ABSTRACT
Lentinula edodes is a popular, cultivated edible and medicinal mushroom. Lentinula edodes is susceptible to postharvest problems, such as gill browning, fruiting body softening, and lentinan degradation. We constructed a de novo assembly draft genome sequence and performed gene prediction for Lentinula edodes. De novo assembly was carried out using short reads from paired-end and mate-paired libraries and by using long reads by PacBio, resulting in a contig number of 1,951 and an N50 of 1 Mb. Furthermore, we predicted genes by Augustus using transcriptome sequencing (RNA-seq) data from the whole life cycle of Lentinula edodes, resulting in 12,959 predicted genes. This analysis revealed that Lentinula edodes lacks lignin peroxidase. To reveal genes involved in the loss of quality of Lentinula edodes postharvest fruiting bodies, transcriptome analysis was carried out using serial analysis of gene expression (SuperSAGE). This analysis revealed that many cell wall-related enzymes are upregulated after harvest, such as β-1,3-1,6-glucan-degrading enzymes in glycoside hydrolase (GH) families GH5, GH16, GH30, GH55, and GH128, and thaumatin-like proteins. In addition, we found that several chitin-related genes are upregulated, such as putative chitinases in GH family 18, exochitinases in GH20, and a putative chitosanase in GH family 75. The results suggest that cell wall-degrading enzymes synergistically cooperate for rapid fruiting body autolysis. Many putative transcription factor genes were upregulated postharvest, such as genes containing high-mobility-group (HMG) domains and zinc finger domains. Several cell death-related proteins were also upregulated postharvest.
IMPORTANCE Our data collectively suggest that there is a rapid fruiting body autolysis system in Lentinula edodes. The genes for the loss of postharvest quality newly found in this research will be targets for the future breeding of strains that keep fresh longer than present strains. De novo Lentinula edodes genome assembly data will be used for the construction of a complete Lentinula edodes chromosome map for future breeding.
KEYWORDS: chitin, fruiting body, glucan, lentinan, senescence
INTRODUCTION
Lentinula edodes, popularly known as the shiitake mushroom, is a widely cultivated edible and medicinal mushroom in Asia. The shiitake is the second most cultivated fungus, and over 1,321,000 tons of Lentinula edodes have been produced in China, Japan, Taiwan, and Korea (1, 2). Lentinula edodes is used as a medicinal mushroom; lentinan, a β-1,3-1,6-glucan with antitumor activity, is present in this mushroom (3). Recently, it was shown that lentinan can be effective for treating gut inflammation (4). It is also reported that polysaccharides extracted from Lentinula edodes can restore age-attenuated immune responses and reverse age-related alterations of gut microbiota compositions in mice (5). Furthermore, Lentinula edodes can be a source of ergosterol and ergothioneine (6). Lentinula edodes has the potential for use not only as a fresh food but also as a source of medicinal compounds. Therefore, it is necessary to breed new Lentinula edodes strains that contain these beneficial compounds. However, Lentinula edodes requires a longer cultivation time than other mushrooms, such as Flammulina velutipes and Pleurotus ostreatus (7), and thus it takes a very long time to breed new strains using typical breeding methods based on mating and evaluating fruiting body traits. Effective breeding methods based on gene function are needed.
Genome sequence data have been published for basidiomycetous fungi such as Phanerochaete chrysosporium (8), Laccaria bicolor (9), Coprinopsis cinerea (10), Agaricus bisporus (11), Schizophyllum commune (12), and Flammulina velutipes (13). Because of the popularity of next-generation sequencing, many genome sequence drafts of basidiomycetous fungi are publicly available, especially from the 1,000 Fungal Genomes Project (14). More recently, the constructions of de novo draft genome sequences of Lentinula edodes were reported (15, 16).
Some transcriptome analyses with genome sequence data have been reported in several mushrooms, such as Agaricus bisporus (11), Schizophyllum commune (12), and others. Transcriptome analyses for fruiting body development in Lentinula edodes were carried out using differential display (17) or serial analysis of gene expression (SAGE) analysis (18), and PCR subtraction was used for analyzing postharvest quality loss (19). More recently, transcriptome analysis was performed by transcriptome sequencing (RNA-seq) during browning film formation on mycelial bags (20) and during cellulose degradation (16). In Lentinula edodes, postharvest quality loss is a very significant problem from an economical viewpoint. In Lentinula edodes, significant quality loss is caused by gill browning and fruiting body softening, resulting in a foul odor 3 to 4 days after harvest if stored at room temperature (21). Lentinula edodes fruiting bodies turn brown owing to melanin synthesis and turn soft because of cell wall degradation. Melanin synthesis occurs from the increases in tyrosinase (22, 23) and laccase (24, 25) after harvest. Cell wall degradation is another significant postharvest issue in Lentinula edodes. Because lentinan, a β-1,3-1,6-glucan in cell walls with antitumor activity (3), is degraded by increased glucanase activity after harvest (26), we identified cell wall degradation-related genes that were upregulated after harvest by PCR subtraction (19). Genome sequence data of Lentinula edodes was not available for PCR subtraction, yet approximately 50 genes, such as putative chitinases (chi1, chi2, and chi3) and a chitosanase (cho1), were identified as upregulated after harvest (19, 21). Several β-1,3-glucanases were purified and characterized from Lentinula edodes fruiting bodies after harvest. EXG2, an exo-β-1,3-glucanase belonging to glycoside hydrolase (GH) family 55 (27), TLG1, an endo-β-1,3-glucanase with high similarity to thaumatin-like protein (28), GLU1, an endo-β-1,3-1,6-gluanase belonging to GH128 (29), and PUS30, an endo-β-1,6-glucanase belonging to GH30 (30), can degrade lentinan. Their expression increased after harvest; therefore, lentinan is degraded by multiple enzymes with synergic effects. The putative transcription factor exp1, which is a homolog of a gene that regulates cap autolysis in Coprinopsis cinerea (31), is also upregulated after harvest (19). This suggests that exp1 in Lentinula edodes regulates genes involved in postharvest quality loss. However, it is highly possible that multiple transcription factors are involved in postharvest quality loss in Lentinula edodes. To establish methods for controlling postharvest quality loss, a more comprehensive transcriptome analysis of Lentinula edodes is needed.
To obtain a basis for constructing a reference sequence for Lentinula edodes transcriptome analysis, we constructed the genome sequence of Lentinula edodes G408PP-4 that was used for linkage mapping (32) using a combination of short-reads from Illumina sequencers and long reads from PacBio. Next, we predicted genes by combining assembled genome sequence data and RNA-seq data. Then, we analyzed transcriptomic data obtained from previously published PCR subtraction data (19) and SuperSAGE data for postharvest quality loss. The results suggest that multiple novel cell wall enzymes, such as putative β-1,3-gluanases, β-1,6-gluanases, and chitinases are upregulated after harvest. We also identified several transcription factors for postharvest quality loss. The results will provide insight for controlling postharvest freshness in Lentinula edodes.
RESULTS AND DISCUSSION
We constructed genome sequence data for G408PP-4, a wild strain isolated on Yakushima Island, Japan, which is crossed with strain D703PP-9 to construct a linkage map (32, 33). Short leads from the Illumina GAIIx and Illumina HiSeq2500 were assembled by the clc genomics workbench, and then we scaffolded the contigs using mate-paired libraries and a PacBio sequencing system. The resulting scaffold number was 1,951, the N50 was 103,018, and the total was 38,944,209 bp (see Table S1 in the supplemental material). The resulting scaffolds were comparable with those from other genome sequence data from mushroom-forming basidiomycetous fungal species, such as from Coprinopsis cinerea (37 Mb) (10), Agaricus bisporus (30 Mb) (11), and Schizophyllum commune (38.5 Mb) (12), but were smaller than that from the mycorrhizal basidiomycetous fungus Laccaria bicolor (65 Mb) (9). Previously, we carried out gene prediction by draft genome sequencing of Lentinula edodes strain D703PP-9 and found 8,271 putative genes (25). On the other hand, the number of predicted genes in Coprinopsis cinerea or Schizophyllum commune is around 15,000, much larger than our prediction for Lentinula edodes. Therefore, to more accurately predict genes, we combined RNA-seq data from whole life cycle and reference genome sequence data. Using RNA-seq data, the number of predicted genes was 12,959, and 100% of recorded genes in Lentinula edodes were predicted (except for pheromone precursors and mitochondrial and partial proteins) (see Table S2). Lentinula edodes belongs to Marasmiaceae, which includes white rot, saprobic, and plant-pathogenic fungi (34). Plant cell wall-degrading enzymes in Lentinula edodes G408PP-4 (see Table S3) were identical to those of strain W1-26 (16) and similar to other species of Marasmiaceae (Table S3) (35–37). Characteristically, no lignin peroxidase (Lip)-encoding genes were found in Lentinula edodes or in Marasmiaceae species whose genome sequences are publicly available (Table S3), suggesting that Lentinula edodes lacks Lip and depends on laccases and manganese peroxidases for lignin degradation.
