identifies the exosome components CD9, annexin 2, and Asp-linked glycosylation protein 2-interacting protein X (ALIX) in affinity-pure preparations of the animal pathogenic bovine vial diarrhea virus (54). They proved by density gradient separation that virus preparations contained exosomal vesicles. This demonstrates the limitation of proteomic methods to determine host protein components of viral particles and calls for validation using orthogonal methods like immunogold labeling or bait–prey swap affinity purification.
In summary, while the viral protein components of flaviviruses and hepaciviruses are well defined, for many of these, the envelope incorporated host proteins remain enigmatic. Technological advancements like highly sensitive mass spectrometers and quantitative analysis methods provide tools, which theoretically allow the analysis of virus proteomes even from small samples (55, 56). However, purification of virus particles, their functional characterization, and the choice of appropriate controls to avoid false positive results is critical for generation of a trustworthy dataset. Importantly, the particle composition of Flaviviridae members differs depending on the host and tissue in which assembly takes place. For instance, viral envelope proteins derived from insect cells often display oligomannose glycans instead of complex glycans as in the case of mammalian-cell-derived envelopes (57, 58). Currently, it is unclear whether these different glycosylation patterns have consequences on PPIs in the viral particle itself. Clearly, it can affect interactions with carbohydrate receptors at the host cell surface. Independently of the glycosylation of envelope, the incorporation of host proteins likely differs between producer cells from distinct hosts and tissues merely due to the fact that each cell type expresses its unique proteome. Possibly, even the subcellular site of virion release might affect particle composition as recently shown for hepatitis A virus (58). This typically nonenveloped virus acquired a host cell envelope when released from the basolateral side of hepatocytes, and the envelope was enriched in host proteins usually found in exosomes. Thus, drawing a complete picture of flavivirus particle composition requires analyzing several producer cell systems and organisms in parallel. Current developments of high-throughput liquid chromatography tandem-MS (LC-MS/MS) machines and protocols may allow such studies in the future (59). Unquestionably, virion proteomics can pave the way for the functional understanding of virus assembly, entry, and possibly escape from host immunity (Fig. 3A) (60).
Viral and Host Protein Interactions during Entry
The entry of flavi- and hepaciviruses involves primary and secondary protein interactions at the plasma membrane and in endosomal compartments. The primary protein interactions at the cell surface either serve to concentrate virus particles in case of attachment factor interactions or trigger the productive virus uptake program in the case of bona fide receptors (29, 61).
The three major attachment factors for flaviviruses are heparin sulfates, dendritic cell-specific intracellular adhesion molecule (ICAM)-3-grabbing nonintegrin 1 (DC-SIGN, CD209 antigen), and DC-SIGNR, which interact with N-linked glycans of the viral E glycoprotein. DENV, JEV, TBEV, WNV, and YFV use heparin sulfates for attachment in vitro (62–66). For DENV, JEV, and WNV, this was confirmed in vivo using a heparin sulfate mimetic that competitively blocks virus entry (65). For DENV, the binding motif was narrowed down to a specific glycochain of the syndecan-2 proteoglycan (67). DC-SIGN and DC-SIGNR are used as an attachment factor by DENV, JEV, WNV, and ZIKV but not by the YFV vaccine strain 17D (68–73). HCV can similarly bind to DC-SIGN and the related molecule liver/lymph node-specific ICAM-3-grabbing non-integrin (L-SIGN), which is expressed on liver sinusoidal endothelial cells. However, these lectins are not expressed on HCV permissive hepatocytes and can therefore only play a role in capturing the virus on the liver endothelium and transmitting it to susceptible hepatocytes (74–76). For DENV, cryo-electron microscopy studies and mutational analyses both confirmed the assumption that the DC-SIGN carbohydrate recognition domain binds to the E protein glycans (34). While most flavivirus attachment factors were identified by genetic and pharmacological methods, the attachment of HIV-1 to DC-SIGN was characterized in detail by MS-based proteomics revealing a role for DC-SIGN tetramers in virus binding (77). Specifically, amine-reactive short range crosslinking in combination with MS-based proteomics elucidated the homo-tetramer formation of DC-SIGN and coimmunoprecipiation (co-IP) revealed HIV-1 glycoprotein binding to the DC-SIGN tetramers. Similar studies could reveal if DC-SIGN clusters upon flavivirus attachment and whether it recruits additional proteins required for flavivirus uptake. Clearly, DC-SIGN itself is not providing an essential internalization signal during DENV entry (78), suggesting that additional yet enigmatic entry factors exist.
HCV, while also using heparin sulfates as attachment factors, does not interact with DC-SIGN but with lipid receptors on hepatocytes. Specifically, the low density lipoprotein receptor, scavenger receptor type B class I and very low density lipoprotein receptor serve to capture HCV particles on liver cells (79–81). This peculiarity is caused by the tight association of HCV virions with serum lipoproteins (reviewed in (51, 82)). Similar to the flavivirus attachment factors, methods alternative to MS-based proteomics, including gene knockout, competition assays, antibody blockage, and crosslinking followed by immunoblotting, led to the identification of HCV attachment factors.
Receptor candidates for flaviviruses have predominantly been characterized by affinity chromatography and virus overlay protein binding assay, a technique first described for the identification of the lymphocytic choriomeningitis virus receptor (83). In a typical virus overlay protein binding assay experiment, whole-cell protein lysates or subcellular fractions of susceptible cells are separated by electrophoresis, transferred to nitrocellulose, and probed with either purified viral glycoproteins or virus particles for binding. MS-based peptide fingerprinting then identifies corresponding candidate proteins. The method relies on the fact that some viral glycoprotein–host receptor interactions tolerate denaturation of the receptor. Denaturation sensitive interactions are not amenable to this technique. DENV and JEV seem to share the usage of laminin receptor and heat shock protein 70 (HSP70) HSP70 (84–87) as entry ports to their human host cells. However, the role of heat shock HSP70 as DENV receptor has been challenged (88). Laminin receptors, on the other hand, have also been implicated in DENV entry into cells of the mosquito vector Aedes albopictus (89). In the second DENV mosquito vector Aedes aegypti, E glycoprotein affinity enrichment and MS identified enolase, beta-adrenergic receptor kinase (beta-ARK), and cadherin as putative entry factors (90). Further postulated receptors for flaviviruses identified by virus overlay protein binding assay followed by MS identification, are the 78 kDa glucose-regulated protein, mosquito 45 kDa, and 64/67 kDa proteins for DENV (91–95) and alpha V beta 3 integrin, the C45 like phosphatase mosPTP-1 and C-type lectin mosGCTL-1 for WNV (Fig. 3B) (96, 97). Glycoprotein-binding receptors for YFV, TBEV, and ZIKV remain elusive to date. Of note, host receptors, which interact with lipids in the virus envelope, are not amenable to virus overlay protein binding assay approaches using purified envelope ectodomains. Consequently, the phosphatidylserine receptors T-cell immunoglobulin mucin receptor 1 (TIM1) and TYRO3, AXL, and MER (TAM) were not discovered as flavivirus entry receptors before cDNA screening methods became more efficient (98–100). Briefly, enveloped viruses can attach to cells through interaction of envelope incorporated phosphatidylserine and lipid receptors of the TIM and TAM family. We refer the reader elsewhere for detailed reading on this apoptotic mimicry mechanism of virus entry (101).
A peculiarity of flaviviruses is the productive internalization of opsonized particles through Fc receptors. In particular for DENV, this antibody-dependent enhancement1 leads significant health risks for patients with secondary infections as reviewed in (102). Notably, DENV antibodies, in particular those targeting the immunodominant fusion loop epitope, can also enhance ZIKV infection through the same mechanism (103). This suggests that previous exposure to DENV not only sensitizes individuals to subsequent challenge with another DENV serotype but also with ZIKV. Clearly, the recent structural and biochemical studies on antiflavivirus antibodies underscore how PPI studies can not only help understand the cell biology of virus infection but also elucidate molecular mechanisms underlying clinical observations.
In contrast to many members of the flavivirus genus, the HCV entry process has been investigated in detail. Essential receptors, typically termed entry factors, include scavenger receptor type B class I, CD81, claudin-1 (CLDN1), and occludin (OCLN), and cDNA complementation screens led to the discovery of the latter three (104–106). Scavenger receptor type B class I was identified using a cell based glycoprotein binding assay in conjunction with biochemical analysis (81). Direct binding of the HCV E2 glycoprotein has been demonstrated for the entry factors scavenger receptor type B class I and low density lipoprotein receptor as well as for CD81 using cell-based binding assays. MS-based proteomics has, however, led to the clarification of secondary interactions during the HCV entry process. We and others have employed quantitative proteomics to identify interaction partners of the CD81 receptor and identified the GTPase HRas, integrin beta1, Ras-related protein Rap2B, and serum response factor binding protein 1 (SRFBP1) as components of the CD81 receptor complex (Fig. 3B) (107, 108). All four proteins are critical cofactors for HCV entry. Importantly, SRFBP1 appears to interact with CD81 only when HCV engages the receptor. This finding fits the notion that HCV triggers its specific penetration program upon receptor binding. Presumably, other Flaviviridae also rely on protein interaction mediated signaling to induce their endocytosis and prepare the cell for invasion (reviewed in (109, 110).
