Abstract
Polymorphisms in X-ray repair cross-complementing (XRCC) genes have been implicated in altering the risk of various urological cancers. However, the results of reported studies are controversial. To ascertain whether polymorphisms in XRCC genes are associated with the risk of urological neoplasms, we conducted present updated meta-analysis and systematic review. Summary odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were used to estimate the association. Finally, 54 publications comprising 129 case-control studies for twelve polymorphisms in five XRCC genes were enrolled. We identified that XRCC1-rs25489 polymorphism was associated with an increased risk of urological neoplasms in heterozygote and dominant models. Moreover, in the subgroup analysis by cancer type, we found that XRCC1-rs25489 polymorphism was associated with an increased risk of bladder cancer (BC) in heterozygote model. Although overall analyses suggested a null result for XRCC1-rs25487 polymorphism, in the stratified analysis by ethnicity, an increased risk of urological neoplasms for Asians in allelic and homozygote models was identified. While for other polymorphisms in XRCC genes, no significant association was uncovered. To sum up, our results indicated that XRCC1-rs25489 polymorphism is a risk factor for urological neoplasms, particularly for BC. Further studies with large sample size are needed to validate these findings.
Keywords: XRCC, Polymorphism, Urological neoplasm, Risk, Meta-analysis
Highlights
-
•
Polymorphisms in XRCC genes have been reported to have potential links with the risk of urological neoplasms.
-
•
XRCC1-rs25489 polymorphism is a risk factor for urological neoplasms, particularly for bladder cancer.
-
•
XRCC1-rs25487 polymorphism is a risk factor particularly for urological neoplasms in Asian population.
Genetic factors play a crucial role in urological neoplasms risk. Polymorphisms in XRCC genes were associated with occurrence or increase of various tumors. But the effects of XRCC genes on urological neoplasms were unclear. Our results identified that XRCC1-rs25489 polymorphism is a risk factor for urological neoplasms, particularly for bladder cancer, while XRCC1-rs25487 polymorphism is a risk factor for urological neoplasms restricted to Asians. By using these variations as biomarkers, it is more feasible to estimate the risk of acquiring urological neoplasms and thus formulate timely preventive strategy.
1. Introduction
Deoxyribonucleic acid (DNA) in a normal cell is capable of withstanding internal and external damage to prevent the damage or death of the cell (Alli et al., 2009, Orlow et al., 2008). The direct reversal, base excision, nucleotide excision in the main DNA repair pathways of human beings’ function as restoring lost gene information and maintaining DNA integrity (Rajaraman et al., 2010). Some research studies have already showed that polymorphisms in DNA-repair genes are an integral part of cancer risk, apart from environmental factors, diet, intake of non-steroidal and anti-inflammatory drugs, and endogenous factors (Spitz et al., 2003). At the cellular level, checkpoints activated by the DNA-repair genes can regulate the cell cycle and transcription to make the choice of the damage or the apoptosis (Vispe et al., 2000). In addition, DNA repair-gene is also critical in defending the cellular genome from the risk of environmental factors (Hoeijmakers, 2001). Therefore, making certain of the genetic mechanisms of DNA repair system might take an insight into the pathogenesis of relevant cancers. X-ray repair cross-complementing (XRCC) genes are members of the family of DNA repair system (Dizdaroglu, 2015), which are polymorphic with several non-synonymous polymorphisms, such as Arg194Trp (rs1799782), Arg280His (rs25489), Arg399Gln (rs25487) in XRCC1, Arg188His (rs3218536) polymorphisms in XRCC2, IVS6-14 (rs1799796) and Thr241Met (rs861539) polymorphisms in XRCC3, rs1805377, rs6869366 and rs28360071 polymorphisms in XRCC4 and rs7003908 in XRCC7. To date, plenty of evidences have indicated that more than one hundred proteins encoded by XRCC genes are implicated in four DNA repair pathways, including nucleotide excision repair (NER), base excision repair (BER), double-strand break repair (DSBR) and mismatch repair (MMR), working as tumor suppressors or oncogenes for the sake of participating in tumorigenesis through posting expression regulation of homologous target genes (Liesegang, 2001). Recently, studies have highlighted the ambivalent association between polymorphisms in XRCC genes and risk of urological neoplasms. In the study conducted by Agalliu et al. (2010), they have proved that there was no significant association between XRCC1 polymorphisms (rs1799782, rs25487, rs25489 and rs915927) and prostate cancer (PCa) risk. Consistent with Agalliu et al.'s conclusion, Lavender et al. (2010) also confirmed that no significant influence of XRCC1-rs25487 polymorphism on PCa risk was identified for African population. While in another population-based case-control dataset, Lan et al. (2006) suggested that XRCC1-rs25487 polymorphism was significantly associated with the development of PCa. Both Matullo (2005) and Nowacka-Zawisza et al. (2015) have not revealed a significant association between XRCC2-rs3218536 polymorphism and urological neoplasms risk in their work, respectively. As for polymorphisms in XRCC3 gene, Wu et al. (2006) indicated that there was no association between XRCC3-rs861539 polymorphism and bladder cancer (BC) risk, while Narter et al. (2009) reported the conflicting results that there was a 4.87-fold protective role of XRCC3 T allele against BC. In 2011, Mandal et al. (2011) conducted a case-control study comprising 192 PCa cases and 224 age-matched healthy controls and obtained a conclusion that XRCC4 promoter-1394 (rs6869366) heterozygote was associated with a lower risk of PCa, a result inconsistent with Chang et al.'s (2008) work. In addition, Mandal et al. (2010) provided a strong supportive evidence that common sequence variants genotype of XRCC7 gene might increase the risk of PCa.
As mentioned above, although many studies have conducted to investigate the associations between one or multiple polymorphism (s) and the risk of urological neoplasms, but there results were not consistent or even contradictory, which was partially due to the heterogeneity within cancer subtypes, the diverse ethnicities of patient cohorts and the small sample sizes. Therefore, we conducted the current updated meta-analysis and systematic review at the aim of precisely determines the association between genetic variants in five XRCC genes and the susceptibility to urological neoplasms.
2. Materials and Methods
2.1. Literature Search
We conducted a systematic literature search on PubMed, Medline, Google Scholar and Web of Science to retrieve all eligible publications on the association between polymorphisms in all XRCC genes and the risk of all urological cancer types (up to December 27, 2016) with the following keywords: (XRCC1-9 OR X-Ray Repair Cross Complementing 1-9) AND (polymorphism OR mutation OR variation OR SNP OR genotype) AND (carcinoma OR cancer OR neoplasm OR adenocarcinoma OR tumor OR malignancy) (Supplementary Table 1). The language of enrolled studies was restricted to English. Moreover, we identified additional articles by screening the references of enrolled eligible articles and Reviews. We would contact authors for critical data not mentioned in the eligible articles. If data or datasets were published in several articles, the publication with largest sample sizes was selected. However, after carefully screening, twelve polymorphisms in five XRCC genes were left for further investigation, and the cancer types were restricted to PCa, BC and renal cell carcinoma (RCC).
2.2. Inclusion Criteria and Exclusion Criteria
Publications satisfied the following inclusion criteria would be enrolled: (1) case-control studies that evaluated the association between polymorphisms in XRCC genes and urological neoplasms risk; (2) publications focusing on population genetic polymorphisms (3) articles with sufficient genotype data to assess ORs and the corresponding 95%CIs; (4) the control subjects satisfied Hardy-Weinberg equilibrium (HWE). The major exclusion criteria were: (1) case-only studies, case reports, or Reviews; (2) studies without raw data for the XRCC genotype (or contacted the corresponding author also cannot obtain the necessary original data): (3) studies that compared the XRCC variants in precancerous lesions and other cancers.
2.3. Data Extraction
Our investigators extracted the data from each study. All the case-control studies satisfied the inclusion criteria and consensus for any controversy was achieved. The data from the eligible articles was composed of the first author's name, year of publication, ethnicity, source of controls, cancer type and numbers of cases and controls in the XRCC1, XRCC2, XRCC3, XRCC4, XRCC7 genotypes. Ethnicity was categorized as “Caucasian”, “Asian”, and “Mixed”. The cancer type was categorized as PCa and BC. With the regard to the sources of controls, all eligible case-control studies were defined as either population-based or hospital-based.
2.4. Statistical Analysis
The strength of association between the polymorphisms in XRCC genes and the risk of urological neoplasms were evaluated using summary ORs and the corresponding 95%CIs in allelic (B vs. A), recessive (BB vs. BA + AA), dominant (BA + BB vs. AA), homozygous (BB vs. AA), and heterozygous (BA vs. AA) models (A: wild allele; B: mutated allele). The P values of our study were adjusted by Bonferroni correction method to compensate for that increased by testing each individual hypothesis at a significance level of a/m (a: the desired overall alpha level; m: the number of the hypothesis), and the Bonferroni correction rejects the null hypothesis with the value of P less than a/m (PA = PZ * 60 < 0.05, was considered as statistical significant) (Bonferroni, 1935). The Cochrane's Q-statistic test was used to assess the heterogeneity between studies (Davey Smith and Egger, 1997), and the inconsistency was quantified with the I2 statistic. The substantial heterogeneity was considered significant when I2 > 50% or PQ ≤ 0.1, then, a random effects model (DerSimonian-Laird method) was used; otherwise, the fixed-effects model (Mantel-Haenszel method) was applied (Mantel and Haenszel, 1959). When it came to the comparison among studies, we performed subgroup analyses categorized by cancer type, ethnicity, HWE and the source of control. Last but not least, we also conducted sensitivity analysis to assess stability of the results by omitting one study each time to exclude studies. HWE was estimated by the asymptotic test using the “sampsi command” in the Stata 12.0 software (version 12.0; State Corporation, College Station, Texas, USA), and deviation was considered when P < 0.05. The potential publication bias of the eligible studies was evaluated by Begg's funnel plots (Begg and Mazumdar, 1995) graphically and Egger's linear regression test (Seagroatt and Stratton, 1998) quantitatively. Moreover, the trim and fill algorithm which trim off the asymmetric outlying part of the funnel and estimate the true center of the funnel further provide effective and relatively powerful tests for evaluating the existence of such publication bias (Sue and Richard, 2000). The data was analyzed using the Stata 12.0 software (version 12.0; State Corporation, College Station, Texas, USA).
2.5. Linkage Disequilibrium (LD) Analysis Across Populations
Data were extracted from the 1000 genomes Project (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r2_B36/) comprising the polymorphisms in XRCC1, XRCC3 and XRCC4 evaluated in present study. Briefly, populations enrolled in the project including CHB (Han Chinese in Beijing, China), CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), JPT (Japanese in Tokyo, Japan) and YRI (Yoruba in Ibadan, Nigeria). Then, Haploview software was applied to conduct analyses and LD was assessed by r2 statistics in each of the above-mentioned populations.
3. Results
3.1. Main Characteristics of the Enrolled Studies
Table 1 showed the characteristics of all the eligible studies and genotype frequency distributions of twelve polymorphisms in five XRCC genes (XRCC1-rs915927, XRCC1-rs25489, XRCC1-rs25487, XRCC1-rs1799782, XRCC1-rs3213245, XRCC2-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs6869366, XRCC4-rs28360071, XRCC4-rs1805377, XRCC7-rs7003908) included in current meta-analysis (Agalliu et al., 2010, Andrew et al., 2015, Andrew et al., 2007, Andrew et al., 2006, Arizono et al., 2008, Berhane et al., 2012, Broberg et al., 2005, Chang et al., 2009, Lan et al., 2006, Lavender et al., 2010, Chang et al., 2008, Dhillon et al., 2009, Figueroa et al., 2007a, Figueroa et al., 2007b, Fontana et al., 2008, Gangwar et al., 2009, Hamano et al., 2008, Hirata et al., 2006, Hirata et al., 2007, Huang et al., 2007, Abe et al., 2011, Mittal et al., 2008, Narter et al., 2009, Nowacka-Zawisza et al., 2015, Ramaniuk et al., 2014, Ritchey et al., 2005, Rybicki et al., 2004, Sak et al., 2007, Sanyal et al., 2004, Shen et al., 2003, Stern et al., 2002, Stern et al., 2001, Van Gils et al., 2002, Wang et al., 2010, Wang et al., 2008, Wen et al., 2009, Wen et al., 2013, Wu et al., 2006, Xu et al., 2007, Zhi et al., 2012, Hao et al., 2008, Zhou et al., 2012, Zhu et al., 2014, Zhu et al., 2012, Kelsey et al., 2004, Kuasne et al., 2011, Luedeke et al., 2009, Mandal et al., 2010, Mandal et al., 2011, Matullo, 2005, Matullo et al., 2006, Matullo et al., 2001, Mittal et al., 2012a, Mittal et al., 2012b). The study selection processes were presented in Supplementary Figs. 1–5.
Table 1.
Characteristics of the enrolled studies.