Transcriptome analysis for fruiting bodies after harvest.
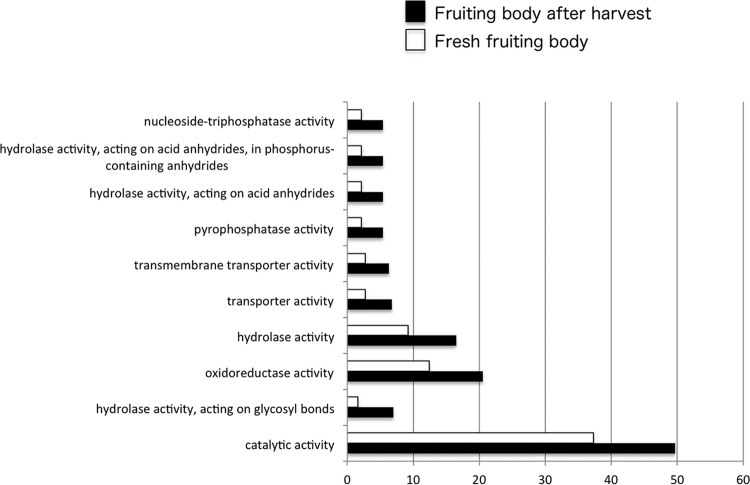
Genes upregulated in the fruiting body after harvest were investigated by SuperSAGE (38). More than 200,000 tags were sequenced from fresh fruiting bodies and postharvest fruiting bodies. Tag counting data are summarized in Fig. S2, and totally, 62% of unique tags were matched with predicted genes. Approximately 95% of the genes that were true-positive upregulated genes after harvest by our previous PCR subtraction analysis (19) were expressed higher than 2-fold in the fruiting body after harvest compared with that in the fresh fruiting body (see Fig. S3, Table S4). This suggests that the results from SuperSAGE are reliable. To identify differentially expressed genes (DEGs), statistical and gene ontology analyses were performed. DEG analysis revealed that 66% of the genes that are true-positive genes upregulated after harvest by our previous PCR subtraction analysis were determined to be DEG in the present SuperSAGE analysis (Fig. S3). We identified an enriched gene ontology in the fruiting body after harvest compared with that in the fresh fruiting body using Blast2GO (Fig. 1). This revealed that catalytic activity and hydrolase activity on glycosyl bonds, transporters, and oxidoreductase activity were upregulated in the fruiting body after harvest (Fig. 1). We identified an enriched gene ontology in the fruiting body after harvest by DAVID and found that dehydrogenases, transporters, oxidoreductases, and transcription factors are found in the fruiting body (see Table S5). On the other hand, we compared the enriched gene ontologies in the fresh fruiting body by Blast2GO and DAVID, and the results suggested that many gene ontologies were enriched in the fresh fruiting body (see Fig. S4, Table S6). In particular, DNA, RNA, protein, and carbohydrate metabolic processes were enriched.
FIG 1.
Comparison of enriched gene ontologies between fruiting bodies after harvest and fresh fruiting bodies (just after harvest) by using Blast2GO enrichment analysis (Fisher's exact test). Black and white bars indicate enriched gene ontologies in the fruiting body after harvest and the fresh fruiting body, respectively. The x axis indicates the number of genes enriched.
Expression of putative β-1,3-glucanases after harvest.
β-1,3-1,6-Glucans are the most abundant cell wall components in the Lentinula edodes fruiting body; therefore, we first analyzed β-1,3-1,6-glucan-degrading enzymes expressed abundantly after harvest (Table 1). Lentinula edodes has three GH55 family exo-β-1,3-glucanases that are mostly conserved in filamentous fungi and also exist in several bacteria (39). EXG2, a member of the GH55 family which is involved in cell wall degradation after harvest (27), has a strong effect on lentinan degradation after harvest (40). The enzyme encoded by exg2 in the GH55 family of exo-β-1,3-glucanases was not determined to be a DEG, but its expression was upregulated more than 15-fold in the fruiting body after harvest (Table 2). Two other genes encoding putative GH55 family enzymes (g7383.t1 and g7404.t2) (Table 1) were slightly increased after harvest (Table 2). We previously characterized another exo-β-1,3-glucanase, EXG1 (g4132.t1), an enzyme of the GH5 family (41). Twenty-one putative GH5 members were found in Lentinula edodes (Table 1). The expression of exg1 decreased after harvest (Table 2), but another GH5 gene (g4131.t1) located in the Lentinula edodes genome in tandem with exg1, similar to the one found in Agaricus bisporus (42), increased after harvest (Table 2). We further identified 3 DEGs that increased after harvest encoding GH5 family members (Table 2). One endo-β-1,3-glucanase in GH128 involved in lentinan degradation in Lentinula edodes, GLU1, was purified and characterized (29). We found 4 putative GH128 family enzymes in Lentinula edodes (Table 1). The expression of glu1 increased significantly after harvest and was deemed a DEG (Table 2), but the other three GH128 family genes were not identified or decreased after harvest (see Table S4). Thaumatin-like protein, TLG1, which also has endo-β-1,3-glucanase activity, was purified from the Lentinula edodes fruiting body after harvest (28). The gene tlg1 was identified as a DEG, and one other thaumatin-like protein was judged to be a DEG as well (g10798.t1) (Table 2). We found four other putative thaumatin-like proteins that were not DEGs (Table 1; see also Table S4), but the expression of one gene (g10969.t1) increased 18.5 times after harvest (Table 2). We also found 6 glycopeptide proteins which are weakly similar to thaumatin-like protein (43), and the expression of three of them (g6764.t1, g6761.t1, and g707.t1) increased significantly after harvest (Table 2). Additionally, we observed that the β-1,6-glucanase LePUS30 that belongs to GH30 is involved in cell wall degradation after harvest (30). Expression of pus30 and ghf30 (19), encoding other GH30 family members, increased after harvest. The other gene encoding a GH30 member enzyme (g1578.t1) was not identified as a DEG (Table 2). Some enzymes belonging to the GH16 family can degrade β-1,3-1,6-glucans, such as laminarin (44), and Lentinula edodes has 28 genes encoding putative GH16 family members (Table 1). We found that 4 of them (including mlg1) are differentially expressed after harvest (Table 2). We expressed mlg1 in Aspergillus oryzae and showed stable expression and secretion. The recombinant protein encoded by mlg1 was purified using ammonium sulfate fractionation, metal affinity resin, and anion-exchange chromatography (see Fig. S5). Purified MLG1 could degrade laminarin (β-1,3-1,6-glucan) and barley glucan (β-1,3-1,4-glucan), a typical feature of mixed-linked glucanases in the GH16 family (Table 3). However, recombinant MLG1 hardly degraded lentinan, cellulose, pustulan, or lichenan. This suggests that MLG1 degrades oligosaccharides degraded from lentinan by endo-β-1,3-glucanases, such as TLG1 and GLU1, and that rapid lentinan degradation after harvest was caused by a synergetic effect between the exo-β-1,3-1,6-glucanase EXG2, endo-β-1,3-glucanases TLG1 and GLU1, β-1,3-glucanase MLG1, and the endo-β-1,6-glucanase PUS30.
TABLE 1.
