Abstract
Streptomyces coelicolor M145 was shown to be able to grow in the presence of high concentrations of polyamines, such as putrescine, cadaverine, spermidine, or spermine, as a sole nitrogen source. However, hardly anything is known about polyamine utilization and its regulation in streptomycetes. In this study, we demonstrated that only one of the three proteins annotated as glutamine synthetase-like protein, GlnA3 (SCO6962), was involved in the catabolism of polyamines. Transcriptional analysis revealed that the expression of glnA3 was strongly induced by exogenous polyamines and repressed in the presence of ammonium. The ΔglnA3 mutant was shown to be unable to grow on defined Evans agar supplemented with putrescine, cadaverine, spermidine, and spermine as sole nitrogen source. HPLC analysis demonstrated that the ΔglnA3 mutant accumulated polyamines intracellularly, but was unable to degrade them. In a rich complex medium supplemented with a mixture of the four different polyamines, the ΔglnA3 mutant grew poorly showing abnormal mycelium morphology and decreased life span in comparison to the parental strain. These observations indicated that the accumulation of polyamines was toxic for the cell. An in silico analysis of the GlnA3 protein model suggested that it might act as a gamma-glutamylpolyamine synthetase catalyzing the first step of polyamine degradation. GlnA3-catalyzed glutamylation of putrescine was confirmed in an enzymatic in vitro assay and the GlnA3 reaction product, gamma-glutamylputrescine, was detected by HPLC/ESI-MS. In this work, the first step of polyamine utilization in S. coelicolor has been elucidated and the putative polyamine utilization pathway has been deduced based on the sequence similarity and transcriptional analysis of homologous genes expressed in the presence of polyamines.
Keywords: nitrogen assimilation, Streptomyces coelicolor, GS-like enzyme, GlnA3, polyamine utilization, gamma-glutamylpolyamine synthetase
Introduction
Streptomyces coelicolor is a Gram-positive, non-motile, soil-dwelling bacterium that belongs to genus Streptomyces, phylum Actinobacteria. It can assimilate nitrogen from a variety of mineral (e.g., ammonium, nitrate, and nitrite) and organic sources (e.g., urea, amino acids, peptides, and amino sugars). However, nitrogen sources such as polyamines, which are also omnipresent in nature, have never been reported to be utilized by S. coelicolor. Polyamines are widespread in all life kingdoms. Common polyamines include cadaverine, putrescine, spermidine, and spermine. Polyamines can be synthesized de novo or taken up from the environment. They are predominantly derived from the following amino acids: ornithine, arginine, and lysine (Kusano and Suzuki, 2015; Miller-Fleming et al., 2015). The canonical biosynthesis pathway begins with decarboxylation of either ornithine or arginine to form putrescine or decarboxylation of lysine to generate cadaverine (Kusano and Suzuki, 2015; Miller-Fleming et al., 2015). Putrescine serves as a substrate for spermine and spermidine biosynthesis via addition of aminopropyl groups donated by the decarboxylated S-adenosylmethionine (Kusano and Suzuki, 2015; Miller-Fleming et al., 2015).
Polyamines have been implicated in a wide range of biological processes, thus their intracellular concentration as well as metabolism is tightly regulated (Miller-Fleming et al., 2015). Extensive studies have demonstrated that intracellular polyamine concentrations accumulate during exposure to several stress conditions (reviewed in Miller-Fleming et al., 2015). In bacterial cells, the role of polyamines is less clear and depends on ecological and physiological context. In bacteria, some studies suggest that polyamines might be involved in cell growth stimulation (Yoshida et al., 2004; Igarashi and Kashiwagi, 2006), biofilm formation (Wortham et al., 2007; Lee et al., 2009; Nesse et al., 2015) as well as regulation of translation (Algranati et al., 1976; Nastri et al., 1993; Higashi et al., 2006; Terui et al., 2012) and transcription (Jain and Tyagi, 1987; Huang et al., 1990; Sarkar et al., 1995; Chattopadhyay et al., 2015). Some studies also suggest that they are involved in acid resistance (Jung and Kim, 2003a; Chattopadhyay and Tabor, 2013; Schneider et al., 2013) and the response to oxidative stress (Chattopadhyay et al., 2003; Jung and Kim, 2003b; Tkachenko and Nesterova, 2003; Sakamoto et al., 2015), biosynthesis of siderophores (Griffiths et al., 1984; Brickman and Armstrong, 1996; Oves-Costales et al., 2008; Burrell et al., 2012), SOS system activation (Oh and Kim, 1999) and antibiotic resistance (Nastri and Algranati, 1996; Kwon and Lu, 2006; Tkachenko et al., 2006; Sarathy et al., 2013).
In eukaryotes, polyamines have been described to be implicated in cell growth and development (Pirnes-Karhu et al., 2015; Reis et al., 2016), maturation (Zhou et al., 2009; Tsaniklidis et al., 2016) and proliferation (Heby, 1981; Dudkowska et al., 2003; Maeda et al., 2006; Bercovich et al., 2011; Weiger and Hermann, 2014). They were also reported to be involved in environmental stress response (Valdes-Santiago and Ruiz-Herrera, 2013; Alcazar and Tiburcio, 2014; Minocha et al., 2014; Liu et al., 2015; Pal et al., 2015), defense response (Walters, 2003; Wimalasekera et al., 2011; Zeier, 2013; Hatmi et al., 2014; Mo et al., 2015; Rossi et al., 2015) and apoptosis (Moschou and Roubelakis-Angelakis, 2014; Parhamifar et al., 2014; Cai et al., 2015). Polyamines are positively charged molecules able to interact with negatively charged molecules like RNA, DNA, proteins, polyphosphate, and phospholipids (Norris et al., 2015). Consequently, an imbalance in polyamine metabolism can deeply affect cellular homeostasis. Abolished polyamine biosynthesis leads to a reduction in growth rate in E. coli. (Cunningham-Rundles and Maas, 1975) and to complete arrest of cell proliferation in eukaryotic cells (Pitkin and Davis, 1990; Ray et al., 1999; Chattopadhyay et al., 2002; Odenlund et al., 2009). Polyamine excess is toxic for prokaryotic and eukaryotic organisms (Davis and Ristow, 1991; Tome et al., 1997; Pegg, 2013; Miller-Fleming et al., 2015), hence polyamine uptake and utilization as well as biosynthesis are under strict regulation (Kusano and Suzuki, 2015; Miller-Fleming et al., 2015).
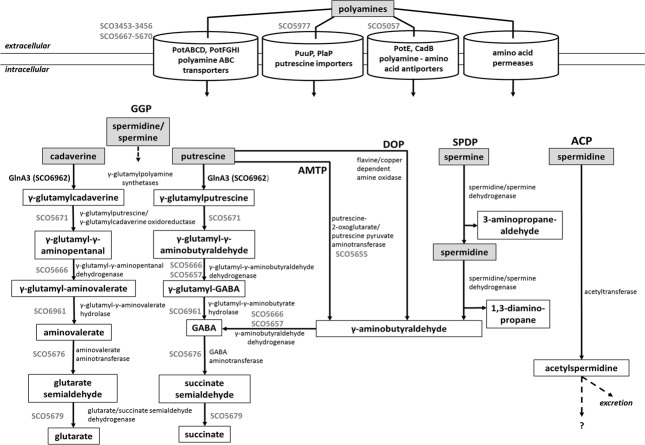
Although polyamine biosynthesis has been broadly studied in evolutionary diverse bacteria and archaea (Tabor and Tabor, 1985; Sekowska et al., 1998; Lee et al., 2009; Nguyen et al., 2015), polyamine catabolism has been mostly studied in gram negative bacteria (Foster et al., 2013; Kusano and Suzuki, 2015). Polyamine utilization pathway is not universal for all bacteria. The currently known polyamine degradative pathways in prokaryotes are summarized in Figure 1. These include the γ-glutamylation pathway, aminotransferase pathway, spermine/spermidine dehydrogenase pathway, direct oxidation pathway and acetylation pathway (Figure 1). However, concerning the acetylation pathway it remains unclear whether acetylated polyamines are further metabolized or excreted (Forouhar et al., 2005; Bai et al., 2011; Planet et al., 2013; Campilongo et al., 2014; Nguyen et al., 2015). Gram-negative bacteria such as E. coli and P. aeruginosa degrade putrescine via GABA to succinate through two pathways: the γ-glutamylation pathway (Kurihara et al., 2005; Yao et al., 2011; Schneider and Reitzer, 2012) and the aminotransferase pathway (Shaibe et al., 1985; Schneider and Reitzer, 2012) (Figure 1). The initial step of the γ-glutamylation pathway involves the glutamylation of putrescine by the γ-glutamylpolyamine synthetase (PuuA) first reported in E. coli (Kurihara et al., 2005). Remarkably, P. aeruginosa possess expanded polyamine catabolic pathway involving seven related γ-glutamylpolyamine synthetases (PauA1–PauA7), each specific for polyamines, monoamines, or other substrates (Yao et al., 2011).
FIGURE 1.
Combined model of polyamine utilization pathways within prokaryotes based on published studies (Yao et al., 2011; Schneider and Reitzer, 2012; Foster et al., 2013; Campilongo et al., 2014; Kusano and Suzuki, 2015). GGP, gamma-glutamylation pathway; AMTP, aminotransferase pathway; DOP, direct oxidation pathway; SPDP, spermine/spermidine dehydrogenase pathway; ACP, acetylation pathway. Dashed arrows represent not entirely investigated metabolic pathways. In gray, deduced steps of the polyamine degradation pathway in S. coelicolor.
Interestingly, the reaction catalyzed by γ-glutamylpolyamine synthetase is comparable to the reaction catalyzed by glutamine synthetase (GS). Both enzymes are able to ligate L-glutamate with an amino group of the specific substrate (polyamine or ammonium, respectively) using energy from ATP. Moreover, both enzyme types share structural similarities (Ladner et al., 2012). Consequently, some proteins annotated as glutamine synthetase-like enzymes (GS-like) may represent still not recognized γ-glutamylpolyamine synthetases. Indeed, Streptomyces coelicolor A3(2) harbors two genes, glnA (SCO2198) and glnII (SCO2210), encoding GSI and GSII, whose function and regulation was extensively studied (Hillemann et al., 1993; Fink et al., 1999; Weisschuh et al., 2000) as well as three other genes, glnA2 (SCO2241), glnA3 (SCO6962), and glnA4 (SCO1613), annotated as encoding putative GS-like enzymes (Rexer et al., 2006). In silico analysis of all glnA-like genes: glnA2, glnA3, and glnA4 across the actinobacterial genomes revealed that glnA has evolved to specialized glnA-like genes encoding proteins that might be involved in the colonization and survival in many diverse habitats (Hayward et al., 2009). Distinct ecological niches occupied by actinobacteria exerted specific evolutional pressure on GS genes resulting in diverse, but so far uncharacterized genes (Hayward et al., 2009). Actinobacterial GSI-like enzymes, whose function is unknown (Rexer et al., 2006) were reported to be poorly expressed and their presence in the cell was judged as not essential for bacterial homeostasis at least under laboratory conditions (Harth et al., 2005). In this work, we demonstrated the involvement of the GSI-like enzyme – GlnA3, a gamma-glutamylpolyamine synthetase, in the polyamine degradation in S. coelicolor M145.