Receptor-mediated endocytosis is a prerequisite for fusion and uncoating of all known Flaviviridae family members. Clathrin-dependent uptake has been described as the major endocytosis mechanism (111–115), but alternative entry routes seem to exist and may be used in a strain-specific manner (116). Mostly, pharmacological assays and genetic silencing methods revealed critical endocytosis factors so that the actual protein networks during uptake of flaviviruses and hepaciviruses remain largely elusive. For HCV, the clathrin adapter protein Huntingtin-interacting protein 1-related protein, originally identified as HCV entry co-factor by RNA interference (117), was found by quantitative proteomics to bind to the HCV receptor CD81 (107). Notably, the Huntingtin-interacting protein 1-related protein –CD81 interaction is enhanced upon ligation of the receptor by the virus. Recently, silencing and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 knockout screens identified RAB5C and Rab5 GDP/GTP exchange factor (RABGEF) as DENV and ZIKV endocytosis factors and endoplasmatic reticulum membrane complex proteins as early flavivirus host factors (98). While in the same study interaction proteomics clarified the endoplasmatic reticulum membrane complex composition in uninfected cells, the mechanistic role of the endoplasmatic reticulum membrane complex in flavivirus infection remains to be clarified.
The second to last step of the flavivirus and hepacivirus entry pathway, the membrane fusion, occurs in endosomal compartments and requires a low pH trigger (reviewed in (118)). For HCV, initial interaction with the receptor CD81 primes the viral E1/E2 glycoprotein complex for later pH-dependent fusion in the early endosomal compartment (119). The Flaviviridae fusion process itself is typically independent of postendocytosis protein interactions. Flaviviruses use the type II fusion protein E to penetrate the limiting endosomal membrane (118). For hepaciviruses, in contrast, the fusion mechanism is unknown and the recent crystallographic models of the E2 glycoprotein core domain and the N-terminal E1 domain do not reveal any classical fusion loop (45, 46, 120). Thus, further structural analyses in conjunction with mutational analyses and cell fusion assays are needed to solve the enigma of hepacivirus membrane fusion.
The final step of Flaviviridae entry is the uncoating, leading to release of the RNA genome into the cytoplasm. This is by far the least-studied step of the entry pathway. Recent evidence suggests a role for ubiquitination of the viral capsid protein in DENV uncoating. Interestingly, proteasomal degradation of the capsid occurs under physiological conditions, but even if the proteasome is blocked pharmacologically, the viral genome gets released (121). This suggests that ubiquitination destabilizes the DENV capsid in a degradation-independent manner. A similar uncoating mechanism requiring association of free ubiquitin with the capsid, but no proteasome activity was recently described for influenza virus (122). Proteomics methods for the quantification of posttranslational modifications and protein interactions become more sensitive and amenable to study virus-infected cells (55, 56, 123). Thus, we might soon learn, whether ubiquitination or free ubiquitin association is a shared uncoating mechanism for flaviviruses and possibly even hepaciviruses.
Lastly, interactions of flavivirus capsid proteins with host proteins may manipulate cellular processes, which are independent of virus entry. For instance, interaction proteomics identified an association of DENV and WNV capsid proteins with the peroxisome biogenesis factor Pex19 (124). Importantly, the capsid-dependent sequestration of Pex19 leads to a loss of peroxisomes in the infected cell, and this, in turn, dampens early antiviral signaling and interferon production. Thus, protein interactions of viral structural proteins may directly reveal strategies of viruses to antagonize innate immune recognition.
In summary, despite the description of some receptors and entry factors of flavi- and hepaciviruses through genetic methods, secondary PPIs, the composition of cell surface receptor complexes, endocytosis complexes and fusion mechanisms are to a large extent unknown today. This calls for the application of the current highly sensitive and high-throughput compatible quantitative interaction proteomics methods to draw a clearer picture of the molecular events during host cell penetration by members of the Flaviviridae family (125).
Viral and Host protein Interactions during Translation and Replication
Translation, genome replication, and particle assembly of Flaviviridae family members take place in the cytoplasm, more precisely at the ER. Here, all Flaviviridae viruses follow the same strategy: Translation results in a single polyprotein that is recruited to the rough ER via signal peptides and signal particle routing to the ER. Subsequently, cellular and viral proteases catalyze cleavage of the polyprotein into functional protein subunits. The viral proteins then induce rearrangement of ER membranes to form membranous compartments, which serve as sites of viral replication and assembly (as reviewed e.g. in (126). At these sites, membrane-associated multiprotein complexes coordinate RNA replication. Many of the RNA replication steps are strictly cis-active; that is, viral proteins act on the RNA molecules that were used to translate them. Due to the restricted coding capacity of their small RNA genomes, viruses of the Flaviviridae family need host cell proteins to complete their life cycle (as reviewed e.g. in (127). Different proteomic approaches identified host proteins recruited by Flaviviridae and supporting translation and replication. In particular co-IP or tandem affinity purification followed by MS-elucidated virus–host PPIs (as reviewed e.g. in (56). This is typically achieved by overexpression of affinity tagged versions of viral proteins or by reverse genetic construction of viruses encoding tagged proteins and infecting cells with these. NS3, NS4, and NS5 are the major viral components of the Flaviviridae replication complex (RC).
Affinity tagged versions of NS3, NS4, and NS5 in conjunction with interaction proteomics can be used to identify PPIs important for virus translation and replication. Accordingly, Carpp et al. used transiently expressed DENV GFP-NS3 and -NS5 to identify interacting proteins by MS after metabolic isotope labeling (128). The authors found 53 proteins interacting with NS5 and 41 proteins interacting with NS3. Interestingly, 13 of these proteins were overlapping, indicating a tight association of proteins in the DENV RC. Formation of a viral RC associated with ER-derived membranes is crucial for the replication of Flaviviridae members. Thus, host cell proteins interacting with the viral RC might play a role in ER structural rearrangements induced by the viruses. Consistently, Carpp et al. found a significant number of NS5-interactors with a role in retrograde Golgi-ER transport. In particular, the DENV NS5 protein interacted with GBF1, which is involved in ER membrane remodeling and RC formation early in the DENV infection process (Fig. 3C).
Similarly, prolactin regulatory element binding, a protein regulating anterograde ER-Golgi transport, was found to interact with HCV NS4B and relocalize to the viral RC upon interaction (129). Silencing of prolactin regulatory element binding decreased the size of membrane webs and reduced the number of double- and multimembrane vesicles, the site of HCV replication. This led to RC destabilization and HCV RNA and protein degradation by cellular RNases and proteases.
Interaction proteomics was not only used to detect proteins regulating the formation of the Flaviviridae replication site but also to identify proteins important for the stabilization of the RC: by overexpression of flag-HA-tagged JEV NS5 in HEK293T cells followed by tandem affinity purification-MS, Ye at al. found that the viral NS5 protein directly binds to cellular HSP70 (130). Further analysis revealed that, in JEV-infected cells, HSP70 not only binds NS5 but also NS3 and the replication intermediate dsRNA. A knockdown of HSP70 led to the ubiquitination and subsequent degradation of NS3 and NS5, which indicates that the interaction with HSP70 stabilizes protein complexes in the JEV RC. Interestingly, proteomics also helped to identified HSP70 as an interaction partner of HCV NS5A (131). In contrast to JEV, this interaction only moderately influenced the replication of the HCV genome but seemed to play a major role in NS5A-regulated IRES-mediated translation. Collectively, these findings underline the importance of virus–host PPIs for the translation and replication of viruses of the Flaviviridae family. In particular, mechanisms of membranous web formation and RC preservation were further elucidated by the identification of PPIs through proteomic approaches.
Of note, interaction proteomics can not only reveal host dependence factors but also mechanisms of innate immune escape. DENV NS4B, for example, was shown to interact with 12 mitochondrial proteins, and these interactions induced changes of the mitochondria architecture necessary to prevent viral RNA recognition by the innate immune system (132). While the regulation of innate immune sensing goes beyond the scope of this review, this study nonetheless highlights that interaction proteomics of virus–host interactions is a suitable unbiased method to reveal novel aspects of virus infection and pathogenesis.
Apart from virus–host PPIs, proteomics approaches also revealed interactions of viral proteins with each other, which is also crucial for replication. For instance, Chatel-Chaix et al. used a DENV-encoding hemagglutinin-tagged NS4B to infect hepatoma cells (132, 133). IP followed by gel purification, and LC-MS-MS detected viral NS4B interactors. NS3 was the main interaction partner of NS4B. The interaction did not require formation of the RC but could be found in cells transfected with full length virus as well as with subgenomic replicons. This indicates that the NS3–NS4B interplay is critical for DENV RNA replication. For WNV, co-IP and subsequent mass spectrometry detected the interaction of NS1 with NS3 and NS4B (134). Interestingly, WNV NS1 resides in the ER lumen and is not part of the WNV RC, which is located on the cytosolic site of the ER. However, NS1 is critical for early RNA replication. The authors therefore speculated that WNV NS1 regulates viral replication by interacting with the transmembrane protein N4B, which is part of the WNV RC.
In conclusion, interaction proteomics helped to elucidate PPIs important for the replication of viruses of the Flaviviridae family, in particular of DENV, WNV, and HCV and shed light on the virus-induced remodeling of cellular organelles to serve as replication sites for the viruses and/or shield the virus from innate immune recognition.
Viral and Host Protein Interactions during Particle Assembly and Release
Virus assembly, budding, and release is an intricate, multistep process that among enveloped RNA viruses generally involves numerous PPI, protein–RNA, and protein–lipid interactions. Collectively, these dynamic interactions coordinate the assembly of RNA-containing capsids, the engagement of cellular membranes, the budding and transport of nascent viruses, particle maturation and processing steps, and ultimately the release of particles from the infected cells. A variety of techniques, including genetic approaches, imaging methods, and biochemical assays, are critical to dissect these steps, to unravel distinct phases of virus assembly, and to define the function of specific protein complexes mediating these steps. Clearly, proteome-based approaches have recently provided major contributions to our understanding of Flaviviridae member particle production. Combined with the other above-mentioned approaches, they have helped to derive a picture of key steps during flavivirus egress that is growing in detail.
