Abstract
Vicinal oxygen chelate proteins (VOC) are members of the metalloenzyme superfamily, which plays roles in many biological reactions. Some members of the VOC superfamily have been systematically characterized but not in Brassica napus. In this study, 38 VOC genes were identified based on their conserved domains. The present results revealed that most of the BnaVOC genes have few introns, and all contained the typical VOC structure of βαβββ modules. The BnaVOC genes are distributed unevenly across 15 chromosomes in B. napus and occur as gene clusters on chromosomes C5 and A6. The synteny and phylogenetic analyses revealed that the VOC gene family is a consequence of mesopolyploidy events that occurred in Brassica evolution, and whole-genome duplication and segmental duplication played a major role in the expansion of the BnaVOC gene family. The expression profile analysis indicated that the expression of most BnaVOCs was increased in the leaves and late stage seeds. Further results indicated that seeds of B. napus with a high oil content show higher expression levels under drought stress conditions, suggesting that BnaVOCs not only respond to abiotic stress but may also affect lipid metabolism in drought stress. This present study provides a comprehensive overview of the VOC gene family and provides new insights into their biological function in B. napus evolution.
Keywords: vicinal oxygen chelate proteins (VOC), B. napus, gene duplication, gene evolution, drought stress, gene expression
Introduction
Vicinal oxygen chelate proteins (VOC) are members of an enzyme superfamily that could catalyse reactions with a common mechanistic attribute that is enabled by certain conserved active site residues. These residues perform the same functions in all members of the superfamily (Gerlt and Babbitt, 2001). The bidentate coordination to a divalent metal center through vicinal oxygen atoms is essential for activation or stabilization and is necessary for the functional mechanism of the VOC superfamily (He and Moran, 2011). In this superfamily, the coordination is mediated by a topological structure (βαβββ modules) that offers a ligand that shields a metal ion (Armstrong, 2000).
Based on the chemistry of the catalyzing enzyme, the VOC superfamily was divided into non-dioxygenase and dioxygenase groups. The non-dioxygenase group includes two families (isomerase and nucleophilic addition) that can catalyse non-redox active reactions. Because the metal center of dioxygenase has many function, such as activating dioxygen and organizing the co-substrates, dioxygenase also contain two families: extradiol dioxygenase and α-keto acid oxygenase (He and Moran, 2011). The first studied members of the VOC superfamily were glyoxalase I (GLYI), fosfomycin resistance protein (FosA), the related fosfomycin resistance protein X (FosX) and extradiol dioxygenases (EXDXs); EXDXs include 2,3-dihydroxybiphenyl1,2-dioxygenase (DHBD), catechol 2,3-dioxygenase (C23O), and homoprotocatechuate 2,3-dioxygenase (HPCD; Bernat et al., 1997). In 2000, methylmalonyl-CoA epimerase (MMCE) was added to the VOC superfamily based on its sequence and biochemical analysis (Armstrong, 2000). Then, the structures of 4-hydroxyphenylpyruvate dioxygenase (HPPD) and hydroxymandelate synthase (HMS) were also identified, and it was recognized that they should be considered members of the VOC superfamily (McCarthy et al., 2001; Brownlee et al., 2008).
GLYI is an important metalloenzyme that participates in the glyoxalase system, which has been reported to be a major pathway for detoxification of methylglyoxal (MG) in living organisms (Thornalley, 1990). MG is formed as a cytotoxic α-ketoaldehyde by-product of carbohydrate and lipid metabolism. MG reacts with nucleic acid molecules and protein to form adducts and can be harmful to organisms. GLYI can use one molecule of glutathione (GSH) to convert MG to S-Dlactoylglutathione (Singla-Pareek et al., 2003). The over-expression of GLYI cloned from Brassica juncea could lead to improved abiotic stress tolerance in transgenic tobacco (Reddy and Sopory, 1999). Gly I is a gene that is induced by drought and cold stress in Arabidopsis (Seki et al., 2001). Nineteen and twenty-two GLYIs have been identified in rice and Arabidopsis thaliana, respectively, and further research studies have indicated that these GLYIs were highly expressed under abiotic stress (Mustafiz et al., 2011). HPPD is another member of the VOC superfamily, and the constitutive over-expression of the barley HPPD gene could enhance the vitamin E content in transgenic tobacco seeds (Falk et al., 2003). The expression of the barley HPPD gene during senescence is most likely related to oxidative stress (Falk et al., 2002). In addition, a member of the VOC superfamily has been reported to be up-regulated during desiccation in the leaves, roots, and seeds of the resurrection plant Xerophyta humilis (Mulako et al., 2008). In addition, the desiccation induced-1VOC (dsi-1VOC) protein was also observed in Brassica napus with a high oil content, suggesting that it might protect the embryonic developmental process from harm under drought conditions (Gan et al., 2013).
B. napus (AACC, 2n = 38) is an allopolyploid species with a triplicated genome structure and many duplicated genes, which originated from Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18; Allender and King, 2010). B. napus is the third largest oil seed crop in the world. However, few studies have focused on the VOC superfamily in B. napus, except for the reported appearance of the VOC protein in high oil content B. napus lines (Gan et al., 2013), suggesting that the VOC protein may contribute to dehydration tolerance during the oil-accumulation period and help increase the oil content in B. napus. Because B. napus originated from hybridization, its genome contains many inversions or translocations and duplications (Chalhoub et al., 2014). Mesopolyploidization events were identified in Brassicaceae evolution, and increasing drought and transient glaciation events coincided with the Brassicaceae major evolutionary splits. Genomic differentiation events resulted in whole genome triplication (WGT), which formed B. napus (Cheng et al., 2014). Structural and functional divergence of duplicated genes within gene families were also found in other gene families, such as the mega-6 fatty acid desaturase (FAD2) gene family, the 12-oxo-phytodienoic acid reductases (OPRs) gene family, and the NBS-encoding gene family (Schlueter et al., 2007; Li et al., 2009; Yu et al., 2014). The characteristics of the duplicated genes suggest that evolution could have caused an adaptive structural diversification, and this process is pervasive and could have contributed to the biological novelty in plants (Li et al., 2009). Thus, many duplicated genes have similar gene sequences but different functional performances, and the large number of duplicated genes in polyploid plants complicates phylogenetic and evolutionary analyses. In addition, the structural and functional divergence of duplicated genes within a gene family could offer support for the gene expansion pattern in the species evolution (Liang et al., 2016). A systemic analysis of the VOC superfamily in B. napus has not been reported. In this study, the VOC superfamily in B. napus was identified, and its structure, evolution and synteny relationship with BnaGLYIs and BnaHPPDs were analyzed. In addition, the expression pattern of VOC superfamily members in different tissues of B. napus were also analyzed. This research study provides a foundation for future studies regarding the VOC superfamily in B. napus.
Materials and methods
Identification of VOC family genes in B. napus and other species
The VOC genes were identified in B. napus based on their homology with the 22 GLYI protein (Mustafiz et al., 2011) and 1 HPPD sequences from Arabidopsis using the BLAT search program in the CNS-Genoscope database (http://www.genoscope.cns.fr/brassicanapus/; Chalhoub et al., 2014). Redundant sequences were removed manually. The VOC genes in B. rapa, B. oleracea, Brassica nigra were obtained from the Brad database (http://brassicadb.org/brad/; Wang et al., 2011). All BnaVOC gene candidates were analyzed using the Hidden Markov Model (HMM), which is a statistical Markov model in which the modeled system is assumed to be a Markov process with unobserved (hidden) states; for this analysis, the following three databases were used for confirmation: the Pfam database (http://pfam.sanger.ac.uk/search; Finn et al., 2010), SMART database (http://smart.embl-heidelberg.de/; Letunic et al., 2004), and NCBI Conserved Domain Search database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; Marchler-Bauer et al., 2015).
A univocal name consisting of two italic letters denoting the source organism, the family name, and subfamily numeral of each gene was assigned to each VOC gene (e.g., BnaGLYI1; Ostergaard and King, 2008). The number of amino acids, CDS lengths, and chromosome locations of the BnaVOC genes were obtained from the B. napus database.
The physicochemical parameters, including the molecular weight (kDa) and pI, of each BnaVOC protein were calculated using the compute pI/Mw tool in ExPASy (http://www.expasy.org/tools/). GRAVY (grand average of hydropathy) values were calculated using the PROTPARAM tool (http://web.expasy.org/protparam/; Gasteiger et al., 2003). The subcellular location predictions were conducted using the TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/) server (Emanuelsson et al., 2007) and Protein Prowler Subcellular Localization Predictor version 1.2 (http://bioinf.scmb.uq.edu.au:8080/; Bodén and Hawkins, 2005).
Multiple alignment and phylogenetic analysis of the VOC family genes
Multiple sequence alignment of all predicted BnaVOC, BrVOC, BoVOC protein sequences was performed using ClustalW software. An unrooted phylogenetic tree of these full-length VOC protein sequences was constructed using MEGA 6 with the Neighbor Joining (NJ) method, and a bootstrap analysis was conducted using 1,000 replicates (Higgins and Sharp, 1988; Tamura et al., 2013).
Gene structure analysis of the BnaVOC family genes
The exon-intron structures of the BnaVOC family genes were determined based on the alignments of their coding sequences with the corresponding genomic sequences, and a diagram was created using GSDS (Gene structure display server: http://gsds.cbi.pku.edu.cn/; Hu et al., 2015). MEME (Multiple Expectation Maximization for Motif Elicitation; http://alternate.meme-suite.org/) was used to identify the conserved motif structures encoded by the BnaVOC family genes (Bailey et al., 2009). A secondary structure analysis was carried out using the following two different tools: PBIL GOR4 (Jones, 1999) and PSIPRED (Buchan et al., 2013). The Tertiary structures of the BnaVOCs were predicted using Phyre2 (Kelley and Sternberg, 2009), and they were analyzed using VAST.