Number of CAZy enzymes among cell wall-degrading enzymes in basidiomycetous fungia
| Classification | Fungal polysaccharide degraded | Lentinula edodes | Gymnopus luxurians | Omphalotus olearius | Moniliophthora perniciosa | Coprinopsis cinereab | Schizophyllum communeb | Laccaria bicolorb | Phanerochaete chrysosporiumb | Auricularia subglabrab | Cryptococcus neoformansb | Ustilago maydisb | Reported gene(s) in Lentinula edodes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH5 | β-1,3-Glucosidase | 21 | 21 | 14 | 5 | 26 | 16 | 23 | 19 | 43 | 10 | 12 | exg1 |
| GH16 | Endo-1,3-β-glucanase | 28 | 8 | 6 | 2 | 32 | 35 | 31 | 23 | 44 | 14 | 20 | mlg1 |
| GH17 | Endo-1,3-β-glucosidase | 5 | 1 | 2 | 0 | 4 | 3 | 4 | 2 | 7 | 1 | 2 | |
| GH18 | Chitinase | 14 | 17 | 11 | 12 | 9 | 16 | 11 | 11 | 26 | 4 | 4 | chi1, chi2, chi3 |
| GH20 | β-Hexosaminidase | 3 | 4 | 3 | 2 | 2 | 3 | 2 | 3 | 4 | 1 | 2 | hex20A, hex20B |
| GH30 | β-1,6-Glucanase | 3 | 6 | 2 | 1 | 3 | 5 | 9 | 2 | 1 | 0 | 2 | pus30 |
| GH55 | Exo-β-1,3-glucanase | 3 | 2 | 3 | 12 | 1 | 2 | 2 | 2 | 5 | 0 | 1 | exg2 |
| GH75 | Chitosanase | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | Cho |
| GH128 | Endo-1,3-β-glucanase | 4 | 2 | 0 | 0 | 4 | 5 | 5 | 5 | 9 | 6 | 5 | glu1 |
| Thaumatin-like protein | Endo-1,3-β-glucanase | 6 | 11 | 3 | 13 | 6 | 7 | 2 | 6 | 10 | 1 | 0 | tlg1 |
CAZy, Carbohydrate-Active Enzymes database.
Data were based on genome sequences in MycoCosm (http://genome.jgi.doe.gov/programs/fungi/index.jsf).
TABLE 2.
β-Glucan-degrading enzymes involved in fruiting body softening after harvest
| Classification | IDa | Day 0 meanb | Day 4 meanc | Fold change (day 4/day 0) | P value | DEGd |
|---|---|---|---|---|---|---|
| GH55 | g7384.t1 (exg2) | 1.67 | 25.33 | 15.2 | 9.82E−03 | 0 |
| GH55 | g7383.t1 | 9 | 12.67 | 1.41 | 9.40E−01 | 0 |
| GH55 | g7404.t2 | 10.67 | 18 | 1.69 | 1.00E+00 | 0 |
| GH5 | g4132.t1 (exg1) | 28.67 | 21.67 | 0.76 | 1.14E−01 | 0 |
| GH5 | g4131.t1 | 3 | 17.33 | 5.78 | 7.62E−02 | 0 |
| GH5 | g3633.t1 | 0 | 19 | ∞ | 1.98E−05 | 1 |
| GH5 | g1981.t1 | 0 | 10.33 | ∞ | 6.98E−04 | 1 |
| GH5 | g1273.t1 | 17 | 152 | 8.94 | 4.26E−04 | 1 |
| GH128 | g8562.t1 (glu1) | 9 | 269.33 | 29.93 | 1.33E−08 | 1 |
| Thaumatin-like protein | g9371.t1 (tlg1) | 27.33 | 560 | 20.49 | 5.15E−08 | 1 |
| Thaumatin-like protein | g10798.t1 | 4.67 | 2328.33 | 498.93 | 1.27E−07 | 1 |
| Thaumatin-like protein | g10969.t1 | 0.67 | 12.33 | 18.5 | 1.44E−02 | 0 |
| Glycopeptide | g707.t1 | 0 | 8.33 | ∞ | 3.18E−03 | 1 |
| Glycopeptide | g6764.t1 | 3 | 3716.33 | 1238.78 | 1.23E−24 | 1 |
| Glycopeptide | g6761.t1 | 0.67 | 239.67 | 359.5 | 1.18E−10 | 1 |
| GH30 | g10719.t2 (ghf30) | 0 | 37.67 | ∞ | 4.37E−08 | 1 |
| GH30 | g1693.t1 (pus30) | 0 | 30 | ∞ | 3.50E−06 | 1 |
| GH30 | g1578.t1 | 251 | 333 | 1.46 | 8.56E−01 | 0 |
| GH16 | g1113.t1 | 0 | 297 | ∞ | 1.49E−09 | 1 |
| GH16 | g6709.t1 | 0 | 120.33 | ∞ | 5.59E−13 | 1 |
| GH16 | g1786.t1 (mlg1) | 159.67 | 6197 | 38.81 | 8.96E−12 | 1 |
| GH16 | g3753.t1 | 9 | 123.33 | 13.7 | 5.67E−06 | 1 |
ID, identification number. The gene names are shown in parentheses.
Mean number of Super-SAGE tag counts (n = 3) of fresh fruiting body (day 0).
Mean number of Super-SAGE tag counts (n = 3) of fruiting body after harvest (day 4).
1, estimated as DEG; 0, not estimated as DEG.
TABLE 3.
Substrate specificity of MLG1
| Substrate | Glycosidic bond | Relative activity (%)a |
|---|---|---|
| Laminarin | β-1,3-1,6 | 100 |
| Pachyman | β-1,3-1,6 | 40.2 |
| Cardran | β-1,3 | 32.0 |
| Lentinan | β-1,3-1,6 | NDb |
| Barley glucan | β-1,3-1,4 | 29.2 |
| Lichenan | β-1,3-1,4 | ND |
| Cellulose | β-1,4 | ND |
| Pustulan | β-1,6 | ND |
Released reducing sugar after incubation of MLG1 and each substrate was matured by the PAHBAH (4-hydroxybenzoic acid hydrazide) method. Relative activity of each substrate was indicated as a ratio (%) of absorbance of each substrate toward that of laminarin.
ND, not detected.
Expression of chitin-related genes after harvest.
Chitin is a major cell wall component in Lentinula edodes, and chitin-degrading-enzyme-encoding genes (chi1, chi2, and chi3) and chitinase activity increased after harvest (19, 21). chi3 is a homolog of the chitinase-encoding gene in Coprinopsis cinerea, chiIII, involved in cap autolysis (45). Therefore, the chitin degradation process is important for postharvest cell wall degradation in Lentinula edodes; however, there is little information on chitin degradation processes in basidiomycetous fungi. We identified 14 putative chitinases with GH18 domains in the Lentinula edodes genome (Table 1). Three of them (chi1, chi2, and chi7) showed significantly increased expression after harvest (Table 4). The chi7 gene is a newly identified putative endochitinase that has a chitin-binding module (CBM5). Other putative chitinases were not determined to be DEGs, but the expression of three other chitinases (chi4, chi5, chi6) increased after harvest (Table 4). These putative chitinases do not have CBM5, but several chitinases without CBM5 have chitin-degrading activity in basidiomycetous fungi (46). In particular, the Coprinellus congregatus chitinase Chi2 purified from droplets during cap autolysis, which has a high similarity to that encoded by chi5, can degrade chitin oligosaccharides (47). These results collectively suggest that multiple proteins with GH18 domains are coordinately involved in chitin degradation in the fruiting body after harvest. One chitin-degrading enzyme family is the hexosaminidases in GH20, namely, Hex20A (48) and Hex20B (49). These two hexosaminidases can degrade crystalized chitin in an exo-manner. Hex20A increased after harvest, and Hex20B decreased after harvest (Table 4). We also identified another GH20 family gene (g9894.t1), and it was not judged as a DEG but its expression increased 6.75-fold after harvest (Table 4). We identified other putative chitin-related proteins that have carbohydrate-binding module family 50 domains (CBM50, known as the LysM domain). Several LysM domains have chitin-binding ability, and several chitinases contain LysM domains (50). We found 7 proteins containing CMB50 domains; 5 of 7 such proteins were increased after harvest, and 2 of 5 were identified as DEGs in the fruiting body after harvest (g6653.t1 and g8231.t1) (Table 4). Expansin is involved in loosening cellulose crystal structures for cellulose degradation (51), and an expansin-like protein in Schizophyllum commune enhances chitin degradation (52). The expression of several expansin-like proteins, such as those encoded by baw28 (g7005.t1) and g6097.t1, increased after harvest. Chitosan is found in Lentinula edodes in addition to chitin (53, 54). Previous PCR subtraction data suggested the chitosanase cho1 is upregulated after harvest (19), and the SuperSAGE data were in agreement (see g3293.t2 in Table 4). The gene cho1 has high similarity to chitosanases in the GH75 family in ascomycetous fungi, but no similar sequence was found in basidiomycetous genomes except for that in Marasmiaceae (Table 1) and Auriculariales (55) species. We also found that the putative chitin deacetylase chd1 (g7070.t1) that encodes a protein that can catalyze chitin to chitosan increases after harvest (19) (Table 4). These data collectively suggest that there are rapid chitin and chitosan degradation systems for cell wall lysis in Lentinula edodes fruiting bodies after harvest.
