Abstract
Plant mitochondrial (mt) genomes have long been known to evolve slowly in sequence. Here we show remarkable departure from this pattern of conservative evolution in a genus of flowering plants. Substitution rates at synonymous sites vary substantially among lineages within Plantago. At the extreme, rates in Plantago exceed those in exceptionally slow plant lineages by ≈4,000-fold. The fastest Plantago lineages set a new benchmark for rapid evolution in a DNA genome, exceeding even the fastest animal mt genome by an order of magnitude. All six mt genes examined show similarly elevated divergence in Plantago, implying that substitution rates are highly accelerated throughout the genome. In contrast, substitution rates show little or no elevation in Plantago for each of four chloroplast and three nuclear genes examined. These results, combined with relatively modest elevations in rates of nonsynonymous substitutions in Plantago mt genes, indicate that major, reversible changes in the mt mutation rate probably underlie the extensive variation in synonymous substitution rates. These rate changes could be caused by major changes in any number of factors that control the mt mutation rate, from the production and detoxification of oxygen free radicals in the mitochondrion to the efficacy of mt DNA replication and/or repair.
Keywords: genome evolution, plant mitochondria, Plantago, rate variation, synonymous substitution rates
In a pioneering study 25 years ago, Brown et al. (1) discovered that primate mitochondrial (mt) DNA evolves rapidly at the sequence level compared with nuclear DNA. With rare exception (2), most animal mt DNAs have been found to evolve rapidly in sequence (3–6). Rapid mt evolution may be the rule in other groups of eukaryotes, although this conclusion must be tempered by the scanty data and distant comparisons available for most groups (7, 8). Plants are the most glaring exception to the general rule that mt DNA evolves rapidly in sequence. In 1987, Wolfe et al. (9) showed that rates of synonymous substitution in angiosperm mt genes are anomalously low, a few-fold lower than in chloroplast genes, ≈10- to 20-fold lower than in nuclear genes of both angiosperms and mammals, and ≈50- to 100-fold lower than in mammalian mt genes. A year later, Palmer and Herbon (10) extended the inference of low rates of sequence change to the entire plant mt genome (most of which is noncoding) and showed that rates of sequence and structural evolution are dramatically uncoupled in plant mt DNA.
All subsequent studies have confirmed that nucleotide substitution rates are in general quite low in land plant mt genomes (11, 12). At the same time, moderate variation in synonymous substitution rates (RS) (up to 7-fold) has been found in comparing several groups of plants (13–16). In most cases, correlated rate changes are seen for chloroplast and/or nuclear genes. Forces operating across the two organelle genomes or all three genomes, such as paternal transmission of organelles or generation-time effects, respectively, have been invoked to explain these patterns, whereas a shift to heterotrophy has been invoked to explain correlated increases in rRNA substitution rates in parasitic plants (17). Phylogenetic studies suggest rate variation in other groups of land plants (18–23), although these studies have lacked a quantitative focus.
We now report unprecedented levels and variation in mt RS, much of which is confined to a single genus of flowering plants (Plantago, a large group of cosmopolitan weeds). Moreover, unlike previous cases of (modest) rate variation in plant mitochondria, rate variation in Plantago is specific to the mt genome. Radical and reversible changes in the underlying mt mutation rate are probably responsible for this rate variation.
Materials and Methods
Most Plantago DNAs were provided by the Royal Botanic Gardens (Kew, U.K.). All other DNAs are as described in ref. 24. PCR products were generated, purified, and sequenced as described in refs. 25 and 26. Taxon names and GenBank accession numbers for all sequences used in this study are listed in Tables 2–4, which are published as supporting information on the PNAS web site. PCR primer sequences and aligned data sets are available upon request.
Phylogenetic relationships within Plantaginaceae (see Fig. 2A) were determined from a 4,730-nt data set consisting of portions of four chloroplast regions (ndhF, rbcL, and intergenic spacers atpB/rbcL and trnL/trnF). Relationships within Plantago subgenus Plantago (see Fig. 2B) were analyzed from a 9,845-nt data set containing two additional chloroplast regions (intergenic spacers psaA/trnS and trnC/trnD). Maximum likelihood (ML) trees were constructed with paup* by using the general time-reversible model, a gamma distribution with four rate categories, and an estimate of the proportion of invariant sites. The rate matrix, base frequencies, shape of the gamma distribution, and proportion of invariant sites were estimated before the ML analysis from a neighbor-joining tree constructed from the data.
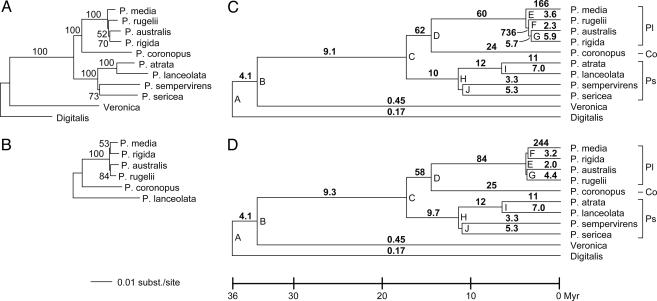
Fig. 2.
