Mutation in the mitochondrial HEMN1 gene of Arabidopsis impairs tetrapyrrole biosynthesis in flower buds, which generates ROS and prevents polar cell fusion in the embryo sac and also affects pollen development.
Abstract
Tetrapyrrole biosynthesis is one of the most essential metabolic pathways in almost all organisms. Coproporphyrinogen III oxidase (CPO) catalyzes the conversion of coproporphyrinogen III into protoporphyrinogen IX in this pathway. Here, we report that mutation in the Arabidopsis (Arabidopsis thaliana) CPO-coding gene At5g63290 (AtHEMN1) adversely affects silique length, ovule number, and seed set. Athemn1 mutant alleles were transmitted via both male and female gametes, but homozygous mutants were never recovered. Plants carrying Athemn1 mutant alleles showed defects in gametophyte development, including nonviable pollen and embryo sacs with unfused polar nuclei. Improper differentiation of the central cell led to defects in endosperm development. Consequently, embryo development was arrested at the globular stage. The mutant phenotype was completely rescued by transgenic expression of AtHEMN1. Promoter and transcript analyses indicated that AtHEMN1 is expressed mainly in floral tissues and developing seeds. AtHEMN1-green fluorescent protein fusion protein was found targeted to mitochondria. Loss of AtHEMN1 function increased coproporphyrinogen III level and reduced protoporphyrinogen IX level, suggesting the impairment of tetrapyrrole biosynthesis. Blockage of tetrapyrrole biosynthesis in the AtHEMN1 mutant led to increased reactive oxygen species (ROS) accumulation in anthers and embryo sacs, as evidenced by nitroblue tetrazolium staining. Our results suggest that the accumulated ROS disrupts mitochondrial function by altering their membrane polarity in floral tissues. This study highlights the role of mitochondrial ROS homeostasis in gametophyte and seed development and sheds new light on tetrapyrrole/heme biosynthesis in plant mitochondria.
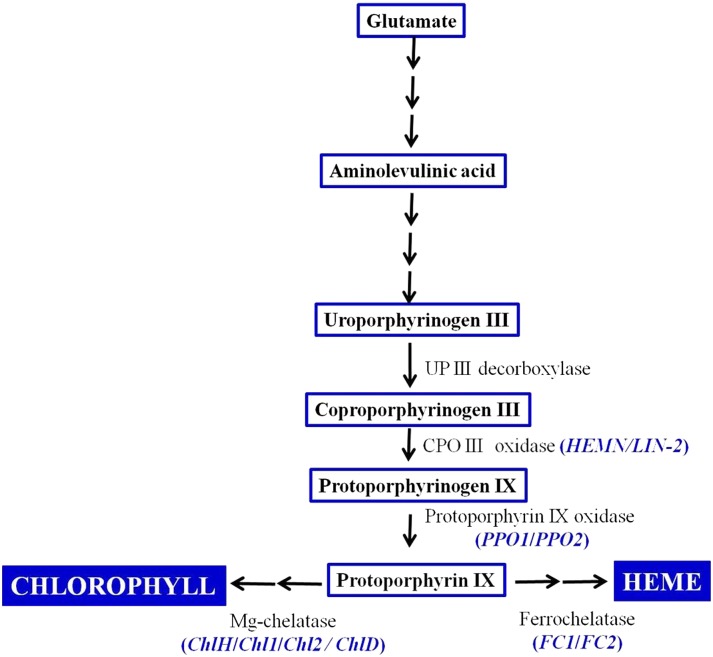
Heme molecules are important components of the electron transport chain that drive aerobic and anaerobic respiration in all organisms and photosynthesis in plants. Heme serves as a prosthetic group of various proteins, such as hemoglobin, myoglobin, cytochrome, and catalase, and also acts as a signaling molecule to coordinate and modulate several molecular and cellular processes ranging from signal transduction to the assembly of protein complexes (Mense and Zhang, 2006). These molecules also are required for the generation of cellular energy, the transport and storage of oxygen, the synthesis and degradation of sterols, lipids, and neurotransmitters, and to control oxidative damage (Sassa and Nagai, 1996; Kumar and Bandyopadhyay, 2005). All organisms including plants share the tetrapyrrole biosynthesis pathway for heme, siroheme, chlorophyll, etc., beginning from Glu via 5-aminolevulinate. This, in turn, is condensed to synthesize a porphyrin ring (Dailey, 1990; Grimm, 1998). This cyclic tetrapyrrole is subsequently modified by three enzymes: uroporphyrinogen III decarboxylase, coproporphyrinogen III oxidase (CPO), and protoporphyrinogen IX oxidase (PPO). CPO (EC 1.3.3.3) catalyzes the oxidative decarboxylation of coproporphyrinogen III (coprogen) to yield protoporphyrinogen IX (protogen). A schematic illustration of the tetrapyrrole/heme biosynthesis pathway is shown in Figure 1. The synthesis and degradation of porphyrins are highly compartmentalized and regulated, as their precursors and breakdown products are extremely cytotoxic. Inactivation of any of the enzymes of the tetrapyrrole biosynthetic pathway leads to the accumulation of porphyrin compounds and causes cell death in plants through reactive oxygen species (ROS) production, as evidenced in lin2, acd1, and acd2 mutants of Arabidopsis (Arabidopsis thaliana; Ishikawa et al., 2001; Tanaka et al., 2003) and the les22 mutant of maize (Zea mays; Hu et al., 1998). Likewise, hereditary coproporphyria, a disease of humans, is attributed to loss of function of the CPO gene (Martásek, 1998).
Figure 1.
Schematic illustration of the tetrapyrrole biosynthesis pathway in plants.
The differentiation and development of male and female gametophytes and genes governing these developmental pathways show a remarkable conservation among angiosperms. In Arabidopsis, the diploid microspore mother cells in the anther undergo meiosis to produce haploid cells. Subsequent rounds of mitosis produce mature pollen grains or male gametophytes that contain two sperm cells and a vegetative cell (McCormick, 2004). Likewise, megaspore mother cells in the ovule undergo meiosis to produce four haploid megaspores. Functional haploid megaspore, after three rounds of mitotic division followed by cellularization, produces a mature seven-celled embryo sac consisting of three antipodals, two synergids, an egg cell, and one central cell (Drews and Yadegari, 2002). Prior to fertilization in Arabidopsis and other species, two polar nuclei, one each from the micropylar end and the chalazal end, migrate to the center and fuse to form a diploid central cell nucleus. During sexual reproduction, one of the sperm cells fertilizes the egg cell and the other fuses with the central cell to produce the embryo and the endosperm, respectively.
Several mutants showing defects in polar nuclei fusion have been identified (Maruyama et al., 2010, 2016). Recent studies have shown that the DIANA/AGAMOUS LIKE61 (AGL61), AGL80 (Portereiko et al., 2006; Bemer et al., 2008; Steffen et al., 2008), and RETINOBLASTAMA RELATED (Johnston et al., 2008, 2010) genes are required for both central cell identity and the fusion of polar nuclei. Furthermore, MAGATAMA3, a gene homologous to the yeast SPLICING ENDONUCLEASE1 involved in RNA metabolism, was found to be essential for polar nuclei fusion and pollen tube guidance (Shimizu et al., 2008). Kägi et al. (2010) reported a slight reduction in polar nuclei size accompanied by the failure of polar nuclei fusion in the fiona mutant. A defect in BiP gene function yields mature female gametophytes with unfused polar nuclei, and the embryonic development is arrested at the globular stage (Maruyama et al., 2010). Likewise, in glauce (Ngo et al., 2007; Leshem et al., 2012) and capulet2 (Grini et al., 2002) mutants where the central cell remains unfertilized, postfertilization embryo development proceeds only up to the globular stage. Schreiber et al. (2004) reported that MADS2 is required for pollen maturation and dehiscence. Recent studies suggest that ROS plays a crucial role in microsporogenesis and megagametogenesis and during embryogenesis in plants (Hu et al., 2011; Martin et al., 2013). ROS homeostasis is regulated through MT-1-4b, where MADS3 acts as a key transcriptional regulator at the late anther development stage in rice (Hu et al., 2011). The expression of MSD1 in the embryo sac restricts central cell fate and modulates ROS homeostasis in Arabidopsis (Martin et al., 2013), establishing the importance of the localization, maintenance, and expression of ROS for proper female gametophyte development.
Here, we report the results of the characterization of a mutation in the Arabidopsis gene At5g63290 that is orthologous to bacterial and mammalian CPO. As this gene shows greater homology with HemN-like CPO, we named it AtHEMN1. Mutations in the AtHEMN1 gene affected the fusion of polar nuclei that led to defects in postfertilization development of the embryo. Disruption of AtHEMN1 function also affected anther/microspore development, leading to nonviable pollen. AtHEMN1 protein was targeted to mitochondria. Impairment of AtHEMN1 function caused the accumulation of coprogen and led to ROS accumulation in developing gametophytes. To the best of our knowledge, this is the first report indicating that the tetrapyrrole/heme biosynthesis pathway operates in mitochondria and that its impairment disturbs ROS homeostasis in flower buds and thereby adversely affects male and female gametophyte development in Arabidopsis.
RESULTS
T-DNA Insertions in the Arabidopsis HEMN1 Gene Cause Seed Sterility
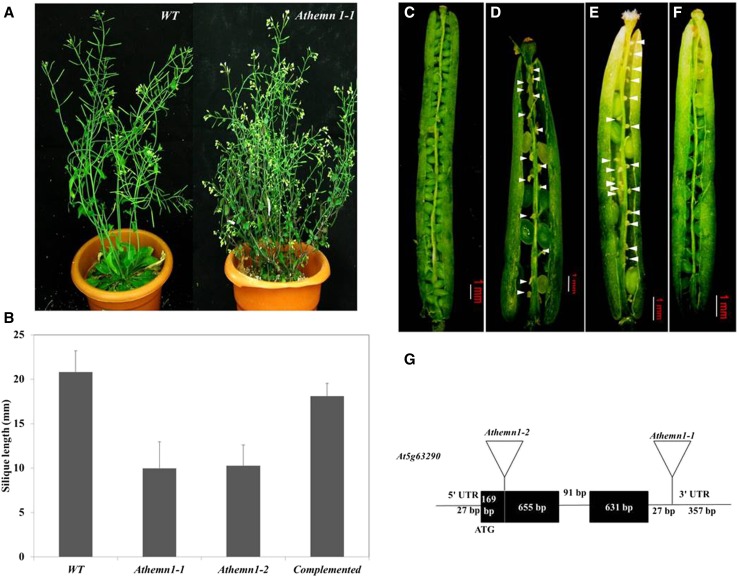
While screening in-house-developed T-DNA insertion mutant lines of Arabidopsis, a mutant with bushy habit and short siliques was identified (Fig. 2A). The mutant had a significantly shorter silique (Fig. 2B) that contained fewer ovules (34 ± 2 versus 46 in the wild type). This mutant, initially named GFP-868, had a low frequency of seed set (Fig. 2D). The mutant plants showed 47.5% (327/688) seed sterility that included 18.8% (130/688) aborted seeds and 28.6% (197/688) aborted ovules compared with the wild-type siliques (Fig. 2C), which had only 1.7% (16/915) aborted seeds and 1% (9/915) aborted ovules (Table I).
