Summary
Many important Plasmodium falciparum genes show clonally variant expression, but little is known about how these genes are used during human malarial infections. This article reports the expression in human infections of the clonally variant clag3 genes linked to solute uptake.
Keywords: Malaria, Plasmodium falciparum, transcription, epigenetics, adaptation, bet-hedging, controlled human malaria infection (CHMI), mutually exclusive gene expression, transcriptional variation, clag3.
Abstract
Background.
Many genes of the malaria parasite Plasmodium falciparum show clonally variant expression regulated at the epigenetic level. These genes participate in fundamental host-parasite interactions and contribute to adaptive processes. However, little is known about their expression patterns during human infections. A peculiar case of clonally variant genes are the 2 nearly identical clag3 genes, clag3.1 and clag3.2, which mediate nutrient uptake and are linked to resistance to some toxic compounds.
Methods.
We developed a procedure to characterize the expression of clag3 genes in naturally infected patients and in experimentally infected human volunteers.
Results.
We provide the first description of clag3 expression during human infections, which revealed mutually exclusive expression and identified the gene predominantly expressed. Adaptation to culture conditions or selection with a toxic compound resulted in isolate-dependent changes in clag3 expression. We also found that clag3 expression patterns were reset during transmission stages.
Conclusions.
Different environment conditions select for parasites with different clag3 expression patterns, implying functional differences between the proteins encoded. The epigenetic memory is likely erased before parasites start infection of a new human host. Altogether, our findings support the idea that clonally variant genes facilitate the adaptation of parasite populations to changing conditions through bet-hedging strategies.
Plasmodium falciparum is responsible for the most severe forms of malaria. Asexual growth of the parasites in the human blood is responsible for all clinical symptoms and also for chronic infection. During the approximately 48-hour asexual multiplication cycle, parasites live inside of human erythrocytes, except for the short time between bursting of parasites at the schizont stage and invasion of new erythrocytes [1]. While the human blood is a relatively stable environment, parasites need to adapt to fluctuating conditions, such as changing nutrient concentrations, presence of drugs, occurrence of fever episodes, or immune responses. Recent studies have demonstrated that populations of genetically identical parasites show extensive transcriptional heterogeneity [2], which potentially allows adaptation by dynamic natural selection of parasites with transcriptional patterns associated with increased fitness as the environment changes. This adaptive strategy, known as bet hedging, is used by many microbial organisms [3–5]. Genes that can be found in either an active or a silenced state in genetically identical parasites at the same stage of cycle progression, known as clonally variant genes, participate in multiple biological pathways involved in fundamental host-parasite interactions [2, 6, 7]. The silenced or active state of these genes is transmitted from one generation to the next by epigenetic mechanisms [8–10]. Switches between the 2 alternative states of these genes occur spontaneously, albeit with low frequency, allowing for the constant generation of transcriptional diversity within parasite populations.
Despite the large number of families of clonally variant genes identified in P. falciparum, there are few for which an adaptive role or an association between the transcriptional state of specific genes and the resulting phenotypes has been clearly established. The best characterized family of clonally variant genes is var, which consists of about 60 genes per genome that encode PfEMP-1, a major virulence factor linked to cytoadherence and antigenic variation. var genes show mutually exclusive expression, such that a single parasite typically expresses only 1 var gene at a time and keeps all of the others silenced [11]. Spontaneous switches in the expression of var genes play an adaptive role, mediating immune evasion and altering the sequestration tropism of infected erythrocytes [12].
A second case of P. falciparum clonally variant genes for which transcriptional switches have been associated with specific phenotypes and adaptation to changes in the environment is clag3. The 2 clag3 genes, clag3.1 (PF3D7_0302500) and clag3.2 (PF3D7_0302200), are separated by only 10 kb and show 95% sequence coincidence. The clag3.1 or clag3.2 identity is determined by the relative position of each gene in the chromosome and by the conserved clag3.1- or clag3.2-specific flanking regions [13]. These genes are part of the 5-member clag family, which encodes the CLAG/RhopH1 component of the RhopH complex. While early reports linked CLAG proteins with erythrocyte invasion or cytoadherence [14], more-recent research has provided strong genetic and biochemical evidence for a key role of CLAG3 proteins in the formation of the plasmodial surface anion channel (PSAC), a broad selectivity channel that mediates the uptake of nutrients and several other solutes at the infected erythrocyte membrane [14–17]. CLAG3 proteins are validated drug targets [16]. Epigenetic silencing of clag3 genes is mediated by formation of heterochromatin, similar to other clonally variant genes [10, 18]. Together with the var family, clag3 is the only known example of mutually exclusive expression in P. falciparum [19]. However, while under normal conditions the vast majority of parasites express only 1 of the 2 clag3 genes at a time, mutually exclusive expression is not strict, which allows for the occurrence of small selectable subpopulations of parasites with alternative expression patterns that enable additional phenotypic plasticity [20]. We and others have recently demonstrated that a compound that is toxic for the parasite, blasticidin S, can select for low abundance subpopulations of parasites with both clag3 genes simultaneously silenced [21, 22]. However, lower concentrations of the drug select for parasites that express a specific paralog, which is suggestive of phenotypic differences associated with expression of one or the other clag3 gene [22]. Altogether, these results indicate that clag3 expression patterns determine the permeability phenotype of infected erythrocytes and can mediate drug resistance at the epigenetic level. Variant expression of these genes needs to fulfill 2 competing requirements: efficient acquisition of nutrients and restriction of the entrance of harmful compounds.