There are some common themes of flavivirus and hepacivirus assembly and in addition numerous unique aspects and distinct assembly pathways of individual members of this large virus family. For instance, for all Flaviviridae, membrane-associated multiprotein complexes coordinate assembly. In contrast to RNA replication, virus assembly functions in trans and nascent virions are composed of structural (core or capsid) and envelope proteins (E1, E2, or prM, E) that do not necessarily package the same RNA that they were expressed from. As a consequence, trans-complementation of structural proteins in flavivivirus assembly is readily possible and a well-described phenomenon (135–137). This also shows that there are no essential RNA packaging signals within the coding regions of these structural proteins. Another common feature of flavivirus assembly is the tight coupling between RNA replication and assembly, which probably ensures that only replicating and thus active viral RNA is packaged into viruses. In addition to viral structural proteins also various NS proteins participate in virus assembly, although they are not packaged into virions (138–142). While it is not fully clear which specific roles these NS proteins play during assembly, it is thought that these functions help to shuttle viral RNA from RCs into nascent particles and to mediate the switch between RNA replication and later stages of the replication cycle. Finally, flaviviruses and hepaciviruses assemble at intracellular membranes most likely the ER or ER-derived membranes. During intracellular transport these particles mature and they are then secreted from the infected cells most likely through the constitutive secretory pathway.
In addition to these common themes there are also unique features to individual viruses of this family. While all Flaviviridae produce particles that infect host cells in a low pH-triggered manner, members of the genus flavivirus (e.g. DENV, TBEV, YFV) and members of the genus hepacivirus (e.g. HCV, non-primate hepacivirus) have evolved divergent mechanisms to protect their envelope proteins from low-pH-triggered fusion during export through the secretory pathway. For instance, HCV encodes an ion channel protein p7 that dissipates the pH in the secretory compartment (143). In contrast, flaviviruses, have evolved a stepwise maturation of particles concomitant with processing of the prM protein to allow export of fully infectious virus (144, 145). Moreover, a unique feature of HCV, the type member of the genus hepacivirus, is the tight link to the lipoprotein synthesis machinery in the infected cells (as reviewed e.g. in (146)). Notably, this tight interaction was first recognized based on a total proteome analysis of the replication compartment (44), thus highlighting the close interplay between replication and assembly complexes and the importance of proteomic approaches to define these.
A number of different and complementary approaches were used to examine individual assembly factors or assembly protein complexes and their function and have provided major insights that shape our understanding of these virus replication steps. First, several yeast two hybrid (Y2H) interaction screens involving various viral proteins or protein fragments as bait were conducted and combined with network analysis and literature mining. Collectively, these studies revealed a wide landscape of several hundred putative cellular interaction partners with HCV proteins (147–150) and highlighted their interconnection in various cellular pathways and networks. In a similar manner, Y2H screens with flaviviruses yielded numerous viral interactions with host proteins (151–153). Moreover, these studies identified several key viral assembly dependence factors. For instance, Xu et al. discovered the interaction between WNV capsid protein and the cellular RNA helicase DDX56/NOH61. They were able to show that DDX-56 is not involved in RNA replication, but it is critical for incorporation of viral RNA into virus particles and for production of highly infectious progeny (151). A Y2H study identified the interaction of DENV E protein with cellular ER chaperones like 78 kDa glucose-regulated protein (GRP-78, BiP), calnexin, and calreticulin and confirmed that these chaperones are important for production of infectious DENV particles. In the case of HCV, Y2H implicated Tip47 in HCV virus production and RNA replication (150, 154, 155). In parallel, key interactions of HCV NS4B, NS5A, and core proteins with various cellular apolipoproteins (ApoB, ApoA1, ApoE) were identified (147, 149), and it is now well recognized that these apolipoproteins facilitate virus assembly (146, 156, 157), likely at a step after capsid envelopment (158). Thus, these examples advocate the utility and relevance of Y2H interaction screening for identification and subsequent characterization of flaviviral assembly co-factors.
Second, the classical method of antibody-based affinity purification was used for identification of interaction partners and their relevance for assembly. Protein co-precipitation from cellular extracts coupled to MS detection of co-purifying proteins successfully revealed flavivirus assembly dependence factors. For instance, Salloum et al. used an infectious HCV variant with strep-tagged NS5A protein combined with a stable isotope labeling by amino acids in cell culture (SILAC) labeling approach to identify the GTPase Rab18 as novel HCV replication and assembly dependence factor (159) (Fig. 3D). Notably, Rab18, which is involved in vesicle trafficking and resides on the ER and lipid droplet surface, is also important for DENV infection. It was proposed that Rab18 functions to recruit enzymes needed for fatty acid synthesis to promote DENV replication (160). Also, nucleic acid-based affinity purification strategies were used to dissect flavivirus assembly co-factors. Using the highly structured DENV 5′and 3′UTR region as bait in conjunction with 2′Fluoro-UTP and 2′Fluoro-CTP to increase resistance to RNAses, Ward et al. successfully identified UTR-binding host proteins (161). Among them, the RNA helicase DDX6 was shown to modulate secretion of infectious viral progeny. Although it is not clear yet, how DDX6 affects production of infectious virus, the approach is interesting and may be particularly suited to identify host factors that facilitate viral RNA trafficking between RNA replication and assembly sites. Finally, a co-IP approach first implicated the ESCRT machinery in YFV particle production (162) (Fig. 3D). The ESCRT pathway is conserved in all eukaryotes, consists of a set of multi-protein complexes that coordinate membrane fission in various cellular budding processes and multiple viruses use the ESCRT machinery for varying replication cycle steps (163). In case of YFV, NS3 interacts with ALG-2-interacting protein X, thereby facilitating virus release (162).
Meanwhile, much more refined proteomic approaches, based on proteomic analysis of cellular membrane compartments or protein complementation assays in mammalian cells have implicated additional ESCRT components in flavivirus budding and release. SILAC and partial purification of cellular membranes identified host proteins specifically recruited to JEV membranous replication sites. In combination with siRNA-based modulation of ESCRT proteins, not only was the involvement of ALG-2-interacting protein X confirmed, but also TSG101 and various CHIMP proteins were implicated in JEV and DENV particle formation (164) (Fig. 3D). Notably, a mammalian cell-based luciferase protein-complementation assay was recently used to show the interaction between ESCRT and HCV proteins (165). This confirms previous proteome-independent studies implicating the ESCRT machinery in HCV virus production (166–168). Recruitment of ESCRT proteins to HCV was independent of overt viral late domains that frequently mediate this in retroviruses and other viral families (169). As shown for YFV, JEV nonstructural proteins (NS2 and NS5A) seem to mediate the ESCRT interaction unlike in retroviruses where typically viral structural components recruit the cellular membrane budding machinery. Again, this finding underscores the relevance of viral NS proteins in Flaviviridae assembly. Notably, proteome-based interrogation of viral RNA RCs led to the identification of several viral assembly co-factors (44, 164, 170). As mentioned above, the involvement of apolipoproteins in HCV assembly was first recognized after their copurification with HCV RCs. Moreover, the relevance of annexin A2 for HCV assembly, an HCV NS5A binding protein, was also discovered upon its identification in HCV RCs (170).
More recently, quantitative proteomic approaches were also employed to dissect specific cellular compartments needed for virus assembly. Cellular lipid droplets are key cellular lipid storage organelles that serve as HCV assembly platforms. Rösch et al. used SILAC labeling and partial purification of lipid droplets (171) to explore HCV-dependent remodeling of the lipid droplet proteome. They showed dramatic changes in lipid droplet protein association. In particular, HCV infection causes recruitment of annexin A3 to these organelles and this relocalization is necessary for a virus particle maturation step after capsid envelopment (172) (Fig. 3D). Collectively, these studies highlight common assembly host factor usage, as well as unique assembly pathways of specific Flaviviridae members. At the same time, they show how proteomic techniques ranging from Y2H and split protein interaction screens to various affinity-based and organelle-specific purification methods in conjunction with MS-based fingerprinting have complemented each other to dissect Flaviviridae assembly steps.
CONCLUSION AND PERSPECTIVE
As detailed above, proteomics studies of the Flaviviridae life cycle led to the discovery of important host factors for virus infection and served to better understand how these viruses shape host cell mechanisms to replicate. Here, we highlighted PPIs relevant for the propagation of the most prominent human pathogenic Flaviviridae family members, namely DENV, WNV, JEV, TBEV, YFV, ZIKV, and HCV. MS-based proteomics revealed receptor candidates for several flaviviruses and interaction proteomics shed light on HCV entry as well as on flavivirus replication mechanisms, including induction of ER membrane alterations and RC stabilization. Moreover, the technique unraveled Flaviviridae assembly factors, in particular host proteins regulating lipid biogenesis, particle maturation,' and membrane fission during particle budding. While the replication mechanisms of DENV and HCV are understood in great detail, we lack knowledge of the molecular events during infection with flaviviruses such as ZIKV, which caused the recent pandemic in the Americas (1). Future work characterizing PPIs and engagement of host factors during infection with ZIKV and other members of the Flaviviridae family might help to rationally design therapeutics and vaccines. In the case of HCV, the establishment of a cell culture model in conjunction with the detailed mechanistic understanding of the HCV NS proteins, including NS3/4A protease, the assembly factor NS5A, and the RNA-dependent RNA polymerase NS5B, indeed led to the discovery of small inhibitory compounds targeting the three viral proteins (173). Several of these direct-acting drugs against HCV are successfully used in the clinics and cure more than 90% of all patients with minimal side effects and after 12 weeks treatment. Time will tell whether, for the other Flaviviridae members, biochemical analysis of their proteins and PPIs can in a similar manner help develop yet-lacking therapeutics.