Materials and Methods
Strains and Growth Conditions
Streptomyces coelicolor M145 (parental strain) and mutants were cultured for 4–6 days at 30°C on the defined Evans-agar base (modified after Evans et al., 1970) supplemented with different nitrogen sources: 50 mM monosodium glutamate, L-glutamine, ammonium chloride, sodium nitrate, urea, cadaverine dihydrochloride, putrescine dihydrochloride, spermidine trihydrochloride and spermine tetrahydrochloride in appropriate concentrations. Growth experiments in liquid culture were performed using either complex S-medium (Okanishi et al., 1974) or YEME:TSB (1:1) (Kieser et al., 2000) and chemically defined medium Evans-medium (modified after Evans et al., 1970). If appropriate, media were supplemented with polyamines or ammonium chloride as a sole nitrogen source. Strains were cultivated at 30°C, on the rotary shaker (180 rpm) for 4–6 days. Genetic manipulation of S. coelicolor M145 was performed as described by Kieser et al. (2000) and Gust et al. (2003). For preparation of the genomic DNA, S. coelicolor M145 was grown for 4 days in S-medium and DNA was isolated with the NucleoSpin Tissue Kit (Macherey-Nagel). All strains and plasmids used in this study are listed in the Table 1.
Table 1.
Strains and plasmids used in this study.
| Strains | Genotype | Reference |
|---|---|---|
| E. coli BW25113 | K-12 derivative (ΔaraBAD ΔrhaBAD) carrying plasmid pIJ790, CmR | Datsenko and Wanner, 2000 |
| E. coli ET12567 | dam-13::Tn9, dcm-6, hsdM, CmR, carrying helper plasmid pUZ8002 | MacNeil et al., 1992 |
| E. coli XL1-Blue | recA1, endA1 gyrA96, thi-1, hsdR17, supE44, relA1, lac [F′ proAB lacIqZΔM15 Tn10 TetR] | Bullock et al., 1987 |
| S. coelicolor M145 | S. coelicolor A3(2) without native plasmids: spc1- and spc2- | Kieser et al., 2000 |
| S. coelicolor M145 ΔglnA | glnA mutant strain of S. coelicolor M145; glnA replaced by an aac(3)IV cassette, AprR | Fink et al., 1999 |
| S. coelicolor M145 ΔglnII | glnII mutant strain of S. coelicolor M145; glnII replaced by an aac(3)IV cassette, AprR | Tiffert et al., 2011 |
| S. coelicolor M145 ΔglnAΔglnII | glnAglnII double deletion mutant strain of S. coelicolor M145; AprR and HygR | This work |
| S. coelicolor M145 ΔglnA2 | glnA2 mutant strain of S. coelicolor M145; insertional inactivation of glnA2 by an aac(3)IV cassette, AprR | Nentwich, 2010 |
| S. coelicolor M145 ΔglnA3 | glnA3 mutant strain of S. coelicolor M145; insertional inactivation of glnA3 by an aac(3)IV cassette, AprR | Rexer et al., 2006 |
| S. coelicolor M145 ΔglnA4 | glnA4 mutant strain of S. coelicolor M145; glnA4 replaced by an aac(3)IV cassette, AprR | This work |
| S. coelicolor M145 ΔglnA3pRM4glnA3 | Complemented glnA3 mutant strain of S. coelicolor M145; AprR and KmR | This work |
| pRM4 | pSET152ermEp∗ with artificial RBS, AprR | Menges et al., 2007 |
| pIJ10700 | pBluescript II KS(+) containing hyg-oriT cassette, HygR | Gust et al., 2003 |
| pJOE2775 | pBR322-derived vector with Prha expression cassette | Volff et al., 1996 |
| cosmid St3H12 | AmpR, KanR | Redenbach et al., 1996 |
Construction of the ΔglnA3 Complementation Mutant
To perform a complementation of the mutant, glnA3 gene without its native promoter was amplified by PCR using CglnA3f and CglnA3r primers (Table 2) and cloned into the multiple cloning site of pRM4 plasmid. The correct construct was confirmed by sequencing and subsequently introduced into the ΔglnA3 mutant by conjugation using the E. coli S17 strain. Clones were then selected on resistant phenotype against kanamycin and apramycin. The correct integration of the pRM4-glnA3 was confirmed by PCR and sequencing.
Table 2.
Oligonucleotides used in this study.
| Oligonucleotides | Sequences 5′–3′ | Reference |
|---|---|---|
| RT_hrdB1 (control) | GAGTCCGTCTCTGTCATGGCG | This work |
| RT_hrdB2 (control) | TCGTCCTCGTCGGACAGCACG | This work |
| RT_glnA3F | CAGGTGGAGCTGAGCGACTG | Nentwich, 2010 |
| RT_glnA3R | AGGCGGAGAGGTGGAGGTG | Nentwich, 2010 |
| RT3453F | CTGGCAGTGACCACCGTTCT | This work |
| RT3453R | TGTCGGGCAGGTCCAGTTCT | This work |
| RT3456F | ACCCTGGGCAACACCAGCTATC | This work |
| RT3456R | AAGTGTCGTCTCCGGCGTTGTC | This work |
| RT5057F | CGTCTACGGGCTGCTGTTCATC | This work |
| RT5057R | GTAGTCCAGCATCGCCATCCAC | This work |
| RT5651F | CAACGCGACGCCGAACTACTAC | This work |
| RT5651R | GTCAACAAGGGCGATTCGGGAC | This work |
| RT5655F | CAACGGCTTCTTCTACGG | This work |
| RT5655R | CAGGATCTGCTCGATCTC | This work |
| RT5656F | CCATCATCGAGCAGCTCCAG | This work |
| RT5656R | CAGACGACCTCCACCATCAG | This work |
| RT5657F | GAAGGCCCTGCTGAAGATCG | This work |
| RT5657R | CGGCCATCATCATCGGGTAG | This work |
| RT5658F | TCAGCCCGTCCCTGATGAAC | This work |
| RT5658R | TCGCAGATCCGGTGGAAGTC | This work |
| RT5666F | AACTACCCGCTCCAGATG | Thiswork |
| RT5666R | GCTGACGACGTTGATCAC | This work |
| RT5667F | TCACGTACACCGAGGACATC | This work |
| RT5667R | CCAGCGCCTTCTTGTTGTAG | This work |
| RT5671F | GCGCCAACCAGTTCCACTAC | This work |
| RT5671R | GACAGCAGGTCGAGCATCAC | This work |
| RT5676F | CTTCACCCACACCTGTTTC | This work |
| RT5676R | TGCTTGTACGGCATGTTC | This work |
| RT5679F | AGAACGGCAAGCCCGTCAAG | This work |
| RT5679R | TTGTGGGCGCACAGGTTGAG | This work |
| RT5977F | TTCCAGGACGGCAACCTCAC | This work |
| RT5977R | AAGACCACGCCGACGATCAG | This work |
| RT6960F | ATGCCGATCCTGTACGTC | Thiswork |
| RT6960R | CGAGGTCCTTCTCCAGATG | This work |
| RT6961F | TATCTCTGCGCGGTCTTC | This work |
| RT6961R | AACTCGGCCAGTCCATAG | This work |
| His-glnA3F | ACATATGCATCATCATCATCATCATAGCGAGAGCGACCCCGTGCC | This work |
| His-glnA3R | AAAGCTTTCAGTACTTCCAGCGGTACG | This work |
| glnII_hygr_P1 | ACGTGCGCCGAGACCCACCCCGAAGGATGTGGCCCCGTGATTCCGGGGATCCGTCGACC | This work |
| glnII_hygr_P2 | CCACTGACAACGCGGCCGGGCGGCGGTGGCCCGGGCTCATGTAGGCTGGAGCTGCTTC | This work |
| CglnA3f | ACGAATTCCCGGACGGCACAATG | This work |
| CglnA3r | AGAAAGCTTGCAGGGCCTCGAAAC | This work |
Construction of ΔglnAΔglnII Mutant
REDIRECT gene-replacement procedure (Gust et al., 2003) was employed to generate in-frame deletion of the glnII gene (SCO2210) in previously generated ΔglnA mutant (Fink et al., 1999). The glnII gene on the cosmid St3H12 was replaced by a hygromycin resistance cassette in E. coli BW25113. The hygromycin resistance cassette was amplified by PCR using glnII_hygr_P1 and glnII_hygr_P2 primers. The obtained cosmid St3H12ΔglnII was then transferred into E. coli ET12567/pUZ8002 by electroporation and subsequently transferred into S. coelicolor M145 ΔglnA by conjugation. Hygromycin and apramycin resistant and kanamycin sensitive trans-conjugants were isolated from MS-plates supplemented with L-glutamine. Correct marker-less deletion of glnII was confirmed by PCR followed by sequencing as well as Southern blot analysis.
Estimation of the Intracellular or Extracellular Polyamine Level Using HPLC
Level of the intracellular polyamines was analyzed using high-performance liquid chromatography (HPLC), as described by Potter and Paton (2014). The method was modified and optimized for S. coelicolor. Cells were harvested by centrifugation (6,000 ×g, 10 min at 4°C), washed three times in phosphate-buffered saline (PBS). The 500 mg of the wet cells were resuspended in morpholinepropanesulfonic acid (MOPS) lysis buffer (100 mM MOPS, 50 mM NaCl, 20 mM MgCl2, pH 8.0) and disrupted using glass beads (150–212 μm, Sigma) and a Precellys homogenizer (6500 rpm, 20 – 30 s; Peqlab). Samples were centrifuged (13,000 ×g, 10 min at 4°C), the pellet was discarded and supernatant was transferred into new tube. Trichloroacetic acid then was added to a final concentration of 10%, and the mixture was incubated on ice for 5 min. After incubation the mixture was cleared by centrifugation (13,000 ×g, 10 min at 4°C), the pH of each sample was neutralized using HCl and the samples were stored at −20°C until analysis. Polyamines were derivatized using pre-column derivatization with ortho-phthalaldehyde (OPA)/mercaptoethanol (MCE) (Dr. Maisch GmbH, Ammerbuch). OPA-derivatized polyamines were separated on a Reprosil OPA column (150 mm × 4.0 mm, 3 μm) fitted with a precolumn (10 mm × 4 mm, same stationary phase) (Dr. Maisch GmbH, Ammerbuch) using a HP1090 Liquid Chromatograph equipped with a diode-array detector, a thermostated autosampler and a HP Kayak XM 600 ChemStation (Agilent, Waldbronn). UV detection was performed at 340 nm. The following gradient was used at a flow rate of 1.1 ml/min with solvent A (25 mM sodium phosphate buffer, pH 7.2 containing 0.75% tetrahydrofuran) and solvent B [25 mM sodium phosphate buffer, pH 7.2 (50%), methanol (35%), acetonitrile (15%) by volume]: t0 = 60% B, t2.5 = 70%, t12 = t22 = 100% B (time in minutes). Cadaverine dihydrochloride, putrescine dihydrochloride, spermidine trihydrochloride, and spermine tetrahydrochloride were purchased from Sigma and used as standards. Standard solutions of polyamines were prepared with distillated water. HPLC detection limit for different polyamines was 0.1 mM.