Figure 2.
Phenotypes of and T-DNA insertion sites in Athemn1-1 and Athemn1-2 mutants of Arabidopsis. The mutant plant refers to heterozygous plants (+/Athemn1) only, as the homozygous mutant is nonviable. A, Mutant plant (right) showing a bushy habit with short siliques. B, Bar chart showing silique length (mean ± sd) in wild-type (WT), Athemn1-1 and Athemn1-2 mutant, and complemented plants. C, Dissected silique from a wild-type plant showing viable seeds. D, Silique from an Athemn1-1 mutant plant showing aborted ovules (arrowheads). E, Silique from an Athemn1-2 mutant plant showing aborted ovules (arrowheads). F, Silique from an Athemn1-1 plant complemented with the p35S::AtHEMN1 transgene showing almost complete seed set, confirming the rescue of the mutant to the wild-type phenotype. G, Diagrammatic representation showing T-DNA insertion in the 3′ UTR and the first exon of the AtHEMN1 gene in Athemn1-1 and Athemn1-2 mutant lines, respectively.
Table I. Seed sterility in wild-type and Athemn1 mutant plants.
| Plant | Ovules Examined | Ovules with Normal Seeds (%) | Aborted Ovules/Seeds (%) |
|---|---|---|---|
| AtHEMN1/AtHEMN1 (wild-type) | 915 | 890 (97.2) | 25 (2.7) |
| AtHEMN1/Athemn1-1 | 689 | 362 (52.5) | 327 (47.5) |
| AtHEMN1/Athemn1-2 | 722 | 346 (47.9) | 376 (52.06) |
| Complemented AtHEMN1/Athemn1-1 | 884 | 806 (91.2) | 78 (8.8) |
The genome walking approach was used to localize the T-DNA insertion in the mutant (Supplemental Fig. S1A), which identified the T-DNA insertion in the 3′ untranslated region (UTR) of At5g63290 (Supplemental Fig. S1, B and C) that encodes CPO. T-DNA insertion in the GFP-868 mutant is diagrammatically represented in Figure 2G, and the mutant is hereafter referred to as the Athemn1-1 mutant. Another T-DNA mutant line (SALK_100305) having T-DNA insertion in the first exon of AtHEMN1 (at 169 bp downstream of the translation initiation codon ATG) was obtained from the Arabidopsis Biological Resource Center and is referred to here as Athemn1-2. This mutant also showed high degrees of ovule abortion and seed sterility (Table I; Fig. 2E). The locations of the T-DNA insertion and position of primers used for genotyping and genome walking are shown in Supplemental Figure S1D.
The kanamycin-positive progeny of the Athemn1-1 and Athemn1-2 mutant plants were analyzed by PCR to identify and isolate homozygous mutant plants (Supplemental Fig. S2). The PCR results confirmed that both the mutant lines are heterozygous for T-DNA insertion. Additional PCR screening of the progeny also failed to identify homozygous mutant individuals. This strongly suggested the nonviability of one of the gametophytes or homozygous mutant embryos/seeds. As we could not recover any homozygous Athemn1 plants, further characterization of the mutant was carried out with the heterozygous plants only.
Both Male and Female Gametes Transmit the Athemn1 Mutant Allele, albeit at Reduced Frequency
The occurrence of Athemn1 mutant plants in progeny of the mutant but absence of homozygous mutant plants indicated that the mutant allele is transmitted via at least one of the gametes. To determine the same, reciprocal crosses were made between wild-type and Athemn1 mutant plants. When selfed progeny of the mutant were tested for kanamycin reaction, kanamycin-resistant (KanR) and kanamycin-susceptible (KanS) plants were obtained in a 0.87:1 ratio. This significantly deviated from the expected 2:1 ratio assuming the lethality of homozygous mutant progeny. When the mutant plant was used as female and pollinated with wild-type pollen, the F1 progeny showed a KanR:KanS ratio of 0.42:1, whereas in the reciprocal cross the ratio was 0.28:1 (Table II). These results deviated significantly from the expected 0:1 KanR:KanS ratio expected assuming that the Athemn1 mutation is lethal to either of the gametes and confirmed that the mutant allele is transmitted via both male and female gametes. Male transmission via pollen was significantly lower (28.4%) than that from the female side (42.4%; Table II). The transmission frequency of the mutant allele observed in the selfed progeny deviated significantly from the 0.71:1 KanR:KanS ratio calculated based on male and female transmission frequencies observed in backcross progeny. The absence of homozygous mutants in selfed progeny clearly indicated that AtHEMN1 function is essential after fertilization for normal seed development, which is ensured by the wild-type allele in crosses between the mutant and the wild type.
Table II. Transmission efficiency (TE) of Athemn1-1 and Athemn1-2 alleles.
| Cross (Female × Male) | Seedlings | KanR | KanS | Ratio | TE |
|---|---|---|---|---|---|
| % | |||||
| AtHEMN1/Athemn1-1 (self) | 144 | 67 | 77 | 0.87:1 | ND |
| Wild type × AtHEMN1/Athemn1-1 | 217 | 48 | 169 | 0.28:1 | 28.4 |
| AtHEMN1/Athemn1-1 × wild type | 235 | 70 | 165 | 0.42:1 | 42.4 |
| Wild type × AtHEMN1/Athemn1-2 | 220 | 47 | 173 | 0.27:1 | 27.1 |
| AtHEMN1/Athemn1-2 × wild type | 214 | 63 | 151 | 0.41:1 | 41.7 |
ND, Not determined.
Transgenic Overexpression of AtHEMN1 Rescues the Athemn1 Mutation
A complementation study was undertaken to confirm that the observed mutant phenotype is indeed due to the Athemn1 mutation. For this, Athemn1 mutant plants were transformed with the binary plasmid containing the p35S::AtHEMN1-GFP gene cassette and the hygromycin selection marker gene (Supplemental Fig. S3A). T1 progeny were selected on hygromycin and were further tested by PCR for the presence of the Athemn1-1 mutant allele in these plants. A 1.5-kb PCR product confirmed the presence of the mutant allele Athemn1-1 in hygromycin-positive plants (Supplemental Fig. S3B). When these plants were tested for the expression of the AtHEMN1-GFP transgene by reverse transcription (RT)-PCR, the expected 1.8-kb amplicon was detected in inflorescence and silique but no amplicon was found in wild-type plants. All five such complemented plants examined showed normal siliques (Fig. 2, B and C) and nearly full seed set (Table II) comparable with the wild type. These results confirmed that the Athemn1 mutation is the cause of the observed mutant phenotype.
The AtHEMN1 Mutation Affects Male Gametophyte Development
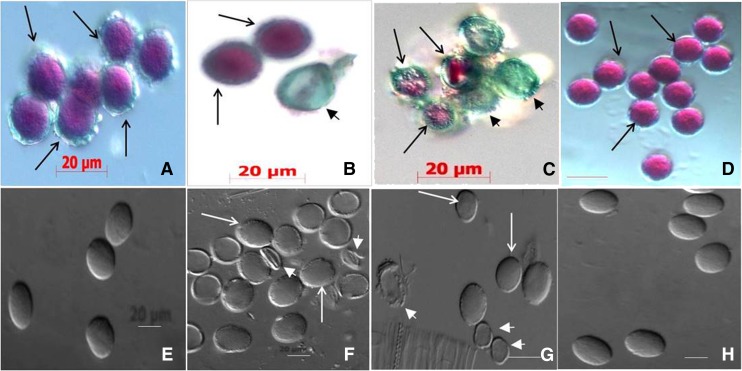
Genetic analysis showed that the Athemn1-1 mutant allele is transmitted at a low frequency from the male side. Therefore, we made a detailed examination of pollen development to identify the nature of the defect in pollen development. Upon Alexander’s staining, 51.4% (n = 1,519) and 49.04% (n = 1,309) pollen grains from Athemn1-1 and Athemn1-2 plants, respectively, did not take any stain and were inferred to be nonviable (Fig. 3, B and C; Table III). On the other hand, pollen of wild-type plants (Fig. 3A) and AtHEMN1-GFP-overexpressing complemented plants (Fig. 3D) showed over 95% viable pollen. DIC microscopic analysis revealed that, compared with the wild type (Fig. 3E), most of the pollen in the mutant plants was collapsed and abnormal in shape (Fig. 3, F and G), whereas pollen of the complemented plants (Fig. 3H) was normal. In mutant plants, morphological defects were observed from pollen development stage 8 (Supplemental Fig. S4). At stage 7, the mutants showed well-developed tapetal layer and normal tetrad similar to wild-type plants (Supplemental Fig. S4, A, F, and K), but at stage 8, the tetrads failed to release the microspores in the mutant (Supplemental Fig. S4, G and L). Even those released from the tetrads failed to form an exine wall (Supplemental Fig. S4, H and M). As a result, collapsed pollen was observed in mature anthers of the mutant plants (Supplemental Fig. S4, I, J, N, and O).
Figure 3.
Assessment of pollen viability by Alexander’s staining (A–D) and differential interference contrast (DIC) microscopy (E–H) of pollen in wild-type and Athemn1 mutant plants. A, Pollen from a wild-type plant showing nearly 100% viable grains (pink/purple). B, Pollen from an Athemn1-1 plant showing both viable (pink) and nonviable (green) grains. C, Pollen from an Athemn1-2 plant showing both viable (pink) and nonviable (green) grains. D, Pollen from a complemented plant showing nearly 100% viable grains (pink). E, Pollen from a wild-type plant showing a normal elliptical shape by DIC microscopy. F, Pollen from an Athemn1-1 mutant plant showing collapsed, empty grains (arrowheads). G, Pollen from an Athemn1-2 mutant plant showing collapsed, empty grains (arrowheads). H, Pollen from a complemented plant showing normal wild-type-like grains.