Estimates of phylogeny, divergence times, and RS values in the Plantaginaceae. (A and B) ML trees of Plantaginaceae (A) and subgenus Plantago (B) were based on four and six chloroplast regions, respectively. Numbers indicate bootstrap support from 1,000 ML replicates. Seven outgroup species were included in the analysis in A but are not shown. (C and D) Chronograms of Plantaginaceae based on the topology found in A (C), and the topology found in B (D) (see Materials and Methods). Internal nodes are labeled A–J. Values above each branch indicate mean RS in SSB from Table 1 for that branch.
Divergence times outside Plantaginaceae were taken from ref. 27. Those within the family were calculated by using a penalized likelihood approach (28) as implemented in the r8s program (29) and a time constraint of 48 million years (27) for the Antirrhinum/Plantago split. The ML tree shown in Fig. 2A was used as the starting tree for the divergence time analysis shown in Fig. 2C. For Fig. 2D, the starting tree was constructed by first constraining the taxa in the 4,730-nt data set to incorporate the alternative relationships within subgenus Plantago from Fig. 2B and then estimating branch lengths for this topology in paup*. A smoothing factor of three was determined by using the r8s cross-validation procedure. Different starting points of initial age estimates and reanalysis after perturbation of the final age estimates had no effect on the results. Standard errors for each time were calculated by rerunning the divergence time analyses on 500 bootstrapped data sets as described in ref. 29.
Branch lengths for the gene trees in Fig. 1 and Fig. 3, which is published as supporting information on the PNAS web site, were estimated by using paml 3.13d (30) and topologically constrained trees. Relationships within Plantaginaceae were constrained according to the analyses shown in Fig. 2 A and B. For all other taxa, topologies were constrained according to ref. 31. Branch lengths, representing the number of substitutions per synonymous site (dS) or number of substitutions per nonsynonymous site (dN), were determined for protein genes by using a simplified Goldman–Yang (GY94) codon model (32) with separate dN/dS ratios (ω) for each branch. Codon frequencies were computed by using the F3 × 4 method (32). The transition/transversion rate ratio (κ) and dN/dS ratios were estimated during the analysis with initial values of 2 and 0.4, respectively. Standard errors for total branch lengths were reported by paml, and these values were propagated to calculate standard errors for their corresponding dS and dN branch lengths. Branch lengths representing total substitutions per site were estimated for rRNA genes by using the general time-reversible nucleotide model and a gamma distribution for rate variation with four categories. The rate matrix, nucleotide frequencies, and shape of the gamma parameter (initial value of 0.5) were estimated during the analysis.
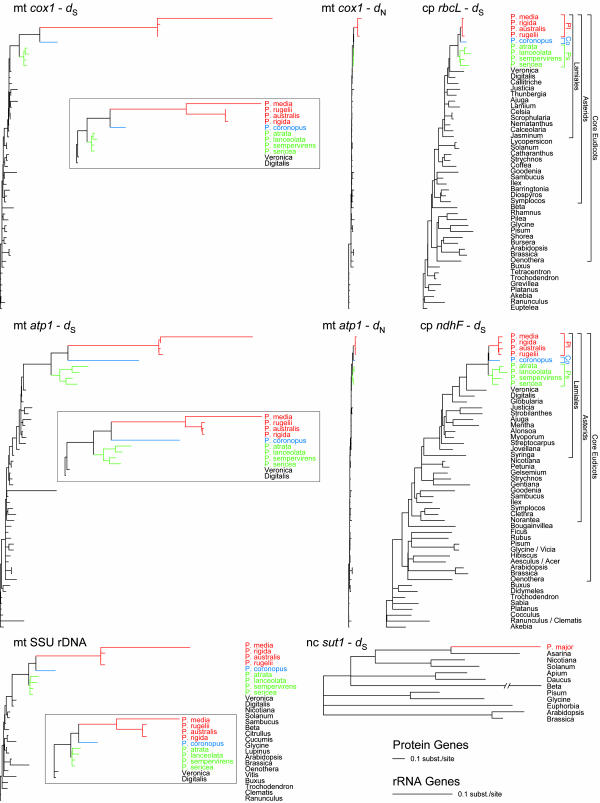
Fig. 1.
Substitution rates in Plantago are highly elevated and variable in mt genes compared with chloroplast (cp) and nuclear (nc) genes. Shown are topologically constrained ML trees based on synonymous (dS) or nonsynonymous (dN) sites for protein genes (all drawn to the same scale) or all sites for SSU rDNA. The three mt trees at left differ from the three boxed (Top Inset, Middle Inset, and Bottom Inset) trees only according to the constraints placed on subgenus Plantago (constrained as shown in Fig. 2 B and A, respectively). Note that the picture of highly elevated and variable RS in Plantago evident in these trees was observed for both unconstrained and constrained trees (Fig. 6) indicating that our findings are not an artifact of topological constraints. Taxa within Plantago are color-coded by subgenus. Pl, subgenus Plantago; Co, Coronopus; Ps, Psyllium. Mt gene trees were based on 1,341 (cox1), 1,272 (atp1), and 1,401 (SSU rDNA) nt; chloroplast gene trees were based on 1,317 (rbcL) and 1,674 (ndhF) nt; and the nuclear sut1 tree was based on 1,293 nt.
