Abstract
Biophysical stimuli presented to cells via microenvironmental properties (e.g., alignment and stiffness) or external forces have a significant impact on cell function and behavior. Recently, the cell nucleus has been identified as a mechanosensitive organelle that contributes to the perception and response to mechanical stimuli. However, the specific mechanotransduction mechanisms that mediate these effects have not been clearly established. Here, we offer a comprehensive review of the evidence supporting (and refuting) three hypothetical nuclear mechanotransduction mechanisms: physical reorganization of chromatin, signaling at the nuclear envelope, and altered cytoskeletal structure/tension due to nuclear remodeling. Our goal is to provide a reference detailing the progress that has been made and the areas that still require investigation regarding the role of nuclear mechanotransduction in cell biology. Additionally, we will briefly discuss the role that mathematical models of cell mechanics can play in testing these hypotheses and in elucidating how biophysical stimulation of the nucleus drives changes in cell behavior. While force-induced alterations in signaling pathways involving lamina-associated polypeptides (LAPs) (e.g., emerin and histone deacetylase 3 (HDAC3)) and transcription factors (TFs) located at the nuclear envelope currently appear to be the most clearly supported mechanism of nuclear mechanotransduction, additional work is required to examine this process in detail and to more fully test alternative mechanisms. The combination of sophisticated experimental techniques and advanced mathematical models is necessary to enhance our understanding of the role of the nucleus in the mechanotransduction processes driving numerous critical cell functions.
Introduction
Over the last decade, there has been a growing appreciation of the importance of mechanical stimuli on cellular behavior. Substrate properties (i.e., alignment and stiffness) modulate changes in cytoskeletal organization and cell contractility, which ultimately drive important biological processes such as stem cell differentiation, cancer progression, and fibrosis [1–5]. Active mechanical inputs (i.e., substrate stretch and fluid shear stress) have similar effects on cell behavior and pathogenesis [6–8]. Understanding how cells sense these biophysical stimuli and ultimately translate them into specific biological outcomes is essential for advancing the field and for developing new clinical therapies.
Recent interest in the field of mechanobiology has improved our understanding of the mechanotransduction mechanisms that underlie these effects. Much of this work has been focused on mechanosensing at focal adhesions and their downstream signaling pathways [9,10]. Focal adhesions are plaques of integrins and other proteins that interface with the extracellular matrix and receive mechanical stimuli from the microenvironment (Fig. 1) [11,12]. Force at focal adhesions is required for their maturation and growth, which is driven by enhanced protein interactions induced by physical unfolding and exposure of cryptic binding sites as well as tyrosine phosphorylation [13–15]. Several actin binding partners (e.g., filamin, α-actinin, and 14-3-3 proteins) that help organize the actin cytoskeleton also mediate mechanotransduction at focal adhesions or cell–cell junctions and possibly throughout the cytoplasm [16–19]. Signaling downstream of focal adhesion maturation involves numerous pathways and is important for cell survival, proliferation, differentiation, and migration [20]. Furthermore, mechanical stimuli can open stretch-activated ion channels within the plasma membrane, which alters the electrochemical potential of the cell leading to oscillations in ion concentrations both locally and throughout the cytoplasm [21]. These ion channels can be activated by stretch of the lipid bilayer itself (e.g., TRAAK and TREK1) or via tension within the actin cytoskeleton (e.g., TRPV4) [22–24]. In particular, calcium ions that pass through mechanosensitive calcium channels (e.g., TRPV4 and Piezo1/2) act as secondary messengers to initiate several signaling processes that mediate the cellular response to mechanical loading [25–28]. Additionally, increased calcium concentrations sensitize the cell to further mechanical stimuli by increasing cell contractility [29,30].
Fig. 1.
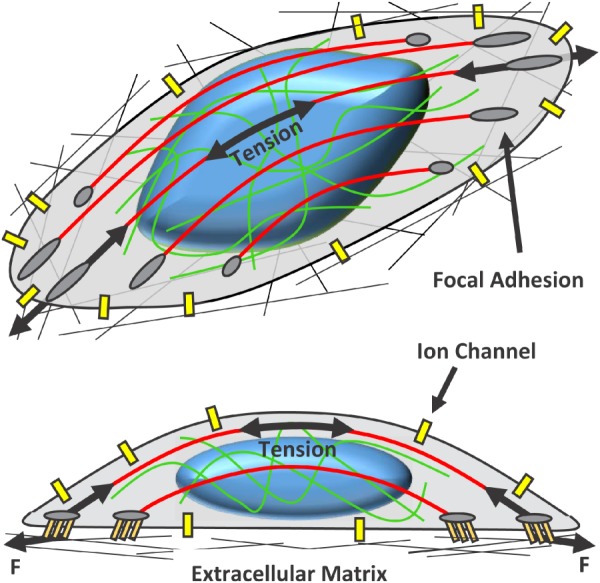
Primary sites of cellular mechanotransduction. Cells attach to the extracellular matrix via integrins and other associated proteins that form focal adhesions. Forces (F) generated by extracellular strain or active cell contraction are produced at the focal adhesions and transmitted through the cell cytoskeleton primarily by actin stress fibers (shown in red) and intermediate filaments (shown in green). Tension within the cytoskeleton transmits forces to the nucleus, which initiates nuclear remodeling and potential mechanotransduction mechanisms. Additionally, stretch of the plasma membrane and cytoskeletal tension may open stretch-activated ion channels. The resulting influx of ions alters the electrochemical potential of the cell and mediates downstream signaling. Color figures are available online.
Mechanotransduction events may also occur at the nuclear envelope and within the nucleoplasm. Forces applied at focal adhesions propagate through the cytoskeleton and are transmitted to the nucleus primarily by actin stress fibers and intermediate filaments [31–34]. Extracellular strains and loads deform the nucleus as a whole and redistribute intranuclear structures, including nucleoli and Cajal bodies [34–38] (Fig. 2). Such changes in nuclear shape are associated with changes in gene expression, even between cells with similar spread areas [39,40]. Furthermore, recent studies demonstrate that isolated nuclei remodel and stiffen in response to mechanical stimulation, clearly demonstrating that the nucleus itself is a mechanoresponsive organelle independent of the cytoplasm [41]. However, it is still unclear whether such nuclear mechanotransduction is causally responsible for alterations in gene expression and cell behavior in response to biophysical inputs.
Fig. 2.
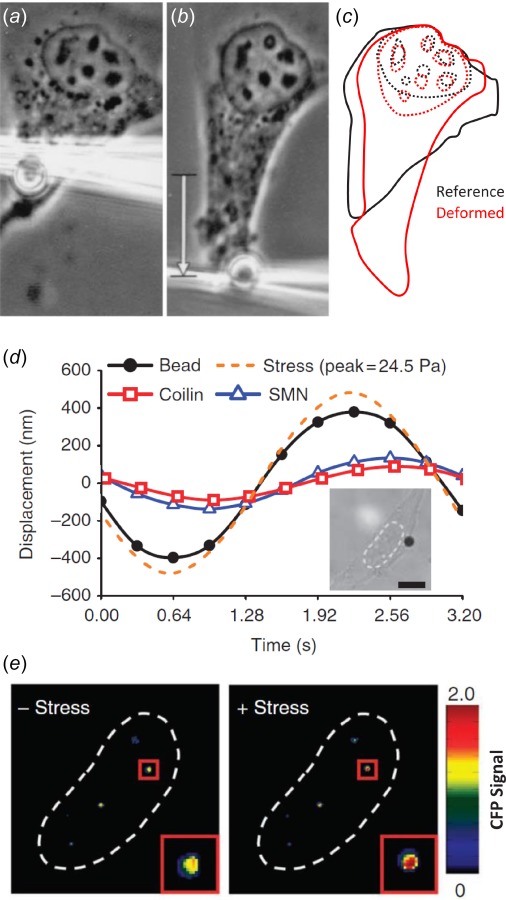
Extracellular forces deform the nucleus. (a)–(c) Force applied to endothelial cells by displacing an RGD-coated bead bound to integrins on the cell surface produces nuclear deformation and displacement of intranuclear nucleoli. (d) Extracellular loading also displaces fluorescently labeled YFP-coilin and CFP-SMN proteins, which are markers for Cajal bodies within the nucleus. Inset: Bright-field image of HeLa cell with RGD-coated bead in black and nucleus outlined with dotted line. Scale bar: 10 μm. (e) Prior to loading, the CFP signal is quenched by fluorescence resonance energy transfer (FRET) due to the association between coilin and SMN. With applied stress, the coilin and SMN proteins separate, resulting in an increase in CFP fluorescence. Adapted with permission from Refs. [34] and [37]. Refer to electronic document for color images.
Several (likely nonexclusive) nuclear mechanotransduction mechanisms have been proposed [42]. Load applied to the nucleus may be transmitted to chromatin positioned at the nuclear envelope, which could alter gene expression by affecting chromatin condensation, topological organization, or positioning of specific gene loci. Alternatively, nuclear loading/deformation could alter the conformation and binding affinities of various proteins located at the nuclear periphery. This could induce structural remodeling of the nucleus and alter the sequestration of genes and associated transcription factors, which would lead to changes in gene transcription. Finally, load-induced nuclear remodeling and stiffening could alter cytoskeletal organization and tension, thereby affecting mechanotransduction processes within the cytoplasm and at the plasma membrane (e.g., at focal adhesions). While several excellent reviews have been recently written discussing the possibility of nuclear mechanotransduction [43–50], few have critically evaluated which of these mechanotransduction mechanisms, either alone or in combination, mediate cellular responses to biophysical stimuli. In this review, we offer a comprehensive synthesis of the existing data supporting (and refuting) each of these three hypotheses regarding nuclear mechanotransduction mechanisms. Our goal is to provide a reference detailing the progress that has been made and the areas that still require investigation regarding the role of nuclear mechanotransduction in cell biology. Additionally, we will briefly discuss the potential contribution of biomechanical engineering to this field through the development of mathematical models of cell mechanics to test these hypotheses and elucidate how biophysical stimulation of the nucleus drives changes in cell behavior.
Nuclear Structure and Connection With Cytoskeleton
The nucleus is physically separated from the cytoplasm by a nuclear envelope composed of an inner and outer nuclear membrane (Fig. 3). On the internal surface of the nuclear envelope is the nuclear lamina, a thin meshwork of intermediate filaments (i.e., lamins A/C, B1, and B2), which provides the primary structural support for the nucleus [51–54]. In particular, lamin A/C (which are isoforms produced from the same LMNA gene) exists in dynamic equilibrium as soluble dimers within the nuclear interior (i.e., nucleoplasm) and as insoluble network assemblies within the lamina meshwork at the nuclear periphery [46,55]. Mechanical load changes the conformation of lamin A/C, which in turn alters accessibility to binding and phosphorylation sites that control the assembly, disassembly, and degradation of lamin A/C [56–60]. Such remodeling of the nuclear lamina is the primary mechanism by which the cell modulates the stiffness of its nucleus in response to changes in microenvironmental stiffness and mechanical loading [56,61]. In addition to their structural role, lamins also bind chromatin and numerous other proteins present within the nucleus (including transcription factors) [62,63]. As will be discussed, given the varied functions of lamin A/C, the nuclear lamina plays an important role in nuclear mechanotransduction [64,65], stem cell differentiation [47,56,66,67], and pathology [68,69].
Fig. 3.
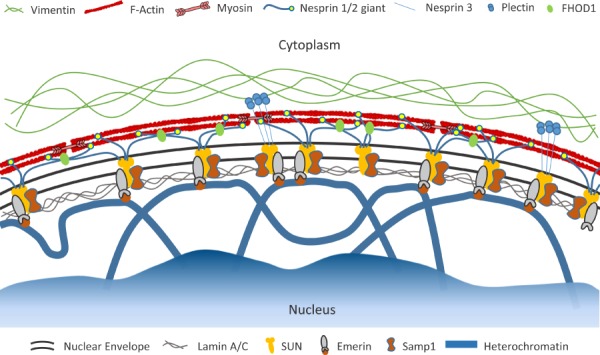
Schematic illustrating the structure of the nuclear envelope. Adjacent to the inner nuclear membrane is the nuclear lamina, which is a meshwork of intermediate filaments that are the primary structural support for the nucleus. Heterochromatic lamina-associated domains (LADs) bind to the lamina and other proteins associated with the nuclear envelope (e.g., emerin). Linker of the nucleoskeleton and cytoskeleton (LINC) complexes are composed of nesprins and SUN proteins as well as other associated molecules (e.g., emerin, FHOD1, and Samp1) and connect the nuclear lamina with the cytoplasmic cytoskeleton. Color figures are available online.
Chromatin is the ensemble of deoxyribonucleic acid (DNA) and associated proteins (e.g., histones), which fills the nuclear volume. The linear DNA molecules are wrapped around core histone complexes to form nucleosomes [70]. These nucleosomes are compacted via internucleosomal interactions into 30 nm chromatin fibers, which are condensed further into topologically associating domains (TADs) and chromosome territories [71–73]. Chromatin condensation, organization, and positioning are highly coordinated and strongly correlated with gene density and expression [74–76]. For example, chromatin containing actively transcribed genes exists in a less condensed state (i.e., euchromatin) compared to the more compact regions (i.e., heterochromatin) that contain silent genes. As will be discussed below, the lamina-associated domains (LADs) within the DNA sequence that bind to the nuclear lamina are generally heterochromatic [77]. Upon activation, these gene loci are decondensed and repositioned to specific intranuclear locations away from the nuclear periphery (i.e., transcription factories), which contain multiple genes, ribonucleic acid (RNA) polymerase, and numerous transcription factors [78–81]. Such reorganization of the genome is a characteristic feature of stem cell differentiation and may be linked to nuclear mechanotransduction processes [44].
The nucleus is physically connected to the cytoskeleton via linker of nucleoskeleton and cytoskeleton (LINC) complexes present within the nuclear envelope (see Ref. [82], and Refs. [83] and [84] for reviews) (Fig. 3). Embedded within the inner nuclear membrane, SUN proteins attach to the nuclear lamina and other membrane-bound proteins. In the perinuclear space between the outer and inner nuclear membranes, these SUN molecules interact with the KASH domain of proteins attached to the outer nuclear membrane. These KASH proteins (in particular, nesprin-1 giant, nesprin-2 giant, and nesprin-3) complete the LINC complex by connecting with actin filaments, intermediate filaments, and microtubules in the cytoplasm. Recently, several other proteins have been discovered to play important roles in maintaining and reinforcing these connections [41,85–87], suggesting that the LINC complex may be the nuclear envelope analog to the more traditional focal adhesions that are present within the plasma membrane [88]. Furthermore, depending on the specific combinations of subcomponents involved in their assembly, LINC complexes play a role in numerous cell functions, including nuclear positioning, migration, morphology, cytoskeletal organization, and intracellular force transmission [84].
Of particular interest for cellular mechanotransduction is the connection between the nucleus and the contractile actomyosin stress fibers within the cytoplasm. While the exact manner in which LINC complexes interface with the actin cytoskeleton is not fully understood, certain details have become clear over the last several years. Specific actin stress fibers have been found to wrap over and around the nucleus to form the so-called perinuclear actin cap [31]. While these stress fibers terminate at focal adhesions within the plasma membrane at both ends [31,89], they are physically connected to the apical surface of the nucleus via nesprins and form transmembrane actin-associated nuclear (TAN) lines, which consist of linear arrays of LINC complex proteins at the nuclear surface [90–92]. As discussed below, these actin cap stress fibers transmit forces to the apical nuclear surface, which can deform and concentrate intranuclear DNA [58,59,92–94]. Therefore, they are thought to serve an important role in transmitting load to the nucleus, determining nuclear shape, and driving potential nuclear mechanotransduction processes [31,32,89,95].
Mechanical Loading of the Nucleus
Following the seminal studies of Guilak [35] and Maniotis et al. [34], numerous groups have demonstrated that extracellular forces applied to cells via ligand-coated beads, microneedles, or substrate strain are transmitted to the nucleus, producing changes in nuclear shape and intranuclear deformations [36–38, 40,93,96–99]. The activation and transcription of many mechanosensitive genes appear to depend on effective transmission of load to the nucleus and the resulting nuclear deformations [39,40,64,99,100]. However, the primary structures that transmit load to the nucleus and the specific nuclear deformations that induce changes in gene expression are still debated.
As mentioned above, substantial evidence exists to suggest that actin stress fibers transmit tensile and compressive loads to the apical surface of the nucleus via LINC complexes. In nearly all cases, depolymerizing actin filaments eliminates nuclear deformations in response to extracellular loads [34–37]. The presence of an intact actin cap has also been shown to flatten and elongate the nucleus with cell spreading [31,32]. This suggests that tension in the actin cap stress fibers produces a downward compressive force onto the nucleus. Indeed, indentations on the order of a few microns are observed in the apical surface of the nucleus, which disappear with inhibition of myosin activity [59,91,92,94] (Fig. 4). Furthermore, nuclear compression due to tension in the actin cap induces structural reorganization of the nuclear lamina and prevents degradation of lamin A [58,59]. This suggests that force transmitted by the actin cap to the nucleus is sufficient to drive load-induced remodeling of the nuclear lamina. Disruption of the LINC complex leads to a loss of actin cap stress fibers [31,89,101,102] and reduced nuclear deformation in response to extracellular loads [96,99], suggesting that the LINC complex is necessary to transmit load from the perinuclear actin cap to the nucleus. Additionally, fluorescence resonance energy transfer (FRET)-based tension sensors have demonstrated that nesprins are under tension at the nuclear surface [93]. Together, this suggests that one potential mediator of nuclear mechanotransduction is via nuclear deformations generated by loads transmitted by actin stress fibers through LINC complexes.
Fig. 4.
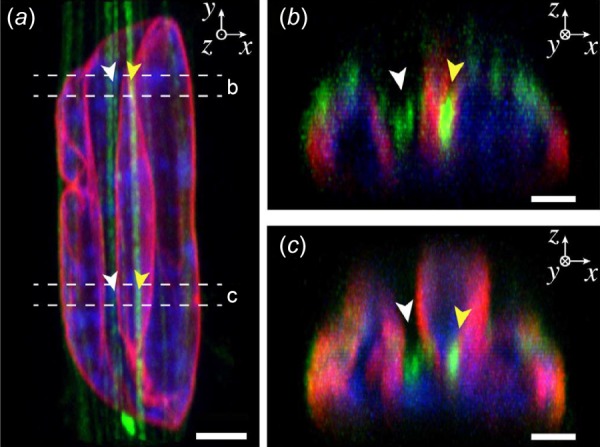
Compressive loading of the apical nuclear surface. (a) Apical actin stress fibers (green) form deep indentations within the nucleus, which deform the nuclear lamina (red) and intranuclear chromatin (blue). Scale bar: 3 μm. (b) and (c) Cross-sectional images of nucleus showing actin stress fibers within nuclear indentations and substantial local nuclear deformations. Scale bars: 1.5 μm. Adapted with permission from Ref. [92]. Color figures are available online.
However, recent findings have also questioned the importance of apical actin stress fibers in determining nuclear shape. Li et al. visualized nuclear deformations within fibroblasts as they were seeded and adhered onto glass substrates [103]. Surprisingly, they found that eliminating cell contractility, actin stress fibers, LINC complex components, intermediate filaments, and microtubules had a minimal effect on nuclear deformation. In fact, the small effect these treatments had on nuclear deformation could be explained simply by changes in cell spread area. Only fully depolymerizing the actin cytoskeleton and stopping cell spreading prevented the initial flattening and elongation of the nucleus, which is consistent with additional data in the literature [31,58,102,104,105]. Furthermore, while cell contractility and intact LINC complexes are generally required to transmit extracellular loads to the nucleus [37,96,99], this is not universally true. For example, knockdown of nesprin-1 in endothelial cells actually increases nuclear strains in response to extracellular loading [106]. Finally, an exogenous compressive force applied to well-spread cells lacking a perinuclear actin cap induced remodeling of the nuclear lamina only when the cells were seeded on stiff substrates [58], suggesting that nuclear forces generated by both cell spreading and the actin cap are necessary to initiate nuclear mechanotransduction.