Table III. Pollen viability in Athemn1 mutants of Arabidopsis.
| Plant | Total Pollen Examined | Viable Pollen | Nonviable Pollen | Percentage Viability |
|---|---|---|---|---|
| AtHEMN1/AtHEMN1 (wild type) | 2,321 | 2,208 | 113 | 95.13 |
| AtHEMN1/Athemn1-1 | 1,519 | 781 | 738 | 51.40 |
| AtHEMN1/Athemn1-2 (SALK_100305) | 1,309 | 642 | 667 | 49.04 |
| Complemented AtHEMN1/Athemn1-1 | 1,139 | 1,100 | 39 | 96.57 |
The AtHEMN1 Mutation Prevents the Fusion of Polar Nuclei in the Female Gametophyte and Affects Endosperm Proliferation
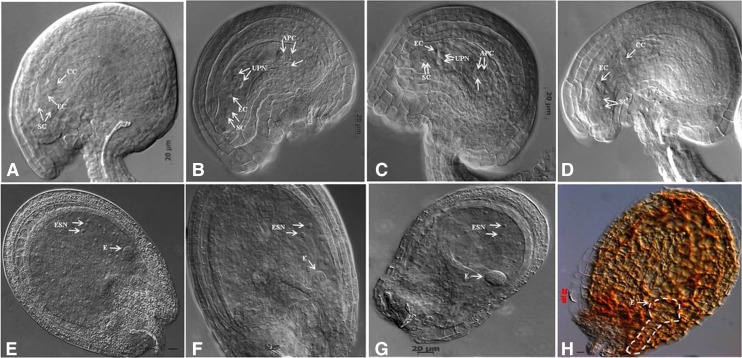
To find the exact defects in female gametophyte development, the tissue-cleared ovules from Athemn1-1 and Athemn1-2 mutants were examined with a DIC microscope. At anthesis, almost all ovules in the wild type contained one central cell nucleus, one egg cell nucleus, and two synergid cell nuclei (Fig. 4A). In both the Athemn1 mutant flowers at this stage, 47.6% to 58.1% (40/84 in Athemn1-1 and 68/117 in Athemn1-2) of ovules displayed a normal wild-type-like phenotype, whereas 40.04% to 38.46% (34/84 in Athemn1-1 and 45/117 in Athemn1-2) of ovules showed a pair of unfused polar nuclei (Fig. 4, B and C; Table IV). Furthermore, 12% (10/84) and 3.4% (4/117) of Athemn1-1 and Athemn1-2 ovules, respectively, showed arrest at the FG1, FG2, or FG4 stage of female gametophyte development (Table IV). In transgene-complemented mutant plants, 94.39% (101/107) of ovules appeared normal (Fig. 4D). These data indicated that the Athemn1 mutation mainly impairs the fusion of the polar nuclei to form the central cell nucleus of the embryo sac. In contrast to the wild type, the Athemn1 embryo sac at stage FG7/8 had comparatively smaller egg cells and persistent antipodal cells (Fig. 4, B and C).
Figure 4.
Abnormalities in prefertilized and postfertilized ovules of Athemn1 mutants of Arabidopsis. A, Ovule from a wild-type plant at anthesis showing a fully differentiated embryo sac at the FG7 stage. B, An Athemn1-1 mutant ovule at a comparable stage showing unfused polar nuclei in the center and nondegenerated antipodals at the chalazal end. C, An Athemn1-2 mutant ovule at a comparable stage showing unfused polar nuclei at the center and nondegenerated antipodals at the chalazal end. D, An Athemn1-1 mutant ovule after complementation with the cDNA of the AtHEMN1 gene showing a large fused polar nucleus. E, Postfertilized ovule of a wild-type plant showing a preglobular stage embryo and proliferating endosperm nuclei. F and G, Postfertilized ovules from the Athemn1-1 mutant at a comparable stage to the wild type showing an abnormal preglobular stage embryo and poorly proliferating endosperm nuclei. H, Aborted seed in a mature silique from an Athemn1-1 mutant showing an embryo arrested at the globular stage (the degenerated globular embryo is marked with dotted lines). APC, Antipodal cell; CC, central cell; E, embryo; EC, egg cell; ESN, endosperm nucleus; SC, synergid cell; UPN, unfused polar nuclei.
Table IV. Effects of the AtHEMN1 mutation on ovule development in Arabidopsis.
| Plant | Stage of Ovule Development at Anthesis |
Total Ovules Examined | ||||||
|---|---|---|---|---|---|---|---|---|
| FG7/8 | FG6 | FG5 | FG4 | FG3 | FG2 | FG1 | ||
| AtHEMN1/AtHEMN1 (wild type) | 115 | 2 | 0 | 0 | 0 | 2 | 1 | 120 |
| AtHEMN1/Athemn1-1 | 40 | 34 | 0 | 4 | 0 | 4 | 2 | 84 |
| AtHEMN1/Athemn1-2 (SALK_100305) | 68 | 45 | 0 | 2 | 0 | 1 | 1 | 117 |
To define the stage and nature of seed abortion in the mutant, whole mounts of cleared ovules sampled at different times after fertilization were examined with a DIC microscope. No differences were discernible at the zygote stage of development between the wild type and the mutant. The defects in seed development became evident at the early globular stage (Fig. 4, E–G). A number of prominent doughnut-shaped nuclei without any cellularization and with defective embryos were observed in the center of the embryo sac of mutant ovules (Fig. 4F). Some of the ovules showed a lack of proper endosperm proliferation where developing embryos were arrested at the globular stage (Fig. 4G). In the Athem1-1 mutant, about 28% (32/115) of the aborting seeds had embryos arrested at the globular stage (Fig. 4H), whereas in 58% (67/115) of aborting seeds, embryo development proceeded beyond the globular stage and about 31% reached the torpedo or cotyledon stage (Table V).
Table V. Frequency of embryo arrest at different stages of development in the Arabidopsis Athemn1-1 mutant.
Values in parentheses are total ovules present in the silique.
| Stage of Embryo | Silique No. |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Preglobular stage | 3 | 0 | 1 | 3 | 0 | 2 | 0 | 0 | 9 |
| Globular stage | 6 | 0 | 9 | 0 | 2 | 2 | 2 | 2 | 23 |
| Initial heart stage | 4 | 5 | 7 | 0 | 0 | 0 | 0 | 0 | 16 |
| Heart stage | 4 | 11 | 14 | 0 | 1 | 0 | 0 | 1 | 31 |
| Torpedo stage | 0 | 0 | 0 | 4 | 13 | 0 | 0 | 0 | 17 |
| Cotyledon stage | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 10 | 19 |
| Total | 17 (34) | 16 (34) | 31 (35) | 7 (35) | 16 (34) | 8 (32) | 7 (35) | 13 (33) | 115 (272) |
Cell Specification Is Not Affected in Athemn1-1 Mutant Embryo Sacs
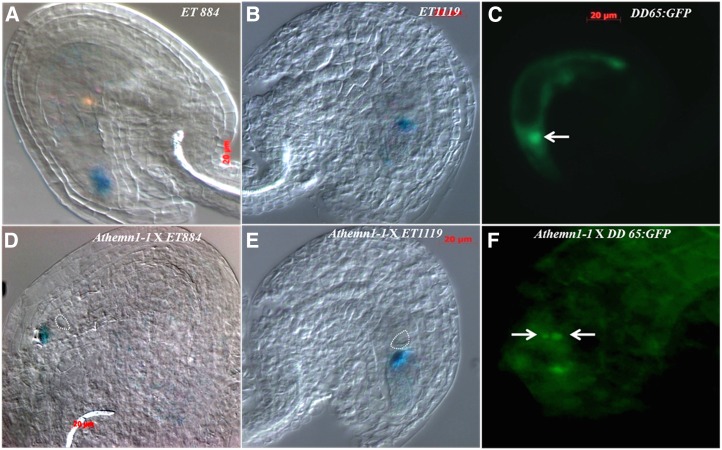
To gain further understanding of the defects in female gametophyte development, the expression of embryo sac cell-specific molecular markers was analyzed in the mutant background. The mutant line Athemn1-1 was crossed separately with different cell-specific marker lines, namely ET884 (synergid cell specific), ET1119 (egg cell specific), and DD65:GFP (central cell specific). The F1 plants carrying the Athemn1-1 mutant allele were identified, and the ovules were examined for GUS or GFP expression (Fig. 5). Since the gynoecium of the heterozygous mutant plant (+/Athemn1-1) contains both normal and defective ovules, we tested the expression of marker genes in defective ovules. GUS expression (conferred by the marker ET884) was observed in synergids of 42.05% (n = 140) of ovules (Fig. 5, A and D). Similarly, the marker ET1119 showed GUS expression in egg cells of 40.3% (n = 176) of ovules (Fig. 5, B and E). These results suggest that both egg cells and synergid cells are properly specified in ovules carrying the Athemn1-1 mutant allele. Expression of the marker DD65:GFP was checked in wild-type and Athemn1-1 ovules. While wild-type ovules with fused polar nuclei showed GFP expression restricted to the central cell of the embryo sac (Fig. 5C), those with unfused polar nuclei displayed green fluorescence in the two polar nuclei of the embryo sacs (Fig. 5F). Thus, the polar nuclei appear to have acquired central cell specification but failed to form a diploid central cell nucleus due to the Athemn1-1 mutation.
Figure 5.
Expression patterns of different embryo sac cell-specific marker genes in wild-type (A–C) and Athemn1-1 mutant (D–F) plants. An Athemn1-1 mutant plant was separately crossed with ET884 (synergid cell-specific), ET1119 (egg cell-specific), and DD65:GFP (central cell-specific) marker lines. F1 plants carrying the Athemn1-1 allele were isolated. Ovules showing defects in polar nuclei fusion were examined for reporter gene (GUS or GFP) expression. A, A wild-type ovule showing ET884 marker expression (blue GUS stain) in the micropylar region corresponding to the synergid cells. B, A wild-type ovule showing ET1119 marker expression (blue GUS stain) in the embryo sac corresponding to the location of the egg cell. C, A wild-type ovule showing DD65:GFP marker expression (green GFP fluorescence) in the central cell (arrow). D, An Athemn1-1 mutant ovule showing ET884 marker expression (blue GUS stain) at the micropylar region corresponding to the location of the synergid cells. Unfused polar nuclei are marked with the dotted circle. E, An Athemn1-1 mutant ovule showing ET1119 marker expression (blue GUS stain) in the embryo sac corresponding to the location of the egg cell. Unfused polar nuclei are marked with the dotted circle. F, An Athemn1-1 mutant ovule showing DD65:GFP marker expression (green GFP fluorescence) in unfused polar nuclei (arrows).
ROS Accumulates around the Central Cell in the Female Gametophytes
The AtHEMN1 gene product is expected to play a role in the porphyrin biosynthetic pathway, which is a bifurcated pathway of chlorophyll biosynthesis (Fig. 1). Available information shows that any defect affecting the synthesis of porphyrins would result in the accumulation of ROS (Busch and Montgomery, 2015). To investigate whether disruption of the AtHEMN1 gene leads to ROS accumulation, nitroblue tetrazolium (NBT) staining of anthers and ovules of wild-type and Athemn1-1 mutant plants was tested. No NBT staining was found in mature anthers and pollen of the wild type (Fig. 6A), whereas the mutant anthers showed strong NBT staining of pollen grains (Fig. 6, B and C). In ovules, no NBT staining was detected in the mutant or the wild-type until the FG5 stage. However, at the FG6-7 stage, dark NBT staining was detected in Athemn1-1 ovules around the unfused polar nuclei and in the micropylar region (Fig. 6, E and F), whereas in the wild type, faint staining was observed in the micropylar region only (Fig. 6D). Some of the ovules of Athemn1-1 that were collapsed (∼32%) showed staining in the entire embryo sac (Supplemental Fig. S5). Thus, NBT staining showed that disruption of AtHEMN1 function leads to high ROS production in developing gametophytes.
Figure 6.