Absolute RS values per branch were calculated by dividing the synonymous branch length by the length of time for that branch. Standard errors for RS were determined by propagating the errors associated with branch length and time. Additional uncertainties in RS stem from the calibration point used in the divergence time estimates (see Fig. 2; see also Supporting Results, Figs. 3–6, and Tables 2–8, which are published as supporting information on the PNAS web site) and from uncertainty in the phylogenetic position of Plantago media (see Results).
Results
In a preliminary report (33), we provided evidence of potentially rapid mt evolution in one species of Plantago (Plantago rugelii) based on anomalously poor mt hybridization in Southern blots and partial sequencing of two mt genes. Nearly full-length sequences have now been obtained for six mt genes from P. rugelii. Phylogenetic analysis of synonymous sites for the four protein genes and of all positions for the two rRNA genes illustrates extraordinary divergence of all six P. rugelii genes when compared with each of several other diverse eudicots examined (Fig. 3). In contrast, sequences from either P. rugelii or the closely related Plantago major for four chloroplast genes and three nuclear genes show unexceptional divergence compared with other eudicots (Fig. 3).
To obtain a fuller picture of the tempo and pattern of mt DNA evolution in Plantago and related taxa, nearly complete sequences were determined from eight additional Plantago species for three mt genes [atp1, cox1, and small subunit (SSU) rRNA-encoding DNA (rDNA)] and two chloroplast genes (rbcL and ndhF). Phylogenetic trees of synonymous and nonsynonymous sites (or of all sites for SSU rDNA) were used to illustrate variation in substitution rates. The most notable feature of these trees (Fig. 1) is the extraordinary divergence within Plantago at synonymous sites for both mt protein genes and at all sites for the mt SSU rDNA. Furthermore, the nine Plantago species fall more or less into the same four groups according to their degree of enhanced mt sequence divergence (Fig. 1; see also the report on relative rate tests in Supporting Results).
To quantify the evolution of mt RS within Plantago and on selected branches elsewhere in the trees shown in Fig. 1, absolute substitution rates were calculated for a given tree branch by dividing its synonymous branch length by the estimated divergence time along that branch. Divergence times within Plantaginaceae were calculated by using two different phylogenetic estimates for the group owing to the uncertain placement of P. media, which shows the greatest mt divergence (Fig. 1). The first estimate was based on a 4,730-nt chloroplast data set. Apart from Plantago subgenus Plantago, and especially the placement of P. media, relationships are well supported in this tree (Fig. 2A) and in good agreement with previous molecular studies of the family (34, 35). To achieve more robust placement of P. media, the number of chloroplast characters was doubled (to 9,845 nt) for subgenus Plantago and two outgroups. This increase in chloroplast characters gave a topology (Fig. 2B) in which P. media is sister to P. rigida but with bootstrap support that is equally as low (53% versus 52%) as its placement in Fig. 2 A as sister to the other three species of subgenus Plantago. The failure to resolve relationships within subgenus Plantago by using nearly 10,000 nt, together with the short internodes within the subgenus, implies a rapid radiation for these four species (see Fig. 4 for the single-gene chloroplast trees).
All estimates of Plantaginaceae divergence times and most estimates of substitution rates are similar between the two analyses (Fig. 2 C and D and Table 1), with the only major differences occurring, as expected, within subgenus Plantago. Consequently, we discuss below only the rate estimates from Fig. 2 B and D, except where noted otherwise. The absolute rate of dS, designated RS, is relatively low in the terminal lineages leading to Veronica and Digitalis, the sister and near-sister of Plantago. For Veronica, RS is 0.28 substitutions per site per billion years (SSB) for cox1 and 0.63 SSB for atp1 (mean RS weighted by gene length = 0.45), whereas the Digitalis values are 0.25 for cox1 and 0.09 for atp1 (mean RS = 0.17). These rates are slightly lower to a few-fold lower than the average rates among the broad array of eudicots analyzed in Fig. 1, as averaged among them from the tips of the trees to either the common ancestor of Lamiales (mean RS for both genes = 1.0), of asterids (mean RS = 0.73), or of all core eudicots (mean RS = 0.66) (Table 1). In striking contrast, synonymous rates are much higher on the stem branch leading to Plantago (mean RS = 9.3) and on most branches within the genus (Table 1 and Fig. 2 C and D).