Of note, ultrasensitive MS methods nowadays allow analyzing virus particle composition and cellular PPIs from sparse biological material, e.g. from arthropod vector species. As flaviviruses are typically transmitted by mosquitoes or ticks, proteomic studies in the respective vector species are critical to understand transmission events as well as vector species tropism. Clearly, expression of essential host factors is—apart from immune restriction—a key mechanism underlying virus tropism for a particular host species. For instance, the HCV entry factors CD81 and OCLN are responsible for the human tropism of HCV (104, 174, 175), and expression of both human proteins in mouse liver can render mice susceptible to HCV (176). In contrast, for many flaviviruses it is currently unclear which factors govern reservoir host and vector species tropism. Proteomics in combination with orthogonal approaches such as gene knockout screens might reveal species-specific host factors and ultimately help evaluate the risk of zoonotic transmission to the human population.
Despite the power of genome-editing techniques, proteomics research should serve as a complementary method in the virologist's toolbox. Importantly, interaction proteomics provides different information as genome-wide CRISPR/Cas9 or other knockout and knockdown screens. Five aspects seem particularly important when comparing both methods for the identification of host factors.
First, viruses often use essential host cell pathways to replicate, and gene knockout of essential pathway components may lead to compensatory mechanisms masking the knockout phenotype. Similarly, certain host factors display redundancy, and, thus, knocking out a single component may reveal false negative results. In contrast, label-free interaction proteomics methods allow the characterization of protein complexes in nonmanipulated cells (55). Obviously, functional follow up analyses using genetic methods will suffer from the same limitations regarding functional redundancy of host factors as an a priori gene editing screen. Nonetheless, the elucidation of protein complexes might already allow hypothesizing whether or not a functionally similar member of the same protein family can replace a specific protein component functionally.
Second, nonredundant proteins with essential cellular functions are poorly amenable to knockout studies as their gene disruption would cause cell death. Here, proteomics studies are clearly advantageous. Examples of essential cellular pathways usurped during virus infection are growth factor signaling, clathrin-mediated endocytosis, and the secretory route (28, 177, 178). In this context, gene silencing still seems the best-suited complementary method to interaction proteomics.
Third, while proteomics does not reveal per se whether a protein has a function during a particular life cycle step, it uncovers whether a physical association or, in the case of near distance crosslinking, a physical interaction with a viral protein exists. In contrast, genomic screens only reveal genetic network linkages, e.g. transcription factors of essential virus host factors score positive despite their indirect role in infection (179). Whether virus–host interaction partners identified by interaction proteomics play a direct role in supporting virus replication or indirectly affect the virus by regulating host cell metabolism, cell death, or innate immune sensing is part of follow up work. In this regard, interaction proteomics should be considered as a hypothesis-generating machine, which opens new avenues for mechanistic in depth studies.
A fourth aspect is the importance of posttranslational modifications and splice variants for the function of a given gene product. While splice variants are accessible through transcriptomics and next generation sequencing approaches, only proteomics can decipher posttranslational modifications. Various proteins carry posttranslational modifications essential for their function as virus host factors. Examples are the palmitate lipid anchor of the HCV receptor CD81 and its interaction partners Glu-Trp-Ile motif-containing protein 2 (EWI-2) and its cleavage product EWI-2wint (180, 181), the phosphorylation of HCV NS5A as a critical replication prerequisite (182), and maturation of flavivirus glycoproteins during particle egress, including ER signal peptidase processing and proteolytic furin processing during GOLGI transit and glycosylation (183–185).
Lastly, quantitative interaction proteomics in combination with the acquisition of whole cell proteomes allows estimation of the stoichiometry of a given protein complex. In addition the proteomic ruler technique, which uses histones as reference proteins, seems to accurately determine absolute protein copy numbers in a whole-cell proteome (186). Thus, it is possible to estimate how many copies of a given host factor are recruited by a virus. This will have important implications in hypothesizing whether or not usage of a specific protein during infection alters endogenous cellular functions. Thereby, proteomics studies of PPIs during virus infection may also reveal important aspects of viral pathogenesis.
Obviously, proteomics methods have their limitations in deciphering virus particle composition and host cell interactions. As flaviviruses seem to display pleiomorphic phenotypes, these can only be investigated with single particle methods, like e.g. cryo-electron microscopy or crystallographic analysis. Also, host protein interaction studies have specific caveats. In particular, false positive interaction partners can occur due to unspecific binding to affinity matrix and due to disruption of the cellular compartmentalization during cell lysis. Careful experimental design with appropriate negative controls, extensive data analysis including comparison to common contaminants (187), and confirmation of interactions with orthogonal methods ideally in the context of an intact cell can reduce the risk of identification of false positives to a minimum. Subcellular fractionation and/or usage of membrane permeable crosslinking reagents can further avoid that interactions of proteins, which are localizing to different cellular compartments, are falsely detected. Of course the choice of cell lysis buffer and IP buffer needs to fit the physiological protein environment. In our experience, acquisition of multiple parallel datasets with different buffer conditions, e.g. different detergents in the case of membrane protein complexes, allows to draw a clearer picture of the stability and order of detected protein interactions.
Taken together, interaction proteomics is an essential method to uncover the molecular details of how viruses enter, translate, and proliferate their genomes; assemble; and finally egress the host cell to spread to other cells, tissues, and organisms. The method serves to generate hypotheses, which require experimental validation with orthogonal techniques. While interaction proteomics has already revealed important aspects of Flaviviridae infection, the rapid improvement of LC-MS/MS sensitivity and high-throughput compatibility will likely lead to an increased usage of this important technology in basic and applied virology research in the future.
Acknowledgments
We thank Dr. Julie Sheldon for critical reading of the manuscript. We apologize to those colleagues whose work we were unable to cite owing to space limitation.
Footnotes
Author contributions: G.G., J.B., and T.P. wrote the paper and G.G. and B.W. prepared the figures.
* This work was supported by fellowships from the German Research Foundation (DFG, GE 2145/3-1 and SFB 900, project A6) and the Helmholtz Association SO-024. Authors declare no conflict of interest.
1 The abbreviations used are:
- ADE
- antibody-dependent enhancement
- ALIX
- Asp-linked glycosylation protein-2-interacting protein X
- APOBEC3G
- apolipoprotein B mRNA editing enzyme complex 3G
- CLDN
- claudin
- co-IP
- co-immunoprecipiation
- CRISPR
- Clustered Regularly Interspaced Short Palindromic Repeats
- DC-SIGN
- dendritic cell-specific ICAM-3-grabbing non-integrin 1
- DENV
- dengue virus
- ECM
- endoplasmatic reticulum membrane complex
- ER
- endoplasmatic reticulum
- ESCRT
- endosomal sorting complexes required for sorting
- EWI-2
- Glu-Trp-Ile motif-containing protein 2
- HIV
- human immunodeficiency virus
- HSC70
- heat shock cognate protein 70
- HSP
- heat shock protein
- HCV
- hepatitis C virus
- HIP1R
- Huntingtin-interacting protein 1-related protein
- ICAM
- intracellular adhesion molecule
- JEV
- Japanese encephalitis virus
- LDLR
- low density lipoprotein receptor
- L-SIGN
- liver/lymph node-specific ICAM-3-grabbing non-integrin
- NS
- non-structural
- Nup
- nucleoporin
- LC-MS/MS
- liquid chromatography tandem-mass spectrometry
- OCLN
- occludin
- PTMs
- posttranslational modifications
- PREB
- prolactin regulatory element binding
- PPI
- protein-protein interaction
- RABGEF
- Rab5 GDP/GTP exchange factor
- RC
- replication complex
- SR-BI
- scavenger receptor type B class I
- SILAC
- stable isotope labeling by amino acids in cell culture
- SRFBP1
- serum response factor binding protein 1
- TAM
- TYRO3, AXL, and MER
- TAP
- tandem affinity purification
- TIM
- T-cell immunoglobulin mucin receptor 1
- TBEV
- Tick-borne encephalitis virus
- VLDLR
- very low density lipoprotein receptor
- VOPBA
- virus overlay protein binding assay
- WNV
- West Nile virus
- Y2H
- yeast two hybrid
- YFV
- Yellow fever virus
- ZIKV
- Zika virus.