| Gene-polymorphism | First author | Year | Ethnicity | Source of control | Cancer type | Case |
Control |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | Y (HWE) | ||||||
| XRCC1-rs25487 | Figueroa et al. | 2007 | Caucasian | H-B | BC | 434 | 494 | 133 | 433 | 453 | 110 | Y |
| Matullo et al. | 2001 | Mixed | H-B | BC | 53 | 58 | 13 | 31 | 41 | 12 | Y | |
| Stern et al. | 2001 | African | H-B | BC | 9 | 10 | 0 | 9 | 4 | 0 | Y | |
| Stern et al. | 2001 | Caucasian | H-B | BC | 87 | 106 | 21 | 79 | 92 | 26 | Y | |
| Mittal et al. | 2012 | Asian | H-B | BC | 67 | 106 | 39 | 102 | 109 | 39 | Y | |
| Arizono et al. | 2008 | Asian | H-B | BC | 139 | 102 | 10 | 140 | 90 | 21 | Y | |
| Mittal et al. | 2008 | Asian | H-B | BC | 37 | 76 | 27 | 73 | 81 | 36 | Y | |
| Sak et al. | 2007 | Caucasian | H-B | BC | 218 | 248 | 66 | 226 | 259 | 75 | Y | |
| Sanyal et al. | 2003 | Caucasian | H-B | BC | 124 | 155 | 32 | 113 | 110 | 23 | Y | |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 136 | 135 | 40 | 120 | 145 | 47 | Y | |
| Fontana et al. | 2008 | Caucasian | H-B | BC | 21 | 25 | 5 | 18 | 18 | 9 | Y | |
| Broberg et al. | 2005 | Caucasian | H-B | BC | 26 | 31 | 4 | 80 | 62 | 13 | Y | |
| Matullo et al. | 2006 | Caucasian | P-B | BC | 54 | 53 | 17 | 484 | 482 | 128 | Y | |
| Shen et al. | 2003 | Caucasian | H-B | BC | 93 | 87 | 21 | 92 | 98 | 24 | Y | |
| Ramaniuk et al. | 2014 | Caucasian | H-B | BC | 141 | 154 | 37 | 151 | 165 | 48 | Y | |
| Zhi et al. | 2012 | Asian | P-B | BC | 121 | 151 | 30 | 148 | 143 | 20 | Y | |
| Wang et al. | 2010 | Asian | P-B | BC | 113 | 102 | 19 | 105 | 126 | 22 | Y | |
| Andrew et al. | 2006 | Mixed | H-B | BC | 412 | 456 | 122 | 533 | 536 | 184 | N | |
| Kelsey et al. | 2004 | Mixed | P-B | BC | 132 | 187 | 36 | 228 | 230 | 86 | N | |
| Huang et al. | 2007 | Caucasian | H-B | BC | 266 | 276 | 71 | 267 | 264 | 65 | Y | |
| Gao et al. | 2010 | Mixed | H-B | PCa | 145 | 151 | 56 | 49 | 47 | 10 | Y | |
| Hamano et al. | 2008 | Asian | H-B | PCa | 16 | 54 | 72 | 11 | 50 | 58 | Y | |
| Berhane et al. | 2012 | Asian | H-B | PCa | 50 | 60 | 40 | 62 | 64 | 24 | Y | |
| Mittal et al. | 2012 | Asian | P-B | PCa | 84 | 62 | 49 | 105 | 102 | 43 | N | |
| Abe et al. | 2010 | Caucasian | P-B | PCa | 326 | 329 | 98 | 154 | 161 | 44 | Y | |
| vanGils et al. | 2002 | Caucasian | P-B | PCa | 37 | 30 | 9 | 77 | 78 | 27 | Y | |
| Xu et al. | 2009 | Asian | P-B | PCa | 108 | 85 | 14 | 153 | 72 | 10 | Y | |
| Ritchey et al. | 2005 | Asian | P-B | PCa | 85 | 53 | 17 | 132 | 99 | 12 | Y | |
| Hirata et al. | 2007 | Asian | H-B | PCa | 87 | 63 | 15 | 86 | 69 | 10 | Y | |
| Dhillon et al. | 2011 | Caucasian | H-B | PCa | 38 | 49 | 28 | 37 | 60 | 33 | Y | |
| Kuasne et al. | 2010 | Caucasian | P-B | PCa | 73 | 52 | 47 | 65 | 73 | 34 | Y | |
| Zhu et al. | 2015 | Asian | H-B | PCa | 249 | 245 | 78 | 276 | 243 | 53 | Y | |
| Chen et al. | 2005 | African | H-B | PCa | 90 | 30 | 3 | 84 | 28 | 3 | Y | |
| Chen et al. | 2005 | Caucasian | H-B | PCa | 95 | 104 | 29 | 109 | 87 | 21 | Y | |
| Rybicki et al. | 2003 | Caucasian | H-B | PCa | 245 | 257 | 70 | 179 | 203 | 55 | Y | |
| Rybicki et al. | 2003 | Mixed | H-B | PCa | 291 | 274 | 72 | 216 | 208 | 56 | Y | |
| Agalliu et al. | 2010 | Caucasian | H-B | PCa | 522 | 576 | 159 | 481 | 590 | 169 | Y | |
| Agalliu et al. | 2010 | Mixed | H-B | PCa | 103 | 37 | 4 | 53 | 27 | 2 | Y | |
| Hirata et al. | 2006 | Asian | H-B | RCC | 64 | 32 | 16 | 102 | 68 | 10 | Y | |
| XRCC1-rs1799782 | Huang et al. | 2007 | Caucasian | H-B | BC | 539 | 73 | 2 | 524 | 74 | 2 | Y |
| Andrew et al. | 2006 | Mixed | H-B | BC | 857 | 115 | 6 | 1041 | 152 | 10 | Y | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 967 | 124 | 5 | 906 | 115 | 1 | Y | |
| Matullo et al. | 2007 | Caucasian | P-B | BC | 108 | 16 | 0 | 951 | 141 | 2 | Y | |
| Stern et al. | 2001 | African | H-B | BC | 18 | 1 | 0 | 10 | 3 | 0 | Y | |
| Stern et al. | 2001 | Caucasian | H-B | BC | 189 | 24 | 0 | 163 | 34 | 0 | Y | |
| Mittal et al. | 2008 | Asian | H-B | BC | 111 | 27 | 2 | 159 | 30 | 1 | Y | |
| Mittal et al. | 2012 | Asian | H-B | BC | 172 | 37 | 3 | 207 | 41 | 2 | Y | |
| Fontana et al. | 2008 | Caucasian | H-B | BC | 0 | 4 | 47 | 0 | 5 | 40 | Y | |
| Wang et al. | 2010 | Asian | H-B | BC | 109 | 102 | 23 | 142 | 102 | 9 | Y | |
| Sak et al. | 2007 | Caucasian | H-B | BC | 476 | 56 | 3 | 498 | 61 | 3 | Y | |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 275 | 40 | 0 | 260 | 51 | 0 | Y | |
| Agalliu et al. | 2010 | Mixed | H-B | PCa | 131 | 15 | 0 | 72 | 9 | 0 | Y | |
| Hamano et al. | 2008 | Asian | H-B | PCa | 70 | 62 | 10 | 79 | 32 | 8 | Y | |
| Hirata et al. | 2007 | Asian | H-B | PCa | 70 | 74 | 21 | 85 | 62 | 18 | Y | |
| Zhu et al. | 2015 | Asian | H-B | PCa | 310 | 208 | 54 | 340 | 203 | 29 | Y | |
| vanGils et al. | 2002 | Caucasian | P-B | PCa | 67 | 9 | 0 | 152 | 28 | 0 | Y | |
| Xu et al. | 2007 | Asian | P-B | PCa | 103 | 84 | 20 | 92 | 117 | 26 | Y | |
| Agalliu et al. | 2010 | Caucasian | H-B | PCa | 1098 | 143 | 5 | 1071 | 158 | 6 | Y | |
| Mittal et al. | 2012 | Asian | P-B | PCa | 157 | 29 | 9 | 203 | 43 | 4 | Y | |
| XRCC1-rs25489 | Sak et al. | 2007 | Caucasian | H-B | BC | 456 | 54 | 3 | 516 | 41 | 3 | N |
| Stern et al. | 2001 | Caucasian | H-B | BC | 198 | 16 | 0 | 180 | 13 | 0 | Y | |
| Stern et al. | 2001 | African | H-B | BC | 17 | 2 | 0 | 13 | 0 | 0 | N | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 955 | 122 | 4 | 911 | 101 | 4 | Y | |
| Mittal et al. | 2012 | Asian | H-B | BC | 112 | 58 | 42 | 146 | 41 | 63 | N | |
| Mittal et al. | 2008 | Asian | H-B | BC | 72 | 39 | 29 | 105 | 28 | 57 | N | |
| Wang et al. | 2010 | Asian | P-B | BC | 140 | 88 | 6 | 201 | 52 | 0 | Y | |
| Xu et al. | 2008 | Asian | P-B | PCa | 165 | 40 | 2 | 193 | 39 | 3 | Y | |
| vanGils et al. | 2002 | Caucasian | P-B | PCa | 66 | 10 | 0 | 164 | 18 | 0 | Y | |
| Agalliu et al. | 2010 | Caucasian | H-B | PCa | 1120 | 121 | 3 | 1145 | 106 | 2 | Y | |
| Agalliu et al. | 2010 | Mixed | H-B | PCa | 137 | 9 | 0 | 76 | 7 | 0 | Y | |
| Mittal et al. | 2012 | Asian | H-B | PCa | 82 | 76 | 37 | 131 | 47 | 72 | N | |
| Zhu et al. | 2015 | Asian | H-B | PCa | 380 | 120 | 73 | 394 | 116 | 62 | N | |
| XRCC1-rs915927 | Sak et al. | 2007 | Caucasian | H-B | BC | 162 | 260 | 93 | 170 | 270 | 105 | Y |
| Matullo et al. | 2006 | Caucasian | P-B | BC | 27 | 56 | 41 | 243 | 508 | 342 | N | |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 87 | 139 | 60 | 116 | 125 | 49 | Y | |
| Agalliu et al. | 2010 | Caucasian | H-B | PCa | 238 | 622 | 400 | 220 | 618 | 409 | Y | |
| Agalliu et al. | 2010 | Mixed | H-B | PCa | 29 | 54 | 62 | 11 | 38 | 30 | Y | |
| XRCC1-rs3213245 | Wang et al. | 2010 | Asian | P-B | BC | 174 | 56 | 4 | 178 | 73 | 2 | Y |
| Sak et al. | 2007 | Caucasian | H-B | BC | 90 | 266 | 174 | 94 | 275 | 187 | Y | |
| Zhi et al. | 2012 | Asian | P-B | BC | 232 | 61 | 9 | 229 | 76 | 6 | Y | |
| Nowacka-Zawisza et al. | 2015 | Caucasian | H-B | PCa | 90 | 11 | 0 | 196 | 20 | 0 | Y | |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 133 | 22 | 1 | 94 | 13 | 2 | Y | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 924 | 208 | 6 | 908 | 208 | 13 | Y | |
| XRCC2-rs3218536 | Nowacka-Zawisza et al. | 2015 | Caucasian | H-B | PCa | 90 | 11 | 0 | 196 | 20 | 0 | Y |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 133 | 22 | 1 | 94 | 13 | 2 | Y | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 924 | 208 | 6 | 908 | 208 | 13 | Y | |
| XRCC3-rs861539 | Narter et al. | 2009 | Caucasian | H-B | BC | 23 | 5 | 27 | 5 | 2 | 32 | N |
| Fontana et al. | 2008 | Caucasian | H-B | BC | 8 | 28 | 15 | 4 | 23 | 18 | Y | |
| Matullo et al. | 2001 | Caucasian | H-B | BC | 33 | 64 | 27 | 42 | 27 | 16 | N | |
| Zhu et al. | 2012 | Asian | H-B | BC | 91 | 44 | 15 | 96 | 49 | 5 | Y | |
| Andrew et al. | 2008 | Mixed | P-B | BC | 397 | 477 | 172 | 482 | 617 | 176 | Y | |
| Gangwar et al. | 2009 | Asian | H-B | BC | 135 | 68 | 9 | 159 | 80 | 11 | Y | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 392 | 524 | 167 | 398 | 468 | 144 | Y | |
| Mittle et al. | 2012 | Asian | H-B | BC | 134 | 68 | 9 | 154 | 79 | 11 | Y | |
| Matullo et al. | 2005 | Caucasian | H-B | BC | 99 | 155 | 63 | 117 | 148 | 52 | Y | |
| Sanyal et al. | 2004 | Caucasian | H-B | BC | 131 | 129 | 51 | 107 | 109 | 30 | Y | |
| Shen et al. | 2003 | Caucasian | H-B | BC | 89 | 87 | 25 | 71 | 116 | 27 | Y | |
| Narter et al. | 2009 | Caucasian | H-B | BC | 23 | 5 | 27 | 5 | 2 | 32 | N | |
| Wu et al. | 2006 | Caucasian | H-B | BC | 230 | 290 | 92 | 250 | 261 | 85 | Y | |
| Broberg et al. | 2005 | Caucasian | P-B | BC | 23 | 33 | 5 | 60 | 72 | 21 | Y | |
| Stern et al. | 2002 | Mixed | H-B | BC | 90 | 110 | 33 | 94 | 91 | 24 | Y | |
| Matullo et al. | 2006 | Caucasian | P-B | BC | 46 | 61 | 17 | 383 | 544 | 167 | Y | |
| Hao et al. | 2008 | Asian | H-B | BC | 268 | 37 | 2 | 292 | 23 | 1 | Y | |
| Nowacka-Zawisza et al. | 2015 | Caucasian | H-B | PCa | 54 | 34 | 13 | 119 | 75 | 52 | N | |
| Ritchey et al. | 2005 | Asian | P-B | PCa | 139 | 17 | 3 | 214 | 31 | 2 | Y | |
| Dhillon et al. | 2011 | Mixed | H-B | PCa | 60 | 44 | 12 | 54 | 72 | 6 | N | |
| Mandal et al. | 2010 | Caucasian | H-B | PCa | 103 | 77 | 12 | 137 | 78 | 9 | Y | |
| Hamano et al. | 2008 | Asian | H-B | PCa | 121 | 18 | 3 | 97 | 20 | 2 | Y | |
| Dhillon et al. | 2011 | Mixed | H-B | PCa | 60 | 44 | 12 | 54 | 72 | 6 | N | |
| XRCC3-rs1799796 | Matullo et al. | 2005 | Caucasian | H-B | BC | 171 | 117 | 21 | 166 | 126 | 19 | Y |
| Mittle et al. | 2012 | Asian | H-B | BC | 122 | 83 | 6 | 160 | 77 | 7 | Y | |
| Wu et al. | 2006 | Caucasian | H-B | BC | 279 | 258 | 63 | 256 | 261 | 75 | Y | |
| Broberg et al. | 2005 | Caucasian | P-B | BC | 25 | 30 | 3 | 57 | 74 | 21 | Y | |
| Matullo et al. | 2006 | Caucasian | P-B | BC | 60 | 47 | 17 | 554 | 447 | 91 | Y | |
| XRCC4-rs6869366 | Chang et al. | 2008 | Asian | H-B | PCa | 113 | 21 | 0 | 126 | 8 | 0 | Y |
| Mandal et al. | 2011 | Asian | H-B | PCa | 117 | 70 | 5 | 112 | 98 | 14 | Y | |
| Mittal et al. | 2011 | Asian | H-B | BC | 120 | 83 | 8 | 121 | 106 | 17 | Y | |
| Chang et al. | 2009 | Asian | H-B | BC | 105 | 53 | 0 | 127 | 31 | 0 | Y | |
| XRCC4-rs28360071 | Mandal et al. | 2011 | Asian | H-B | PCa | 124 | 49 | 19 | 168 | 48 | 8 | Y |
| Mittal et al. | 2011 | Asian | H-B | BC | 153 | 47 | 11 | 188 | 50 | 6 | Y | |
| Chang et al. | 2009 | Asian | H-B | BC | 95 | 61 | 2 | 98 | 57 | 3 | Y | |
| XRCC4-rs1805377 | Mandal et al. | 2011 | Asian | H-B | PCa | 131 | 55 | 6 | 149 | 65 | 10 | Y |
| Luedeke et al. | 2009 | Caucasian | H-B | PCa | 8 | 107 | 422 | 8 | 89 | 410 | Y | |
| Broberg et al. | 2005 | Caucasian | H-B | BC | 44 | 9 | 1 | 103 | 23 | 1 | Y | |
| Mittal et al. | 2011 | Asian | H-B | BC | 140 | 70 | 1 | 156 | 79 | 9 | Y | |
| Figueroa et al. | 2007 | Caucasian | H-B | BC | 13 | 232 | 841 | 12 | 168 | 852 | Y | |
| XRCC7-rs7003908 | Hirata et al. | 2006 | Asian | H-B | PCa | 74 | 79 | 12 | 86 | 67 | 12 | Y |
| Mandal et al. | 2010 | Asian | H-B | PCa | 48 | 82 | 62 | 75 | 105 | 44 | Y | |
| Wang et al. | 2008 | Asian | H-B | BC | 129 | 80 | 4 | 118 | 103 | 14 | Y | |
| Gangwar et al. | 2009 | Asian | H-B | BC | 32 | 81 | 99 | 80 | 116 | 54 | Y | |
| Zhi et al. | 2012 | Asian | H-B | BC | 185 | 105 | 12 | 152 | 134 | 25 | Y | |
| Hirata et al. | 2006 | Asian | H-B | RCC | 57 | 40 | 15 | 90 | 76 | 14 | Y | |
PCa: prostate cancer; BC: bladder cancer; RCC: renal cell carcinoma; H-B: hospital-based; P-B: population-based; HWE: Hardy Weinberg equilibrium; Y: controls conformed to HWE; N: controls were not conformed to HWE; Mixed: more than two ethnicities.
For polymorphisms in XRCC1 gene (XRCC1-rs915927, XRCC1-rs25489, XRCC1-rs25487, XRCC1-rs1799782, XRCC1-rs3213245), a total of 80 case-control studies with 28,095 cases and 31,363 controls met the inclusion criteria. 37 studies of them were performed in Caucasians, 29 studies in Asians, four in Africans and the others were in mixed ethnic groups (including at least one race). Controls of 60 studies were hospital-based controls, and the others were population-based controls. Additionally, the distributions of polymorphisms in XRCC1 for control groups were consistent with HWE, except for ten studies (Andrew et al., 2006, Mittal et al., 2008, Sak et al., 2007, Stern et al., 2001, Zhu et al., 2014, Kelsey et al., 2004, Matullo et al., 2006, Mittal et al., 2012b). For XRCC2-rs3218536 polymorphism, three eligible studies comprising1395 cases and 1454 controls were enrolled. All the studies were performed on subjects in Caucasians. Controls of studies were hospital-based. All of the studies were consistent with HWE. For polymorphisms in XRCC3 (XRCC3-rs1799796 and XRCC3-rs861539), we analyzed 28 studies with 7283 cases and 9773 controls, which were published between 2002 and 2016. 17 of the studies were performed in Caucasians, seven studies in Asians and the other four in Mixed group. Controls of 23 studies were hospital-based controls, and others were population-based controls. There are six case-control studies that were not consistent with HWE (Nowacka-Zawisza et al., 2015, Narter et al., 2009, Dhillon et al., 2009, Matullo et al., 2001). For polymorphisms in XRCC4 (XRCC4-rs6869366, XRCC4-rs28360071 and XRCC4-rs1805377), 12 case-control studies comprising 3336 cases and 3520 controls were considered eligible. Nine studies were conducted in Asians and the others were in Caucasians. Controls of all studies were hospital-based controls and no study was deviated from HWE. For XRCC7-rs7003908, six studies with 1196 cases and 1365 controls were enrolled. All the six studies were performed in Asians. Source of control of all enrolled studies were hospital-based controls and no study was deviated from HWE. In addition, we applied a Newcastle-Ottawa scale (NOS) to evaluate the quality of these enrolled studies (Wells et al., 2000), which was presented in Table 2, and employed a PRISMA 2009 checklist to present our meta-analysis work (Supplementary Table 4).
Table 2.