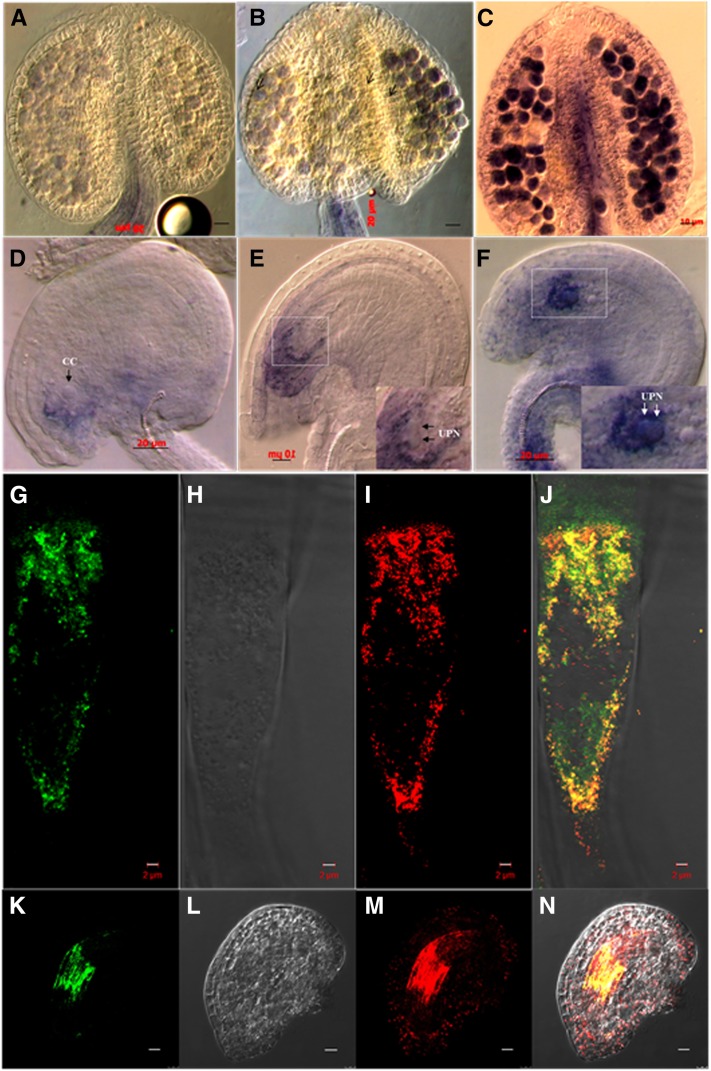
Patterns of ROS accumulation in anther (A–C) and ovule (D–F) of wild-type and Athemn1 mutant plants of Arabidopsis visualized through NBT staining. A and D, Anther (A) and ovule (D) from a wild-type plant showing no or faint NBT staining (blue) in mature pollen and toward the micropylar end in the ovule. B and E, Anther (B) and ovule (E) from an Athemn1-1 plant showing NBT staining (blue) in pollen and around the egg cell and the central cell (CC) in the ovule. The inset in E shows a magnified view of the embryo sac with unfused polar nuclei (UPN; arrows). C and F, Anther (C) and ovule (F) from an Athemn1-2 plant showing intense NBT (blue) staining in the pollen and around the egg cell and the central cell in the ovule. The inset in F shows a magnified view of the embryo sac with unfused polar nuclei (arrows). G to N, Subcellular localization of the AtHEMN1-GFP fusion protein. Root cells and ovules of an Athemn1-1 mutant plant complemented with the AtHEMN1-GFP fusion construct were examined by confocal microscopy after staining with MitoSOX Red. G, GFP fluorescence in a root cell overexpressing AtHEMN1-GFP. H, DIC image of the cell shown in G. I, MitoSOX Red fluorescence of the cell shown in G. J, Merged image of G to I showing colocalization (yellow) of GFP and MitoSOX Red signals, confirming that AtHEMN1 is targeted to the mitochondria. K, GFP fluorescence in an ovule overexpressing AtHEMN1-GFP. L, DIC image of the ovule shown in K. M, MitoSOX Red fluorescence of the ovule shown in K. N, Merged image of K to M showing colocalization (yellow) of GFP and MitoSOX Red signals corresponding to the mitochondrial cloud around the central cell.
AtHEMN1 Is Targeted to Mitochondria
As per TAIR annotation, At5g63290 has two exons (824 and 631 bp) and a 91-bp intron and is capable of encoding a 484-amino acid polypeptide. Furthermore, its transcript has a 27-bp 5′ UTR and a 384-bp 3′ UTR. The polypeptide sequence possesses a mitochondrial targeting signal, MLKTTISPIFSSFTGKPKCSSKLFFRAFSKVVLQDTPPSARRN, at the N-terminal end (Supplemental Fig. S6). A phylogenetic tree of CPO proteins prepared from different organisms like bacteria, cyanobacteria, mammals, and plants, including Arabidopsis, is shown in Supplemental Figure S7 and Supplemental Table S1. On the basis of homology, CPO proteins were found to be conserved across different kingdoms. To determine the similarity of AtHEMN1 with the two available analogs of CPOs (i.e. hemF and hemN), amino acid sequences were compared using homology search. The AtHEMN1 protein showed 44% similarity with plastid-localized hemF CPO and 91% similarity with mitochondria-localized hemN CPO (Fig. 7A; Supplemental Table S2). It also has three Elongator protein (Elp) domains and a C-terminal hemN domain (Fig. 7B). The Elp domain shares significant sequence homology with the radical S-adenosyl-Met superfamily. Members of this family of bacterial proteins contain an FeS cluster and use S-adenosyl-Met to catalyze a variety of radical reactions (Goto et al., 2010).
Figure 7.
Comparison of the AtHEMN amino acid sequence with CPO polypeptides of Arabidopsis and other plant species. A, Alignment of AtHEMN1 (At5g63290) with CPOs (hemF) of Arabidopsis (i.e. LIN2 and HEMF2). AtHEMN1 shows low homology with hemF. B, Alignment of AtHEMN1 (At5g63290) with hemN polypeptides of Camelina sativa, Brassica rapa, Citrus, Oryza sativa, Glycine, Ricinus communis, and maize. Identical residues are marked in gray. The Elp3 and hemN domains are marked with thick lines and labeled. AtHEMN1 shows high homology with the known hemN sequences.
Tetrapyrrole biosynthesis in plants is known to occur in the plastids. However, AtHEMN1 contains a mitochondrial targeting signal. Therefore, the cellular localization of AtHEMN1 was tested in Athemn1-1 mutant plants complemented with the 35S::AtHEMN1-GFP gene construct. To localize the mitochondria, tissue was stained with MitoSOX Red, a mitochondrial superoxide indicator that selectively moves into mitochondria, becomes oxidized, and turns fluorescent (Robinson et al., 2006). Stained root tip tissues were examined for GFP and MitoSOX Red fluorescence. A strong overlap was observed between AtHEMN1/GFP and MitoSOX Red signals (Fig. 6, G–J). A similar type of colocalization was observed in the mitochondrial cloud around the central cell of the mature embryo sac (Fig. 6, K–N). These results confirmed the targeting of AtHEMN1 to mitochondria.
Mitochondrial Function Is Compromised in the Female Gametophytes of the Athemn1 Mutant
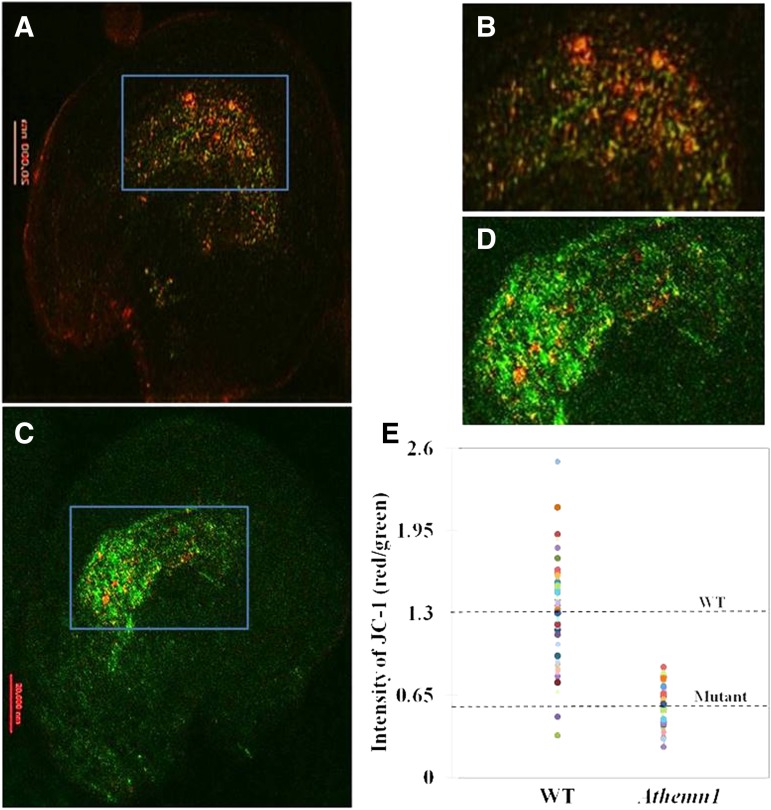
Documentation of the unfused polar nuclei phenotype by DIC and strong NBT staining around these unfused polar nuclei in the mutant clearly establish the critical role of AtHEMN1 function for polar cell fusion during gametophyte development. These results also confirmed that the Athemn1 mutation has disturbed ROS homeostasis in both anthers and ovules and led to defects in pollen and embryo sac development. Since AtHEMN1 encodes an enzyme involved in tetrapyrrole biosynthesis, it is likely that this accumulation of ROS is a consequence of disruption in the tetrapyrrole biosynthesis in the mitochondria, which ultimately affects the functionality of these mitochondria around the central cell in this mutant. Interestingly, the fiona and oiwa mutants with defects in polar nuclei fusion also display altered mitochondrial ultrastructure in the central cell (Kägi et al., 2010; Martin et al., 2013). To verify the status of mitochondrial function within the embryo sac, we used JC-1 dye to monitor the mitochondrial membrane potential. JC-1 is a lipophilic dye that selectively enters into mitochondria and reversibly changes color from green to red as the membrane potential increases (Martin et al., 2013). In normal cells showing high mitochondrial membrane potential, JC-1 spontaneously forms complexes giving intense red fluorescence. On the other hand, in cells having mitochondria with low membrane potential, the dye remains in the monomeric form and exhibits green fluorescence only (Wang et al., 2010; Martin et al., 2013). Ovules of the Athemn1-1 mutant and wild-type plants were stained with JC-1 dye and examined for its fluorescence to assess the functionality of the mitochondria within the embryo sac. Embryo sacs of wild-type plants showed comparatively more red fluorescence than green (Fig. 8, A and B), whereas in mutant embryo sacs, green fluorescence was predominant over red (Fig. 8, C and D). The ratio of red to green fluorescence in wild-type embryo sacs was about 1.3 (Fig. 8E), whereas it was significantly lower (i.e. 0.58) in the Athemn1-1 mutant. These results confirmed that the embryo sacs of Athemn1 mutants have dysfunctional mitochondria around the central cell because of the accumulated ROS, which induces the damage to the organelle.