TABLE 4.
Chitin- and chitosan-degrading enzymes involved in fruiting body softening after harvest
| Classification | IDa | Day 0 meanb | Day 4 meanc | Fold change (day 4/day 0) | P value | DEGd |
|---|---|---|---|---|---|---|
| GH18 | g976.t2 (chi5) | 0 | 5.67 | ∞ | 1.41E−02 | 0 |
| GH18 | g10886.t1 (chi7) | 1 | 116 | 116 | 4.64E−05 | 1 |
| GH18 | g1744.t1 (chi4) | 1.33 | 10.67 | 8 | 3.56E−01 | 0 |
| GH18 | g10746.t4 (chi6) | 3 | 9.33 | 3.11 | 5.54E−01 | 0 |
| GH18 | g2804.t1 (chi8) | 104 | 146 | 1.4 | 7.87E−01 | 0 |
| GH18 | g5859.t1 (chi3) | 0 | 5 | ∞ | 1.65E−01 | 0 |
| GH18 | g8924.t1 (chi2) | 1 | 1110 | 1110 | 8.91E−17 | 1 |
| GH18 | g8040.t1 (chi1) | 8.33 | 405.67 | 48.68 | 3.40E−06 | 1 |
| GH20 | g10209.t2 (Hex20A) | 0 | 2.67 | ∞ | 2.24E−01 | 0 |
| GH20 | g10237.t1 (Hex20B) | 19 | 11.33 | 0.6 | 5.24E−02 | 0 |
| GH20 | g9894.t1 | 1.33 | 9 | 6.75 | 8.10E−02 | 0 |
| CBM50 | g6653.t1 | 0 | 18.33 | ∞ | 8.30E−05 | 1 |
| CBM50 | g8231.t1 | 0.67 | 104.67 | 157 | 3.74E−05 | 1 |
| CBM50 | g6766.t1 | 205.67 | 1024.67 | 4.98 | 1.04E−02 | 0 |
| CBM50 | g6268.t1 | 43.33 | 136 | 3.14 | 1.31E−01 | 0 |
| CBM50 | g11628.t1 | 89.33 | 270 | 3.02 | 2.58E−01 | 0 |
| Expansin family protein | g6097.t1 | 2 | 60 | 23.67 | 4.70E−04 | 1 |
| Expansin family protein | g7005.t1 (baw1) | 27.67 | 3210.5 | 122.76 | 9.33E−17 | 1 |
| GH75 | g3293.t2 (cho1) | 0 | 39 | ∞ | 8.52E−09 | 1 |
| CE4 | g7070.t1 (chd1) | 242.33 | 613 | 3.59 | 5.28E−02 | 0 |
ID, identification number. The gene names are shown in parentheses.
Mean number of Super-SAGE tag counts (n = 3) of fresh fruiting body (day 0).
Mean number of Super-SAGE tag counts (n = 3) of fruiting body after harvest (day 4).
1, estimated as DEG; 0, not estimated as DEG.
Other genes upregulated in the fruiting body after harvest.
Many transcription factor (TF) genes were found in the fruiting bodies after harvest using DAVID (see Table S5); especially, TF genes containing zinc finger domains were upregulated after harvest (Table 5). Some genes containing Zn2Cy6 in their N termini, designated GAL4 domains, are involved in fruiting body development, such as priB (56) and ftf1, ftf2, and ftf3 (19, 21), and are upregulated during fruiting body formation. We found 7 genes (including ftf2) that have GAL4 domains in their N termini that were upregulated after harvest. The transcription factor exp1 regulates cap autolysis during spore diffusion (31), and its homolog in Lentinula edodes is upregulated after harvest (19). Two novel genes that contain high-mobility-group (HMG) domains were upregulated after harvest (g11706.t1 and g429.t2) (Table 5). Some of the genes with HMG domains have a wide gene regulation ability (57, 58) by interacting with other transcription factors to regulate transcription in a variety of genes (59). Therefore, these genes might be more effective in controlling fruiting body quality after harvest. We found other putative transcription factor genes that contain basic leucine zipper (bzip [g5086.t1 and g6148.t1]) and helix-loop-helix (bHLH [g271.t1]) domains (Table 5) that were upregulated after harvest. We found chromatin remodeling-related genes, such as the SWI/SNF complex genes (g872.t1, g5556.t1, g11151.t1, and g2328.t1), and histone deacetylases containing Sin3 domains (g143.t1) in fruiting bodies after harvest (Table 5). These results collectively suggest that several transcription factor genes and chromatin remodeling are coordinated to transcribe genes related to quality loss after harvest.
TABLE 5.
Transcription factor genes that were upregulated in the fruiting body after harvest
| Sequence IDa | Domain | Day 0 meanb | Day 4 meanc | Fold change (day 4/day 0) | P value |
|---|---|---|---|---|---|
| g3094.t1 | Zn2Cys6 (GAL4:ftf2) | 5.67 | 64 | 11.29 | 9.88E−04 |
| g9959.t1 | Zn2Cys6 (GAL4) | 2.33 | 27.67 | 11.86 | 6.51E−04 |
| g5145.t1 | Zn2Cys6 (GAL4) | 5 | 661 | 132.2 | 1.24E−08 |
| g4572.t3 | Zn2Cys6 (GAL4) | 0 | 31.67 | ∞ | 2.89E−07 |
| g5092.t1 | Zn2Cys6 (GAL4) | 0 | 30.67 | ∞ | 1.30E−06 |
| g250.t1 | Zn2Cys6 (GAL4) | 0 | 22.33 | ∞ | 8.63E−05 |
| g5558.t1 | Zn2Cys6 (GAL4) | 0 | 10.67 | ∞ | 3.94E−04 |
| g7227.t1 | Zn2Cys6 | 1 | 19 | 19 | 4.47E−03 |
| g9056.t2 | Zn2Cys6 | 0 | 16 | ∞ | 2.02E−05 |
| g721.t1 | Zn2Cys6 | 0 | 13.67 | ∞ | 1.21E−03 |
| g5340.t1 | Zn2Cys6 | 0 | 11.33 | ∞ | 9.82E−04 |
| g2567.t1 | Zn2Cys6 | 0 | 10.33 | ∞ | 2.40E−03 |
| g8104.t1 | Znf-CCHC | 0.67 | 17.67 | 26.5 | 1.37E−03 |
| g9472.t1 | Znf-CCHC | 0 | 40 | ∞ | 9.53E−08 |
| g7860.t1 | Znf-C2H2 | 3 | 34 | 11.33 | 7.75E−04 |
| g9975.t1 | Znf-C2H2 | 0 | 38.33 | ∞ | 4.86E−08 |
| g10687.t1 | Znf-C2H2 | 0 | 25 | ∞ | 4.32E−05 |
| g10687.t1 | Znf-C2H2 | 0 | 25 | ∞ | 4.32E−05 |
| g2176.t3 | Znf-C2H2 | 0 | 18.33 | ∞ | 1.35E−04 |
| g9593.t1 | HMG (exp1) | 1.33 | 90.67 | 68 | 7.23E−06 |
| g10925.t1 | HMG | 9.67 | 147 | 15.21 | 7.38E−04 |
| g429.t2 | HMG | 1 | 42.33 | 42.33 | 2.66E−03 |
| g6148.t1 | bZIP | 6.67 | 53 | 7.95 | 3.43E−03 |
| g5086.t1 | bZIP | 2.67 | 137.33 | 51.5 | 2.03E−06 |
| g271.t1 | bHLH (hlh) | 3.67 | 49.33 | 13.45 | 9.48E−04 |
| g5556.t1 | SWI | 1.67 | 52 | 31.2 | 1.03E−03 |
| g872.t1 | SNF5 | 2.67 | 74.33 | 27.88 | 5.52E−06 |
| g11151.t1 | SNF5 | 0.67 | 31 | 46.5 | 4.77E−03 |
| g2328.t1 | SNF2 | 1.33 | 106.33 | 79.75 | 1.85E−08 |
| g143.t1 | Homeodomain-like | 0.67 | 80.67 | 121 | 5.77E−05 |
ID, identification number.
Mean number of Super-SAGE tag counts (n = 3) of fresh fruiting body (day 0).
Mean number of Super-SAGE tag counts (n = 3) of fruiting body after harvest (day 4).