Table 1. Absolute synonymous substitution rates (RS) for atp1 and cox1.
| atp1
|
cox1
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Branch | Time, Myr | dS × 100 | RS, SSB | dN/dS | dS × 100 | RS, SSB | dN/dS | Mean RS, SSB |
| Group means | ||||||||
| Lamiales | 64 ± 4 | 10 ± 4 | 1.6 ± 0.6 | 0.095 | 3.2 ± 2.5 | 0.50 ± 0.39 | 0.17 | 1.0 |
| Asterids | 107 ± 5 | 10 ± 6 | 0.96 ± 0.58 | 0.096 | 5.5 ± 2.7 | 0.51 ± 0.26 | 0.13 | 0.73 |
| Core eudicots | 114 ± 5 | 9.3 ± 6.0 | 0.81 ± 0.52 | 0.11 | 5.8 ± 2.6 | 0.51 ± 0.23 | 0.21 | 0.66 |
| Selected Non-Plantaginaceae | ||||||||
| Goodenia | 94 ± 5 | 41 ± 5 | 4.4 ± 0.5 | 0.033 | 10 ± 2 | 1.1 ± 0.2 | 0.14 | 2.7 |
| Sambucus | 94 ± 5 | 0.95 ± 0.37 | 0.10 ± 0.04 | 0.45 | 0.25 ± 0.09 | 0.027 ± 0.010 | 3.0 | 0.063 |
| Trochodendron | 124 ± 5 | 0.97 ± 0.43 | 0.078 ± 0.035 | 0.23 | 0.59 ± 0.29 | 0.048 ± 0.024 | 0.54 | 0.062 |
| Platanus | 130 ± 6 | 0.98 ± 0.57 | 0.075 ± 0.044 | 0 | 1.05 ± 0.4 | 0.081 ± 0.027 | 0.65 | 0.078 |
| Plantaginaceae† | ||||||||
| A to Digitalis | 36 ± 1 | 0.33 ± 0.19 | 0.091 ± 0.052 | 0.64 | 0.90 ± 0.41 | 0.25 ± 0.11 | 0.23 | 0.17 |
| A to B | 3 ± 2 | 1.4 ± 0.6 | 5.1 ± 4.0 | 0.16 | 0.84 ± 0.55 | 3.1 ± 2.8 | 0 | 4.1 |
| B to Veronica | 33 ± 1 | 2.1 ± 0.8 | 0.63 ± 0.24 | 0.099 | 0.92 ± 0.48 | 0.28 ± 0.14 | 0.11 | 0.45 |
| B to C | 16 ± 2 | 23 ± 4 | 14 ± 3 | 0.037 | 7.8 ± 1.6 | 4.7 ± 1.1 | 0.13 | 9.3 |
| C to D | 3 ± 1 | 14 ± 4 | 52 ± 29 | 0.045 | 17 ± 5 | 64 ± 35 | 0.016 | 58 |
| D to P. coronopus | 14 ± 1 | 59 ± 8 | 41 ± 6 | 0.022 | 15 ± 4 | 10 ± 3 | 0.022 | 25 |
| D to E | 10 ± 1 | 76 ± 9 | 73 ± 11 | 0.022 | 99 ± 29 | 95 ± 29 | 0.034 | 84 |
| F to P. media | 3.6 ± 0.4 | 77 ± 9 | 215 ± 35 | 0.014 | 97 ± 11 | 271 ± 45 | 0.030 | 244 |
| F to P. rigida | 3.6 ± 0.4 | 0 ± 0 | 0 | — | 2.2 ± 1.7 | 6.3 ± 4.7 | 0.14 | 3.2 |
| G to P. australis | 3.7 ± 0.4 | 1.2 ± 0.6 | 3.2 ± 1.7 | 0.36 | 0.31 ± 0.22 | 0.84 ± 0.60 | 0.35 | 2.0 |
| G to P. rugelii | 3.7 ± 0.4 | 3.1 ± 1.2 | 8.3 ± 3.4 | 0 | 0.29 ± 0.08 | 0.79 ± 0.24 | 4.8 | 4.4 |
| C to H | 5.7 ± 1.1 | 7.3 ± 2.8 | 13 ± 6 | 0.044 | 3.8 ± 1.9 | 6.7 ± 3.5 | 0.026 | 9.7 |
| H to I | 4.8 ± 0.9 | 10 ± 2 | 21 ± 6 | 0.10 | 1.2 ± 0.6 | 2.6 ± 1.3 | 0.081 | 12 |
| I to P. atrata | 6.4 ± 0.6 | 13 ± 2 | 21 ± 4 | 0.018 | 1.6 ± 0.6 | 2.4 ± 0.9 | 0.22 | 11 |
| I to P. lanceolata | 6.4 ± 0.6 | 5.9 ± 1.6 | 9.2 ± 2.7 | 0 | 3.2 ± 1.0 | 5.0 ± 1.5 | 0.084 | 7.0 |
| J to P. sempervirens | 11 ± 1 | 5.3 ± 1.5 | 4.9 ± 1.4 | 0.023 | 2.0 ± 0.6 | 1.8 ± 0.6 | 0.19 | 3.3 |
| J to P. sericea | 11 ± 1 | 9.2 ± 1.9 | 8.5 ± 1.8 | 0.038 | 2.4 ± 0.9 | 2.2 ± 0.8 | 0.057 | 5.3 |
| Subgenus Plantago‡ | ||||||||
| D to E | 10 ± 1 | 62 ± 9 | 60 ± 11 | 0.023 | 63 ± 15 | 61 ± 16 | 0.037 | 60 |
| E to P. media | 3.8 ± 0.5 | 64 ± 8 | 168 ± 30 | 0.009 | 62 ± 10 | 163 ± 33 | 0.029 | 166 |
| E to F | 0.3 ± 0.6 | 14 ± 6 | 435 ± 886 | 0.031 | 33 ± 9 | 1,020 ± 2,053 | 0.035 | 736 |
| F to P. rugelii | 3.5 ± 0.4 | 2.3 ± 1.0 | 6.5 ± 3.0 | 0 | 0.29 ± 0.08 | 0.83 ± 0.25 | 4.4 | 3.6 |
| G to P. australis | 3.2 ± 0.4 | 1.2 ± 0.9 | 3.8 ± 2.7 | 0 | 0.31 ± 0.22 | 0.96 ± 0.69 | 0.35 | 2.3 |
| G to P. rigida | 3.2 ± 0.4 | 1.6 ± 0.9 | 4.9 ± 2.8 | 0 | 2.2 ± 0.7 | 6.9 ± 2.3 | 0.14 | 5.9 |
Mean RS is the mean of atp1 (1,272 nt) and cox1 (1,341 nt) values weighted by gene length. Group means are the average root to tip distances for taxa within that group (Fig. 1), excluding Goodenia and Plantago (P.). Myr, Million years; —, denominator (dS) = 0.