While the expression patterns of var genes in field isolates have been the subject of intensive investigation [12, 23–26], very little is known about the expression of clag3 genes during human infections. Previous studies of clag3 expression were conducted with culture-adapted parasites. Genome-wide transcriptomic analysis of P. falciparum field isolates [27–29] could not reliably characterize the expression patterns of these genes because the sequences of clag3.1 and clag3.2 are almost identical and the regions that are more distinct between the 2 genes are highly polymorphic between parasite isolates [13]. Here we developed a procedure to analyze clag3 expression in natural human infections and used it to study the expression of these genes in clinical malaria cases and after parasite adaptation to culture conditions or to blasticidin S pressure. We also studied clag3 expression in experimental human malarial infections.
METHODS
Ethics Approval
This study was approved by the Institute of Tropical Medicine (ITM) Institutional Review Board (IRB; protocol ITG913/13), the University Hospital of Antwerp IRB (protocol B300201319284), and the Gambian government/Medical Research Council Joint Ethics Committee (protocol SCC1392). Approval for the controlled human malaria infection (CHMI) trial has been previously described [30]. All participants provided written informed consent before enrollment. The study was conducted according to the principles stated in the Declaration of Helsinki.
Sample Collection
Blood samples were obtained from returning travelers (≥18 years old) with clinical P. falciparum malaria who attended clinics in Antwerp (Belgium) and from children (≤12 years old) with clinical P. falciparum malaria who attended health centers in the Gambia. We also analyzed blood samples collected from volunteers participating in a CHMI study [30]. Additional details of sample collection and processing are provided in the Supplementary Methods.
Genetic and Transcriptional Analysis
Multiplicity of infection was estimated by genotyping the msp1 and msp2 loci [31]. To assess for recombination events between the 2 clag3 genes, the clag3 loci were analyzed by long polymerase chain reaction (PCR) [13]. For each isolate, we sequenced the hypervariable region (HVR) [13] of each clag3 gene from the long PCR products to design gene- and isolate-specific primers.
To prepare RNA for clag transcriptional analysis, cultures were harvested when the majority of parasites were at the schizont stage. For natural infections, parasites were cultured only until they reached the schizont stage, with the exception of 2 samples (Supplementary Methods). In the CHMI study, samples collected on day 9 after sporozoite injection and on the day of malaria diagnosis were cryopreserved, and, after thawing, parasites were cultured for 2–3 weeks and for approximately 1 week, respectively, until they reached a parasitemia level of ≥0.02%.
RNA was purified approximately as described elsewhere [32]. The protocol was validated for use at very low parasitemia levels (Supplementary Figure 1). Quantitative PCR analysis was performed using the standard curve method approximately as described previously [10], with primers listed in Supplementary Table 1.
All new sequences obtained in this study have been deposited to GenBank as accession numbers KY092485-KY092488 (full sequences) and KY364642-KY364689 (HVR sequences). Additional details of the methods for the genetic and transcriptional analysis and also for the CLAG3 sequence analysis can be found in the Supplementary Methods.