Together these data suggest that the overall nuclear shape is primarily dictated by passive forces generated within the actin cytoskeleton with cell spreading and that forces transmitted by the actin cap or LINC complexes contribute to a lesser degree. Specifically, it appears that nuclear height is largely determined by cell spread area, whereas lateral compression of the nucleus in elongated cells is driven by tension in the actin cap [32]. However, it is unclear what specific nuclear deformations are important for activating mechanotransduction mechanisms. That is, while cell spreading may determine global nuclear shape, the additional load and local deformations generated by actin cap stress fibers may be critical for nuclear mechanotransduction. For example, in mesenchymal stem cells and fibroblasts cytoskeletal connection to the nucleus and actomyosin contractility are necessary to transmit extracellular loads to the nucleus [37,96,99], which is a prerequisite for any nuclear mechanotransduction process. Beyond changes in nuclear shape induced by cell spreading, forces generated by actin cap stress fibers compact chromatin by reducing nuclear volume, which is associated with changes in cell proliferation [32]. Furthermore, nuclear loading by actin cap stress fibers produces significant local deformations in the apical nuclear surface and is necessary to induce remodeling of the nuclear lamina [58,59,92], which may be an important mediator of downstream signaling and gene transcription (discussed below). Finally, actin cap stress fibers are more sensitive than basal stress fibers to changes in substrate stiffness [89], suggesting that they are one of the primary actin structures involved in cell mechanotransduction.
Mechanisms of Nuclear Mechanotransduction
Mechanotransduction Via Physical Reorganization of Chromatin
Load-Induced Gene Activation and Repositioning.
The spatial organization of the genome has a strong influence on gene expression [76,81,107,108]. In particular, chromatin contained in LADs is generally heterochromatic and repressed [51,77,109,110]. Decondensation and detachment of heterochromatic gene loci from the nuclear periphery and their translocation to the nuclear interior are associated with elevated transcription. For example, during stem cell commitment and differentiation, lineage-specific genes are untethered from the nuclear lamina and are relocated to the nucleoplasm (Fig. 5) where they form intra- and interchromosomal interactions at transcription factories within the nucleus [78–80,111–113]. Furthermore, genes associated with pluripotency are repressed and also repositioned with differentiation [78,80,114].
Fig. 5.
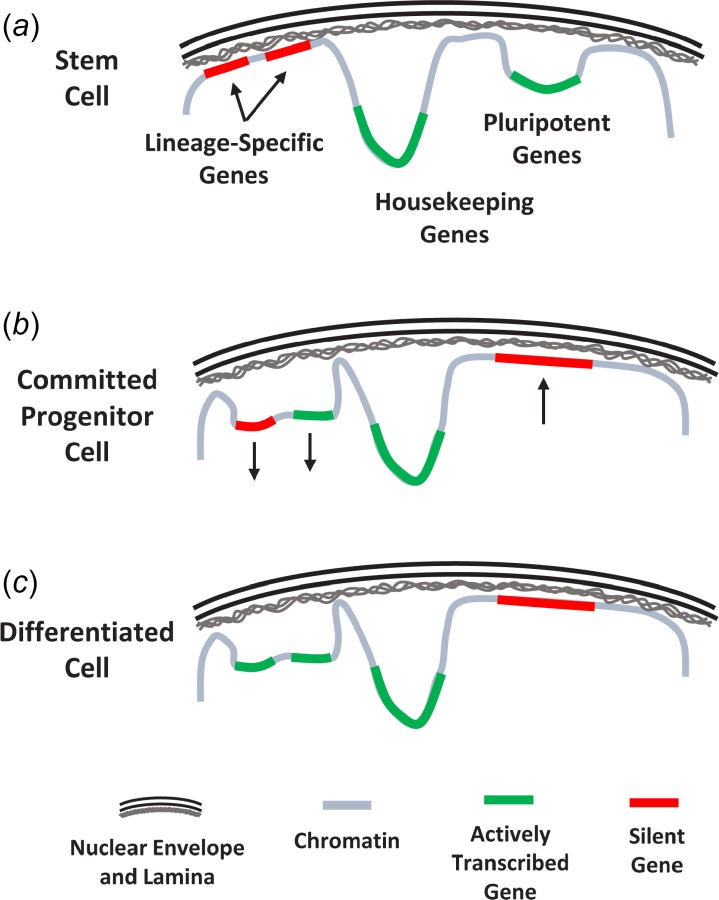
Relocation of gene loci within the nucleus during stem cell differentiation. (a) In stem cells, pluripotent and housekeeping genes are actively transcribed within the nuclear interior while lineage-specific genes are silenced at the nuclear periphery. (b) With lineage commitment, lineage-specific genes are detached from the nuclear lamina and relocated to the nuclear interior. In contrast, pluripotent genes are silenced and attach to the nuclear lamina. (c) Additionally, some lineage-specific genes remain inactive despite being displaced from the nuclear envelope and are transcribed only after terminal differentiation. Adapted with permission from Ref. [78]. Color figures are available online.
Based on these findings, it has been postulated that mechanical force might dislodge chromatin from the nuclear periphery and initiate gene activation. Numerous proteins attached to the inner nuclear membrane (including lamins) directly bind chromatin and act in a cooperative fashion to maintain LADs at the nuclear periphery [62,63,77,109,115–118]. Experiments utilizing fluorescence resonance energy transfer (FRET) demonstrated that extracellular forces can separate protein complexes within Cajal bodies inside the nucleoplasm [37]. Similar load-induced dissociation of protein complexes at the nuclear envelope could result in untethering LADs from the nuclear lamina, thereby initiating gene repositioning and/or transcription.
Rather than directly untethering gene loci, mechanical forces could also decondense silent heterochromatic genes at the nuclear lamina, which could allow better access for transcription machinery and initiate changes in gene transcription. Indeed, the forces required to alter chromatin condensation and potentially initiate transcription are physically reasonable. A force of approximately 5 pN is required to decondense a 30 nm chromatin fiber [119], which is the same as that required to unfold talin at a focal adhesion within the plasma membrane [120]. Additionally, the level of decondensation necessary to initiate transcription is likely small, given that gene loci expand only 1.5–3 times upon artificially induced transcription [121]. For context, this is about half the level of compaction present within a 30 nm chromatin fiber [122]. Furthermore, chromatin decondensation by itself is capable of untethering gene loci from the nuclear periphery [123].
An excellent recent study in this area demonstrated that chromatin stretching induced by exogenous forces is indeed capable of activating gene transcription [124]. Using magnetic beads, researchers from the labs of Wang and Belmont applied shear forces to the cell surface, which generated strains throughout the nucleus. More importantly, by inserting multiple copies of a fluorescently labeled bacterial artificial chromosome (BAC) into the same chromatin locus, they were able to directly observe the stretching of a single chromatin fiber. Furthermore, this stretching enabled the binding of RNA polymerase II to dihydrofolate reductase (DHFR) genes within the BAC construct and upregulated DHFR transcription. Fluorescent in situ hybridization demonstrated that the generated messenger RNA colocalized with the DHFR transgene and that upregulation occurred within 15 s, suggesting that the response was indeed due to the mechanical stretching of the chromatin and not signaling cascades. Finally, these effects required active cytoskeletal tension and chromatin tethering to the nuclear membrane via lamin A/C, SUN 1/2, and other proteins associated with the nuclear lamina. These results demonstrate for the first time that forces applied to the cell surface can physically stretch chromatin within the nucleus and that such stretching can activate local gene transcription. However, a major caveat is that this work focused on a transgene that was not under endogenous transcriptional regulation. That is, the inserted loci were not part of highly condensed heterochromatin attached to the nuclear periphery, but were rather likely pre-activated and located near the nuclear interior.
An example supporting gene activation of endogenous heterochromatin is seen with induced pluripotent stem cell (iPSC) reprograming. While heterochromatin is associated with gene repression, gene loci within these condensed regions are still generally accessible to transcription factors [125–127]. However, heterochromatin with trimethylation of histone 3 (i.e., H3K9me3) is impenetrable to the transcription factors Oct-4, Sox2, and Klf4, which poses a barrier for fibroblast reprograming [128]. Removal of these repressive marks, and presumably decondensation of the affected genes, is necessary for iPSC reprograming [129,130]. Interestingly, H3K9 methylation is particularly enriched in LADs and is associated with the nuclear lamina [79,130,131]. If mechanical force could promote the decondensation of these regions and allow transcription factor penetration, then increased nuclear loading should improve reprograming efficiency. Intriguingly, increasing nuclear elongation by culturing cells on patterned substrates improves reprograming efficiency fourfold [132]. Importantly, this effect required tension within actin cap stress fibers, suggesting that mechanical force transmitted to the nucleus mediated this improved reprograming. However, encapsulation of fibroblasts in soft hydrogels, which causes cell rounding and presumably reduced contractility and nuclear deformation, also improves reprograming [133]. Clearly, additional data are necessary to determine whether direct loading of intranuclear chromatin may contribute to iPSC reprograming and to cellular mechanotransduction in general.
Protection of Chromatin From Load Transmission.
There is also evidence suggesting that force-induced untethering or decondensation of chromatin at the nuclear lamina is an unlikely mechanism for driving changes in gene transcription and ultimately cell behavior. First of all, gene association with the nuclear lamina or the nuclear periphery is not uniformly repressive [76,107,114, 134–137]. Recruitment of RNA polymerase II and transcription can occur at the nuclear periphery and precede locus repositioning [111,138]. Conversely, gene silencing often precedes relocation to the periphery [139] and ectopic tethering to the nuclear envelope does not always impede expression [140–142]. Second, physically detaching genes from the nuclear envelope does not necessarily enhance their expression. In a study where inactive gene loci were artificially untethered from the nuclear lamina, the genes remained silent [143]. Additionally, genes naturally relocated away from the periphery do not become active until after additional modifications involving hyperacetylation and promoter–enhancer interactions [78,144]. Finally, translocation of gene loci from the nuclear periphery toward the nuclear interior is a highly coordinated process involving numerous factors and is not simply due to untethering from the nuclear envelope. For example, intranuclear actin polymerization and myosin activity is necessary to reposition gene loci, suggesting that translocation events are active processes driven by nuclear actomyosin motor complexes [145–148]. Furthermore, recurrent repositioning of gene loci to and from the nuclear periphery in embryonic stem cells is observed with circadian oscillation of gene expression and in the absence of mechanical stimulation [116].
Despite the complex processes involved in repositioning genes, mechanical stimuli applied to chromatin regions containing specific gene loci may still initiate such processes. This would imply that cell-specific mechanosensitive genes are positioned in a “mechanically poised” state at the nuclear periphery. Interestingly, while knocking out LMNA increased nuclear deformation, the expression of mechanosensitive genes was reduced in response to extracellular strains [64]. This suggests that widespread chromatin deformation alone is not sufficient for gene activation and that specific interactions between gene loci and the nuclear lamina (which are lost with lamin A/C knockout) are necessary to induce transcription of mechanically activated genes. This would further suggest that the genes associated with the nuclear lamina are cell-type specific. However, this does not appear to be the case.
After mitosis, the LADs that associate with the nuclear lamina are not conserved from mother to daughter cells [149]. Instead, a proportion of LADs randomly associate with the surface of nucleoli, which is also a site for heterochromatin positioning [150]. In fact, LADs share a significant overlap with nucleoli-associated domains (NADs) across the genome [151,152]. Although nuclear loading does deform intranuclear chromatin and structures (e.g., nucleoli) [34,37,124], the gene loci that assemble on the nucleoli surface are presumably partially insulated from mechanical forces applied at the nuclear periphery. The stochastic nature of the positioning of heterochromatin suggests that mechanosensitive genes are not targeted to the nuclear envelope for the purpose of mechanotransduction. This is consistent with the finding that disruption of the LINC complex, which reduced nuclear deformations in response to extracellular loading, did not alter fibroblast expression of numerous mechanosensitive genes [96]. However, in cardiomyocytes, disruption of the LINC complex via knockout of nesprins 1 and 2 does eliminate the expression of the same mechanosensitive genes [100]. This discrepancy may be due to the fact that, besides reducing nuclear loading, loss of nesprins 1 and 2 also alters the localization of other proteins associated with LINC complexes, including lamin A/C [100,153]. Therefore, it is unclear whether the different effects produced by LINC disruption between cell types are due to altered nuclear loading or altered tissue-specific signaling pathways involving the mislocalized proteins (discussed in detail below).
Consistent with this evidence against load-induced gene activation is the alternative hypothesis that the nuclear lamina serves to prevent forces from being transmitted to the nuclear interior and protect the delicate chromatin structure and organization from force-induced disruption. As mentioned above, the nuclear lamina is the primary structural support for the nuclear envelope. In cells lacking lamin A/C, large nuclear deformations and chromatin displacements are observed as a result of applied extracellular forces or active actomyosin contraction [33,64,154,155] (Fig. 6). Such disturbances of the nucleus may negatively impact cell behavior. For example, mutations in the gene coding for lamin A/C result in numerous disorders (termed laminopathies) that primarily affect mechanically loaded tissues like muscle and bone [68,69]. This suggests a mechanical origin to these diseases resulting from defects in the nuclear lamina and alterations in nuclear mechanics or mechanotransduction. Indeed, lamin mutations that produce muscular disorders also reduce the stiffness of cell nuclei, whereas laminopathies that lack muscular phenotypes do not affect nuclear mechanics [156]. Recent data also suggest that nuclear lamins help prevent nuclear rupture, DNA breakage, and chromatin damage induced during cell migration through tight pores, which may be related to cancer development and metastasis [157–160].
Fig. 6.
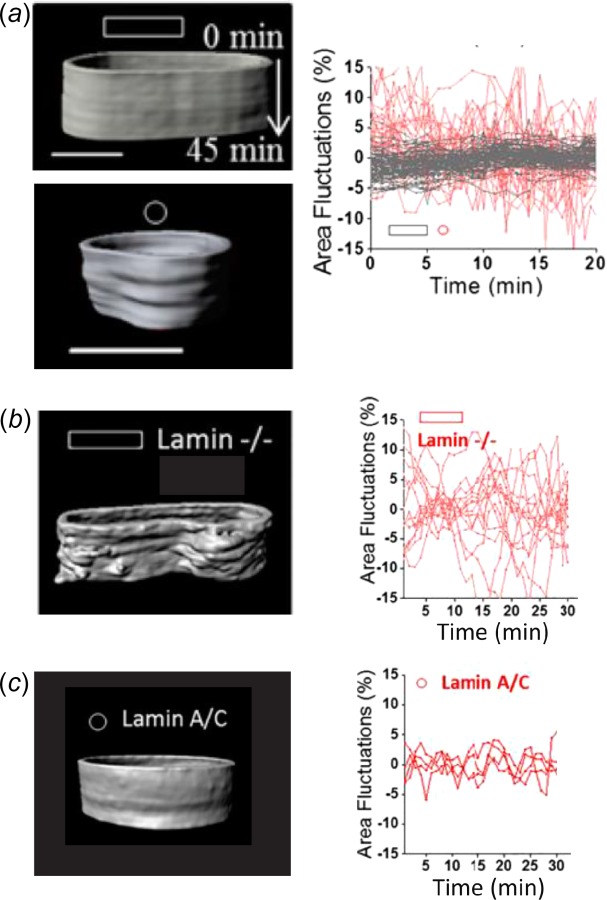
A stiff nuclear lamina prevents large deformations of the nucleus. (a) Kymographs of the nuclear envelope demonstrate that elongated cells on rectangular micropatterned islands (gray lines) have significantly smaller local perturbations of the nuclear surface compared to round cells on circular islands (red lines). These differences in the stability of the nuclear envelope are due to remodeling of the nuclear lamina, since (b) knockout of lamin A/C in elongated cells produces larger fluctuations in the nuclear surface, whereas (c) overexpression of lamin A/C in round cells has the opposite effect. Adapted with permission from Ref. [154]. Color figures are available online.
To prevent such damage, the nuclear lamina actively remodels itself in response to forces applied to the nucleus. When loaded, lamin A/C undergoes a conformational change that enhances its assembly at the nuclear envelope and protects it from being phosphorylated and ultimately degraded [56–60]. This self-assembly process is fully coordinated by the nucleus itself, as mechanical stimulation of isolated nuclei is sufficient to initiate lamin A/C assembly at the nuclear envelope [41]. This demonstrates that one potential outcome of nuclear mechanotransduction is a stiffening of the nuclear membrane, which may protect the nucleus and its contents from further loading [161]. In fact, lamin A/C content in primary cells from various tissues scales with the in situ tissue stiffness, suggesting that a stiffer lamina is necessary to protect cells from a stiffer microenvironment [56]. However, it is unclear if cells in a stiff extracellular matrix experience increased loads/strains since the stiffer microenvironment would shield the cells from mechanical forces [162,163]. As discussed below, it is also possible that such nuclear stiffening serves to enhance tension in the cytoskeleton and increase the sensitivity of mechanotransduction mechanisms at the nuclear envelope, within the cytoplasm, or at the plasma membrane.
Mechanotransduction at the Nuclear Envelope
Signaling at the Inner Nuclear Membrane.
While load may not directly reposition chromatin or alter endogenous gene expression, it may initiate a remodeling process at the nuclear envelope that plays an important role in downstream signaling pathways. This distinction is best exemplified by the numerous laminopathies that result from defects in the LMNA gene [68,69]. One of the most well-studied laminopathies is Hutchinson–Gilford progeria syndrome (HGPS), which is characterized by accelerated aging and early death [55]. This disease is caused by a C1824T mutation that alters the splicing of LMNA mRNA, preventing the removal of the farnesylated end of the protein. In cells from HGPS patients, this results in increased lamin A at the nuclear periphery and increased nuclear stiffness, which is similar to the effects of lamin A overexpression [164–166]. However, mouse models of progeria, which exhibit remarkably similar phenotypes to HGPS patients, have softer nuclei and reduced nuclear loading due to the lack of actin cap stress fibers as well as a lack of properly organized LINC complexes [31,167,168]. The fact that both nuclear stiffening and softening produce similar disease states suggests that altered nuclear mechanics and mechanotransduction are not necessarily the cause of disease.
Instead, the tissue-specificity of laminopathies may be due to the elimination of binding sites in the mutated lamin A protein for specific gene loci, chromatin remodeling complexes, or other nuclear proteins. For example, chromosome positioning is altered within HGPS cells and the actomyosin machinery that actively repositions chromosomes is disabled [169]. HGPS cells also display a loss of heterochromatin, with specifically fewer H3K27me3 and H3K9me3 marks as well as altered localization and chromatin association of heterochromatin protein 1 (HP1) [170]. Furthermore, the LMNA mutation responsible for Emery–Dreifuss muscular dystrophy (EDMD) blocks muscle-specific gene relocation [171]. Finally, mutations associated with both HGPS and EDMD affect F-actin bundling by lamin A, which may be the cause of the inhibited gene relocation [172]. These findings suggest that the diversity and tissue-specificity of laminopathies may be due to altered binding of the nuclear lamina with chromatin and tissue-specific nuclear proteins [173] rather than altered mechanotransduction.