Chromosomal location and gene duplication of the BnaVOC family genes
The chromosomal locations of the BnaVOC genes were determined based on the positional information obtained from the B. napus database. Tandemly duplicated VOC genes were defined as adjacent to homologous VOC genes on B. napus chromosomes or within a sequence distance of 50 kb (Yu et al., 2014). The synteny relationships between the BnaVOCs and AtVOCs, BrVOCs, and BoVOCs were evaluated using the search syntenic genes tool in BRAD (http://brassicadb.org/brad/; Wang et al., 2011) and the synteny tools in the B. napus Genome Browser (Chalhoub et al., 2014).
Calculation of the Ka/Ks values in the BnaVOC family genes
The VOC gene sequences of each paralogous pair were first aligned using ClustalW. Files containing the multiple sequence alignments of the VOC gene sequences were then converted to a PHYLIP alignment using MEGA. Then, the converted sequence alignments were imported into the YN00 program in PAML to calculate the non-synonymous and synonymous substitution rates (Yang, 2007).
Plant materials, drought treatment, RNA extraction, and qRT-PCR analysis
The late grown plants (40 days after pollination) from the high oil content (55.19%) and low oil content (36.49%) B. napus lines were moved into a green chamber at 25°C with a 16-h light/8-h dark photoperiod. After 10 days of adaption, they were treated. For the drought stress treatment, water was withheld from these plants for 30 days. After 10, 20, or 30 days of treatment, RNA was extracted from the siliques. An RNAprep Pure Plant Kit (Tiangen) was used to isolate the total RNA from each frozen sample, and first-strand cDNA was synthesized from the RNA using a PrimeScriptTM RT Master Mix Kit (TaKaRa) according to the manufacturer's instructions.
Gene-specific primers were designed using Primer5.0 (Table S3). Each reaction was carried out in triplicate with a reaction volume of 20 μl containing 1.6 μl of gene-specific primers (1.0 μM), 1.0 μl of cDNA, 10 μl of SYBR green (TaKaRa), and 7.4 μl of sterile distilled water. The PCR conditions were as follows: Stage 1: 95°C for 3 min; stage 2: 40 cycles of 15 s at 95°C and 45 s at 60°C; and stage 3: 95°C for 15 s, 60°C for 1 min, 95°C for 15 s. At stage 3, a melting curve was generated to estimate the specificity of the reactions. A housekeeping gene (actin) that is constitutively expressed in B. napus was used as a reference for the normalization and analyzed using an ABI3100 DNA sequencer (Applied Biosystems; Quantitation-Comparative: ΔΔCT); three biological replicates were performed for the qRT-PCR analysis (Kagale et al., 2007).
Results
Genome-wide identification of the VOC superfamily genes in the B. napus genome
In total, 38 genes in the B. napus genome were identified as VOC superfamily genes using the CNS-Genoscope database based on their homology with GLYI genes and HPPD genes from Arabidopsis (Table 1). The GLYI and HPPD families were observed, while the other seven subfamilies were not found in B. napus, and no homologous genes were reported in Arabidopsis (FosA, FosX, DHBD, C23O, HPCD, MMCE, and HMS). The GLYI family and HPPD family in B. napus contains 34 members and 4 members, and the members were named BnaGLYI1-BnaGLYI34 and BnaHPPD1-BnaHPPD4, respectively. The physicochemical parameters of each VOC gene were calculated using ExPASy. Except for BnaGLYI11 and BnaGLYI12, the VOC (BnaVOC) proteins in B. napus had a molecular mass <50 KDa. In total, 31 of the BnaVOC proteins had relatively low isoelectric points (pI < 7). Nearly all the BnaVOCs had a GRAVY value < 0, indicating that a large proportion of the BnaVOC proteins are hydrophilic. Low hydrophobicity is a feature observed in other drought stress induced proteins (Beck et al., 2007), suggesting that the BnaVOC proteins may play a role under drought stress conditions. PProwler and TargetP were used to predict the subcellular location of the 38 BnaVOC proteins; the subcellular locations of the different BnaVOC proteins were diverse, and some proteins are predicted to be located in secretory pathways and the nucleus (Table S1).
Table 1.
VOC genes in B. napus genome and their sequence characteristics and subcellular location prediction.
| Name | Gene ID | Family | Chr. | Gene position | Gene length (bp) | protein length (aa) | Mol.Wt. (KD) | pI | GRAVY | Intron number | Subcellular location PProwler TargetP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||||||
| BnaGLYI1 | BnaA09g49270D | Glyoxalase | A9 | 32,814,199 | 32,815,336 | 1,138 | 137 | 15.3942 | 5.84 | −0.4635036 | 2 | Other | S |
| BnaGLYI2 | BnaA06g04170D | Glyoxalase | A6 | 2,547,123 | 2,548,306 | 1,184 | 171 | 19.2747 | 7.77 | −0.4046783 | 3 | Other | S |
| BnaGLYI3 | BnaC05g05340D | Glyoxalase | A5 | 2,609,787 | 2,610,529 | 743 | 138 | 15.5964 | 5.94 | −0.4688405 | 2 | Other | S |
| BnaGLYI4 | BnaCnng38880D | Glyoxalase | Cn-R | 37,521,585 | 37,522,696 | 1,112 | 137 | 15.5023 | 6.2 | −0.5124087 | 2 | Other | S |
| BnaGLYI5 | BnaC05g05770D | Glyoxalase | C5 | 2,848,240 | 2,850,316 | 2,077 | 235 | 26.4389 | 8.74 | −0.4885106 | 7 | C | C |
| BnaGLYI6 | BnaA06g04580D | Glyoxalase | A6 | 2,708,014 | 2,710,119 | 2,106 | 237 | 26.6351 | 8.33 | −0.4206751 | 7 | C | C |
| BnaGLYI7 | BnaC03g51010D | Glyoxalase | C3 | 35,479,070 | 35,480,133 | 1,064 | 150 | 16.9921 | 5.25 | −0.4893333 | 4 | Other | O |
| BnaGLYI8 | BnaCnng47290D | Glyoxalase | Cn-R | 46,764,251 | 46,765,432 | 1,182 | 143 | 16.1803 | 5.39 | −0.4349650 | 5 | Other | |
| BnaGLYI9 | BnaC08g15100D | Glyoxalase | C8 | 19,658,835 | 19,661,229 | 2,395 | 283 | 31.8142 | 5.26 | −0.3279151 | 7 | Other | O |
| BnaGLYI10 | BnaA08g25110D | Glyoxalase | A8 | 17,357,805 | 17,359,871 | 2,067 | 283 | 31.8724 | 5.26 | −0.3215547 | 7 | Other | O |
| BnaGLYI11 | BnaC05g08770D | Glyoxalase | C5 | 4,659,651 | 4,665,014 | 5,364 | 724 | 80.6725 | 7.49 | −0.2138121 | 19 | M | M |
| BnaGLYI12 | BnaA06g07360D | Glyoxalase | A6 | 3,915,659 | 3,923,909 | 8,251 | 1215 | 134.9846 | 8.86 | −0.2618930 | 26 | M | M |
| BnaGLYI13 | BnaC05g11680D | Glyoxalase | C5 | 6,800,739 | 6,802,181 | 1,443 | 174 | 19.7337 | 5.68 | −0.1994252 | 2 | Other | O |
| BnaGLYI14 | BnaA08g23870D | Glyoxalase | A8 | 1,684,3513 | 16,844,540 | 1,028 | 174 | 19.8469 | 5.88 | −0.2264367 | 2 | Other | O |
| BnaGLYI15 | BnaA06g10060D | Glyoxalase | A6 | 5,342,612 | 5,343,786 | 1,175 | 174 | 19.7787 | 5.68 | −0.2086206 | 2 | Other | O |
| BnaGLYI16 | BnaC08g16660D | Glyoxalase | C8 | 20,576,966 | 20,578,018 | 1,053 | 174 | 19.8218 | 5.89 | −0.2862068 | 2 | Other | O |
| BnaGLYI17 | BnaA09g56790D | Glyoxalase | A9-R | 3,889,968 | 3,891,223 | 1,256 | 167 | 18.9438 | 5.86 | −0.2449101 | 2 | M | |
| BnaGLYI18 | BnaC08g38920D | Glyoxalase | C8 | 34,930,459 | 34,931,660 | 1,202 | 173 | 19.5686 | 5.86 | −0.2150289 | 2 | ||
| BnaGLYI19 | BnaA02g19970D | Glyoxalase | A2 | 12,337,272 | 12,338,438 | 1,167 | 167 | 18.8186 | 5.81 | −0.2095808 | 2 | Other | O |
| BnaGLYI20 | BnaC02g46640D | Glyoxalase | C2-R | 2,394,713 | 2,395,834 | 1,122 | 167 | 18.8046 | 5.66 | −0.2071856 | 2 | Other | O |
| BnaGLYI21 | BnaC02g23290D | Glyoxalase | C2 | 20,281,012 | 20,282,133 | 1,122 | 167 | 18.7625 | 5.66 | −0.2347305 | 2 | Other | O |