RESULTS
Parasites Predominantly Express clag3.2 in Natural Malaria Infections
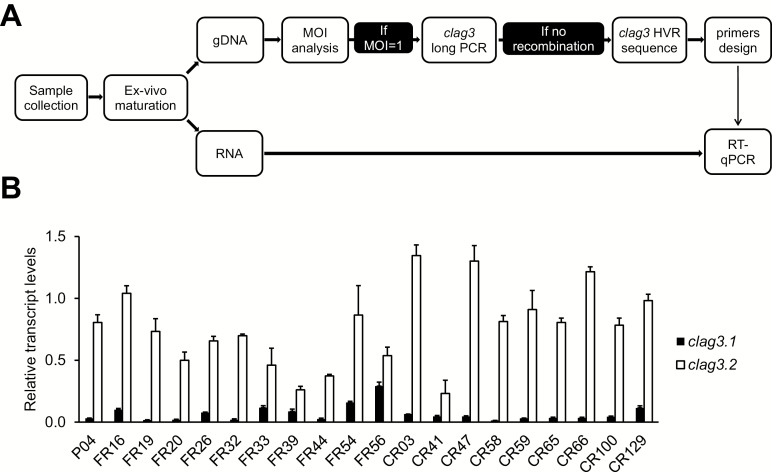
We analyzed clag3 expression patterns in blood specimens from P. falciparum–infected symptomatic patients. Parasites were cultured ex vivo until they reached the schizont stage, when clag3 genes are expressed, and were harvested for genomic DNA and RNA extraction (Figure 1A). To reduce the complexity of the analysis, only samples with a single clone or a clearly predominant clone were retained for clag3 expression characterization. Isolates presenting a single clag3 gene in their genome as a consequence of recombination [13] were excluded. For the 20 remaining samples, we sequenced the HVR of the 2 genes to design isolate-specific primers for clag3.1 and for clag3.2 and used them to analyze clag3 expression by reverse transcription followed by quantitative PCR. All isolates showed predominant expression of 1 of the 2 clag3 paralogs (Figure 1B), consistent with the mutually exclusive expression observed in culture-adapted parasite lines [15, 16, 18, 19]. However, in contrast to the majority of culture-adapted lines, which predominantly express clag3.1 [15, 18], we observed predominant clag3.2 expression in all isolates (Figure 1B).
Figure 1.
Transcriptional analysis of clag3.1 and clag3.2 in Plasmodium falciparum natural human infections. A, Schematic of the work flow. Forty field isolates were cultured ex vivo until they reached the schizont stage and were harvested for genomic DNA (gDNA) and RNA extraction. In samples with a multiplicity of infection (MOI) of 1 (single infections) or a clear predominant clone and without recombination between the 2 clag3 genes, the hypervariable region (HVR) of clag3.1 and clag3.2 was sequenced to design paralog- and isolate-specific primers for the analysis of clag3 expression by reverse transcription followed by quantitative polymerase chain reaction analysis (RT-PCR). B, Expression of clag3.1 and clag3.2 in 20 field isolates. Pxx are samples collected in Belgium, FRxx are samples collected in the Gambia and analyzed directly without freezing, and CRxx are cryopreserved isolates collected in the Gambia. Of note, clag3.1 has a stronger promoter than clag3.2 [10, 20], implying that the actual proportion of clag3.1-expressing parasites in each isolate is even lower than the proportion of clag3.1 transcripts relative to total clag3 transcripts. The significance of variation in total clag3 transcript levels (clag3.1 plus clag3.2 transcripts) among these samples is unclear (Supplementary Methods); the focus of this analysis is on the relative transcript levels between the 2 clag3 genes. Transcript levels are normalized against rhoph2. Error bars are SD.
Adaptation of Field Isolates to Culture Conditions or to Low Drug Pressure Is Associated With Isolate-Dependent Changes in clag3 Expression
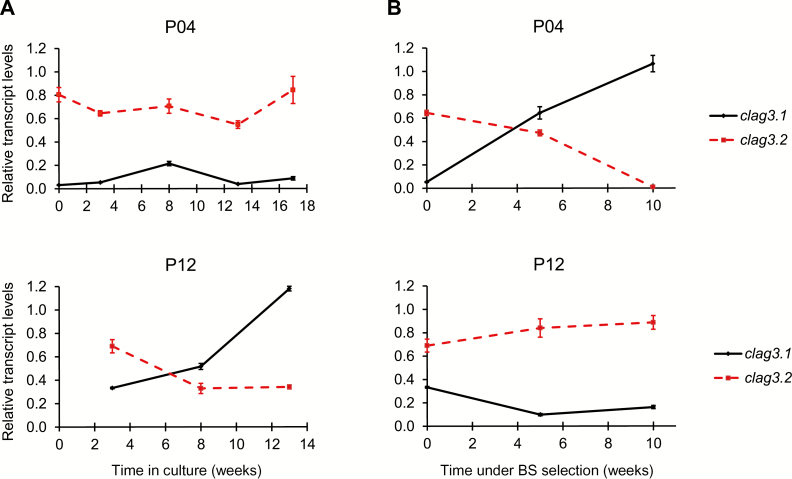
Two of the field isolates, P04 and P12, were maintained under culture conditions for 17 and 13 weeks, respectively, with regular monitoring of clag3 expression (Figure 2A). In the P12 isolate, parasites expressing clag3.2 were progressively replaced by parasites expressing clag3.1. In contrast, in the P04 isolate, the majority of parasites maintained clag3.2 expression throughout the experiment. Next we selected the same isolates with a sublethal concentration of blasticidin S that, in the 3D7 genetic background, selects for parasites that express clag3.1 [22]. In the P04 isolate, parasites expressing clag3.1 were quickly selected, and by 10 weeks of selection they had almost completely displaced clag3.2-expressing parasites (Figure 2B). However, blasticidin S had the opposite effect on the P12 isolate by instead favoring survival of parasites expressing clag3.2: while normal culturing of this isolate resulted in selection of clag3.1-expressing parasites (Figure 2A), this did not occur in the presence of blasticidin S (Figure 2B). These results indicate that the selective advantage conferred by expression of one or the other clag3 paralog under different conditions is isolate specific and likely depends on the clag3 genes sequences.