We previously identified tyrosinase and laccase (22–25) as involved in melanin synthesis in the Lentinula edodes fruiting body after harvest. The putative phenylalanine ammonia-lyase (PAL) that produces several phenolic compounds (60) for pigment synthesis was upregulated after harvest. This suggests that PAL is coordinately involved in pigment formation with tyrosinase and laccase after harvest (see Table S7). Oxidative stress affects postharvest quality in horticultural crops (61, 62). We found that 7 glutathione S-transferase genes were DEGs in the fruiting body after harvest (Table S7), and glutathione reductase (g3721.t1) and catalase (g2239.t2) were not DEGs but were expressed at 3-fold higher levels in the fruiting body after harvest compared with those in the fresh fruiting body (Table S4). These proteins are involved in antioxidant defense (63). Furthermore, the expression of ascorbate oxidase (g9281.t1) also increased after harvest (Table S7). The ascorbate glutathione pathway has an antioxidant role in oxidative stress (64). These genes are involved in reducing oxidation stress. Thioredoxin and cytochromes that are also involved in the reduction of oxidation stress (63) were upregulated in the fruiting body postharvest (Table S7). These antioxidant enzyme activities increased after harvest (65) in Lentinula edodes, collectively suggesting that the antioxidant defense system is induced after harvest. Protein kinases have a critical role for signal transduction in stress responses (66), and many protein kinases were upregulated after harvest (Table S7). This suggests that some signal transduction systems are newly functioning after harvest. We found that several programmed cell death-related genes were upregulated after harvest (Table S7). The autophagy-related genes encoding inositol hexakisphosphate kinase 1 (InsP6K1 [g615.t1]) and a cullin domain-containing protein (g11374.t1) were upregulated after harvest (Table S7). InsP6K forms diphosphoinositol pentakisphosphate [InsP(7)], which induces autophagy (67). Furthermore, InsP6K mediates the assembly/disassembly of the cullin-RING ubiquitin ligases (CRL)-signalosome complex to regulate cell death (68). The vacuolar assembling sorting protein VPS16 (g9687.t2) (Table S7), which is involved in vacuolar assembling for biosynthetic, endocytic, and autophagic pathways (69), was also upregulated after harvest. Typical autophagy-related genes, such as those encoding ATG8 and APG6 (g3164.t2 and g3678.t1, respectively) (70), were expressed in the fruiting body after harvest (Table S4), suggesting that autophagy occurs in fruiting bodies postharvest. We also found that genes encoding proteases, such as subtilisin-like protease and metacaspase (g9815.t1 and g485.t1, respectively), involved in autophagy or programmed cell death (71, 72), were upregulated after harvest (Table S7). These results collectively suggest that programmed cell death occurs after harvest in the Lentinula edodes fruiting body. Nutrient limitation by harvesting will induce postharvest autophagy or programmed cell death.
Conclusion.
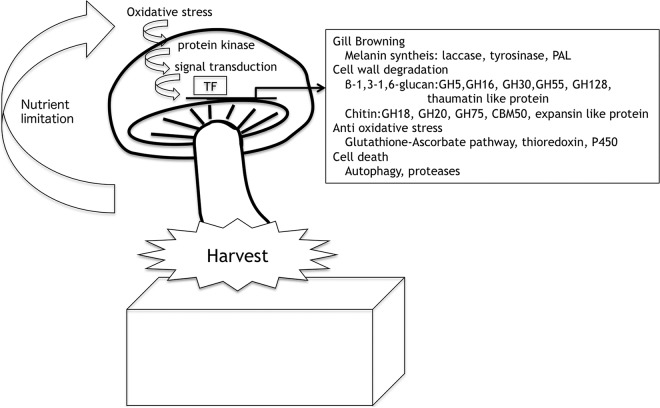
We constructed a de novo assembly of genome sequence data of Lentinula edodes strain G408PP-4 and predicted 12,959 genes from the genome sequence. We also constructed a predicted gene set of Lentinula edodes as a reference sequence for transcriptome analysis. This analysis provided significant insight into freshness control in the Lentinula edodes fruiting body after harvest. We identified novel cell wall-associated genes, putative transcription factors, and putative cell death-related genes (Fig. 2). RNA interference tools are available in Lentinula edodes (40, 73), and TILLING, a method for screening mutants that have a mutation in a target gene (74), will be applicable for breeding. Gene expression for wood decay, biofilm formation in sawdust, medium cultivation, and fruiting body primordia in Lentinula edodes has been reported. Therefore, to carefully identify postharvest-specific genes and mutate the genes by the above techniques, we will construct strains that retain freshness long after harvest. These results collectively suggest that genome sequence data, predicted gene sets, and transcriptome data will be applicable for breeding Lentinula edodes in the near future.
FIG 2.
Working model illustrating the modification of the postharvest quality loss of a Lentinula edodes fruiting body. Oxidative stress will occur after harvest, as will sequential signal transduction via protein kinases and de novo gene expression via transcription factors. De novo gene expression includes genes for gill browning, cell wall degradation, antioxidative stress, and cell death.
MATERIALS AND METHODS
Strains and culture conditions.
Lentinula edodes monokaryotic strain G408PP-4 (NBRC 111202), which was a mating partner of D703PP-9 (ICMP 20921) for linkage map construction (32), was used for genome sequence analysis. The dikaryotic commercially cultivated strain Lentinula edodes KRCF1660 (NBRC 111652, designated H600 [19, 75]) was used for transcriptome analysis. Mycelial cultures on sawdust medium were prepared as described previously (41). Fruiting body growth was performed as described previously (41), and for postharvest analysis, harvested mature fruiting bodies were immediately transferred to a desiccator at 25°C (41) and sampled daily from day 0 (fresh) to day 4. Upon sampling, mushrooms were separated into the pileus, gill, and stipe, and were immediately frozen in liquid nitrogen.
Genome assembling using pyrosequence data.
Genomic DNA was extracted from 2-week-old liquid cultures after crushing the mycelia in liquid nitrogen and using a MasterPure yeast DNA extraction kit (Epicentre Biotechnologies, WI, USA) in accordance wtih the manufacturer's instructions. Libraries for genome sequencing were prepared using a TruSeq DNA sample Prep kit v2 (Illumina, CA, USA), and 76-bp paired-end sequencing was performed with an Illumina Genome Analyzer IIx system. Furthermore, 76-bp paired-end sequencing was performed with a HiSeq 2500 through a custom service provided by Genaris, Inc. (Kanagawa, Japan). The mate pair library was constructed using a Mate Pair library prep kit v2 (Illumina, CA USA) and was sequenced using an Illumina Genome Analyzer IIx system. PacBio sequencing was performed using the custom service provided by Filgen, Inc. (Aichi, Japan). De novo sequences were assembled using Velvet assembler version 0.7.34 (http://www.ebi.ac.uk/~zerbino/velvet/) by varying several parameters and by the clc genomics workbench 8.5.1 (Filgen Inc., Aichi, Japan). We chose a set of contigs created under the conditions that generated the longest N50 for further analyses. Scaffolding of assembled contigs was performed using SSPACE (76) (BaseClear B.V., Netherlands) with sequence reads from the mate pair library. Further scaffolding was carried out using sequence data from PacBio sequence with Pbjelly (77) (http://sourceforge.net/p/pb-jelly/wiki/Home/).
Gene prediction with RNA-seq data.
For RNA-seq analysis, we used RNA from mycelia cultivated for 2 weeks in liquid medium, mycelia grown for 3 weeks on sawdust medium, mycelia grown for 3 months just before fruiting body production, primodium, stipe, and pileus of a young fruiting body, stipe, pileus, and gills of a mature fruiting body, and gills at 4 days after harvest of the fruiting body. For RNA extraction, mycelia were cultured in malt extract-yeast extract-peptone-glucose (MYPG) liquid medium at 25°C with shaking as described previously (41). To extract RNA from mycelia grown on sawdust medium, a membrane filter was placed on the sawdust and covered with 1.5% agar. Mycelia from sawdust cultures were harvested from the surface of the filter membrane 2 weeks after inoculation (19). To extract RNA from fruiting bodies, primordia and fruiting bodies were prepared after harvest, as described previously (27, 78). Equal amounts of RNA were mixed and used for RNA-seq analysis. The construction of a library for RNA-seq and subsequent sequencing by a HiSeq 2500 instrument (Illumina, CA, USA) were carried out using a custom service (Genaris, Inc., Kanagawa, Japan). Gene prediction was performed using Augustus 3.0.1 (79) (http://augustus.gobics.de) using training files from Coprinopsis cinerea and a mixture of RNA from mycelia (grown in liquid medium and sawdust medium), from young fruiting bodies, from mature fruiting bodies, and from fruiting bodies after harvest. Library construction and sequencing using the HiSeq 2500 (Illumina) were performed through a custom service from Genaris, Inc. (Kanagawa, Japan). RNA-seq reads were mapped onto the reference genome using Bowtie2 (80) (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) and TopHat2 (81) (https://ccb.jhu.edu/software/tophat/index.shtml). Intron regions were extracted by filterBam in Augustus; hint files for gene prediction were constructed, and genes were predicted by Augustus (3.0.1) with the hint file. The annotation of predicted genes was performed using Blast2GO Pro (82) in the clc genomics workbench plug-in.