Gene Expression Analysis by Reverse Transcriptase/PCR
For the transcriptional analysis experiments, the S. coelicolor M145 wild type and the glnA3 mutant were grown in S-medium. After 4 days, cells were washed twice with defined Evans medium and cultivated for 24 h in defined Evans medium supplemented either with 25 mM ammonium chloride or 25 mM polyamine (cadaverine, spermidine, or putrescine). RNA isolation was performed with an RNeasy kit (Qiagen). RNA preparations were treated twice with DNase (Fermentas). First, an on-column digestion was carried out for 30 min at 24°C, and afterward RNA samples were treated with DNase for 1.5 h at 37°C. RNA concentrations and quality were checked using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). cDNA from 3 μg RNA was generated with random nonamer primers (Sigma), reverse transcriptase and cofactors (Fermentas). PCR reactions were performed with the primers listed in Table 2. The PCR conditions were 95°C for 5 min; 35 cycles of 95°C for 15 s, 55–60°C for 30 s and 72°C for 30 s; and 72°C for 10 min. As a positive control, cDNA was amplified from the major vegetative sigma factor (hrdB) transcript, which is produced constitutively. To exclude DNA contamination, negative controls were carried out by using total RNA as a template for each RT-PCR.
Survival Assay
The ability of the S. coelicolor M145 and ΔglnA3 mutant to survive in the presence of polyamines was examined by the estimation of the dry weight biomass after 72 and 168 h of incubation in the YEME–TSB (1:1) rich complex medium. Both strains, M145 and ΔglnA3 mutant were inoculated into YEME–TSB supplemented with putrescine, cadaverine, spermidine as well as spermine (25 mM of each) and incubated for 3 days at 30°C. As a control M145 and ΔglnA3 mutant were cultivated YEME:TSB without polyamines supplementation and incubated as described above. Samples (1 ml) were taken every day and cells were harvested by centrifugation (16,200 ×g, 15 min at 4°C), the residual liquid was removed and pellets were dried for 24 h. The middle value of three technical replicated was compared with the middle value of three biological replicates and the standard error was calculated.
Light Microscopy and Viability Staining
Morphology and viability of S. coelicolor M145 and ΔglnA3 mutant cells grown in a rich complex YEME–TSB (1:1) medium was analyzed in presence of polyamines. Samples were taken from every culture after 72 and168 h of growth, and obtained perpetrates were observed under phase-contrast microscope under 400× enlargement. To detect live and dead cells, SYTO9 and PI (propidium iodide) stains of the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes) were used. The SYTO 9 green fluorescent stain labels cells with intact membranes, as well as those with damaged ones. PI penetrates cells with damaged membranes, decreasing SYTO 9 stain fluorescence when both dyes are present. Thus, in the presence of both SYTO9 and PI, cells with intact cell membranes appear fluorescent green whereas cells with damaged membranes appear red. The staining solution was prepared by mixing 0.75 μl of component A and B in 500 μl of water. Stained cells were analyzed by the fluorescence microscopy using Olympus BX60 microscope with an Olympus UPlanFl 100 × oil objective and an Olympus BX-FLA reflected light fluorescence attachment. Images were taken with the F-view II camera (Olympus), using TxRed and eGFP filter sets for detection of the fluorescent markers. The ImageJ was used for image processing. Significant number of images (10) was analyzed in a minimum of three independent culture analyses.
Cloning, Expression, and Purification of His-GlnA3
The gene encoding GlnA3 (SCO6962) was amplified by PCR form S. coelicolor genomic DNA and ligated into the expression vector pJOE2775 (Volff et al., 1996) under the control of rhamnose inducible promoter Prha. The restriction sites NdeI and HindIII were used for cloning purposes. His-GlnA3 was overexpressed in E. coli strain BL21 (DE3) using the auto-induction method for protein production (Li et al., 2011; Studier, 2014). Cells were grown initially at 37°C until the culture density reached an optical density of ∼0.6 at 600 nm at which time the temperature of the shaking incubator was reduced to 20°C. The bacterial cells were then harvested by centrifugation after 48 h cultivation and stored at −20°C until needed. His-GlnA3 was purified by nickel ion affinity chromatography essentially as directed by the resin manufacturer (GE-Healthcare). Purified His-GlnA3 was dialyzed against 20 mM Tris, 100 mM NaCl (pH 8) and immediately used for further analysis.
GlnA3 In Vitro Assay and HPLC/ESI-MS Detection of the Glutamylated-product
HPLC/ESI-MS was used to evaluate putrescine and glutamate as GlnA3 substrates and generated gamma-glutamylated putrescine as a product. Standard reactions typically contained 20 mM HEPES (pH 7.2), 150 mM glutamate sodium monohydrate, 150 mM putrescine dihydrochloride, 20 mM MgCl2 × 6H2O, and 10 mM ATP were mixed with 10 μg of the purified His-GlnA3 (or without GlnA3 as a control) and incubated at 30°C for 5 min. The reaction was stopped by incubation of the reaction mixture at 100°C for 5 min. HPLC/ESI-MS analysis of the glutamylated product generated by GlnA3 was done on an Agilent 1200 HPLC series using a Reprosil 120 C18 AQ column, 5 μm, 200 mm × 2 mm fitted with a precolumn 10 mm × 2 mm (Dr. Maisch GmbH, Ammerbuch, Germany) coupled to an Agilent LC/MSD Ultra Trap System XCT 6330 (Agilent, Waldbronn, Germany). Analysis was carried out using 0.1% formic acid as solvent A and acetonitrile with 0.06% formic acid as solvent B at a flow rate of 0.4 ml min−1. The gradient was as follows: t0 = t5 = 0% B, t20 = 40% B (time in minutes). Injection volume was 2.5 μl, column temperature was 40°C. ESI ionization was done in positive mode with a capillary voltage of 3.5 kV and a drying gas temperature of 350°C.
Results
GlnA2, GlnA3, and GlnA4 Annotated as GSI-Like Do Not Have a Glutamine Synthase Function in S. coelicolor M145
In order to determine whether GlnA2, GlnA3, and GlnA4 were involved or not involved in glutamine biosynthesis in S. coelicolor M145, single and double mutants of genes glnA and glnII encoding proven GSs, GSI and GSII, respectively, were deleted in the parental strain M145. REDIRECT gene-replacement procedure (Gust et al., 2003) was employed to delete glnII gene (SCO2210) in a previously generated ΔglnA mutant (Fink et al., 1999) as described in Section “Materials and Methods.” Independent clones of the single ΔglnA and ΔglnII mutants, of the double ΔglnAΔglnII mutant and of the parental strain M145 were tested for their ability to grow on defined Evans-agar supplemented with ammonium chloride (50 mM) as sole nitrogen source. Glutamine synthetases, GSI and GSII catalyze condensation of L-glutamate and ammonia to form L-glutamine in the ATP dependent reaction. Consequently, growth reveals the ability of the strains to assimilate ammonium and to synthesize glutamine whereas absence of growth indicates absence of this ability. S. coelicolor M145 and the single deletion mutants, ΔglnA and ΔglnII, were able to grow on the plates, demonstrating that both glnA and glnII encode functional GSs as reported previously (Reuther and Wohlleben, 2007). In contrast, the ΔglnAΔglnII double mutant was unable to grow on this medium (Figure 2) and was auxotrophic for glutamine. Addition of L-glutamine restored growth of the ΔglnAΔglnII double mutant on this medium. These results indicated that GlnA2, GlnA3, and GlnA4 do not have a GS function that can substitute for that of GSI and GSII and avoid glutamine auxotrophy.
FIGURE 2.
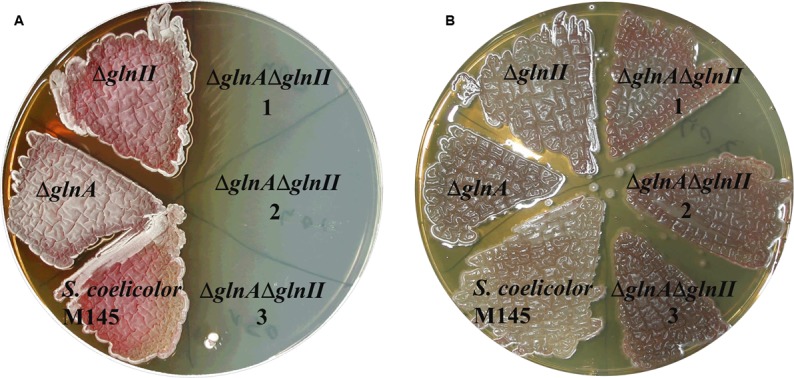
Phenotypic analysis of parental strain S. coelicolor M145 and mutants: ΔglnA, ΔglnII and ΔglnAΔglnII (1, 2, 3 – independent clones) on defined Evans medium supplemented with 50 mM ammonium chloride (A) or with 50 mM ammonium chloride and 50 mM L-glutamine (B). Deletion of both genes, glnA and glnII, for GSs, resulted in a glutamine auxotrophic phenotype indicating that GlnA2, GlnA3, and GlnA4 were incapable to substitute GSI and GSII metabolic function in the cell.
The Deletion of GlnA3, But Not that of GlnA2 and GlnA4 Abolished Polyamine Utilization
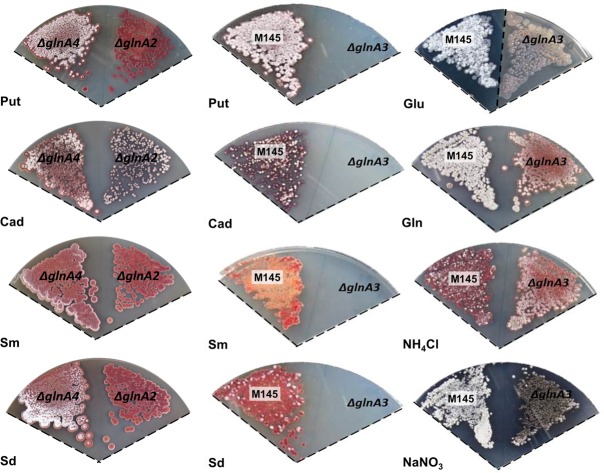
In an attempt to decipher the function of GS-like enzymes, the genes glnA2, glnA3, and glnA4 were deleted in the parental strain S. coelicolor M145. The ΔglnA2 (Nentwich, 2010), ΔglnA3 (Rexer et al., 2006), and the ΔglnA4 (this work) mutants were tested for their ability to utilize different nitrogen sources. Phenotypic analysis was performed on defined Evans-agar supplemented with the following sole nitrogen sources: ammonium chloride, sodium nitrate, monosodium L-glutamate, L-glutamine, putrescine dihydrochloride, cadaverine dihydrochloride, spermidine trihydrochloride, and spermine tetrahydrochloride. Growth and morphology of the parental strain M145 and the ΔglnA2, ΔglnA3, and ΔglnA4 mutants were monitored on defined Evans-agar after 3 to 12 days of incubation at 30°C. Remarkably, S. coelicolor M145 could grow on all tested nitrogen sources (Figure 3). Interestingly, growth of the S. coelicolor M145 on polyamines resulted in delayed aerial mycelium and spore formation as well as abolition of actinorhodin (blue antibiotic) and enhanced prodigiosin (red antibiotic) production (Figure 3). Growth of the glnA2, glnA3, and glnA4 mutants was similar to that of the parental strain in the presence of ammonium chloride, sodium nitrate, L-glutamine, and monosodium L-glutamate. Growth of the glnA3 mutant, but not that of the glnA2 and glnA4 mutants, was inhibited in the presence of putrescine, cadaverine, spermidine, and spermine (Figure 3). The complementation of the glnA3 mutant by the glnA3 gene under control of the constitutive promoter (PermE) restored the growth of this mutant on polyamine plates (Supplementary Figure S1). Altogether these results indicated that the disruption of glnA3, but not that of glnA2 and glnA4, completely abolished the utilization of polyamines as a nitrogen source. GlnA2 and GlnA4 have obviously a function different from that of GlnA3 since their deletion did not influence polyamine utilization. Interestingly, the glnA3 mutant showed defect in formation of aerial mycelium as well as sporulation on defined medium supplemented with glutamate (50 mM). Moreover, enhanced actinorhodin and prodigiosin production was observed on the nitrate, glutamine and ammonium plates, respectively (Figure 3), indicating an impact of glnA3 deletion on the secondary metabolites production and morphological differentiation in S. coelicolor.