Figure 2.
Expression of clag3.1 and clag3.2 in field isolates of Plasmodium falciparum maintained under culture conditions or under drug pressure. A, Transcript levels of clag3.1 and clag3.2 in the P04 and P12 isolates maintained under culture conditions. B, Transcript levels of clag3.1 and clag3.2 in the same isolates selected with a low concentration of blasticidin S (BS; 0.3 µg/mL). Transcript levels are normalized against rhoph2. Error bars are SD.
Analysis of clag3 Sequences Identifies Paralog-Specific and Promiscuous Sequence Features
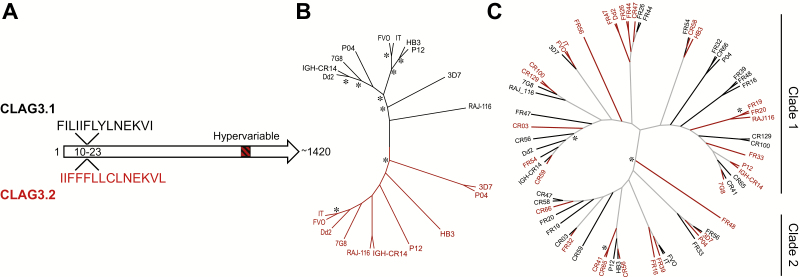
To gain insight into the sequence determinants of the fitness advantages conferred by expression of one or the other protein, we analyzed publicly available CLAG3 sequences and the newly obtained sequences from the P04 and P12 isolates. We identified a sequence feature at the N-terminal end of the protein (positions 10–23) that was different between CLAG3.1 and CLAG3.2 but conserved for each protein among all isolates analyzed (Figure 3A). This hydrophobic region, which likely corresponds to either a signal peptide or a transmembrane domain, is a candidate for determining general CLAG3 properties that, under normal human blood conditions, confer a selective advantage to parasites expressing CLAG3.2. At other positions, polymorphisms are not paralog specific, such that the same sequence can be found in CLAG3.1 or in CLAG3.2 in different isolates. This is probably attributable to frequent recombination and gene conversion events between the 2 paralogs (Supplementary Figure 2A). However, clustering analysis of the full CLAG3 sequences confirmed that CLAG3.1 and CLAG3.2 separate into discrete clades (Figure 3B), as previously reported [21]. This is a consequence of several polymorphisms occurring at very different frequencies between the 2 paralogs. In contrast, clustering analysis of the HVR did not show separation of CLAG3.1 and CLAG3.2 sequences (Figure 3C), which indicates that a CLAG3.1 HVR can be as similar to a CLAG3.2 HVR as to another CLAG3.1 HVR, and vice versa. HVR sequences separated in 2 distinct clades, both including CLAG3.1 and CLAG3.2 sequences (Figure 3C and Supplementary Figure 2B). Phenotypic traits such as resistance to blasticidin S that are associated with clag3.1 expression in some isolates and with clag3.2 expression in others likely depend on polymorphism at the HVR or other positions where polymorphism is not paralog specific.
Figure 3.
Analysis of CLAG3 sequences. A, Schematic of CLAG3 sequences, showing the single paralog-specific conserved sequence feature identified (positions 10–23) and the hypervariable region located near the C-terminal end of the protein (starting at approximately position 1110). The analysis is based on publicly available full CLAG3 sequences and the newly obtained sequences from the P04 and P12 isolates (10 sequences for each gene in total). B, Cladogram of full CLAG3 sequences. C, Cladogram of hypervariable region sequences, including publicly available sequences and new sequences from 24 patient isolates. In panels B and C, CLAG3.1 and CLAG3.2 sequences are represented as black and red branches, respectively (black and dark gray, respectively, in the printed edition). Asterisks indicate bootstrap values of >70%.