Similarly, the mechanisms by which nuclear loading affects gene transcription may be mediated by remodeling of the nuclear lamina, which then changes the binding properties of nuclear envelope proteins and transcription factors. Numerous lamina-associated polypeptides (LAPs) directly bind lamins at the nuclear envelope and play important roles in chromatin binding, histone modification, assembly of protein complexes, and transcription factor sequestration [63,174] (Fig. 7). For example, LAP2α mediates soluble lamin A/C binding to actively transcribed euchromatin within the nuclear interior [175], whereas lamin-B receptor (LBR) mediates binding of heterochromatin to the nuclear periphery [176]. LAP2β, emerin, and MAN1 each contain LEM domains that bind barrier-to-autointegration factor (BAF), which mediates chromatin binding to the nuclear envelope [63]. The CCCTC-binding factor (CTCF) is an insulator that spatially organizes the genome into TADs, controls heterochromatin spreading, and connects LADs to the nuclear lamina [77,116–118]. Many of these LAPs also bind and sequester numerous transcription factors at the nuclear periphery, including c-Fos, sterol regulatory element-binding protein 1 (SREBP1), retinoblastoma protein, germ cell-less (GCL), octamer-binding transcription factor 1 (Oct-1), β-catenin, and Smads [45,174]. It is also worth noting that small isoforms of nesprins 1 and 2 exist within the nucleoplasm and directly bind lamin A, emerin, and β-catenin [177–179]. Force-induced remodeling of the nuclear lamina may alter the affinity of these LAPs with the nuclear envelope, thereby affecting chromatin binding, enhancer–promoter interactions, and transcription factor accessibility, ultimately altering gene expression.
Fig. 7.
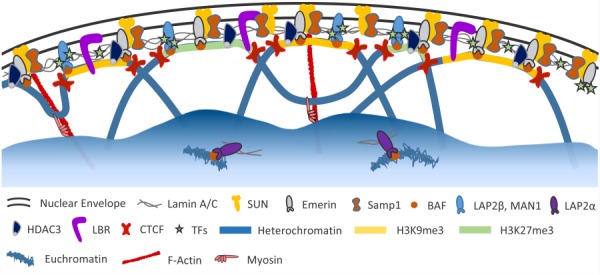
Proteins associated with the nuclear lamina. In addition to the components of the LINC complex, numerous lamina-associated polypeptides (LAPs) bind to the nuclear lamina and serve various functions. Emerin, LAP2β, and MAN1 help connect heterochromatic LADs to the nuclear lamina via their interaction with BAF. In addition, HDAC3 and LBR directly bind chromatin. These proteins, as well as the nuclear lamina itself, also bind various transcription factors (TFs) involved in important signaling pathways (e.g., c-Fos, SREB1, β-catenin, and Smads). CTCF helps position chromatin at the nuclear envelope and also flanks regions of histone modifications associated with gene silencing (i.e., H3K9me3 and H3K27me3). Soluble lamin A/C dimers associate with actively transcribed euchromatin within the nuclear interior via LAP2α. Lamin A/C and emerin are also likely involved in the actomyosin machinery potentially responsible for relocating gene loci to the nuclear interior upon activation. Color figures are available online.
Two particularly compelling examples of mechanical loading influencing factor binding to the nuclear lamina involve emerin and histone deacetylase 3 (HDAC3). As mentioned above, force applied to isolated nuclei via nesprins increases lamin A assembly at the nuclear envelope and stiffens the nucleus. This is dependent on emerin phosphorylation by Src, which likely alters emerin binding with lamin A and reinforces binding between SUN and the nuclear lamina [41]. In living cells, while emerin knockout does not change nuclear deformation in response to extracellular strain, the expression of mechanosensitive genes is repressed [155]. This may be linked to the role emerin plays in sequestering various transcription factors at the nuclear periphery. For example, binding of β-catenin to emerin negatively regulates Wnt signaling by restricting its accumulation within the nucleoplasm [180]. Furthermore, emerin maintains the proper position of myosin IIB at the nuclear membrane [181] and nucleates actin polymerization [182], which suggests that it could be a critical component of the actomyosin machinery used to relocate gene loci and activate transcription [183].
Like other HDACs, HDAC3 inactivates gene transcription by removing acetyl groups from lysines on H3 and H4 core histones, which compacts chromatin in a more condensed state [184]. Beyond its catalytic activity, HDAC3 also mediates heterochromatin binding to the nuclear lamina [185,186]. Furthermore, HDAC3 nuclear localization is dependent on cell contractility, with greater contractility leading to less nuclear retention and increased histone acetylation [187]. Interestingly, histone hyperacetylation resulting from tension in actin cap stress fibers is localized to the nuclear lamina [59] and lamin A/C knockdown eliminates the mechanically mediated repression of HDAC activity [188]. Decreased HDAC activity and increased histone hyperacetylation were also shown to contribute to the increased iPSC reprograming efficiency as a result of nuclear loading and elongation on microgrooved substrates [132]. Finally, emerin also directly binds HDAC3 at the nuclear envelope and together they coordinate the nuclear positioning and expression of the myogenic regulatory factors Myf5, MyoD, and Pax7 during myogenesis [80,189]. These findings strongly suggest that binding of emerin and HDAC3 to the nuclear lamina is sensitive to nuclear loading and that force-induced alterations of their interaction with the lamina affect histone modifications, chromatin condensation, gene positioning, transcription, and cell fate.
Signaling at the Outer Nuclear Membrane.
Mechanical stimuli may also alter protein complexes located on the outer nuclear membrane. The LINC complex is slowly being appreciated as an organizational structure on par with focal adhesions at the plasma membrane, with numerous interacting subcomponents that dynamically assemble and disassemble in response to various signals, including mechanical load [45,88]. While components of the LINC complex are distributed across the nuclear envelope [58,59], nesprins 1 and 2 (and the corresponding SUN proteins within the inner nuclear membrane) are enriched at sites along actin cap stress fibers running over the nuclear surface [90,92,94]. This suggests that LINC complexes may form higher-order structures in response to mechanical force similar to focal adhesions. Indeed, nesprins can interact directly and through supporting proteins like formin homology 2 domain containing 1 (FHOD1) [86,190–192], which could potentially form interconnected assemblies of individual LINC complexes. Additionally, despite the concentration of LINC complex proteins at actin cap stress fibers, tension within these structures at the nuclear envelope is uniform across the nuclear surface, suggesting that LINC complex enrichment may be driven by achieving an optimal tensional load [93]. Formation and maintenance of LINC complexes at the nuclear envelope are also likely influenced by the remodeling of the nuclear lamina, given that numerous LINC complex proteins are improperly localized with lamin A/C mutations and knockouts in some cell types [82,168,178,193–195]. Finally, the outer nuclear membrane contains mechanosensitive ion channels and functions as an activation scaffold for numerous enzymes, suggesting that stretch of the nuclear envelope itself may initiate mechanotransduction [196].
Mechanotransduction at the Plasma Membrane Resulting From Nuclear Remodeling.
In addition to possible mechanotransduction mechanisms that may take place within the nucleus or at the nuclear envelope, recent evidence suggests that nuclear remodeling in response to load may also influence mechanotransduction events throughout the cytoplasm and even at focal adhesions [197]. As the stiffest organelle in the cell, the mechanical properties of the nucleus strongly affect the mechanics of the entire cell [198]. For example, embryonic stem cells, which lack lamin A/C, have nuclei and cytoplasms that are more compliant than terminally differentiated cells [67]. Furthermore, knocking out lamin A in differentiated cells not only reduces the nuclear stiffness to that of naive stem cells [199] but also reduces the cytoplasmic stiffness of the cell [64,168]. Similar reductions in cytoplasmic stiffness are also seen with LINC complex disruption [200].
Such changes in cytoplasmic stiffness are likely mediated by alterations in cytoskeletal organization in response to remodeling of the nuclear structure. As mentioned above, alterations in the nuclear lamina can disrupt the integrity and function of the LINC complex [168,178,193–195], which interfaces with the cytoskeleton. This then disrupts or eliminates the actin cap stress fibers [31,101,102], which are a primary contractile element of the actin cytoskeleton [89], as well as inhibits F-actin polymerization [201,202]. Furthermore, loss of lamin A/C or LINC complex components can also disrupt the architecture of intermediate filaments and microtubules within the cytoplasm [54,96,203]. These changes in cytoskeletal organization and tension affect mechanotransduction at focal adhesions and potentially throughout the cytoplasm via mechanosensitive actin binding partners, including filamin, α-actinin, and 14-3-3 proteins [16–19]. For example, disruption of the LINC complex leads to alterations in focal adhesion size and number [89,168,204] as well as altered nuclear localization of important transcription factors associated with the cytoskeleton, like yes-associated protein (YAP) and megakaryoblastic leukemia (translocation) 1 (MKL1) [99,205,206]. Finally, cytoskeletal architecture is further modulated by downstream changes in expression of numerous proteins involved in the organization of actomyosin and intermediate filaments [57,207]. These data suggest the existence of a dynamic feedback loop (Fig. 8), where force-induced nuclear remodeling leads to reorganization of the cytoskeletal structure, which in turn alters the loads transmitted to the nucleus and affects mechanotransduction processes acting within the cytoplasm and at the plasma membrane.
Fig. 8.
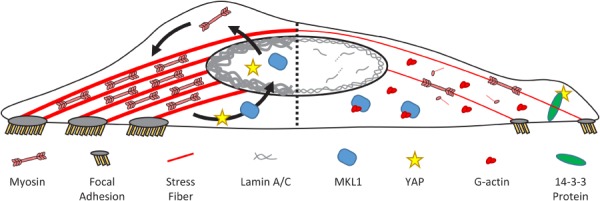
Changes in biophysical stimuli dynamically modulate nuclear and cytoskeletal structure. (Left) Force applied to the nucleus promotes assembly of the lamina and nuclear stiffening. This in turn increases the forces at focal adhesions generated by actomyosin contractility, which leads to further growth of focal adhesions and stress fibers. Increased actin polymerization and nuclear loading induce import of transcription factors (e.g., MKL1 and YAP), which drive further structural remodeling in the cytoplasm via upregulation of several cytoskeletal proteins (e.g., myosin-IIA). (Right) Loss of nuclear loading causes disassembly and degradation of lamin A/C. This softens the nucleus and disrupts existing LINC complexes, causing reductions in stress fiber and focal adhesion size as well as cytoskeletal tension. Increased levels of G-actin and loss of nuclear loading lead to sequestration of MKL1 and YAP within the cytoplasm and downregulation of cytoskeletal proteins.
Mathematical Models of Nuclear Mechanotransduction
Clearly, future work is necessary to conclusively determine the specific mechanisms underlying nuclear mechanotransduction and to decipher their interplay with one another as well as with additional mechanotransduction processes located in the cytoplasm or at the plasma membrane. Use of cutting edge experimental techniques on living cells, including molecular force probes [93], labeling of individual gene loci [124,145,208], super-resolution and fluorescence-lifetime imaging microscopy (FLIM) [209,210], and methods for applying localized force [33,211], will further our understanding of how load transmitted to the nucleus alters gene expression [45]. In addition, the development and application of new mechanical models that capture load transmission through the cytoskeleton and to the nucleus, nuclear remodeling, and the changing mechanics of the nuclear lamina and chromatin can be a powerful tool for testing specific hypotheses concerning the mechanisms of nuclear mechanotransduction.
Numerous mechanical models have been used to successfully replicate cell mechanics and various cell behaviors at the whole cell level as well as the mechanical behavior of individual intracellular structures [212–214], including focal adhesions [215–223] and the actin cytoskeleton [224–232]. Several models have additionally included force transmission through the cytoskeleton and to the nucleus [225,227,233]. While these models incorporate phenomenological representations describing the mechanics of the nucleus [103,220,233–238], they generally neglect the mechanical response of specific nuclear substructures, like the nuclear lamina and intranuclear chromatin. However, mechanical models of these nuclear components currently exist. For example, network models have been used to explain the behavior of the lamina during nuclear swelling [239,240] and even the formation of nuclear blebs observed in cells from HGPS patients [241]. Models based on polymer physics and molecular dynamics have also successfully replicated key features of chromatin organization and mechanics, including nucleosome folding and chromatin condensation [242], forced chromatin decondensation with the application of load [119,243], and even looping out of gene loci from their chromosomal territories [244]. Furthermore, the self-organization of replication, transcription, and splicing factories within the nucleus has been explained by entropy-driven processes [245,246].
Incorporating the mechanics of nuclear substructures into whole cell models is required in order to test the hypotheses regarding nuclear mechanotransduction described above. To demonstrate the power and need for such modeling approaches, consider a simple calculation regarding how force is transmitted to a lamina-associated chromatin fiber due to a local force applied at the nuclear envelope and the likelihood that this could initiate chromatin decondensation (Fig. 9). Micropipette aspiration has provided an estimate of the network elastic modulus of the nuclear lamina as approximately 25 pN/nm in differentiated fibroblasts and mesenchymal stem cells [56,199,239]. This measurement was based on the Young-Laplace equation [239], which can be converted to a force–displacement relation
Fig. 9.
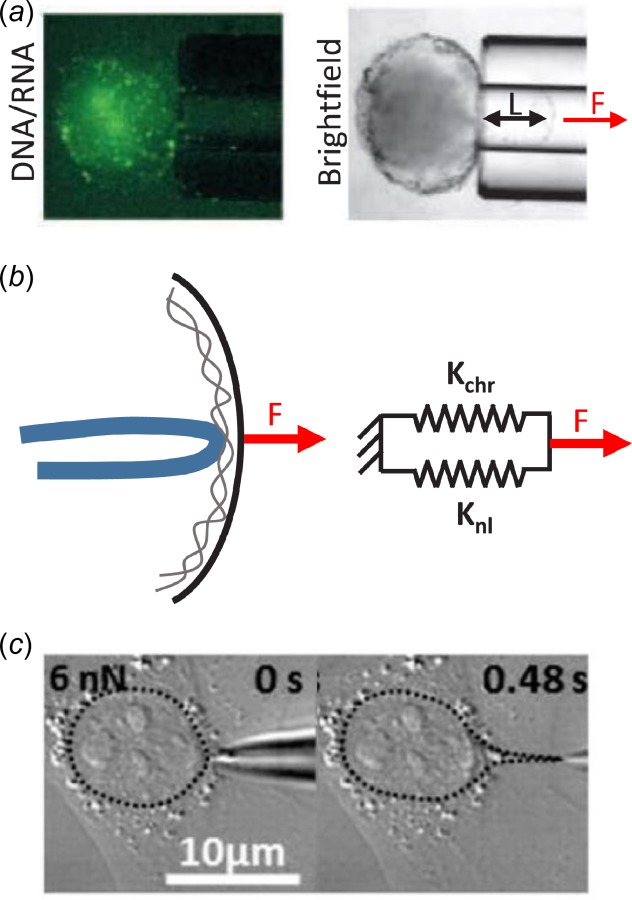
Simple calculation of forces required to locally deform nuclear lamina and decondense chromatin. (a) Application of local force (F) via micropipette aspiration of isolated nuclei displaces the nuclear lamina a distance L, while the intranuclear chromatin is excluded from the pipette lumen. This provides an estimate of the network elastic modulus of the nuclear lamina. (b) Force applied to the nuclear envelope is transmitted to the nuclear lamina and attached chromatin fiber, which act in parallel and have stiffnesses Knl and Kchr, respectively. (c) Micropipette aspiration of live adherent fibroblasts produces substantial local deformation of the nucleus, which is more than sufficient to decondense chromatin and potentially initiate gene transcription. Adapted with permission from Refs. [33] and [239].
| (1) |
where F is the force applied at a point on the nuclear lamina, L is the radial displacement of that point, and E is the network elastic modulus of the nuclear lamina. The term 2πE represents the stiffness of the lamina in response to a force applied perpendicular to its surface and is approximately 157 pN/nm. Additionally, tensile testing of condensed individual chromatin fibers demonstrates that they have a stiffness of approximately 0.0125 pN/nm and decondense at about 5 pN [119]. Assuming that the far-field displacement of the nucleus is zero and that the chromatin fiber and nuclear lamina are loaded in parallel, these estimates suggest that only about 0.008% of the load applied to the nuclear lamina is transmitted to the chromatin. Furthermore, if we assume that an actin stress fiber terminates at the nuclear surface, applying the full 5 nN of force generated at a typical focal adhesion to a point on the nucleus [247,248], this would produce a displacement of about 32 nm at the nuclear envelope and a force on the chromatin fiber of about 0.4 pN, which are both about an order-of-magnitude too low to induce decondensation of the chromatin fiber [119]. This would contradict the hypothesis that nuclear mechanotransduction is mediated by direct manipulation of chromatin organization and supports the idea that assembly of lamin A/C at the nuclear periphery and nuclear stiffening in response to load protects chromatin from forces transmitted by the cytoskeleton. However, recent data demonstrate that 5 nN applied to the nuclear surface of live adherent fibroblasts is sufficient to produce localized nuclear protrusions of about 4 μm in length [33], which is 100 times larger than the estimate calculated above and more than sufficient to decondense (or possibly untether) gene loci from the nuclear periphery. Clearly, improved mechanical models and additional measurements of the dynamic mechanical properties of the nucleus and subnuclear structures are needed to investigate how loads impact nuclear mechanics and alter cellular mechanotransduction.
Finally, inclusion of active remodeling of the nuclear lamina and the cytoskeleton would elucidate the mechanical interplay between the two cell compartments. Such an approach would enable the modeling of dynamic mechanotransduction processes, similar to the benefits provided by cell models incorporating active cytoskeletal contraction [214,219,220,222,225–227,230,231, 249–255]. Furthermore, these models could capture the influence of load-induced nuclear remodeling on mechanotransduction processes that occur within the cytoplasm and plasma membrane. For example, we recently showed that stiffening of the nucleus leads to the formation of larger focal adhesions [220]. Additionally, Zemel demonstrated that nuclear stiffness and the entropic swelling forces of chromatin can affect actomyosin organization [234]. The complexity of representing numerous dynamic structures across a wide range of length and time scales is of course a large obstacle for developing such models [45,213]. Novel approaches, like the cytoskeleton divided medium (CDM) model, can overcome such obstacles by representing the cell and intracellular structures with dynamic particles that interact via simple force–displacement relations [256]. This approach has recently been used to successfully recapitulate the dynamic remodeling of the cytoskeleton and nuclear deformations in response to cell spreading and contraction [257,258]. Such advances in mechanical models are essential for determining the role of nuclear loading and mechanotransduction on gene expression, cell behavior, and differentiation.
Conclusions
Substantial evidence suggests that nuclear loading plays an important role in cellular mechanotransduction. However, the specific mechanisms that mediate these effects remain elusive. Of the three main hypotheses discussed in this review, alterations in intranuclear signaling pathways as a result of force-induced remodeling of the nuclear lamina appear to be the most clearly supported mechanism for nuclear mechanotransduction. The inner nuclear membrane and nuclear lamina provide a scaffolding for a vast array of important proteins and transcription factors (Fig. 7), and the interactions between these elements are highly sensitive to lamina assembly and remodeling. Furthermore, mutations in lamins and inner nuclear membrane proteins (e.g., emerin) alter cell function and response to mechanical loading, even in cases where the nuclear mechanics remain unaffected [155,156]. Still, additional research is necessary to identify the means by which nuclear loading alters protein interactions at the nuclear envelope and to clearly define the intranuclear signaling pathways that lead to changes in gene transcription.
Less is known about the potential effects of load transmission to intranuclear chromatin or the effects of nuclear mechanics on mechanotransduction processes outside the nucleus. Directly observing load-induced changes in chromatin organization or positioning is difficult, and isolating the effects of such physical manipulations from the myriad other known (and unknown) mechanosensitive pathways has been a challenge. While a recent study demonstrated that stretching of transgenic chromatin due to forces applied at the cell surface can alter gene expression [124], it is unclear if similar mechanisms regulate the activation of endogenous genes, particularly those silenced within heterochromatin at the nuclear periphery. Additionally, significant evidence exists to refute this hypothesis and suggests that nuclear remodeling is an attempt to protect chromatin from extracellular forces or cell-mediated contraction [161]. Alternatively, recent data suggest that remodeling of the nuclear lamina and the resulting changes in nuclear stiffness affect cytoskeletal tension and mechanotransduction processes within the cytoplasm. Beyond additional experiments, improved mechanical models incorporating subnuclear structures as well as cytoskeletal and focal adhesion organization would be extremely valuable to test these hypotheses and elucidate the mechanisms of nuclear mechanotransduction.