SuperSAGE analysis.
RNA from the gills of fresh fruiting bodies and fruiting bodies 4 days after harvest was used for SuperSAGE according to the method previously described (38). Purified and mixed PCR products for SuperSAGE were applied to cluster formation on the flow cell of the Illumina Genome Analyzer IIx (illumina, CA, USA) and then were sequenced. Matching of tag sequences to Lentinula edodes genes was performed with the blastn algorithm (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), and tag counts were calculated following a scheme described in Fig. S1 in the supplemental material. Statistical analysis was performed using TCC in the R package (83). Gene ontology analysis was performed by Blast2GO (BioBam Bioinformatics S.L., Valencia, Spain) and DAVID (84). Transcription factor genes were analyzed using the Fungal Transcription Factor Database (85) (http://ftfd.snu.ac.kr/index.php?a=view).
Expression of putative cell wall-related enzymes in A. oryzae.
Several putative glucanases containing GH16 domains were heterologously expressed by using the vector pPPAMYBsp in Aspergillus oryzae according to the methods described previously (86). Transformants were cultured for 3 days in dextrin-polypeptone-yeast extract (DPY) medium with shaking, and proteins were expressed and secreted according to the methods described previously (28). Recombinant proteins secreted in the medium were concentrated by precipitation with 80% saturated sulfur ammonium. Recombinant proteins were detected by Western blot analysis and visualized by a penta-His horseradish peroxidase (HRP) conjugate kit (Qiagen GmbH, Germany). Recombinant proteins were purified by a Talon metal affinity resin (TaKaRa Bio, Inc., Tokyo, Japan) and further purified by anion-exchange chromatography using Mono Q (GE Healthcare, UK). Glucanase activity was measured using the methods described previously (87).
Accession number(s).
Scaffold data were deposited in the DDBJ (accession no. BDGU01000001 to BDGU01001951).
Supplementary Material
ACKNOWLEDGMENTS
We thank Akiko Uchidate, Junko Kawaguchi, Ayumi Obara, and Miyuki Ito for their help with the experiments.
Y. Sakamoto, K. Miyazaki, and K. Nakade contributed to the scaffolding of the genome sequence and transcriptome analysis, K. Yoshida and S. Natsume contributed to the de novo assembly of short reads, K. Nakade and S. Sato contributed to the library construction for genome sequencing, and N. Konno contributed to the analysis of cell wall-degrading enzymes. All authors declare no conflict of interest.
This study was funded by the Iwate Biotechnology Research Center, Research Center Forestry and Forest Products Research Institute, and Agriculture, Forestry, Fisheries (scientific technique research promotion program for agriculture, forestry, fisheries, and the food industry, no. 23051).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02990-16.
REFERENCES
- 1.Lin FC, Yang XM, Wang ZW. 2000. Cultivation of the black oak mushroom Lentinula edodes in China, p 955–958. In Proc 15th Int Congr Sci Cultiv Edible Fungi, Maastricht, Netherlands, 15–19 May 2000. [Google Scholar]
- 2.Chang ST, Miles PG. 2004. Mushrooms: cultivation, nutritional value, medicinal effect, and environmental impact, 2nd ed CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 3.Chihara G, Maeda Y, Hamuro J, Sasaki T, Fukuoka F. 1969. Inhibition of mouse sarcoma 180 by polysaccharides from Lentinus edodes (Berk.) sing. Nature 222:687–688. doi: 10.1038/222687a0. [DOI] [PubMed] [Google Scholar]
- 4.Nishitani Y, Zhang L, Yoshida M, Azuma T, Kanazawa K, Hashimoto T, Mizuno M. 2013. Intestinal anti-inflammatory activity of lentinan: influence on IL-8 and TNFR1 expression in intestinal epithelial cells. PLoS One 8:e62441. doi: 10.1371/journal.pone.0062441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X, Yang J, Ning Z, Zhang X. 2015. Lentinula edodes-derived polysaccharide rejuvenates mice in terms of immune responses and gut microbiota. Food Funct 6:2653–2663. doi: 10.1039/C5FO00689A. [DOI] [PubMed] [Google Scholar]
- 6.Feeney MJ, Dwyer J, Hasler-Lewis CM, Milner JA, Noakes M, Rowe S, Wach M, Beelman RB, Caldwell J, Cantorna MT, Castlebury LA, Chang S-T, Cheskin LJ, Clemens R, Drescher G, Fulgoni VL, Haytowitz DB, Hubbard VS, Law D, Myrdal Miller A, Minor B, Percival SS, Riscuta G, Schneeman B, Thornsbury S, Toner CD, Woteki CE, Wu D. 2014. Mushrooms and health summit proceedings. J Nutr 144:1128S–1136S. doi: 10.3945/jn.114.190728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamets P. 2000. Growing gourmet and medicinal mushrooms, 3rd ed, p 201–422. Ten Speed Press, Berkeley, CA. [Google Scholar]
- 8.Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 9.Martin F, Aerts A, Ahrén D, Brun A, Danchin EGJ, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, et al. 2008. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 10.Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, Deng J, Dietrich FS, Fargo DC, Farman ML, Gathman AC, Goldberg J, Guigó R, Hoegger PJ, Hooker JB, Huggins A, James TY, Kamada T, Kilaru S, Kodira C, Kües U, Kupfer D, Kwan HS, Lomsadze A, Li W, Lilly WW, Ma L-J, Mackey AJ, Manning G, Martin F, Muraguchi H, Natvig DO, Palmerini H, Ramesh MA, Rehmeyer CJ, Roe BA, Shenoy N, Stanke M, Ter-Hovhannisyan V, Tunlid A, Velagapudi R, Vision TJ, Zeng Q, Zolan ME, Pukkila PJ. 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci U S A 107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hildén K, Kües U, Labutti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HA, Xu J, Eastwood DC, Foster GD, Sonnenberg ASM, Cullen D, de Vries RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F. 2012. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci U S A 109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FWMR, VanKuyk PA, Horton JS, Grigoriev IV, Wösten HAB. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 13.Park Y-J, Baek JH, Lee S, Kim C, Rhee H, Kim H, Seo J-S, Park H-R, Yoon D-E, Nam J-Y, Kim H-I, Kim J-G, Yoon H, Kang H-W, Cho J-Y, Song E-S, Sung G-H, Yoo Y-B, Lee C-S, Lee B-M, Kong W-S. 2014. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS One 9:e93560. doi: 10.1371/journal.pone.0093560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shim D, Park S-G, Kim K, Bae W, Lee GW, Ha B-S, Ro H-S, Kim M, Ryoo R, Rhee S-K, Nou I-S, Koo C-D, Hong CP, Ryu H. 2016. Whole genome de novo sequencing and genome annotation of the world popular cultivated edible mushroom, Lentinula edodes. J Biotechnol 223:24–25. doi: 10.1016/j.jbiotec.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Gong Y, Cai Y, Liu W, Zhou Y, Xiao Y, Xu Z, Liu Y, Lei X, Wang G, Guo M, Ma X, Bian Y. 2016. Genome sequence of the edible cultivated mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PLoS One 11:e0160336. doi: 10.1371/journal.pone.0160336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki Y, Nakamura M, Babasaki K. 2005. Molecular cloning of developmentally specific genes by representational difference analysis during the fruiting body formation in the basidiomycete Lentinula edodes. Fungal Genet Biol 42:493–505. doi: 10.1016/j.fgb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Chum WWY, Ng KTP, Shih RSM, Au CH, Kwan HS. 2008. Gene expression studies of the dikaryotic mycelium and primordium of Lentinula edodes by serial analysis of gene expression. Mycol Res 112:950–964. doi: 10.1016/j.mycres.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Sakamoto Y, Nakade K, Sato T. 2009. Characterization of the post-harvest changes in gene transcription in the gill of the Lentinula edodes fruiting body. Curr Genet 55:409–423. doi: 10.1007/s00294-009-0255-9. [DOI] [PubMed] [Google Scholar]