Taken from Fig. 2 B and D topology.
Taken from Fig. 2 A and C topology.
At the extreme, in the topology with P. media sister to P. rigida (Fig. 2D), RS is 244 SSB on the terminal branch leading to P. media. In the alternative topology (Fig. 2C), RS on this branch is somewhat lower (166), whereas an even more spectacular RS of 736 is estimated on the topology-specific, short E–F internode. Compared with the Digitalis and Veronica terminals, the mean RS on the P. media terminal in Fig. 2D is estimated to be 1,400- and 540-fold higher, respectively, and that on the superfast E–F internode of Fig. 2C is estimated to be 4,300- and 1,600-fold higher.
Our analyses (Fig. 1 and Fig. 5) show evidence for synonymous rate variation elsewhere in eudicots beyond the extreme variation seen in Plantaginaceae. Examples include ≈43-fold faster evolution in Goodenia relative to its sister Sambucus (see also Table 1) and rapid evolution in Justicia and Ajuga relative to their respective sisters. The relatively long internodes subtending short termini leading to the crucifers Arabidopsis and Brassica (Fig. 1), a pattern seen for many other genes in these genomes (36), raise the possibility of relatively rapid evolution early in this group followed by a rate decrease. Exceptionally low RS values over long periods of time are estimated for the “basal” eudicots Trochodendron and Platanus (0.062 and 0.078 SSB, respectively), and for Sambucus (0.063 SSB) (Fig. 1 and Table 1). With the Trochodendron rate as one extreme and the fastest Plantago rate as the other, RS among the eudicots examined in this study ranges by a factor of 3,900 (for the topology in Fig. 2D) or 12,000 (for the topology in Fig. 2C).
Discussion
We have discovered unprecedented variation in rates of mt synonymous substitution within a genus of flowering plants. To the best of our knowledge, the greatest previously measured disparity in mt RS is ≈7-fold, in comparisons of grasses and palms (14), whereas a 10- to 30-fold range of overall substitution rates was reported for a mt intron in comparisons of diverse monocots and eudicots (15). In contrast, we have uncovered on the order of 4,000-fold (and possibly even 12,000-fold) variation in mt synonymous substitution across eudicots. Most of this range is found within a single family (Plantaginaceae) and even within a single genus (Plantago), but significant rate heterogeneity is also evident in a number of other lineages and comparisons (Fig. 1 and Table 1).
Considering these findings and the number of cases of plant mt rate variation already reported (13–17) or indicated by phylogenetic analysis (18–23), we suspect that, as more plants are sampled, more and more evidence will be found for widespread heterogeneity in rates of synonymous substitutions among plant mt DNAs. Denser taxon sampling can only reveal more peaks and valleys (now hidden by averaging across relatively long, poorly sampled branches) in what seems to be an emerging pattern of episodic evolution of mt gene sequences in plants. Discovery of additional cases of dramatic Plantago-like accelerated evolution can be expected, and indeed, we have found a second such case (Geraniaceae, especially Pelargonium; ref. 33 and unpublished data).
Rates of synonymous substitutions in chloroplast genes are much less variable than in mt genes of the same plants (Fig. 1). Chloroplast rates are almost uniformly a few-fold higher than mt rates, and only in Plantago (and Pelargonium) do mt rates significantly exceed chloroplast rates. Thus the long-standing generalization (9, 10) that sequence evolution proceeds more slowly in plant mitochondria compared with chloroplasts is still true, albeit now with spectacular exception.
mt Sequence Evolution in Plants Versus Animals. Animal mt DNA has reigned for 25 years (1) as the exemplar of rapid sequence evolution among cellular genomes, with only certain RNA viruses, replicated by using polymerases that lack proofreading ability, known to evolve more rapidly. However, even the fastest recorded animal mt DNAs (3, 4) pale at 37 SSB compared with the 244 SSB (and possibly even 736 SSB) estimated for the fastest Plantago lineage. Two recent studies have identified lineages of lice (5) and gastropods (6) that may prove to set new speed records for animal mt DNA, however absolute rates have not yet been estimated. Plants further eclipse animals in terms of the known range of mt substitution rates. Even with the discovery of relatively low mt rates in anthozoans, rates that may be ancestrally low for animals (2), the range of mt RS values among all metazoans is only ≈40–100 (2), compared with 4,000 (and possibly as high as 12,000) among just eudicots.