REFERENCES
- 1. Valentine G, Marquez L., and Pammi M. (2016) Zika virus epidemic: An update. Expert Rev. Anti Infect. Ther. 14, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 2. Monath T. P. (2008) Treatment of yellow fever. Antiviral Res. 78, 116–124 [DOI] [PubMed] [Google Scholar]
- 3. Guzman M. G., Halstead S. B., Artsob H., Buchy P., Farrar J., Gubler D. J., Hunsperger E., Kroeger A., Margolis H. S., Martínez E., Nathan M. B., Pelegrino J. L., Simmons C., Yoksan S., Peeling, and R. W. (2010) Dengue: A continuing global threat. Nat. Rev. Microbiol. 8, S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Halstead S. B., and Thomas S. J. (2010) Japanese encephalitis: New options for active immunization. Clin. Infect. Dis. 50, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 5. Gubler D. J. (2002) The global emergence/resurgence of arboviral diseases as public health problems. Arch. Med. Res. 33, 330–342 [DOI] [PubMed] [Google Scholar]
- 6. Hayes E. B., Sejvar J. J., Zaki S. R., Lanciotti R. S., Bode A. V., Campbell G. L. (2005) Virology, pathology, and clinical manifestations of West Nile virus disease. Emerging Infect. Dis. 11, 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gower E., Estes C., Blach S., Razavi-Shearer K., and Razavi H. (2014) Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 61, S45–S57 [DOI] [PubMed] [Google Scholar]
- 8. Staples J. E., and Monath T. P. (2008) Yellow fever: 100 years of discovery. JAMA 300, 960–962 [DOI] [PubMed] [Google Scholar]
- 9. Dick G. W., Kitchen S. F., and Haddow A. J. (1952) Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 46, 509–520 [DOI] [PubMed] [Google Scholar]
- 10. Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., and Houghton M. (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 [DOI] [PubMed] [Google Scholar]
- 11. Ishikawa T., Yamanaka A., and Konishi E. (2014) A review of successful flavivirus vaccines and the problems with those flaviviruses for which vaccines are not yet available. Vaccine 32, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 12. Pawlotsky J. M., Feld J. J., Zeuzem S., and Hoofnagle J. H. (2015) From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 62, S87–S99 [DOI] [PubMed] [Google Scholar]
- 13. Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., and Rice C. M. (1993) A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. U.S.A. 90, 10583–10587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., and Rice C. M. (1993) Characterization of the hepatitis C virus-encoded serine proteinase: Determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67, 2832–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartenschlager R., Ahlborn-Laake L., Mous J., and Jacobsen H. (1993) Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67, 3835–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., and Rice C. M. (2005) Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 17. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., and Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aubry F., Nougairède A., Gould E. A., and de Lamballerie X. (2015) Flavivirus reverse genetic systems, construction techniques and applications: A historical perspective. Antiviral Res. 114, 67–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerold G., and Pietschmann T. (2014) The HCV life cycle: In vitro tissue culture systems and therapeutic targets. Dig Dis. 32, 525–537 [DOI] [PubMed] [Google Scholar]
- 20. Scheel T. K., Rice C. M. (2013) Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19, 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chatel-Chaix L., and Bartenschlager R. (2014) Dengue virus- and hepatitis C virus-induced replication and assembly compartments: The enemy inside-caught in the web. J. Virol. 88, 5907–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shan C., Xie X., Muruato A. E., Rossi S. L., Roundy C. M., Azar S. R., Yang Y., Tesh R. B., Bourne N., Barrett A. D., Vasilakis N., Weaver S. C., and Shi P. Y. (2016) An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19, 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsetsarkin K. A., Kenney H., Chen R., Liu G., Manukyan H., Whitehead S. S., Laassri M., Chumakov K., and Pletnev A. G. (2016) A full-length infectious cDNA clone of Zika virus from the 2015 epidemic in Brazil as a genetic platform for studies of virus-host interactions and vaccine development. MBio 7, e01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mukhopadhyay S., Kuhn R. J., and Rossmann M. G. (2005) A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 [DOI] [PubMed] [Google Scholar]
- 25. Voisset C., and Dubuisson J. (2004) Functional hepatitis C virus envelope glycoproteins. Biol. Cell 96, 413–420 [DOI] [PubMed] [Google Scholar]
- 26. Tan J. L., and Lok S. M. (2014) Dengue virus purification and sample preparation for cryo-electron microscopy. Methods Mol. Biol. 1138, 41–52 [DOI] [PubMed] [Google Scholar]
- 27. Catanese M. T., Uryu K., Kopp M., Edwards T. J., Andrus L., Rice W. J., Silvestry M., Kuhn R. J., and Rice C. M. (2013) Ultrastructural analysis of hepatitis C virus particles. Proc. Natl. Acad. Sci. U.S.A. 110, 9505–9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pierson T. C., and Diamond M. S. (2012) Degrees of maturity: The complex structure and biology of flaviviruses. Curr. Opin. Virol. 2, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perera-Lecoin M., Meertens L., Carnec X., Amara A. (2013) Flavivirus entry receptors: An update. Viruses 6, 69–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Potk;4levka P., Battisti A. J., Junjhon J., Winkler D. C., Holdaway H. A., Keelapang P., Sittisombut N., Kuhn R. J., Steven A. C., and Rossmann M. G. (2011) Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12, 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuhn R. J., Zhang W., Rossmann M. G., Pletnev S. V., Corver J., Lenches E., Jones C. T., Mukhopadhyay S., Chipman P. R., Strauss E. G., Baker T. S., and Strauss J. H. (2002) Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Modis Y., Ogata S., Clements D., and Harrison S. C. (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 100, 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Modis Y., Ogata S., Clements D., and Harrison S. C. (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 [DOI] [PubMed] [Google Scholar]
- 34. Pokidysheva E., Zhang Y., Battisti A. J., Bator-Kelly C. M., Chipman P. R., Xiao C., Gregorio G. G., Hendrickson W. A., Kuhn R. J., and Rossmann M. G. (2006) Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124, 485–493 [DOI] [PubMed] [Google Scholar]
- 35. Ferlenghi I., Clarke M., Ruttan T., Allison S. L., Schalich J., Heinz F. X., Harrison S. C., Rey F. A., and Fuller S. D. (2001) Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7, 593–602 [DOI] [PubMed] [Google Scholar]
- 36. Rey F. A., Heinz F. X., Mandl C., Kunz C., and Harrison S. C. (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375, 291–298 [DOI] [PubMed] [Google Scholar]
- 37. Kanai R., Kar K., Anthony K., Gould L. H., Ledizet M., Fikrig E., Marasco W. A., Koski R. A., and Modis Y. (2006) Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80, 11000–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mukhopadhyay S., Kim B. S., Chipman P. R., Rossmann M. G., and Kuhn R. J. (2003) Structure of West Nile virus. Science 302, 248. [DOI] [PubMed] [Google Scholar]
- 39. Dai L., Song J., Lu X., Deng Y. Q., Musyoki A. M., Cheng H., Zhang Y., Yuan Y., Song H., Haywood J., Xiao H., Yan J., Shi Y., Qin C. F., Qi J., and Gao G. F. (2016) Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 [DOI] [PubMed] [Google Scholar]
- 40. Barba-Spaeth G., Dejnirattisai W., Rouvinski A., Vaney M. C., Medits I., Sharma A., Simon-Lorière E., Sakuntabhai A., Cao-Lormeau V. M., Haouz A., England P., Stiasny K., Mongkolsapaya J., Heinz F. X., Screaton G. R., and Rey F. A. (2016) Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53 [DOI] [PubMed] [Google Scholar]
- 41. Sirohi D., Chen Z., Sun L., Klose T., Pierson T. C., Rossmann M. G., and Kuhn R. J. (2016) The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kostyuchenko V. A., Lim E. X., Zhang S., Fibriansah G., Ng T. S., Ooi J. S., Shi J., and Lok S. M. (2016) Structure of the thermally stable Zika virus. Nature 533, 425–428 [DOI] [PubMed] [Google Scholar]
- 43. Merz A., Long G., Hiet M. S., Brügger B., Chlanda P., Andre P., Wieland F., Krijnse-Locker J., and Bartenschlager R. (2011) Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286, 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang H., Sun F., Owen D. M., Li W., Chen Y., Gale M. Jr., and Ye J. (2007) Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khan A. G., Whidby J., Miller M. T., Scarborough H., Zatorski A. V., Cygan A., Price A. A., Yost S. A., Bohannon C. D., Jacob J., Grakoui A., Marcotrigiano J. (2014) Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kong L., Giang E., Nieusma T., Kadam R. U., Cogburn K. E., Hua Y., Dai X., Stanfield R. L., Burton D. R., Ward A. B., Wilson I. A., and Law M. (2013) Hepatitis C virus E2 envelope glycoprotein core structure. Science 342, 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciczora Y., Callens N., Montpellier C, Bartosch B, Cosset F. L., Op de Beeck A, Dubuisson J. (2005) Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J. Gen. Virol. 86, 2793–2798 [DOI] [PubMed] [Google Scholar]
- 48. Ciczora Y., Callens N., Penin F., Pécheur E. I., and Dubuisson J. (2007) Transmembrane domains of hepatitis C virus envelope glycoproteins: Residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 81, 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rychlowska M., Owsianka A. M., Foung S. K., Dubuisson J., Bienkowska-Szewczyk K., and Patel A. H. (2011) Comprehensive linker-scanning mutagenesis of the hepatitis C virus E1 and E2 envelope glycoproteins reveals new structure-function relationships. J. Gen. Virol. 92, 2249–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lussignol M., Kopp M., Molloy K., Vizcay-Barrena G., Fleck R. A., Dorner M., Bell K. L., Chait B. T., Rice C. M., and Catanese M. T. (2016) Proteomics of HCV virions reveals an essential role for the nucleoporin Nup98 in virus morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 113, 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lindenbach B. D., and Rice C. M. (2013) The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 11, 688–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alvisi G., Madan V., and Bartenschlager R. (2011) Hepatitis C virus and host cell lipids: An intimate connection. RNA Biol. 8, 258–269 [DOI] [PubMed] [Google Scholar]