Methodological quality of the included studies according to the Newcastle-Ottawa scale.
| Gene-polymorphism | Author | Ethnicity | Adequacy of case definition | Representativeness of the cases | Selection of controls | Definition of controls | Comparability cases/controls | Ascertainment of exposure | Same method of ascertainment | Non-response rate |
|---|---|---|---|---|---|---|---|---|---|---|
| XRCC1-rs25487 | Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * |
| Matullo et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Arizono et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | * | NA | * | * | |
| Sak et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Sanyal et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| Matullo et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Fontana et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Broberg et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Matullo et al. | Caucasian | * | * | NA | * | * | * | * | * | |
| Shen et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Ramaniuk et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Zhi et al. | Asian | * | * | * | * | ** | * | * | * | |
| Wang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Andrew et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Kelsey et al. | Mixed | * | * | * | * | ** | * | * | * | |
| Huang et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Gao et al. | Mixed | NA | * | NA | * | * | NA | * | - | |
| Hamano et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Berhane et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Abe et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| vanGils et al. | Caucasian | * | * | * | * | ** | NA | * | * | |
| Xu et al. | Asian | * | * | * | * | ** | * | * | * | |
| Ritchey et al. | Asian | * | * | * | * | ** | * | * | * | |
| Hirata et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Dhillon et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| Kuasne et al. | Caucasian | * | * | * | * | ** | NA | * | * | |
| Zhu et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Chen et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Chen et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Rybicki et al. | Mixed | * | * | NA | * | * | * | * | * | |
| Rybicki et al. | Mixed | * | * | NA | * | * | * | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Hirata et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| XRCC1-rs1799782 | Huang et al. | Caucasian | * | * | NA | * | ** | * | * | * |
| Andrew et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Matullo et al. | Caucasian | * | * | * | * | ** | * | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | * | NA | * | * | |
| Fontana et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Wang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Sak et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Matullo et al. | Caucasian | * | * | NA | * | * | * | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Hamano et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Hirata et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Zhu et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| vanGils et al. | Caucasian | * | * | * | * | ** | NA | * | * | |
| Xu et al. | Asian | * | * | * | * | ** | * | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Mittal et al. | Asian | * | * | NA | * | * | NA | * | * | |
| XRCC1-rs25489 | Sak et al. | Mixed | * | * | NA | * | ** | * | * | * |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | * | NA | * | * | |
| Wang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Xu et al. | Asian | * | * | * | * | ** | * | * | * | |
| vanGils et al. | Caucasian | * | * | * | * | ** | NA | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Zhu et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| XRCC1-rs915927 | Sak et al. | Caucasian | * | * | NA | * | ** | * | * | * |
| Matullo et al. | Caucasian | * | * | * | * | ** | * | * | * | |
| Matullo et al. | Caucasian | * | * | NA | * | * | * | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| Agalliu et al. | Mixed | * | * | NA | * | ** | NA | * | * | |
| XRCC1-rs3213245 | Wang et al. | Asian | * | * | NA | * | ** | * | * | * |
| Sak et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Zhi et al. | Asian | * | * | * | * | ** | * | * | * | |
| XRCC2-rs3218536 | Nowacka-Zawisza et al. | Caucasian | NA | * | NA | * | ** | * | * | * |
| Matullo et al. | Caucasian | * | * | NA | * | * | * | * | * | |
| Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| XRCC3-rs861539 | Narter et al. | Caucasian | * | * | NA | * | ** | * | * | * |
| Fontana et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Matullo et al. | Caucasian | * | * | * | * | ** | * | * | * | |
| Zhu et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Andrew et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Gangwar et al. | Asian | * | * | NA | * | * | * | * | * | |
| Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| Mittle et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Matullo et al. | Caucasian | * | * | NA | * | * | * | * | * | |
| Sanyal et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| Shen et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Narter et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Wu et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Broberg et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Stern et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Matullo et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| Yang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Hao et al. | Asian | NA | * | NA | * | ** | NA | * | * | |
| XRCC3-rs861539 | Nowacka-Zawisza et al. | Caucasian | NA | * | NA | * | ** | * | * | * |
| Ritchey et al. | Asian | * | * | * | * | ** | * | * | * | |
| Dhillon et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| Mandal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Hamano et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Dhillon et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| XRCC3-rs1799796 | Matullo et al. | Caucasian | * | * | * | * | ** | * | * | * |
| Mittle et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Wu et al. | Caucasian | * | * | NA | * | ** | * | * | * | |
| Broberg et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Matullo et al. | Mixed | * | * | NA | * | ** | * | * | * | |
| XRCC4-rs1805377 | Mandal et al. | Asian | * | * | NA | * | * | * | * | * |
| Luedeke et al. | Caucasian | * | * | NA | * | ** | NA | * | * | |
| Broberg et al. | Asian | * | * | NA | * | ** | NA | * | * | |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Figueroa et al. | Caucasian | * | * | NA | * | * | NA | * | * | |
| XRCC4-rs6869366 | Chang et al. | Asian | * | * | NA | * | ** | * | * | * |
| Mandal et al. | Asian | * | * | NA | * | * | * | * | * | |
| Mittal et al. | Asian | * | * | NA | * | * | NA | * | * | |
| Chang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| XRCC4-rs28360071 | Mandal et al. | Asian | * | * | NA | * | ** | * | * | * |
| Mittal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Chang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| XRCC7-rs7003908 | Hirata et al. | Asian | * | * | NA | * | ** | NA | * | * |
| Mandal et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Wang et al. | Asian | * | * | NA | * | ** | * | * | * | |
| Gangwar et al. | Asian | * | * | NA | * | * | * | * | * | |
| Zhi et al. | Asian | * | * | * | * | ** | * | * | * | |
| Hirata et al. | Asian | * | * | NA | * | ** | NA | * | * |
H′ quality choices with a ‘star’. A study can be awarded a maximum of one star for each numbered item within the Selection and Exposure categories. A maximum of two stars can be given for Comparability.
3.2. Quantitative Synthesis
Table 3 listed the main results of the meta-analysis of polymorphisms in XRCC genes and risk of urological neoplasm.
Table 3.
Results of meta-analysis for polymorphisms in XRCC genes and risk of urological neoplasms.
| SNP | Comparison | Subgroup | N | Cases | Controls | PH | PZ | PA | Random | Fixed |
|---|---|---|---|---|---|---|---|---|---|---|
| XRCC1-rs25487 | B vs. A | Overall | 39 | 12,565 | 13,362 | 0.068 | 0.103 | 1.000 | 1.040 (0.992–1.090) | 1.031 (0.993–1.070) |
| B vs. A | African | 2 | 142 | 128 | 0.330 | 0.718 | 1.000 | 1.084 (0.676–1.738) | 1.090 (0.682–1.743) | |
| B vs. A | Asian | 13 | 2837 | 3169 | 0.168 | 0.000 | 0.000 | 1.174 (1.069–1.288) | 1.176 (1.089–1.271) | |
| B vs. A | Caucasian | 18 | 6984 | 7516 | 0.582 | 0.795 | 1.000 | 0.993 (0.945–1.044) | 0.993 (0.945–1.044) | |
| B vs. A | Mixed | 6 | 2602 | 2549 | 0.515 | 0.617 | 1.000 | 0.978 (0.900–1.064) | 0.979 (0.900–1.064) | |
| B vs. A | H-B | 29 | 9992 | 9719 | 0.120 | 0.317 | 1.000 | 1.029 (0.977–1.084) | 1.022 (0.980–1.066) | |
| B vs. A | P-B | 10 | 2573 | 3643 | 0.125 | 0.123 | 1.000 | 1.072 (0.966–1.191) | 1.066 (0.983–1.157) | |
| B vs. A | N | 3 | 1540 | 2047 | 0.508 | 0.902 | 1.000 | 0.994 (0.902–1.096) | 0.994 (0.902–1.095) | |
| B vs. A | Y | 36 | 11,025 | 11,315 | 0.051 | 0.093 | 1.000 | 1.046 (0.993–1.101) | 1.037 (0.996–1.080) | |
| B vs. A | BC | 20 | 6438 | 7928 | 0.266 | 0.535 | 1.000 | 1.016 (0.959–1.075) | 1.016 (0.966–1.068) | |
| B vs. A | PCa | 18 | 6015 | 5254 | 0.046 | 0.090 | 1.000 | 1.072 (0.989–1.160) | 1.046 (0.988–1.107) | |
| BA vs. AA | Overall | 39 | 12,565 | 13,362 | 0.095 | 0.331 | 1.000 | 1.033 (0.967–1.104) | 1.033 (0.978–1.090) | |
| BA vs. AA | African | 2 | 142 | 128 | 0.261 | 0.638 | 1.000 | 1.217 (0.582–2.542) | 1.141 (0.659–1.974) | |
| BA vs. AA | Asian | 13 | 2837 | 3169 | 0.024 | 0.339 | 1.000 | 1.082 (0.920–1.272) | 1.093 (0.979–1.221) | |
| BA vs. AA | Caucasian | 18 | 6984 | 7516 | 0.568 | 0.747 | 1.000 | 0.988 (0.919–1.062) | 0.988 (0.919–1.062) | |
| BA vs. AA | Mixed | 6 | 2602 | 2549 | 0.256 | 0.186 | 1.000 | 1.074 (0.924–1.249) | 1.084 (0.962–1.223) | |
| BA vs. AA | H-B | 29 | 9992 | 9719 | 0.493 | 0.278 | 1.000 | 1.034 (0.973–1.099) | 1.034 (0.973–1.099) | |
| BA vs. AA | P-B | 10 | 2573 | 3643 | 0.008 | 0.934 | 1.000 | 0.992 (0.820–1.200) | 1.027 (0.914–1.154) | |
| BA vs. AA | N | 3 | 1540 | 2047 | 0.061 | 0.515 | 1.000 | 1.095 (0.832–1.442) | 1.121 (0.971–1.293) | |
| BA vs. AA | Y | 36 | 11,025 | 11,315 | 0.172 | 0.532 | 1.000 | 1.022 (0.955–1.094) | 1.019 (0.961–1.080) | |
| BA vs. AA | BC | 20 | 6438 | 7928 | 0.227 | 0.014 | 0.840 | 1.097 (1.008–1.193) | 1.095 (1.019–1.177) | |
| BA vs. AA | PCa | 18 | 6015 | 5254 | 0.271 | 0.401 | 1.000 | 0.968 (0.881–1.063) | 0.966 (0.890–1.048) | |
| BA + BB vs. AA | Overall | 39 | 12,565 | 13,362 | 0.145 | 0.134 | 1.000 | 1.044 (0.983–1.109) | 1.040 (0.988–1.094) | |
| BA + BB vs. AA | African | 2 | 142 | 128 | 0.255 | 0.663 | 1.000 | 1.211 (0.576–2.543) | 1.125 (0.662–1.914) | |
| BA + BB vs. AA | Asian | 13 | 2837 | 3169 | 0.076 | 0.036 | 1.000 | 1.160 (1.010–1.333) | 1.165 (1.050–1.293) | |
| BA + BB vs. AA | Caucasian | 18 | 6984 | 7516 | 0.577 | 0.760 | 1.000 | 0.989 (0.924–1.059) | 0.989 (0.924–1.059) | |
| BA + BB vs. AA | Mixed | 6 | 2602 | 2549 | 0.473 | 0.525 | 1.000 | 1.037 (0.926–1.162) | 1.038 (0.926–1.162) | |
| BA + BB vs. AA | H-B | 29 | 9992 | 9719 | 0.315 | 0.249 | 1.000 | 1.038 (0.974–1.105) | 1.034 (0.977–1.095) | |
| BA + BB vs. AA | P-B | 10 | 2573 | 3643 | 0.066 | 0.540 | 1.000 | 1.048 (0.901–1.219) | 1.060 (0.949–1.183) | |