| BnaGLYI22 | BnaC09g12330D | Glyoxalase | C9 | 8,747,895 | 8,748,643 | 749 | 118 | 12.9659 | 6.4 | 0.09152542 | 2 | M | M |
| BnaGLYI23 | BnaA09g12000D | Glyoxalase | A9 | 6,301,614 | 6,302,406 | 793 | 118 | 12.9839 | 6.4 | 0.04745762 | 2 | M | M |
| BnaGLYI24 | BnaC09g11920D | Glyoxalase | C9 | 8,321,491 | 8,322,355 | 865 | 118 | 12.9839 | 6.4 | 0.02966101 | 2 | M | M |
| BnaGLYI25 | BnaC06g28360D | Glyoxalase | C6 | 29,576,211 | 29,578,606 | 2,396 | 345 | 38.3689 | 6.19 | −0.2481159 | 8 | C | C |
| BnaGLYI26 | BnaA07g26290D | Glyoxalase | A7 | 19,358,162 | 19,361,477 | 3,316 | 341 | 37.8944 | 6.48 | −0.2533724 | 8 | C | C |
| BnaGLYI27 | BnaC04g15790D | Glyoxalase | C4 | 13,701,067 | 13,701,624 | 558 | 34 | 3.8642 | 6.7 | −1.6470588 | 1 | Other | O |
| BnaGLYI28 | BnaA07g13890D | Glyoxalase | A7 | 12,263,440 | 12,263,994 | 555 | 184 | 2.0887 | 4.78 | −0.6913043 | 0 | Other | O |
| BnaGLYI29 | BnaA05g35240D | Glyoxalase | A5-R | 670,044 | 672,049 | 2,006 | 138 | 15.2522 | 5.46 | −0.2255474 | 3 | Other | O |
| BnaGLYI30 | BnaCnng59150D | Glyoxalase | Cn-R | 58,862,580 | 58,863,862 | 1,283 | 137 | 15.2531 | 5.45 | −0.2788321 | 3 | Other | O |
| BnaGLYI31 | BnaA09g11460D | Glyoxalase | A9 | 5,894,872 | 5,895,565 | 694 | 118 | 12.9378 | 6.4 | 0.07288135 | 2 | M | M |
| BnaGLYI32 | BnaA10g11070D | Glyoxalase | A10 | 9,346,288 | 9,347,612 | 1,325 | 195 | 21.959 | 6.71 | −0.4492307 | 3 | O | |
| BnaGLYI33 | BnaC03g13130D | Glyoxalase | C3 | 6,313,042 | 6,314,254 | 1,213 | 193 | 21.7128 | 7.77 | −0.4398963 | 3 | O | |
| BnaGLYI34 | BnaA03g10440D | Glyoxalase | A3 | 4,700,661 | 4,701,804 | 1,144 | 193 | 21.7859 | 8.43 | −0.4880829 | 3 | C | |
| BnaHPPD1 | BnaC05g04530D | HPPD | C5 | 2,231,485 | 2,233,489 | 2,005 | 445 | 48.8579 | 5.65 | −0.2617977 | 1 | Other | O |
| BnaHPPD2 | BnaA09g49870D | HPPD | A9 | 33,093,200 | 33,095,241 | 2,042 | 440 | 48.0721 | 5.72 | −0.2372727 | 1 | Other | O |
| BnaHPPD3 | BnaC08g44820D | HPPD | C8 | 37,849,659 | 37,851,160 | 1,502 | 440 | 48.1622 | 5.46 | −0.2440909 | 1 | Other | O |
| BnaHPPD4 | BnaA10g04310D | HPPD | A10 | 2,268,795 | 2,272,272 | 3,478 | 587 | 48.973 | 5.45 | −0.2878651 | 11 | Other | O |
Sequence alignment and phylogenetic analysis of the VOC genes in B. napus, B.rapa, B. oleracea, B. nigra, and A. thaliana
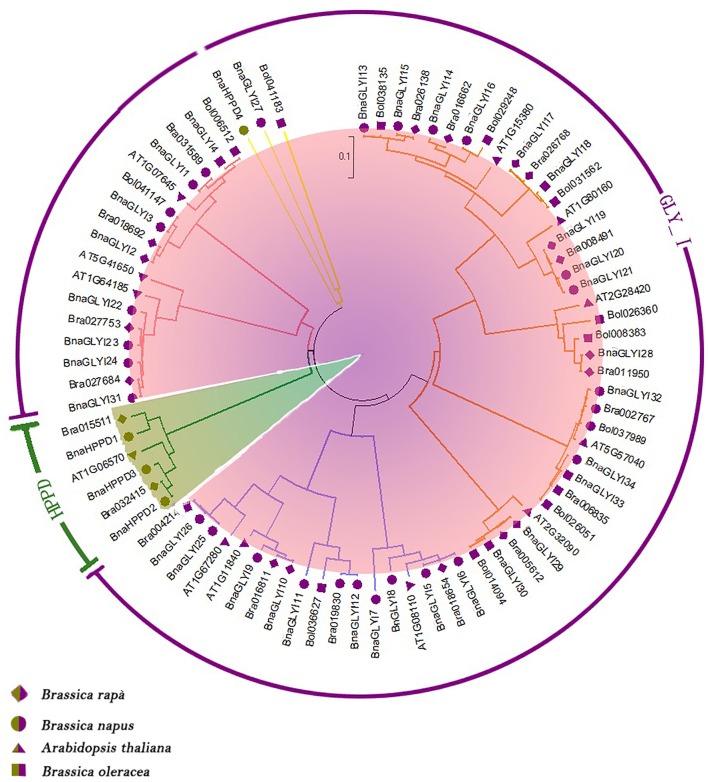
To determine the similarity and homology of the BnaVOC genes in B. napus and other brassica species, sequence alignments and a phylogenetic analysis were performed. The GLYI genes and HPPD genes in B. rapa, B. oleracea, and A. thaliana were used to construct an unrooted phylogenetic tree (Figure 1). Eighteen and ten homologous genes were identified in B. rapa and B. oleracea, respectively (Figure 1, Table S4). All VOC gene sequences clustered into five main clades, and almost half of the GLYI genes were clustered in one branch. Furthermore, the VOC genes in another diploid Brassica species, Brassica nigra (B. nigra), were also analyzed. Fourteen homologous VOC genes were detected in B. nigra (Table S4). Interestingly, BnaGLYI27, Bol041183, and BnaHPPD4 were in the same clade, but this clade has a long branch length; on the one hand, this clade might result from the characteristics of their sequences because they contain consensus sequences; on the other hand, this result suggested that the divergence has not occurred in recent years. Almost all the B. napus VOC genes appeared as pair-wises with the VOC genes in B. oleracea or B. rapa in the phylogenetic relationship, and they contained long branch lengths, indicating that they have been evolved for a long time. For the phylogenetic analysis of the Brassica species, the VOC genes in B. nigra were usually in a different clade from the pair-wise genes, except for BniB014868, BniB034245, and BniB003628 (Figure S4). This result suggested that B. nigra had a farther relationship with B. rapa, B. oleracea, and B. napus, but BnaHPPD1, BnaGLYI7, and Bol026360 may be more closely related to B. nigra. While the AtVOC genes were in the same clade, they were on opposite sides, which is consistent with the evolutionary history of these Brassica plants. Altogether, these results indicated that in the VOC gene family, BnaVOCs, BoVOCs, and BrVOCs have a high homology. Some genes (BnaGLYI 27 and BnaHPPD4) were in the same clade, but they had different conserved domains, suggesting that the other parts of these genes have evolved a closer phylogenetic relationship.
Figure 1.
Phylogenetic analysis of the B. napus, B. rapa, B. oleracea, and A. thaliana VOC genes. VOC gene families are distinguished by different colors. The unrooted tree was generated by using ClustalW in MEGA with the full-length amino acid sequences of the VOC proteins.
Chromosomal location and expansion pattern analysis of the VOC genes in B. napus
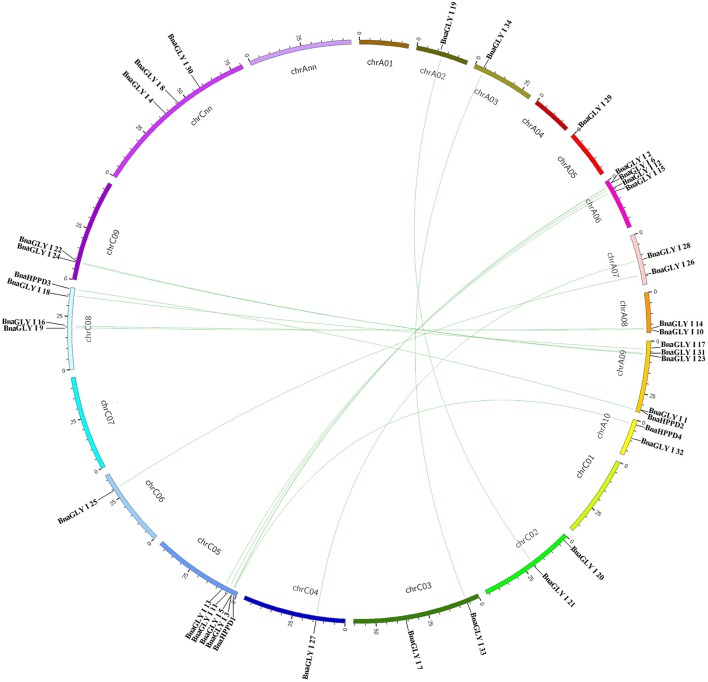
The chromosomal location of the BnaVOC genes was analyzed, and the positions and chromosome locations of 35 BnVOC genes were clearly identified on the 14 chromosomes in B. napus (Table 1, Figure 2). The number of BnaVOC genes varies among the different chromosomes, and no BnaVOC genes were observed on chromosome A1, A4, C1, and C7. Chromosome C5 contains the greatest number of BnaVOC genes, and the five BnaVOC genes that are located on chromosome C5 appear as a gene cluster. The final chromosomal location may result from VOC gene duplication in the long evolutionary history. Compared with A. thaliana, Brassica species experienced an extra WGT event, and the WGT event contributed to a gene-level evolution and drove the diversification of the Brassica plants (Cheng et al., 2014). In addition, the homology synteny and chromosomal gene location analysis revealed that the BnaVOC genes are closely phylogenetically related to other VOC genes in Brassicaceae species (B. oleracea, B. rapa, and A. thaliana; Figures 1, 3). No tandemly duplicated genes were identified in the BnaVOC gene family, and 31 BnaVOC genes are associated with segmental duplications (Figure 2). Two loci (At1g11840 and At1g06570) had four copies that are involved in segmental duplications (Table 2). One loci, At1g15380, contains eight copies that are associated with segmental duplications (Table 2); these results indicate that segmental duplication played an important role in the gene evolution of this loci. Interestingly, the BnaVOC genes that clustered on chromosome A6 were closely linked with the genes located on chromosome C5, suggesting that the fragment in this region experienced segmental duplication. In addition, all the VOC genes in the Brassica species have a syntenic relationship with the chromosome of translocation Proto-Calepineae Karyotype (tPCK), which is an ancestral genome of the Brassica species. AT1G07645, AT1G08110, AT1G11840, AT1G15380, AT1G06570 and the related Brassica genes were identified from tPCK1, and genes from AT1G80160 and AT1G67280 were linked with tPCK6, AT2G28420, and AT5G57040, which had synteny relationship with tPCK3 and tPCK5. The synteny relationship between tPCK and the Brassica species indicated that the VOC genes evolved from different ancestral genomes and expanded to diverse chromosomes through segmental duplications.