clag3 Expression in Parasites Obtained From Experimentally Infected Humans
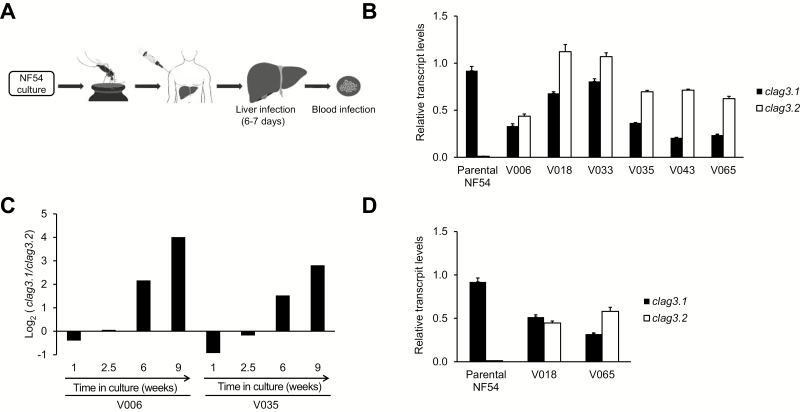
We analyzed clag3 expression patterns in parasites collected from human volunteers participating in a CHMI trial [30] in which cryopreserved sporozoites of the culture-adapted line NF54 (from which 3D7 was derived) were used for infection (Figure 4A). In samples collected from 6 different volunteers when parasites were first detected by microscopy (ie, on the day of malaria diagnosis [12–15 days after injection]), we observed higher levels of clag3.2 transcripts, compared with clag3.1, in contrast to the parental NF54 line used to infect the mosquitoes that almost exclusively expresses clag3.1 (Figure 4B). Next we readapted to culture conditions the parasites obtained from 2 volunteers, and in both cases we observed a progressive increase in the ratio of clag3.1 to clag3.2 transcripts, reflecting selection of parasites expressing clag3.1 (Figure 4C). This result indicates that clag3.1 expression confers a fitness advantage under culture conditions in the NF54 genetic background. Similar levels of transcripts for both clag3 genes in volunteer samples (Figure 4B) imply that they likely consist of a mixture of parasites expressing clag3.2 and parasites expressing clag3.1, rather than a homogeneous population of individual parasites expressing the 2 genes simultaneously. This idea is based on previous results with culture-adapted parasite lines with similar transcript levels of clag3.1 and clag3.2 [19, 20] and is also supported by the analysis of clag3 expression in subclones of one of the volunteer samples (Supplementary Figure 3).
Figure 4.
Expression of clag3.1 and clag3.2 in controlled human malaria infections. A, Schematic of the controlled human malaria infection trial. Sporozoites obtained from mosquitoes fed with NF54-infected blood were cryopreserved and injected into healthy volunteers. B, Expression of clag3.1 and clag3.2 in the NF54 parental line used to infect the mosquitoes and in parasites collected from volunteers on the day of malaria diagnosis by light microscopy. The significance of variation in total clag3 transcript levels (clag3.1 plus clag3.2 transcripts) among these samples is unclear (Supplementary Methods); the focus of this analysis is on the relative transcript levels between the 2 clag3 genes. C, Relative transcript levels of clag3.1 and clag3.2, expressed as the log2 of the ratio of clag3.1/clag3.2 transcript levels, in parasites obtained from 2 volunteers and maintained under culture conditions for 9 weeks. D, Expression of clag3.1 and clag3.2 in parasites collected from 2 volunteers at day 9 after sporozoite injection (approximately 1 cycle in the blood circulation). In all panels, transcript levels are normalized against rhoph2. Error bars are SD.
The Expression Patterns of clag3 Genes Are Reset During Transmission Stages
The clag3 expression patterns observed in parasites obtained from experimentally infected volunteers can be explained by 2 nonexclusive scenarios: clag3.2-expressing parasites are selected under the conditions of the human circulation, or there is a reset of clag3 expression patterns during transmission stages. To distinguish between the 2 possibilities, we analyzed clag3 expression in blood samples collected 9 days after sporozoite injection from 2 volunteers. Considering that parasite liver stage development takes 6–7 days [33], parasites collected at day 9 had been in the peripheral blood for only about 1 multiplication cycle. Despite this, we observed similar levels of transcripts for both clag3 genes at day 9 (Figure 4D), a pattern similar to that observed on the day of malaria diagnosis. This result is inconsistent with a blood-stage selection–only scenario and supports the idea that clag3 expression patterns are reset when parasites go through transmission stages. Because parasite densities were extremely low at day 9 after injection, parasites had to be cultured for 2–3 weeks before we could obtain sufficient material for transcriptional analysis. However, this is unlikely to be a confounder for these results because, in parasites of the NF54 genetic background, culture conditions progressively selected for parasites that express clag3.1 (Figure 4C). Hence, at day 9 after injection, the parasites population contained a large proportion of parasites expressing clag3.2, a composition that was clearly distinct from the parental NF54 line.