| BA + BB vs. AA | N | 3 | 1540 | 2047 | 0.517 | 0.335 | 1.000 | 1.068 (0.934–1.222) | 1.068 (0.934–1.222) | |
| BA + BB vs. AA | Y | 36 | 11,025 | 11,315 | 0.106 | 0.221 | 1.000 | 1.041 (0.974–1.114) | 1.035 (0.980–1.093) | |
| BA + BB vs. AA | BC | 20 | 6438 | 7928 | 0.210 | 0.066 | 1.000 | 1.067 (0.984–1.157) | 1.066 (0.996–1.142) | |
| BA + BB vs. AA | PCa | 18 | 6015 | 5254 | 0.170 | 0.831 | 1.000 | 1.020 (0.928–1.122) | 1.008 (0.934–1.089) | |
| BB vs. AA | Overall | 39 | 12,565 | 13,362 | 0.040 | 0.176 | 1.000 | 1.077 (0.967–1.199) | 1.053 (0.970–1.143) | |
| BB vs. AA | African | 2 | 142 | 128 | 0.968 | 0.942 | 1.000 | 0.949 (0.229–3.933) | 0.949 (0.229–3.933) | |
| BB vs. AA | Asian | 13 | 2837 | 3169 | 0.131 | 0.000 | 0.000 | 1.456 (1.170–1.812) | 1.464 (1.232–1.740) | |
| BB vs. AA | Caucasian | 18 | 6984 | 7516 | 0.778 | 0.729 | 1.000 | 0.981 (0.880–1.094) | 0.981 (0.880–1.094) | |
| BB vs. AA | Mixed | 6 | 2602 | 2549 | 0.354 | 0.212 | 1.000 | 0.890 (0.725–1.093) | 0.890 (0.742–1.068) | |
| BB vs. AA | H-B | 29 | 9992 | 9719 | 0.072 | 0.443 | 1.000 | 1.049 (0.929–1.184) | 1.031 (0.940–1.130) | |
| BB vs. AA | P-B | 10 | 2573 | 3643 | 0.121 | 0.150 | 1.000 | 1.171 (0.928–1.478) | 1.140 (0.954–1.363) | |
| BB vs. AA | N | 3 | 1540 | 2047 | 0.116 | 0.314 | 1.000 | 0.930 (0.667–1.297) | 0.900 (0.733–1.105) | |
| BB vs. AA | Y | 36 | 11,025 | 11,315 | 0.074 | 0.097 | 1.000 | 1.101 (0.983–1.233) | 1.085 (0.992–1.186) | |
| BB vs. AA | BC | 20 | 6438 | 7928 | 0.247 | 0.610 | 1.000 | 0.971 (0.853–1.105) | 0.971 (0.869–1.086) | |
| BB vs. AA | PCa | 18 | 6015 | 5254 | 0.111 | 0.038 | 1.000 | 1.194 (1.015–1.403) | 1.138 (1.007–1.287) | |
| BB vs. BA + AA | Overall | 39 | 12,565 | 13,362 | 0.022 | 0.233 | 1.000 | 1.064 (0.961–1.179) | 1.039 (0.963–1.121) | |
| BB vs. BA + AA | African | 2 | 142 | 128 | 0.853 | 0.842 | 1.000 | 0.865 (0.211–3.546) | 0.866 (0.211–3.548) | |
| BB vs. BA + AA | Asian | 13 | 2837 | 3169 | 0.087 | 0.002 | 0.120 | 1.378 (1.120–1.695) | 1.376 (1.176–1.609) | |
| BB vs. BA + AA | Caucasian | 18 | 6984 | 7516 | 0.828 | 0.941 | 1.000 | 0.997 (0.900–1.104) | 0.996 (0.900–1.103) | |
| BB vs. BA + AA | Mixed | 6 | 2602 | 2549 | 0.161 | 0.049 | 1.000 | 0.860 (0.666–1.109) | 0.843 (0.710–0.999) | |
| BB vs. BA + AA | H-B | 29 | 9992 | 9719 | 0.140 | 0.758 | 1.000 | 1.025 (0.922–1.139) | 1.014 (0.931–1.104) | |
| BB vs. BA + AA | P-B | 10 | 2573 | 3643 | 0.021 | 0.166 | 1.000 | 1.202 (0.926–1.560) | 1.145 (0.969–1.353) | |
| BB vs. BA + AA | N | 3 | 1540 | 2047 | 0.006 | 0.701 | 1.000 | 0.910 (0.563–1.473) | 0.857 (0.709–1.037) | |
| BB vs. BA + AA | Y | 36 | 11,025 | 11,315 | 0.170 | 0.074 | 1.000 | 1.088 (0.986–1.200) | 1.079 (0.993–1.172) | |
| BB vs. BA + AA | BC | 20 | 6438 | 7928 | 0.343 | 0.129 | 1.000 | 0.924 (0.824–1.035) | 0.922 (0.831–1.024) | |
| BB vs. BA + AA | PCa | 18 | 6015 | 5254 | 0.180 | 0.006 | 0.360 | 1.215 (1.057–1.397) | 1.172 (1.047–1.312) | |
| XRCC1-rs25489 | B vs. A | Overall | 13 | 4854 | 5050 | 0.040 | 0.028 | 1.000 | 1.168 (1.017–1.343) | 1.156 (1.053–1.268) |
| B vs. A | Asian | 6 | 1561 | 1750 | 0.001 | 0.204 | 1.000 | 1.168 (0.919–1.484) | 1.141 (1.017–1.280) | |
| B vs. A | Caucasian | 5 | 3128 | 3204 | 0.900 | 0.028 | 1.000 | 1.197 (1.020–1.405) | 1.197 (1.020–1.405) | |
| B vs. A | H-B | 10 | 4337 | 4380 | 0.820 | 0.089 | 1.000 | 1.090 (0.987–1.204) | 1.090 (0.987–1.204) | |
| B vs. A | P-B | 3 | 517 | 670 | 0.030 | 0.097 | 1.000 | 1.580 (0.921–2.711) | 1.715 (1.322–2.223) | |
| B vs. A | N | 6 | 1652 | 1835 | 0.539 | 0.262 | 1.000 | 1.071 (0.949–1.209) | 1.071 (0.950–1.208) | |
| B vs. A | Y | 7 | 3202 | 3215 | 0.027 | 0.054 | 1.000 | 1.289 (0.995–1.670) | 1.291 (1.116–1.494) | |
| B vs. A | BC | 7 | 2413 | 2475 | 0.003 | 0.118 | 1.000 | 1.246 (0.946–1.641) | 1.206 (1.054–1.381) | |
| B vs. A | PCa | 6 | 2441 | 2575 | 0.899 | 0.102 | 1.000 | 1.112 (0.979–1.264) | 1.112 (0.979–1.264) | |
| BA vs. AA | Overall | 13 | 4854 | 5050 | 0.010 | 0.000 | 0.000 | 1.455 (1.198–1.768) | 1.388 (1.233–1.563) | |
| BA vs. AA | Asian | 6 | 1561 | 1750 | 0.003 | 0.001 | 0.060 | 1.738 (1.244–2.428) | 1.615 (1.364–1.912) | |
| BA vs. AA | Caucasian | 5 | 3128 | 3204 | 0.870 | 0.025 | 1.000 | 1.213 (1.025–1.437) | 1.213 (1.025–1.437) | |
| BA vs. AA | H-B | 10 | 4337 | 4380 | 0.035 | 0.002 | 0.120 | 1.393 (1.133–1.712) | 1.323 (1.162–1.506) | |
| BA vs. AA | P-B | 3 | 517 | 670 | 0.076 | 0.050 | 1.000 | 1.653 (1.000–2.735) | 1.764 (1.322–2.354) | |
| BA vs. AA | N | 6 | 1652 | 1835 | 0.031 | 0.002 | 0.120 | 1.667 (1.211–2.295) | 1.526 (1.272–1.830) | |
| BA vs. AA | Y | 7 | 3202 | 3215 | 0.061 | 0.037 | 1.000 | 1.309 (1.016–1.686) | 1.294 (1.107–1.513) | |
| BA vs. AA | BC | 7 | 2413 | 2475 | 0.082 | 0.000 | 0.000 | 1.611 (1.242–2.090) | 1.540 (1.298–1.827) | |
| BA vs. AA | PCa | 6 | 2441 | 2575 | 0.032 | 0.077 | 1.000 | 1.304 (0.971–1.750) | 1.260 (1.068–1.485) | |
| BA + BB vs. AA | Overall | 13 | 4854 | 5050 | 0.153 | 0.000 | 0.000 | 1.297 (1.129–1.491) | 1.281 (1.148–1.428) | |
| BA + BB vs. AA | Asian | 6 | 1561 | 1750 | 0.019 | 0.010 | 0.600 | 1.392 (1.083–1.790) | 1.352 (1.168–1.564) | |
| BA + BB vs. AA | Caucasian | 5 | 3128 | 3204 | 0.882 | 0.024 | 1.000 | 1.211 (1.025–1.431) | 1.211 (1.025–1.431) | |
| BA + BB vs. AA | H-B | 10 | 4337 | 4380 | 0.918 | 0.002 | 0.120 | 1.207 (1.073–1.359) | 1.207 (1.073–1.359) | |
| BA + BB vs. AA | P-B | 3 | 517 | 670 | 0.034 | 0.080 | 1.000 | 1.666 (0.942–2.947) | 1.802 (1.357–2.394) | |
| BA + BB vs. AA | N | 6 | 1652 | 1835 | 0.791 | 0.004 | 0.240 | 1.258 (1.078–1.468) | 1.258 (1.078–1.468) | |
| BA + BB vs. AA | Y | 7 | 3202 | 3215 | 0.025 | 0.051 | 1.000 | 1.316 (0.999–1.733) | 1.304 (1.118–1.521) | |
| BA + BB vs. AA | BC | 7 | 2413 | 2475 | 0.052 | 0.008 | 0.480 | 1.403 (1.091–1.804) | 1.378 (1.177–1.614) | |
| BA + BB vs. AA | PCa | 6 | 2441 | 2575 | 0.718 | 0.020 | 1.000 | 1.197 (1.029–1.392) | 1.197 (1.029–1.392) | |
| BB vs. AA | Overall | 13 | 4854 | 5050 | 0.773 | 0.933 | 1.000 | 0.978 (0.790–1.209) | 0.991 (0.803–1.223) | |
| BB vs. AA | Asian | 6 | 1561 | 1750 | 0.202 | 0.879 | 1.000 | 0.959 (0.716–1.285) | 0.983 (0.790–1.223) | |
| BB vs. AA | Caucasian | 5 | 3128 | 3204 | 0.975 | 0.678 | 1.000 | 1.194 (0.524–2.719) | 1.190 (0.524–2.703) | |
| BB vs. AA | H-B | 10 | 4337 | 4380 | 0.936 | 0.664 | 1.000 | 0.954 (0.769–1.184) | 0.954 (0.769–1.182) | |
| BB vs. AA | P-B | 3 | 517 | 670 | 0.232 | 0.094 | 1.000 | 2.33 (0.508–10.683) | 2.554 (0.852–7.657) | |
| BB vs. AA | N | 6 | 1652 | 1835 | 0.674 | 0.647 | 1.000 | 0.950 (0.762–1.186) | 0.950 (0.762–1.184) | |
| BB vs. AA | Y | 7 | 3202 | 3215 | 0.640 | 0.243 | 1.000 | 1.360 (0.638–2.898) | 1.527 (0.751–3.107) | |
| BB vs. AA | BC | 7 | 2413 | 2475 | 0.548 | 0.561 | 1.000 | 0.878 (0.635–1.215) | 0.910 (0.662–1.251) | |
| BB vs. AA | PCa | 6 | 2441 | 2575 | 0.779 | 0.688 | 1.000 | 1.060 (0.800–1.405) | 1.059 (0.800–1.401) | |
| BB vs. BA + AA | Overall | 13 | 4854 | 5050 | 0.361 | 0.096 | 1.000 | 0.830 (0.657–1.048) | 0.843 (0.689–1.031) | |
| BB vs. BA + AA | Asian | 6 | 1561 | 1750 | 0.036 | 0.277 | 1.000 | 0.811 (0.556–1.183) | 0.828 (0.671–1.020) | |
| BB vs. BA + AA | Caucasian | 5 | 3128 | 3204 | 0.976 | 0.715 | 1.000 | 1.169 (0.513–2.661) | 1.165 (0.513–2.646) | |
| BB vs. BA + AA | H-B | 10 | 4337 | 4380 | 0.454 | 0.048 | 1.000 | 0.815 (0.662–1.003) | 0.812 (0.661–0.998) | |
| BB vs. BA + AA | P-B | 3 | 517 | 670 | 0.298 | 0.150 | 1.000 | 2.034 (0.515–8.037) | 2.260 (0.744–6.863) | |
| BB vs. BA + AA | N | 6 | 1652 | 1835 | 0.143 | 0.042 | 1.000 | 0.779 (0.574–1.057) | 0.804 (0.651–0.992) | |
| BB vs. BA + AA | Y | 7 | 3202 | 3215 | 0.742 | 0.321 | 1.000 | 1.298 (0.609–2.765) | 1.436 (0.703–2.935) | |
| BB vs. BA + AA | BC | 7 | 2413 | 2475 | 0.537 | 0.083 | 1.000 | 0.740 (0.542–1.010) | 0.763 (0.562–1.036) | |
| BB vs. BA + AA | PCa | 6 | 2441 | 2575 | 0.215 | 0.492 | 1.000 | 0.892 (0.578–1.378) | 0.910 (0.696–1.191) | |
| XRCC1-rs1799782 | B vs. A | Overall | 20 | 7280 | 8577 | 0.013 | 0.468 | 1.000 | 1.044 (0.930–1.171) | 1.055 (0.977–1.139) |
| B vs. A | Asian | 8 | 1867 | 2034 | 0.036 | 0.020 | 1.000 | 1.227 (1.033–1.458) | 1.223 (1.096–1.365) | |
| B vs. A | Caucasian | 9 | 4270 | 5246 | 0.756 | 0.193 | 1.000 | 0.923 (0.816–1.043) | 0.922 (0.816–1.042) | |
| B vs. A | Mixed | 2 | 1124 | 1248 | 0.964 | 0.383 | 1.000 | 0.903 (0.719–1.135) | 0.903 (0.719–1.135) | |
| B vs. A | H-B | 16 | 6678 | 6818 | 0.019 | 0.260 | 1.000 | 1.075 (0.948–1.220) | 1.086 (1.000–1.179) | |
| B vs. A | P-B | 4 | 602 | 1759 | 0.324 | 0.229 | 1.000 | 0.893 (0.707–1.127) | 0.881 (0.716–1.083) | |
| B vs. A | BC | 12 | 4531 | 5740 | 0.123 | 0.661 | 1.000 | 1.017 (0.881–1.174) | 1.024 (0.920–1.141) | |
| B vs. A | PCa | 8 | 2749 | 2837 | 0.011 | 0.440 | 1.000 | 1.081 (0.887–1.319) | 1.086 (0.974–1.210) | |
| BA vs. AA | Overall | 20 | 7280 | 8577 | 0.080 | 0.833 | 1.000 | 0.987 (0.878–1.111) | 0.986 (0.902–1.077) | |
| BA vs. AA | Asian | 8 | 1867 | 2034 | 0.020 | 0.230 | 1.000 | 1.153 (0.914–1.455) | 1.132 (0.983–1.305) | |
| BA vs. AA | Caucasian | 9 | 4270 | 5246 | 0.862 | 0.111 | 1.000 | 0.901 (0.791–1.026) | 0.900 (0.790–1.025) | |
| BA vs. AA | Mixed | 2 | 1124 | 1248 | 0.994 | 0.503 | 1.000 | 0.919 (0.717–1.177) | 0.919 (0.717–1.177) | |
| BA vs. AA | H-B | 16 | 6678 | 6818 | 0.097 | 0.653 | 1.000 | 1.029 (0.907–1.168) | 1.019 (0.926–1.120) | |
| BA vs. AA | P-B | 4 | 602 | 1759 | 0.592 | 0.054 | 1.000 | 0.774 (0.597–1.002) | 0.774 (0.596–1.004) | |
| BA vs. AA | BC | 12 | 4531 | 5740 | 0.555 | 0.607 | 1.000 | 0.971 (0.861–1.094) | 0.969 (0.860–1.092) | |
| BA vs. AA | PCa | 8 | 2749 | 2837 | 0.011 | 0.836 | 1.000 | 1.026 (0.802–1.314) | 1.007 (0.880–1.151) | |
| BA + BB vs. AA | Overall | 20 | 7280 | 8577 | 0.023 | 0.783 | 1.000 | 1.018 (0.897–1.154) | 1.018 (0.934–1.110) | |
| BA + BB vs. AA | Asian | 8 | 1867 | 2034 | 0.018 | 0.083 | 1.000 | 1.218 (0.975–1.523) | 1.207 (1.055–1.381) | |
| BA + BB vs. AA | Caucasian | 9 | 4270 | 5246 | 0.819 | 0.136 | 1.000 | 0.908 (0.798–1.032) | 0.907 (0.798–1.031) | |
| BA + BB vs. AA | Mixed | 2 | 1124 | 1248 | 0.984 | 0.435 | 1.000 | 0.908 (0.713–1.157) | 0.908 (0.713–1.157) | |
| BA + BB vs. AA | H-B | 16 | 6678 | 6818 | 0.032 | 0.399 | 1.000 | 1.060 (0.925–1.215) | 1.051 (0.958–1.153) | |
| BA + BB vs. AA | P-B | 4 | 602 | 1759 | 0.398 | 0.103 | 1.000 | 0.813 (0.634–1.042) | 0.813 (0.634–1.043) | |
| BA + BB vs. AA | BC | 12 | 4531 | 5740 | 0.290 | 0.886 | 1.000 | 0.992 (0.868–1.134) | 0.991 (0.882–1.114) | |
| BA + BB vs. AA | PCa | 8 | 2749 | 2837 | 0.006 | 0.621 | 1.000 | 1.064 (0.832–1.360) | 1.051 (0.924–1.196) | |
| BB vs. AA | Overall | 20 | 7280 | 8577 | 0.459 | 0.001 | 0.060 | 1.486 (1.158–1.908) | 1.502 (1.178–1.916) | |
| BB vs. AA | Asian | 8 | 1867 | 2034 | 0.099 | 0.014 | 0.840 | 1.676 (1.111–2.530) | 1.659 (1.259–2.187) | |
| BB vs. AA | Caucasian | 9 | 4270 | 5246 | 0.907 | 0.424 | 1.000 | 1.278 (0.657–2.486) | 1.303 (0.681–2.492) | |
| BB vs. AA | Mixed | 2 | 1124 | 1248 | 0.852 | 0.476 | 1.000 | 0.705 (0.271–1.832) | 0.706 (0.271–1.839) | |
| BB vs. AA | H-B | 16 | 6678 | 6818 | 0.749 | 0.000 | 0.000 | 1.625 (1.227–2.154) | 1.648 (1.252–2.170) | |
| BB vs. AA | P-B | 4 | 602 | 1759 | 0.107 | 0.826 | 1.000 | 1.583 (0.563–4.450) | 1.061 (0.625–1.802) | |
| BB vs. AA | BC | 12 | 4531 | 5740 | 0.653 | 0.016 | 0.960 | 1.706 (1.066–2.731) | 1.735 (1.106–2.722) | |
| BB vs. AA | PCa | 8 | 2749 | 2837 | 0.198 | 0.019 | 1.000 | 1.357 (0.918–2.006) | 1.415 (1.060–1.890) | |