- 53. Cantin R., Méthot S., and Tremblay M. J. (2005) Plunder and stowaways: Incorporation of cellular proteins by enveloped viruses. J. Virol. 79, 6577–6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Callens N., Brügger B., Bonnafous P., Drobecq H., Gerl M. J., Krey T., Roman-Sosa G., Rümenapf T., Lambert O., Dubuisson J., and Rouillé Y. (2016) Morphology and molecular composition of purified bovine viral diarrhea virus envelope. PLoS Pathog. 12, e1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lum K. K., and Cristea I. M. (2016) Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection. Expert Rev. Proteomics 13, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mason P. W. (1989) Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169, 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feng Z., Hensley L., McKnight K. L., Hu F., Madden V., Ping L., Jeong S. H., Walker C., Lanford R. E., and Lemon S. M. (2013) A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hosp F., Scheltema R. A., Eberl H. C., Kulak N. A., Keilhauer E. C., Mayr K., and Mann M. (2015) A double-barrel liquid chromatography-tandem mass spectrometry (LC-MS/MS) system to quantify 96 interactomes per day. Mol. Cell. Proteomics 14, 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maxwell K. L., and Frappier L. (2007) Viral proteomics. Microbiol. Mol. Biol. Rev. 71, 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pierson T. C., and Kielian M. (2013) Flaviviruses: Braking the entering. Curr. Opin. Virol. 3, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Y., Maguire T., Hileman R. E., Fromm J. R., Esko J. D., Linhardt R. J., Marks R. M. (1997). Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature Medicine 3, 866–871 [DOI] [PubMed] [Google Scholar]
- 63. Germi R., Crance J. M., Garin D., Guimet J., Lortat-Jacob H., Ruigrok R. W., Zarski J. P., and Drouet E. (2002) Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292, 162–168 [DOI] [PubMed] [Google Scholar]
- 64. Hilgard P., and Stockert R. (2000) Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology 32, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 65. Lee E., Pavy M., Young N., Freeman C., and Lobigs M. (2006) Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antiviral Res. 69, 31–38 [DOI] [PubMed] [Google Scholar]
- 66. Kroschewski H., Allison S. L., Heinz F. X., and Mandl C. W. (2003) Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology 308, 92–100 [DOI] [PubMed] [Google Scholar]
- 67. Okamoto K., Kinoshita H., Parquet M. de L., Raekiansyah M., Kimura D., Yui K., Islam M. A., Hasebe F., and Morita K. (2012) Dengue virus strain DEN2 16681 utilizes a specific glycochain of syndecan-2 proteoglycan as a receptor. J. Gen. Virol. 93, 761–770 [DOI] [PubMed] [Google Scholar]
- 68. Barba-Spaeth G., Longman R. S., Albert M. L., and Rice C. M. (2005) Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J. Exp. Med. 202, 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Davis C. W., Nguyen H. Y., Hanna S. L., Sánchez M. D., Doms R. W., and Pierson T. C. (2006) West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 80, 1290–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Navarro-Sanchez E., Altmeyer R., Amara A., Schwartz O., Fieschi F., Virelizier J. L., Arenzana-Seisdedos F., and Desprès P. (2003) Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tassaneetrithep B., Burgess T. H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M. A., Pattanapanyasat K., Sarasombath S., Birx D. L., Steinman R. M., Schlesinger S., and Marovich M. A. (2003) DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hamel R., Dejarnac O., Wichit S., Ekchariyawat P., Neyret A., Luplertlop N., Perera-Lecoin M., Surasombatpattana P., Talignani L., Thomas F., Cao-Lormeau V. M., Choumet V., Briant L., Desprès P., Amara A., Yssel H., and Missé D. (2015) Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 89, 8880–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shimojima M., Takenouchi A., Shimoda H., Kimura N., and Maeda K. (2014) Distinct usage of three C-type lectins by Japanese encephalitis virus: DC-SIGN, DC-SIGNR, and LSECtin. Arch. Virol. 159, 2023–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lozach P. Y., Lortat-Jacob H., de Lacroix de Lavalette A., Staropoli I., Foung S., Amara A., Houles C., Fieschi F., Schwartz O., Virelizier J. L., Arenzana-Seisdedos F., and Altmeyer R. (2003) DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278, 20358–20366 [DOI] [PubMed] [Google Scholar]
- 75. Pöhlmann S., Zhang J., Baribaud F., Chen Z., Leslie G. J., Lin G., Granelli-Piperno A., Doms R. W., Rice C. M., and McKeating J. A. (2003) Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77, 4070–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gardner J. P., Durso R. J., Arrigale R. R., Donovan G. P., Maddon P. J., Dragic T., and Olson W. C. (2003) L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 100, 4498–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bernhard O. K., Lai J., Wilkinson J., Sheil M. M., and Cunningham A. L. (2004) Proteomic analysis of DC-SIGN on dendritic cells detects tetramers required for ligand binding but no association with CD4. J. Biol. Chem. 279, 51828–51835 [DOI] [PubMed] [Google Scholar]
- 78. Lozach P. Y., Burleigh L., Staropoli I., Navarro-Sanchez E., Harriague J., Virelizier J. L., Rey F. A., Desprès P., Arenzana-Seisdedos F., and Amara A. (2005) Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 280, 23698–23708 [DOI] [PubMed] [Google Scholar]
- 79. Yamamoto S., Fukuhara T., Ono C., Uemura K., Kawachi Y., Shiokawa M., Mori H., Wada M., Shima R., Okamoto T., Hiraga N., Suzuki R., Chayama K., Wakita T., and Matsuura Y. (2016) Lipoprotein receptors redundantly participate in entry of hepatitis C virus. PLoS Pathog. 12, e1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Agnello V., Abel G., Elfahal M., Knight G. B., and Zhang Q. X. (1999) Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 12766–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Scarselli E., Ansuini H., Cerino R., Roccasecca R. M., Acali S., Filocamo G., Traboni C., Nicosia A., Cortese R., and Vitelli A. (2002) The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Paul D., Madan V., and Bartenschlager R. (2014) Hepatitis C virus RNA replication and assembly: Living on the fat of the land. Cell Host Microbe 16, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Borrow P., and Oldstone M. B. (1992) Characterization of lymphocytic choriomeningitis virus-binding protein (s): A candidate cellular receptor for the virus. J. Virol. 66, 7270–7281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Thepparit C., and Smith D. R. (2004) Serotype-specific entry of dengue virus into liver cells: Identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J. Virol. 78, 12647–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Reyes-Del Valle J., Chávez-Salinas S., Medina F., and Del Angel R. M. (2005) Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 79, 4557–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thongtan T., Wikan N., Wintachai P., Rattanarungsan C., Srisomsap C., Cheepsunthorn P., and Smith D. R. (2012) Characterization of putative Japanese encephalitis virus receptor molecules on microglial cells. J. Med. Virol. 84, 615–623 [DOI] [PubMed] [Google Scholar]
- 87. Das S., Laxminarayana S. V., Chandra N., Ravi V., and Desai A. (2009) Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology 385, 47–57 [DOI] [PubMed] [Google Scholar]
- 88. Cabrera-Hernandez A., Thepparit C., Suksanpaisan L., and Smith D. R. (2007) Dengue virus entry into liver (HepG2) cells is independent of hsp90 and hsp70. J. Med. Virol. 79, 386–392 [DOI] [PubMed] [Google Scholar]
- 89. Sakoonwatanyoo P., Boonsanay V., and Smith D. R. (2006) Growth and production of the dengue virus in C6/36 cells and identification of a laminin-binding protein as a candidate serotype 3 and 4 receptor protein. Intervirology 49, 161–172 [DOI] [PubMed] [Google Scholar]
- 90. Muñoz M. de L., Limón-Camacho G., Tovar R., Diaz-Badillo A., Mendoza-Hernández G., and Black W. C. 4th (2013) Proteomic identification of dengue virus binding proteins in Aedes aegypti mosquitoes and Aedes albopictus cells. BioMed. Res. Int. 2013, 875958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jindadamrongwech S., Thepparit C., and Smith D. R. (2004) Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch. Virol. 149, 915–927 [DOI] [PubMed] [Google Scholar]
- 92. Salas-Benito J., Reyes-Del Valle J., Salas-Benito M., Ceballos-Olvera I., Mosso C., and del Angel R. M. (2007) Evidence that the 45-kD glycoprotein, part of a putative dengue virus receptor complex in the mosquito cell line C6/36, is a heat-shock related protein. Am. J. Trop. Med. Hyg. 77, 283–290 [PubMed] [Google Scholar]
- 93. Salas-Benito J. S, and del Angel R. M. (1997) Identification of two surface proteins from C6/36 cells that bind dengue type 4 virus. J. Virol. 71, 7246–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mercado-Curiel R. F., Black W. C. 4th, and Muñoz M. de L. (2008) A dengue receptor as possible genetic marker of vector competence in Aedes aegypti. BMC Microbiol. 8, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Smith D. R. (2012) An update on mosquito cell expressed dengue virus receptor proteins. Insect Mol. Biol. 21, 1–7 [DOI] [PubMed] [Google Scholar]
- 96. Cheng G., Cox J., Wang P., Krishnan M. N., Dai J., Qian F., Anderson J. F., and Fikrig E. (2010) A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell 142, 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chu J. J., and Ng M. L. (2003) Characterization of a 105-kDa plasma membrane associated glycoprotein that is involved in West Nile virus binding and infection. Virology 312, 458–469 [DOI] [PubMed] [Google Scholar]
- 98. Savidis G., McDougall W. M., Meraner P., Perreira J. M., Portmann J. M., Trincucci G., John S. P., Aker A. M., Renzette N., Robbins D. R., Guo Z., Green S., Kowalik T. F., and Brass A. L. (2016) Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 16, 232–246 [DOI] [PubMed] [Google Scholar]