FIGURE 3.
Physiological role of the glnA3 gene product in S. coelicolor M145 cells grown in the presence of polyamines. (A) Phenotypic comparison of parental strain S. coelicolor M145 and ΔglnA2, ΔglnA3, and ΔglnA4 mutants grown on defined Evans medium supplemented with Put – putrescine dihydrochloride (200 mM), Cad – cadaverine dihydrochloride (50 mM), Sm – spermine tetrahydrochloride (25 mM), Sd – spermidine trihydrochloride (25 mM) as well as on Glu – monosodium glutamate (50 mM), Gln – glutamine (50 mM), NH4Cl – ammonium chloride (50 mM) and NaNO3 – sodium nitrate (50 mM) as sole nitrogen source. Each panel represents observations on a single agar plate, except the phenotypic analysis of the glnA3 mutant and parental strain in the presence of glutamate has been documented on two separate agar plates as indicated by the dotted line. Deletion of glnA3 resulted in the no-growth phenotype in the presence of high polyamine concentrations.
Expression of glnA3 Is Induced by Polyamines and Starvation Conditions
Reverse transcription/PCR experiments were performed as described in Section “Materials and Methods” to determine whether glnA3 expression was influenced by N- and/or C-concentrations as reported for glnA and glnII (Amin et al., 2016) as well as to investigate the expression of glnA3 in the presence of polyamines. For this purpose, parental strain M145 was grown in complex S-medium subsequently transferred into defined Evans medium supplemented either with polyamines (25 mM) or ammonium chloride (25 mM) as a sole nitrogen source. Total RNA isolated after 24 h from S. coelicolor M145 was used to generate cDNA. Subsequently, reverse transcription/PCR analysis was performed using primers internal to glnA3. HrdB, encoding the main sigma factor of RNA polymerase, that has relatively constant levels of expression throughout growth (Buttner et al., 1990) was used as internal control. Expression of genes encoding GSs, glnA and glnII, was shown to be strongly enhanced in condition of ammonium limitation and reduced or completely abolished in condition of ammonium proficiency in S. coelicolor, irrespective of the glucose concentration (Amin et al., 2016). Interestingly, the expression of glnA3 was also strongly enhanced under ammonium limitation, but only in combination with low glucose concentration suggesting that its expression does not require polyamines. Furthermore, transcriptional analysis of glnA3 revealed strong induction of the expression of this gene in the presence of putrescine, cadaverine and spermidine (Figure 4), putative substrates of the encoded enzyme.
FIGURE 4.
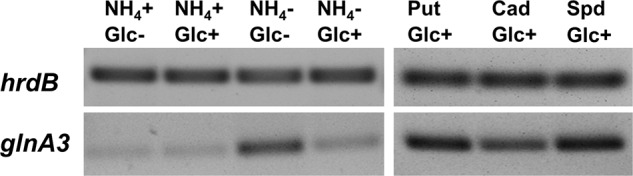
Transcriptional analysis of glnA3 in the presence of polyamines and ammonium as a sole nitrogen sources. Reverse transcriptase/PCR of glnA3 and hrdB (control) from S. coelicolor M145 cultivated in defined Evans medium with low (5 mM) or high (50 mM) concentrations of ammonium chloride or polyamines (Put – putrescine; Cad – cadaverine and Spd – spermidine, 25 mM of each) and glucose as a sole carbon source high Glc+ (25 g/l) or low Glc– (2.5 g/l). Total RNA was isolated from mycelium harvested after 24 h of cultivation in the defined Evans medium.
Transcriptional Analysis-Based Identification of Orthologs of the Putative Polyamine Transport and Polyamine Utilization Genes in S. coelicolor M145
Polyamine uptake and utilization pathways are largely uncharacterized in streptomycetes. BlastP analysis was used to predict uptake systems and putative enzymes involved in polyamine utilization in S. coelicolor, whose amino acid sequences are closely related to the amino acid sequences of the characterized proteins from E. coli and P. aeruginosa (Kusano and Suzuki, 2015). Similarity based prediction revealed 18 strong homologs in S. coelicolor (genome accession number AL645882). Their amino acid identities ranged from 24 to 50%, while their similarities ranged from 41 to 65% (Figure 5). To demonstrate that these genes were induced by polyamines, their expression patterns were analyzed in the presence of putrescine, cadaverine, and spermidine as sole N-source as well as ammonium as control.
FIGURE 5.
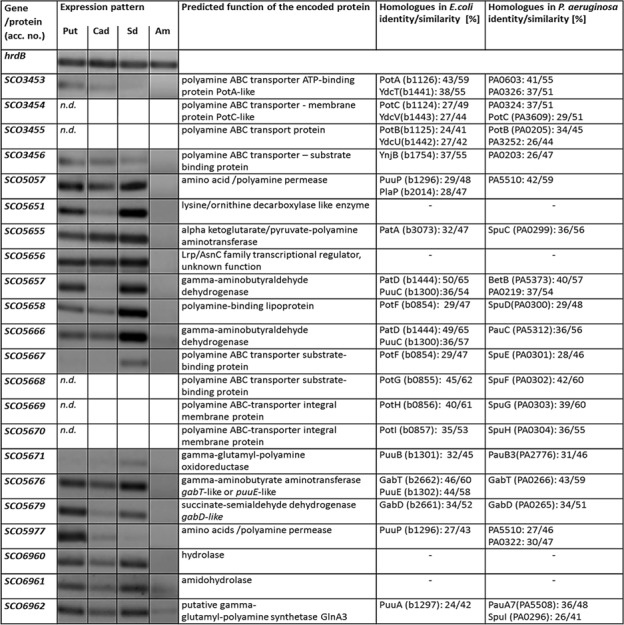
Transcriptional analyses of putative gene homologs encoding predicted proteins involved in polyamine degradation in S. coelicolor. Reverse transcriptase/PCR of genes and hrdB (control) from S. coelicolor M145 cultivated in defined Evans medium with polyamines (Put – putrescine; Cad – cadaverine or Spd – spermidine, 25 mM of each) and glucose as a sole carbon source Glc (25 g/l). Total RNA was isolated from mycelium harvested after 24 h of cultivation in the defined Evans medium.
Analysis of the glnA3 genomic region revealed two genes SCO6960 and SCO6961 located downstream of glnA3 and likely co-transcribed with the latter. SCO6961 encodes an uncharacterized protein belonging to the amidohydrolase 2 family, whereas SCO6960 encodes a small protein of unknown function conserved among streptomycetes. However, no orthologs of these genes could be found in E. coli and P. aeruginosa, suggesting that the polyamine utilization pathway might be different in S. coelicolor. Transcriptional analysis of the SCO6960 and SCO6961 indicated that the expression of these genes was inducible by polyamine as that of glnA3 (Figure 5). In contrast, the expression of the genes SCO6963, SCO6964, and SCO6965 located upstream of this putative operon and encoding uncharacterized lipoprotein and two small proteins of unknown function were not induced in conditions of ammonium limitation nor in the presence of polyamine (data not shown).
Furthermore, BlastP analysis predicted two ABC transporters and two permeases potentially involved in the uptake of polyamines. The SCO5667-70 and SCO3453-56 predicted gene products resemble the ABC transport systems PotFGHI or SpuEFGH from E. coli and P. aeruginosa, respectively. The expression of these genes was only weakly inducible in the presence of polyamines (Figure 5). In contrast, the expression of the genes encoding the putative polyamine permeases SCO5057 and SCO5977 was strongly induced in the presence of all polyamines or only putrescine, respectively (Figure 5). The expression of SCO5658 (encoding predicted polyamine binding protein) was strongly induced in the presence of spermidine (Figure 5).
Prediction of genes encoding enzymes involved in the post-glutamylation steps in S. coelicolor revealed orthologs for gamma-glutamylpolyamine oxidoreductase (SCO5671), polyamine-pyruvate/2-oxoglutarate transaminase (SCO5655) as well as close orthologs of (gamma-glutamyl-) gamma-aminobutyraldehyde dehydrogenases PatD and PuuC (SCO5657 and SCO5666) and SCO5676 encoding predicted GabT-like gamma-aminobutyrate transaminase and finally SCO5676, encoding predicted succinate semialdehyde dehydrogenase (GabD-like). The expression of all these genes (except SCO5671) was strongly induced in the presence of polyamines (Figure 5). Altogether, putative homologs of genes encoding predicted polyamine uptake systems and proteins involved in the polyamine utilization in S. coelicolor showed enhanced expression in the presence of polyamines whereas their expression was abolished in the presence of ammonium. To summarize, the search for proteins orthologs of those involved in the catabolism of polyamine in other bacteria using BlastP and the analysis of gene expression patterns, led us to predict, so far unknown, polyamine degradation pathway in S. coelicolor (Figure 1).
GlnA3 Deletion Mutant Accumulates Polyamines Intracellularly
In an attempt to understand the basis of polyamine toxicity in the glnA3 mutant grown in the presence of polyamine, the intracellular polyamine content of S. coelicolor M145 and glnA3 mutant was analyzed by HPLC. To do so, the parental strain M145 and the glnA3 deletion mutant were cultivated in a complex S-medium for 4 days at 30°C. Cells were harvested and washed with Evans medium without N-source to remove remaining complex N-sources. Washed cells were transferred into defined Evans medium containing polyamines as a sole N-source and incubated at 30°C for further 4 days. Samples for the biomass determination and extraction of intracellular polyamines were collected every day. Cell pellets and supernatants were used to extract intracellular and extracellular polyamines, respectively. Comparison of intracellular polyamine levels of both strains revealed three, six and twenty fold higher levels of putrescine, cadaverine, and spermidine in ΔglnA3 mutant, respectively (Figure 6) than in the parental strain after 4 days of cultivation. The intracellular concentration of polyamines decreased over time in the parental strain M145, but not in the mutant strain. After 4 days of cultivation, intracellular polyamine concentrations ranged between 0.5 and 0.7 μmol/g of biomass, whereas that of the glnA3 mutant ranged between 3.1 and 5.1 μmol/g. Furthermore, comparison of extracellular polyamine levels in the parental strain culture and glnA3 culture revealed strong differences. The residual level of polyamines in the glnA3 mutant culture supernatant remained high from the first to the fourth day of incubation (Figure 7), whereas after 4 days of incubation no peak for polyamines could be detected in the culture supernatant of the parental strain. Altogether, these data indicated that the glnA3 mutant was able to uptake polyamines, but was unable to degrade them. The absence of polyamine degradation in the glnA3 mutant obviously resulted in high concentration of intracellular polyamine, likely to be responsible for the inhibition of further uptake of external polyamines (Figure 7).