| BB vs. BA + AA | Overall | 20 | 7280 | 8577 | 0.673 | 0.002 | 0.120 | 1.443 (1.134–1.837) | 1.461 (1.155–1.850) | |
| BB vs. BA + AA | Asian | 8 | 1867 | 2034 | 0.212 | 0.001 | 0.060 | 1.570 (1.108–2.226) | 1.583 (1.210–2.071) | |
| BB vs. BA + AA | Caucasian | 9 | 4270 | 5246 | 0.910 | 0.321 | 1.000 | 1.335 (0.723–2.467) | 1.355 (0.743–2.470) | |
| BB vs. BA + AA | Mixed | 2 | 1124 | 1248 | 0.851 | 0.489 | 1.000 | 0.712 (0.274–1.850) | 0.714 (0.274–1.857) | |
| BB vs. BA + AA | H-B | 16 | 6678 | 6818 | 0.781 | 0.002 | 0.120 | 1.509 (1.150–1.980) | 1.534 (1.176–2.001) | |
| BB vs. BA + AA | P-B | 4 | 602 | 1759 | 0.195 | 0.453 | 1.000 | 1.615 (0.683–3.820) | 1.217 (0.729–2.032) | |
| BB vs. BA + AA | BC | 12 | 4531 | 5740 | 0.758 | 0.017 | 1.000 | 1.660 (1.061–2.598) | 1.688 (1.097–2.598) | |
| BB vs. BA + AA | PCa | 8 | 2749 | 2837 | 0.356 | 0.027 | 1.000 | 1.340 (0.977–1.838) | 1.374 (1.037–1.821) | |
| XRCC1-rs915927 | B vs. A | Overall | 5 | 2330 | 3254 | 0.170 | 0.895 | 1.000 | 1.028 (0.915–1.154) | 1.005 (0.927–1.090) |
| B vs. A | Caucasian | 4 | 2185 | 3175 | 0.094 | 0.584 | 1.000 | 1.038 (0.908–1.188) | 1.007 (0.927–1.094) | |
| B vs. A | H-B | 4 | 2206 | 2161 | 0.097 | 0.675 | 1.000 | 1.031 (0.893–1.191) | 1.001 (0.920–1.090) | |
| B vs. A | Y | 4 | 2206 | 2161 | 0.097 | 0.675 | 1.000 | 1.031 (0.893–1.191) | 1.001 (0.920–1.090) | |
| B vs. A | BC | 3 | 925 | 1928 | 0.108 | 0.254 | 1.000 | 1.093 (0.904–1.322) | 1.074 (0.950–1.214) | |
| B vs. A | PCa | 2 | 1405 | 1326 | 0.926 | 0.408 | 1.000 | 0.956 (0.858–1.064) | 0.956 (0.858–1.064) | |
| BA vs. AA | Overall | 5 | 2330 | 3254 | 0.133 | 0.897 | 1.000 | 1.019 (0.823–1.261) | 1.010 (0.874–1.165) | |
| BA vs. AA | Caucasian | 4 | 2185 | 3175 | 0.198 | 0.676 | 1.000 | 1.054 (0.869–1.280) | 1.032 (0.891–1.194) | |
| BA vs. AA | H-B | 4 | 2206 | 2161 | 0.070 | 0.878 | 1.000 | 1.021 (0.785–1.327) | 1.011 (0.870–1.175) | |
| BA vs. AA | Y | 4 | 2206 | 2161 | 0.070 | 0.878 | 1.000 | 1.021 (0.785–1.327) | 1.011 (0.870–1.175) | |
| BA vs. AA | BC | 3 | 925 | 1928 | 0.223 | 0.236 | 1.000 | 1.139 (0.882–1.471) | 1.129 (0.924–1.379) | |
| BA vs. AA | PCa | 2 | 1405 | 1326 | 0.201 | 0.298 | 1.000 | 0.819 (0.520–1.288) | 0.896 (0.729–1.102) | |
| BA + BB vs. AA | Overall | 5 | 2330 | 3254 | 0.104 | 0.853 | 1.000 | 1.032 (0.836–1.273) | 1.013 (0.885–1.160) | |
| BA + BB vs. AA | Caucasian | 4 | 2185 | 3175 | 0.099 | 0.561 | 1.000 | 1.066 (0.860–1.320) | 1.029 (0.897–1.181) | |
| BA + BB vs. AA | H-B | 4 | 2206 | 2161 | 0.053 | 0.810 | 1.000 | 1.032 (0.797–1.336) | 1.011 (0.878–1.166) | |
| BA + BB vs. AA | Y | 4 | 2206 | 2161 | 0.053 | 0.810 | 1.000 | 1.032 (0.797–1.336) | 1.011 (0.878–1.166) | |
| BA + BB vs. AA | BC | 3 | 925 | 1928 | 0.128 | 0.194 | 1.000 | 1.152 (0.867–1.530) | 1.133 (0.938–1.367) | |
| BA + BB vs. AA | PCa | 2 | 1405 | 1326 | 0.378 | 0.277 | 1.000 | 0.898 (0.739–1.093) | 0.897 (0.738–1.091) | |
| BB vs. AA | Overall | 5 | 2330 | 3254 | 0.238 | 0.913 | 1.000 | 1.017 (0.822–1.258) | 0.991 (0.841–1.168) | |
| BB vs. AA | Caucasian | 4 | 2185 | 3175 | 0.158 | 0.992 | 1.000 | 1.045 (0.820–1.332) | 1.001 (0.846–1.184) | |
| BB vs. AA | H-B | 4 | 2206 | 2161 | 0.145 | 0.831 | 1.000 | 1.018 (0.779–1.331) | 0.981 (0.825–1.167) | |
| BB vs. AA | Y | 4 | 2206 | 2161 | 0.145 | 0.831 | 1.000 | 1.018 (0.779–1.331) | 0.981 (0.825–1.167) | |
| BB vs. AA | BC | 3 | 925 | 1928 | 0.167 | 0.348 | 1.000 | 1.153 (0.821–1.618) | 1.125 (0.880–1.439) | |
| BB vs. AA | PCa | 2 | 1405 | 1326 | 0.743 | 0.322 | 1.000 | 0.895 (0.717–1.116) | 0.895 (0.717–1.116) | |
| BB vs. BA + AA | Overall | 5 | 2330 | 3254 | 0.596 | 0.974 | 1.000 | 1.002 (0.882–1.138) | 1.002 (0.882–1.139) | |
| BB vs. BA + AA | Caucasian | 4 | 2185 | 3175 | 0.517 | 0.897 | 1.000 | 0.991 (0.870–1.130) | 0.991 (0.870–1.130) | |
| BB vs. BA + AA | H-B | 4 | 2206 | 2161 | 0.457 | 0.919 | 1.000 | 0.993 (0.868–1.136) | 0.993 (0.868–1.136) | |
| BB vs. BA + AA | Y | 4 | 2206 | 2161 | 0.457 | 0.919 | 1.000 | 0.993 (0.868–1.136) | 0.993 (0.868–1.136) | |
| BB vs. BA + AA | BC | 3 | 925 | 1928 | 0.422 | 0.617 | 1.000 | 1.055 (0.855–1.302) | 1.055 (0.855–1.302) | |
| BB vs. BA + AA | PCa | 2 | 1405 | 1326 | 0.408 | 0.734 | 1.000 | 0.972 (0.828–1.142) | 0.973 (0.828–1.142) | |
| XRCC1-rs3213245 | B vs. A | Overall | 3 | 1066 | 1120 | 0.835 | 0.504 | 1.000 | 0.954 (0.830–1.096) | 0.954 (0.830–1.096) |
| B vs. A | Asian | 2 | 536 | 564 | 0.891 | 0.388 | 1.000 | 0.899 (0.706–1.145) | 0.899 (0.706–1.145) | |
| B vs. A | P-B | 2 | 536 | 564 | 0.891 | 0.388 | 1.000 | 0.899 (0.706–1.145) | 0.899 (0.706–1.145) | |
| BA vs. AA | Overall | 3 | 1066 | 1120 | 0.537 | 0.212 | 1.000 | 0.873 (0.705–1.081) | 0.873 (0.705–1.081) | |
| BA vs. AA | Asian | 2 | 536 | 564 | 0.973 | 0.095 | 1.000 | 0.789 (0.597–1.042) | 0.789 (0.597–1.042) | |
| BA vs. AA | P-B | 2 | 536 | 564 | 0.973 | 0.095 | 1.000 | 0.789 (0.597–1.042) | 0.789 (0.597–1.042) | |
| BA + BB vs. AA | Overall | 3 | 1066 | 1120 | 0.697 | 0.297 | 1.000 | 0.897 (0.730–1.101) | 0.896 (0.730–1.101) | |
| BA + BB vs. AA | Asian | 2 | 536 | 564 | 0.916 | 0.180 | 1.000 | 0.831 (0.635–1.089) | 0.831 (0.635–1.089) | |
| BA + BB vs. AA | P-B | 2 | 536 | 564 | 0.916 | 0.180 | 1.000 | 0.831 (0.635–1.089) | 0.831 (0.635–1.089) | |
| BB vs. AA | Overall | 3 | 1066 | 1120 | 0.555 | 0.795 | 1.000 | 1.042 (0.749–1.449) | 1.045 (0.752–1.451) | |
| BB vs. AA | Asian | 2 | 536 | 564 | 0.752 | 0.288 | 1.000 | 1.617 (0.661–3.955) | 1.622 (0.664–3.958) | |
| BB vs. AA | P-B | 2 | 536 | 564 | 0.752 | 0.288 | 1.000 | 1.617 (0.661–3.955) | 1.622 (0.664–3.958) | |
| BB vs. BA + AA | Overall | 3 | 1066 | 1120 | 0.454 | 0.946 | 1.000 | 1.006 (0.789–1.283) | 1.008 (0.791–1.285) | |
| BB vs. BA + AA | Asian | 2 | 536 | 564 | 0.743 | 0.234 | 1.000 | 1.711 (0.702–4.172) | 1.715 (0.705–4.175) | |
| BB vs. BA + AA | P-B | 2 | 536 | 564 | 0.743 | 0.234 | 1.000 | 1.711 (0.702–4.172) | 1.715 (0.705–4.175) | |
| XRCC2-rs3218536 | B vs. A | Overall | 3 | 1395 | 1454 | 0.815 | 0.524 | 1.000 | 0.943 (0.788–1.130) | 0.943 (0.787–1.130) |
| B vs. A | BC | 2 | 1294 | 1238 | 0.856 | 0.446 | 1.000 | 0.930 (0.773–1.120) | 0.930 (0.773–1.120) | |
| BA vs. AA | Overall | 3 | 1395 | 1454 | 0.798 | 0.925 | 1.000 | 1.010 (0.828–1.230) | 1.010 (0.828–1.230) | |
| BA vs. AA | BC | 2 | 1294 | 1238 | 0.615 | 0.984 | 1.000 | 0.998 (0.813–1.224) | 0.998 (0.814–1.224) | |
| BA + BB vs. AA | Overall | 3 | 1395 | 1454 | 0.815 | 0.796 | 1.000 | 0.975 (0.803–1.184) | 0.975 (0.803–1.184) | |
| BA + BB vs. AA | BC | 2 | 1294 | 1238 | 0.728 | 0.703 | 1.000 | 0.962 (0.787–1.175) | 0.962 (0.787–1.175) | |
| BB vs. AA | Overall | 3 | 1395 | 1454 | 0.552 | 0.116 | 1.000 | 0.510 (0.216–1.203) | 0.507 (0.217–1.183) | |
| BB vs. AA | BC | 2 | 1294 | 1238 | 0.851 | 0.073 | 1.000 | 0.438 (0.178–1.079) | 0.438 (0.178–1.080) | |
| BB vs. BA + AA | Overall | 3 | 1395 | 1454 | 0.556 | 0.115 | 1.000 | 0.509 (0.216–1.199) | 0.506 (0.217–1.180) | |
| BB vs. BA + AA | BC | 2 | 1294 | 1238 | 0.835 | 0.073 | 1.000 | 0.438 (0.178–1.078) | 0.438 (0.178–1.079) | |
| XRCC3-rs861539 | B vs. A | Overall | 23 | 5979 | 7382 | 0.000 | 0.914 | 1.000 | 0.994 (0.890–1.110) | 1.044 (0.988–1.102) |
| B vs. A | Asian | 6 | 1181 | 1326 | 0.314 | 0.265 | 1.000 | 1.102 (0.916–1.326) | 1.099 (0.931–1.297) | |
| B vs. A | Caucasian | 13 | 3287 | 4308 | 0.000 | 0.326 | 1.000 | 0.914 (0.765–1.093) | 1.031 (0.961–1.106) | |
| B vs. A | Mixed | 4 | 1511 | 1748 | 0.463 | 0.351 | 1.000 | 1.049 (0.948–1.161) | 1.049 (0.948–1.161) | |
| B vs. A | H-B | 19 | 4589 | 4613 | 0.000 | 0.807 | 1.000 | 0.983 (0.855–1.130) | 1.051 (0.986–1.121) | |
| B vs. A | P-B | 4 | 1390 | 2769 | 0.801 | 0.643 | 1.000 | 1.025 (0.924–1.137) | 1.025 (0.924–1.137) | |
| B vs. A | N | 6 | 567 | 673 | 0.000 | 0.094 | 1.000 | 0.637 (0.376–1.081) | 0.788 (0.663–0.936) | |
| B vs. A | Y | 17 | 5412 | 6709 | 0.301 | 0.011 | 0.660 | 1.076 (1.007–1.150) | 1.077 (1.017–1.141) | |
| B vs. A | BC | 17 | 5153 | 6282 | 0.000 | 0.918 | 1.000 | 1.007 (0.884–1.146) | 1.056 (0.997–1.119) | |
| B vs. A | PCa | 6 | 826 | 1100 | 0.277 | 0.534 | 1.000 | 0.945 (0.784–1.139) | 0.950 (0.807–1.117) | |
| BA vs. AA | Overall | 23 | 5979 | 7382 | 0.005 | 0.823 | 1.000 | 1.014 (0.894–1.151) | 1.031 (0.952–1.116) | |
| BA vs. AA | Asian | 6 | 1181 | 1326 | 0.392 | 0.830 | 1.000 | 1.020 (0.830–1.253) | 1.022 (0.837–1.249) | |
| BA vs. AA | Caucasian | 13 | 3287 | 4308 | 0.031 | 0.304 | 1.000 | 1.093 (0.922–1.296) | 1.108 (0.997–1.232) | |
| BA vs. AA | Mixed | 4 | 1511 | 1748 | 0.021 | 0.244 | 1.000 | 0.815 (0.577–1.150) | 0.896 (0.772–1.040) | |
| BA vs. AA | H-B | 19 | 4589 | 4613 | 0.002 | 0.764 | 1.000 | 1.024 (0.876–1.198) | 1.064 (0.970–1.167) | |
| BA vs. AA | P-B | 4 | 1390 | 2769 | 0.882 | 0.473 | 1.000 | 0.945 (0.811–1.102) | 0.945 (0.811–1.102) | |
| BA vs. AA | N | 6 | 567 | 673 | 0.001 | 0.686 | 1.000 | 0.880 (0.473–1.637) | 0.878 (0.676–1.141) | |
| BA vs. AA | Y | 17 | 5412 | 6709 | 0.226 | 0.273 | 1.000 | 1.046 (0.947–1.156) | 1.048 (0.964–1.138) | |
| BA vs. AA | BC | 17 | 5153 | 6282 | 0.029 | 0.271 | 1.000 | 1.076 (0.944–1.227) | 1.065 (0.978–1.160) | |
| BA vs. AA | PCa | 6 | 826 | 1100 | 0.068 | 0.184 | 1.000 | 0.808 (0.590–1.106) | 0.839 (0.678–1.039) | |
| BA + BB vs. AA | Overall | 23 | 5979 | 7382 | 0.000 | 0.997 | 1.000 | 1.000 (0.873–1.145) | 1.041 (0.966–1.122) | |
| BA + BB vs. AA | Asian | 6 | 1181 | 1326 | 0.412 | 0.508 | 1.000 | 1.065 (0.878–1.292) | 1.067 (0.881–1.292) | |
| BA + BB vs. AA | Caucasian | 13 | 3287 | 4308 | 0.000 | 0.906 | 1.000 | 0.987 (0.796–1.224) | 1.075 (0.974–1.186) | |
| BA + BB vs. AA | Mixed | 4 | 1511 | 1748 | 0.061 | 0.492 | 1.000 | 0.905 (0.681–1.203) | 0.963 (0.837–1.109) | |
| BA + BB vs. AA | H-B | 19 | 4589 | 4613 | 0.000 | 0.940 | 1.000 | 0.994 (0.838–1.178) | 1.063 (0.975–1.160) | |
| BA + BB vs. AA | P-B | 4 | 1390 | 2769 | 0.969 | 0.808 | 1.000 | 0.982 (0.849–1.135) | 0.982 (0.849–1.136) | |
| BA + BB vs. AA | N | 6 | 567 | 673 | 0.000 | 0.173 | 1.000 | 0.646 (0.345–1.211) | 0.768 (0.609–0.970) | |
| BA + BB vs. AA | Y | 17 | 5412 | 6709 | 0.228 | 0.061 | 1.000 | 1.077 (0.979–1.184) | 1.078 (0.997–1.167) | |
| BA + BB vs. AA | BC | 17 | 5153 | 6282 | 0.000 | 0.535 | 1.000 | 1.050 (0.899–1.227) | 1.070 (0.987–1.159) | |
| BA + BB vs. AA | PCa | 6 | 826 | 1100 | 0.157 | 0.203 | 1.000 | 0.855 (0.658–1.110) | 0.877 (0.716–1.073) | |
| BB vs. AA | Overall | 23 | 5979 | 7382 | 0.004 | 0.479 | 1.000 | 1.076 (0.878–1.318) | 1.119 (0.995–1.258) | |
| BB vs. AA | Asian | 6 | 1181 | 1326 | 0.527 | 0.140 | 1.000 | 1.397 (0.853–2.289) | 1.434 (0.888–2.315) | |
| BB vs. AA | Caucasian | 13 | 3287 | 4308 | 0.000 | 0.485 | 1.000 | 0.900 (0.669–1.210) | 1.035 (0.894–1.198) | |
| BB vs. AA | Mixed | 4 | 1511 | 1748 | 0.729 | 0.035 | 1.000 | 1.263 (1.014–1.572) | 1.265 (1.016–1.574) | |
| BB vs. AA | H-B | 19 | 4589 | 4613 | 0.002 | 0.554 | 1.000 | 1.079 (0.839–1.387) | 1.123 (0.978–1.291) | |
| BB vs. AA | P-B | 4 | 1390 | 2769 | 0.420 | 0.363 | 1.000 | 1.111 (0.890–1.388) | 1.108 (0.888–1.381) | |
| BB vs. AA | N | 6 | 567 | 673 | 0.000 | 0.474 | 1.000 | 0.728 (0.306–1.733) | 0.751 (0.535–1.056) | |
| BB vs. AA | Y | 17 | 5412 | 6709 | 0.617 | 0.009 | 0.540 | 1.181 (1.041–1.341) | 1.182 (1.043–1.340) | |
| BB vs. AA | BC | 17 | 5153 | 6282 | 0.002 | 0.710 | 1.000 | 1.044 (0.833–1.307) | 1.117 (0.988–1.264) | |