Transcript Levels of clag2, clag8, and clag9 Did Not Show Major Differences Among Isolates or Between Different Growth Conditions
We also analyzed the expression of clag2 (PF3D7_0220800), clag8 (PF3D7_0831600), and clag9 (PF3D7_0935800) in all samples described in this study. There was little variation in the transcript levels of these genes (Supplementary Figures 4–8). Even clag2, which shows clonally variant expression in culture-adapted lines [2, 19], was expressed at similar levels in all samples.
DISCUSSION
Variantly expressed malarial genes play key roles in host-parasite interactions and contribute to parasites’ adaptation to changes in their environment, but little is known about the expression patterns of these genes during human infections. This is an important limitation because the expression of clonally variant genes in a population of parasites is shaped by the environment, and the environment is different between culture conditions and the natural conditions of the human blood circulation. Here we characterized the expression in human infections of the clonally variant P. falciparum clag3 genes, which provide one of the best models in malaria to study functional variation linked to epigenetic switches. We observed differences from the clag3 expression patterns commonly observed under culture conditions, but expression conformed with the mutual exclusion principle previously described in cultured parasites. By comparing expression patterns in the same parasite lines between human circulation and culture conditions or by challenging parasites with a toxic compound, we observed that different environments dynamically select for parasites with different patterns of clag3 expression in an isolate-dependent manner. These results support the idea that transcriptional variation and bet-hedging strategies play an important role in malarial parasite adaptation.
In 20 clinical malarial infections, parasites predominantly expressed clag3.2, in contrast to most culture-adapted parasite lines that predominantly express clag3.1 [15, 18]. This result suggests that, under the conditions of the human circulation, with lower concentrations of most nutrients than in the regular parasite culture medium [16], expression of clag3.2 confers a growth advantage. Whether different clinical presentation (eg, asymptomatic or severe malaria), host malnutrition, exposure to drugs, or other conditions are associated with different clag3 expression patterns remains to be determined. Together with previous observations showing that, in the 3D7 genetic background, expression of clag3.1 appears to restrict the entrance of the toxic compound blasticidin S [22], this result may suggest that the PSAC resulting from clag3.1 expression mediates less efficient solute uptake than the PSAC resulting from clag3.2 expression. However, we found that blasticidin S pressure or growth under culture conditions select for parasites expressing a different clag3 gene in isolates of different genetic background, revealing a more complex scenario. Predominant expression of clag3.2 during clinical malarial infections was the only observation common to all isolates, which suggests that the advantage conferred by clag3.2 expression in this type of infection depends on characteristics unique to the CLAG3.2 protein, such as the conserved sequence feature identified at its N-terminus. On the other hand, phenotypic characteristics that in different isolates are associated with the expression of a different clag3 paralog may depend on nonconserved CLAG3 sequence features that in some isolates occur in CLAG3.1 and, in others, in CLAG3.2. In any case, considering that natural selection only operates on phenotypes, our culture adaptation and blasticidin S selection experiments (together with our previous studies using blasticidin S selection of a culture-adapted line [22]) clearly demonstrate that expression of one or the other clag3 paralog results in phenotypic differences. This is remarkable considering that the 2 CLAG3 proteins have nearly identical sequences. These phenotypic differences are likely linked to infected erythrocyte permeability, although we cannot exclude the possibility that they also involve processes such as cytoadherence or erythrocyte invasion in which CLAG3 proteins may also play a role [14].
Mutually exclusive expression is a phenomenon that affects gene families of utmost importance in several pathogens [34]. In P. falciparum, it has been observed for var [11] and clag3 [15, 16, 18, 19] genes in culture-adapted parasites, although it was found not to be strict: for both gene families, single-cell analysis or strong selection applied to cultures revealed the existence of small parasite subpopulations that do not conform with mutually exclusive expression patterns [20, 22, 35–37]. Considering that the selective pressures operating on parasites in the human blood circulation are different from those under culture conditions, this raises the formal possibility that mutual exclusion may not be the most common pattern in human infections. By focusing only on single infections and using isolate- specific clag3.1 and clag3.2 primers, here we provide evidence for predominant mutually exclusive expression in P. falciparum genes during natural infections. In the majority of isolates, clag3.2 transcript levels were >10-fold higher than clag3.1 levels, although residual expression of the latter was observed in all cases. Residual expression of the silenced paralog is also observed in clonal culture-adapted parasite lines and likely corresponds to small subpopulations of parasites that spontaneously switch the active clag3 at each cycle of multiplication. The existence of these subpopulations of parasites with alternative expression patterns is essential to enable natural selection when changes in host conditions occur.