- 20.Tang L-H, Jian H-H, Song C-Y, Bao D-P, Shang X-D, Wu D-Q, Tan Q, Zhang X-H. 2013. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Appl Microbiol Biotechnol 97:4977–4989. doi: 10.1007/s00253-013-4832-y. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto Y, Nakade K, Konno K, Sato T. 2012. Senescence of the Lentinula edodes fruiting body after harvesting, p 84–110. In Kapiris K. (ed), Food quality. InTech, Croatia. [Google Scholar]
- 22.Kanda K, Sato T, Ishii S, Enei H, Ejiri S. 1996. Purification and properties of tyrosinase isozymes from the gill of Lentinus edodes fruiting body. Biosci Biotechnol Biochem 60:1273–1278. doi: 10.1271/bbb.60.1273. [DOI] [PubMed] [Google Scholar]
- 23.Sato T, Kanda K, Okawa K, Takahashi M, Watanabe H, Hirano T, Yaegashi K, Sakamoto Y, Uchimiya H. 2009. The tyrosinase-encoding gene of Lentinula edodes, Letyr, is abundantly expressed in the gills of the fruit-body during post-harvest preservation. Biosci Biotechnol Biochem 73:1042–1047. doi: 10.1271/bbb.80810. [DOI] [PubMed] [Google Scholar]
- 24.Nagai M, Kawata M, Watanabe H, Ogawa M, Saito K, Takesawa T, Kanda K, Sato T. 2003. Important role of fungal intracellular laccase for melanin synthesis: purification and characterization of an intracellular laccase from Lentinula edodes fruit bodies. Microbiology 149:2455–2462. doi: 10.1099/mic.0.26414-0. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto Y, Nakade K, Yoshida K, Natsume S, Miyazaki K, Sato S, van Peer AF, Konno N. 2015. Grouping of multicopper oxidases in Lentinula edodes by sequence similarities and expression patterns. AMB Express 5:63. doi: 10.1186/s13568-015-0151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minato K, Mizuno M, Terai H, Tsuchida H. 1999. Autolysis of lentinan, an antitumor polysaccharide, during storage of Lentinus edodes, shiitake mushroom. J Agric Food Chem 47:1530–1532. doi: 10.1021/jf981022w. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto Y, Minato K, Nagai M, Mizuno M, Sato T. 2005. Characterization of the Lentinula edodes exg2 gene encoding a lentinan-degrading exo-beta-1,3-glucanase. Curr Genet 48:195–203. doi: 10.1007/s00294-005-0002-9. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto Y, Watanabe H, Nagai M, Nakade K, Takahashi M, Sato T. 2006. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol 141:793–801. doi: 10.1104/pp.106.076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto Y, Nakade K, Konno N. 2011. Endo-β-1,3-glucanase GLU1, from the fruiting body of Lentinula edodes, belongs to a new glycoside hydrolase family. Appl Environ Microbiol 77:8350–8354. doi: 10.1128/AEM.05581-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konno N, Sakamoto Y. 2011. An endo-β-1,6-glucanase involved in Lentinula edodes fruiting body autolysis. Appl Microbiol Biotechnol 91:1365–1373. doi: 10.1007/s00253-011-3295-2. [DOI] [PubMed] [Google Scholar]
- 31.Muraguchi H, Fujita T, Kishibe Y, Konno K, Ueda N, Nakahori K, Yanagi SO, Kamada T. 2008. The exp1 gene essential for pileus expansion and autolysis of the inky cap mushroom Coprinopsis cinerea (Coprinus cinereus) encodes an HMG protein. Fungal Genet Biol 45:890–896. doi: 10.1016/j.fgb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Miyazaki K, Huang F, Zhang B, Shiraishi S, Sakai M, Shimaya C, Shishido K. 2008. Genetic map of a basidiomycete fungus, Lentinula edodes (shiitake mushroom), constructed by tetrad analysis. Breed Sci 58:23–30. doi: 10.1270/jsbbs.58.23. [DOI] [Google Scholar]
- 33.Miyazaki K, Sakai M, Miyazaki Y. 2010. Mapping of genes abundantly expressed during fruiting body formation of Lentinula edodes. Breed Sci 60:81–86. doi: 10.1270/jsbbs.60.81. [DOI] [Google Scholar]
- 34.Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo J-M, Ge Z-W, Slot JC, Ammirati JF, Baroni TJ, Bougher NL, Hughes KW, Lodge DJ, Kerrigan RW, Seidl MT, Aanen DK, DeNitis M, Daniele GM, Desjardin DE, Kropp BR, Norvell LL, Parker A, Vellinga EC, Vilgalys R, Hibbett DS. 2006. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia 98:982–995. doi: 10.3852/mycologia.98.6.982. [DOI] [PubMed] [Google Scholar]
- 35.Wawrzyn GT, Quin MB, Choudhary S, López-Gallego F, Schmidt-Dannert C. 2012. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem Biol 19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mondego JMC, Carazzolle MF, Costa GGL, Formighieri EF, Parizzi LP, Rincones J, Cotomacci C, Carraro DM, Cunha AF, Carrer H, Vidal RO, Estrela RC, García O, Thomazella DPT, de Oliveira BV, Pires AB, Rio MCS, Araújo MRR, de Moraes MH, Castro LAB, Gramacho KP, Gonçalves MS, Neto JPM, Neto AG, Barbosa LV, Guiltinan MJ, Bailey BA, Meinhardt LW, Cascardo JC, Pereira GAG. 2008. A genome survey of Moniliophthora perniciosa gives new insights into Witches' Broom Disease of cacao. BMC Genomics 9:548. doi: 10.1186/1471-2164-9-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Doré J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Högberg N, Johansson T, Khouja H-R, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Tunlid A, Grigoriev IV, Hibbett DS, Martin F. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 38.Matsumura H, Yoshida K, Luo S, Kimura E, Fujibe T, Albertyn Z, Barrero RA, Krüger DH, Kahl G, Schroth GP, Terauchi R. 2010. High-throughput SuperSAGE for digital gene expression analysis of multiple samples using next generation sequencing. PLoS One 5:e12010. doi: 10.1371/journal.pone.0012010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bianchetti CM, Takasuka TE, Deutsch S, Udell HS, Yik EJ, Bergeman LF, Fox BG. 2015. Active site and laminarin binding in glycoside hydrolase family 55. J Biol Chem 290:11819–11832. doi: 10.1074/jbc.M114.623579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konno N, Nakade K, Nishitani Y, Mizuno M, Sakamoto Y. 2014. Lentinan degradation in the Lentinula edodes fruiting body during postharvest preservation is reduced by downregulation of the exo-β-1,3-glucanase EXG2. J Agric Food Chem 62:8153–8157. doi: 10.1021/jf501578w. [DOI] [PubMed] [Google Scholar]
- 41.Sakamoto Y, Irie T, Sato T. 2005. Isolation and characterization of a fruiting body-specific exo-beta-1,3-glucanase-encoding gene, exg1, from Lentinula edodes. Curr Genet 47:244–252. doi: 10.1007/s00294-005-0563-7. [DOI] [PubMed] [Google Scholar]
- 42.van de Rhee MD, Mendes O, Werten MWT, Huizing HJ, Mooibroek H. 1996. Highly efficient homologous integration via tandem exo-β-1,3-glucanase genes in the common mushroom, Agaricus bisporus. Curr Genet 30:166–173. doi: 10.1007/s002940050116. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka H, Yoshida G, Baba Y, Matsumura K, Wasada H, Murata J, Agawa M, Itakura S, Enoki A. 2007. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol 128:500–511. doi: 10.1016/j.jbiotec.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Kawai R, Igarashi K, Yoshida M, Kitaoka M, Samejima M. 2006. Hydrolysis of beta-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-beta-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 71:898–906. doi: 10.1007/s00253-005-0214-4. [DOI] [PubMed] [Google Scholar]
- 45.Niu X, Liu C-C, Xiong Y-J, Yang M-M, Ma F, Liu Z-H, Yuan S. 2016. The modes of action of ChiIII, a chitinase from mushroom Coprinopsis cinerea, shift with changes in the length of GlcNAc oligomers. J Agric Food Chem 64:6958–6968. doi: 10.1021/acs.jafc.6b03086. [DOI] [PubMed] [Google Scholar]
- 46.Galante RS, Taranto AG, Koblitz MGB, Góes-Neto A, Pirovani CP, Cascardo JCM, Cruz SH, Pereira GAG, De Assis SA. 2012. Purification, characterization and structural determination of chitinases produced by Moniliophthora perniciosa. An Acad Bras Cienc 84:469–486. doi: 10.1590/S0001-37652012000200016. [DOI] [PubMed] [Google Scholar]
- 47.Kang Y, Kim H, Choi HT. 2013. Biochemical characterization of chitinase 2 expressed during the autolytic phase of the inky cap, Coprinellus congregatus. J Microbiol 51:189–193. doi: 10.1007/s12275-013-2535-9. [DOI] [PubMed] [Google Scholar]