Figure 8.
Determination of the membrane potential of mitochondria in wild-type and Athemn1 embryo sacs using confocal microscopy following JC-1 dye staining. A, Image of a wild-type ovule stained with JC-1 dye showing reddish fluorescence in the central cell region of the embryo sac. B, Enlarged view of the rectangular area in A. C, Image of an Athemn1-1 ovule stained with JC-1 dye showing green fluorescence in the central cell region of the embryo sac. D, Enlarged view of the rectangular area in C. E, Dispersion graph depicting the red-green JC-1 fluorescence ratio values recorded in wild-type (WT) and Athemn1 mutant embryo sacs. The horizontal dotted lines indicate mean values for wild-type and Athemn1-1 embryo sacs.
AtHEMN1 Is Expressed Mainly in Anthers, Ovules, and Endosperm of Developing Seeds
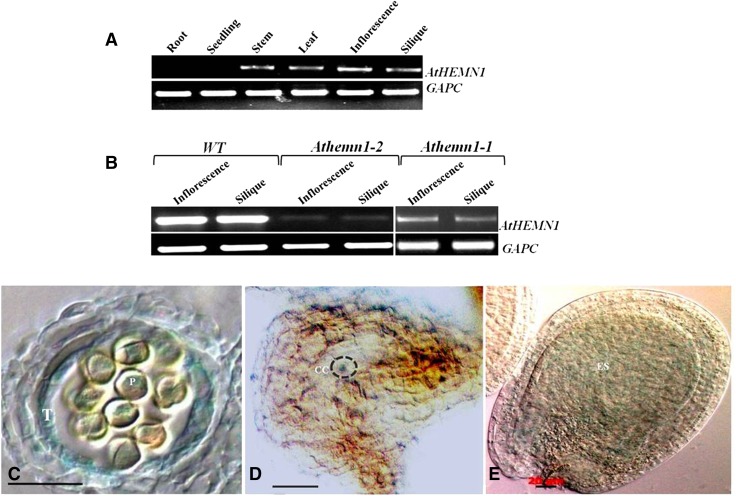
The Arabidopsis eFP browser database (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) shows that AtHEMN1 is expressed in all plant parts, with the highest expression in dry and imbibed seeds, whereas the GENVESTIGATOR (Zimmermann et al., 2004) expression pattern shows the highest expression of AtHEMN1 in pollen and seed. RT-PCR expression analysis confirmed the presence of AtHEMN1 transcripts in stems, leaves, inflorescences, and siliques, whereas no transcripts were detected in roots and seedlings of wild-type plants (Fig. 9A). However, AtHEMN1 transcript levels were reduced drastically in the mutant plants (Fig. 9B). Since mutant plants are heterozygous, these results indicate that haploinsufficiency of AtHEMN1 leads to the observed mutant phenotype.
Figure 9.
Assessment of the expression pattern of the AtHEMN1 gene by RT-PCR and reporter gene assay. A, RT-PCR analysis of the AtHEMN1 gene in different tissues of a wild-type plant (the bottom gel shows amplification of the GAPC transcript used as a control). B, RT-PCR analysis of the AtHEMN1 gene in inflorescences and siliques of wild-type (WT) and Athemn1-1 and Athemn1-2 mutant plants (the bottom gel shows amplification of the GAPC transcript used as a control). C to E, Histochemical GUS assay of tissues from the transgenic plant carrying the pAtHEMN::uidA construct. C, Cross section of an anther showing GUS expression in tapetum (T) and microspores (P). D, Cross section of an FG7-stage ovule showing GUS expression in the central cell (CC) region of the mature embryo sac. E, Developing seed showing GUS expression in the endosperm (ES).
To determine the temporal and spatial expression patterns of the AtHEMN1 gene, a 1.2-kb fragment upstream of the AtHEMN1 translation start site was PCR amplified and cloned upstream of the reporter gene uidA in PORE-R2 vector and used to transform wild-type Arabidopsis plants. GUS expression was examined in five independent transgenic lines. Weak GUS expression was observed in stems, leaves (trichomes), inflorescences, and developing seeds (Supplemental Fig. S8). GUS activity was detected specifically in anthers. In anthers, GUS expression was found in the tapetum, developing microspores (Fig. 9C), and mature pollen. GUS activity also was found in the central cell region of the mature embryo sacs of the ovules and in the endosperm of the developing seeds (Fig. 9, D and E). Thus, based on in silico data of transcript abundance in different tissues and promoter activity determined using GUS expression, we infer that the AtHEMN1 gene is expressed primarily in anther, embryo sac, and endosperm during seed maturation.
The Tetrapyrrole Biosynthesis Pathway Is Disturbed in Athemn1 Mutants
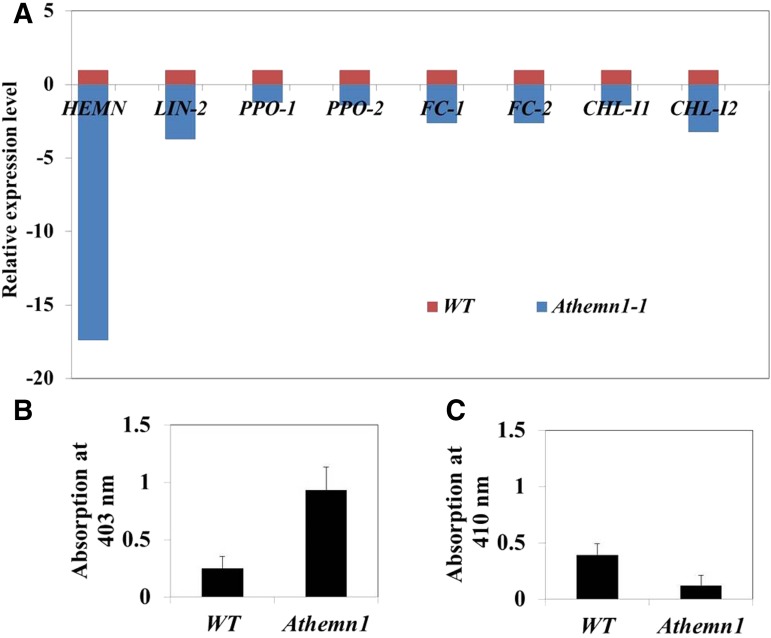
To determine whether the Athemn1 mutation causes disturbance in the tetrapyrrole pathway, we assessed the expression levels of different genes of the pathway, namely coproporphyrinogen oxidase (AtHEMN1 [At5g63290] and LIN2 [At1g03475]), protoporphyrinogen IX oxidase (PPO1 [At5g14220] and PPO2 [At4g01690]), ferrochelatase (FC1 [At5g26030] and FC2 [At2g30390]), and Mg-chelatase (CHL-I1 [At4g18490] and CHL-I2 [At5g45930]). Quantitative reverse transcription (qRT)-PCR analysis using RNA samples from the inflorescence revealed 17-fold down-regulation of AtHEMN1 transcripts in the mutant (Fig. 10A). The plastidial CPO gene (LIN2) also showed 3.7-fold down-regulation. Likewise, FC1, FC2, and CHL-I2 also showed 2- to 3-fold down-regulation in the Athemn1-1 mutant. In contrast, the expression of PPO1, PPO2, and CHL-I1 in the mutant was comparable to that in the wild type. These results show that the Athemn1 mutation affects the expression of other downstream genes of the pathway. To further understand the consequences of these gene expression changes on the accumulation of products of the pathway, relative levels of coproporphyrin and protoporphyrin were estimated in the mutant and wild-type plants through spectrophotometry (Supplemental Fig. S9). A 4-fold increase in coproporphyrin level was found in ovules of Athemn1-1 mutant plants compared with the wild type (Fig. 10B), whereas the protoporphyrin level was reduced by 3-fold (Fig. 10C). Thus, the AtHEMN1 mutation disrupted the tetrapyrrole/heme biosynthesis pathway and caused the accumulation of intermediates.
Figure 10.
Expression analysis of genes of the tetrapyrrole biosynthesis pathway in the Athemn1-1 mutant by qRT-PCR. A, Relative expression analysis of different genes in inflorescences of the Athemn1-1 mutant taking expression level in the wild type (WT) as the calibrator. CPO1, LIN2, FC1, FC2, and CHL-I2 showed significant down-regulation in the Athemn1-1 mutant, whereas the expression of PPO1, PPO2, and CHL-I1 was comparable to that of the wild type. B and C, Relative quantification of tetrapyrrole biosynthesis pathway intermediates in inflorescences of wild-type and Athemn1-1 mutant plants. Accumulated intermediates were extracted from the inflorescence, and their quantities were estimated using spectrometry. Values indicate means ± sd of three biological replicates. B, Bar chart showing a significantly higher amount of coproporphyrin in the Athemn1-1 mutant compared with the wild type. C, Bar chart showing a significantly lower level of protoporphyrin in the Athemn1-1 mutant compared with the wild type.
DISCUSSION
The differentiation and development of male and female gametophytes is governed by ROS (Martin et al., 2013). Analyzing oiwa mutants of Arabidopsis, Martin et al. (2013) reported that mitochondrial MnSOD modulation of ROS plays a key role in female gametophyte development. Similarly, nuclear gene mutations affecting mitochondrial functions such as transcription, protein synthesis, and oxidative phosphorylation have been shown to affect polar nuclei fusion (Maruyama et al., 2016). This study shows, to our knowledge for the first time, that disturbance of the heme biosynthesis pathway leads to increased ROS accumulation in developing gametophytes, thereby adversely affecting gametophyte and seed development. Furthermore, our study sheds new light on tetrapyrrole biosynthesis in plant mitochondria.
The T-DNA insertion mutant in the AtHEMN1 locus described in this study was identified as a mutant with short siliques, fewer ovules, and high frequency of pollen abortion and seed sterility. Failure of the recovery of homozygous mutant plants and the appearance of a mutant phenotype in plants heterozygous for the mutation suggest that it is an essential gene displaying a dosage effect. Furthermore, qRT-PCR results showed a drastic reduction in AtHEMN1 transcripts in the heterozygous mutant plants. This could possibly also be due to an RNA interference effect arising from the instability of the mutant transcript. Similar effects have been reported for uroporphyrinogen III decarboxylase mutants in maize (Hu et al., 1998) and human (Moore, 1993). Despite the nonrecovery of homozygous mutant plants, mutant alleles were transmitted from both the male and female sides. Furthermore, the mutant allele was less efficiently transmitted from the male side. The heterozygous mutant plants produce pollen that will carry either wild-type or mutant alleles. Therefore, the low-frequency transmission of the mutant allele via pollen could be attributed either to poor viability or the reduced competitive ability of pollen carrying the mutant allele. The AtHEMN1 promoter was found to drive uidA expression in anthers, ovules, and endosperm. Thus, the above results point to the critical role of AtHEMN1 during gametophyte and seed development and account for the absence of homozygous mutant progeny.