We compared clag3 expression between blood-stage parasites obtained from infected volunteers and the parental cultured parasite line used for the infections. An analogous approach has been previously used to study the expression of var genes in the context of a human infection, which revealed a reset of the expression patterns of this gene family during transmission stages [38–40]. Here we show that the expression of clag3 genes is also reset. This result strongly suggests that the epigenetic memory for the expression of clag3 genes is erased during gametocyte, mosquito, or liver stages and stochastically reestablished before the onset of a new blood infection, thus providing support to the idea that the epigenetic memory for the expression of clonally variant genes in general is erased during transmission stages, rather than only the epigenetic memory for the peculiar var family. An alternative explanation would be that the reset of clag3 expression depends on selection of parasites expressing a specific clag3 gene during transmission stages, but we consider this an unlikely possibility because such selection seems incompatible with the relatively small parasite population sizes and few multiplications cycles occurring during transmission stages [41]. Furthermore, no function has been described for CLAG3 proteins outside the asexual cycle. The idea that mosquito passage resets the epigenetic patterns for virulence genes has also been proposed for Plasmodium chabaudi [42]. Erasing the epigenetic memory and releasing a transcriptionally diverse population of parasites at the onset of a blood infection is an intuitively advantageous strategy for the parasite to ensure the survival of the population in a new human host with unpredictable conditions.
Altogether, our results support the idea that variant expression of clag3 genes plays an important adaptive role and provide the first insight into how these genes are used under the natural conditions of a human infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank the patients and clinical staff at the ITM, Antwerp University Hospital, and the Medical Research Council Unit, Gambia, for their cooperation; the volunteers and clinical staff who participated in the CHMI in Barcelona; José Muñoz (Barcelona Institute for Global Health [ISGlobal]), for clinical support in the CHMI trial; Alfredo Mayor and Ariel Magallón-Tejada (ISGlobal), for their contribution to the collection of samples for transcriptional analysis in the CHMI study; Stephen L. Hoffman and Kim Lee Sim (Sanaria), for providing cryopreserved P. falciparum sporozoites (for P. falciparum sporozoite challenge); Conor Meehan (ITM), for assistance with phylogenetic analysis; and Jacqueline E. Broerse (Vrije University Amsterdam), for useful discussion and cosupervision of S. M. M.
Financial support. This work was supported by the Spanish Ministry of Economy and Competitiveness (SAF2013-43601-R to A. C.), cofunded by the European Regional Development Fund, European Union; the Secretary for Universities and Research, Department of Economy and Knowledge, Government of Catalonia (2014 SGR 485 to A. C.); the Institute of Tropical Medicine, Antwerp (funding to A. R. U.); ISGlobal is a member of the CERCA Programme, Government of Catalonia; the National Institute of Allergy and Infectious Diseases (R44AI058375 to Sanaria); and the Trans Global Health–Erasmus Mundus Joint Doctorate Programme, European Union (scholarship to S. M. M.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature 2002; 415:673–9. [DOI] [PubMed] [Google Scholar]
- 2. Rovira-Graells N, Gupta AP, Planet E, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res 2012; 22:925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 2008; 62:193–210. [DOI] [PubMed] [Google Scholar]
- 4. Starrfelt J, Kokko H. Bet-hedging–a triple trade-off between means, variances and correlations. Biol Rev Camb Philos Soc 2012; 87:742–55. [DOI] [PubMed] [Google Scholar]
- 5. Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol 2012; 10:e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voss TS, Bozdech Z, Bártfai R. Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol 2014; 20:88–95. [DOI] [PubMed] [Google Scholar]
- 7. Cortés A, Deitsch K. Malaria Epigenetics. Cold Spring Harb Perspect Med 2017; in press, doi:10. 1101/CSHPERSPECT.a025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5’ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol 2007; 66:1296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang L, López-Barragán MJ, Jiang H, et al. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci U S A 2010; 107:2224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crowley VM, Rovira-Graells N, Ribas de Pouplana L, Cortés A. Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol Microbiol 2011; 80:391–406. [DOI] [PubMed] [Google Scholar]
- 11. Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 2008; 62:445–70. [DOI] [PubMed] [Google Scholar]