Figure 2.
Distribution of BnVOC gene family members on B. napus chromosomes. The BnVOC genes that chromosomal information was available in the database were mapped to the B. napus chromosomes and synteny relationship were lined.
Figure 3.
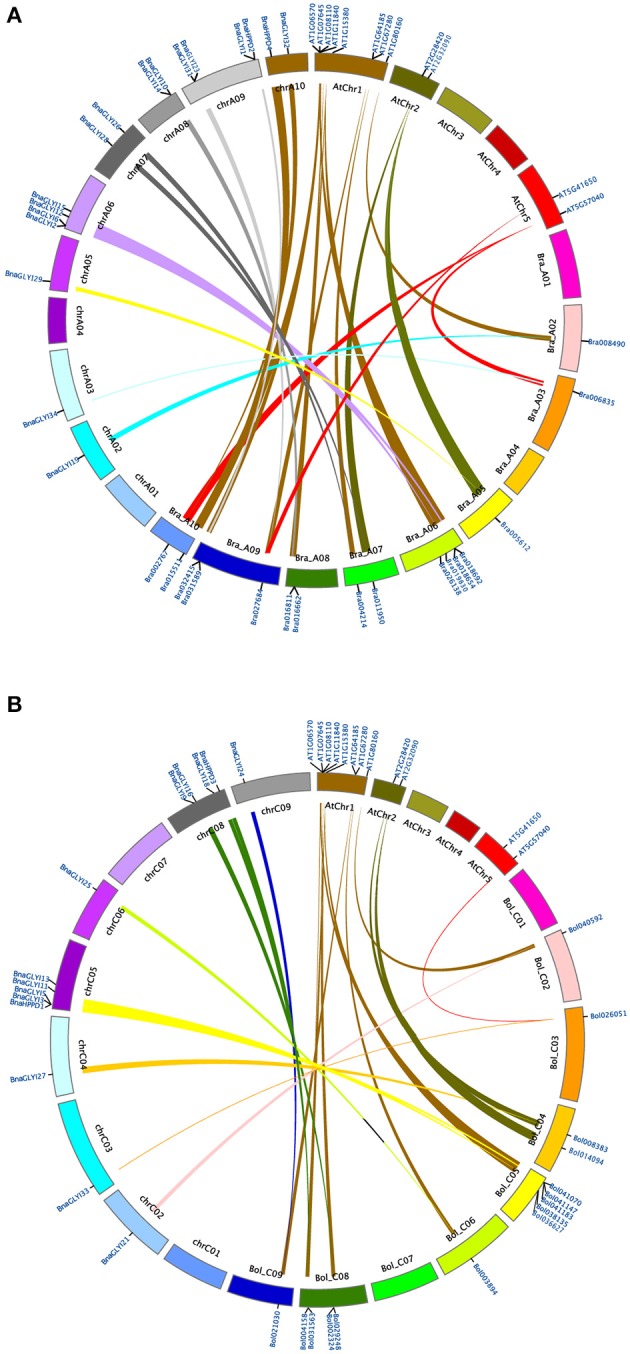
Synteny analysis map of VOC gene in B. napus, B. rapa, B. oleracea, and A. thaliana chromosomes. (A) Genes located on B. napus A genome are syntenic with genes of B. rapa and A. thaliana. (B) Genes located on B. napus C genome are syntenic with genes of B. oleracea and A. thaliana. The different degrees of thickness of lines indicate their synteny relationship (thick, strong; thin, weak).
Table 2.
Non-synonymous (Ka) and synonymous (Ks) nucleotide substitution rates for Arabidopsis thaliana and B. napus VOC gene coding loci.
| A. thaliana ID | B. napus gene | B. napus ID | VOC family | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|
| ONE COPY LOCI | ||||||
| AT5G41650 | BnaGLYI31 | BnaA09g11460D | Glyoxalase | 0.1318 | 0.0949 | 0.7202 |
| TWO-COPY LOCI | ||||||
| AT1G67280 | BnaGLYI25 | BnaC06g28360D | Glyoxalase | 0.1413 | 0.0598 | 0.4232 |
| BnaGLYI26 | BnaA07g26290D | Glyoxalase | 0.1499 | 0.0605 | 0.4039 | |
| AT2G28420 | BnaGLYI27 | BnaC04g15790D | Glyoxalase | 0.3604 | 0.2398 | 0.6654 |
| BnaGLYI28 | BnaA07g13890D | Glyoxalase | 0.3669 | 0.2534 | 0.6907 | |
| AT2G32090 | BnaGLYI29 | BnaA05g35240D | Glyoxalase | 0.0607 | 0.0328 | 0.5402 |
| BnaGLYI30 | BnaCnng59150D | Glyoxalase | 0.071 | 0.0327 | 0.4612 | |
| THREE-COPY LOCI | ||||||
| AT1G64185 | BnaGLYI22 | BnaC09g12330D | Glyoxalase | 0.0669 | 0.0306 | 0.4579 |
| BnaGLYI23 | BnaA09g12000D | Glyoxalase | 0.0829 | 0.0346 | 0.417 | |
| BnaGLYI24 | BnaC09g11920D | Glyoxalase | 0.0745 | 0.0386 | 0.5178 | |
| AT5G57040 | BnaGLYI32 | BnaA10g11070D | Glyoxalase | 0.1294 | 0.0564 | 0.4361 |
| BnaGLYI33 | BnaC03g13130D | Glyoxalase | 0.1193 | 0.0599 | 0.5023 | |
| BnaGLYI34 | BnaA03g10440D | Glyoxalase | 0.1183 | 0.0649 | 0.5486 | |
| FOUR-COPY LOCI | ||||||
| AT1G07645 | BnaGLYI1 | BnaA09g49270D | Glyoxalase | 0.0534 | 0.0274 | 0.5133 |
| BnaGLYI2 | BnaA06g04170D | Glyoxalase | 0.0963 | 0.0494 | 0.5124 | |
| BnaGLYI3 | BnaC05g05340D | Glyoxalase | 0.1078 | 0.0493 | 0.4571 | |
| BnaGLYI4 | BnaCnng38880D | Glyoxalase | 0.0606 | 0.0291 | 0.4807 | |
| AT1G08110 | BnaGLYI5 | BnaC05g05770D | Glyoxalase | 0.0491 | 0.0206 | 0.4196 |
| BnaGLYI6 | BnaA06g04580D | Glyoxalase | 0.0546 | 0.0206 | 0.3771 | |
| BnaGLYI7 | BnaC03g51010D | Glyoxalase | 0.4269 | 0.2508 | 0.5874 | |
| BnaGLYI8 | BnaCnng47290D | Glyoxalase | 0.1701 | 0.0679 | 0.3992 | |
| AT1G11840 | BnaGLYI9 | BnaC08g15100D | Glyoxalase | 0.0986 | 0.0341 | 0.3459 |
| BnaGLYI10 | BnaA08g25110D | Glyoxalase | 0.0357 | 0.0056 | 0.1561 | |
| BnaGLYI11 | BnaC05g08770D | Glyoxalase | 0.4815 | 0.1503 | 0.312 | |
| BnaGLYI12 | BnaA06g07360D | Glyoxalase | 0.2625 | 0.0884 | 0.3368 | |
| AT1G06570 | BnaHPPD1 | BnaC05g04530D | HPPD | 0.0607 | 0.0553 | 0.9118 |
| BnaHPPD2 | BnaA09g49870D | HPPD | 0.048 | 0.0461 | 0.9598 | |
| BnaHPPD3 | BnaC08g44820D | HPPD | 0.0531 | 0.0507 | 0.9549 | |
| BnaHPPD4 | BnaA10g04310D | HPPD | 0.6695 | 1.767 | 2.6391 | |
| NINE-COPY LOCI | ||||||
| AT1G15380 | BnaGLYI13 | BnaC05g11680D | Glyoxalase | 0.1089 | 0.0543 | 0.4983 |
| BnaGLYI14 | BnaA08g23870D | Glyoxalase | 0.1361 | 0.0583 | 0.4285 | |
| BnaGLYI15 | BnaA06g10060D | Glyoxalase | 0.1152 | 0.0598 | 0.5195 | |
| BnaGLYI16 | BnaC08g16660D | Glyoxalase | 0.1445 | 0.0613 | 0.424 | |
| BnaGLYI17 | BnaA09g56790D | Glyoxalase | 0.1976 | 0.0887 | 0.449 | |
| BnaGLYI18 | BnaC08g38920D | Glyoxalase | 0.2202 | 0.0917 | 0.4162 | |
| BnaGLYI19 | BnaA02g19970D | Glyoxalase | 0.0747 | 0.0864 | 1.1556 | |
| BnaGLYI20 | BnaC02g46640D | Glyoxalase | 0.0764 | 0.0922 | 1.2066 | |
| BnaGLYI21 | BnaC02g23290D | Glyoxalase | 0.0819 | 0.0952 | 1.1624 | |
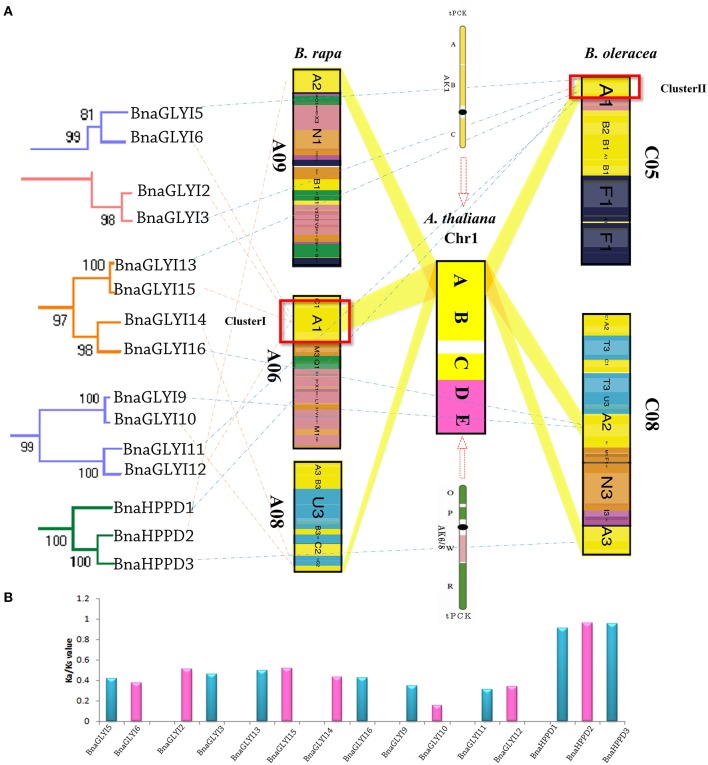
By comparing the distributions of the genes around the VOC genes in the genomes of A. thaliana, B. rapa, B. oleracea, and B. napus, the synteny of the GLY I and HPPD families was revealed to be preserved, and some genes were either duplicated or lost. Furthermore, the synteny maps of the homologous genes in A. thaliana, B. rapa and the BnaVOC genes in B. napus genome A and the homologous genes in A. thaliana, B. oleracea and the BnaVOC genes in B. napus genome C were analyzed (Figure 3); it was revealed that most of homologous VOC genes were clustered on top of Arabidopsis chromosome 1; these AtVOC genes had synteny relationships with the BrVOC and BoVOC genes; and the AtVOC genes were duplicated and distributed to the BrVOCs or BoVOCs located on different B. rapa or B. oleracea chromosomes. In addition, nearly all of these homologous BrVOCs or BoVOCs genes maintained a synteny relationship with BnaVOCs (Figure 3). Interestingly, most of the BnaVOCs appear as pair-wises for more than 90% of the AtVOCs that have synteny relationships