The examination of developing anthers showed an impairment of microspore release from the tetrad. Furthermore, nearly 50% of the pollen was nonviable. Mutant studies have demonstrated that loss-of-function HEMN mutant plants accumulate coprogen, which triggers a series of uncontrolled reactions and generates ROS (Smith, 1987; Ishikawa et al., 2001). NBT staining of anthers of Athemn1-1 also showed a large number of deeply stained pollen grains compared with the wild type. Thus, the Athemn1-1 mutation appears to have affected ROS equilibrium in anthers. Recently, Zafra et al. (2012) reported that SOD regulates ROS homeostasis during the late tetrad/early microspore stage and in mature pollen to produce viable pollen. In a separate study, Matveyeva et al. (2012) revealed the role of ROS in the deposition of sporopollenin on the microspore wall. The anthers with disturbed ROS released microspores that were smaller and shrunken and that collapsed after entering the mitotic phase. Our histological observations of anther development support the above findings and show that AtHEMN1 disruption affecting microspore development and pollen maturation is accompanied by elevated ROS production.
The Athemn1-1 mutation affected the fusion of polar nuclei during female gametophyte development. The expression of embryo sac cell-specific markers in the Athemn1-1 background showed that egg and synergid cells are normal in the mutant embryo sacs. Although unfused polar nuclei of the mutant embryo sac showed expression of the central cell-specific marker DD65:GFP at anthesis, the staining pattern deviated markedly from that of the wild type. These results stress the need for a cell-autonomous synthesis of heme in the central cell. A high level of ROS is characteristic of the central cell and is regulated by mitochondrial MnSOD (Martin et al., 2013). Interestingly, the central cell is enriched with mitochondria, whereas the egg cell shows relatively fewer mitochondria (Kägi et al., 2010). Loss of HEMN function in Athemn1 mutant embryo sacs resulted in even higher ROS accumulation around the central cell and in the micropylar region. This might account for the observed mutant phenotypes in Athemn1. The Arabidopsis fiona mutant defective in mitochondrial cysteinyl t-RNA synthetase has deformed mitochondrial cristae and shows a failure of polar nuclear fusion (Kägi et al., 2010). Similarly, ectopic expression of the dominant aac2A199D allele in the central cell led to the deformation of mitochondrial cristae and the failure of polar nuclear fusion (Kägi et al., 2010). Thus, a defect in mitochondrial function in the embryo sac appears to hinder the fusion of polar nuclei. In Athemn1 mutants, the disruption of tetrapyrrole synthesis seems to trigger ROS production and thereby impair mitochondrial function. These results attest to the need for the proper functioning of mitochondria in and around the central cell to ensure polar nuclear fusion and would explain the observed defects in polar nuclear fusion in Athemn1 mutants. Mutants with unfused polar nuclei often show defects in egg and antipodal cells (Kägi et al., 2010; Krohn et al., 2012; Wu et al., 2013). This implies cross talk among the egg cell, the central cell, and the synergids. Intercommunication among the cells of the embryo sac also was reported in the fiona/syco-1 mutants (Wu et al., 2013). Athemn1 mutant ovules also displayed a similar kind of phenotype, namely, small egg cell and persistent antipodals, which suggests that genes responsible for these mutants operate in a common developmental pathway.
Biochemical and molecular studies have established that tetrapyrrole biosynthesis in plants occurs in the chloroplasts (Joyard et al., 2009; Tanaka et al., 2011). This is in contrast to animals, where tetrapyrrole biosynthesis leading to heme production occurs sequentially in mitochondria (up to 5-aminolevulinic acid), cytoplasm (5-aminolevulinic acid to coprogen), and mitochondria (Yin and Bauer, 2013). Chlorophyll and heme, the main tetrapyrroles found in plants, share a common pathway until protoporphyrin IX (Fig. 1). The Arabidopsis At1g03475 (LIN2) gene has been identified to code for the CPO enzyme of the protoporphyrin IX synthesis pathway, and this protein is targeted to chloroplasts (Ishikawa et al., 2001). The lin2 mutant of Arabidopsis displays the characteristic ROS-induced lesions on leaves and siliques (Ishikawa et al., 2001), confirming the impairment of tetrapyrrole synthesis in the plastids. Another CPO-coding gene in Arabidopsis is At4g03205, annotated as chloroplast-localized hemf2 (uncharacterized). Thus, there are two plastid-localized CPO-coding genes (At4g03205 and At1g03475) in Arabidopsis. Homology analysis revealed that AtHEMN1 shares 91% and 44% similarity (amino acid level) to the mitochondrial and plastidic isoforms, respectively, of CPOs of different organisms (Fig. 7). Furthermore, a domain search analysis of the AtHEMN protein revealed a mitochondrial targeting sequence at the N-terminal end (Supplemental Fig. S6). Therefore, we tested the subcellular localization of this protein. Our confocal colocalization study of the AtHEMN1:GFP fusion protein and MitoSOX Red confirmed that the AtHEMN1 gene product is indeed targeted to mitochondria. PPO is targeted to both chloroplasts and mitochondria in tobacco (Nicotiana tabacum; Lermontova et al., 1997) and spinach (Spinacia oleracea; Watanabe et al., 2001). Williams et al. (2006) have reported two isoforms of CPO in maize encoded by CPX1 and CPX2 genes. CPX1 was targeted to plastids, whereas CPX2 was localized in the mitochondria. In agreement with these observations, homozygous cpx1 mutants were albino and developed necrotic lesions under light, but no clear phenotypes were observed in homozygous cpx2 mutants. Our results strongly support the findings of Williams et al., (2006) that duplicated CPO genes have undergone subfunctionalization in plants during evolution.
Considering that Arabidopsis carries a single gene, At2g26540, that codes for uroporphyrinogen III synthase, which is targeted to plastids (Tanaka et al., 2011), it is reasonable to suppose that the tetrapyrrole pathway at least up to uroporphyrinogen III operates in the plastids. On the other hand, the occurrence of downstream enzymes CPO and PPO in both plastids and mitochondria suggests that the pathway downstream of coprogen also operates in the mitochondria. More studies are needed to clearly establish the cellular location of tetrapyrrole/heme biosynthesis in plants. Comparative qRT-PCR analysis indicated significant reductions in HEMN1 transcript levels in Athemn1 mutants compared with the wild type, whereas PPO1, PPO2, and CHL-I1 levels were not affected. Furthermore, LIN2, FC1, FC2, and CHL-I2 levels were reduced 2- to 3-fold in the mutant. The cross talk between genes of the heme and chlorophyll synthesis pathways has not been clearly worked out. Some of the observed changes in gene expression could be due to overall poor mitochondrial function leading to reduced ATP synthesis, as evidenced by the membrane polarity changes observed in the studies with JC-1 dye.
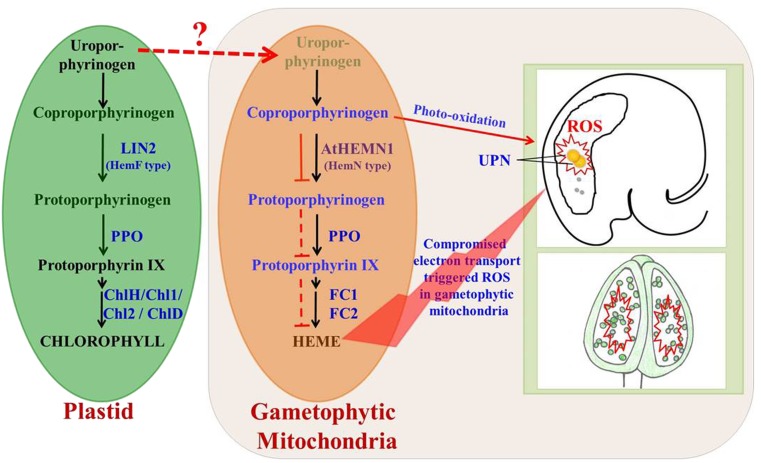
According to Goto et al. (2010), there are two analogs of CPOs in various organisms, including plants: oxygen-dependent hemF CPO and oxygen-independent hemN CPO. hemF is essential for the aerobic growth of photosynthetic bacteria and plants (Kruse et al., 1995). In tobacco, growth retardation and necrosis were reported by the overexpression of antisense RNA of CPO (hemF; Kruse et al., 1995). Similarly, lesion formation was reported by the loss of CPO (LIN2; hemF) activity in Arabidopsis (Ishikawa et al., 2001). Athemn1 mutant plants were normal during the vegetative stage but displayed abnormalities at flowering. The flowering stage is said to be highly energy demanding, and to cope with this, floral tissues increase the mitochondrial copies per cell (Huang et al., 1994; Martin et al., 2014). Considering the fact that AtHEMN1 is targeted to mitochondria, it appears that plants have recruited HEMN1 copies to meet the mitochondrial tetrapyrrole demand in floral tissues. Thus, loss of AtHEMN1 function would be evident in floral tissues and developing seeds. Based on the results obtained, a mechanistic model is proposed (Fig. 11) to explain the role of AtHEMN1 in male and female gametophyte development and in subsequent seed formation in Arabidopsis. LIN2 and AtHEMN1 perform the same function in different locations (i.e. in plastids and mitochondria, respectively). Our proposed mechanism does not rule out the transfer of tetrapyrrole biosynthesis pathway substrates/products from plastids to mitochondria but emphasizes the need for local production of the same in mitochondria when the energy demand is high. Quantitative estimation revealed increased coproporphyrin and decreased protoporphyrin levels in accordance with the reduced expression of Athemn1 in the heterozygous mutant. The oxidization of accumulated coproporphyrin leads to an increased generation of ROS. Furthermore, it is likely that the decreased availability of pathway intermediates and the down-regulation of ferro-chelatases might have affected the overall tetrapyrrole synthesis in mitochondria. Based on the available evidence, it can be concluded that the Athemn1 mutation affects the conversion of coprogen to protogen and, thus, impairs the synthesis of tetrapyrrole molecules and subsequent heme synthesis in the mitochondria. This leads to a shortage of heme molecules for the synthesis of cytochromes that affect the electron transport chain and oxygen metabolism. The oxidation of accumulated tetrapyrrole biosynthesis pathway intermediates generates ROS and impairs male and female gametophyte development, including the fusion of polar nuclei in Arabidopsis. To our knowledge, this is the first report linking mitochondrial tetrapyrrole biosynthesis and ROS homeostasis during the reproductive development of plants.
Figure 11.