- 99. Jemielity S., Wang J. J., Chan Y. K., Ahmed A. A., Li W., Monahan S., Bu X., Farzan M., Freeman G. J., Umetsu D. T., Dekruyff R. H., and Choe H. (2013) TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 9, e1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Meertens L., Carnec X., Lecoin M. P, Ramdasi R., Guivel-Benhassine F., Lew E., Lemke G., Schwartz O., and Amara A. (2012) The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12, 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Amara A., and Mercer J. (2015) Viral apoptotic mimicry. Nat. Rev. Microbiol. 13, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Guzman M. G., Alvarez M., and Halstead S. B. (2013) Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 158, 1445–1459 [DOI] [PubMed] [Google Scholar]
- 103. Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V. M., Malasit P., Rey F. A., Mongkolsapaya J., and Screaton G. R. (2016) Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ploss A., Evans M. J., Gaysinskaya V. A., Panis M., You H., de Jong Y. P., and Rice C. M. (2009) Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Evans M. J., von Hahn T., Tscherne D. M., Syder A. J., Panis M., Wölk B., Hatziioannou T., McKeating J. A., Bieniasz P. D., and Rice C. M. (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 [DOI] [PubMed] [Google Scholar]
- 106. Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., and Abrignani S. (1998) Binding of hepatitis C virus to CD81. Science 282, 938–941 [DOI] [PubMed] [Google Scholar]
- 107. Gerold G., Meissner F., Bruening J., Welsch K., Perin P. M., Baumert T. F., Vondran F. W., Kaderali L., Marcotrigiano J., Khan A. G., Mann M., Rice C. M., and Pietschmann T. (2015) Quantitative proteomics identifies serum response factor binding protein 1 as a host factor for hepatitis C virus entry. Cell Rep. 12, 864–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zona L., Lupberger J., Sidahmed-Adrar N., Thumann C., Harris H. J., Barnes A., Florentin J., Tawar R. G., Xiao F., Turek M., Durand S. C., Duong F. H., Heim M. H., Cosset F. L., Hirsch I., Samuel D., Brino L., Zeisel M. B., Le Naour F., McKeating J. A., and Baumert T. F. (2013) HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13, 302–313 [DOI] [PubMed] [Google Scholar]
- 109. Mercer J., Schelhaas M., and Helenius A. (2010) Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 [DOI] [PubMed] [Google Scholar]
- 110. Grove J., and Marsh M. (2011) The cell biology of receptor-mediated virus entry. J. Cell Biol. 195, 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Acosta E. G., Castilla V., and Damonte E. B. (2008) Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J. Gen. Virol. 89, 474–484 [DOI] [PubMed] [Google Scholar]
- 112. Van der Schaar H. M., Rust M. J., Chen C., van der Ende-Metselaar H., Wilschut J., Zhuang X., and Smit J. M. (2008) Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4, e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nawa M., Takasaki T., Yamada K., Kurane I., and Akatsuka T. (2003) Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J. Gen. Virol. 84, 1737–1741 [DOI] [PubMed] [Google Scholar]
- 114. Chu J. J., and Ng M. L. (2004) Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78, 10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Blanchard E., Belouzard S., Goueslain L., Wakita T., Dubuisson J., Wychowski C., and Rouillé Y. (2006) Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80, 6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fernandez-Garcia M. D., Meertens L., Chazal M., Hafirassou M. L., Dejarnac O., Zamborlini A., Despres P., Sauvonnet N., Arenzana-Seisdedos F., Jouvenet N., and Amara A. (2016) Vaccine and wild-type strains of yellow fever virus engage distinct entry mechanisms and differentially stimulate antiviral immune responses. MBio 7, e01956-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Coller K. E., Berger K. L., Heaton N. S., Cooper J. D., Yoon R., and Randall G. (2009) RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kielian M., and Rey F. A. (2006) Virus membrane-fusion proteins: More than one way to make a hairpin. Nat. Rev. Microbiol. 4, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sharma N. R., Mateu G., Dreux M., Grakoui A., Cosset F. L., and Melikyan G. B. (2011) Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J. Biol. Chem. 286, 30361–30376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. El Omari K., Iourin O., Kadlec J., Sutton G., Harlos K., Grimes J. M., and Stuart D. I. (2014) Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat. Commun. 5, 4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Byk L. A., Iglesias N. G., De Maio F. A., Gebhard L. G., Rossi M., and Garmarnik A. V. (2016) Dengue virus genome uncoating requires ubiquitination. MBio 7, e00804-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Banerjee I., Miyake Y., Nobs S. P., Schneider C., Horvath P., Kopf M., Matthias P., Helenius A., and Yamauchi Y. (2014) Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346, 473–477 [DOI] [PubMed] [Google Scholar]
- 123. Cox J., and Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 124. You J., Hou S., Malik-Soni N., Xu Z., Kumar A., Rachubinski R. A., Frappier L., and Hobman T. C. (2015) Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J. Virol. 89, 12349–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gerold G., Bruening J., and Pietschmann T. (2016) Decoding protein networks during virus entry by quantitative proteomics. Virus Res. 218, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Paul D., and Bartenschlager R. (2015) Flaviviridae replication organelles: Oh, what a tangled web we weave. Annu. Rev. Virol. 2, 289–310 [DOI] [PubMed] [Google Scholar]
- 127. Fernandez-Garcia M. D., Mazzon M., Jacobs M., and Amara A. (2009) Pathogenesis of flavivirus infections: Using and abusing the host cell. Cell Host Microbe 5, 318–328 [DOI] [PubMed] [Google Scholar]
- 128. Carpp L. N., Rogers R. S., Moritz R. L., Aitchison JD. (2014) Quantitative proteomic analysis of host-virus interactions reveals a role for Golgi brefeldin A resistance factor 1 (GBF1) in dengue infection. Mol. Cell. Proteomics 13, 2836–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kong L., Fujimoto A, Nakamura M, Aoyagi H, Matsuda M, Watashi K, Suzuki R., Arita M, Yamagoe S, Dohmae N, Suzuki T, Sakamaki Y, Ichinose S, Suzuki T, Wakita T., Aizaki H. (2016) Prolactin Regulatory Element Binding Protein Is Involved in Hepatitis C Virus Replication by Interaction with NS4B. J. Virol. 90, 3093–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ye J., Chen Z., Zhang B, Miao H, Zohaib A, Xu Q, Chen H, Cao S. (2013) Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS ONE 8, e75188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gonzalez O, Fontanes V, Raychaudhuri S, Loo R, Loo J, Arumugaswami V, Sun R, Dasgupta A, French SW. (2009) The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology 50, 1756–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chatel-Chaix L., Cortese M., Romero-Brey I., Bender S., Neufeldt C. J., Fischl W., Scaturro P., Schieber N., Schwab Y., Fischer B., Ruggieri A., and Bartenschlager R. (2016) Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe 20, 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chatel-Chaix L., Fischl W., Scaturro P., Cortese M., Kallis S., Bartenschlager M., Fischer B., and Bartenschlager R. (2015) A combined genetic-proteomic approach identifies residues within dengue virus NS4B critical for interaction with NS3 and viral replication. J. Virol. 89, 7170–7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Youn S., Li T., McCune B. T., Edeling M. A., Fremont D. H., Cristea I. M., and Diamond M. S. (2012) Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 86, 7360–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Khromykh A. A., Kenney M. T., and Westaway E. G. (1998) Trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72, 7270–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Tautz N., Thiel H. J., Dubovi E. J., and Meyers G. (1994) Pathogenesis of mucosal disease: A cytopathogenic pestivirus generated by an internal deletion. J. Virol. 68, 3289–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Steinmann E., Brohm C., Kallis S., Bartenschlager R., and Pietschmann T. (2008) Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 82, 7034–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Liu W. J., Chen H. B., and Khromykh A. A. (2003) Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77, 7804–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Leung J. Y., Pijlman G. P., Kondratieva N., Hyde J., Mackenzie J. M., and Khromykh A. A. (2008) Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 82, 4731–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kümmerer B. M., and Rice C. M. (2002) Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76, 4773–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Patkar C. G., and Kuhn R. J. (2008) Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82, 3342–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lindenbach B. D. (2013) Virion assembly and release. Curr. Top. Microbiol Immunol. 369, 199–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wozniak A. L., Griffin S., Rowlands D., Harris M., Yi M., Lemon S. M., and Weinman S. A. (2010) Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6, e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yu I. M., Zhang W., Holdaway H. A., Li L., Kostyuchenko V. A., Chipman P. R., Kuhn R. J., Rossmann M. G., and Chen J. (2008) Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 [DOI] [PubMed] [Google Scholar]
- 145. Perera R., and Kuhn R. J. (2008) Structural proteomics of dengue virus. Curr. Opin. Microbiol. 11, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fukuhara T., Ono C., Puig-Basagoiti F., and Matsuura Y. (2015) Roles of lipoproteins and apolipoproteins in particle formation of hepatitis C virus. Trends Microbiol. 23, 618–629 [DOI] [PubMed] [Google Scholar]
- 147. De Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugué S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., André P., Rabourdin-Combe C., and Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Tripathi L. P., Kambara H., Chen Y.A, Nishimura Y., Moriishi K., Okamoto T., Morita E., Abe T., Mori Y., Matsuura Y., and Mizuguchi K. (2013) Understanding the biological context of NS5A-host interactions in HCV infection: A network-based approach. J. Proteome Res. 12, 2537–2551 [DOI] [PubMed] [Google Scholar]
- 149. Tripathi L. P., Kataoka C., Taguwa S., Moriishi K., Mori Y., Matsuura Y., and Mizuguchi K. (2010) Network based analysis of hepatitis C virus core and NS4B protein interactions. Mol. Biosyst. 6, 2539–2553 [DOI] [PubMed] [Google Scholar]