Apart from the specific questions addressed in this review, significant other open areas remain to be investigated. For example, besides assembly of lamin A/C at the nuclear periphery, the nuclear envelope also undergoes numerous remodeling activities including nuclear pore complex formation, release of large ribonucleoprotein particles too big to pass through nuclear pores, microautophagy of the nucleus, and nuclear envelope dissolution during mitosis [259]. Each of these processes requires local breakdown of the nuclear lamina and reorganization of the nuclear envelope as a whole. Interestingly, much of the same machinery involved in such remodeling is also necessary for the repair of nuclear rupture resulting from migration through constricted micropores [157,158]. Still, how these processes work in concert to establish or interrupt nuclear mechanotransduction processes is completely unknown [259,260]. Additionally, while apical stress fibers have been shown to be critical elements of nuclear mechanotransduction, much of this research has been performed with cells on two-dimensional substrates. Other than in the endo- or epithelium, cells in native three-dimensional contexts lack this apical–basal polarization [261], and it is unclear whether such apical stress fibers (or stress fibers generally) even form [3,262,263]. While recent work shows that deep nuclear invaginations produced by actin and intermediate filaments exist in three-dimensional organoid cultures [264], additional work is still needed to evaluate the role of nuclear mechanotransduction in native cell environments.
Finally, the persistence of changes in nuclear structure and mechanics may play an important role in the establishment of stable cellular phenotypes. For example, nuclei in cells that migrated through small micropores or cells exposed to shear loading remain deformed and stiffened even after isolation from the cell [265,266]. Similarly, the effects of substrate stiffness or extracellular loading persist even after cells are exposed to new environments [1,267–269]. It is possible that such “mechanical memory” is a result of permanent alterations in nuclear structure and mechanical properties. Clearly, many open questions remain in the rapidly changing field of nuclear mechanotransduction with new and exciting discoveries constantly emerging. Indeed, during the publication of this review article, two particularly relevant studies were released. One investigates the role of nuclear mechanics on cellular mechanosensation and mesenchymal stem cell differentiation [270]. The other provides a chemomechanical model to describe nuclear deformation and rupture during cell migration through small constrictions [271]. The combination of such sophisticated experimental techniques and advanced mathematical models will continue to enhance our understanding of the role of the nucleus in the mechanotransduction processes driving numerous critical cell functions.
Acknowledgment
This work was supported by the National Institutes of Health (NIBIB R01 EB002425, NIAMS R01 AR056624, NIAMS T32 AR053461, and NIAMS F32 AR070562) and by the National Science Foundation Science and Technology Center for Engineering Mechanobiology (CMMI-1548571).
Contributor Information
Spencer E. Szczesny, Department of Orthopaedic Surgery, , University of Pennsylvania, , 424 Stemmler Hall, , 36th Street and Hamilton Walk, , Philadelphia, PA 19104; , Translational Musculoskeletal Research Center, , Corporal Michael J. Crescenz Veterans Affairs , Medical Center, , 3900 Woodland Avenue, , Philadelphia, PA 19104
Robert L. Mauck, Department of Orthopaedic Surgery, , University of Pennsylvania, , 424 Stemmler Hall, , 36th Street and Hamilton Walk, , Philadelphia, PA 19104; , Translational Musculoskeletal Research Center, , Corporal Michael J. Crescenz Veterans Affairs , Medical Center, , 3900 Woodland Avenue, , Philadelphia, PA 19104;; Department of Bioengineering, , University of Pennsylvania, , 240 Skirkanich Hall, , 210 South 33rd Street, , Philadelphia, PA 19104 , e-mail: lemauck@mail.med.upenn.edu
Nomenclature
- BAC =
bacterial artificial chromosome
- BAF =
barrier-to-autointegration factor
- CDM =
cytoskeleton divided medium
- CFP =
cyan fluorescent protein
- CTCF =
CCCTC-binding factor
- DHFR =
dihydrofolate reductase
- DNA =
deoxyribonucleic acid
- E =
network elastic modulus of the nuclear lamina
- EDMD =
Emery–Dreifuss muscular dystrophy
- F =
force
- F-actin =
filamentous actin
- FHOD1 =
formin homology 2 domain containing 1
- FLIM =
fluorescence-lifetime imaging microscopy
- FRET =
fluorescence resonance energy transfer
- G-actin =
globular actin
- GCL =
germ cell-less
- HDAC =
histone deacetylase
- HGPS =
Hutchinson–Gilford progeria syndrome
- HP1 =
heterochromatin protein 1
- iPSC =
induced pluripotent stem cell
- Kchr =
stiffness of chromatin fiber
- Knl =
stiffness of nuclear lamina
- Klf4 =
Kruppel-like factor 4
- KASH =
Klarsicht, ANC-1, SYNE/nesprin homology
- L =
radial displacement of the nuclear lamina
- LAD =
lamina-associated domain
- LAP =
lamina-associated polypeptide
- LBR =
lamin-B receptor
- LEM =
LAP2, emerin, MAN1
- LINC =
linker of nucleoskeleton and cytoskeleton
- mRNA =
messenger ribonucleic acid
- MKL1 =
megakaryoblastic leukemia (translocation) 1
- Myf5 =
myogenic factor 5
- MyoD =
myogenic differentiation 1
- NAD =
nucleoli-associated domain
- Oct-1/4 =
octamer-binding transcription factor 1/4
- Pax7 =
paired box 7
- PRC1 =
polycomb-group repressive complex 1
- RGD =
arginylglycylaspartic acid
- SMN =
survival motor neuron
- SOX2 =
SRY (sex determining region Y)-box 2
- SREBP1 =
sterol regulatory element-binding protein 1
- SUN =
Sad1, UNC-84
- TAD =
topologically associating domain
- TAN =
transmembrane actin-associated nuclear
- TRAAK =
TWIK related arachidonic acid activated K+ channel
- TREK =
TWIK related K+ channel
- TRPV4 =
transient receptor potential cation channel, subfamily V, member 4
- YAP =
yes-associated protein
- YFP =
yellow fluorescent protein
References
- [1]. Engler, A. J. , Sen, S. , Sweeney, H. L. , and Discher, D. E. , 2006, “ Matrix Elasticity Directs Stem Cell Lineage Specification,” Cell, 126(4), pp. 677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- [2]. McBeath, R. , Pirone, D. M. , Nelson, C. M. , Bhadriraju, K. , and Chen, C. S. , 2004, “ Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment,” Dev. Cell, 6(4), pp. 483–495. 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- [3]. Baker, B. M. , Trappmann, B. , Wang, W. Y. , Sakar, M. S. , Kim, I. L. , Shenoy, V. B. , Burdick, J. A. , and Chen, C. S. , 2015, “ Cell-Mediated Fibre Recruitment Drives Extracellular Matrix Mechanosensing in Engineered Fibrillar Microenvironments,” Nat. Mater., 14(12), pp. 1262–1268. 10.1038/nmat4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Przybyla, L. , Muncie, J. M. , and Weaver, V. M. , 2016, “ Mechanical Control of Epithelial-to-Mesenchymal Transitions in Development and Cancer,” Annu. Rev. Cell Dev. Biol., 32, pp. 527–554. 10.1146/annurev-cellbio-111315-125150 [DOI] [PubMed] [Google Scholar]
- [5]. Duscher, D. , Maan, Z. N. , Wong, V. W. , Rennert, R. C. , Januszyk, M. , Rodrigues, M. , Hu, M. , Whitmore, A. J. , Whittam, A. J. , Longaker, M. T. , and Gurtner, G. C. , 2014, “ Mechanotransduction and Fibrosis,” J. Biomech., 47(9), pp. 1997–2005. 10.1016/j.jbiomech.2014.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Cui, Y. , Hameed, F. M. , Yang, B. , Lee, K. , Pan, C. Q. , Park, S. , and Sheetz, M. , 2015, “ Cyclic Stretching of Soft Substrates Induces Spreading and Growth,” Nat. Commun., 6, p. 6333. 10.1038/ncomms7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Kurpinski, K. , Chu, J. , Hashi, C. , and Li, S. , 2006, “ Anisotropic Mechanosensing by Mesenchymal Stem Cells,” Proc. Natl. Acad. Sci. U.S.A., 103(44), pp. 16095–16100. 10.1073/pnas.0604182103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Johnson, B. D. , Mather, K. J. , and Wallace, J. P. , 2011, “ Mechanotransduction of Shear in the Endothelium: Basic Studies and Clinical Implications,” Vasc. Med., 16(5), pp. 365–377. 10.1177/1358863X11422109 [DOI] [PubMed] [Google Scholar]
- [9]. Schwartz, M. A. , 2010, “ Integrins and Extracellular Matrix in Mechanotransduction,” Cold Spring Harbor Perspect. Biol., 2(12), p. a005066. 10.1101/cshperspect.a005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Janmey, P. A. , Wells, R. G. , Assoian, R. K. , and McCulloch, C. A. , 2013, “ From Tissue Mechanics to Transcription Factors,” Differ. Res. Biol. Diversity, 86(3), pp. 112–120. 10.1016/j.diff.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Zaidel-Bar, R. , Itzkovitz, S. , Ma'ayan, A. , Iyengar, R. , and Geiger, B. , 2007, “ Functional Atlas of the Integrin Adhesome,” Nat. Cell Biol., 9(8), pp. 858–867. 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Kanchanawong, P. , Shtengel, G. , Pasapera, A. M. , Ramko, E. B. , Davidson, M. W. , Hess, H. F. , and Waterman, C. M. , 2010, “ Nanoscale Architecture of Integrin-Based Cell Adhesions,” Nature, 468(7323), pp. 580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Geiger, B. , Spatz, J. P. , and Bershadsky, A. D. , 2009, “ Environmental Sensing Through Focal Adhesions,” Nat. Rev. Mol. Cell Biol., 10(1), pp. 21–33. 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- [14]. Yan, J. , Yao, M. , Goult, B. T. , and Sheetz, M. P. , 2015, “ Talin Dependent Mechanosensitivity of Cell Focal Adhesions,” Cell. Mol. Bioeng., 8(1), pp. 151–159. 10.1007/s12195-014-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Harburger, D. S. , and Calderwood, D. A. , 2009, “ Integrin Signalling at a Glance,” J. Cell Sci., 122(2), pp. 159–163. 10.1242/jcs.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Zhou, A.-X. , Hartwig, J. H. , and Akyürek, L. M. , 2010, “ Filamins in Cell Signaling, Transcription and Organ Development,” Trends Cell Biol., 20(2), pp. 113–123. 10.1016/j.tcb.2009.12.001 [DOI] [PubMed] [Google Scholar]
- [17]. Craig, D. H. , Haimovich, B. , and Basson, M. D. , 2007, “ α-Actinin-1 Phosphorylation Modulates Pressure-Induced Colon Cancer Cell Adhesion Through Regulation of Focal Adhesion Kinase-Src Interaction,” Am. J. Physiol. Cell Physiol., 293(6), pp. C1862–C1874. 10.1152/ajpcell.00118.2007 [DOI] [PubMed] [Google Scholar]
- [18]. Sluchanko, N. N. , and Gusev, N. B. , 2010, “ 14-3-3 Proteins and Regulation of Cytoskeleton,” Biochem. Biokhimiia, 75(13), pp. 1528–1546. 10.1134/S0006297910130031 [DOI] [PubMed] [Google Scholar]
- [19]. Schlegelmilch, K. , Mohseni, M. , Kirak, O. , Pruszak, J. , Rodriguez, J. R. , Zhou, D. , Kreger, B. T. , Vasioukhin, V. , Avruch, J. , Brummelkamp, T. R. , and Camargo, F. D. , 2011, “ Yap1 Acts Downstream of α-Catenin to Control Epidermal Proliferation,” Cell, 144(5), pp. 782–795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Jaalouk, D. E. , and Lammerding, J. , 2009, “ Mechanotransduction Gone Awry,” Nat. Rev. Mol. Cell Biol., 10(1), pp. 63–73. 10.1038/nrm2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Martinac, B. , 2004, “ Mechanosensitive Ion Channels: Molecules of Mechanotransduction,” J. Cell Sci., 117(12), pp. 2449–2460. 10.1242/jcs.01232 [DOI] [PubMed] [Google Scholar]
- [22]. Anishkin, A. , Loukin, S. H. , Teng, J. , and Kung, C. , 2014, “ Feeling the Hidden Mechanical Forces in Lipid Bilayer Is an Original Sense,” Proc. Natl. Acad. Sci., 111(22), pp. 7898–7905. 10.1073/pnas.1313364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Matthews, B. D. , Thodeti, C. K. , Tytell, J. D. , Mammoto, A. , Overby, D. R. , and Ingber, D. E. , 2010, “ Ultra-Rapid Activation of TRPV4 Ion Channels by Mechanical Forces Applied to Cell Surface Beta1 Integrins,” Integr. Biol. Quant. Biosci. Nano Macro, 2(9), pp. 435–442. 10.1039/c0ib00034e [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Hayakawa, K. , Tatsumi, H. , and Sokabe, M. , 2008, “ Actin Stress Fibers Transmit and Focus Force to Activate Mechanosensitive Channels,” J. Cell Sci., 121(Pt. 4), pp. 496–503. 10.1242/jcs.022053 [DOI] [PubMed] [Google Scholar]
- [25]. Clapham, D. E. , 2007, “ Calcium Signaling,” Cell, 131(6), pp. 1047–1058. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- [26]. O'Conor, C. J. , Leddy, H. A. , Benefield, H. C. , Liedtke, W. B. , and Guilak, F. , 2014, “ TRPV4-Mediated Mechanotransduction Regulates the Metabolic Response of Chondrocytes to Dynamic Loading,” Proc. Natl. Acad. Sci., 111(4), pp. 1316–1321. 10.1073/pnas.1319569111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Lee, W. , Leddy, H. A. , Chen, Y. , Lee, S. H. , Zelenski, N. A. , McNulty, A. L. , Wu, J. , Beicker, K. N. , Coles, J. , Zauscher, S. , Grandl, J. , Sachs, F. , Guilak, F. , and Liedtke, W. B. , 2014, “ Synergy Between Piezo1 and Piezo2 Channels Confers High-Strain Mechanosensitivity to Articular Cartilage,” Proc. Natl. Acad. Sci. U.S.A., 111(47), pp. E5114–5122. 10.1073/pnas.1414298111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Pathak, M. M. , Nourse, J. L. , Tran, T. , Hwe, J. , Arulmoli, J. , Le, D. T. T. , Bernardis, E. , Flanagan, L. A. , and Tombola, F. , 2014, “ Stretch-Activated Ion Channel Piezo1 Directs Lineage Choice in Human Neural Stem Cells,” Proc. Natl. Acad. Sci., 111(45), pp. 16148–16153. 10.1073/pnas.1409802111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Lierop, J. E. V. , Wilson, D. P. , Davis, J. P. , Tikunova, S. , Sutherland, C. , Walsh, M. P. , and Johnson, J. D. , 2002, “ Activation of Smooth Muscle Myosin Light Chain Kinase by Calmodulin ROLE OF LYS30 and GLY40,” J. Biol. Chem., 277(8), pp. 6550–6558. 10.1074/jbc.M111404200 [DOI] [PubMed] [Google Scholar]
- [30]. Munevar, S. , Wang, Y.-L. , and Dembo, M. , 2004, “ Regulation of Mechanical Interactions Between Fibroblasts and the Substratum by Stretch-Activated Ca2+ Entry,” J. Cell Sci., 117(Pt. 1), pp. 85–92. 10.1242/jcs.00795 [DOI] [PubMed] [Google Scholar]
- [31]. Khatau, S. B. , Hale, C. M. , Stewart-Hutchinson, P. J. , Patel, M. S. , Stewart, C. L. , Searson, P. C. , Hodzic, D. , and Wirtz, D. , 2009, “ A Perinuclear Actin Cap Regulates Nuclear Shape,” Proc. Natl. Acad. Sci. U.S.A., 106(45), pp. 19017–19022. 10.1073/pnas.0908686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Versaevel, M. , Grevesse, T. , and Gabriele, S. , 2012, “ Spatial Coordination Between Cell and Nuclear Shape Within Micropatterned Endothelial Cells,” Nat. Commun., 3, p. 671. 10.1038/ncomms1668 [DOI] [PubMed] [Google Scholar]
- [33]. Neelam, S. , Chancellor, T. J. , Li, Y. , Nickerson, J. A. , Roux, K. J. , Dickinson, R. B. , and Lele, T. P. , 2015, “ Direct Force Probe Reveals the Mechanics of Nuclear Homeostasis in the Mammalian Cell,” Proc. Natl. Acad. Sci. U.S.A., 112(18), pp. 5720–5725. 10.1073/pnas.1502111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Maniotis, A. J. , Chen, C. S. , and Ingber, D. E. , 1997, “ Demonstration of Mechanical Connections Between Integrins, Cytoskeletal Filaments, and Nucleoplasm That Stabilize Nuclear Structure,” Proc. Natl. Acad. Sci. U.S.A., 94(3), pp. 849–854. 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Guilak, F. , 1995, “ Compression-Induced Changes in the Shape and Volume of the Chondrocyte Nucleus,” J. Biomech., 28(12), pp. 1529–1541. 10.1016/0021-9290(95)00100-X [DOI] [PubMed] [Google Scholar]
- [36]. Nathan, A. S. , Baker, B. M. , Nerurkar, N. L. , and Mauck, R. L. , 2011, “ Mechano-Topographic Modulation of Stem Cell Nuclear Shape on Nanofibrous Scaffolds,” Acta Biomater., 7(1), pp. 57–66. 10.1016/j.actbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Poh, Y.-C. , Shevtsov, S. P. , Chowdhury, F. , Wu, D. C. , Na, S. , Dundr, M. , and Wang, N. , 2012, “ Dynamic Force-Induced Direct Dissociation of Protein Complexes in a Nuclear Body in Living Cells,” Nat. Commun., 3, p. 866. 10.1038/ncomms1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Booth-Gauthier, E. A. , Alcoser, T. A. , Yang, G. , and Dahl, K. N. , 2012, “ Force-Induced Changes in Subnuclear Movement and Rheology,” Biophys. J., 103(12), pp. 2423–2431. 10.1016/j.bpj.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Thomas, C. H. , Collier, J. H. , Sfeir, C. S. , and Healy, K. E. , 2002, “ Engineering Gene Expression and Protein Synthesis by Modulation of Nuclear Shape,” Proc. Natl. Acad. Sci., 99(4), pp. 1972–1977. 10.1073/pnas.032668799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Heo, S.-J. , Nerurkar, N. L. , Baker, B. M. , Shin, J.-W. , Elliott, D. M. , and Mauck, R. L. , 2011, “ Fiber Stretch and Reorientation Modulates Mesenchymal Stem Cell Morphology and Fibrous Gene Expression on Oriented Nanofibrous Microenvironments,” Ann. Biomed. Eng., 39(11), pp. 2780–2790. 10.1007/s10439-011-0365-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Guilluy, C. , Osborne, L. D. , Van Landeghem, L. , Sharek, L. , Superfine, R. , Garcia-Mata, R. , and Burridge, K. , 2014, “ Isolated Nuclei Adapt to Force and Reveal a Mechanotransduction Pathway in the Nucleus,” Nat. Cell Biol., 16(4), pp. 376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Wang, N. , Tytell, J. D. , and Ingber, D. E. , 2009, “ Mechanotransduction at a Distance: Mechanically Coupling the Extracellular Matrix With the Nucleus,” Nat. Rev. Mol. Cell Biol., 10(1), pp. 75–82. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- [43]. Shivashankar, G. V. , 2011, “ Mechanosignaling to the Cell Nucleus and Gene Regulation,” Annu. Rev. Biophys., 40, pp. 361–378. 10.1146/annurev-biophys-042910-155319 [DOI] [PubMed] [Google Scholar]