A proposed model showing tetrapyrrole biosynthesis in mitochondria of gametophyte cells and the effect of AtHEMN1 disruption on male and female gametophyte development in Arabidopsis. LIN2 (hemF) and AtHEMN1 (hemN) convert coproporphyrinogen to protoporphyrinogen in plastids and in mitochondria, respectively. The source of uroporphyrinogen in mitochondria is currently unknown (locally synthesized or acquired from plastids) and is denoted with the dotted red arrow. We propose that the AtHEMN1 (hemN)-mediated conversion of coproporphyrinogen to protoporphyrinogen is affected in mitochondria by the Athemn1 mutation without disturbing the same in plastids. This leads to the accumulation of coproporphyrinogen and a drop in the levels of protoporphyrinogen and subsequent downstream pathway intermediates in the mitochondria in the Athemn1 mutant. Black arrows indicate normal tetrapyrrole biosynthesis in plastids and mitochondria. In mitochondria, the red solid line indicates inhibition of the conversion of coproporphyrinogen to protoporphyrinogen. Red dotted lines between protoporphyrinogen and heme indicate reduced accumulation of these products in the Athemn1 mutant. Accumulated coproporphyrinogen leads to an increased generation of ROS partly attributed to its photooxidation. The inadequate supply of heme in the mitochondria affects the synthesis of cytochrome c, affects the electron transport chain, and contributes to the generation of ROS. The ROS-mediated oxidative damage of mitochondria impairs microspore development, leading to pollen abortion. Increased ROS production in the mitochondria around the central cell of the female gametophyte prevents the fusion of polar nuclei and thereby affects ovule and seed development in Arabidopsis. UPN, Unfused polar nuclei.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The development and screening of the T-DNA insertion population of Arabidopsis (Arabidopsis thaliana ecotype Columbia) has been described (Pratibha et al., 2013). Briefly, the T-DNA promoter trap lines were generated by floral dip transformation (Clough and Bent, 1998) of Arabidopsis with Agrobacterium tumefaciens strain GV3101 harboring a promoter trap vector carrying a promoterless GFP gene at the right border of the T-DNA. T1 seeds were screened for T-DNA insertion by selection on a medium containing kanamycin (50 mg L−1). The green seedlings were transferred to pots and allowed to grow under controlled conditions (20°C ± 1°C, 60% relative humidity, 16 h of light/8 h of dark, under fluorescent illumination of 100 μmol m−2 s−1), and T2 seeds were collected. Each KanR T1 plant was considered as an independent trap. T2 plants of each line were examined for reporter gene expression or seed sterility. One of the lines showing seed sterility (designated as GFP-868) was identified and analyzed in this study.
Identification and Confirmation of the T-DNA Insertion Site
The T-DNA insertion site was determined through the genome walking approach (Pratibha et al., 2013; Sharma et al., 2015). Briefly, genome walking libraries were constructed by digesting DNA with DraI, EcoRV, PvuII, or StuI restriction enzyme followed by adapter ligation. PCR amplification was carried out according to the manufacturer’s instructions (Clontech). Primary PCR product (Supplemental Fig. S1A) was diluted (1:250) and used as a template for nested PCR amplification using a nested adapter primer (AP2) and a nested GFP gene-specific primer (NGWGS; Supplemental Table S3). Resolved nested PCR product (Supplemental Fig. S1B) was eluted using the GFX PCR DNA Gel Band Purification kit (GE Healthcare), cloned into pGEM-T Easy vector (Promega), and sequenced. The sequence retrieved after removal of the vector backbone was BLAST searched against the Arabidopsis genome database (TAIR; www.arabidopsis.org) to identify the T-DNA insertion site.
T-DNA insertion in the GFP-868 mutant is diagrammatically represented in Supplemental Figure S1D. The insertion was further confirmed by PCR amplification with the primer pairs P1-P2 and P1-P3. Amplification of the expected size fragments (i.e. 2.5 and 1.8 kb, respectively) confirmed the location of the T-DNA in At5g63290 (Supplemental Fig. S1C). Similarly, location of the T-DNA insertion in the Arabidopsis Biological Resource Center line SALK_100305 was confirmed using the primer pairs P4-P5 and P1-P6 targeting T-DNA and T-DNA flanking region-specific primers (Supplemental Fig. S1D).
Hybridization and Segregation Studies
For segregation analysis of Athemn1 mutations, T1 plants were allowed to self-pollinate and the seeds were collected. T2 seeds were germinated on Murashige and Skoog agar (MSA) medium (Murashige and Skoog, 1962) containing 50 mg L−1 kanamycin. The number of KanR and KanS plants was scored. For transmission studies, reciprocal crosses were made between heterozygous mutant plants and wild-type plants. The seeds from the crosses were collected, and progeny were tested for kanamycin sensitivity on MSA plates.
Egg cell-specific marker ET1119 and synergid cell-specific marker ET884 lines were obtained from Ueli Grossniklaus’s laboratory and analyzed according to Kirioukhova et al. (2011). The central cell-specific marker DD65:GFP was obtained from G. Pagnussat’s laboratory and analyzed according to Steffen et al. (2007) and Leshem et al. (2012). These marker lines were individually crossed with the Athemn1-1 mutant lines, and F1 plants carrying the Athemn1-1 mutant allele and the GFP/GUS marker gene were identified through PCR. The gametophytes of such F1 plants were analyzed for marker gene expression.
RT-PCR Analysis
Total RNA from different parts of both mutant and wild-type plants was isolated using the Total RNA Extraction kit (Real Genomics). Total RNA was quantified with a Nanodrop spectrophotometer (ND-1000; Thermo Scientific) and treated with DNase (Invitrogen). SuperScript III reverse transcriptase (Invitrogen) with oligo(dT) primer was used for first-strand cDNA synthesis followed by the amplification of double-stranded cDNA with gene-specific forward and reverse primers. The constitutively expressed GAPC gene was used as the normalization control. Primer sequences used for the amplification of different DNA sequences are given in Supplemental Table S3.
Phenotypic Characterization of Pollen and Ovules
The inflorescences were collected and fixed in ethanol:acetic acid (9:1) for 24 h, dehydrated in an ethanol series (50% [v/v], 70% [v/v], and 90% [v/v]), followed by dissection of the ovary and anthers in Hoyer’s solution (Anderson, 1954) using a stereo zoom microscope (SMZ1500; Nikon), then mounted on a glass slide for 2 h under a cover slip and observed after 2 h using DIC optics. Pollen viability was examined using Alexander’s staining (Alexander, 1969). After staining, the number of viable and nonviable pollen grains was counted.
For histological studies, the inflorescence was fixed in 3.7% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol, dehydrated in an ethanol series, infiltrated, and embedded in paraffin wax. Tissues were sectioned (5–10 µm thickness) with a microtome (Shadon Finnse) and observed after staining with 0.05% (w/v) Toluidine Blue using a microscope (AXIO imager.M1; Carl Zeiss) with bright-field illumination. The anther development stages were determined according to Sanders et al. (1999).
GUS staining was performed as described by Jefferson et al. (1987). The GUS-stained samples were rinsed three times with 70% (v/v) ethanol, cleared in Hoyer’s solution, and observed with a microscope (AXIO imager.M1; Carl Zeiss).
In Situ ROS Detection and Imaging
NBT staining was performed as described by Martin et al. (2013) to detect in situ ROS. The inflorescence was vacuum infiltrated in 10 mm sodium phosphate buffer (pH 7.8) containing 10 mm NaN3 and 0.1% (w/v) NBT stain (SRL Diagnostics) twice for 5 min each and incubated in complete darkness for 30 min at 37°C. After that, the anthers and ovules were dissected using a microscope (SMZ1500; Nikon) in Hoyer’s solution and left overnight for tissue clearance. NBT-stained anthers and ovules were examined with a microscope (AXIO imager.M1; Carl Zeiss), and images were taken.
Complementation Analysis
Full-length cDNA of the AtHEMN1 gene (1,455 bp) was amplified using gene-specific primers P1 and P6 (Supplemental Table S3; Supplemental Fig. S1D) with additional sequences for BglII and SpeI restriction sites, respectively, at the 5′ end. The amplified product was cloned into pGEM-T Easy vector and subcloned into the binary vector pCAMBIA1302 between BglII and SpeI sites upstream of the GFP reporter gene to get the AtHEMN1-GFP fusion transcript. The pCAMBIA1302 construct with the 35S:AtHEMN1-GFP gene cassette containing hygromycin as a selectable marker (Supplemental Fig. S3) was introduced into Athemn1-1 mutant plants, and transgenic plants were selected on MSA medium supplemented with hygromycin (20 mg mL−1). Out of 10 independent transformants carrying p35S::AtHEMN1-GFP, two lines were taken for further phenotypic analysis. All these transformants were confirmed for the presence of the AtHEMN1-GFP gene (Supplemental Fig. S3B) and transcripts (Supplemental Fig. S3C) using appropriate primers.
Cellular Localization of AtHEMN1 Gene Expression
A 1.2-kb fragment upstream of the start codon of the AtHEMN1 gene was PCR amplified with the primers P11 and P12 (Supplemental Table S3) having SacII and NotI restriction sites at the 5′ end, respectively, using Phusion high-fidelity DNA polymerase (Finnzymes). The amplified product was cloned into pGEM-T Easy vector and subcloned into the binary vector pORE R2 (Coutu et al., 2007) between SacII and NotI sites upstream of the uidA reporter gene. The recombinant binary vector was mobilized into A. tumefaciens strain GV 3101 and used to transform Arabidopsis plants. T2 plants were examined to localize GUS expression in vegetative and floral parts.
For colocalization analysis, root tips from 7-d-old seedlings of complemented plants (T3 generation) were dissected in 5 µm MitoSOX Red (from a stock of 5 mm diluted 1,000 times in 20 mm HEPES buffer, pH 7.2) on a microscope slide and incubated for 30 min at 37°C in the dark. The root tips were washed with HEPES buffer (20 mm), pH 7.2, and immediately examined using a confocal laser-scanning microscope (750 Meta; Zeiss) with excitation/emission at 514/580 nm (Li et al., 2013). Ovules were dissected from pistils in 5 µm MitoSOX Red on a microscope slide using a dissecting microscope and allowed to incubate for 30 min at room temperature (Martin et al., 2013). Ovules were then washed with HEPES buffer (20 mm), pH 7.2, and examined with a confocal laser-scanning microscope with excitation/emission at 514/580 nm.
Isolation and Estimation of Coproporphyrin and Protoporphyrin
Porphyrins were isolated according to Marsh et al. (1963) with some modifications. One gram of inflorescence was homogenized in 4 mL of acetic acid (1.3 n) with a mortar and pestle for 2 min. Blending was continued for another 2 min while adding 5 mL of ethyl acetate, the homogenate was taken into a collection tube, the volume was made up to 25 mL with ethyl acetate, and the pH was adjusted to 3.1 with glacial acetic acid. The resulting slurry was allowed to stand overnight to form an emulsion. The emulsion was centrifuged, and after removing the organic fraction, the aqueous fraction was extracted in 10 mL of ethyl acetate saturated with HCl (15% [v/v]) and then concentrated to 4 mL. The acid extracts were combined and adjusted to pH 3.1 with a saturated solution of sodium acetate. The porphyrins were quantitatively separated from the aqueous solution into peroxide-free ether. From the ether extracts, coproporphyrin and protoporphyrin were separated using 0.2% (v/v) and 10% (v/v) HCl, respectively (Dresel and Falk, 1956). The absorbance spectra of coproporphyrin and protoporphyrin were measured over the Soret band 400 to 403 nm and 408 to 410 nm, respectively, using a scanning UV-Visible Spectrophotometer (Shimadzu) equipped with a chart recorder (Supplemental Fig. S9). The experiment was performed with three biological replicates. The mean absorbance value (at 400–403 nm for coproporphyrin and 408–410 nm for protoporphyrin) was used for histogram preparation. Error bars indicate sd.
qRT-PCR Analysis
Single-stranded cDNA was prepared from 1 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. qRT-PCR was performed in an Applied Biosystems real-time PCR system (Life Technologies) using Power SYBR Green PCR Master mix (Life Technologies). Total cDNA was diluted to ∼25 ng μL−1, and a total of 100 ng was used in a 10-μL reaction mixture. Three technical replicates were taken for each reaction along with a no-template control to check for contaminants. The thermal cycling program for qRT-PCR was as follows: 3 min at 95°C, followed by 40 cycles of 3 s at 95°C and 30 s at 60°C, which included data acquisition. Dissociation curve analysis was performed from 65°C to 95°C in increments of 0.5°C, each lasting for 5 s, to confirm the presence of a specific product. qRT-PCR primers were designed using standard parameters available at http://eu.idtdna.com/scitools/Applications/RealTimePCR. The concentration of ACT2 (At3g18780) was used to normalize the gene expression in different samples. The 2-ΔΔCT method was used for the calculation of change in expression according to Livak and Schmittgen (2001).