FIGURE 6.
Effect of glnA3 deletion on intracellular polyamine concentration. The intracellular polyamine level of parental strain S. coelicolor M145 and ΔglnA3 mutant was monitored after 24 and 96 h of cultivation in defined Evans medium supplemented with polyamines (Put – putrescine; Cad – cadaverine or Sd – spermidine, 25 mM of each). The mean value of three biological replicates was calculated in μmol per 1 g of wet cells. Error bars indicate standard error of three biological replicates.
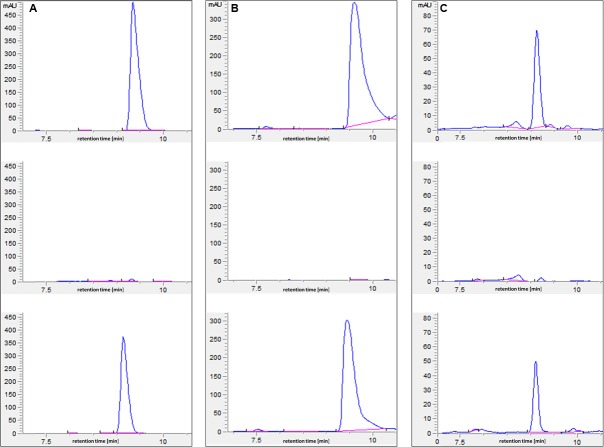
FIGURE 7.
HPLC-based detection of polyamines in supernatant from the 96 h culture of parental strain M145 and ΔglnA3 mutant. Both strains were grown in defined Evans medium supplemented with either (A) putrescine (25 mM), (B) cadaverine (25 mM) or (C) spermidine (25 mM). Upper HPLC chromatogram: polyamine standard, middle HPLC chromatogram: detection of polyamines remained in the supernatant of the M145 culture, lower HPLC chromatogram: detection of polyamines remained in the supernatant of the ΔglnA3 mutant culture. Results indicate that only parental strain S. coelicolor M145 was able to completely utilize whole polyamines from the medium after 96 h of incubation.
The ΔglnA3 Mutant Grown in Complex Medium with Polyamines Showed Atypical Mycelial Morphology and Short Lifetime Span
The ΔglnA3 mutant and its parental strain S. coelicolor M145 were cultivated in a rich complex liquid medium (YEME:TSB) supplemented or not (control) with polyamines (putrescine, cadaverine, spermidine, and spermine, 25 mM of each). Biomass yields as well as morphology of both strains were monitored over 7 days. The ΔglnA3 mutant yielded significantly lower biomass than the parental strain after 72 and 168 h of cultivation in the complex medium supplemented with polyamines (Figure 8). Prolonged incubation of the ΔglnA3 mutant culture did not result in any significant biomass increase. In this medium, S. coelicolor M145 showed its typical pellet growth whereas growth of the ΔglnA3 mutant was dispersed. This indicated that despite the presence of alternative nitrogen sources in this complex medium, besides polyamines, growth of ΔglnA3 mutant was impaired most likely because of intracellular accumulation of polyamines.
FIGURE 8.
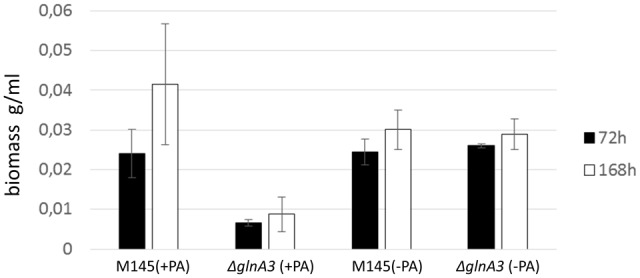
Effect of the glnA3 deletion on a biomass accumulation in a rich complex medium supplemented with and without polyamines. Monitoring of changes in biomass accumulation of the parental strain S. coelicolor M145 and ΔglnA3 mutant in the YEME:TSB (1:1) medium supplemented with polyamines total concentration 100 mM (putrescine, cadaverine, spermidine, and spermine, 25 mM of each) after 72 and 168 h of incubation. Error bars indicate standard error of three biological replicates.
The mycelium of the ΔglnA3 mutant grown in the complex medium with polyamines was observed under microscope. After 3 days of cultivation, the parental strain formed a long and compact, branched mycelium constituting pellets whereas the ΔglnA3 mutant formed non-branched mycelium (Figure 9). The mycelial fragments of the ΔglnA3 mutant were significantly shorter and thicker than those of the parental strain M145 after 3 or 7 days of cultivation in the medium containing polyamines (Figure 10).
FIGURE 9.
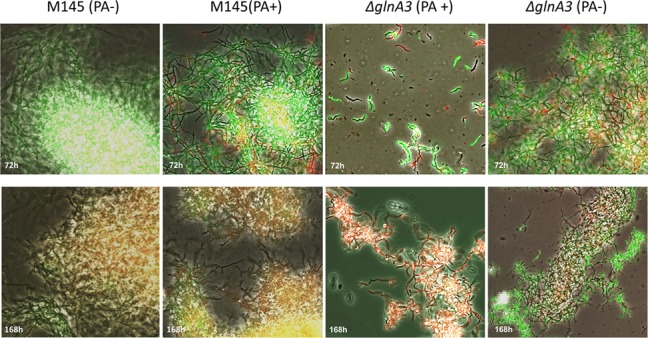
Effect of the glnA3 deletion on cell morphology and its viability in the presence of polyamines. Parental strain S. coelicolor M145 and ΔglnA3 mutant were cultivated in YEME:TSB (1:1) medium with or without polyamines total concentration 100 mM (putrescine, cadaverine, spermidine, and spermine, 25 mM of each). Phase contrast microscopic pictures of parental strain M145 and ΔglnA3 mutant mycelium stained with SYTO9/PI were taken after 72 and 168 h of growth (under 400× enlargement).
FIGURE 10.

Comparison of the hyphae thickness of parental strain S. coelicolor M145 and ΔglnA3 mutant grown in the presence of polyamines. Parental strain S. coelicolor M145 and ΔglnA3 mutant were cultivated in YEME:TSB (1:1) medium with or without polyamines. The hyphae thickness was measured after 72 and 168 h of growth. Error bars indicate standard error of n = 100 biological replicates.
In order to assess more precisely the toxic effect of polyamines, the viability of the ΔglnA3 mutant was assessed with SYTO9/PI staining. The SYTO 9 green fluorescent stain labels all cells, whereas PI penetrates only cells with damaged membranes, decreasing SYTO 9 stain fluorescence when both dyes are present. In the presence of both, SYTO9 and PI, cells with intact cell membranes appear fluorescent green and dead cells with damaged membranes appear red. Whereas the parental strain M145 appeared green (alive) in the presence of polyamines, most filaments in the culture of the ΔglnA3 mutant were red (dead) after 7 days of incubation (Figure 9). These results demonstrated that the absence of GlnA3 led to the toxic accumulation of polyamine that impaired growth and viability and suggested that GlnA3 plays a major role in polyamine degradation.
GlnA3 a Predicted Gamma-Glutamylpolyamine Synthetase in S. coelicolor M145
The GlnA3 protein belongs to large class of ligases which form carbon-nitrogen bonds using energy from ATP (class 6.3.1). This class contains twenty different subclasses of enzymes including GS (class 6.3.1.2), glutamate – ethylamine ligase (class 6.3.1.6) and glutamate-putrescine ligase (6.3.1.11). GlnA3 protein sequence analysis by InterProScan1 predicted two enzymatic domains: N-terminal GS, beta-Grasp domain (IPR008147) and the C-terminal catalytic domain of GS (IPR008146). Searching for functionally and structurally characterized GlnA3 homologs from the Protein Data Bank (PDB) revealed a gamma-glutamylmonoamine/polyamine synthetase PauA7 (PA5508) as a likely. PauA7 is involved in monoamine/polyamine gamma-glutamylation in P. aeruginosa (Ladner et al., 2012). The available crystal structure of Pau7 (Protein Data Bank entry: 4HPP) and GS GSISt from S. typhimurium (Protein Data Bank entry: 1FPY) were used as the templates to generate a structural identical conserved binding residues for glutamate and ATP as well as two divalent metal ions (Mg++/Mn++). However, the binding pocket for ammonium and monoamine differ significantly. The GSISt (as well as other eukaryotic and bacterial GS) and the PauA7 require two divalent metal ions per one subunit for activity. These ions have a structural as well as a catalytic role. Comparison of the conserved catalytic residues for the binding of divalent metal ions revealed four conserved glutamate residues E151, E153, E207, E214 and one conserved histidine residue H263 in the GlnA3 model structure (corresponding to E131, E133, E180, E187, and H236 in PauA7, as well to E129, E131, E212, E220, and H269 in GSISt, respectively). Moreover, three conserved residues involved in coordination of the glutamate binding in GSISt and PauA7, were found in the GlnA3 model structure. These GlnA3 residues (N260, G261, and R316) correspond to the N233, 258 G234, and R290 in PauA7 as well as to N264, G265, and R321 in GSISt. Finally, two conserved residues (H265 and R339 both corresponding to H271 und R344 in GSISt) important for coordination of the beta-phosphate and alpha-phosphate group of ADP were found in the GlnA3 model. This comparative in silico analysis demonstrates that GlnA3 possess conserved residues for binding of two metal ions, L-glutamate and ATP (Supplementary Figure S2). An essential element of GSISt catalytic pocket is a loop, termed “the E327 flap” (GSISt). The GSI active site is located between two subunits, and the E327-flap closes the catalytic site by interaction of the E335 with D50 from an adjacent subunit within the same ring, shielding the γ-glutamyl phosphate intermediate from hydrolysis. The second conserved residue D50 is involved in the deprotonation of ammonium for attack on the γ-glutamyl phosphate. Interestingly, these key acidic residues (E327 and D50) essential for the catalytic synthesis of glutamine in GSISt are not conserved in PauA7 nor in GlnA3 (Supplementary Figure S2). In PauA7 structure and GlnA3 model these loops are much bigger and instead E327, a non-polar W327 residue occupies analogous position in the GlnA3 model structure (corresponding to W296 in PauA7). The conserved residue D50 that increases the affinity for ammonium binding in GSISt is replaced by G40 and G63 in PauA7 and 273 GlnA3, respectively. Moreover, the conserved Y179 in GSISt that coordinates the ammonium binding pocket is substituted by A147 in PauA7 and A169 in GlnA3, providing much more space for a bulky substrate such as polyamine (Supplementary Figure S2). The significant overall similarity of GlnA3 to the gamma-glutamylmonoamine/polyamine synthetase PauA7, the lack of the conserved residues for ammonium binding as well as our observations strongly suggest that GlnA3 may function as a gamma-glutamylpolyamine synthetase catalyzing the first step of polyamine degradation.