Unlike animal mt DNA, whose generally high substitution rate is usually accompanied by a large excess of transitions relative to transversions (37), there is no evidence for an elevated transition/transversion ratio in Plantago mt DNAs. For cox1 and atp1, this ratio is generally low (≈1.0 in most cases), both within Plantago and across eudicots, and, if anything, it is somewhat depressed in Plantago, especially for cox1 (Table 5). We have limited evidence (unpublished data) for an elevated rate of insertions/deletions in Plantago mt DNAs. More data are needed, however, before it can be concluded, as in the case of mammal mt and nuclear genomes (38, 39), that rates of point and length mutations are closely coupled.
Number and Directionality of Changes in mt Substitution Rates in Plantago. It is clear that the rate of mt synonymous substitutions changed at least several times during the evolution of the Plantaginaceae (Figs. 1 and 2). How many changes, on what branches, with what magnitude, and in what direction are all less clear issues. Certainly, the rate must have increased in a common ancestor of Plantago (Fig. 1), but exactly when and at what magnitude cannot be discerned from the present data. A second rate change is needed to account for the difference in rates between subgenus Psyllium and subgenus Plantago (Fig. 2), with the directionality and timing of this second change dependent on the magnitude of the initial rate increase. In one scenario, a very large initial increase late along branch B–C was perpetuated along the branches leading to subgenus Plantago, followed by a rate decrease along the C–H branch leading to subgenus Psyllium. In a second scenario, a smaller initial increase early on the B–C branch was perpetuated throughout subgenus Psyllium, followed by an additional rate increase along branch C–D. A decrease in RS in the terminal lineage leading to Plantago coronopus is likely under either scenario, although the magnitude and/or timing of this seems to be different for cox1 than for atp1 (Table 1).
Interpretation of the magnitude and timing of rate changes in subgenus Plantago depends on the topology analyzed (Figs. 2, compare C with D). The most parsimonious interpretation of Fig. 2D postulates a major rate decrease in the common ancestor of subgenus Plantago (presumably near the end of the D–E branch), followed by an even greater rate increase on the terminal branch leading to P. media. The topology of Fig. 2C implies a huge, transient change in RS (an increase to 736 SSB, followed by >100-fold decrease) on the very short E–F branch, although the huge error associated with this value (Table 1) makes this change difficult to assess with any confidence, and a nearly 3-fold rate increase on the P. media terminal branch.
These scenarios for the evolution of RS in Plantaginaceae mt DNAs are necessarily speculative and tentative. No matter how one views it, however, it seems clear that mt sequences in Plantaginaceae have been riding a wild rollercoaster, one with multiple major speed-ups and slow-downs. More refined and accurate substitution-rate scenarios should emerge with the generation of further sequence data of three kinds: (i) from mt genes of additional species of Plantago (>200 have yet to be examined), (ii) from mt genes besides atp1 and cox1, and (iii) from additional non-mt markers to better resolve the phylogeny of subgenus Plantago. Denser taxon sampling within Plantago may identify still further episodes of RS change, as some of the relatively long internodes in our current trees may average different periods with significantly different substitution rates. By the same token, we may discover relatively intense but brief periods of even faster evolution than those documented thus far. Further sampling of taxa and mt genes will also allow us to determine whether synonymous rates are relatively homogenous across a mt genome, or, for example, whether rates are occasionally or even consistently higher in certain genes and regions of the mt genome than in others. In this respect, it is worth noting that RS is often 2- to 4-fold higher for atp1 than for cox1 and is rarely much lower (Table 1 and data not shown).
Underlying Basis of Extreme Molecular Divergence in Plantago. The exceptional divergence of Plantago mt genes could potentially be explained if they had been relocated to the (generally) much higher mutational environment of the nucleus (9, 11–13). This possibility can be ruled out because (i) four of the six analyzed mt genes (cox1, cob, and both SSU and large subunit rDNA) have never been found functionally transferred to nuclear DNA of any eukaryote, whereas transfer of the other two in plants is extremely rare (24); (ii) this scenario would not explain the subsequent major rate decrease(s) inferred in subgenus Plantago without invoking unprecedented reverse transfer of genes back to the mitochondrion or major decreases in nuclear substitution rates; (iii) probes for two genes (cob and cox1) have been shown (25) to hybridize preferentially to P. rugelii mt DNA relative to total DNA; (iv) cDNA sequences from these two P. rugelii genes undergo mt-characteristic (40) C→U editing [at one and two sites, respectively (25)]; and (v) both genes lack any obvious mt targeting sequences within 300 nt upstream of their presumptive start codons. The recovery of RNA-edited cDNA sequences also indicates that these two genes are probably functional. Functionality is indicated more generally because all mt protein genes reported in this study have intact reading frames despite their extreme divergence and because dN/dS is not close to 1.0 on any Plantago branch (Table 1).