- [44]. Martins, R. P. , Finan, J. D. , Guilak, F. , and Lee, D. A. , 2012, “ Mechanical Regulation of Nuclear Structure and Function,” Annu. Rev. Biomed. Eng., 14, pp. 431–455. 10.1146/annurev-bioeng-071910-124638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Fedorchak, G. R. , Kaminski, A. , and Lammerding, J. , 2014, “ Cellular Mechanosensing: Getting to the Nucleus of It All,” Prog. Biophys. Mol. Biol., 115(2–3), pp. 76–92. 10.1016/j.pbiomolbio.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Osmanagic-Myers, S. , Dechat, T. , and Foisner, R. , 2015, “ Lamins at the Crossroads of Mechanosignaling,” Genes Dev., 29(3), pp. 225–237. 10.1101/gad.255968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Uzer, G. , Fuchs, R. K. , Rubin, J. , and Thompson, W. R. , 2016, “ Concise Review: Plasma and Nuclear Membranes Convey Mechanical Information to Regulate Mesenchymal Stem Cell Lineage,” Stem Cells, 34(6), pp. 1455–1463. 10.1002/stem.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Navarro, A. P. , Collins, M. A. , and Folker, E. S. , 2016, “ The Nucleus Is a Conserved Mechanosensation and Mechanoresponse Organelle,” Cytoskeleton, 73(2), pp. 59–67. 10.1002/cm.21277 [DOI] [PubMed] [Google Scholar]
- [49]. Graham, D. M. , and Burridge, K. , 2016, “ Mechanotransduction and Nuclear Function,” Curr. Opin. Cell Biol., 40, pp. 98–105. 10.1016/j.ceb.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Belaadi, N. , Aureille, J. , and Guilluy, C. , 2016, “ Under Pressure: Mechanical Stress Management in the Nucleus,” Cells, 5(2), p. 27. 10.3390/cells5020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Gruenbaum, Y. , and Foisner, R. , 2015, “ Lamins: Nuclear Intermediate Filament Proteins With Fundamental Functions in Nuclear Mechanics and Genome Regulation,” Annu. Rev. Biochem., 84, pp. 131–164. 10.1146/annurev-biochem-060614-034115 [DOI] [PubMed] [Google Scholar]
- [52]. Davidson, P. M. , and Lammerding, J. , 2014, “ Broken Nuclei: Lamins, Nuclear Mechanics, and Disease,” Trends Cell Biol., 24(4), pp. 247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Shimi, T. , Pfleghaar, K. , Kojima, S. , Pack, C.-G. , Solovei, I. , Goldman, A. E. , Adam, S. A. , Shumaker, D. K. , Kinjo, M. , Cremer, T. , and Goldman, R. D. , 2008, “ The A- and B-Type Nuclear Lamin Networks: Microdomains Involved in Chromatin Organization and Transcription,” Genes Dev., 22(24), pp. 3409–3421. 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Broers, J. L. V. , Peeters, E. A. G. , Kuijpers, H. J. H. , Endert, J. , Bouten, C. V. C. , Oomens, C. W. J. , Baaijens, F. P. T. , and Ramaekers, F. C. S. , 2004, “ Decreased Mechanical Stiffness in LMNA−/− Cells Is Caused by Defective Nucleo-Cytoskeletal Integrity: Implications for the Development of Laminopathies,” Hum. Mol. Genet., 13(21), pp. 2567–2580. 10.1093/hmg/ddh295 [DOI] [PubMed] [Google Scholar]
- [55]. Broers, J. L. V. , Ramaekers, F. C. S. , Bonne, G. , Yaou, R. B. , and Hutchison, C. J. , 2006, “ Nuclear Lamins: Laminopathies and Their Role in Premature Ageing,” Physiol. Rev., 86(3), pp. 967–1008. 10.1152/physrev.00047.2005 [DOI] [PubMed] [Google Scholar]
- [56]. Swift, J. , Ivanovska, I. L. , Buxboim, A. , Harada, T. , Dingal, P. C. D. P. , Pinter, J. , Pajerowski, J. D. , Spinler, K. R. , Shin, J.-W. , Tewari, M. , Rehfeldt, F. , Speicher, D. W. , and Discher, D. E. , 2013, “ Nuclear Lamin-A Scales With Tissue Stiffness and Enhances Matrix-Directed Differentiation,” Science, 341(6149), p. 1240104. 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Buxboim, A. , Swift, J. , Irianto, J. , Spinler, K. R. , Dingal, P. C. D. P. , Athirasala, A. , Kao, Y.-R. C. , Cho, S. , Harada, T. , Shin, J.-W. , and Discher, D. E. , 2014, “ Matrix Elasticity Regulates Lamin-A,C Phosphorylation and Turnover With Feedback to Actomyosin,” Curr. Biol., 24(16), pp. 1909–1917. 10.1016/j.cub.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Ihalainen, T. O. , Aires, L. , Herzog, F. A. , Schwartlander, R. , Moeller, J. , and Vogel, V. , 2015, “ Differential Basal-to-Apical Accessibility of Lamin A/C Epitopes in the Nuclear Lamina Regulated by Changes in Cytoskeletal Tension,” Nat. Mater., 14(12), pp. 1252–1261. 10.1038/nmat4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Kim, D.-H. , and Wirtz, D. , 2015, “ Cytoskeletal Tension Induces the Polarized Architecture of the Nucleus,” Biomaterials, 48, pp. 161–172. 10.1016/j.biomaterials.2015.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Machowska, M. , Piekarowicz, K. , and Rzepecki, R. , 2015, “ Regulation of Lamin Properties and Functions: Does Phosphorylation Do It All?,” Open Biol., 5(11), p. 150094. 10.1098/rsob.150094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Lammerding, J. , Fong, L. G. , Ji, J. Y. , Reue, K. , Stewart, C. L. , Young, S. G. , and Lee, R. T. , 2006, “ Lamins A and C But Not Lamin B1 Regulate Nuclear Mechanics,” J. Biol. Chem., 281(35), pp. 25768–25780. 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- [62]. Taniura, H. , Glass, C. , and Gerace, L. , 1995, “ A Chromatin Binding Site in the Tail Domain of Nuclear Lamins That Interacts With Core Histones,” J. Cell Biol., 131(1), pp. 33–44. 10.1083/jcb.131.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Wilson, K. L. , and Foisner, R. , 2010, “ Lamin-Binding Proteins,” Cold Spring Harbor Perspect. Biol., 2(4), p. a000554. 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64]. Lammerding, J. , Schulze, P. C. , Takahashi, T. , Kozlov, S. , Sullivan, T. , Kamm, R. D. , Stewart, C. L. , and Lee, R. T. , 2004, “ Lamin A/C Deficiency Causes Defective Nuclear Mechanics and Mechanotransduction,” J. Clin. Invest., 113(3), pp. 370–378. 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Cupesi, M. , Yoshioka, J. , Gannon, J. , Kudinova, A. , Stewart, C. L. , and Lammerding, J. , 2010, “ Attenuated Hypertrophic Response to Pressure Overload in a Lamin A/C Haploinsufficiency Mouse,” J. Mol. Cell. Cardiol., 48(6), pp. 1290–1297. 10.1016/j.yjmcc.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Akter, R. , Rivas, D. , Geneau, G. , Drissi, H. , and Duque, G. , 2009, “ Effect of Lamin A/C Knockdown on Osteoblast Differentiation and Function,” J. Bone Miner. Res., 24(2), pp. 283–293. 10.1359/jbmr.081010 [DOI] [PubMed] [Google Scholar]
- [67]. Mao, X. , Gavara, N. , and Song, G. , 2015, “ Nuclear Mechanics and Stem Cell Differentiation,” Stem Cell Rev., 11(6), pp. 804–812. 10.1007/s12015-015-9610-z [DOI] [PubMed] [Google Scholar]
- [68]. Capell, B. C. , and Collins, F. S. , 2006, “ Human Laminopathies: Nuclei Gone Genetically Awry,” Nat. Rev. Genet., 7(12), pp. 940–952. 10.1038/nrg1906 [DOI] [PubMed] [Google Scholar]
- [69]. Burke, B. , and Stewart, C. L. , 2006, “ The Laminopathies: The Functional Architecture of the Nucleus and Its Contribution to Disease,” Annu. Rev. Genomics Hum. Genet., 7, pp. 369–405. 10.1146/annurev.genom.7.080505.115732 [DOI] [PubMed] [Google Scholar]
- [70]. McGinty, R. K. , and Tan, S. , 2015, “ Nucleosome Structure and Function,” Chem. Rev., 115(6), pp. 2255–2273. 10.1021/cr500373h [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Cremer, T. , and Cremer, M. , 2010, “ Chromosome Territories,” Cold Spring Harbor Perspect. Biol., 2(3), p. a003889. 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72]. Woodcock, C. L. , and Ghosh, R. P. , 2010, “ Chromatin Higher-Order Structure and Dynamics,” Cold Spring Harbor Perspect. Biol., 2(5), p. a000596. 10.1101/cshperspect.a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Phillips-Cremins, J. E. , 2014, “ Unraveling Architecture of the Pluripotent Genome,” Curr. Opin. Cell Biol., 28, pp. 96–104. 10.1016/j.ceb.2014.04.006 [DOI] [PubMed] [Google Scholar]
- [74]. Parada, L. A. , McQueen, P. G. , and Misteli, T. , 2004, “ Tissue-Specific Spatial Organization of Genomes,” Genome Biol., 5, p. R44. 10.1186/gb-2004-5-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Bolzer, A. , Kreth, G. , Solovei, I. , Koehler, D. , Saracoglu, K. , Fauth, C. , Müller, S. , Eils, R. , Cremer, C. , Speicher, M. R. , and Cremer, T. , 2005, “ Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes,” PLoS Biol., 3(5), p. e157. 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Hübner, M. R. , and Spector, D. L. , 2010, “ Chromatin Dynamics,” Annu. Rev. Biophys., 39, pp. 471–489. 10.1146/annurev.biophys.093008.131348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77]. Guelen, L. , Pagie, L. , Brasset, E. , Meuleman, W. , Faza, M. B. , Talhout, W. , Eussen, B. H. , de Klein, A. , Wessels, L. , de Laat, W. , and van Steensel, B. , 2008, “ Domain Organization of Human Chromosomes Revealed by Mapping of Nuclear Lamina Interactions,” Nature, 453(7197), pp. 948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- [78]. Peric-Hupkes, D. , Meuleman, W. , Pagie, L. , Bruggeman, S. W. M. , Solovei, I. , Brugman, W. , Gräf, S. , Flicek, P. , Kerkhoven, R. M. , van Lohuizen, M. , Reinders, M. , Wessels, L. , and van Steensel, B. , 2010, “ Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions During Differentiation,” Mol. Cell, 38(4), pp. 603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Lund, E. , Oldenburg, A. R. , Delbarre, E. , Freberg, C. T. , Duband-Goulet, I. , Eskeland, R. , Buendia, B. , and Collas, P. , 2013, “ Lamin A/C-Promoter Interactions Specify Chromatin State-Dependent Transcription Outcomes,” Genome Res., 23(10), pp. 1580–1589. 10.1101/gr.159400.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Demmerle, J. , Koch, A. J. , and Holaska, J. M. , 2013, “ Emerin and Histone Deacetylase 3 (HDAC3) Cooperatively Regulate Expression and Nuclear Positions of MyoD, Myf5, and Pax7 Genes During Myogenesis,” Chromosome Res., 21(8), pp. 765–779. 10.1007/s10577-013-9381-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81]. Misteli, T. , 2007, “ Beyond the Sequence: Cellular Organization of Genome Function,” Cell, 128(4), pp. 787–800. 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- [82]. Crisp, M. , Liu, Q. , Roux, K. , Rattner, J. B. , Shanahan, C. , Burke, B. , Stahl, P. D. , and Hodzic, D. , 2006, “ Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex,” J. Cell Biol., 172(1), pp. 41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Chang, W. , Worman, H. J. , and Gundersen, G. G. , 2015, “ Accessorizing and Anchoring the LINC Complex for Multifunctionality,” J. Cell Biol., 208(1), pp. 11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Rothballer, A. , and Kutay, U. , 2013, “ The Diverse Functional LINCs of the Nuclear Envelope to the Cytoskeleton and Chromatin,” Chromosoma, 122(5), pp. 415–429. 10.1007/s00412-013-0417-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Borrego-Pinto, J. , Jegou, T. , Osorio, D. S. , Auradé, F. , Gorjánácz, M. , Koch, B. , Mattaj, I. W. , and Gomes, E. R. , 2012, “ Samp1 Is a Component of TAN Lines and Is Required for Nuclear Movement,” J. Cell Sci., 125(Pt. 5), pp. 1099–1105. 10.1242/jcs.087049 [DOI] [PubMed] [Google Scholar]
- [86]. Kutscheidt, S. , Zhu, R. , Antoku, S. , Luxton, G. W. G. , Stagljar, I. , Fackler, O. T. , and Gundersen, G. G. , 2014, “ FHOD1 Interaction With Nesprin-2G Mediates TAN Line Formation and Nuclear Movement,” Nat. Cell Biol., 16(7), pp. 708–715. 10.1038/ncb2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Bone, C. R. , Tapley, E. C. , Gorjánácz, M. , and Starr, D. A. , 2014, “ The Caenorhabditis Elegans SUN Protein UNC-84 Interacts With Lamin to Transfer Forces From the Cytoplasm to the Nucleoskeleton During Nuclear Migration,” Mol. Biol. Cell, 25(18), pp. 2853–2865. 10.1091/mbc.E14-05-0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Antoku, S. , Zhu, R. , Kutscheidt, S. , Fackler, O. T. , and Gundersen, G. G. , 2015, “ Reinforcing the LINC Complex Connection to Actin Filaments: The Role of FHOD1 in TAN Line Formation and Nuclear Movement,” Cell Cycle, 14(14), pp. 2200–2205. 10.1080/15384101.2015.1053665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89]. Kim, D.-H. , Khatau, S. B. , Feng, Y. , Walcott, S. , Sun, S. X. , Longmore, G. D. , and Wirtz, D. , 2012, “ Actin Cap Associated Focal Adhesions and Their Distinct Role in Cellular Mechanosensing,” Sci. Rep., 2, p. 555. 10.1038/srep00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90]. Luxton, G. W. G. , Gomes, E. R. , Folker, E. S. , Vintinner, E. , and Gundersen, G. G. , 2010, “ Linear Arrays of Nuclear Envelope Proteins Harness Retrograde Actin Flow for Nuclear Movement,” Science, 329(5994), pp. 956–959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Nagayama, K. , Yahiro, Y. , and Matsumoto, T. , 2011, “ Stress Fibers Stabilize the Position of Intranuclear DNA Through Mechanical Connection With the Nucleus in Vascular Smooth Muscle Cells,” FEBS Lett., 585(24), pp. 3992–3997. 10.1016/j.febslet.2011.11.006 [DOI] [PubMed] [Google Scholar]
- [92]. Versaevel, M. , Braquenier, J.-B. , Riaz, M. , Grevesse, T. , Lantoine, J. , and Gabriele, S. , 2014, “ Super-Resolution Microscopy Reveals LINC Complex Recruitment at Nuclear Indentation Sites,” Sci. Rep., 4, p. 7362. 10.1038/srep07362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93]. Arsenovic, P. T. , Ramachandran, I. , Bathula, K. , Zhu, R. , Narang, J. D. , Noll, N. A. , Lemmon, C. A. , Gundersen, G. G. , and Conway, D. E. , 2016, “ Nesprin-2G: A Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension,” Biophys. J., 110(1), pp. 34–43. 10.1016/j.bpj.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94]. Nagayama, K. , Yahiro, Y. , and Matsumoto, T. , 2013, “ Apical and Basal Stress Fibers Have Different Roles in Mechanical Regulation of the Nucleus in Smooth Muscle Cells Cultured on a Substrate,” Cell. Mol. Bioeng., 6(4), pp. 473–481. 10.1007/s12195-013-0294-7 [DOI] [Google Scholar]
- [95]. Khatau, S. B. , Kusuma, S. , Hanjaya-Putra, D. , Mali, P. , Cheng, L. , Lee, J. S. H. , Gerecht, S. , and Wirtz, D. , 2012, “ The Differential Formation of the LINC-Mediated Perinuclear Actin Cap in Pluripotent and Somatic Cells,” PLoS One, 7(5), p. e36689. 10.1371/journal.pone.0036689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. Lombardi, M. L. , Jaalouk, D. E. , Shanahan, C. M. , Burke, B. , Roux, K. J. , and Lammerding, J. , 2011, “ The Interaction Between Nesprins and Sun Proteins at the Nuclear Envelope Is Critical for Force Transmission Between the Nucleus and Cytoskeleton,” J. Biol. Chem., 286(30), pp. 26743–26753. 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Han, W. M. , Heo, S.-J. , Driscoll, T. P. , Smith, L. J. , Mauck, R. L. , and Elliott, D. M. , 2013, “ Macro- to Microscale Strain Transfer in Fibrous Tissues Is Heterogeneous and Tissue-Specific,” Biophys. J., 105(3), pp. 807–817. 10.1016/j.bpj.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98]. Li, Q. , Makhija, E. , Hameed, F. M. , and Shivashankar, G. V. , 2015, “ Micropillar Displacements by Cell Traction Forces Are Mechanically Correlated With Nuclear Dynamics,” Biochem. Biophys. Res. Commun., 461(2), pp. 372–377. 10.1016/j.bbrc.2015.04.041 [DOI] [PubMed] [Google Scholar]
- [99]. Driscoll, T. P. , Cosgrove, B. D. , Heo, S.-J. , Shurden, Z. E. , and Mauck, R. L. , 2015, “ Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells,” Biophys. J., 108(12), pp. 2783–2793. 10.1016/j.bpj.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Banerjee, I. , Zhang, J. , Moore-Morris, T. , Pfeiffer, E. , Buchholz, K. S. , Liu, A. , Ouyang, K. , Stroud, M. J. , Gerace, L. , Evans, S. M. , McCulloch, A. , and Chen, J. , 2014, “ Targeted Ablation of Nesprin 1 and Nesprin 2 From Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response,” PLoS Genet., 10(2), p. e1004114. 10.1371/journal.pgen.1004114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101]. Chambliss, A. B. , Khatau, S. B. , Erdenberger, N. , Robinson, D. K. , Hodzic, D. , Longmore, G. D. , and Wirtz, D. , 2013, “ The LINC-Anchored Actin Cap Connects the Extracellular Milieu to the Nucleus for Ultrafast Mechanotransduction,” Sci. Rep., 3, p. 1087. 10.1038/srep01087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102]. Lovett, D. B. , Shekhar, N. , Nickerson, J. A. , Roux, K. J. , and Lele, T. P. , 2013, “ Modulation of Nuclear Shape by Substrate Rigidity,” Cell. Mol. Bioeng., 6(2), pp. 230–238. 10.1007/s12195-013-0270-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Li, Y. , Lovett, D. , Zhang, Q. , Neelam, S. , Kuchibhotla, R. A. , Zhu, R. , Gundersen, G. G. , Lele, T. P. , and Dickinson, R. B. , 2015, “ Moving Cell Boundaries Drive Nuclear Shaping During Cell Spreading,” Biophys. J., 109(4), pp. 670–686. 10.1016/j.bpj.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104]. Mazumder, A. , and Shivashankar, G. V. , 2010, “ Emergence of a Prestressed Eukaryotic Nucleus During Cellular Differentiation and Development,” J. R. Soc. Interface R. Soc., 7(Suppl. 3), pp. S321–330. 10.1098/rsif.2010.0039.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105]. Jean, R. P. , Gray, D. S. , Spector, A. A. , and Chen, C. S. , 2004, “ Characterization of the Nuclear Deformation Caused by Changes in Endothelial Cell Shape,” ASME J. Biomech. Eng., 126(5), pp. 552–558. 10.1115/1.1800559 [DOI] [PubMed] [Google Scholar]