with two or more BrVOCs or BoVOCs. Further analysis revealed that this phenomenon was an evolutionary result of genome rearrangement after WGT (Figure 4). Most phylogenetic pair-wise BnaVOCs share an ancestral gene on A. thaliana chromosome 1, and these AtVOC genes were syntenic linked with tPCK1 or tPCK6 (Figure 4). Abundant genome rearrangements caused triplicated ancestral genomic blocks in the A and C genomes of B. rapa and B. oleracea (Liu et al., 2014), and B. napus was formed by allopolyploidy; the VOC genes in B. napus were syntenically linked with the genes in B. rapa and B. oleracea. This pattern can be observed not only in the VOC gene clusters that are located on the A6 and C5 chromosomes but also in the VOC genes that are located on other chromosomes. These findings suggested that the VOC gene family expansion pattern is a consequence of mesopolyploidy in Brassica evolution. Then, the non-synonymous (Ka) and synonymous (Ks) value were used to explore the selective pressure on the duplicated BnaVOC genes. Most VOC genes in the A genome have a higher Ka/Ks ratio than the homologous genes in the C genome (Figure 4, Table 2), suggesting that different evolution pressures existed in the Brassica species divergence evolution. In general, the Ka/Ks ratio indicates the different evolution pressures; a Ka/Ks ratio that is >1 indicates positive selection, while a Ka/Ks ratio that is <1 indicates a functional constraint, and a Ka/Ks ratio equal to 1 indicates neutral selection (Nekrutenko et al., 2002). The results revealed that most of the BnaVOC genes have Ka/Ks ratios >0.1 and <1. However, BnaGLYI19, BnaGLYI20, and BnaGLYI21 had Ka/Ks ratios that were >1, suggesting that these three genes experienced positive selection pressure in evolution. The genes in the GLYI families exhibit relatively lower Ka/Ks ratios, whereas the HPPD gene families have higher Ka/Ks ratios. These findings indicated that most BnaVOC genes might preferentially conserve function and structure under selective pressure. BnaGLYI19, BnaGLYI20, and BnaGLYI21 show positive selection, suggesting that they might develop a relatively functional formation. These results indicated that WGD and segmental duplication play a role in the VOC gene family evolution, which is consistent with the Brassica evolutionary process.
Figure 4.
Evolutionary pattern analysis of pair-wised BnaVOC genes. (A) Phylogenetic relationships and evolutionary progress of the VOC genes in Brassica evolution. (B) Ka/Ks value comparison of the pair-wised BnaVOC genes.
Structure analysis in the BnaVOC genes
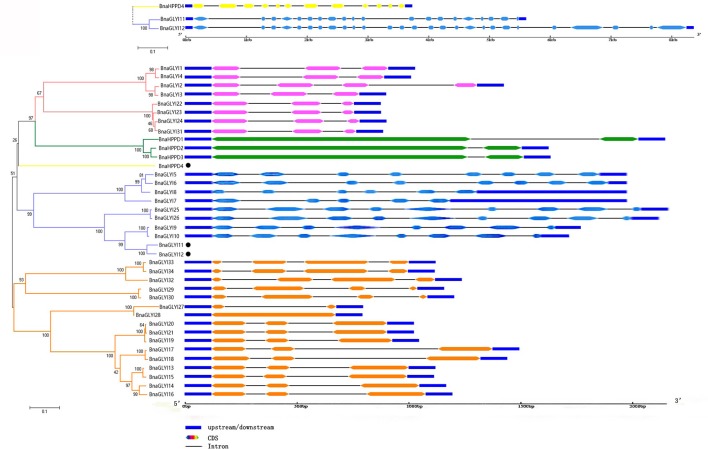
The phylogenetic tree of the B. napus VOC genes shows four main clades, and BnaHPPD4 is in a special clade. The lengths of most BnaVOC genes were shorter than 3 kb, except for BnaHPPD4, BnaGLYI11 and BnaGLYI12 (Figure 5). In the three main clades of the BnaVOC genes, the different clades contain different intron-exon structural features. Most of the BnaVOC genes have two or three introns, and more than three introns were observed in the BnaVOC genes that were longer than 1.5 kb. BnaGLYI11 and BnaGLYI12 were both longer than 5 kb, and it was revealed that these two genes have similar intron-exon structural characteristics with more than 10 introns; their intron-exon organization also reflects their close phylogenetic relationship. The same phenomenon is also observed in BnaHPPD4. Most BnaVOC genes were pair-wises, and their intron-exon structures are similar to that in the pair-wise BnaVOCs.
Figure 5.
Exon-intron organization of the BnaVOC genes. Double-sided wedge boxes represent exons, and different colors indicate sub-clade families. Black lines represent introns, and untranslated regions (UTRs) are indicated by mazarine boxes. The lengths of the proteins and motifs can be estimated using the scale at the bottom.
Because all the BnaVOCs genes have a low similarity, the genes in each clade were submitted to MEME for a motif structure analysis. Six motifs were identified to be conserved motifs, except for BnaGLYI32, BnaGLYI33, BnaGLYI34, and BnaGLYI 27. Motif 1 was present in each clade and encoded a conserved VOC family domain as indicated by the Pfam codes (Pfam000903 and Pfam 14696) and WebLogo (Figure S1). Most of the closely related genes in each phylogenetic branch exhibited similar motif compositions, suggesting the presence of functional similarities in the VOC family. BnaGLYI11 and BnaGLYI12, which are longer than 2 kb, contain nine and eleven motifs, respectively. BnaGLYI9, BnaGLYI10, BnaGLYI25, and BnaGLYI26 are in the same clade and have shorter lengths, but they contain a higher number of motifs than the genes in the other clades (Figure S1). These results imply that the composition of the structural motifs varies among the different VOC genes but is similar within the same phylogenetic branch and that the motifs encoding the VOC domains are conserved.