- 150. Vogt D. A., Camus G., Herker E., Webster B. R., Tsou C. L., Greene W. C., Yen T. S., and Ott M. (2013) Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 9, e1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Xu Z., Anderson R., and Hobman T. C. (2011) The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J. Virol. 85, 5571–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Limjindaporn T., Wongwiwat W., Noisakran S., Srisawat C., Netsawang J., Puttikhunt C., Kasinrerk W., Avirutnan P., Thiemmeca S., Sriburi R., Sittisombut N., Malasit P., and Yenchitsomanus P. T. (2009) Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production. Biochem. Biophys. Res. Commun. 379, 196–200 [DOI] [PubMed] [Google Scholar]
- 153. Le Breton M., Meyniel-Schicklin L., Deloire A., Coutard B., Canard B., de Lamballerie X., Andre P., Rabourdin-Combe C., Lotteau V., and Davoust N. (2011) Flavivirus NS3 and NS5 proteins interaction network: A high-throughput yeast two-hybrid screen. BMC Microbiol. 11, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ploen D., Hafirassou M. L., Himmelsbach K., Schille S. A., Biniossek M. L., Baumert T. F., Schuster C., and Hildt E. (2013) TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur. J. Cell Biol. 92, 374–382 [DOI] [PubMed] [Google Scholar]
- 155. Ploen D., Hafirassou M. L., Himmelsbach K., Sauter D, Biniossek M. L., Weiss TS, Baumert T. F., Schuster C., and Hildt E. (2013) TIP47 plays a crucial role in the life cycle of hepatitis C virus. J. Hepatol. 58, 1081–1088 [DOI] [PubMed] [Google Scholar]
- 156. Fukuhara T., Wada M., Nakamura S., Ono C., Shiokawa M., Yamamoto S., Motomura T., Okamoto T., Okuzaki D., Yamamoto M., Saito I., Wakita T., Koike K., and Matsuura Y. (2014) Amphipathic α-helices in apolipoproteins are crucial to the formation of infectious hepatitis C virus particles. PLoS Pathog. 10, e1004534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chang K. S., Jiang J., Cai Z., and Luo G. (2007) Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hueging K., Doepke M., Vieyres G., Bankwitz D., Frentzen A., Doerrbecker J., Gumz F., Haid S., Wölk B., Kaderali L., and Pietschmann T. (2014) Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J. Virol. 88, 1433–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Salloum S., Wang H., Ferguson C., Parton R. G., and Tai A. W. (2013) Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 9, e1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tang W. C., Lin R. J., Liao C. L., and Lin Y. L. (2014) Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J. Virol. 88, 6793–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ward A. M., Bidet K., Yinglin A., Ler S. G., Hogue K., Blackstock W., Gunaratne J., and Garcia-Blanco M. A. (2011) Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 8, 1173–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Carpp L. N., Galler R., and Bonaldo M. C. (2011) Interaction between the yellow fever virus nonstructural protein NS3 and the host protein Alix contributes to the release of infectious particles. Microbes Infect. 13, 85–95 [DOI] [PubMed] [Google Scholar]
- 163. Schöneberg J., Lee I. H., Iwasa J. H., Hurley, and J. H. (2017) Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tabata K., Arimoto M., Arakawa M., Nara A., Saito K., Omori H., Arai A., Ishikawa T., Konishi E., Suzuki R., Matsuura Y., and Morita E. (2016) Unique requirement for ESCRT factors in flavivirus particle formation on the endoplasmic reticulum. Cell Rep. 16, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 165. Barouch-Bentov R., Neveu G., Xiao F., Beer M., Bekerman E., Schor S., Campbell J., Boonyaratanakornkit J., Lindenbach B., Lu A., Jacob Y., and Einav S. (2016) Hepatitis C virus proteins interact with the endosomal sorting complex required for transport (ESCRT) machinery via ubiquitination to facilitate viral envelopment. MBio 7, e01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Tamai K., Shiina M., Tanaka N., Nakano T., Yamamoto A., Kondo Y., Kakazu E., Inoue J., Fukushima K., Sano K., Ueno Y., Shimosegawa T., and Sugamura K. (2012) Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 422, 377–385 [DOI] [PubMed] [Google Scholar]
- 167. Ariumi Y., Kuroki M., Maki M., Ikeda M., Dansako H., Wakita T., and Kato N. (2011) The ESCRT system is required for hepatitis C virus production. PLoS ONE 6, e14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Corless L., Crump C. M., Griffin S. D., and Harris M. (2010) Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 91, 362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lee S., Joshi A., Nagashima K., Freed E. O., and Hurley J. H. (2007) Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Backes P., Quinkert D., Reiss S., Binder M., Zayas M., Rescher U., Gerke V., Bartenschlager R., and Lohmann V. (2010) Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84, 5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 172. Rösch K., Kwiatkowski M., Hofmann S., Schöbel A., Grüttner C., Wurlitzer M., Schlüter H., and Herker E. (2016) Quantitative lipid droplet proteome analysis identifies annexin A3 as a cofactor for HCV particle production. Cell Rep. 16, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 173. Rice C., and Bartenschlager R. (2016) Bringing the hepatitis C virus to life. Cell 167, 39–42 [DOI] [PubMed] [Google Scholar]
- 174. Flint M., von Hahn T., Zhang J., Farquhar M., Jones C. T., Balfe P., Rice C. M., and McKeating J. A. (2006) Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 80, 11331–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Vogt A., Scull M. A., Friling T., Horwitz J. A., Donovan B. M., Dorner M., Gerold G., Labitt R. N., Rice C. M., and Ploss A. (2013) Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology 444, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Dorner M., Horwitz J. A., Robbins J. B., Barry W. T., Feng Q., Mu K., Jones C. T., Schoggins J. W., Catanese M. T., Burton D. R., Law M., Rice C. M., and Ploss A. (2011) A genetically humanized mouse model for hepatitis C virus infection. Nature 474, 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lupberger J., Zeisel M. B., Xiao F., Thumann C., Fofana I., Zona L., Davis C., Mee C. J., Turek M., Gorke S., Royer C., Fischer B., Zahid M. N., Lavillette D., Fresquet J., Cosset F. L., Rothenberg S. M., Pietschmann T., Patel A. H., Pessaux P., Doffoël M., Raffelsberger W., Poch O., McKeating J. A., Brino L., and Baumert T. F. (2011) EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Kaufmann B., and Rossmann M. G. (2011) Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 13, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Law G. L., Korth M. J., Benecke A. G., and Katze M. G. (2013) Systems virology: Host-directed approaches to viral pathogenesis and drug targeting. Nat. Rev. Microbiol. 11, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhu Y. Z., Luo Y., Cao M. M., Liu Y., Liu X. Q., Wang W., Wu D. G., Guan M., Xu Q. Q., Ren H., Zhao P., and Qi Z. T. (2012) Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology 429, 112–123 [DOI] [PubMed] [Google Scholar]
- 181. Montpellier C., Tews B. A., Poitrimole J., Rocha-Perugini V., D'Arienzo V, Potel J., Zhang X. A., Rubinstein E., Dubuisson J., and Cocquerel L. (2011) Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 286, 13954–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ross-Thriepland D., Mankouri J., and Harris M. (2015) Serine phosphorylation of the hepatitis C virus NS5A protein controls the establishment of replication complexes. J. Virol. 89, 3123–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Zhang R., Miner J. J., Gorman M. J., Rausch K., Ramage H., White J. P., Zuiani A., Zhang P., Fernandez E., Zhang Q., Dowd K. A., Pierson T. C., Cherry S., and Diamond M. S. (2016) A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Marceau C. D., Puschnik A. S., Majzoub K., Ooi Y. S., Brewer S. M., Fuchs G., Swaminathan K., Mata M. A., Elias J. E., Sarnow P., and Carette J. E. (2016) Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Roby J. A., Setoh Y. X., Hall R. A., and Khromykh A. A. (2015) Post-translational regulation and modifications of flavivirus structural proteins. J. Gen. Virol. 96, 1551–1569 [DOI] [PubMed] [Google Scholar]
- 186. Wiśniewski J. R., Hein M. Y., Cox J., and Mann M. (2014) A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics 13, 3497–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Acosta E. G., and Bartenschlager R. (2016) The quest for host targets to combat dengue virus infections. Curr. Opin. Virol. 20, 47–54 [DOI] [PubMed] [Google Scholar]
- 189. Moradpour D., Grakoui A., and Manns M. P. (2016) Future landscape of hepatitis C research—Basic, translational and clinical perspectives. J. Hepatol. 65, S143–S155 [DOI] [PubMed] [Google Scholar]
- 190. Bukh J. (2016) The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J. Hepatol. 65, S2-S21 [DOI] [PubMed] [Google Scholar]
- 191. Mackenzie J. S., Gubler D. J., and Petersen L. R. (2004) Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10, S98–S109 [DOI] [PubMed] [Google Scholar]
- 192. Süss J. (2011) Tick-borne encephalitis, epidemiology, risk areas, and virus strains in Europe and Asia-an overview. Ticks Tick Borne Dis. 2, 2–15 [DOI] [PubMed] [Google Scholar]
- 193. Mansfield K. L., Johnson N., Phipps L. P., Stephenson J. R., Fooks A. R., and Solomon T. (2009) Tick-borne encephalitis virus—A review of an emerging zoonosis. J. Gen. Virol. 90, 1781–1794 [DOI] [PubMed] [Google Scholar]
- 194. Colpitts T. M., Conway M. J., Montgomery R. R., and Fikrig E. (2012) West Nile virus: Biology, transmission, and human infection. Clin. Microbiol. Rev. 25, 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Chancey C., Grinev A., Volkova E., and Rios M. (2015) The global ecology and epidemiology of West Nile virus. Biomed. Res. Int. 2015, 376230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Monath T. P, and Vasconcelos P. F. (2015) Yellow fever. J. Clin. Virol. 64, 160–173 [DOI] [PubMed] [Google Scholar]
- 197. Beasley D. W., McAuley A. J., and Bente D. A. (2015) Yellow fever virus: Genetic and phenotypic diversity and implications for detection, prevention and therapy. Antiviral Res. 115, 48–70 [DOI] [PubMed] [Google Scholar]
- 198. Petersen L. R., Jamieson D. J., Powers A. M., and Honein M. A. (2016) Zika virus. N. Engl. J. Med. 374, 1552–1563 [DOI] [PubMed] [Google Scholar]
- 199. Musso D., and Gubler D. J. (2016) Zika virus. Clin. Microbiol. Rev. 29, 487–524 [DOI] [PMC free article] [PubMed] [Google Scholar]