GlnA3 Catalyzes Gamma-Glutamylation of Putrescine In Vitro
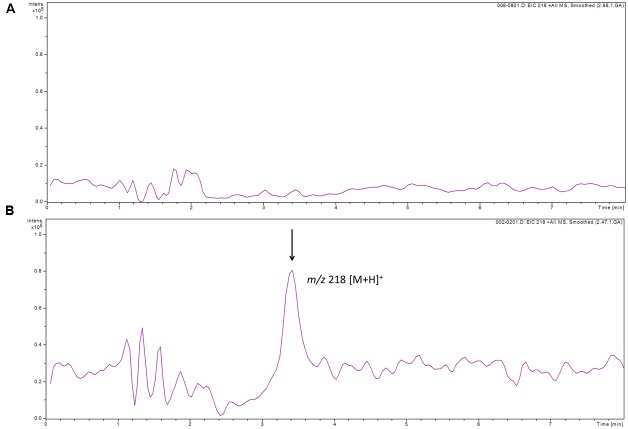
To determine whether GlnA3 was able to catalyze the predicted glutamylation reaction we developed an HPLC/ESI-MS based assay designed to detect a gamma-glutamyl product formed by GlnA3. For this, GlnA3 was overexpressed in E. coli BL21 (DE3) and purified by nickel ion affinity chromatography. The purified His-GlnA3 was used in an in vitro assay as described in Section “Materials and Methods.” The reaction conditions were as follows: 10 μg of His-GlnA3 were incubated at 30°C with glutamate, putrescine, MgCl2 and ATP in HEPES buffer (pH 7.2). The reaction was stopped after 5 min. by incubation at 100°C for 5 min. HPLC analysis of generated product was performed in a positive MS mode. Result indicated that GlnA3 was able to use glutamate and putrescine as substrates in the ATP dependent reaction, generating a product with the mass to charge ratio of 218 m/z, corresponding to calculated mass of gamma-glutamylputrescine, confirming the function of GlnA3 as a gamma-glutamylpolyamine synthetase (Figure 11).
FIGURE 11.
HPLC/ESI-MS analysis of the glutamylated product generated by GlnA3 in in vitro assay. Two samples were analyzed in the MS positive mode: reaction mixtures without addition of GlnA3 (A) and with addition of GlnA3 (B). Extracted ion chromatograms for the GlnA3 reaction product corresponding to gamma-glutamylputrescine with charge to mass ratio of 218 m/z was shown (B), and no product in the sample without addition of GlnA3 could be detected (A).
Discussion
In this study, we demonstrated that S. coelicolor M145 is able to utilize polyamines such as putrescine, cadaverine, spermidine, and spermine as a sole nitrogen source and it is able to withstand high polyamine concentrations. Our results indicate that this ability is due to GlnA3 since the deletion of glnA3 resulted in intracellular polyamine accumulation, abnormal morphology of the mycelial fragments as well as a polyamine sensitive phenotype resulting in cell death. Our in vivo data concerning the deletion of glnA and glnII encoding GSs, GSI and GSII, respectively, indicated that GlnA3 doesn’t fulfill a function of GS. Comparative in silico analysis of PauA7, GSI and GlnA3 as well as the in vitro GlnA3 enzymatic assay revealed that GlnA3 bear features of a gamma-glutamylpolyamine synthetase, an enzyme catalyzing the first step of polyamines catabolism in Streptomyces.
Using BlastP for comparative analyses of Streptomyces proteome sets (taxid: 1883), 116 homologs of the predicted GlnA3 protein were identified with 75–99% sequence identity and 95–100% query cover in Streptomyces sp. Frequent occurrence of GlnA3 homologs in Streptomyces genomes as well as its highly conserved protein sequence in all strains indicates its evolutionary importance. Homologs of the predicted GlnA3 protein were also found in other actinobacteria such as: Nocardia spp., Rhodococcus spp., Frankia spp., Amycolatopsis spp., Kitasatospora spp., Saccharopolyspora spp. and Mycobacterium spp. (45–76% identity and 92–100% query cover).
Since orthologs of GlnA3 were found also in some pathogenic actinobacteria, GlnA3 might ensure colonization and persistence in the host by conferring resistance against toxic concentrations of polyamines produced during infection. Indeed, the polyamine content was reported to increase dramatically in both proliferating cells and extracellular tissue fluids during inflammation, tissue regeneration and cell damage (Yatin, 2011). Increased polyamines levels have been demonstrated at inflammatory sites of bacterial and viral infections (Hirsch and Dubos, 1952; Colombatto et al., 1989; Clarke and Tyms, 1991; Lasbury et al., 2007). Interestingly, pathogens belonging to the actinobacteria group are able to survive in a tissue in presence of high polyamine concentrations produced by defense system in a response to infection and stress (Liao et al., 2009; Grasemann et al., 2012; Jimenez-Bremont et al., 2014). Symbiotic actinobacteria can survive in the nitrogen fixing nodules where they are constantly confronted with fluctuating polyamine concentrations (Fujihara et al., 1994). Free living S. coelicolor and other non-motile actinobacteria might have to cope with high concentration of polyamines that are released during the decomposition of animal cadaver and plant tissues in the soil.
The search for proteins orthologs of those involved in the catabolism of polyamine in other bacteria using BlastP and the analysis of the expression patterns of these genes, led us to predict, polyamine glutamylation pathway in S. coelicolor (Figure 1). We were able to show that the gamma-glutamylpolyamine synthetase GlnA3 is important for polyamine catabolism in S. coelicolor under the conditions tested. This implies that further post-glutamylation steps of the so far known polyamine glutamylation pathway in E. coli and P. aeruginosa (or a variation of this pathway) are most likely present in streptomycetes. The second step of glutamylation pathway requires a gamma-glutamyl polyamine oxidoreductase (PuuB in E. coli/pauB1-B4 in P. aeruginosa). The SCO5671 (predicted ortholog of the PuuB) showed no expression in the presence of putrescine and cadaverine, but a weak expression has been observed in the presence of spermidine. It remains to be elucidated whether other potential homologs (SCO1281, SCO6051) with lower sequence similarity might represent gamma-glutamylpolyamine oxidoreductase PuuB in streptomycetes. The next step of the polyamine glutamylation pathway involves predicted PuuC dehydrogenase homologs (SCO5666, SCO5657), followed by the predicted hydrolase SCO6961. Last two steps of polyamine glutamylation pathway lead to the formation of succinate and are catalyzed by the predicted GabT and GabD homologs, SCO5676 and SCO5679, respectively. Interestingly, expression of the SCO5676 has been shown to be stimulated by arginine (a precursor of putrescine) in S. coelicolor M145 (Perez-Redondo et al., 2012).
Moreover, the predicted PatA aminotransferase (SCO5655), along with orthologs of additional predicted aminotransferases and dehydrogenases, suggests the possibility of an alternate pathway in streptomycetes. Interestingly, induction of the SCO5655 has been reported to be stimulated by diamide (Kallifidas et al., 2010), however, not by arginine (Perez-Redondo et al., 2012). Further genetic analysis of SCO5655 and other potential aminotransferase homologs (SCO1223, SCO1284, and SCO6769) would be required to determine whether a PatA-dependent pathway is functional. The regulation and importance of glutamylation and aminotransferase pathways may dependent on polyamine concentration or another environmental parameter such as temperature (Schneider and Reitzer, 2012).
A better knowledge of polyamine utilization in actinobacteria is of fundamental importance, since this pathway may be important for the survival strategy of pathogenic, symbiotic, and non-pathogenic actinobacteria living in as diverse habitats as human, animal, and plant tissues or in soil environment, respectively. Our work constitutes the first attempt to decipher polyamine metabolism in the actinobacterial model organism Streptomyces coelicolor via the characterization of a major polyamine resistance factor, the gamma-glutamylpolyamine synthetase GlnA3 that is indispensable for survival under high polyamine concentration.
Author Contributions
AM performed the reverse transcriptase/PCR analysis. AK performed the HPLC and HPLC/MS analysis. JG constructed and analyzed the ΔglnA/glnII mutant. NO constructed the ΔglnA4 mutant as well as was involved in technical assistance. NO and SK performed phenotypic analysis of all mutants and parental strain M145 on Evans medium with different nitrogen sources. SK performed complementation of the ΔglnA3 mutant, survival and viability assays, microscopic analysis of cells, intracellular polyamine analysis and bioinformatics analysis of DNA and protein sequences. SK cloned, overexpressed and purified GlnA3 protein and performed GlnA3 in vitro assay. AB formulated the original problem and provided direction and guidance as well as designed the study and developed the methodology. AB generated and analyzed the model of GlnA3 structure. WW provided helpful feedback on an early draft of the paper and assisted with data analysis. AB contributed to the writing of the manuscript and resolved final approval of the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Manuel Espinosa Padrón (Centro de Investigaciones Biológicas, CSIC, Madrid, Spain) for precious time he spent reading the manuscript and for his helpful and substantial comments – always greatly appreciated. We acknowledge support by Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Tübingen.