Another possible explanation for the extreme divergence of Plantago mt genes is exceptionally high levels of RNA editing, such that only mt genes, per se, but not their encoded cDNAs and proteins, are evolving extremely rapidly. This explanation can be ruled out because all three Plantago cDNA sequences determined show only one or two (see above) or no RNA edits, and because Plantago mt genes show no major bias in base composition (e.g., all T residues are not converted to C residues, as would be expected if the conventional, low-level C→U editing of plant mitochondria had been extended to all possible sites).
Instead, because the dramatic rate variation in Plantago is confined to the mt genome (Figs. 1 and 3) and nonsynonymous rates are proportionately less elevated than synonymous rates in Plantago mt genes (Fig. 1, Table 1, and Table 6), we conclude that rate variation in Plantago mt genes is probably a direct consequence of underlying changes in the mt mutation rate. In contrast, most previously reported cases of (much more modest) synonymous site variation in plant mt DNA have been postulated to reflect generation time effects (13–15), although this interpretation has been questioned (41).
The overall pattern and magnitude of mutation rate change in Plantago—a series of relatively recent, major, and reversible changes specifically in the mt mutation rate—is reminiscent of the well established literature on bacterial mutators and antimutators (42, 43). The most common mutators in bacteria are in genes for DNA replication and DNA mismatch repair, and the limited number of mt mutators characterized in lab strains of yeast (44, 45) also fall into these same categories (note that in plants, as in yeast, all of the genes responsible for mt DNA replication and repair are nuclear genes). Mutations causing error-prone replication and/or defective repair could have led to major increases in the mt mutation rate at the base of genus Plantago and at subsequent times in Plantago phylogeny, whereas direct reversals or compensatory suppressions of these mutations could have led to subsequent slow-downs in the mutation rate in certain descendant Plantago lineages. Although replication and mismatch repair are the most commonly found mutators, many other kinds of mutators have also been characterized in bacteria (42, 43). Major changes in the production and/or detoxification (by superoxide dismutase and catalase) of oxygen free-radicals, thought to be a major cause of damage to mt DNA, could in principle underlie this mutation-rate variation.
Supplementary Material
Acknowledgments
We thank Mark Chase (Royal Botanic Gardens) for providing most of the Plantago DNAs used in this study, Danny Rice for helpful discussion, Na Zhou (Indiana University, Bloomington) for help generating some of the Plantago chloroplast sequences, and Greg Young (Indiana University, Bloomington) for help generating two of the atp1 sequences. This research was supported by National Institutes of Health Grant R01-GM-35087 (to J.D.P.)
Abbreviations: mt, mitochondrial; ML, maximum likelihood; SSB, substitutions per site per billion years; dN, substitutions per nonsynonymous site; dS, substitutions per synonymous site; RS, synonymous substitution rate; SSU, small subunit; rDNA, rRNA-encoding DNA.
Data deposition: The nucleotide sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ389588–AJ389621 and AY818897–AY818951).
References
- 1.Brown, W. M., George, M., Jr., & Wilson, A. C. (1979) Proc. Natl. Acad. Sci. USA 76, 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shearer, T. L., Van Oppen, M. J. H., Romano, S. L. & Worheide, G. (2002) Mol. Ecol. 11, 2475–2487. [DOI] [PubMed] [Google Scholar]
- 3.Pesole, G., Gissi, C., De Chirico, A. & Saccone, C. (1999) J. Mol. Evol. 48, 427–434. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, A. J. (2003) J. Mol. Evol. 57, 636–641. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, K. P., Cruickshank, R. H., Adams, R. J., Smith, V. S., Page, R. D. & Clayton, D. H. (2003) Mol. Phylogenet. Evol. 26, 231–242. [DOI] [PubMed] [Google Scholar]
- 6.Williams, S. T., Reid, D. G. & Littlewood, D. T. J. (2003) Mol. Phylogenet. Evol. 28, 60–86. [DOI] [PubMed] [Google Scholar]
- 7.Bruns, T. D. & Szaro, T. M. (1992) Mol. Biol. Evol. 9, 836–855. [DOI] [PubMed] [Google Scholar]
- 8.Gray, M. W. & Spencer, D. F. (1996) in Evolution of Microbial Life, eds. Roberts, D., Sharp, P., Alderson, G. & Collins, M. A. (Cambridge Univ. Press, Cambridge, U.K.), pp. 109–126.