- [106]. Anno, T. , Sakamoto, N. , and Sato, M. , 2012, “ Role of Nesprin-1 in Nuclear Deformation in Endothelial Cells Under Static and Uniaxial Stretching Conditions,” Biochem. Biophys. Res. Commun., 424(1), pp. 94–99. 10.1016/j.bbrc.2012.06.073 [DOI] [PubMed] [Google Scholar]
- [107]. Sexton, T. , Schober, H. , Fraser, P. , and Gasser, S. M. , 2007, “ Gene Regulation Through Nuclear Organization,” Nat. Struct. Mol. Biol., 14(11), pp. 1049–1055. 10.1038/nsmb1324 [DOI] [PubMed] [Google Scholar]
- [108]. Bickmore, W. A. , and van Steensel, B. , 2013, “ Genome Architecture: Domain Organization of Interphase Chromosomes,” Cell, 152(6), pp. 1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- [109]. Amendola, M. , and van Steensel, B. , 2014, “ Mechanisms and Dynamics of Nuclear Lamina–Genome Interactions,” Curr. Opin. Cell Biol., 28, pp. 61–68. 10.1016/j.ceb.2014.03.003 [DOI] [PubMed] [Google Scholar]
- [110]. Dobrzynska, A. , Gonzalo, S. , Shanahan, C. , and Askjaer, P. , 2016, “ The Nuclear Lamina in Health and Disease,” Nucleus, 7(3), pp. 233–248. 10.1080/19491034.2016.1183848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111]. Ragoczy, T. , Bender, M. A. , Telling, A. , Byron, R. , and Groudine, M. , 2006, “ The Locus Control Region Is Required for Association of the Murine Beta-Globin Locus With Engaged Transcription Factories During Erythroid Maturation,” Genes Dev., 20(11), pp. 1447–1457. 10.1101/gad.1419506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112]. Szczerbal, I. , Foster, H. A. , and Bridger, J. M. , 2009, “ The Spatial Repositioning of Adipogenesis Genes Is Correlated With Their Expression Status in a Porcine Mesenchymal Stem Cell Adipogenesis Model System,” Chromosoma, 118(5), pp. 647–663. 10.1007/s00412-009-0225-5 [DOI] [PubMed] [Google Scholar]
- [113]. Yao, J. , Fetter, R. D. , Hu, P. , Betzig, E. , and Tjian, R. , 2011, “ Subnuclear Segregation of Genes and Core Promoter Factors in Myogenesis,” Genes Dev., 25(6), pp. 569–580. 10.1101/gad.2021411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Udagawa, K. , and Ohyama, T. , 2014, “ Positions of Pluripotency Genes and Hepatocyte-Specific Genes in the Nucleus Before and After Mouse ES Cell Differentiation,” Genet. Mol. Res., 13(1), pp. 1979–1988. 10.4238/2014.March.24.2 [DOI] [PubMed] [Google Scholar]
- [115]. Shachar, S. , Voss, T. C. , Pegoraro, G. , Sciascia, N. , and Misteli, T. , 2015, “ Identification of Gene Positioning Factors Using High-Throughput Imaging Mapping,” Cell, 162(4), pp. 911–923. 10.1016/j.cell.2015.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116]. Zhao, H. , Sifakis, E. G. , Sumida, N. , Millán-Ariño, L. , Scholz, B. A. , Svensson, J. P. , Chen, X. , Ronnegren, A. L. , Mallet de Lima, C. D. , Varnoosfaderani, F. S. , Shi, C. , Loseva, O. , Yammine, S. , Israelsson, M. , Rathje, L.-S. , Németi, B. , Fredlund, E. , Helleday, T. , Imreh, M. P. , and Göndör, A. , 2015, “ PARP1- and CTCF-Mediated Interactions Between Active and Repressed Chromatin at the Lamina Promote Oscillating Transcription,” Mol. Cell, 59(6), pp. 984–997. 10.1016/j.molcel.2015.07.019 [DOI] [PubMed] [Google Scholar]
- [117]. Harr, J. C. , Luperchio, T. R. , Wong, X. , Cohen, E. , Wheelan, S. J. , and Reddy, K. L. , 2015, “ Directed Targeting of Chromatin to the Nuclear Lamina Is Mediated by Chromatin State and A-Type Lamins,” J. Cell Biol., 208(1), pp. 33–52. 10.1083/jcb.201405110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Ottaviani, A. , Schluth-Bolard, C. , Rival-Gervier, S. , Boussouar, A. , Rondier, D. , Foerster, A. M. , Morere, J. , Bauwens, S. , Gazzo, S. , Callet-Bauchu, E. , Gilson, E. , and Magdinier, F. , 2009, “ Identification of a Perinuclear Positioning Element in Human Subtelomeres That Requires A-Type Lamins and CTCF,” EMBO J., 28(16), pp. 2428–2436. 10.1038/emboj.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119]. Cui, Y. , and Bustamante, C. , 2000, “ Pulling a Single Chromatin Fiber Reveals the Forces That Maintain Its Higher-Order Structure,” Proc. Natl. Acad. Sci. U.S.A., 97(1), pp. 127–132. 10.1073/pnas.97.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120]. Yao, M. , Goult, B. T. , Chen, H. , Cong, P. , Sheetz, M. P. , and Yan, J. , 2014, “ Mechanical Activation of Vinculin Binding to Talin Locks Talin in an Unfolded Conformation,” Sci. Rep., 4, p. 4610. 10.1038/srep04610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121]. Hu, Y. , Kireev, I. , Plutz, M. , Ashourian, N. , and Belmont, A. S. , 2009, “ Large-Scale Chromatin Structure of Inducible Genes: Transcription on a Condensed, Linear Template,” J. Cell Biol., 185(1), pp. 87–100. 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122]. Marko, J. F. , 2008, “ Micromechanical Studies of Mitotic Chromosomes,” Chromosome Res., 16(3), pp. 469–497. 10.1007/s10577-008-1233-7 [DOI] [PubMed] [Google Scholar]
- [123]. Therizols, P. , Illingworth, R. S. , Courilleau, C. , Boyle, S. , Wood, A. J. , and Bickmore, W. A. , 2014, “ Chromatin Decondensation Is Sufficient to Alter Nuclear Organization in Embryonic Stem Cells,” Science, 346(6214), pp. 1238–1242. 10.1126/science.1259587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124]. Tajik, A. , Zhang, Y. , Wei, F. , Sun, J. , Jia, Q. , Zhou, W. , Singh, R. , Khanna, N. , Belmont, A. S. , and Wang, N. , 2016, “ Transcription Upregulation Via Force-Induced Direct Stretching of Chromatin,” Nat. Mater., 15(12), pp. 1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125]. Verschure, P. J. , van der Kraan, I. , Manders, E. M. M. , Hoogstraten, D. , Houtsmuller, A. B. , and van Driel, R. , 2003, “ Condensed Chromatin Domains in the Mammalian Nucleus Are Accessible to Large Macromolecules,” EMBO Rep., 4(9), pp. 861–866. 10.1038/sj.embor.embor922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126]. Chen, D. , Dundr, M. , Wang, C. , Leung, A. , Lamond, A. , Misteli, T. , and Huang, S. , 2005, “ Condensed Mitotic Chromatin Is Accessible to Transcription Factors and Chromatin Structural Proteins,” J. Cell Biol., 168(1), pp. 41–54. 10.1083/jcb.200407182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127]. Bancaud, A. , Huet, S. , Daigle, N. , Mozziconacci, J. , Beaudouin, J. , and Ellenberg, J. , 2009, “ Molecular Crowding Affects Diffusion and Binding of Nuclear Proteins in Heterochromatin and Reveals the Fractal Organization of Chromatin,” EMBO J., 28(24), pp. 3785–3798. 10.1038/emboj.2009.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128]. Becker, J. S. , Nicetto, D. , and Zaret, K. S. , 2016, “ H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes,” Trends Genet., 32(1), pp. 29–41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Mattout, A. , Biran, A. , and Meshorer, E. , 2011, “ Global Epigenetic Changes During Somatic Cell Reprogramming to iPS Cells,” J. Mol. Cell Biol., 3(6), pp. 341–350. 10.1093/jmcb/mjr028 [DOI] [PubMed] [Google Scholar]
- [130]. Soufi, A. , Donahue, G. , and Zaret, K. S. , 2012, “ Facilitators and Impediments of the Pluripotency Reprogramming Factors' Initial Engagement With the Genome,” Cell, 151(5), pp. 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131]. Towbin, B. D. , González-Aguilera, C. , Sack, R. , Gaidatzis, D. , Kalck, V. , Meister, P. , Askjaer, P. , and Gasser, S. M. , 2012, “ Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery,” Cell, 150(5), pp. 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- [132]. Downing, T. L. , Soto, J. , Morez, C. , Houssin, T. , Fritz, A. , Yuan, F. , Chu, J. , Patel, S. , Schaffer, D. V. , and Li, S. , 2013, “ Biophysical Regulation of Epigenetic State and Cell Reprogramming,” Nat. Mater., 12(12), pp. 1154–1162. 10.1038/nmat3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133]. Caiazzo, M. , Okawa, Y. , Ranga, A. , Piersigilli, A. , Tabata, Y. , and Lutolf, M. P. , 2016, “ Defined Three-Dimensional Microenvironments Boost Induction of Pluripotency,” Nat. Mater., 15(3), pp. 344–352. 10.1038/nmat4536 [DOI] [PubMed] [Google Scholar]
- [134]. Akhtar, A. , and Gasser, S. M. , 2007, “ The Nuclear Envelope and Transcriptional Control,” Nat. Rev. Genet., 8(7), pp. 507–517. 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- [135]. Mekhail, K. , and Moazed, D. , 2010, “ The Nuclear Envelope in Genome Organization, Expression and Stability,” Nat. Rev. Mol. Cell Biol., 11(5), pp. 317–328. 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136]. Towbin, B. D. , Meister, P. , and Gasser, S. M. , 2009, “ The Nuclear Envelope: A Scaffold for Silencing?,” Curr. Opin. Genet. Dev., 19(2), pp. 180–186. 10.1016/j.gde.2009.01.006 [DOI] [PubMed] [Google Scholar]
- [137]. Jost, K. L. , Haase, S. , Smeets, D. , Schrode, N. , Schmiedel, J. M. , Bertulat, B. , Herzel, H. , Cremer, M. , and Cardoso, M. C. , 2011, “ 3D-Image Analysis Platform Monitoring Relocation of Pluripotency Genes During Reprogramming,” Nucleic Acids Res., 39(17), pp. e113–e113. 10.1093/nar/gkr486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138]. Zink, D. , Amaral, M. D. , Englmann, A. , Lang, S. , Clarke, L. A. , Rudolph, C. , Alt, F. , Luther, K. , Braz, C. , Sadoni, N. , Rosenecker, J. , and Schindelhauer, D. , 2004, “ Transcription-Dependent Spatial Arrangements of CFTR and Adjacent Genes in Human Cell Nuclei,” J. Cell Biol., 166(6), pp. 815–825. 10.1083/jcb.200404107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139]. Brown, K. E. , Baxter, J. , Graf, D. , Merkenschlager, M. , and Fisher, A. G. , 1999, “ Dynamic Repositioning of Genes in the Nucleus of Lymphocytes Preparing for Cell Division,” Mol. Cell, 3(2), pp. 207–217. 10.1016/S1097-2765(00)80311-1 [DOI] [PubMed] [Google Scholar]
- [140]. Reddy, K. L. , Zullo, J. M. , Bertolino, E. , and Singh, H. , 2008, “ Transcriptional Repression Mediated by Repositioning of Genes to the Nuclear Lamina,” Nature, 452(7184), pp. 243–247. 10.1038/nature06727 [DOI] [PubMed] [Google Scholar]
- [141]. Kumaran, R. I. , and Spector, D. L. , 2008, “ A Genetic Locus Targeted to the Nuclear Periphery in Living Cells Maintains Its Transcriptional Competence,” J. Cell Biol., 180(1), pp. 51–65. 10.1083/jcb.200706060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142]. Finlan, L. E. , Sproul, D. , Thomson, I. , Boyle, S. , Kerr, E. , Perry, P. , Ylstra, B. , Chubb, J. R. , and Bickmore, W. A. , 2008, “ Recruitment to the Nuclear Periphery Can Alter Expression of Genes in Human Cells,” PLoS Genet, 4(3), p. e1000039. 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143]. Gartenberg, M. R. , Neumann, F. R. , Laroche, T. , Blaszczyk, M. , and Gasser, S. M. , 2004, “ Sir-Mediated Repression Can Occur Independently of Chromosomal and Subnuclear Contexts,” Cell, 119(7), pp. 955–967. 10.1016/j.cell.2004.11.008 [DOI] [PubMed] [Google Scholar]
- [144]. Schübeler, D. , Francastel, C. , Cimbora, D. M. , Reik, A. , Martin, D. I. , and Groudine, M. , 2000, “ Nuclear Localization and Histone Acetylation: A Pathway for Chromatin Opening and Transcriptional Activation of the Human Beta-Globin Locus,” Genes Dev., 14(8), pp. 940–950. 10.1101/gad.14.8.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145]. Chuang, C.-H. , Carpenter, A. E. , Fuchsova, B. , Johnson, T. , de Lanerolle, P. , and Belmont, A. S. , 2006, “ Long-Range Directional Movement of an Interphase Chromosome Site,” Curr. Biol., 16(8), pp. 825–831. 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- [146]. Dundr, M. , Ospina, J. K. , Sung, M.-H. , John, S. , Upender, M. , Ried, T. , Hager, G. L. , and Matera, A. G. , 2007, “ Actin-Dependent Intranuclear Repositioning of an Active Gene Locus in vivo,” J. Cell Biol., 179(6), pp. 1095–1103. 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147]. Levi, V. , Ruan, Q. , Plutz, M. , Belmont, A. S. , and Gratton, E. , 2005, “ Chromatin Dynamics in Interphase Cells Revealed by Tracking in a Two-Photon Excitation Microscope,” Biophys. J., 89(6), pp. 4275–4285. 10.1529/biophysj.105.066670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148]. Mehta, I. S. , Amira, M. , Harvey, A. J. , and Bridger, J. M. , 2010, “ Rapid Chromosome Territory Relocation by Nuclear Motor Activity in Response to Serum Removal in Primary Human Fibroblasts,” Genome Biol., 11, p. R5. 10.1186/gb-2010-11-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149]. Kind, J. , Pagie, L. , Ortabozkoyun, H. , Boyle, S. , de Vries, S. S. , Janssen, H. , Amendola, M. , Nolen, L. D. , Bickmore, W. A. , and van Steensel, B. , 2013, “ Single-Cell Dynamics of Genome-Nuclear Lamina Interactions,” Cell, 153(1), pp. 178–192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- [150]. Padeken, J. , and Heun, P. , 2014, “ Nucleolus and Nuclear Periphery: Velcro for Heterochromatin,” Curr. Opin. Cell Biol., 28, pp. 54–60. 10.1016/j.ceb.2014.03.001 [DOI] [PubMed] [Google Scholar]
- [151]. Van Koningsbruggen, S. , Gierlinski, M. , Schofield, P. , Martin, D. , Barton, G. J. , Ariyurek, Y. , den Dunnen, J. T. , and Lamond, A. I. , 2010, “ High-Resolution Whole-Genome Sequencing Reveals That Specific Chromatin Domains From Most Human Chromosomes Associate With Nucleoli,” Mol. Biol. Cell, 21(21), pp. 3735–3748. 10.1091/mbc.E10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152]. Németh, A. , Conesa, A. , Santoyo-Lopez, J. , Medina, I. , Montaner, D. , Péterfia, B. , Solovei, I. , Cremer, T. , Dopazo, J. , and Längst, G. , 2010, “ Initial Genomics of the Human Nucleolus,” PLoS Genet., 6(3), p. e1000889. 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153]. Zhang, Q. , Bethmann, C. , Worth, N. F. , Davies, J. D. , Wasner, C. , Feuer, A. , Ragnauth, C. D. , Yi, Q. , Mellad, J. A. , Warren, D. T. , Wheeler, M. A. , Ellis, J. A. , Skepper, J. N. , Vorgerd, M. , Schlotter-Weigel, B. , Weissberg, P. L. , Roberts, R. G. , Wehnert, M. , and Shanahan, C. M. , 2007, “ Nesprin-1 and -2 Are Involved in the Pathogenesis of Emery Dreifuss Muscular Dystrophy and Are Critical for Nuclear Envelope Integrity,” Hum. Mol. Genet., 16(23), pp. 2816–2833. 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- [154]. Makhija, E. , Jokhun, D. S. , and Shivashankar, G. V. , 2016, “ Nuclear Deformability and Telomere Dynamics Are Regulated by Cell Geometric Constraints,” Proc. Natl. Acad. Sci. U.S.A., 113(1), pp. E32–40. 10.1073/pnas.1513189113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155]. Lammerding, J. , Hsiao, J. , Schulze, P. C. , Kozlov, S. , Stewart, C. L. , and Lee, R. T. , 2005, “ Abnormal Nuclear Shape and Impaired Mechanotransduction in Emerin-Deficient Cells,” J. Cell Biol., 170(5), pp. 781–791. 10.1083/jcb.200502148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156]. Zwerger, M. , Jaalouk, D. E. , Lombardi, M. L. , Isermann, P. , Mauermann, M. , Dialynas, G. , Herrmann, H. , Wallrath, L. L. , and Lammerding, J. , 2013, “ Myopathic Lamin Mutations Impair Nuclear Stability in Cells and Tissue and Disrupt Nucleo-Cytoskeletal Coupling,” Hum. Mol. Genet., 22(12), pp. 2335–2349. 10.1093/hmg/ddt079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157]. Denais, C. M. , Gilbert, R. M. , Isermann, P. , McGregor, A. L. , te Lindert, M. , Weigelin, B. , Davidson, P. M. , Friedl, P. , Wolf, K. , and Lammerding, J. , 2016, “ Nuclear Envelope Rupture and Repair During Cancer Cell Migration,” Science, 352(6283), pp. 353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158]. Raab, M. , Gentili, M. , de Belly, H. , Thiam, H. R. , Vargas, P. , Jimenez, A. J. , Lautenschlaeger, F. , Voituriez, R. , Lennon-Duménil, A. M. , Manel, N. , and Piel, M. , 2016, “ ESCRT III Repairs Nuclear Envelope Ruptures During Cell Migration to Limit DNA Damage and Cell Death,” Science, 352(6283), pp. 359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- [159]. Bell, E. S. , and Lammerding, J. , 2016, “ Causes and Consequences of Nuclear Envelope Alterations in Tumour Progression,” Eur. J. Cell Biol., 95(11), pp. 449–464. 10.1016/j.ejcb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160]. Irianto, J. , Pfeifer, C. R. , Ivanovska, I. L. , Swift, J. , and Discher, D. E. , 2016, “ Nuclear Lamins in Cancer,” Cell. Mol. Bioeng., 9(2), pp. 258–267. 10.1007/s12195-016-0437-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161]. Swift, J. , and Discher, D. E. , 2014, “ The Nuclear Lamina Is Mechano-Responsive to ECM Elasticity in Mature Tissue,” J. Cell Sci., 127(14), pp. 3005–3015. 10.1242/jcs.149203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162]. Knight, M. M. , Lee, D. A. , and Bader, D. L. , 1998, “ The Influence of Elaborated Pericellular Matrix on the Deformation of Isolated Articular Chondrocytes Cultured in Agarose,” Biochim. Biophys. Acta, 1405(1–3), pp. 67–77. 10.1016/S0167-4889(98)00102-5 [DOI] [PubMed] [Google Scholar]