The secondary structures and three-dimensional structures of the BnaVOCs were also analyzed. GOR4 and PSIPRED were used for the secondary structure prediction, and similar results were obtained (Figure S2). The BnaVOCs mainly contained α-helixes, extended strands and random coils. The α-helixes accounts for ~20%, and the strand structures account for ~30%. Based on the results from the GOR4 database, the VOC-special structure (βαβββ) was observed in these BnaVOCs sequences. Some BnaVOCs, such as BnaGLYI22 and BnaGLYI24, contain more at least two VOC-special structures (βαβββ). The three-dimensional structures of the BnaVOCs were modeled and predicted using the Phyre2 database. First, all protein sequences were analyzed by VAST, and then, their sequences were compared with the structures obtained from the Protein Data Bank. The predicted domains were separately presented in Cn3D macromolecular structure viewer, and the human glyoxalase I and HPPD structures were selected for the homology modeling because their structures were well-studied, clear, and have been used in modeling the VOC in Arabidopsis (Figure 6). The tertiary structures of the s βαβββ domains. Compared with the BnaGLY I proteins, the BnaHPPDs contain more than one βαβββ repeat. In addition, the divalent metal centers of the BnaGLY I proteins and BnaHPPD proteins were different, and the zinc ions in the BnaGLYI proteins were usually predicted to appear in the metal center; however, the BnaHPPD proteins were predicted to contain ferric ion in the center in the intermediate state through the vicinal oxygen atoms (Figure 6). These results suggest that the BnaVOC proteins have conserved structures but also show some differences, particularly in the divalent metal centers and the number of βαβββ folds. All these differences might contribute to their different functions in diverse environments.
Figure 6.
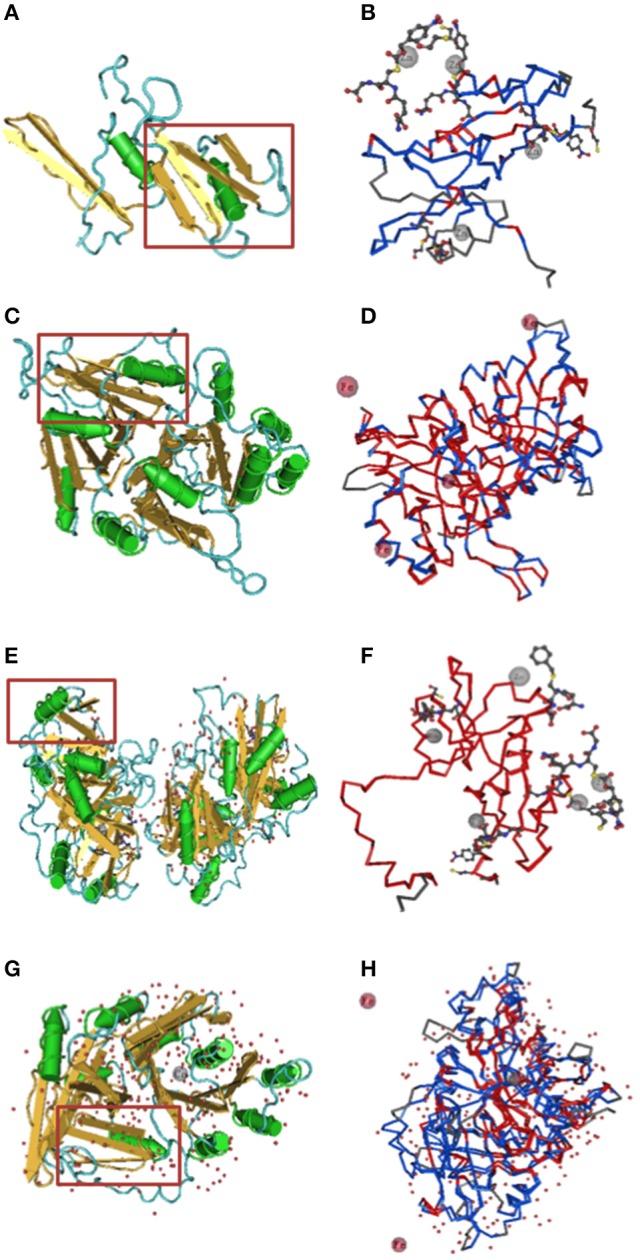
Predictive 3D domain structure of BnaVOCs. BnGLYI19 and BnHPPD2 were selected as examples to show. The models were generated from Phyre2 with 100% of confidence. Conserved domain analysis was highlighted using VAST. (A) Predictive 3D structure of BnGLYI19. (B) Conserved domains and chelated metal ion of BnGLYI19. (C) Predictive 3D structure of BnHPPD2. (D) Conserved domains and chelated metal ion of BnHPPD2. (E) 3D structure of human GLYI. (F) Conserved domains and chelated metal ion of human GLYI. (G) 3D structure of human HPPD. (H) Conserved domains and chelated metal ion of human HPPD. Red boxes indicate a typical structure of βαβββ.
Expression profile analysis of the BnaGLYI and BnaHPPD genes in different tissues
To investigate the expression pattern of the VOC genes in B. napus, the root, stem, leaves, flower, early developmental stage seeds (24 weeks after seeding), and late stage developmental seeds (30 weeks after seeding) were used to perform a gene expression analysis using the qPCR technique. Because homologous VOC genes were mainly related to desiccation and drought stress, there are fewer reports regarding their function under other types of abiotic stress (Mulako et al., 2008; Mustafiz et al., 2014). B. napus were treated with drought stress to study the BnaVOC expression pattern under drought stress conditions; to determine whether the BnaVOCs expression is linked to lipid formation under drought condition, high oil content and low oil content B. napus were used, and the analysis was performed with qPCR analysis.
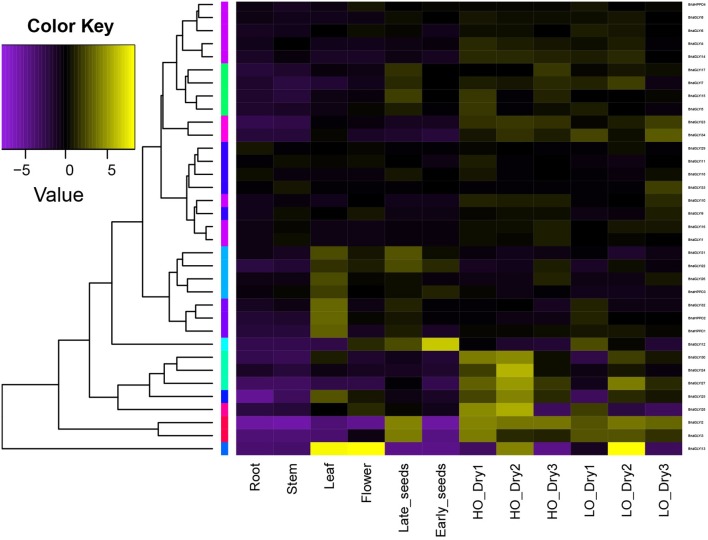
Compared with the other organs, the leaf, and late developmental stage seeds show a higher expression level of BnaVOCs (Figure 7). The leaves are important organs for transpiration and are sensitive tissues under stress conditions (Xiong and Zhu, 2002); late developmental stage seeds frequently experience dehydration, and the high expression level of the BnaVOCs in these two tissues was consistent with reported proteins levels of the VOC gene family and their expression pattern in other plants (Mulako et al., 2008; Mustafiz et al., 2011). After the dry treatment, the expression level of most BnaVOC genes was higher in the high oil content B. napus seeds than that in the low oil content B. napus seeds. For example, BnaGLYI28, BnaGLYI30, BnaGLYI24, BnaGLYI25, BnaGLYI10, and BnaHPPD3 (Figure 7, Table S2). Interestingly, during the dry treatment, different BnaVOC genes presented different expression patterns. Certain BnaVOC genes, such as BnaGLYI12, BnaGLYI32, and BnaHPPD2, showed a higher expression level during the early drought stress stage (HO_Dry1, LO_Dry1) and a lower expression level during the late drought stress stage and intermediate drought stress stage. Certain BnaVOC genes were highly expressed during the late drought stress stage (HO_Dry3, LO_Dry3), such as BnaGLYI17, BnaGLYI26, and BnaGLYI33. Many BnaVOC genes presented a higher expression during the intermediate drought stress stage (HO_Dry2, LO_Dry2), particularly BnGLYI13. Certain BnaVOC genes showed very different expression patterns between the high oil content seeds and low oil content seeds during the dry treatment, such as BnaGLYI18, BnaGLYI10, BnaGLYI32, and BnaGLYI29. These results suggest that these genes share a homologous structure, but they might function differently under drought stress conditions. In addition, because certain BnaVOC genes contain different expression features between the high and low oil content of B. napus, they might affect lipid metabolism and the final oil formation in B. napus. Most of the pair-wise genes showed similar expression patterns, but some phylogenetic gene pairs had different expression patterns (BnaHPPD2/BnaHPPD3, BnaGLYI11/BnaGLYI12, BnaGLYI13/BnaGLYI15, BnaGLYI17/BnaGLYI18, BnaGLYI25/BnaGLYI26, and BnaGLYI29/BnaGLYI30). This finding suggests that these pair-wise genes may have different functions in B. napus. These results indicate that even if the BnaVOC genes are diversely expressed in different tissues, their high expression level in the leaves shows their important roles in drought stress. When the different oil content B. napus lines were under the dry treatment, the BnaVOC genes showed different expression patterns between the two types of B. napus, and certain BnaVOC genes had a higher level of expression in the high oil content B. napus; this result suggests that the BnaVOC protein not only has a biological function in drought resistance but may also affect lipid metabolism and lead to the differences in the final oil content, which is consistent with the results of the proteomic analysis of the different oil content B. napus lines (Gan et al., 2013).
Figure 7.
Hierarchical clustering of the expression profiles of BnaVOC genes in different tissues and the seeds in drought stress. The log-transformed values of the relative expression levels of BnaVOC genes were used for hierarchical cluster analysis (original data shown in Table S2). The color scale represents relative expression levels with increased transcript (yellow) or decreased transcript (purple). Late_stage seeds were drought stress treated (HO_Dry1, HO_Dry2, HO_Dry3 indicate high oil content B. napus lines were treated for 1, 2, 3 weeks, respectively; LO_Dry1, LO_Dry2, LO_Dry3 indicate low oil content B. napus lines were treated for 1, 2, 3 weeks, respectively).