- 12. Smith JD, Rowe JA, Higgins MK, Lavstsen T. Malaria’s deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol 2013; 15:1976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iriko H, Kaneko O, Otsuki H, et al. Diversity and evolution of the rhoph1/clag multigene family of Plasmodium falciparum. Mol Biochem Parasitol 2008; 158:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta A, Thiruvengadam G, Desai SA. The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction. Drug Resist Updat 2015; 18:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguitragool W, Bokhari AA, Pillai AD, et al. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 2011; 145:665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pillai AD, Nguitragool W, Lyko B, et al. Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol 2012; 82:1104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguitragool W, Rayavara K, Desai SA. Proteolysis at a specific extracellular residue implicates integral membrane CLAG3 in malaria parasite nutrient channels. PLoS One 2014; 9:e93759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Comeaux CA, Coleman BI, Bei AK, Whitehurst N, Duraisingh MT. Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth. Mol Microbiol 2011; 80:378–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cortés A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 2007; 3:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rovira-Graells N, Crowley VM, Bancells C, Mira-Martínez S, Ribas de Pouplana L, Cortés A. Deciphering the principles that govern mutually exclusive expression of Plasmodium falciparum clag3 genes. Nucleic Acids Res 2015; 43:8243–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma P, Wollenberg K, Sellers M, et al. An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem 2013; 288:19429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mira-Martínez S, Rovira-Graells N, Crowley VM, Altenhofen LM, Llinás M, Cortés A. Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell Microbiol 2013; 15:1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaestli M, Cortes A, Lagog M, Ott M, Beck HP. Longitudinal assessment of Plasmodium falciparum var gene transcription in naturally infected asymptomatic children in Papua New Guinea. J Infect Dis 2004; 189:1942–51. [DOI] [PubMed] [Google Scholar]
- 24. Rottmann M, Lavstsen T, Mugasa JP, et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun 2006; 74:3904–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lavstsen T, Turner L, Saguti F, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A 2012; 109:E1791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdi AI, Warimwe GM, Muthui MK, et al. Global selection of Plasmodium falciparum virulence antigen expression by host antibodies. Sci Rep 2016; 6:19882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daily JP, Scanfeld D, Pochet N, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 2007; 450:1091–5. [DOI] [PubMed] [Google Scholar]
- 28. Mackinnon MJ, Li J, Mok S, et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog 2009; 5:e1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vignali M, Armour CD, Chen J, et al. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest 2011; 121:1119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gómez-Pérez GP, Legarda A, Muñoz J, et al. Controlled human malaria infection by intramuscular and direct venous inoculation of cryopreserved Plasmodium falciparum sporozoites in malaria-naïve volunteers: effect of injection volume and dose on infectivity rates. Malar J 2015; 14:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schoepflin S, Valsangiacomo F, Lin E, Kiniboro B, Mueller I, Felger I. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar J 2009; 8:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wampfler R, Mwingira F, Javati S, et al. Strategies for detection of Plasmodium species gametocytes. PLoS One 2013; 8:e76316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sauerwein RW, Roestenberg M, Moorthy VS. Experimental human challenge infections can accelerate clinical malaria vaccine development. Nat Rev Immunol 2011; 11:57–64. [DOI] [PubMed] [Google Scholar]
- 34. Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 2009; 7:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duffy MF, Brown GV, Basuki W, et al. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin sulphate a binding phenotype. Mol Microbiol 2002; 43:1285–93. [DOI] [PubMed] [Google Scholar]
- 36. Joergensen L, Bengtsson DC, Bengtsson A, et al. Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathog 2010; 6:e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merrick CJ, Jiang RH, Skillman KM, et al. Functional analysis of sirtuin genes in multiple Plasmodium falciparum strains. PLoS One 2015; 10:e0118865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peters J, Fowler E, Gatton M, Chen N, Saul A, Cheng Q. High diversity and rapid changeover of expressed var genes during the acute phase of Plasmodium falciparum infections in human volunteers. Proc Natl Acad Sci U S A 2002; 99:10689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang CW, Hermsen CC, Sauerwein RW, Arnot DE, Theander TG, Lavstsen T. The Plasmodium falciparum var gene transcription strategy at the onset of blood stage infection in a human volunteer. Parasitol Int 2009; 58:478–80. [DOI] [PubMed] [Google Scholar]
- 40. Bachmann A, Petter M, Krumkamp R, et al. Mosquito passage dramatically changes var gene expression in controlled human Plasmodium falciparum Infections. PLoS Pathog 2016; 12:e1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinden RE. A biologist’s perspective on malaria vaccine development. Hum Vaccin 2010; 6:3–11. [DOI] [PubMed] [Google Scholar]
- 42. Spence PJ, Brugat T, Langhorne J. Mosquitoes reset malaria parasites. PLoS Pathog 2015; 11:e1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.