- 9.Wolfe, K. H., Li, W.-H. & Sharp, P. M. (1987) Proc. Natl. Acad. Sci. USA 84, 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer, J. D. & Herbon, L. A. (1988) J. Mol. Evol. 28, 87–97. [DOI] [PubMed] [Google Scholar]
- 11.Gaut, B. S. (1998) Evol. Biol. 30, 93–120. [Google Scholar]
- 12.Muse, S. V. (2000) Plant Mol. Biol. 42, 25–43. [PubMed] [Google Scholar]
- 13.Laroche, J., Li, P., Maggia, L. & Bousquet, J. (1997) Proc. Natl. Acad. Sci. USA 94, 5722–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyre-Walker, A. & Gaut, B. S. (1997) Mol. Biol. Evol. 14, 455–460. [DOI] [PubMed] [Google Scholar]
- 15.Laroche, J. & Bousquet, J. (1999) Mol Biol. Evol. 16, 441–451. [DOI] [PubMed] [Google Scholar]
- 16.Whittle, C.-A. & Johnston, M. O. (2002) Mol. Biol. Evol. 19, 938–949. [DOI] [PubMed] [Google Scholar]
- 17.Duff, R. J. & Nickrent, D. L. (1997) J. Mol. Evol. 45, 631–639. [DOI] [PubMed] [Google Scholar]
- 18.Beckert, S., Steinhauser, S., Muhle, H. & Knoop, V. (1999) Plant Syst. Evol. 218, 179–192. [Google Scholar]
- 19.Vangerow, S., Teerkorn, T. & Knoop, V. (1999) Plant Biol. 1, 235–243. [Google Scholar]
- 20.Chaw, S.-M., Parkinson, C. L., Cheng, Y., Vincent, T. M. & Palmer, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowe, L. M., Coat, G. & dePamphilis, C. W. (2000) Proc. Natl. Acad. Sci. USA 97, 4092–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkman, T. J., Lim, S.-H., Mat Salleh, K. & Nais, J. (2004) Proc. Natl. Acad. Sci. USA 101, 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis, J. I., Stevenson, D. W., Petersen, G., Seberg, L., Campbell, L. M., Freudenstein, J. V., Goldman, D. H., Hardy, C., R., Michelangeli, F. A., Simmons, M. P., et al. (2004). Syst. Bot. 29, 467–510. [Google Scholar]
- 24.Adams, K. L., Qiu, Y.-L., Stoutemyer, M. & Palmer, J. D. (2002) Proc. Natl. Acad. Sci. USA 99, 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho, Y. (1997). Ph.D. thesis, (Indiana University, Bloomington).
- 26.Mower, J. P., Stefanović, S., Young, G. J. & Palmer, J. D. (2004) Nature 432, 165–166. [DOI] [PubMed] [Google Scholar]
- 27.Wikström, N., Savolainen, V. & Chase, M. W. (2001) Proc. R. Soc. London Ser. B 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson, M. J. (2002) Mol. Biol. Evol. 19, 101–109. [DOI] [PubMed] [Google Scholar]
- 29.Sanderson, M. J. (2003) Bioinformatics 19, 301–302. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 31.Angiosperm Phylogeny Group (2003) Bot. J. Linn. Soc. 141, 399–436. [Google Scholar]
- 32.Yang, Z. & Nielson, R. (1998) J. Mol. Evol. 46, 409–418. [DOI] [PubMed] [Google Scholar]
- 33.Palmer, J. D., Adams, K. L., Cho, Y., Parkinson, C. L., Qiu, Y.-L. & Song, K. (2000) Proc. Natl. Acad. Sci. USA 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olmstead, R. G., dePamphilis, C. W., Wolfe, A. D., Young, N. D., Elisons, W. J. & Reeves, P. A. (2001) Am. J. Bot. 88, 348–361. [PubMed] [Google Scholar]
- 35.Rønsted, N., Chase, M. W., Albach, D. C. & Bello, M. A. (2002) Bot. J. Linn. Soc. 139, 323–338. [Google Scholar]
- 36.Bergthorsson, U., Richardson, A. O., Young, G. J., Goertzen, L. R. & Palmer, J. D. (2004) Proc. Natl. Acad. Sci. USA 101, 17747–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown, W. M., Prager, E. M., Wang, A. & Wilson, A. C. (1982) J. Mol. Evol. 18, 225–239. [DOI] [PubMed] [Google Scholar]
- 38.Cann, R. L. & Wilson, A. C. (1983) Genetics 104, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, R. & Ueda, S. (1994). (1994) Mol. Biol. Evol. 11, 504–512. [DOI] [PubMed] [Google Scholar]
- 40.Maier, R. M., Zeltz, P., Kössel, H., Bonnard, G., Gualberto, J. M. & Grienenberger, J. M. (1996) Plant Mol. Biol. 32, 343–365. [DOI] [PubMed] [Google Scholar]
- 41.Whittle, C.-A. & Johnston, M. O. (2003) J. Mol. Evol. 56, 223–233. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. (1996) Annu. Rev. Microbiol. 50, 625–643. [DOI] [PubMed] [Google Scholar]
- 43.Horst, J.-P., Wu, T. & Marinus, M. G. (1999) Trends Microbiol. 7, 29–36. [DOI] [PubMed] [Google Scholar]
- 44.Foury, F. & Vanderstraeten, S. (1992) EMBO J. 11, 2717–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu, J. P., Vanderstraeten, S. & Foury, F. (1995) Gene 160, 105–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.