Estimation of Mitochondrial Membrane Polarity Changes
Pistils at the FG6-7 stage were dissected and incubated in JC-1 dye (Molecular Probes) for 30 min at room temperature, washed with buffer A (20 mM HEPES buffer [pH 7.2]), and observed with a confocal microscope (Carl Zeiss). The green (excitation/emission wavelength = 485/538 nm) and red (excitation/emission wavelength = 485/590 nm) fluorescence intensities were documented from five different spots of 60 embryo sacs from wild-type and Athemn1-1 mutant plants, respectively. The mean intensity for each embryo sac was scored in both red and green channels. Then, the ratio of red to green fluorescence of JC-1 images was calculated for all the embryo sacs from both wild-type and mutant plants.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification and diagrammatic representation of the T-DNA insertion in the GFP-868 mutant by the genome walking approach.
Supplemental Figure S2. Genotyping of Athemn1-1 and Athemn1-2 mutant plants through PCR.
Supplemental Figure S3. Complementation of the Athemn1-1 mutant by ectopic expression of the AtHEMN1 gene.
Supplemental Figure S4. Stage-wise developmental analysis of pollen in wild-type and Athemn1 mutant plants.
Supplemental Figure S5. NBT staining of an aborted ovule from the Athemn1 mutant showing the accumulation of ROS in the entire ovule.
Supplemental Figure S6. Structure of the AtHEMN1 gene and predicted amino acid sequence of AtHEMN1.
Supplemental Figure S7. Phylogenic tree of HEMN proteins of different genera constructed using the neighbor-joining method.
Supplemental Figure S8. GUS expression in transgenic Arabidopsis plants carrying the pAtHEMN1::uidA construct.
Supplemental Figure S9. UV-visible spectra of purified coproporphyrin and protoporphyrin from wild-type and Athemn1-1 mutant inflorescences.
Supplemental Table S1. Homology of the AtHEMN1 protein with CPOs from different genera.
Supplemental Table S2. Details of the similarity of AtHEMN1 with mitochondria-localized hemN-type CPOs of different organisms.
Supplemental Table S3. Particulars of the primers used in this study.
Supplementary Material
Glossary
- ROS
reactive oxygen species
- UTR
untranslated region
- RT
reverse transcription
- DIC
differential interference contrast
- NBT
nitroblue tetrazolium
- qRT
quantitative reverse transcription
- MSA
Murashige and Skoog agar
Footnotes
This work was supported by the Council of Scientific and Industrial Research (project nos. MLP-072 and BSC-0107 as well as junior and senior research fellowships to P.P. and S.K.S., respectively) and the Indian Council of Agricultural Research through the National Agricultural Innovation Project (grant no. NAIP-4157).
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Anderson LE. (1954) Hoyer’s solution as a rapid permanent mounting medium for bryophytes. Bryologist 57: 242–244 [Google Scholar]
- Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC (2008) The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch AWU, Montgomery BL (2015) Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response. Redox Biol 4: 260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Dailey HA. (1990) Biosynthesis of Heme and Chlorophylls. McGraw-Hill, New York [Google Scholar]
- Dresel EIB, Falk JE (1956) Studies on the biosynthesis of blood pigments. 2. Haem and porphyrin formation in intact chicken erythrocytes. Biochem J 63: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Goto T, Aoki R, Minamizaki K, Fujita Y (2010) Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 51: 650–663 [DOI] [PubMed] [Google Scholar]
- Grimm B. (1998) Novel insights in the control of tetrapyrrole metabolism of higher plants. Curr Opin Plant Biol 1: 245–250 [DOI] [PubMed] [Google Scholar]
- Grini PE, Jürgens G, Hülskamp M (2002) Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Struck F, Matzinger DF, Levings CS III (1994) Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Okamoto H, Iwasaki Y, Asahi T (2001) A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J 27: 89–99 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Kirioukhova O, Barrell PJ, Rutten T, Moore JM, Baskar R, Grossniklaus U, Gruissem W (2010) Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet 6: e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W (2008) A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 18: 1680–1686 [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Kägi C, Baumann N, Nielsen N, Stierhof YD, Gross-Hardt R (2010) The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc Natl Acad Sci USA 107: 22350–22355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirioukhova O, Johnston AJ, Kleen D, Kägi C, Baskar R, Moore JM, Bäumlein H, Gross-Hardt R, Grossniklaus U (2011) Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development 138: 3409–3420 [DOI] [PubMed] [Google Scholar]
- Krohn NG, Lausser A, Juranić M, Dresselhaus T (2012) Egg cell signaling by the secreted peptide ZmEAL1 controls antipodal cell fate. Dev Cell 23: 219–225 [DOI] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B (1995) Reduction of coproporphyrinogen oxidase level by antisense RNA synthesis leads to deregulated gene expression of plastid proteins and affects the oxidative defense system. EMBO J 14: 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Bandyopadhyay U (2005) Free heme toxicity and its detoxification systems in human. Toxicol Lett 157: 175–188 [DOI] [PubMed] [Google Scholar]
- Lermontova I, Kruse E, Mock HP, Grimm B (1997) Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc Natl Acad Sci USA 94: 8895–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y, Johnson C, Wuest SE, Song X, Ngo QA, Grossniklaus U, Sundaresan V (2012) Molecular characterization of the glauce mutant: a central cell-specific function is required for double fertilization in Arabidopsis. Plant Cell 24: 3264–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Liang DD, Li J, Duan YB, Li H, Yang YC, Qin RY, Li L, Wei PC, Yang JB (2013) Unravelling mitochondrial retrograde regulation in the abiotic stress induction of rice ALTERNATIVE OXIDASE 1 genes. Plant Cell Environ 36: 775–788 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Marsh HV Jr, Evans HJ, Matrone G (1963) Investigations of the role of iron in chlorophyll metabolism. I. Effect of iron deficiency on chlorophyll and heme content and on the activities of certain enzymes in leaves. Plant Physiol 38: 632–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martásek P. (1998) Hereditary coproporphyria. Semin Liver Dis 18: 25–32 [DOI] [PubMed] [Google Scholar]
- Martin MV, Distéfano AM, Bellido A, Córdoba JP, Soto D, Pagnussat GC, Zabaleta E (2014) Role of mitochondria during female gametophyte development and fertilization in A. thaliana. Mitochondrion 19b: 350–356 [DOI] [PubMed] [Google Scholar]
- Martin MV, Fiol DF, Sundaresan V, Zabaleta EJ, Pagnussat GC (2013) oiwa, a female gametophytic mutant impaired in a mitochondrial manganese-superoxide dismutase, reveals crucial roles for reactive oxygen species during embryo sac development and fertilization in Arabidopsis. Plant Cell 25: 1573–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Ohtsu M, Higashiyama T (2016) Cell fusion and nuclear fusion in plants. Semin Cell Dev Biol 60: 127–135 [DOI] [PubMed] [Google Scholar]
- Matveyeva NP, Polevova SV, Smirnova AV, Yermakov IP (2012) Sporopollenin accumulation in Nicotiana tabacum L. microspore wall during its development. Cell Tissue Biol 6: 293–301 [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell (Suppl) 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense SM, Zhang L (2006) Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res 16: 681–692 [DOI] [PubMed] [Google Scholar]
- Moore MR. (1993) Biochemistry of porphyria. Int J Biochem 25: 1353–1368 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ngo QA, Moore JM, Baskar R, Grossniklaus U, Sundaresan V (2007) Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134: 4107–4117 [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN (2006) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratibha P, Singh SK, Sharma I, Kumar R, Srinivasan R, Bhat SR, Ahuja PS, Sreenivasulu Y (2013) Characterization of a T-DNA promoter trap line of Arabidopsis thaliana uncovers a cryptic bi-directional promoter. Gene 524: 22–27 [DOI] [PubMed] [Google Scholar]
- Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS (2006) Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci USA 103: 15038–15043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, Mclntire KN, Hsu Y, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sassa S, Nagai T (1996) The role of heme in gene expression. Int J Hematol 63: 167–178 [DOI] [PubMed] [Google Scholar]
- Schreiber DN, Bantin J, Dresselhaus T (2004) The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol 134: 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma I, Srinivasan R, Ahuja PS, Bhat SR, Sreenivasulu Y (2015) Identification and characterization of a T-DNA promoter trap line of Arabidopsis thaliana uncovers an embryo sac-specific bi-directional promoter. Plant Mol Biol Rep 33: 1404–1412 [Google Scholar]
- Shimizu KK, Ito T, Ishiguro S, Okada K (2008) MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana. Plant Cell Physiol 49: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (1987) Mechanisms of toxicity of photoactivated artificial porphyrins: role of porphyrin-protein interactions. Ann N Y Acad Sci 514: 309–322 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Portereiko MF, Lloyd A, Drews GN (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44: 1266–1274 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, doi/10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Qu GZ, Li HY, Wu YJ, Wang C, Liu GF, Yang CP (2010) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol Biol Rep 37: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Che FS, Iwano M, Takayama S, Yoshida S, Isogai A (2001) Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J Biol Chem 276: 20474–20481 [DOI] [PubMed] [Google Scholar]
- Williams P, Hardeman K, Fowler J, Rivin C (2006) Divergence of duplicated genes in maize: evolution of contrasting targeting information for enzymes in the porphyrin pathway. Plant J 45: 727–739 [DOI] [PubMed] [Google Scholar]
- Wu JJ, Peng XB, Sun M (2013) Mitochondria responsive signaling between egg and central cell controls their coordinated maturation. Plant Signal Behav 8: e24076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Bauer CE (2013) Controlling the delicate balance of tetrapyrrole biosynthesis. Philos Trans R Soc Lond B Biol Sci 368: 20120262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra A, Jimenez-Quesada MJ, Traverso JA, Corpas FJ, Rodrguez-Garcia MI, Alche JD (2012) Peroxisomal localization of Cu-Zn superoxide dismutase in the male reproductive tissues of the olive tree. Microsc Microanal 18: 33–34 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.