Discussion
Structural characteristics of the BnaVOC family
The VOC superfamily is a type of metalloenzyme superfamily; therefore, this superfamily contains structurally related proteins (Bergdoll et al., 1998). These VOC proteins can provide a metal coordination environment for the electrophilic participation of the metal ion in catalysis, and these proteins may participate in the evolution of protein folding (Armstrong, 2000; Gerlt and Babbitt, 2001). Some gene structures in the VOC superfamily have been reported in different species (Bernat et al., 1997; McCarthy et al., 2001; Thornalley, 2003); however, a genome-wide identification and annotation of the VOC genes have not been reported in B. napus. In this study, 38 genes were identified as VOC family genes in the B. napus genome. In general, gene families that are associated with stress resistance contain fewer introns (Liang et al., 2016), and our present results confirmed this conclusion to a certain extent in 27 of the BnaVOC genes that have no more than three introns. However, one clade of BnaGLYIs and BnaHPPD4 had more than three introns, and this likely was due to their lengths, which were longer than 1,000 bp, and the number of βαβββ folds; many introns were conserved in evolution (Figure 1). The composition and motif numbers in each clade varied, but the conserved motif of Pfam 00903 or Pfam 14696 was observed in every member of the BnaVOC gene family. The differences in the motifs between BnaGLYIs and BnaHPPDs were likely due to their configurations in the functional form; for instance, glyoxalase I can exist as a dimer or a monomer (Saint-Jean et al., 1998). The three-dimensional structural analysis showed that Fe2+ and Zn2+ were in the metal center (Figure 6), and they were also found in the VOC family in other species (He and Moran, 2011; Mustafiz et al., 2011), supporting that the BnaVOCs can play important roles in catalytic function as metalloenzymes.
Duplication patterns and synteny analysis of the BnaVOC super family
Gene duplication could expand the genome and lead to differential gene functions for optimal adaptability in the evolution of plants (Xu et al., 2012). There are three mechanism contributing to the gene family expansion, i.e., tandem duplication, segmental duplication, and whole-genome duplication (WGD; Xu et al., 2012). WGDs were assumed to have played a major role in the diversification of angiosperms (Soltis et al., 2009). Mesopolyploidization is one category of WGD events, and it has been defined in Brassicaceae (Mandakova et al., 2010). The progenitor diploid genomes of B. napus are ancient polyploids (Schmidt et al., 2001), and B. napus was formed by allopolyploidy (Chalhoub et al., 2014). Many research studies have revealed that the Brassica species has undergone WGD events during their evolution; in addition, several independent lineage-specific WGD events have been identified in Brassicaceae (Rana et al., 2004; Cheng et al., 2014). In this study, the BnaVOC superfamily formation was associated with segmental duplications and WGD. In Arabidopsis and rice, segmental duplication events were also found in the VOC family genes (Mustafiz et al., 2011). The Arabidopsis genome contains 22 GLYI genes; therefore, a WGT event would be expected to produce more than 66 GLYI genes in the B. oleracea or B. rapa genome, ultimately leading to even more GLYI genes in B. napus. However, only 34 genes were observed in the B. napus genome in the present study. This finding implies that more than 50% of the duplicated GLY I genes were lost after WGT, which might be due to the extensive chromosome reshuffling during the rediploidization after WGT (Cheng et al., 2014). Most likely, 34 GLYI genes were sufficient for B. napus during the long natural selection process, and thus, some duplicated GLYI genes did not remain in the B. napus genome. For example, certain genes that are homologous to Arabidopsis (AT5G41650, AT1G67280, AT2G28420, AT2G32090, AT2G32090, AT5G57040) were detected with less than four copies in B. napus; thus, the eleven genes that were not detected might have been lost (Table 2). Similar losses of genes after WGT have also been observed in other gene families in Brassica (Yu et al., 2014; Liang et al., 2016). The synteny analysis demonstrated that most VOC gene family members are located in well-conserved synteny regions, and VOC genes in the A genomes from B. rapa and C genomes from B. oleracea exhibited a greater homology to B. napus than to A. thaliana. WGDs play a major role in Brassicas, particularly the mesopolyploidization events, which are simultaneously accompanied by extensive chromosomal and genetic diploidization processes (Hohmann et al., 2015). After WGT, extensive genome fractionation, block reshuffling and chromosome reduction accompanied by paleocentromere, which was a descendent of the tPCK subgenomes during the rediploidization process, produced stable diploid species (Schranz et al., 2006; Cheng et al., 2014). All the VOC genes in the Brassica species have a syntenic relationship with the tPCK chromosome. The characteristics of the BnaVOC family duplication patterns and synteny analysis were consistent with the Brassicaceae evolutionary history.
Evolution and phylogenetic analysis of the VOC family genes
An evolutionary pathway for the structural scaffolding of the VOC superfamily has been proposed (Bergdoll et al., 1998), and then, the pathway was modified (Armstrong, 2000). Usually, gene fusion and duplication can create a stronger two-motif pseudosymmetric metallomonomer, which enhances the utility of the dimer. This evolutionary step in the VOC family genes was also observed in B. napus. The phylogenetic, evolution and structural analysis results revealed that the BnaVOCs have a closer phylogenetic relationship with a similar structure, and their Ka/Ks ratio confirmed this result (Figures 4, 5, Table 2). Most BnaVOCs do not have closer phylogenetic relationships with B. nigra than with other Brassica species (Figure S4). This result is consistent with the evolutionary history of Brassica (Hohmann et al., 2015). The progenitor of the VOC superfamily is assumed to be a mini gene-encoded single βαβββ motif (Armstrong, 2000); thus, the members of BnaVOCs in the phylogenetic clade that contains more βαβββ motifs likely resulted from this evolution step. As discussed above, the BnaVOC family genes experienced WGD and segmental duplication, these events may have also affected their structural formation in evolution. BnaHPPD4 is in a single phylogenetic clade, and its structure was different with that of the other BnaHPPDs, which likely resulted from certain additional gene duplication-fusion events. Such events can provide four motif monomers as is eventually observed in some of the extradiol dioxygenases and yeast GLYI (Ridderstrom and Mannervik, 1996; Armstrong, 2000).
Expression profile analysis reveals that the BnaVOC genes have diverse expression patterns in normal tissues under dry stress conditions
The BnaVOC genes have different expression patterns both in normal tissues and under dry stress conditions. The expression in late stage seeds was obviously higher than that in the early stage seeds, which indicated that the BnaVOCs accumulated during the dehydration progress, which is a feature that is consistent with certain VOCs in other plants (Mulako et al., 2008). High expression of BnaVOCs was also observed in the leaf, and the leaves were closely connected to abiotic stress. GLYIs in Arabidopsis and rice were highly expressed under abiotic stress conditions (Mustafiz et al., 2011), and GLYIs in rice were reported to function under abiotic stress conditions (Mustafiz et al., 2014). After the dry stress treatment, the B. napus seeds showed a higher expression level (Figure 7), and all these results suggested that the BnaVOCs might also function under certain abiotic stress conditions, particularly dry stress. Dsi-1VOC proteins were found in the high oil content B. napus lines in our previous study (Gan et al., 2013), and certain VOC proteins could also be induced in seed desiccation (Mulako et al., 2008); additionally, the functional networks of the BnaVOCs and dsi-1VOC proteins show that they are linked with genes that are involved in drought stress or lipid metabolism (Figure S3); therefore, the BnaVOCs are likely involved in lipid formation, and the expression profiles of the BnaVOC genes in this study support this assumption. Under the dry stress condition, the high oil content B. napus lines showed a higher level of expression of the BnaVOC genes than that in the low oil content lines, which might be due to their biological function of tolerance to methylglyoxal and their participation in lipid metabolism.
The glyoxalase system plays major roles in the detoxification of methylglyoxal (MG), and MG is a cytotoxic by-product of the glycolytic pathway by catalyzing the reaction forming S-Dlactoylglutathione (GSH; Singla-Pareek et al., 2003). MG was involved in abiotic stress and hormonal responses in plants (Mustafiz et al., 2014), and GSH affects lipid peroxidation (De La Cruz et al., 2000); all these results suggest that GLYI is important in lipid metabolism in plants under abiotic stress conditions. HPPDs catalyse the reaction from 4-hydroxyphenylpyruvate (HPP) to 2,5-dihydroxyphenylacetate (homogentisate, HG), which is at an intersection of certain important biological processes (Falk et al., 2003; He and Moran, 2011). HG is vital for photosynthesis in plants because it is the precursor to tocopherol and plastoquinone, which are both important for photosynthesis systems (Sakuragi et al., 2006). Under the dry stress condition, more efficient photosynthesis helps the formation of carbohydrate (Alonso et al., 2010), and carbohydrate is interrelated with the oil content in B. napus (King et al., 1997). In addition, the displacement of metals from metalloenzymes or metabolites is one of the five mechanisms linked with heavy metal toxicity, which could lead to membrane structural changes (Baker et al., 2006). Fatty acids (FAs) were also connected with the membrane structure and were closely related to heavy metal tolerance and abiotic stress in higher plants (Maksymiec, 1998; Zemanova et al., 2015). These results suggested that the BnaVOCs may affect lipid metabolism and show different expression pattern in high and low oil content B. napus lines under dry stress conditions.
Conclusion
In this study, 38 BnaVOC genes were identified and a comprehensive analysis was performed in which the conserved structures in the VOC superfamily were observed; WGD and segmental duplication contributed to the BnaVOC gene family duplication in the phylogenetic evolution. The expression profile analysis provided novel insight into the biological function of the BnaVOC protein, and BnaVOCs not only respond to abiotic stress but may also affect lipid metabolism and oil formation.