- [163]. Choi, J. B. , Youn, I. , Cao, L. , Leddy, H. A. , Gilchrist, C. L. , Setton, L. A. , and Guilak, F. , 2007, “ Zonal Changes in the Three-Dimensional Morphology of the Chondron Under Compression: The Relationship Among Cellular, Pericellular, and Extracellular Deformation in Articular Cartilage,” J. Biomech., 40(12), pp. 2596–2603. 10.1016/j.jbiomech.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164]. Dahl, K. N. , Scaffidi, P. , Islam, M. F. , Yodh, A. G. , Wilson, K. L. , and Misteli, T. , 2006, “ Distinct Structural and Mechanical Properties of the Nuclear Lamina in Hutchinson-Gilford Progeria Syndrome,” Proc. Natl. Acad. Sci. U.S.A., 103(27), pp. 10271–10276. 10.1073/pnas.0601058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165]. Scaffidi, P. , and Misteli, T. , 2008, “ Lamin A-Dependent Misregulation of Adult Stem Cells Associated With Accelerated Ageing,” Nat. Cell Biol., 10(4), pp. 452–459. 10.1038/ncb1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166]. Verstraeten, V. L. R. M. , Ji, J. Y. , Cummings, K. S. , Lee, R. T. , and Lammerding, J. , 2008, “ Increased Mechanosensitivity and Nuclear Stiffness in Hutchinson-Gilford Progeria Cells: Effects of Farnesyltransferase Inhibitors,” Aging Cell, 7(3), pp. 383–393. 10.1111/j.1474-9726.2008.00382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167]. Mounkes, L. C. , Kozlov, S. , Hernandez, L. , Sullivan, T. , and Stewart, C. L. , 2003, “ A Progeroid Syndrome in Mice Is Caused by Defects in A-Type Lamins,” Nature, 423(6937), pp. 298–301. 10.1038/nature01631 [DOI] [PubMed] [Google Scholar]
- [168]. Hale, C. M. , Shrestha, A. L. , Khatau, S. B. , Stewart-Hutchinson, P. J. , Hernandez, L. , Stewart, C. L. , Hodzic, D. , and Wirtz, D. , 2008, “ Dysfunctional Connections Between the Nucleus and the Actin and Microtubule Networks in Laminopathic Models,” Biophys. J., 95(11), pp. 5462–5475. 10.1529/biophysj.108.139428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169]. Mehta, I. S. , Eskiw, C. H. , Arican, H. D. , Kill, I. R. , and Bridger, J. M. , 2011, “ Farnesyltransferase Inhibitor Treatment Restores Chromosome Territory Positions and Active Chromosome Dynamics in Hutchinson-Gilford Progeria Syndrome Cells,” Genome Biol., 12(8), p. R74. 10.1186/gb-2011-12-8-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170]. Shumaker, D. K. , Dechat, T. , Kohlmaier, A. , Adam, S. A. , Bozovsky, M. R. , Erdos, M. R. , Eriksson, M. , Goldman, A. E. , Khuon, S. , Collins, F. S. , Jenuwein, T. , and Goldman, R. D. , 2006, “ Mutant Nuclear Lamin A Leads to Progressive Alterations of Epigenetic Control in Premature Aging,” Proc. Natl. Acad. Sci. U.S.A., 103(23), pp. 8703–8708. 10.1073/pnas.0602569103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171]. Mattout, A. , Pike, B. L. , Towbin, B. D. , Bank, E. M. , Gonzalez-Sandoval, A. , Stadler, M. B. , Meister, P. , Gruenbaum, Y. , and Gasser, S. M. , 2011, “ An EDMD Mutation in C. Elegans Lamin Blocks Muscle-Specific Gene Relocation and Compromises Muscle Integrity,” Curr. Biol., 21(19), pp. 1603–1614. 10.1016/j.cub.2011.08.030 [DOI] [PubMed] [Google Scholar]
- [172]. Simon, D. N. , Zastrow, M. S. , and Wilson, K. L. , 2010, “ Direct Actin Binding to A- and B-Type Lamin Tails and Actin Filament Bundling by the Lamin A Tail,” Nucleus, 1(3), pp. 264–272. 10.4161/nucl.11799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173]. Worman, H. J. , and Schirmer, E. C. , 2015, “ Nuclear Membrane Diversity: Underlying Tissue-Specific Pathologies in Disease?,” Curr. Opin. Cell Biol., 34, pp. 101–112. 10.1016/j.ceb.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174]. Heessen, S. , and Fornerod, M. , 2007, “ The Inner Nuclear Envelope as a Transcription Factor Resting Place,” EMBO Rep., 8(10), pp. 914–919. 10.1038/sj.embor.7401075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175]. Gesson, K. , Rescheneder, P. , Skoruppa, M. P. , von Haeseler, A. , Dechat, T. , and Foisner, R. , 2016, “ A-Type Lamins Bind Both Hetero- and Euchromatin, the Latter Being Regulated by Lamina-Associated Polypeptide 2 Alpha,” Genome Res., 26(4), pp. 462–473. 10.1101/gr.196220.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176]. Solovei, I. , Wang, A. S. , Thanisch, K. , Schmidt, C. S. , Krebs, S. , Zwerger, M. , Cohen, T. V. , Devys, D. , Foisner, R. , Peichl, L. , Herrmann, H. , Blum, H. , Engelkamp, D. , Stewart, C. L. , Leonhardt, H. , and Joffe, B. , 2013, “ LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation,” Cell, 152(3), pp. 584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- [177]. Mislow, J. M. K. , Holaska, J. M. , Kim, M. S. , Lee, K. K. , Segura-Totten, M. , Wilson, K. L. , and McNally, E. M. , 2002, “ Nesprin-1α Self-Associates and Binds Directly to Emerin and Lamin A in vitro,” FEBS Lett., 525(1–3), pp. 135–140. 10.1016/S0014-5793(02)03105-8 [DOI] [PubMed] [Google Scholar]
- [178]. Zhang, Q. , Ragnauth, C. D. , Skepper, J. N. , Worth, N. F. , Warren, D. T. , Roberts, R. G. , Weissberg, P. L. , Ellis, J. A. , and Shanahan, C. M. , 2005, “ Nesprin-2 Is a Multi-Isomeric Protein That Binds Lamin and Emerin at the Nuclear Envelope and Forms a Subcellular Network in Skeletal Muscle,” J. Cell Sci., 118(Pt. 4), pp. 673–687. 10.1242/jcs.01642 [DOI] [PubMed] [Google Scholar]
- [179]. Neumann, S. , Schneider, M. , Daugherty, R. L. , Gottardi, C. J. , Eming, S. A. , Beijer, A. , Noegel, A. A. , and Karakesisoglou, I. , 2010, “ Nesprin-2 Interacts With {Alpha}-Catenin and Regulates Wnt Signaling at the Nuclear Envelope,” J. Biol. Chem., 285(45), pp. 34932–34938. 10.1074/jbc.M110.119651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180]. Markiewicz, E. , Tilgner, K. , Barker, N. , van de Wetering, M. , Clevers, H. , Dorobek, M. , Hausmanowa-Petrusewicz, I. , Ramaekers, F. C. S. , Broers, J. L. V. , Blankesteijn, W. M. , Salpingidou, G. , Wilson, R. G. , Ellis, J. A. , and Hutchison, C. J. , 2006, “ The Inner Nuclear Membrane Protein Emerin Regulates Beta-Catenin Activity by Restricting Its Accumulation in the Nucleus,” EMBO J., 25(14), pp. 3275–3285. 10.1038/sj.emboj.7601230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181]. Chang, W. , Folker, E. S. , Worman, H. J. , and Gundersen, G. G. , 2013, “ Emerin Organizes Actin Flow for Nuclear Movement and Centrosome Orientation in Migrating Fibroblasts,” Mol. Biol. Cell, 24(24), pp. 3869–3880. 10.1091/mbc.E13-06-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182]. Holaska, J. M. , Kowalski, A. K. , and Wilson, K. L. , 2004, “ Emerin Caps the Pointed End of Actin Filaments: Evidence for an Actin Cortical Network at the Nuclear Inner Membrane,” PLoS Biol., 2(9), p. e231. 10.1371/journal.pbio.0020231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183]. Mehta, I. S. , Elcock, L. S. , Amira, M. , Kill, I. R. , and Bridger, J. M. , 2008, “ Nuclear Motors and Nuclear Structures Containing A-Type Lamins and Emerin: Is There a Functional Link?,” Biochem. Soc. Trans., 36(Pt. 6), pp. 1384–1388. 10.1042/BST0361384 [DOI] [PubMed] [Google Scholar]
- [184]. Eberharter, A. , and Becker, P. B. , 2002, “ Histone Acetylation: A Switch Between Repressive and Permissive Chromatin. Second in Review Series on Chromatin Dynamics,” EMBO Rep., 3(3), pp. 224–229. 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185]. Milon, B. C. , Cheng, H. , Tselebrovsky, M. V. , Lavrov, S. A. , Nenasheva, V. V. , Mikhaleva, E. A. , Shevelyov, Y. Y. , and Nurminsky, D. I. , 2012, “ Role of Histone Deacetylases in Gene Regulation at Nuclear Lamina,” PLoS One, 7(11), p. e49692. 10.1371/journal.pone.0049692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186]. Zullo, J. M. , Demarco, I. A. , Piqué-Regi, R. , Gaffney, D. J. , Epstein, C. B. , Spooner, C. J. , Luperchio, T. R. , Bernstein, B. E. , Pritchard, J. K. , Reddy, K. L. , and Singh, H. , 2012, “ DNA Sequence-Dependent Compartmentalization and Silencing of Chromatin at the Nuclear Lamina,” Cell, 149(7), pp. 1474–1487. 10.1016/j.cell.2012.04.035 [DOI] [PubMed] [Google Scholar]
- [187]. Jain, N. , Iyer, K. V. , Kumar, A. , and Shivashankar, G. V. , 2013, “ Cell Geometric Constraints Induce Modular Gene-Expression Patterns Via Redistribution of HDAC3 Regulated by Actomyosin Contractility,” Proc. Natl. Acad. Sci., 110(28), pp. 11349–11354. 10.1073/pnas.1300801110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188]. Li, Y. , Chu, J. S. , Kurpinski, K. , Li, X. , Bautista, D. M. , Yang, L. , Sung, K.-L. P. , and Li, S. , 2011, “ Biophysical Regulation of Histone Acetylation in Mesenchymal Stem Cells,” Biophys. J., 100(8), pp. 1902–1909. 10.1016/j.bpj.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189]. Demmerle, J. , Koch, A. J. , and Holaska, J. M. , 2012, “ The Nuclear Envelope Protein Emerin Binds Directly to Histone Deacetylase 3 (HDAC3) and Activates HDAC3 Activity,” J. Biol. Chem., 287(26), pp. 22080–22088. 10.1074/jbc.M111.325308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190]. Lu, W. , Schneider, M. , Neumann, S. , Jaeger, V.-M. , Taranum, S. , Munck, M. , Cartwright, S. , Richardson, C. , Carthew, J. , Noh, K. , Goldberg, M. , Noegel, A. A. , and Karakesisoglou, I. , 2012, “ Nesprin Interchain Associations Control Nuclear Size,” Cell. Mol. Life Sci., 69(20), pp. 3493–3509. 10.1007/s00018-012-1034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191]. Taranum, S. , Sur, I. , Müller, R. , Lu, W. , Rashmi, R. N. , Munck, M. , Neumann, S. , Karakesisoglou, I. , and Noegel, A. A. , 2012, “ Cytoskeletal Interactions at the Nuclear Envelope Mediated by Nesprins, Cytoskeletal Interactions at the Nuclear Envelope Mediated by Nesprins,” Int. J. Cell Biol., 2012, p. e736524. 10.1155/2012/736524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192]. Ketema, M. , Wilhelmsen, K. , Kuikman, I. , Janssen, H. , Hodzic, D. , and Sonnenberg, A. , 2007, “ Requirements for the Localization of Nesprin-3 at the Nuclear Envelope and Its Interaction With Plectin,” J. Cell Sci., 120(Pt. 19), pp. 3384–3394. 10.1242/jcs.014191 [DOI] [PubMed] [Google Scholar]
- [193]. Muchir, A. , van Engelen, B. G. , Lammens, M. , Mislow, J. M. , McNally, E. , Schwartz, K. , and Bonne, G. , 2003, “ Nuclear Envelope Alterations in Fibroblasts From LGMD1B Patients Carrying Nonsense Y259X Heterozygous or Homozygous Mutation in Lamin A/C Gene,” Exp. Cell Res., 291(2), pp. 352–362. 10.1016/j.yexcr.2003.07.002 [DOI] [PubMed] [Google Scholar]
- [194]. Libotte, T. , Zaim, H. , Abraham, S. , Padmakumar, V. C. , Schneider, M. , Lu, W. , Munck, M. , Hutchison, C. , Wehnert, M. , Fahrenkrog, B. , Sauder, U. , Aebi, U. , Noegel, A. A. , and Karakesisoglou, I. , 2005, “ Lamin A/C-Dependent Localization of Nesprin-2: A Giant Scaffolder at the Nuclear Envelope,” Mol. Biol. Cell, 16(7), pp. 3411–3424. 10.1091/mbc.E04-11-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195]. Folker, E. S. , Ostlund, C. , Luxton, G. W. G. , Worman, H. J. , and Gundersen, G. G. , 2011, “ Lamin A Variants That Cause Striated Muscle Disease Are Defective in Anchoring Transmembrane Actin-Associated Nuclear Lines for Nuclear Movement,” Proc. Natl. Acad. Sci. U.S.A., 108(1), pp. 131–136. 10.1073/pnas.1000824108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196]. Enyedi, B. , and Niethammer, P. , 2016, “ A Case for the Nuclear Membrane as a Mechanotransducer,” Cell. Mol. Bioeng., 9(2), pp. 247–251. 10.1007/s12195-016-0430-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197]. Wallrath, L. L. , Bohnekamp, J. , and Magin, T. M. , 2016, “ Cross Talk Between the Cytoplasm and Nucleus During Development and Disease,” Curr. Opin. Genet. Dev., 37, pp. 129–136. 10.1016/j.gde.2016.03.007 [DOI] [PubMed] [Google Scholar]
- [198]. McKee, C. T. , Raghunathan, V. K. , Nealey, P. F. , Russell, P. , and Murphy, C. J. , 2011, “ Topographic Modulation of the Orientation and Shape of Cell Nuclei and Their Influence on the Measured Elastic Modulus of Epithelial Cells,” Biophys. J., 101(9), pp. 2139–2146. 10.1016/j.bpj.2011.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199]. Pajerowski, J. D. , Dahl, K. N. , Zhong, F. L. , Sammak, P. J. , and Discher, D. E. , 2007, “ Physical Plasticity of the Nucleus in Stem Cell Differentiation,” Proc. Natl. Acad. Sci. U.S.A., 104(40), pp. 15619–15624. 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200]. Stewart-Hutchinson, P. J. , Hale, C. M. , Wirtz, D. , and Hodzic, D. , 2008, “ Structural Requirements for the Assembly of LINC Complexes and Their Function in Cellular Mechanical Stiffness,” Exp. Cell Res., 314(8), pp. 1892–1905. 10.1016/j.yexcr.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201]. Münter, S. , Enninga, J. , Vazquez-Martinez, R. , Delbarre, E. , David-Watine, B. , Nehrbass, U. , and Shorte, S. L. , 2006, “ Actin Polymerisation at the Cytoplasmic Face of Eukaryotic Nuclei,” BMC Cell Biol., 7(23), p. 23. 10.1186/1471-2121-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202]. González-Granado, J. M. , Silvestre-Roig, C. , Rocha-Perugini, V. , Trigueros-Motos, L. , Cibrián, D. , Morlino, G. , Blanco-Berrocal, M. , Osorio, F. G. , Freije, J. M. P. , López-Otín, C. , Sánchez-Madrid, F. , and Andrés, V. , 2014, “ Nuclear Envelope Lamin-A Couples Actin Dynamics With Immunological Synapse Architecture and T Cell Activation,” Sci. Signalling, 7(322), p. ra37. 10.1126/scisignal.2004872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203]. Morgan, J. T. , Pfeiffer, E. R. , Thirkill, T. L. , Kumar, P. , Peng, G. , Fridolfsson, H. N. , Douglas, G. C. , Starr, D. A. , and Barakat, A. I. , 2011, “ Nesprin-3 Regulates Endothelial Cell Morphology, Perinuclear Cytoskeletal Architecture, and Flow-Induced Polarization,” Mol. Biol. Cell, 22(22), pp. 4324–4334. 10.1091/mbc.E11-04-0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204]. Chancellor, T. J. , Lee, J. , Thodeti, C. K. , and Lele, T. , 2010, “ Actomyosin Tension Exerted on the Nucleus Through Nesprin-1 Connections Influences Endothelial Cell Adhesion, Migration, and Cyclic Strain-Induced Reorientation,” Biophys. J., 99(1), pp. 115–123. 10.1016/j.bpj.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205]. Ho, C. Y. , Jaalouk, D. E. , Vartiainen, M. K. , and Lammerding, J. , 2013, “ Lamin A/C and Emerin Regulate MKL1-SRF Activity by Modulating Actin Dynamics,” Nature, 497(7450), pp. 507–511. 10.1038/nature12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206]. Bertrand, A. T. , Ziaei, S. , Ehret, C. , Duchemin, H. , Mamchaoui, K. , Bigot, A. , Mayer, M. , Quijano-Roy, S. , Desguerre, I. , Lainé, J. , Ben Yaou, R. , Bonne, G. , and Coirault, C. , 2014, “ Cellular Microenvironments Reveal Defective Mechanosensing Responses and Elevated YAP Signaling in LMNA-Mutated Muscle Precursors,” J. Cell Sci., 127(Pt. 13), pp. 2873–2884. 10.1242/jcs.144907 [DOI] [PubMed] [Google Scholar]
- [207]. Foster, C. R. , Robson, J. L. , Simon, W. J. , Twigg, J. , Cruikshank, D. , Wilson, R. G. , and Hutchison, C. J. , 2011, “ The Role of Lamin A in Cytoskeleton Organization in Colorectal Cancer Cells: A Proteomic Investigation,” Nucleus, 2(5), pp. 434–443. 10.4161/nucl.2.5.17775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208]. Chen, B. , Gilbert, L. A. , Cimini, B. A. , Schnitzbauer, J. , Zhang, W. , Li, G.-W. , Park, J. , Blackburn, E. H. , Weissman, J. S. , Qi, L. S. , and Huang, B. , 2013, “ Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System,” Cell, 155(7), pp. 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209]. Godin, A. G. , Lounis, B. , and Cognet, L. , 2014, “ Super-Resolution Microscopy Approaches for Live Cell Imaging,” Biophys. J., 107(8), pp. 1777–1784. 10.1016/j.bpj.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210]. Spagnol, S. T. , and Dahl, K. N. , 2016, “ Spatially Resolved Quantification of Chromatin Condensation Through Differential Local Rheology in Cell Nuclei Fluorescence Lifetime Imaging,” PLoS One, 11(1), p. e0146244. 10.1371/journal.pone.0146244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211]. Fedorchak, G. , and Lammerding, J. , 2016, “ Cell Microharpooning to Study Nucleo-Cytoskeletal Coupling,” Methods Mol. Biol., 1411, pp. 241–254. 10.1007/978-1-4939-3530-7_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212]. Rodriguez, M. L. , McGarry, P. J. , and Sniadecki, N. J. , 2013, “ Review on Cell Mechanics: Experimental and Modeling Approaches,” ASME Appl. Mech. Rev., 65(6), p. 060801. 10.1115/1.4025355 [DOI] [Google Scholar]