| BB vs. AA | PCa | 6 | 826 | 1100 | 0.201 | 0.535 | 1.000 | 1.264 (0.743–2.151) | 1.135 (0.762–1.690) | |
| BB vs. BA + AA | Overall | 23 | 5979 | 7382 | 0.004 | 0.641 | 1.000 | 1.046 (0.867–1.261) | 1.087 (0.976–1.212) | |
| BB vs. BA + AA | Asian | 6 | 1181 | 1326 | 0.505 | 0.126 | 1.000 | 1.407 (0.863–2.295) | 1.447 (0.901–2.324) | |
| BB vs. BA + AA | Caucasian | 13 | 3287 | 4308 | 0.002 | 0.294 | 1.000 | 0.875 (0.683–1.122) | 0.980 (0.857–1.120) | |
| BB vs. BA + AA | Mixed | 4 | 1511 | 1748 | 0.371 | 0.009 | 0.540 | 1.320 (1.053–1.654) | 1.308 (1.069–1.600) | |
| BB vs. BA + AA | H-B | 19 | 4589 | 4613 | 0.002 | 0.711 | 1.000 | 1.044 (0.831–1.313) | 1.068 (0.939–1.214) | |
| BB vs. BA + AA | P-B | 4 | 1390 | 2769 | 0.289 | 0.209 | 1.000 | 1.081 (0.795–1.470) | 1.138 (0.930–1.394) | |
| BB vs. BA + AA | N | 6 | 567 | 673 | 0.000 | 0.468 | 1.000 | 0.743 (0.333–1.658) | 0.725 (0.527–0.997) | |
| BB vs. BA + AA | Y | 17 | 5412 | 6709 | 0.724 | 0.019 | 1.000 | 1.147 (1.021–1.288) | 1.148 (1.023–1.288) | |
| BB vs. BA + AA | BC | 17 | 5153 | 6282 | 0.006 | 0.957 | 1.000 | 1.005 (0.826–1.224) | 1.078 (0.962–1.207) | |
| BB vs. BA + AA | PCa | 6 | 826 | 1100 | 0.075 | 0.250 | 1.000 | 1.437 (0.775–2.665) | 1.203 (0.822–1.761) | |
| XRCC3-rs1799796 | B vs. A | Overall | 5 | 1302 | 2391 | 0.116 | 0.693 | 1.000 | 0.998 (0.845–1.180) | 0.977 (0.872–1.095) |
| B vs. A | Caucasian | 4 | 1091 | 2147 | 0.204 | 0.335 | 1.000 | 0.951 (0.808–1.119) | 0.942 (0.834–1.064) | |
| B vs. A | H-B | 3 | 1120 | 1147 | 0.155 | 0.535 | 1.000 | 0.990 (0.818–1.197) | 0.960 (0.843–1.092) | |
| B vs. A | P-B | 2 | 182 | 1244 | 0.069 | 0.893 | 1.000 | 0.967 (0.597–1.568) | 1.040 (0.819–1.322) | |
| BA vs. AA | Overall | 5 | 1302 | 2391 | 0.396 | 0.828 | 1.000 | 0.984 (0.841–1.151) | 0.983 (0.842–1.148) | |
| BA vs. AA | Caucasian | 4 | 1091 | 2147 | 0.992 | 0.320 | 1.000 | 0.918 (0.775–1.087) | 0.918 (0.775–1.087) | |
| BA vs. AA | H-B | 3 | 1120 | 1147 | 0.133 | 0.909 | 1.000 | 1.018 (0.786–1.317) | 0.990 (0.832–1.178) | |
| BA vs. AA | P-B | 2 | 182 | 1244 | 0.898 | 0.801 | 1.000 | 0.957 (0.682–1.344) | 0.957 (0.682–1.344) | |
| BA + BB vs. AA | Overall | 5 | 1302 | 2391 | 0.276 | 0.780 | 1.000 | 0.991 (0.833–1.179) | 0.979 (0.845–1.135) | |
| BA + BB vs. AA | Caucasian | 4 | 1091 | 2147 | 0.735 | 0.308 | 1.000 | 0.920 (0.784–1.080) | 0.920 (0.784–1.080) | |
| BA + BB vs. AA | H-B | 3 | 1120 | 1147 | 0.118 | 0.736 | 1.000 | 1.006 (0.779–1.299) | 0.972 (0.823–1.148) | |
| BA + BB vs. AA | P-B | 2 | 182 | 1244 | 0.372 | 0.966 | 1.000 | 1.006 (0.732–1.383) | 1.007 (0.733–1.384) | |
| BB vs. AA | Overall | 5 | 1302 | 2391 | 0.088 | 0.913 | 1.000 | 0.976 (0.627–1.518) | 0.927 (0.708–1.214) | |
| BB vs. AA | Caucasian | 4 | 1091 | 2147 | 0.046 | 0.844 | 1.000 | 0.949 (0.565–1.596) | 0.917 (0.694–1.210) | |
| BB vs. AA | H-B | 3 | 1120 | 1147 | 0.611 | 0.329 | 1.000 | 0.856 (0.626–1.170) | 0.856 (0.626–1.170) | |
| BB vs. AA | P-B | 2 | 182 | 1244 | 0.020 | 0.824 | 1.000 | 0.830 (0.160–4.302) | 1.171 (0.694–1.977) | |
| BB vs. BA + AA | Overall | 5 | 1302 | 2391 | 0.082 | 0.974 | 1.000 | 0.993 (0.645–1.529) | 0.948 (0.732–1.228) | |
| BB vs. BA + AA | Caucasian | 4 | 1091 | 2147 | 0.040 | 0.948 | 1.000 | 0.983 (0.592–1.634) | 0.946 (0.725–1.234) | |
| BB vs. BA + AA | H-B | 3 | 1120 | 1147 | 0.668 | 0.408 | 1.000 | 0.881 (0.653–1.189) | 0.881 (0.654–1.189) | |
| BB vs. BA + AA | P-B | 2 | 182 | 1244 | 0.017 | 0.843 | 1.000 | 0.848 (0.167–4.320) | 1.175 (0.712–1.939) | |
| XRCC4-rs1805377 | B vs. A | Overall | 5 | 2080 | 2134 | 0.813 | 0.005 | 0.300 | 0.827 (0.725–0.944) | 0.827 (0.725–0.943) |
| B vs. A | Asian | 2 | 403 | 468 | 0.734 | 0.238 | 1.000 | 0.863 (0.676–1.102) | 0.863 (0.676–1.102) | |
| B vs. A | Caucasian | 3 | 1677 | 1666 | 0.524 | 0.009 | 0.540 | 0.812 (0.694–0.951) | 0.812 (0.694–0.950) | |
| B vs. A | BC | 3 | 1351 | 1403 | 0.693 | 0.005 | 0.300 | 0.789 (0.668–0.931) | 0.789 (0.668–0.931) | |
| B vs. A | PCa | 2 | 729 | 731 | 0.963 | 0.326 | 1.000 | 0.897 (0.721–1.115) | 0.897 (0.721–1.115) | |
| BA vs. AA | Overall | 5 | 2080 | 2134 | 0.969 | 0.949 | 1.000 | 1.009 (0.784–1.298) | 1.008 (0.784–1.297) | |
| BA vs. AA | Asian | 2 | 403 | 468 | 0.931 | 0.869 | 1.000 | 0.976 (0.730–1.305) | 0.976 (0.730–1.305) | |
| BA vs. AA | Caucasian | 3 | 1677 | 1666 | 0.847 | 0.675 | 1.000 | 1.116 (0.672–1.854) | 1.114 (0.672–1.847) | |
| BA vs. AA | BC | 3 | 1351 | 1403 | 0.827 | 0.917 | 1.000 | 1.018 (0.734–1.412) | 1.017 (0.734–1.411) | |
| BA vs. AA | PCa | 2 | 729 | 731 | 0.693 | 0.981 | 1.000 | 0.995 (0.670–1.478) | 0.995 (0.670–1.478) | |
| BA + BB vs. AA | Overall | 5 | 2080 | 2134 | 0.998 | 0.567 | 1.000 | 0.931 (0.729–1.189) | 0.931 (0.729–1.189) | |
| BA + BB vs. AA | Asian | 2 | 403 | 468 | 0.921 | 0.518 | 1.000 | 0.911 (0.687–1.208) | 0.911 (0.687–1.208) | |
| BA + BB vs. AA | Caucasian | 3 | 1677 | 1666 | 0.989 | 0.981 | 1.000 | 0.994 (0.608–1.626) | 0.994 (0.608–1.626) | |
| BA + BB vs. AA | BC | 3 | 1351 | 1403 | 0.975 | 0.618 | 1.000 | 0.922 (0.670–1.269) | 0.922 (0.669–1.270) | |
| BA + BB vs. AA | PCa | 2 | 729 | 731 | 0.803 | 0.766 | 1.000 | 0.944 (0.646–1.380) | 0.944 (0.646–1.380) | |
| BB vs. AA | Overall | 5 | 2080 | 2134 | 0.376 | 0.216 | 1.000 | 0.799 (0.471–1.357) | 0.735 (0.451–1.197) | |
| BB vs. AA | Asian | 2 | 403 | 468 | 0.139 | 0.050 | 1.000 | 0.367 (0.070–1.918) | 0.413 (0.171–1.000) | |
| BB vs. AA | Caucasian | 3 | 1677 | 1666 | 0.814 | 0.984 | 1.000 | 0.996 (0.545–1.821) | 0.994 (0.544–1.816) | |
| BB vs. AA | BC | 3 | 1351 | 1403 | 0.139 | 0.211 | 1.000 | 0.630 (0.151–2.629) | 0.652 (0.333–1.275) | |
| BB vs. AA | PCa | 2 | 729 | 731 | 0.575 | 0.641 | 1.000 | 0.847 (0.414–1.733) | 0.844 (0.414–1.721) | |
| BB vs. BA + AA | Overall | 5 | 2080 | 2134 | 0.338 | 0.001 | 0.060 | 0.764 (0.617–0.946) | 0.753 (0.635–0.894) | |
| BB vs. BA + AA | Asian | 2 | 403 | 468 | 0.136 | 0.051 | 1.000 | 0.370 (0.070–1.948) | 0.418 (0.174–1.006) | |
| BB vs. BA + AA | Caucasian | 3 | 1677 | 1666 | 0.466 | 0.004 | 0.240 | 0.774 (0.650–0.921) | 0.773 (0.649–0.921) | |
| BB vs. BA + AA | BC | 3 | 1351 | 1403 | 0.175 | 0.001 | 0.060 | 0.585 (0.181–1.889) | 0.706 (0.571–0.872) | |
| BB vs. BA + AA | PCa | 2 | 729 | 731 | 0.676 | 0.280 | 1.000 | 0.853 (0.638–1.140) | 0.852 (0.638–1.139) | |
| XRCC4-rs6869366 | B vs. A | Overall | 4 | 695 | 760 | 0.000 | 0.590 | 1.000 | 1.163 (0.672–2.015) | 0.916 (0.758–1.107) |
| B vs. A | BC | 2 | 369 | 402 | 0.002 | 0.731 | 1.000 | 1.165 (0.488–2.780) | 0.987 (0.770–1.265) | |
| B vs. A | PCa | 2 | 326 | 358 | 0.002 | 0.717 | 1.000 | 1.291 (0.324–5.146) | 0.828 (0.618–1.108) | |
| BA vs. AA | Overall | 4 | 695 | 760 | 0.000 | 0.476 | 1.000 | 1.253 (0.674–2.328) | 1.030 (0.819–1.295) | |
| BA vs. AA | BC | 2 | 369 | 402 | 0.003 | 0.634 | 1.000 | 1.258 (0.490–3.230) | 1.120 (0.828–1.514) | |
| BA vs. AA | PCa | 2 | 326 | 358 | 0.002 | 0.683 | 1.000 | 1.345 (0.324–5.585) | 0.918 (0.646–1.306) | |
| BA + BB vs. AA | Overall | 4 | 695 | 760 | 0.000 | 0.553 | 1.000 | 1.216 (0.637–2.320) | 0.974 (0.778–1.218) | |
| BA + BB vs. AA | BC | 2 | 369 | 402 | 0.002 | 0.694 | 1.000 | 1.222 (0.450–3.318) | 1.065 (0.792–1.430) | |
| BA + BB vs. AA | PCa | 2 | 326 | 358 | 0.001 | 0.726 | 1.000 | 1.304 (0.295–5.773) | 0.862 (0.611–1.216) | |
| BB vs. AA | Overall | 4 | 695 | 760 | 0.762 | 0.015 | 0.900 | 0.462 (0.244–0.875) | 0.458 (0.244–0.861) | |
| BB vs. AA | BC | 2 | 369 | 402 | 0.530 | 0.118 | 1.000 | 0.516 (0.224–1.192) | 0.515 (0.224–1.183) | |
| BB vs. AA | PCa | 2 | 326 | 358 | 0.436 | 0.060 | 1.000 | 0.396 (0.148–1.062) | 0.393 (0.149–1.039) | |
| BB vs. BA + AA | Overall | 4 | 695 | 760 | 0.882 | 0.032 | 1.000 | 0.509 (0.271–0.954) | 0.505 (0.271–0.942) | |
| BB vs. BA + AA | BC | 2 | 369 | 402 | 0.666 | 0.159 | 1.000 | 0.557 (0.244–1.267) | 0.555 (0.245–1.260) | |
| BB vs. BA + AA | PCa | 2 | 326 | 358 | 0.546 | 0.100 | 1.000 | 0.449 (0.169–1.188) | 0.445 (0.170–1.166) | |
| XRCC4-rs28360071 | B vs. A | Overall | 3 | 561 | 626 | 0.148 | 0.004 | 0.240 | 1.359 (1.010–1.829) | 1.369 (1.106–1.695) |
| B vs. A | BC | 2 | 369 | 402 | 0.350 | 0.206 | 1.000 | 1.189 (0.909–1.554) | 1.189 (0.909–1.554) | |
| BA vs. AA | Overall | 3 | 561 | 626 | 0.772 | 0.162 | 1.000 | 1.207 (0.927–1.571) | 1.207 (0.927–1.571) | |
| BA vs. AA | BC | 2 | 369 | 402 | 0.890 | 0.458 | 1.000 | 1.130 (0.819–1.558) | 1.130 (0.819–1.558) | |
| BA + BB vs. AA | Overall | 3 | 561 | 626 | 0.406 | 0.027 | 1.000 | 1.325 (1.032–1.701) | 1.325 (1.032–1.700) | |
| BA + BB vs. AA | BC | 2 | 369 | 402 | 0.611 | 0.295 | 1.000 | 1.180 (0.866–1.608) | 1.180 (0.866–1.608) | |
| BB vs. AA | Overall | 3 | 561 | 626 | 0.318 | 0.005 | 0.300 | 2.298 (1.169–4.519) | 2.358 (1.289–4.312) | |
| BB vs. AA | BC | 2 | 369 | 402 | 0.263 | 0.234 | 1.000 | 1.592 (0.553–4.586) | 1.690 (0.712–4.011) | |
| BB vs. BA + AA | Overall | 3 | 561 | 626 | 0.337 | 0.009 | 0.540 | 2.194 (1.146–4.198) | 2.230 (1.224–4.061) | |
| BB vs. BA + AA | BC | 2 | 369 | 402 | 0.259 | 0.262 | 1.000 | 1.533 (0.528–4.454) | 1.635 (0.692–3.863) | |
| XRCC7-rs7003908 | B vs. A | Overall | 6 | 1196 | 1365 | 0.000 | 0.567 | 1.000 | 1.133 (0.738–1.741) | 1.148 (1.020–1.293) |
| B vs. A | BC | 3 | 727 | 796 | 0.000 | 0.978 | 1.000 | 1.012 (0.430–2.383) | 1.044 (0.894–1.219) | |
| B vs. A | PCa | 2 | 357 | 389 | 0.259 | 0.003 | 0.180 | 1.376 (1.081–1.752) | 1.384 (1.119–1.711) | |
| BA vs. AA | Overall | 6 | 1196 | 1365 | 0.004 | 0.979 | 1.000 | 0.996 (0.719–1.379) | 0.937 (0.789–1.112) | |
| BA vs. AA | BC | 3 | 727 | 796 | 0.003 | 0.717 | 1.000 | 0.903 (0.520–1.567) | 0.822 (0.658–1.026) | |
| BA vs. AA | PCa | 2 | 357 | 389 | 0.725 | 0.116 | 1.000 | 1.295 (0.938–1.789) | 1.295 (0.938–1.789) | |
| BA + BB vs. AA | Overall | 6 | 1196 | 1365 | 0.000 | 0.643 | 1.000 | 1.113 (0.708–1.749) | 1.021 (0.868–1.199) | |
| BA + BB vs. AA | BC | 3 | 727 | 796 | 0.000 | 0.994 | 1.000 | 1.003 (0.430–2.341) | 0.881 (0.715–1.086) | |
| BA + BB vs. AA | PCa | 2 | 357 | 389 | 0.698 | 0.023 | 1.000 | 1.423 (1.049–1.929) | 1.423 (1.050–1.929) | |
| BB vs. AA | Overall | 6 | 1196 | 1365 | 0.000 | 0.677 | 1.000 | 1.195 (0.517–2.763) | 1.584 (1.219–2.057) | |
| BB vs. AA | BC | 3 | 727 | 796 | 0.000 | 0.830 | 1.000 | 0.809 (0.117–5.612) | 1.422 (1.000–2.023) | |
| BB vs. AA | PCa | 2 | 357 | 389 | 0.214 | 0.007 | 0.420 | 1.755 (0.964–3.196) | 1.845 (1.178–2.888) | |
| BB vs. BA + AA | Overall | 6 | 1196 | 1365 | 0.000 | 0.628 | 1.000 | 1.180 (0.603–2.308) | 1.630 (1.293–2.054) | |
| BB vs. BA + AA | BC | 3 | 727 | 796 | 0.000 | 0.795 | 1.000 | 0.811 (0.166–3.957) | 1.570 (1.152–2.140) | |
| BB vs. BA + AA | PCa | 2 | 357 | 389 | 0.165 | 0.010 | 0.600 | 1.537 (0.820–2.879) | 1.677 (1.133–2.482) |
PH: P value of Q test for heterogeneity test; PZ: means statistically significant; P (Adjust): Multiple testing P value according to Bonferroni correction (P value less than 0.05/(12 polymorphisms * 5 models) was considered as statistically significant, which was marked with bold font in the PA column); PCa: Prostate cancer; BC: Bladder cancer; H-B: hospital-based; P-B: population-based; HWE: Hardy Weinberg equilibrium; Note: Heterogeneity was considered to be significant when the P-value was less than 0.1. If there was no significant heterogeneity, a fixed effect model (Der-Simonian Laird) was used to evaluate the point estimates and 95% CI; otherwise, a random effects model (Der-Simonian Laird) was used. And the PZ was calculated based on the actual model adopted.