Funding. SK and AB are members of the DFG Research Training Group GRK1708 (I and II). This project as well as the Ph.D. work of SK was funded by DFG Research Training Group GRK1708 (II).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00726/full#supplementary-material
References
- Alcazar R., Tiburcio A. F. (2014). Plant polyamines in stress and development: an emerging area of research in plant sciences. Front. Plant Sci. 5:319 10.3389/fpls.2014.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algranati I. D., Echandi G., Garcia-Patrone M., Gonzalez N. S., Goldemberg S. H. (1976). Polyamines, equilibrium between ribosomal particles and protein synthesis in bacteria. Arch. Biol. Med. Exp. 10 49–60. [PubMed] [Google Scholar]
- Amin R., Franz-Wachtel M., Tiffert Y., Heberer M., Meky M., Ahmed Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 3:38 10.3389/fmolb.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Chang M., Shan J., Jiang R., Zhang Y., Zhang R., et al. (2011). Identification and characterization of a novel spermidine/spermine acetyltransferase encoded by gene ste26 from Streptomyces sp. 139. Biochimie 93 1401–1407. 10.1016/j.biochi.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Bercovich Z., Snapir Z., Keren-Paz A., Kahana C. (2011). Antizyme affects cell proliferation and viability solely through regulating cellular polyamines. J. Biol. Chem. 286 33778–33783. 10.1074/jbc.M111.270637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman T. J., Armstrong S. K. (1996). The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178 54–60. 10.1128/jb.178.1.54-60.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock W. O., Fernandez J. M., Short J. M. (1987). Xl1-blue - a high-efficiency plasmid transforming reca Escherichia-coli strain with beta-galactosidase selection. Biotechniques 5 376–378. [Google Scholar]
- Burrell M., Hanfrey C. C., Kinch L. N., Elliott K. A., Michael A. J. (2012). Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 86 485–499. 10.1111/j.1365-2958.2012.08208.x [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Chater K. F., Bibb M. J. (1990). Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 172 3367–3378. 10.1128/jb.172.6.3367-3378.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Sobieszczuk-Nowicka E., Aloisi I., Fattorini L., Serafini-Fracassini D., Del Duca S. (2015). Polyamines are common players in different facets of plant programmed cell death. Amino Acids 47 27–44. 10.1007/s00726-014-1865-1 [DOI] [PubMed] [Google Scholar]
- Campilongo R., Di Martino M. L., Marcocci L., Pietrangeli P., Leuzzi A., Grossi M., et al. (2014). Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: looking into the gap between commensal E. coli and harmful Shigella. PLoS ONE 9:e106589 10.1371/journal.pone.0106589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M. K., Keembiyehetty C. N., Chen W., Tabor H. (2015). Polyamines stimulate the level of the sigma38 subunit (RpoS) of Escherichia coli RNA polymerase, resulting in the induction of the glutamate decarboxylase-dependent acid response system via the gadE regulon. J. Biol. Chem. 290 17809–17821. 10.1074/jbc.M115.655688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M. K., Tabor C. W., Tabor H. (2002). Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe). Proc. Natl. Acad. Sci. U.S.A. 99 10330–10334. 10.1073/pnas.162362899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M. K., Tabor C. W., Tabor H. (2003). Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U.S.A. 100 2261–2265. 10.1073/pnas.2627990100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M. K., Tabor H. (2013). Polyamines are critical for the induction of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 288 33559–33570. 10.1074/jbc.M113.510552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. R., Tyms A. S. (1991). Polyamine biosynthesis in cells infected with different clinical isolates of human cytomegalovirus. J. Med. Virol. 34 212–216. 10.1002/jmv.1890340403 [DOI] [PubMed] [Google Scholar]
- Colombatto S., De Agostini M., Corsi D., Sinicco A. (1989). Polyamines in lymphocytes from patients infected by human immunodeficiency virus. Biol. Chem. Hoppe Seyler 370 745–748. 10.1515/bchm3.1989.370.2.745 [DOI] [PubMed] [Google Scholar]
- Cunningham-Rundles S., Maas W. K. (1975). Isolation, characterization, and mapping of Escherichia coli mutants blocked in the synthesis of ornithine decarboxylase. J. Bacteriol. 124 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., Ristow J. L. (1991). Polyamine toxicity in Neurospora crassa: protective role of the vacuole. Arch. Biochem. Biophys. 285 306–311. 10.1016/0003-9861(91)90364-O [DOI] [PubMed] [Google Scholar]
- Dudkowska M., Lai J., Gardini G., Stachurska A., Grzelakowska-Sztabert B., Colombatto S., et al. (2003). Agmatine modulates the in vivo biosynthesis and interconversion of polyamines and cell proliferation. Biochim. Biophys. Acta 1619 159–166. 10.1016/S0304-4165(02)00476-2 [DOI] [PubMed] [Google Scholar]
- Evans C. G. T., Herbert D., Tempest D. W. (1970). Chapter XIII the continuous cultivation of micro-organisms: 2. construction of a chemostat. Methods Microbiol. 2 277–327. 10.1016/S0580-9517(08)70227-7 [DOI] [Google Scholar]
- Fink D., Falke D., Wohlleben W., Engels A. (1999). Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology 145(Pt 9), 2313–2322. 10.1099/00221287-145-9-2313 [DOI] [PubMed] [Google Scholar]
- Forouhar F., Lee I. S., Vujcic J., Vujcic S., Shen J., Vorobiev S. M., et al. (2005). Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J. Biol. Chem. 280 40328–40336. 10.1074/jbc.M505332200 [DOI] [PubMed] [Google Scholar]
- Foster A., Barnes N., Speight R., Keane M. A. (2013). Genomic organisation, activity and distribution analysis of the microbial putrescine oxidase degradation pathway. Syst. Appl. Microbiol. 36 457–466. 10.1016/j.syapm.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Fujihara S., Abe H., Minakawa Y., Akao S., Yoneyama T. (1994). Polyamines in nodules from various plant-microbe symbiotic associations. Plant Cell Physiol. 35 1127–1134. 10.1093/oxfordjournals.pcp.a078705 [DOI] [Google Scholar]
- Grasemann H., Shehnaz D., Enomoto M., Leadley M., Belik J., Ratjen F. (2012). L-ornithine derived polyamines in cystic fibrosis airways. PLoS ONE 7:e46618 10.1371/journal.pone.0046618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G. L., Sigel S. P., Payne S. M., Neilands J. B. (1984). Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259 383–385. [PubMed] [Google Scholar]
- Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100 1541–1546. 10.1073/pnas.0337542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G., Maslesa-Galic S., Tullius M. V., Horwitz M. A. (2005). All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58 1157–1172. 10.1111/j.1365-2958.2005.04899.x [DOI] [PubMed] [Google Scholar]
- Hatmi S., Trotel-Aziz P., Villaume S., Couderchet M., Clement C., Aziz A. (2014). Osmotic stress-induced polyamine oxidation mediates defence responses and reduces stress-enhanced grapevine susceptibility to Botrytis cinerea. J. Exp. Bot. 65 75–88. 10.1093/jxb/ert351 [DOI] [PubMed] [Google Scholar]
- Hayward D., Van Helden P. D., Wiid I. J. (2009). Glutamine synthetase sequence evolution in the mycobacteria and their use as molecular markers for Actinobacteria speciation. BMC Evol. Biol. 9:48 10.1186/1471-2148-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heby O. (1981). Role of polyamines in the control of cell proliferation and differentiation. Differentiation 19 1–20. 10.1111/j.1432-0436.1981.tb01123.x [DOI] [PubMed] [Google Scholar]
- Higashi K., Kashiwagi K., Taniguchi S., Terui Y., Yamamoto K., Ishihama A., et al. (2006). Enhancement of +1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli. J. Biol. Chem. 281 9527–9537. 10.1074/jbc.M513752200 [DOI] [PubMed] [Google Scholar]
- Hillemann D., Dammann T., Hillemann A., Wohlleben W. (1993). Genetic and biochemical characterization of the two glutamine synthetases GSI and GSII of the phosphinothricyl-alanyl-alanine producer, Streptomyces viridochromeogenes Tu494. J. Gen. Microbiol. 139 1773–1783. 10.1099/00221287-139-8-1773 [DOI] [PubMed] [Google Scholar]
- Hirsch J. G., Dubos R. J. (1952). The effect of spermine on tubercle bacilli. J. Exp. Med. 95 191–208. 10.1084/jem.95.2.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. C., Panagiotidis C. A., Canellakis E. S. (1990). Transcriptional effects of polyamines on ribosomal proteins and on polyamine-synthesizing enzymes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87 3464–3468. 10.1073/pnas.87.9.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (2006). Polyamine Modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. 139 11–16. 10.1093/jb/mvj020 [DOI] [PubMed] [Google Scholar]
- Jain A., Tyagi A. K. (1987). Role of polyamines in the synthesis of RNA in mycobacteria. Mol. Cell. Biochem. 78 3–8. 10.1007/BF00224418 [DOI] [PubMed] [Google Scholar]
- Jimenez-Bremont J. F., Marina M., Guerrero-Gonzalez M. d. L., Rossi F. R., Sanchez-Rangel D., Rodriguez-Kessler M., et al. (2014). Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 5:95 10.3389/fpls.2014.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I. L., Kim I. G. (2003a). Polyamines and glutamate decarboxylase-based acid resistance in Escherichia coli. J. Biol. Chem. 278 22846–22852. [DOI] [PubMed] [Google Scholar]
- Jung I. L., Kim I. G. (2003b). Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem. Biophys. Res. Commun. 301 915–922. [DOI] [PubMed] [Google Scholar]
- Kallifidas D., Thomas D., Doughty P., Paget M. S. (2010). The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology 156 1661–1672. 10.1099/mic.0.037804-0 [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation. [Google Scholar]
- Kurihara S., Oda S., Kato K., Kim H. G., Koyanagi T., Kumagai H., et al. (2005). A novel putrescine utilization pathway involves gamma-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 280 4602–4608. 10.1074/jbc.M411114200 [DOI] [PubMed] [Google Scholar]
- Kusano T., Suzuki H. (eds). (2015). Polyamines: A Universal Molecular Nexus for Growth, Survival, and Specialized Metabolism. Tokyo: Springer. [Google Scholar]
- Kwon D. H., Lu C. D. (2006). Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 50 1615–1622. 10.1128/AAC.50.5.1615-1622.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Atanasova V., Dolezelova Z., Parsons J. F. (2012). Structure and activity of PA5508, a hexameric glutamine synthetase homologue. Biochemistry 51 10121–10123. 10.1021/bi3014856 [DOI] [PubMed] [Google Scholar]
- Lasbury M. E., Merali S., Durant P. J., Tschang D., Ray C. A., Lee C. H. (2007). Polyamine-mediated apoptosis of alveolar macrophages during Pneumocystis pneumonia. J. Biol. Chem. 282 11009–11020. 10.1074/jbc.M611686200 [DOI] [PubMed] [Google Scholar]
- Lee J., Sperandio V., Frantz D. E., Longgood J., Camilli A., Phillips M. A., et al. (2009). An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284 9899–9907. 10.1074/jbc.M900110200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Kessler W., Van Den Heuvel J., Rinas U. (2011). Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl. Microbiol. Biotechnol. 91 1203–1213. 10.1007/s00253-011-3407-z [DOI] [PubMed] [Google Scholar]
- Liao C. P., Phanstiel O. T., Lasbury M. E., Zhang C., Shao S., Durant P. J., et al. (2009). Polyamine transport as a target for treatment of Pneumocystis pneumonia. Antimicrob. Agents Chemother. 53 5259–5264. 10.1128/AAC.00662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Wang W., Wu H., Gong X., Moriguchi T. (2015). Polyamines function in stress tolerance: from synthesis to regulation. Front. Plant Sci. 6:827 10.3389/fpls.2015.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil D. J., Occi J. L., Gewain K. M., Macneil T., Gibbons P. H., Ruby C. L., et al. (1992). Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115 119–125. 10.1016/0378-1119(92)90549-5 [DOI] [PubMed] [Google Scholar]
- Maeda T., Wakasawa T., Shima Y., Tsuboi I., Aizawa S., Tamai I. (2006). Role of polyamines derived from arginine in differentiation and proliferation of human blood cells. Biol. Pharm. Bull. 29 234–239. 10.1248/bpb.29.234 [DOI] [PubMed] [Google Scholar]