- [213]. Mak, M. , Kim, T. , Zaman, M. H. , and Kamm, R. D. , 2015, “ Multiscale Mechanobiology: Computational Models for Integrating Molecules to Multicellular Systems,” Integr. Biol. Quant. Biosci. Nano Macro, 7(10), pp. 1093–1108. 10.1039/c5ib00043b [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214]. Nava, M. M. , Raimondi, M. T. , and Pietrabissa, R. , 2014, “ Bio-Chemo-Mechanical Models for Nuclear Deformation in Adherent Eukaryotic Cells,” Biomech. Model. Mechanobiol., 13(5), pp. 929–943. 10.1007/s10237-014-0558-8 [DOI] [PubMed] [Google Scholar]
- [215]. Shemesh, T. , Geiger, B. , Bershadsky, A. D. , and Kozlov, M. M. , 2005, “ Focal Adhesions as Mechanosensors: A Physical Mechanism,” Proc. Natl. Acad. Sci. U.S.A., 102(35), pp. 12383–12388. 10.1073/pnas.0500254102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216]. Chan, C. E. , and Odde, D. J. , 2008, “ Traction Dynamics of Filopodia on Compliant Substrates,” Science, 322(5908), pp. 1687–1691. 10.1126/science.1163595 [DOI] [PubMed] [Google Scholar]
- [217]. Hytönen, V. P. , and Vogel, V. , 2008, “ How Force Might Activate Talin's Vinculin Binding Sites: SMD Reveals a Structural Mechanism,” PLoS Comput. Biol., 4(2), p. e24. 10.1371/journal.pcbi.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218]. Paszek, M. J. , Boettiger, D. , Weaver, V. M. , and Hammer, D. A. , 2009, “ Integrin Clustering Is Driven by Mechanical Resistance From the Glycocalyx and the Substrate,” PLoS Comput. Biol., 5(12), p. e1000604. 10.1371/journal.pcbi.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219]. Elosegui-Artola, A. , Oria, R. , Chen, Y. , Kosmalska, A. , Pérez-González, C. , Castro, N. , Zhu, C. , Trepat, X. , and Roca-Cusachs, P. , 2016, “ Mechanical Regulation of a Molecular Clutch Defines Force Transmission and Transduction in Response to Matrix Rigidity,” Nat. Cell Biol., 18(5), pp. 540–548. 10.1038/ncb3336 [DOI] [PubMed] [Google Scholar]
- [220]. Cao, X. , Lin, Y. , Driscoll, T. P. , Franco-Barraza, J. , Cukierman, E. , Mauck, R. L. , and Shenoy, V. B. , 2015, “ A Chemomechanical Model of Matrix and Nuclear Rigidity Regulation of Focal Adhesion Size,” Biophys. J., 109(9), pp. 1807–1817. 10.1016/j.bpj.2015.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221]. Maraldi, M. , and Garikipati, K. , 2015, “ The Mechanochemistry of Cytoskeletal Force Generation,” Biomech. Model. Mechanobiol., 14(1), pp. 59–72. 10.1007/s10237-014-0588-2 [DOI] [PubMed] [Google Scholar]
- [222]. Ronan, W. , Deshpande, V. S. , McMeeking, R. M. , and McGarry, J. P. , 2013, “ Cellular Contractility and Substrate Elasticity: A Numerical Investigation of the Actin Cytoskeleton and Cell Adhesion,” Biomech. Model. Mechanobiol., 13(2), pp. 417–435. 10.1007/s10237-013-0506-z [DOI] [PubMed] [Google Scholar]
- [223]. Jamali, Y. , Jamali, T. , and Mofrad, M. R. K. , 2013, “ An Agent Based Model of Integrin Clustering: Exploring the Role of Ligand Clustering, Integrin Homo-Oligomerization, Integrin–Ligand Affinity, Membrane Crowdedness and Ligand Mobility,” J. Comput. Phys., 244, pp. 264–278. 10.1016/j.jcp.2012.09.010 [DOI] [Google Scholar]
- [224]. Zemel, A. , Rehfeldt, F. , Brown, A. E. X. , Discher, D. E. , and Safran, S. A. , 2010, “ Optimal Matrix Rigidity for Stress Fiber Polarization in Stem Cells,” Nat. Phys., 6(6), pp. 468–473. 10.1038/nphys1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225]. Gouget, C. L. M. , Hwang, Y. , and Barakat, A. I. , 2016, “ Model of Cellular Mechanotransduction Via Actin Stress Fibers,” Biomech. Model. Mechanobiol., 15(2), pp. 331–344. 10.1007/s10237-015-0691-z [DOI] [PubMed] [Google Scholar]
- [226]. Kang, J. , Puskar, K. M. , Ehrlicher, A. J. , LeDuc, P. R. , and Schwartz, R. S. , 2015, “ Structurally Governed Cell Mechanotransduction Through Multiscale Modeling,” Sci. Rep., 5, p. 8622. 10.1038/srep08622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227]. De Santis, G. , Lennon, A. B. , Boschetti, F. , Verhegghe, B. , Verdonck, P. , and Prendergast, P. J. , 2011, “ How Can Cells Sense the Elasticity of a Substrate? An Analysis Using a Cell Tensegrity Model,” Eur. Cell. Mater., 22, pp. 202–213. 10.22203/eCM.v022a16 [DOI] [PubMed] [Google Scholar]
- [228]. Kim, T. , Hwang, W. , Lee, H. , and Kamm, R. D. , 2009, “ Computational Analysis of Viscoelastic Properties of Crosslinked Actin Networks,” PLoS Comput. Biol., 5(7), p. e1000439. 10.1371/journal.pcbi.1000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229]. Huisman, E. M. , van Dillen, T. , Onck, P. R. , and Van der Giessen, E. , 2007, “ Three-Dimensional Cross-Linked F-Actin Networks: Relation Between Network Architecture and Mechanical Behavior,” Phys. Rev. Lett., 99(20), p. 208103. 10.1103/PhysRevLett.99.208103 [DOI] [PubMed] [Google Scholar]
- [230]. Wang, S. , and Wolynes, P. G. , 2012, “ Active Contractility in Actomyosin Networks,” Proc. Natl. Acad. Sci., 109(17), pp. 6446–6451. 10.1073/pnas.1204205109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231]. Alvarado, J. , Sheinman, M. , Sharma, A. , MacKintosh, F. C. , and Koenderink, G. H. , 2013, “ Molecular Motors Robustly Drive Active Gels to a Critically Connected State,” Nat. Phys., 9(9), pp. 591–597. 10.1038/nphys2715 [DOI] [Google Scholar]
- [232]. Åström, J. A. , Kumar, P. B. S. , Vattulainen, I. , and Karttunen, M. , 2008, “ Strain Hardening, Avalanches, and Strain Softening in Dense Cross-Linked Actin Networks,” Phys. Rev. E, 77(5), p. 051913. 10.1103/PhysRevE.77.051913 [DOI] [PubMed] [Google Scholar]
- [233]. Zeng, Y. , Yip, A. K. , Teo, S.-K. , and Chiam, K.-H. , 2012, “ A Three-Dimensional Random Network Model of the Cytoskeleton and Its Role in Mechanotransduction and Nucleus Deformation,” Biomech. Model. Mechanobiol., 11(1–2), pp. 49–59. 10.1007/s10237-011-0292-4 [DOI] [PubMed] [Google Scholar]
- [234]. Zemel, A. , 2015, “ Active Mechanical Coupling Between the Nucleus, Cytoskeleton and the Extracellular Matrix, and the Implications for Perinuclear Actomyosin Organization,” Soft Matter, 11(12), pp. 2353–2363. 10.1039/C4SM02425G [DOI] [PubMed] [Google Scholar]
- [235]. Caille, N. , Thoumine, O. , Tardy, Y. , and Meister, J.-J. , 2002, “ Contribution of the Nucleus to the Mechanical Properties of Endothelial Cells,” J. Biomech., 35(2), pp. 177–187. 10.1016/S0021-9290(01)00201-9 [DOI] [PubMed] [Google Scholar]
- [236]. Ferko, M. C. , Bhatnagar, A. , Garcia, M. B. , and Butler, P. J. , 2007, “ Finite-Element Stress Analysis of a Multicomponent Model of Sheared and Focally Adhered Endothelial Cells,” Ann. Biomed. Eng., 35(2), pp. 208–223. 10.1007/s10439-006-9223-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237]. Slomka, N. , and Gefen, A. , 2010, “ Confocal Microscopy-Based Three-Dimensional Cell-Specific Modeling for Large Deformation Analyses in Cellular Mechanics,” J. Biomech., 43(9), pp. 1806–1816. 10.1016/j.jbiomech.2010.02.011 [DOI] [PubMed] [Google Scholar]
- [238]. Jean, R. P. , Chen, C. S. , and Spector, A. A. , 2005, “ Finite-Element Analysis of the Adhesion-Cytoskeleton-Nucleus Mechanotransduction Pathway During Endothelial Cell Rounding: Axisymmetric Model,” ASME J. Biomech. Eng., 127(4), pp. 594–600. 10.1115/1.1933997 [DOI] [PubMed] [Google Scholar]
- [239]. Dahl, K. N. , Kahn, S. M. , Wilson, K. L. , and Discher, D. E. , 2004, “ The Nuclear Envelope Lamina Network Has Elasticity and a Compressibility Limit Suggestive of a Molecular Shock Absorber,” J. Cell Sci., 117(Pt. 20), pp. 4779–4786. 10.1242/jcs.01357 [DOI] [PubMed] [Google Scholar]
- [240]. Tessier, F. , Boal, D. H. , and Discher, D. E. , 2003, “ Networks With Fourfold Connectivity in Two Dimensions,” Phys. Rev. E, 67(1), p. 011903. 10.1103/PhysRevE.67.011903 [DOI] [PubMed] [Google Scholar]
- [241]. Funkhouser, C. M. , Sknepnek, R. , Shimi, T. , Goldman, A. E. , Goldman, R. D. , and Olvera de la Cruz, M. , 2013, “ Mechanical Model of Blebbing in Nuclear Lamin Meshworks,” Proc. Natl. Acad. Sci. U.S.A., 110(9), pp. 3248–3253. 10.1073/pnas.1300215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242]. Arya, G. , and Schlick, T. , 2006, “ Role of Histone Tails in Chromatin Folding Revealed by a Mesoscopic Oligonucleosome Model,” Proc. Natl. Acad. Sci., 103(44), pp. 16236–16241. 10.1073/pnas.0604817103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243]. Cocco, S. , Marko, J. F. , Monasson, R. , Sarkar, A. , and Yan, J. , 2003, “ Force-Extension Behavior of Folding Polymers,” Eur. Phys. J. E Soft Matter, 10(3), pp. 249–263. 10.1140/epje/i2002-10113-2 [DOI] [PubMed] [Google Scholar]
- [244]. Barbieri, M. , Chotalia, M. , Fraser, J. , Lavitas, L.-M. , Dostie, J. , Pombo, A. , and Nicodemi, M. , 2012, “ Complexity of Chromatin Folding Is Captured by the Strings and Binders Switch Model,” Proc. Natl. Acad. Sci. U.S.A., 109(40), pp. 16173–16178. 10.1073/pnas.1204799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245]. Marenduzzo, D. , Micheletti, C. , and Cook, P. R. , 2006, “ Entropy-Driven Genome Organization,” Biophys. J., 90(10), pp. 3712–3721. 10.1529/biophysj.105.077685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246]. Carrero, G. , Hendzel, M. J. , and de Vries, G. , 2006, “ Modelling the Compartmentalization of Splicing Factors,” J. Theor. Biol., 239(3), pp. 298–312. 10.1016/j.jtbi.2005.07.019 [DOI] [PubMed] [Google Scholar]
- [247]. Stricker, J. , Sabass, B. , Schwarz, U. S. , and Gardel, M. L. , 2010, “ Optimization of Traction Force Microscopy for Micron-Sized Focal Adhesions,” J. Phys. Condens. Matter Inst. Phys. J., 22(19), p. 194104. 10.1088/0953-8984/22/19/194104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248]. Lessey, E. C. , Guilluy, C. , and Burridge, K. , 2012, “ From Mechanical Force to RhoA Activation,” Biochemistry, 51(38), pp. 7420–7432. 10.1021/bi300758e [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249]. Borau, C. , Kamm, R. D. , and García-Aznar, J. M. , 2014, “ A Time-Dependent Phenomenological Model for Cell Mechano-Sensing,” Biomech. Model. Mechanobiol., 13(2), pp. 451–462. 10.1007/s10237-013-0508-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250]. Étienne, J. , Fouchard, J. , Mitrossilis, D. , Bufi, N. , Durand-Smet, P. , and Asnacios, A. , 2015, “ Cells as Liquid Motors: Mechanosensitivity Emerges From Collective Dynamics of Actomyosin Cortex,” Proc. Natl. Acad. Sci., 112(9), pp. 2740–2745. 10.1073/pnas.1417113112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251]. Moreo, P. , García-Aznar, J. M. , and Doblaré, M. , 2008, “ Modeling Mechanosensing and Its Effect on the Migration and Proliferation of Adherent Cells,” Acta Biomater., 4(3), pp. 613–621. 10.1016/j.actbio.2007.10.014 [DOI] [PubMed] [Google Scholar]
- [252]. McGarry, J. P. , Fu, J. , Yang, M. T. , Chen, C. S. , McMeeking, R. M. , Evans, A. G. , and Deshpande, V. S. , 2009, “ Simulation of the Contractile Response of Cells on an Array of Micro-Posts,” Philos. Trans. R. Soc. London Math. Phys. Eng. Sci., 367(1902), pp. 3477–3497. 10.1098/rsta.2009.0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253]. Marcq, P. , Yoshinaga, N. , and Prost, J. , 2011, “ Rigidity Sensing Explained by Active Matter Theory,” Biophys. J., 101(6), pp. L33–L35. 10.1016/j.bpj.2011.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254]. Parameswaran, H. , Lutchen, K. R. , and Suki, B. , 2014, “ A Computational Model of the Response of Adherent Cells to Stretch and Changes in Substrate Stiffness,” J. Appl. Physiol., 116(7), pp. 825–834. 10.1152/japplphysiol.00962.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255]. Vernerey, F. J. , and Farsad, M. , 2011, “ A Constrained Mixture Approach to Mechano-Sensing and Force Generation in Contractile Cells,” J. Mech. Behav. Biomed. Mater., 4(8), pp. 1683–1699. 10.1016/j.jmbbm.2011.05.022 [DOI] [PubMed] [Google Scholar]
- [256]. Milan, J. L. , Wendling-Mansuy, S. , Jean, M. , and Chabrand, P. , 2007, “ Divided Medium-Based Model for Analyzing the Dynamic Reorganization of the Cytoskeleton During Cell Deformation,” Biomech. Model. Mechanobiol., 6(6), pp. 373–390. 10.1007/s10237-006-0057-7 [DOI] [PubMed] [Google Scholar]
- [257]. Milan, J.-L. , Lavenus, S. , Pilet, P. , Louarn, G. , Wendling, S. , Heymann, D. , Layrolle, P. , and Chabrand, P. , 2013, “ Computational Model Combined With in vitro Experiments to Analyse Mechanotransduction During Mesenchymal Stem Cell Adhesion,” Eur. Cell. Mater., 25, pp. 97–113. 10.22203/eCM.v025a07 [DOI] [PubMed] [Google Scholar]
- [258]. Milan, J.-L. , Manifacier, I. , Beussman, K. M. , Han, S. J. , Sniadecki, N. J. , About, I. , and Chabrand, P. , 2016, “ In Silico CDM Model Sheds Light on Force Transmission in Cell From Focal Adhesions to Nucleus,” J. Biomech., 49(13), pp. 2625–2634. 10.1016/j.jbiomech.2016.05.031 [DOI] [PubMed] [Google Scholar]
- [259]. King, M. C. , and Lusk, C. P. , 2016, “ A Model for Coordinating Nuclear Mechanics and Membrane Remodeling to Support Nuclear Integrity,” Curr. Opin. Cell Biol., 41, pp. 9–17. 10.1016/j.ceb.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260]. Saunders, C. A. , and Luxton, G. W. G. , 2016, “ LINCing Defective Nuclear-Cytoskeletal Coupling and DYT1 Dystonia,” Cell. Mol. Bioeng., 9(2), pp. 207–216. 10.1007/s12195-016-0432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261]. Baker, B. M. , and Chen, C. S. , 2012, “ Deconstructing the Third Dimension: How 3D Culture Microenvironments Alter Cellular Cues,” J. Cell Sci., 125(Pt. 13), pp. 3015–3024. 10.1242/jcs.079509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262]. Grinnell, F. , Ho, C.-H. , Tamariz, E. , Lee, D. J. , and Skuta, G. , 2003, “ Dendritic Fibroblasts in Three-Dimensional Collagen Matrices,” Mol. Biol. Cell, 14(2), pp. 384–395. 10.1091/mbc.E02-08-0493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263]. Hakkinen, K. M. , Harunaga, J. S. , Doyle, A. D. , and Yamada, K. M. , 2011, “ Direct Comparisons of the Morphology, Migration, Cell Adhesions, and Actin Cytoskeleton of Fibroblasts in Four Different Three-Dimensional Extracellular Matrices,” Tissue Eng. Part A, 17(5–6), pp. 713–724. 10.1089/ten.tea.2010.0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264]. Jorgens, D. M. , Inman, J. L. , Wojcik, M. , Robertson, C. , Palsdottir, H. , Tsai, W.-T. , Huang, H. , Bruni-Cardoso, A. , López, C. S. , Bissell, M. J. , Xu, K. , and Auer, M. , 2016, “ Deep Nuclear Invaginations Linked to Cytoskeletal Filaments: Integrated Bioimaging of Epithelial Cells in 3D Culture,” J. Cell Sci. (in press). 10.1242/jcs.190967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265]. Deguchi, S. , Maeda, K. , Ohashi, T. , and Sato, M. , 2005, “ Flow-Induced Hardening of Endothelial Nucleus as an Intracellular Stress-Bearing Organelle,” J. Biomech., 38(9), pp. 1751–1759. 10.1016/j.jbiomech.2005.06.003 [DOI] [PubMed] [Google Scholar]
- [266]. Harada, T. , Swift, J. , Irianto, J. , Shin, J.-W. , Spinler, K. R. , Athirasala, A. , Diegmiller, R. , Dingal, P. C. D. P. , Ivanovska, I. L. , and Discher, D. E. , 2014, “ Nuclear Lamin Stiffness Is a Barrier to 3D Migration, But Softness Can Limit Survival,” J. Cell Biol., 204(5), pp. 669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267]. Balestrini, J. L. , Chaudhry, S. , Sarrazy, V. , Koehler, A. , and Hinz, B. , 2012, “ The Mechanical Memory of Lung Myofibroblasts,” Integr. Biol. Quant. Biosci. Nano Macro, 4(4), pp. 410–421. 10.1039/c2ib00149g [DOI] [PubMed] [Google Scholar]
- [268]. Yang, C. , Tibbitt, M. W. , Basta, L. , and Anseth, K. S. , 2014, “ Mechanical Memory and Dosing Influence Stem Cell Fate,” Nat. Mater., 13(6), pp. 645–652. 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269]. Heo, S.-J. , Thorpe, S. D. , Driscoll, T. P. , Duncan, R. L. , Lee, D. A. , and Mauck, R. L. , 2015, “ Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells,” Sci. Rep., 5, p. 16895. 10.1038/srep16895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270]. Heo, S. J. , Driscoll, T. P. , Thorpe, S. D. , Nerurkar, N. L. , Baker, B. M. , Yang, M. T. , Chen, C. S. , Lee, D. A. , and Mauck, R. L. , 2016, “ Differentiation Alters Stem Cell Nuclear Architecture, Mechanics, and Mechano-sensitivity,” eLife, 5, p. e18207. 10.7554/eLife.18207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271]. Cao, X. , Moeendarbary, E. , Isermann, P. , Davidson, P. M. , Wang, X. , Chen, M. B. , Burkart, A. K. , Lammerding, J. , Kamm, R. D. , and Shenoy, V. B. , 2016, “ A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration,” Biophys. J., 111(7), pp. 1541–1552. 10.1016/j.bpj.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]