Author contributions
YL did the identification of VOC family genes in B. napus, duplication pattern analysis and wrote the main manuscript text. NW did multiple alignment of the BnaVOC genes and made the phylogenetic analysis. YL, NW, ZC, YM, and BL did the qRT-PCR, collected VOC genes information of other species, and did structural analysis and prepared Figures 1–7. HL, NR, and YY prepared for supplementary information. ML designed the experiment. All authors reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was supported by the National Key Research and Development Program (2016YFD0101300), the National Natural Science Foundation of China (31671721) and the Program for New Century Excellent Talents in University (NCET110172).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00745/full#supplementary-material
Availability of data and materials
The data set of Figures 1, 5 supporting the results of this article is available in the TreeBase (https://treebase.org/treebase-web/home.html) repository, under the accession URL (http://purl.org/phylo/treebase/phylows/study/TB2:S20148). The genes sequences are available in CNS-Genoscope database (http://www.genoscope.cns.fr/brassicanapus/).
References
- Allender C. J., King G. J. (2010). Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10:54. 10.1186/1471-2229-10-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso D. M., Bond J. Q., Dumesic J. A. (2010). Catalytic conversion of biomass to biofuels. Green Chem. 12, 1493–1513. 10.1039/c004654j [DOI] [Google Scholar]
- Armstrong R. N. (2000). Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39, 13625–13632. 10.1021/bi001814v [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. (2006). Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci. 11, 124–132. 10.1016/j.tplants.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Beck E. H., Fettig S., Knake C., Hartig K., Bhattarai T. (2007). Specific and unspecific responses of plants to cold and drought stress. J. Biosci. 32, 501–510. 10.1007/s12038-007-0049-5 [DOI] [PubMed] [Google Scholar]
- Bergdoll M., Eltis L. D., Cameron A. D., Dumas P., Bolin J. T. (1998). All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 7, 1661–1670. 10.1002/pro.5560070801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernat B. A., Laughlin L. T., Armstrong R. N. (1997). Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry 36, 3050–3055. 10.1021/bi963172a [DOI] [PubMed] [Google Scholar]
- Bodén M., Hawkins J. (2005). Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21, 2279–2286. 10.1093/bioinformatics/bti372 [DOI] [PubMed] [Google Scholar]
- Brownlee J., He P., Moran G. R., Harrison D. H. (2008). Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate. Biochemistry 47, 2002–2013. 10.1021/bi701438r [DOI] [PubMed] [Google Scholar]
- Buchan D. W., Minneci F., Nugent T. C., Bryson K., Jones D. T. (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., Wang X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- Cheng F., Wu J., Wang X. (2014). Genome triplication drove the diversification of Brassica plants. Hortic. Res. 1, 14024. 10.1038/hortres.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz J. P., Quintero L., Villalobos M. A., Sanchez de la Cuesta F. (2000). Lipid peroxidation and glutathione system in hyperlipemic rabbits: influence of olive oil administration. Biochim. Biophys. Acta 1485, 36–44. 10.1016/S1388-1981(00)00027-5 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Falk J., Andersen G., Kernebeck B., Krupinska K. (2003). Constitutive overexpression of barley 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Lett. 540, 35–40. 10.1016/S0014-5793(03)00166-2 [DOI] [PubMed] [Google Scholar]
- Falk J., Krauss N., Dahnhardt D., Krupinska K. (2002). The senescence associated gene of barley encoding 4-hydroxyphenylpyruvate dioxygenase is expressed during oxidative stress. J. Plant Physiol. 159, 1245–1253. 10.1078/0176-1617-00804 [DOI] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L., Zhang C. Y., Wang X. D., Wang H., Long Y., Yin Y. T., et al. (2013). Proteomic and comparative genomic analysis of two Brassica napus lines differing in oil content. J. Proteome Res. 12, 4965–4978. 10.1021/pr4005635 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A., Babbitt P. C. (2001). Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 70, 209–246. 10.1146/annurev.biochem.70.1.209 [DOI] [PubMed] [Google Scholar]
- He P., Moran G. R. (2011). Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J. Inorg. Biochem. 105, 1259–1272. 10.1016/j.jinorgbio.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. (1988). CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73, 237–244. 10.1016/0378-1119(88)90330-7 [DOI] [PubMed] [Google Scholar]
- Hohmann N., Wolf E. M., Lysak M. A., Koch M. A. (2015). A Time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27, 2770–2784. 10.1105/tpc.15.00482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- Kagale S., Divi U. K., Krochko J. E., Keller W. A., Krishna P. (2007). Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225, 353–364. 10.1007/s00425-006-0361-6 [DOI] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J. E. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- King S. P., Lunn J. E., Furbank R. T. (1997). Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol. 114, 153–160. 10.1104/pp.114.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144. 10.1093/nar/gkh088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu B., Yu L., Feng D., Wang H., Wang J. (2009). Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol. Biol. 9:90. 10.1186/1471-2148-9-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Xiong Z., Zheng J., Xu D., Zhu Z., Xiang J., et al. (2016). Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 6:24265. 10.1038/srep24265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. 10.1038/ncomms4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksymiec W. (1998). Effect of copper on cellular processes in higher plants. Photosynthetica 34, 321–342. 10.1023/A:1006818815528 [DOI] [Google Scholar]
- Mandakova T., Joly S., Krzywinski M., Mummenhoff K., Lysak M. A. (2010). Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22, 2277–2290. 10.1105/tpc.110.074526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res 43, D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. A., Baker H. M., Shewry S. C., Patchett M. L., Baker E. N. (2001). Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure 9, 637–646. 10.1016/s0969-2126(01)00622-0 [DOI] [PubMed] [Google Scholar]
- Mulako I., Farrant J. M., Collett H., Illing N. (2008). Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. J. Exp. Bot. 59, 3885–3901. 10.1093/jxb/ern226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafiz A., Ghosh A., Tripathi A. K., Kaur C., Ganguly A. K., Bhavesh N. S., et al. (2014). A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78, 951–963. 10.1111/tpj.12521 [DOI] [PubMed] [Google Scholar]
- Mustafiz A., Singh A. K., Pareek A., Sopory S. K., Singla-Pareek S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11, 293–305. 10.1007/s10142-010-0203-2 [DOI] [PubMed] [Google Scholar]
- Nekrutenko A., Makova K. D., Li W.-H. (2002). The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 12, 198–202. 10.1101/gr.200901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L., King G. J. (2008). Standardized gene nomenclature for the Brassica genus. Plant Methods 4:10. 10.1186/1746-4811-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D., van den Boogaart T., O'Neill C. M., Hynes L., Bent E., Macpherson L., et al. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40, 725–733. 10.1111/j.1365-313X.2004.02244.x [DOI] [PubMed] [Google Scholar]
- Reddy V. S., Sopory S. K. (1999). Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 17, 385–395. 10.1046/j.1365-313X.1999.00390.x [DOI] [PubMed] [Google Scholar]
- Ridderstrom M., Mannervik B. (1996). The primary structure of monomeric yeast glyoxalase I indicates a gene duplication resulting in two similar segments homologous with the subunit of dimeric human glyoxalase I. Biochem. J. 316 (Pt 3), 1005–1006. 10.1042/bj3161005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jean A. P., Phillips K. R., Creighton D. J., Stone M. J. (1998). Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: evidence for 3D domain swapping. Biochemistry 37, 10345–10353. 10.1021/bi980868q [DOI] [PubMed] [Google Scholar]
- Sakuragi Y., Maeda H., DellaPenna D., Bryant D. A. (2006). α-Tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141, 508–521. 10.1104/pp.105.074765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J. A., Vaslenko-Sanders I. F., Deshpande S., Yi J., Siegfried M., Roe B. A., et al. (2007). The FAD2 gene family of soybean: insights into the structural and functional divergence of a paleoplyploid genome. Crop Sci. 47, S14–S26. 10.2135/cropsci2006.06.0382tpg [DOI] [Google Scholar]
- Schmidt R., Acarkan A., Boivin K. (2001). Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 39, 253–262. 10.1016/S0981-9428(01)01239-6 [DOI] [Google Scholar]
- Schranz M. E., Lysak M. A., Mitchell-Olds T. (2006). The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. 10.1016/j.tplants.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., et al. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. 10.1105/tpc.13.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek S. L., Reddy M. K., Sopory S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U.S.A. 100, 14672–14677. 10.1073/pnas.2034667100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Albert V. A., Leebens-Mack J., Bell C. D., Paterson A. H., Zheng C., et al. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. 10.3732/ajb.0800079 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269, 1–11. 10.1042/bj2690001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. (2003). Glyoxalase I–structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 31(Pt 6), 1343–1348. 10.1042/bst0311343 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Xiong L., Zhu J. K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. 10.1046/j.1365-3040.2002.00782.x [DOI] [PubMed] [Google Scholar]
- Xu G., Guo C., Shan H., Kong H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. 10.1073/pnas.1109047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yu J. Y., Tehrim S., Zhang F. Q., Tong C. B., Huang J. Y., Cheng X. H., et al. (2014). Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics 15:3. 10.1186/1471-2164-15-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanova V., Pavlik M., Kyjakova P., Pavlikova D. (2015). Fatty acid profiles of ecotypes of hyperaccumulator Noccaea caerulescens growing under cadmium stress. J. Plant Physiol. 180, 27–34. 10.1016/j.jplph.2015.02.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data set of Figures 1, 5 supporting the results of this article is available in the TreeBase (https://treebase.org/treebase-web/home.html) repository, under the accession URL (http://purl.org/phylo/treebase/phylows/study/TB2:S20148). The genes sequences are available in CNS-Genoscope database (http://www.genoscope.cns.fr/brassicanapus/).