- 48.Konno N, Takahashi H, Nakajima M, Takeda T, Sakamoto Y. 2012. Characterization of β-N-acetylhexosaminidase (LeHex20A), a member of glycoside hydrolase family 20, from Lentinula edodes (shiitake mushroom). AMB Express 2:29. doi: 10.1186/2191-0855-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konno N, Obara A, Sakamoto Y. 2015. Molecular cloning, characterization, and expression analysis of a β-N-acetylhexosaminidase (LeHex20B) from the shiitake mushroom, Lentinula edodes. J Wood Sci 61:178–184. doi: 10.1007/s10086-015-1459-x. [DOI] [Google Scholar]
- 50.Akcapinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V. 2015. Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet 61:103–113. doi: 10.1007/s00294-014-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. 2002. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci U S A 99:9055–9060. doi: 10.1073/pnas.132080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tovar-Herrera OE, Batista-García RA, Sánchez-Carbente M del R, Iracheta-Cárdenas MM, Arévalo-Niño K, Folch-Mallol JL. 2015. A novel expansin protein from the white-rot fungus Schizophyllum commune. PLoS One 10:e0122296. doi: 10.1371/journal.pone.0122296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pochanavanich P, Suntornsuk W. 2002. Fungal chitosan production and its characterization. Lett Appl Microbiol 35:17–21. doi: 10.1046/j.1472-765X.2002.01118.x. [DOI] [PubMed] [Google Scholar]
- 54.Crestini C, Kovac B, Giovannozzi-Sermanni G. 1996. Production and isolation of chitosan by submerged and solid-state fermentation from Lentinus edodes. Biotechnol Bioeng 50:207–210. doi: 10.1002/bit.260500202. [DOI] [PubMed] [Google Scholar]
- 55.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyazaki Y, Tsunoka O, Shishido K. 1997. Determination of the DNA-binding sequences of the Zn(II)2Cys6 zinc-cluster-containing PRIB protein, derived from the basidiomycete Lentinus edodes gene. J Biochem 122:1088–1091. doi: 10.1093/oxfordjournals.jbchem.a021866. [DOI] [PubMed] [Google Scholar]
- 57.Thomas JO, Travers AA. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26:167–174. doi: 10.1016/S0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 58.Antosch M, Mortensen SA, Grasser KD. 2012. Plant proteins containing high mobility group box DNA-binding domains modulate different nuclear processes. Plant Physiol 159:875–883. doi: 10.1104/pp.112.198283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wissmüller S, Kosian T, Wolf M, Finzsch M, Wegner M. 2006. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res 34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weijn A, Bastiaan-Net S, Wichers HJ, Mes JJ. 2013. Melanin biosynthesis pathway in Agaricus bisporus mushrooms. Fungal Genet Biol 55:42–53. doi: 10.1016/j.fgb.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Hodges DM. 2003. Overview: oxidative stress and postharvest produce, p 1–12. In Hodges DM. (ed), Postharvest oxidative stress in horticultural crops. Food Products Press, New York, NY. [Google Scholar]
- 62.Lum GB, Shelp BJ, DeEll JR, Bozzo GG. 2016. Oxidative metabolism is associated with physiological disorders in fruits stored under multiple environmental stresses. Plant Sci 245:143–152. doi: 10.1016/j.plantsci.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Blokhina O, Fagerstedt KV. 2010. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol Biochem 48:359–373. doi: 10.1016/j.plaphy.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Anjum NA, Umar S, Chan MT (ed). 2010. Ascorbate-glutathione pathway and stress tolerance in plants. Springer Netherlands, Houten, Netherlands. [Google Scholar]
- 65.Jiang T, Jahangir MM, Jiang Z, Lu X, Ying T. 2010. Influence of UV-C treatment on antioxidant capacity, antioxidant enzyme activity and texture of postharvest shiitake (Lentinus edodes) mushrooms during storage. Postharvest Biol Technol 56:209–215. doi: 10.1016/j.postharvbio.2010.01.011. [DOI] [Google Scholar]
- 66.Hirt H. 2000. Connecting oxidative stress, auxin, and cell cycle regulation through a plant mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A 97:2405–2407. doi: 10.1073/pnas.97.6.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J, Shibata M, Takizawa S, Takagi S. 2010. Inositol hexakisphosphate kinases promote autophagy. Int J Biochem Cell Biol 42:2065–2071. doi: 10.1016/j.biocel.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 68.Rao F, Xu J, Khan AB, Gadalla MM, Cha JY, Xu R, Tyagi R, Dang Y, Chakraborty A, Snyder SH. 2014. Inositol hexakisphosphate kinase-1 mediates assembly/disassembly of the CRL4-signalosome complex to regulate DNA repair and cell death. Proc Natl Acad Sci U S A 111:16005–16010. doi: 10.1073/pnas.1417900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wartosch L, Günesdogan U, Graham SC, Luzio JP. 2015. Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic 16:727–742. doi: 10.1111/tra.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Yang Y, Ming M, Liu B. 2011. Mitochondrial ROS generation for regulation of autophagic pathways in cancer. Biochem Biophys Res Commun 414:5–8. doi: 10.1016/j.bbrc.2011.09.046. [DOI] [PubMed] [Google Scholar]
- 71.van Diepeningen AD, Engelmoer DJP, Sellem CH, Huberts DHEW, Slakhorst SM, Sainsard-Chanet A, Zwaan BJ, Hoekstra RF, Debets AJM. 2014. Does autophagy mediate age-dependent effect of dietary restriction responses in the filamentous fungus Podospora anserina? Philos Trans R Soc Lond B Biol Sci 369:20130447. doi: 10.1098/rstb.2013.0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV. 2011. Metacaspases. Cell Death Differ 18:1279–1288. doi: 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakade K, Watanabe H, Sakamoto Y, Sato T. 2011. Gene silencing of the Lentinula edodes lcc1 gene by expression of a homologous inverted repeat sequence. Microbiol Res 166:484–493. doi: 10.1016/j.micres.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 74.McCallum CM, Comai L, Greene EA, Henikoff S. 2000. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol 123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakamoto Y, Nakade K, Yano A, Nakagawa Y, Hirano T, Irie T, Watanabe H, Nagai M, Sato T. 2008. Heterologous expression of lcc1 from Lentinula edodes in tobacco BY-2 cells results in the production an active, secreted form of fungal laccase. Appl Microbiol Biotechnol 79:971–980. doi: 10.1007/s00253-008-1507-1. [DOI] [PubMed] [Google Scholar]
- 76.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 77.English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, Gibbs RA. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirano T, Sato T, Okawa K, Kanda K, Yaegashi K, Enei H. 1999. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Lentinus edodes. Biosci Biotechnol Biochem 63:1223–1227. doi: 10.1271/bbb.63.1223. [DOI] [PubMed] [Google Scholar]
- 79.Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conesa A, Götz S. 2008. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun J, Nishiyama T, Shimizu K, Kadota K. 2013. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 85.Park J, Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Kim S, Park B, Jung K, Kim S, Kang S, Lee Y-H. 2008. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24:1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 86.Gomi K, Iimura Y, Hara S. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric Biol Chem 51:2549–2555. doi: 10.1271/bbb1961.51.2549. [DOI] [Google Scholar]
- 87.Takeda T, Nakano Y, Takahashi M, Sakamoto Y, Konno N. 2013. Polysaccharide-inducible endoglucanases from Lentinula edodes exhibit a preferential hydrolysis of 1,3-1,4-β-glucan and xyloglucan. J Agric Food Chem 61:7591–7598. doi: 10.1021/jf401543m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.