3.2.1. XRCC1-rs25489
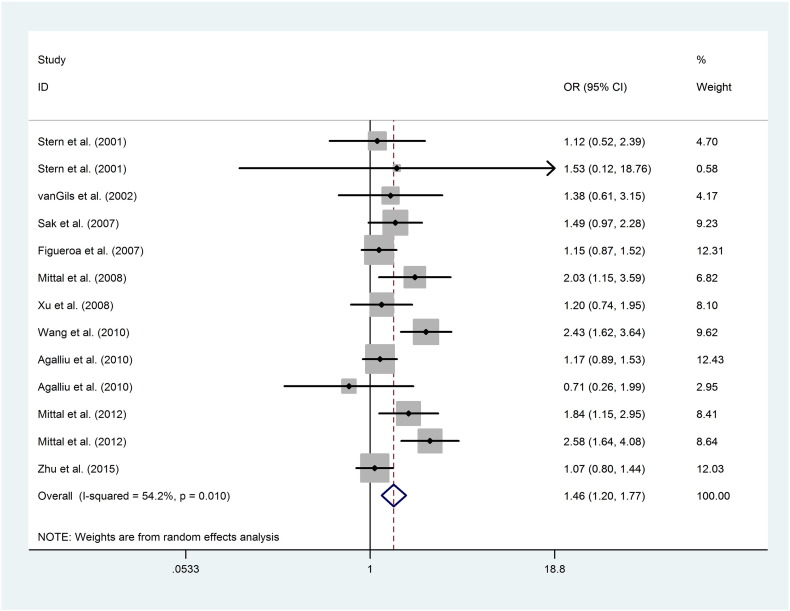
The pooled results based on 13 included studies indicated that the XRCC1-rs25489 polymorphism conferred a significantly increased overall risk to urological neoplasms in heterozygote (BA vs. AA: OR = 1.455, 95%CI = 1.198–1.768, PA < 0.001, Fig. 1) and dominant models (BA + BB vs. AA: OR = 1.281, 95%CI = 1.148–1.428, PA < 0.001), respectively. Further subgroup analysis by cancer type indicated that the ‘B’ allele was significantly related to an increased risk of BC in heterozygote model (BA vs. AA: OR = 1.611, 95%CI = 1.242–2.090, PA < 0.001). Moreover, when the subgroup analyses were performed based on source of controls, ethnicity and HWE status, null result was uncovered (Table 3).
Fig. 1.
Forest plots of the association between XRCC1-rs25489 polymorphism and the risk of urological neoplasms (BA vs. AA). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. CI = confidence interval, OR = odds ratio.
3.2.2. XRCC1-rs1799782
Overall, no significant association was uncovered for the association between XRCC1-rs1799782 polymorphism and urological neoplasms risk. However, in the stratification analysis by source of control, we observed hospital-based controls groups were one of the heterogeneity sources in homozygote model (BB vs. AA: OR = 1.648, 95%CI = 1.252–2.170, PA < 0.001) instead of population-based controls groups.
3.2.3. XRCC1-rs25487
With regard to the XRCC1-rs25487 polymorphism, overall results revealed a null association between the polymorphism and risk of urological neoplasms (Fig. 2). However, in the stratification analysis by ethnicity, a significant increased risk of urological neoplasms risk was uncovered for Asians in allelic (B vs. A: OR = 1.176, 95%CI = 1.089–1.271, PA < 0.001) and homozygote models (BB vs. AA: OR = 1.464, 95%CI = 1.232–1.740, PA < 0.001). However, in the stratified analysis by HWE status, source of controls and cancer type, null result was obtained.
Fig. 2.
Forest plots of the association between XRCC1-rs25487 polymorphism and the risk of urological neoplasms (B vs. A). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. CI = confidence interval, OR = odds ratio.
3.2.4. XRCC1-rs3213245, XRCC1-rs915927, XRCC2-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs1805377, XRCC4-rs28360071, XRCC4-rs6869366 and XRCC7-rs7003908
There was no significant association between XRCC1-rs3213245, XRCC1-rs915927, XRCC2-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs1805377, XRCC4-rs28360071 and XRCC4-rs6869366, XRCC7-rs7003908 polymorphisms and risk of urological neoplasms. Furthermore, in the subgroup analysis by ethnicity, HWE status, source of controls and cancer type, similar results were also obtained (Table 3).
3.3. Sensitivity Analysis and Publication Bias
Sensitivity analyses were performed to evaluate the influence of the separate case-control study on the integrated data. The results showed that there was no material alteration in corresponding pooled ORs for XRCC1-rs915927, XRCC1-rs25489, XRCC1-rs25487, XRCC1-rs1799782, XRCC1-rs3213245, XRCC2-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs6869366, XRCC4-rs28360071, XRCC4-rs1805377, XRCC7-rs7003908 polymorphisms (Supplementary Table 2 and Figs. 6–17). Additionally, Begg's funnel plot and Egger's regression test were performed to evaluate the publication bias. If the tests indicated significant publication bias existed in several genetic models, it might reflect differences in the selection of controls, age distributions, and some other lifestyle characteristics. As for XRCC1-rs915927, XRCC1-rs25489, XRCC1-rs3213245, XRCC1-rs25487, XRCC1-rs1799782, XRCC2-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs1805377, XRCC4-rs6869366, and XRCC7-rs7003908 polymorphisms, no evidence of publication bias was identified by viewing the shape of Begg's funnel plot, which was further validated by Egger's regression test (Supplementary Table 3 and Figs. 18–29). However, for XRCC4-rs28360071 polymorphism in overall (P > | t | = 0.043), publication bias was existed. Therefore, we conducted a sensitivity analysis using the trim and fill method (Sue and Richard, 2000). The imputed results provide a symmetrical funnel plot, which indicated that no publication bias for XRCC4-rs28360071 polymorphism was identified after adjusting.
3.4. LD Analyses Across Populations
In order to better understand the quantitative synthesis, LD analysis was performed to test for the existence of bins in the region comprising these polymorphisms in each XRCC genes, respectively (polymorphisms including XRCC1-rs915927, XRCC1-rs25489, XRCC1-rs25487, XRCC1-rs1799782, XRCC1-rs3213245, XRCC1-rs3218536, XRCC3-rs1799796, XRCC3-rs861539, XRCC4-rs6869366, XRCC4-rs28360071, XRCC4-rs1805377, XRCC7-rs7003908). Finally, only XRCC4-rs28360071 polymorphism cannot be matched from the database. LD plots for polymorphisms in each gene were presented in Supplementary Figs. 30–31. Highlighted, for the two significant risk factors (XRCC1-rs25489 and XRCC1-rs25487), no significant LD was identified in all the four populations (CHB: r2 = 0.03; CEU: r2 = 0.02; JPT: r2 = 0.04; YRI: r2 = 0).
4. Discussion
Tumors of the urinary system were reported to make significant threaten to the overall human cancer burden (Parkin, 2008). A wide variable incidence of urological neoplasms indicates its multi-factorial aetiology that involves the interactions between genetic and ethnic backgrounds, as well as the environmental factors. In human beings, XRCC genes that are relevant to DNA repair and damage prevention pathways are critical for preventing cancer initiation and progression. The XRCC1, situated at chromosome 19q13.3, can produce XRCC1 enzyme that involved in BER pathway. It may be particularly important for urological neoplasms, functioning as repairing uracil and oxidative DNA damage (Taylor et al., 2002). The XRCC2 protein encoded by the XRCC2 genes, one of homologue of the RecA protein, displaces replication protein A (RPA) on the exposed single-stranded DNA, which takes responsibility for repairing the DNA double-strand breaks (DBS) (Riha et al., 2006). Similarly, the XRCC3 protein is involved in the homologous recombination repair (HR) pathway and the XRCC4, XRCC7 protein in the non-homologous end joining (NHEJ), also responsible for repairing DBS.
It is hypothesized that polymorphisms in XRCC genes of BER, NER, DSBR and MMR pathways may be important risk factors for the development of urological neoplasms. Some investigators have conducted case-control studies to evaluate the association between polymorphisms in XRCC genes and the risk of urological tumors. However, most of former studies stressed on limited polymorphisms in XRCC genes while neglected potential multiple genes' influence on carcinogenesis. In current study, we presented a comprehensive meta-analysis and systematic review for five DNA repair genes (XRCC1, XRCC2, XRCC3, XRCC4 and XRCC7) to examine the association between these polymorphisms and the risk of urological neoplasms. Overall, our findings suggested that XRCC1-rs25489 polymorphism was associated with an increased risk of urological neoplasms in heterozygote and dominant models, a result consistent with Mittal et al.'s (2012b) work. However, in the research conducted by Zhu et al. (2014), they did not uncover a significant association between XRCC1 polymorphisms and urological neoplasms risk. In further subgroup analyses categorized by cancer type, XRCC1-rs25489 polymorphism was identified as a risk factor for BC in heterozygote model. In addition, significantly increased risk of urological neoplasms in Asians was also identified for XRCC1-rs25487 polymorphism in allelic and homozygote models, while no significant association was revealed for the overall, a result consistent with previous study conducted by Fontana et al. (2008). Furthermore, we also performed LD analysis to find the potential LD association between the two significant risk factors in XRCC1 (rs25487 and rs25489), however, an extremely lower LD was identified for them in all the four commonly researched populations (r2 < 0.10). Moreover, subgroup analysis based on source of controls suggests a significant association between XRCC1-1799782 polymorphism and the risk of urological neoplasms in homozygote model for hospital-based group. The existence of this phenomenon may be due to the inconsistencies in control groups. Although majority of the controls were selected from healthy populations, many individuals may have suffered from other non-cancer diseases. While for other polymorphisms, no significant association was found.
It is worth noting that our data for XRCC3-rs861539 was not consistent with several previously published studies. In the study performed by Shen et al. (2003), they found that the XRCC3 rs861539 variant genotype exhibited a protective effect against BC (OR = 0.63; 95% CI = 0.42–0.93), which was further validated by Narter et al. (2009). On the contrary, Zhu et al. (2012) genotyped a comprehensive case-control studies of 150 BC cases and 150 controls and identified an elevated BC risk among individuals who carry at least one mutated variant allele (OR = 3.22, 95% CI = 1.14–9.11, P = 0.030), and similar results was also obtained by Andrew et al. (2007) Moreover, the frequency of XRCC3-rs861539 genotype distributions in some of the control groups were departed from HWE and thus we cannot rule out the possibility that such an association occurred as a result of bias. Then, we conducted a subgroup analysis by HWE status, and identified that HWE status did not give rise to the bias of results. In addition, the stability of meta-results was further enhanced by sensitivity analysis.
In the present study, we have put considerable effort on carefully searching for published studies, setting strict criteria for study inclusion. There are some advantages that should be illustrated. Firstly, we have conducted a comprehensive search to identify more eligible studies thus, makes our analysis more persuasive and substantial. Secondly, we assessed quality of enrolled studies by NOS, excluding low quality studies to raise the overall quality. Thirdly, we performed various subgroup analysis by ethnicity, source of controls and so on, in order to provide the sources of heterogeneity and the tumor and/or race markers. Fourthly, results were adjusted according to the recognized formula, ensuring the accuracy of the results. In addition, the stability of these studies was further confirmed by sensitivity analysis, and publication bias was tested by Egger's test and Begg's funnel plot. However, several drawbacks in our study should also be noted. Firstly, for XRCC1-rs1799782 polymorphism, relatively heterogeneity existed between some studies, although we conducted this analysis with severe inclusion criteria and explicit extraction for data. Therefore, after stratified analysis by source of control, we observed that the subgroup heterogeneity reduced significantly. It can be assumed that the heterogeneity possibly derived from differenced of ethnicity, source of control, HWE status and cancer type. Secondly, we did not obtain sufficient published studies for several polymorphisms, and some small sample sizes studies may not have enough statistical power to prove authentic associations. Thirdly, all of the studies were published in English, exclusion of studies in other languages may influence effects of polymorphisms tested here. Fourthly, although we want to explore the association between all eligible polymorphisms in XRCC genes and the risk of all urological cancers, however, eligible studies were only identified for the three most commonly researched cancer types (PCa, BC and RCC). Follow-up studies will continue to focus on this issue. Last but not least, our unadjusted estimated results were lacking in information for data analysis, which might lead to failure to confirm marginal association. Hence, result presented here should be interpreted with care, and future studies with more covariates are required.
In conclusion, our meta-analysis suggests that XRCC1-rs25489 polymorphism is a risk factor for urological neoplasms, particularly for BC. Further studies with larger sample size are needed to validate our findings.
Conflicts of Interests
The authors declare no competing financial interests.
Funding Sources
This work was supported by the Clinical Key Subjects Program of the Ministry of Public Health (Urology) and National Natural Science Foundation of China (81370856 and 81401518).
Author Contributions
M.Z., W.L., Z.H., J.Z., L.Z. and C.L. contributed to the conception and design of the study, or acquisition of data, or analysis and interpretation of data; M.Z., W.L. and L.Z. drafting the article or revising it critically for important intellectual content; C.L. final approval of the version to be submitted.
Acknowledgements
We are grateful to Dr. Michael J. Hackett at Seoul National University for participating in the critical revision of this meta-analysis and systematic review.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.03.009.
Contributor Information
Li Zhang, Email: lzhang@ahmu.edu.cn.