- Menges R., Muth G., Wohlleben W., Stegmann E. (2007). The ABC transporter Tba of Amycolatopsis balhimycina is required for efficient export of the glycopeptide antibiotic balhimycin. Appl. Microbiol. Biotechnol. 77 125–134. 10.1007/s00253-007-1139-x [DOI] [PubMed] [Google Scholar]
- Miller-Fleming L., Olin-Sandoval V., Campbell K., Ralser M. (2015). Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427 3389–3406. 10.1016/j.jmb.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Minocha R., Majumdar R., Minocha S. C. (2014). Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 5:175 10.3389/fpls.2014.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H., Wang X., Zhang Y., Zhang G., Zhang J., Ma Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83 962–975. 10.1111/tpj.12941 [DOI] [PubMed] [Google Scholar]
- Moschou P. N., Roubelakis-Angelakis K. A. (2014). Polyamines and programmed cell death. J. Exp. Bot. 65 1285–1296. 10.1093/jxb/ert373 [DOI] [PubMed] [Google Scholar]
- Nastri H. G., Algranati I. D. (1996). Effect of polyamines on plasmid-mediated kanamycin resistance and kanamycin phosphotransferase gene expression in Escherichia coli. Cell. Mol. Biol. 42 711–717. [PubMed] [Google Scholar]
- Nastri H. G., Fastame I. G., Algranati I. D. (1993). Polyamines modulate streptomycin-induced mistranslation in Escherichia coli. Biochim. Biophys. Acta 1216 455–459. 10.1016/0167-4781(93)90014-5 [DOI] [PubMed] [Google Scholar]
- Nentwich M. (2010). Die Funktion von GlnA2 in der Transkriptionellen und Posttranslationalen Kontrolle des Stickstoffmetabolismus in Streptomyces coelicolor M145. Doctoral Dissertation, Eberhard Karls Universität Tübingen, Tübingen. [Google Scholar]
- Nesse L. L., Berg K., Vestby L. K. (2015). Effects of norspermidine and spermidine on biofilm formation by potentially pathogenic Escherichia coli and Salmonella enterica wild-type strains. Appl. Environ. Microbiol. 81 2226–2232. 10.1128/AEM.03518-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. Q., Schneider J., Wendisch V. F. (2015). Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J. Biotechnol. 201 75–85. 10.1016/j.jbiotec.2014.10.035 [DOI] [PubMed] [Google Scholar]
- Norris V., Reusch R. N., Igarashi K., Root-Bernstein R. (2015). Molecular complementarity between simple, universal molecules and ions limited phenotype space in the precursors of cells. Biol. Direct 10 28 10.1186/s13062-014-0028-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenlund M., Holmqvist B., Baldetorp B., Hellstrand P., Nilsson B. O. (2009). Polyamine synthesis inhibition induces S phase cell cycle arrest in vascular smooth muscle cells. Amino Acids 36 273–282. 10.1007/s00726-008-0060-7 [DOI] [PubMed] [Google Scholar]
- Oh T. J., Kim I. G. (1999). The expression of Escherichia coli SOS genes recA and uvrA is inducible by polyamines. Biochem. Biophys. Res. Commun. 264 584–589. 10.1006/bbrc.1999.1553 [DOI] [PubMed] [Google Scholar]
- Okanishi M., Suzuki K., Umezawa H. (1974). Formation and reversion of Streptomycete protoplasts: cultural condition and morphological study. J. Gen. Microbiol. 80 389–400. 10.1099/00221287-80-2-389 [DOI] [PubMed] [Google Scholar]
- Oves-Costales D., Kadi N., Fogg M. J., Song L., Wilson K. S., Challis G. L. (2008). Petrobactin biosynthesis: AsbB catalyzes condensation of spermidine with N8-citryl-spermidine and its N1-(3,4-dihydroxybenzoyl) derivative. Chem. Commun. (Camb.) 4034–4036. 10.1039/b809353a [DOI] [PubMed] [Google Scholar]
- Pal M., Szalai G., Janda T. (2015). Speculation: polyamines are important in abiotic stress signaling. Plant Sci. 237 16–23. 10.1016/j.plantsci.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Parhamifar L., Andersen H., Wu L., Hall A., Hudzech D., Moghimi S. M. (2014). Polycation-mediated integrated cell death processes. Adv. Genet. 88 353–398. 10.1016/B978-0-12-800148-6.00012-2 [DOI] [PubMed] [Google Scholar]
- Pegg A. E. (2013). Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 26 1782–1800. 10.1021/tx400316s [DOI] [PubMed] [Google Scholar]
- Perez-Redondo R., Rodriguez-Garcia A., Botas A., Santamarta I., Martin J. F., Liras P. (2012). ArgR of Streptomyces coelicolor is a versatile regulator.PLoS ONE 7:e32697. 10.1371/journal.pone.0032697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirnes-Karhu S., Maatta J., Finnila M., Alhonen L., Uimari A. (2015). Overexpression of spermidine/spermine N1-acetyltransferase impairs osteoblastogenesis and alters mouse bone phenotype. Transgenic Res. 24253–265. 10.1007/s11248-014-9836-6 [DOI] [PubMed] [Google Scholar]
- Pitkin J., Davis R. H. (1990). The genetics of polyamine synthesis in Neurospora crassa. Arch. Biochem. Biophys. 278 386–391. 10.1016/0003-9861(90)90275-4 [DOI] [PubMed] [Google Scholar]
- Planet P. J., Larussa S. J., Dana A., Smith H., Xu A., Ryan C., et al. (2013). Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4:e00889-13 10.1128/mBio.00889-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter A. J., Paton J. C. (2014). Spermidine biosynthesis and transport modulate pneumococcal autolysis. J. Bacteriol. 196 3556–3561. 10.1128/JB.01981-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R. M., Zimmerman B. J., Mccormack S. A., Patel T. B., Johnson L. R. (1999). Polyamine depletion arrests cell cycle and induces inhibitors p21(Waf1/Cip1), p27(Kip1), and p53 in IEC-6 cells. Am. J. Physiol. 276C684–C691. [DOI] [PubMed] [Google Scholar]
- Redenbach M., Kieser H. M., Denapaite D., Eichner A., Cullum J., Kinashi H., et al. (1996). A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21 77–96. 10.1046/j.1365-2958.1996.6191336.x [DOI] [PubMed] [Google Scholar]
- Reis R. S., Vale Ede M., Heringer A. S., Santa-Catarina C., Silveira V. (2016). Putrescine induces somatic embryo development and proteomic changes in embryogenic callus of sugarcane. J. Proteomics 130 170–179. 10.1016/j.jprot.2015.09.029 [DOI] [PubMed] [Google Scholar]
- Reuther J., Wohlleben W. (2007). Nitrogen metabolism in streptomyces coelicolor: transcriptional and post-translational regulation. J. Mol. Microbiol. Biotechnol. 12 139–146. 10.1159/000096469 [DOI] [PubMed] [Google Scholar]
- Rexer H. U., Schaberle T., Wohlleben W., Engels A. (2006). Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 186 447–458. 10.1007/s00203-006-0159-8 [DOI] [PubMed] [Google Scholar]
- Rossi F. R., Marina M., Pieckenstain F. L. (2015). Role of Arginine decarboxylase (ADC) in Arabidopsis thaliana defence against the pathogenic bacterium Pseudomonas viridiflava. Plant Biol. 17 831–839. 10.1111/plb.12289 [DOI] [PubMed] [Google Scholar]
- Sakamoto A., Terui Y., Yoshida T., Yamamoto T., Suzuki H., Yamamoto K., et al. (2015). Three members of polyamine modulon under oxidative stress conditions: two transcription factors (SoxR and EmrR) and a glutathione synthetic enzyme (GshA). PLoS ONE 10:e0124883 10.1371/journal.pone.0124883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy J. P., Lee E., Dartois V. (2013). Polyamines inhibit porin-mediated fluoroquinolone uptake in mycobacteria. PLoS ONE 8:e65806 10.1371/journal.pone.0065806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar N. K., Shankar S., Tyagi A. K. (1995). Polyamines exert regulatory control on mycobacterial transcription: a study using RNA polymerase from Mycobacterium phlei. Biochem. Mol. Biol. Int. 351189–1198. [PubMed] [Google Scholar]
- Schneider B. L., Hernandez V. J., Reitzer L. (2013). Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol. Microbiol. 88 537–550. 10.1111/mmi.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B. L., Reitzer L. (2012). Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J. Bacteriol. 194 4080–4088. 10.1128/JB.05063-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A., Bertin P., Danchin A. (1998). Characterization of polyamine synthesis pathway in Bacillus subtilis 168. Mol. Microbiol. 29 851–858. 10.1046/j.1365-2958.1998.00979.x [DOI] [PubMed] [Google Scholar]
- Shaibe E., Metzer E., Halpern Y. S. (1985). Metabolic pathway for the utilization of L-arginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J. Bacteriol. 163 933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. (2014). Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol. 1091 17–32. 10.1007/978-1-62703-691-7_2 [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. (1985). Polyamines in microorganisms. Microbiol. Rev. 49 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y., Akiyama M., Sakamoto A., Tomitori H., Yamamoto K., Ishihama A., et al. (2012). Increase in cell viability by polyamines through stimulation of the synthesis of ppGpp regulatory protein and omega protein of RNA polymerase in Escherichia coli. Int. J. Biochem. Cell Biol. 44 412–422. 10.1016/j.biocel.2011.11.017 [DOI] [PubMed] [Google Scholar]
- Tiffert Y., Franz-Wachtel M., Fladerer C., Nordheim A., Reuther J., Wohlleben W., et al. (2011). Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 89 1149–1159. 10.1007/s00253-011-3086-9 [DOI] [PubMed] [Google Scholar]
- Tkachenko A. G., Nesterova L. Y. (2003). Polyamines as modulators of gene expression under oxidative stress in Escherichia coli. Biochemistry 68 850–856. [DOI] [PubMed] [Google Scholar]
- Tkachenko A. G., Pozhidaeva O. N., Shumkov M. S. (2006). Role of polyamines in formation of multiple antibiotic resistance of Escherichia coli under stress conditions. Biochemistry 71 1042–1049. 10.1134/s0006297906090148 [DOI] [PubMed] [Google Scholar]
- Tome M. E., Fiser S. M., Payne C. M., Gerner E. W. (1997). Excess putrescine accumulation inhibits the formation of modified eukaryotic initiation factor 5A (eIF-5A) and induces apoptosis. Biochem. J. 328(Pt 3), 847–854. 10.1042/bj3280847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaniklidis G., Kotsiras A., Tsafouros A., Roussos P. A., Aivalakis G., Katinakis P., et al. (2016). Spatial and temporal distribution of genes involved in polyamine metabolism during tomato fruit development. Plant Physiol. Biochem. 100 27–36. 10.1016/j.plaphy.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Valdes-Santiago L., Ruiz-Herrera J. (2013). Stress and polyamine metabolism in fungi. Front. Chem. 1:42 10.3389/fchem.2013.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff J. N., Eichenseer C., Viell P., Piendl W., Altenbuchner J. (1996). Nucleotide sequence and role in DNA amplification of the direct repeats composing the amplifiable element AUD1 of Streptomyces lividans 66. Mol. Microbiol. 21 1037–1047. 10.1046/j.1365-2958.1996.761428.x [DOI] [PubMed] [Google Scholar]
- Walters D. R. (2003). Polyamines and plant disease. Phytochemistry 64 97–107. 10.1016/S0031-9422(03)00329-7 [DOI] [PubMed] [Google Scholar]
- Weiger T. M., Hermann A. (2014). Cell proliferation, potassium channels, polyamines and their interactions: a mini review. Amino Acids 46 681–688. 10.1007/s00726-013-1536-7 [DOI] [PubMed] [Google Scholar]
- Weisschuh N., Fink D., Vierling S., Bibb M. J., Wohlleben W., Engels A. (2000). Transcriptional analysis of the gene for glutamine synthetase II and two upstream genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 264 461–469. 10.1007/s004380000315 [DOI] [PubMed] [Google Scholar]
- Wimalasekera R., Tebartz F., Scherer G. F. (2011). Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181 593–603. 10.1016/j.plantsci.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Wortham B. W., Patel C. N., Oliveira M. A. (2007). Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 603 106–115. 10.1007/978-0-387-72124-8_9 [DOI] [PubMed] [Google Scholar]
- Yao X., He W., Lu C. D. (2011). Functional characterization of seven gamma-Glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and beta-Alanine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 193 3923–3930. 10.1128/JB.05105-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatin M. (2011). Polyamines in living organisms. J. Cell Mol. Biol. 1 57–67. [Google Scholar]
- Yoshida M., Kashiwagi K., Shigemasa A., Taniguchi S., Yamamoto K., Makinoshima H., et al. (2004). A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 279 46008–46013. 10.1074/jbc.M404393200 [DOI] [PubMed] [Google Scholar]
- Zeier J. (2013). New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 36 2085–2103. 10.1111/pce.12122 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ma C., Karmouch J., Katbi H. A., Liu X. J. (2009). Antiapoptotic role for ornithine decarboxylase during oocyte maturation. Mol. Cell. Biol. 29 1786–1795. 10.1128/MCB.01815-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.