Chaozhao Liang, Email: liang_chaozhao@ahmu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- Abe M., Xie W., Regan M.M., King I.B., Stampfer M.J., Kantoff P.W., Oh W.K., Chan J.M. Single-nucleotide polymorphisms within the antioxidant defence system and associations with aggressive prostate cancer. BJU Int. 2011;107:126–134. doi: 10.1111/j.1464-410X.2010.09344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalliu I., Kwon E.M., Salinas C.A., Koopmeiners J.S., Ostrander E.A., Stanford J.L. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli E., Sharma V.B., Sunderesakumar P., Ford J.M. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 2009;69:3589–3596. doi: 10.1158/0008-5472.CAN-08-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew A.S., Gui J., Hu T., Wyszynski A., Marsit C.J., Kelsey K.T., Schned A.R., Tanyos S.A., Pendleton E.M., Ekstrom R.M. Genetic polymorphisms modify bladder cancer recurrence and survival in a USA population-based prognostic study. BJU Int. 2015;115:238–247. doi: 10.1111/bju.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew A.S., Karagas M.R., Nelson H.H., Guarrera S., Polidoro S., Gamberini S., Sacerdote C., Moore J.H., Kelsey K.T., Demidenko E. DNA Repair Polymorphisms Modify Bladder Cancer Risk: A Multi-factor Analytic Strategy. Hum. Hered. 2007;65:105–118. doi: 10.1159/000108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew A.S., Nelson H.H., Kelsey K.T., Moore J.H., Meng A.C., Casella D.P., Tosteson T.D., Schned A.R., Karagas M.R. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- Arizono K., Osada Y., Kuroda Y. DNA repair gene hOGG1 Codon 326 and XRCC1 Codon 399 polymorphisms and bladder cancer risk in a Japanese population. Jpn. J. Clin. Oncol. 2008;38:186–191. doi: 10.1093/jjco/hym176. [DOI] [PubMed] [Google Scholar]
- Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1995;50:1088–1101. [PubMed] [Google Scholar]
- Berhane N., Sobti R.C., Mahdi S.A. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol. Biol. Rep. 2012;39:2471–2479. doi: 10.1007/s11033-011-0998-5. [DOI] [PubMed] [Google Scholar]
- Bonferroni C.E. Teoria Statistica Delle Classi e Calcolo Delle Probabilità. Comm. Firenze. 1935:216–218. [Google Scholar]
- Broberg K., Björk J., Paulsson K., Höglund M., Albin M. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis. 2005;26:1263–1271. doi: 10.1093/carcin/bgi063. [DOI] [PubMed] [Google Scholar]
- Chang C.H., Chang C.L., Tsai C.W., Wu H.C., Chiu C.F., Wang R.F., Liu C.S., Lin C.C., Bau D.T. Significant association of an XRCC4 single nucleotide polymorphism with bladder cancer susceptibility in Taiwan. Anticancer Res. 2009;29:1777–1782. [PubMed] [Google Scholar]
- Chang C.H., Chiu C.F., Wu H.C., Tseng H.C., Wang C.H., Lin C.C., Tsai C.W., Liang S.Y., Wang C.L., Bau D.T. Significant association of XRCC4 single nucleotide polymorphisms with prostate cancer susceptibility in Taiwanese males. Mol. Med. Rep. 2008;1:525–530. [PubMed] [Google Scholar]
- Davey Smith G., Egger M. Meta-analyses of randomised controlled trials. Lancet. 1997;350(9085):1182. doi: 10.1016/s0140-6736(05)63833-0. (PubMed PMID: 9343537) [DOI] [PubMed] [Google Scholar]
- Dhillon V.S., Yeoh E., Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia—results of a pilot study. Urol. Oncol. 2009;29:641–646. doi: 10.1016/j.urolonc.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Oxidatively induced DNA damage and its repair in cancer. Mutat. Res. Rev. Mutat. Res. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Figueroa J.D., Malats N., Real F.X., Silverman D., Kogevinas M., Chanock S., Welch R., Dosemeci M., Tardón A., Serra C. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum. Genet. 2007;121:233–242. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- Figueroa J.D., Malats N., Rothman N., Real F.X., Silverman D., Kogevinas M., Chanock S., Yeager M., Welch R., Dosemeci M. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28:1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- Fontana L., Bosviel R., Delort L., Guy L., Chalabi N., Kwiatkowski F., Satih S., Rabiau N., Boiteux J.P., Chamoux A. DNA repair gene ERCC2, XPC, XRCC1, XRCC3 polymorphisms and associations with bladder cancer risk in a French cohort. Anticancer Res. 2008;28:1853–1856. [PubMed] [Google Scholar]
- Gangwar R., Ahirwar D., Mandhani A., Mittal R.D. Do DNA repair genes OGG1, XRCC3 and XRCC7 have an impact on susceptibility to bladder cancer in the North Indian population? Mutat. Res. 2009;680:56–63. doi: 10.1016/j.mrgentox.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Hamano T., Matsui H., Ohtake N., Nakata S., Suzuki K. Polymorphisms of DNA repair genes, XRCC1 and XRCC3, and susceptibility to familial prostate cancer in a Japanese population. Asia-Pacific J. Clin. Oncol. 2008;4:21–26. [Google Scholar]
- Hao G.Y., Zhang Y.Y., Zhang W.D., Yang M.S., Jia Q. Relationship between XRCC3 gene polymorphism and bladder cancer in the Han population. J. Shandong Univ. 2008;46:612–615. [Google Scholar]
- Hirata H., Hinoda Y., Matsuyama H., Tanaka Y., Okayama N., Suehiro Y., Zhao H., Urakami S., Kawamoto K., Kawakami T. Polymorphisms of DNA repair genes are associated with renal cell carcinoma. Biochem. Biophys. Res. Commun. 2006;342:1058–1062. doi: 10.1016/j.bbrc.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Hirata H., Hinoda Y., Tanaka Y., Okayama N., Suehiro Y., Kawamoto K., Kikuno N., Majid S., Vejdani K., Dahiya R. Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur. J. Cancer. 2007;43:231–237. doi: 10.1016/j.ejca.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H.J. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Huang M., Dinney C.P., Lin X., Lin J., Grossman H.B., Wu X. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer Epidemiol. Biomark. Prev. 2007;16:84–91. doi: 10.1158/1055-9965.EPI-06-0712. [DOI] [PubMed] [Google Scholar]
- Kelsey K.T., Park S., Nelson H.H., Karagas M.R. A population-based case-control study of the XRCC1 Arg399Gln polymorphism and susceptibility to bladder cancer. Cancer Epidemiol. Biomark. Prev. 2004;13:1337–1341. [PubMed] [Google Scholar]
- Kuasne H., Rodrigues I.S., Losi-Guembarovski R., Reis M.B., Fuganti P.E., Gregório E.P., Libos J.F., Matsuda H.M., Rodrigues M.A., Kishima M.O. Base excision repair genes XRCC1 and APEX1 and the risk for prostate cancer. Mol. Biol. Rep. 2011;38:1585–1591. doi: 10.1007/s11033-010-0267-z. [DOI] [PubMed] [Google Scholar]
- Lan C., Ambrosone C.B., Lee J., Sellers T.A., Pow-Sang J., Park J.Y. Association between polymorphisms in the DNA repair genes XRCC1 and APE1, and the risk of prostate cancer in white and black Americans. J. Urol. 2006;175:108–112. doi: 10.1016/S0022-5347(05)00042-X. [DOI] [PubMed] [Google Scholar]
- Lavender N.A., Komolafe O.O., Benford M., Brock G., Moore J.H., Vancleave T.T., States J.C., Kittles R.A., Kidd L.C.R. No association between variant DNA repair genes and prostate cancer risk among men of African descent †. Prostate. 2010;70:113–119. doi: 10.1002/pros.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesegang T.J. Human DNA repair genes. Am J. Ophthalmol. 2001;132:298. [Google Scholar]
- Luedeke M., Linnert C.M., Hofer M.D., Surowy H.M., Rinckleb A.E., Hoegel J., Kuefer R., Rubin M.A., Vogel W., Maier C. Predisposition for TMPRSS2-ERG fusion in prostate cancer by variants in DNA repair genes. Cancer Epidemiol. Biomark. Prev. 2009;18:3030–3035. doi: 10.1158/1055-9965.EPI-09-0772. [DOI] [PubMed] [Google Scholar]
- Mandal R.K., Kapoor R., Mittal R.D. Polymorphic variants of DNA repair gene XRCC3 and XRCC7 and risk of prostate cancer: a study from North Indian population. DNA Cell Biol. 2010;29:669–674. doi: 10.1089/dna.2010.1047. [DOI] [PubMed] [Google Scholar]
- Mandal R.K., Singh V., Kapoor R., Mittal R.D. Do polymorphisms in XRCC4 influence prostate cancer susceptibility in North Indian population? Biomarkers. 2011;16:236–242. doi: 10.3109/1354750X.2010.547599. [DOI] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natil Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Matullo G. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol. Biomark. Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- Matullo G., Dunning A.M., Guarrera S., Baynes C., Polidoro S., Garte S., Autrup H., Malaveille C., Peluso M., Airoldi L. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- Matullo G., Guarrera S., Carturan S., Peluso M., Malaveille C., Davico L., Piazza A., Vineis P. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int. J. Cancer. 2001;92:562–567. doi: 10.1002/ijc.1228. [DOI] [PubMed] [Google Scholar]
- Mittal R.D., Gangwar R., Mandal R.K., Srivastava P., Ahirwar D.K. Gene variants of XRCC4 and XRCC3 and their association with risk for urothelial bladder cancer. Mol. Biol. Rep. 2012;39:1667–1675. doi: 10.1007/s11033-011-0906-z. [DOI] [PubMed] [Google Scholar]
- Mittal R.D., Mandal R.K., Gangwar R. Base excision repair pathway genes polymorphisminprostate and bladder cancer risk in North Indian population. Mech. Ageing Dev. 2012;133(4):127–132. doi: 10.1016/j.mad.2011.10.002. (PubMed PMID: 22019847) [DOI] [PubMed] [Google Scholar]
- Mittal R.D., Singh R., Manchanda P.K., Ahirwar D., Gangwar R., Kesarwani P., Mandhani A. XRCC1 codon 399 mutant allele: a risk factor for recurrence of urothelial bladder carcinoma in patients on BCG immunotherapy. Cancer Biol. Ther. 2008;7:645–650. doi: 10.4161/cbt.7.5.5763. [DOI] [PubMed] [Google Scholar]
- Narter K.F., Ergen A., Ağaçhan B., Görmüş U., Timirci Ö., Isbir T. Bladder Cancer and Polymorphisms of DNA Repair Genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1) Anticancer Res. 2009;29:1389–1393. [PubMed] [Google Scholar]
- Nowacka-Zawisza M., Wiśnik E., Wasilewski A., Skowrońska M., Forma E., Bryś M., Różański W., Krajewska W.M. Polymorphisms of Homologous Recombination RAD51, RAD51B, XRCC2, and XRCC3 Genes and the Risk of Prostate Cancer. Anal. Cell. Pathol. 2015;2015:2087–2095. doi: 10.1155/2015/828646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlow I., Park B.J., Mujumdar U., Patel H., Siu-Lau P., Clas B.A., Downey R., Flores R., Bains M., Rizk N. DNA damage and repair capacity in patients with lung cancer: prediction of multiple primary tumors. J. Clin. Oncol. 2008;26:3560–3566. doi: 10.1200/JCO.2007.13.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin D.M. The global burden of urinary bladder cancer. Scand. J. Urol. 2008;42:1–9. doi: 10.1080/03008880802285032. [DOI] [PubMed] [Google Scholar]
- Rajaraman P., Hutchinson A., Wichner S., Black P.M., Fine H.A., Loeffler J.S., Selker R.G., Shapiro W.R., Rothman N., Linet M.S. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro-Oncology. 2010;12:37–48. doi: 10.1093/neuonc/nop012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaniuk V.P., Nikitchenko N.V., Savina N.V., Kuzhir T.D., Rolevich A.I., Krasny S.A., Sushinsky V.E., Goncharova R.I. Polymorphism of DNA repair genes OGG1, XRCC1, XPD and ERCC6 in bladder cancer in Belarus. Biomarkers. 2014;19:1–8. doi: 10.3109/1354750X.2014.943291. [DOI] [PubMed] [Google Scholar]
- Riha K., Heacock M.L., Shippen D.E. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Genetics. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Ritchey J.D., Huang W.Y., Chokkalingam A.P., Gao Y.T., Deng J., Levine P., Stanczyk F.Z., Hsing A.W. Genetic variants of DNA repair genes and prostate cancer: a population-based study. Cancer Epidemiol. Biomark. Prev. 2005;14:1703–1709. doi: 10.1158/1055-9965.EPI-04-0809. [DOI] [PubMed] [Google Scholar]
- Rybicki B.A., Conti D.V., Moreira A., Cicek M., Casey G., Witte J.S. DNA repair gene XRCC1 and XPD polymorphisms and risk of prostate cancer. Cancer Epidemiol. Biomark. Prev. 2004;13:23–29. doi: 10.1158/1055-9965.epi-03-0053. [DOI] [PubMed] [Google Scholar]
- Sak S.C., Barrett J.H., Paul A.B., Bishop D.T., Kiltie A.E. DNA repair gene XRCC1 polymorphisms and bladder cancer risk. BMC Genet. 2007;8:1–8. doi: 10.1186/1471-2156-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S., Festa F., Sakano S., Zhang Z., Steineck G., Norming U., Wijkström H., Larsson P., Kumar R., Hemminki K. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- Seagroatt V., Stratton I. Bias in meta-analysis detected by a simple, graphical test. Test had 10% false positive rate. BMJ. 1998;315:629–634. [PMC free article] [PubMed] [Google Scholar]
- Shen M., Hung R.J., Brennan P., Malaveille C., Donato F., Placidi D., Carta A., Hautefeuille A., Boffetta P., Porru S. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in northern Italy. Cancer Epidemiol. Biomark. Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- Spitz M.R., Wei Q., Dong Q., Amos C.I., Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol. Biomark. Prev. 2003;12:689–698. [PubMed] [Google Scholar]
- Stern M.C., Umbach D.M., Lunn R.M., Taylor J.A. DNA repair gene XRCC3 codon 241 polymorphism, its interaction with smoking and XRCC1 polymorphisms, and bladder cancer risk. Cancer Epidemiol. Biomark. Prev. 2002;11:939–943. [PubMed] [Google Scholar]
- Stern M.C., Umbach D.M., Van Gils C.H., Lunn R.M., Taylor J.A. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol. Biomark. Prev. 2001;10:125–131. [PubMed] [Google Scholar]
- Sue D., Richard T. Trim and fill: a simple funnel-plot–based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Taylor R.M., Thistlethwaite A., Caldecott K.W. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gils C.H., Bostick R.M., Stern M.C., Taylor J.A. Differences in base excision repair capacity may modulate the effect of dietary antioxidant intake on prostate cancer risk: an example of polymorphisms in the XRCC1 gene. Cancer Epidemiol. Biomark. Prev. 2002;11:1279–1284. [PubMed] [Google Scholar]
- Vispe S., Yung T.M., Ritchot J., Serizawa H., Satoh M.S. A cellular defense pathway regulating transcription through poly(ADP-ribosyl)ation in response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2000;97:185–189. doi: 10.1073/pnas.170280397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Qin C., Zhu J., Yuan L., Fu G., Zhang Z., Yin C. Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010;29:303–311. doi: 10.1089/dna.2009.0969. [DOI] [PubMed] [Google Scholar]
- Wang S.Y., Peng L., Li C.P., Li A.P., Zhou J.W., Zhang Z.D., Liu Q.Z. Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int. J. Urol. 2008;15:534–539. doi: 10.1111/j.1442-2042.2008.02049.x. [DOI] [PubMed] [Google Scholar]
- Wells G.A., Shea B.J., O'connell D., Peterson J., Welch V., Losos M., Tugwell P. The Newcastle–Ottawa Scale (NOS) for assessing the quality of non-randomized studies in meta-analysis. Appl. Eng. Agric. 2000;18:727–734. [Google Scholar]
- Wen H., Ding Q., Fang Z.J., Xia G.W., Fang J. Population study of genetic polymorphisms and superficial bladder cancer risk in Han-Chinese smokers in Shanghai. Int. Urol. Nephrol. 2009;41:855–864. doi: 10.1007/s11255-009-9560-y. [DOI] [PubMed] [Google Scholar]
- Wen H., Feng C.C., Fang Z.J., Xia G.W., Jiang H.W., Xu G., Huang X.D., Ding Q. Estudio sobre susceptibilidad de cáncer de vejiga y polimorfismos genéticos de XPC, XPG y CYP en fumadores y no fumadores. Actas Urol. Esp. 2013;37:259–265. doi: 10.1016/j.acuro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Wu X., Gu J., Grossman H.B., Amos C.I., Etzel C., Huang M., Zhang Q., Millikan R.E., Lerner S., Dinney C.P. Bladder cancer predisposition: a multigenic approach to dna-repair and cell-cycle–control genes. Am. J. Hum. Genet. 2006;78:464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Qian H.L.X., Yang J., Wang X.R., Zhang W., Wu H.F. Relationship between XRCC1 polymorphisms and susceptibility to prostate cancer in men from Han, Southern China. Asian J. Androl. 2007;9:331–338. doi: 10.1111/j.1745-7262.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- Zhi Y., Yu J., Liu Y., Wei Q., Yuan F., Zhou X., Song B., Chen Z., Yang J. Interaction between polymorphisms of DNA repair genes significantly modulated bladder cancer risk. Int. J. Med. Sci. 2012;9:498–505. doi: 10.7150/ijms.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.F., Zhang G.B., Qu P., Zhou J., Pan H.X., Hou J.Q. Association between single nucleotide polymorphisms in the XRCC1 gene and susceptibility to prostate cancer in Chinese men. Asian Pac. J. Cancer Prev. 2012;13:5241–5243. doi: 10.7314/apjcp.2012.13.10.5241. [DOI] [PubMed] [Google Scholar]
- Zhu H., Jiu T., Wang D. Impact of polymorphisms of the DNA repair gene XRCC1 and their role in the risk of prostate cancer. Pak. J. Med. Sci. 2014;31 doi: 10.12669/pjms.312.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhong Z., Zhang X., Zhao X., Xu R., Ren W., Li S. DNA repair gene XRCC3 T241M polymorphism and bladder cancer risk in a Chinese population. Genet. Test. Mol. Biomarkers. 2012;16:640–643. doi: 10.1089/gtmb.2011.0334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material