Abstract
An insertion in the promoter of the operon that encodes the molecular chaperone GroE was isolated as an antimutator for stationary-phase or adaptive mutation. The groE operon consists of two genes, groES and groEL; point mutations in either gene conferred the same phenotype, reducing Lac+ adaptive mutation 10- to 20-fold. groE mutant strains had 1/10 the amount of error-prone DNA polymerase IV (Pol IV). In recG+ strains, the reduction in Pol IV was sufficient to account for their low rate of adaptive mutation, but in recG mutant strains, a deficiency of GroE had some additional effect on adaptive mutation. Pol IV is induced as part of the SOS response, but the effect of GroE on Pol IV was independent of LexA. We were unable to show that GroE interacts directly with Pol IV, suggesting that GroE may act indirectly. Together with previous results, these findings indicate that Pol IV is a component of several cellular stress responses.
Members of the recently discovered Y family of DNA polymerases are highly error prone and poorly processive (22, 63). Escherichia coli has two members of this family, DNA polymerase IV (Pol IV), encoded by the dinB gene (59), and DNA Pol V, encoded by the umuD and umuC genes (48, 54, 56). Both of these polymerases are induced by DNA damage as part of the SOS response (2, 8, 28). Pol V is able to replicate past a variety of DNA lesions and is rightly called a lesion bypass polymerase; however, Pol IV has only a limited ability to replicate damaged DNA (30, 44, 55) and its true role in the cell remains speculative.
When populations of microorganisms are placed under certain nonlethal selective conditions, mutations that relieve the selective pressure arise, a process called adaptive mutation (7, 16). Most of the research on adaptive mutation has focused on E. coli strain FC40 (6). FC40 cannot utilize lactose because of a frameshift mutation that affects the lacZ gene; when FC40 cells are plated with lactose as the sole carbon and energy source, the Lac− population remains stable but Lac+ revertants arise at a constant rate for about a week (6). Unlike growth-dependent mutations, adaptive mutations in FC40 require functions for recombination (6, 17, 20, 23, 24). In addition, the production of Lac+ adaptive mutations in FC40 is enhanced when the mutational target is on a conjugal plasmid and conjugal functions are expressed (19, 21, 46). The last result probably reflects the fact that DNA nicking at the conjugal origin stimulates the mutational process (50).
In FC40, loss of Pol IV reduces the rate of adaptive mutation three- to fivefold (18). Recently we reported that Pol IV is induced late in stationary phase under the positive control of RpoS, the stationary-phase sigma factor. After induction, high levels of Pol IV are maintained in the starving cells for at least 3 days, but only if the cells are rpoS+ (34). Other researchers have shown that the dinB gene is transcribed in a 5-day-old culture (63). In an rpoS mutant strain, the rate of Lac+ adaptive mutation is reduced 5- to 10-fold (34, 39). About half of this reduction is due to the decreased amount of Pol IV in the rpoS mutant strain, and the rest is due to some other effect that RpoS has on adaptive mutation (34).
The results summarized above indicate that Pol IV is induced as part of two stress responses: the SOS response to DNA damage, under the control of the LexA repressor, and the starvation response, positively regulated by RpoS. Both of these responses are also induced under a variety of other conditions. The SOS response is induced at the end of growth in liquid rich medium (11) and in aging colonies on solid rich medium (52). RpoS is responsive to stresses in addition to starvation that tend to inhibit active cell growth (25). Thus, cells respond to a variety of difficult situations by increasing their levels of Pol IV, which would tend to increase the error rate of DNA synthesis during repair or replication. Here we report that levels of Pol IV are also positively affected by the heat shock-induced molecular chaperone GroE, bringing to three the number of stress responses that influence Pol IV.
MATERIALS AND METHODS
Bacterial strains.
The strains used are listed in Table 1. Genetic manipulations were performed using standard techniques (43). The original isolation of the groES::Cm mutant was as previously described for the rpoS::Cm mutant (34). To make groEL100 and groES30 derivatives, parental strains were transduced to tetracycline resistance (Tetr) with bacteriophage P1vir lysates of strains GW7503 (= AB1157 groEL100 purA::Tn10) and GW7504 (= AB1157 groES30 purA::Tn10) (9). Tetr, adenine-requiring (Ade−) candidates were tested for resistance to λvir bacteriophage to identify sensitive (groE+) and resistant(groE[minus]) isogenic pairs. The rpoS::Cm and groES::Cm alleles were transferred by P1vir transduction, with selection for chloramphenicol resistance (Cmr). Tets Arg+ derivatives of TE8197 and TE8222 were constructed by transducing them to arginine prototrophy with a P1vir lysate of FC36 and then screening Arg+ isolates for sensitivity to tetracycline. Strains that have dinB deleted on both the chromosome and the episome were constructed as previously described (34) except that the nonpolar dinB::Zeo allele was used (5) and selection was for resistance to zeomycin (Zeor). F′ Φ(lacI33-lacZ) Pro+ was mated into F− Pro− strains by conjugation with a Met−/F′ Φ(lacI33-lacZ) Pro+ donor, with selection for proline and methionine prototrophy.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain(s) | Relevant phenotype or genotype | No. of copies of dinB+ | Reference |
|---|---|---|---|
| FC29 | ara Δ(gpt-lac)5 thi/F′ Δ(lacIZ) Pro+ | 2 | 6 |
| FC36 | ara Δ(gpt-lac)5 thi Rifr | 1 | 6 |
| FC40 | FC36/F′ Φ(lacI33-lacZ) Pro+ | 2 | 6 |
| FC526 | FC40 ΔrecG263::Kn | 2 | 20 |
| FC1230 | FC40/pYG768 | Many | This study and reference 29 |
| FC1240 | FC722 ΔdinB::Kn on chromosome and episome | None | 34 |
| PF2244 | TE8197a Arg+ Tets | 1 | This study and reference 26 |
| PF2245 | TE8222b Arg+ Tets | 1 | This study and reference 26 |
| PF2252 | WAM106c PL-lacZ | 1 | 61 |
| PF2253 | WAM106c PgroE-lacZ | 1 | 61 |
| PF2258 | PF2252 rpoS::Cm | 1 | This study and reference 34 |
| PF2259 | PF2253 rpoS::Cm | 1 | This study and reference 34 |
| PFG60, PFG64d | FC40 groEL+purA::Tn10 | 2 | This study |
| PFG61 | FC40 groEL100 purA::Tn10 | 2 | This study and reference 66 |
| PFG62, PFG66d | FC526 groEL+purA::Tn10 | 2 | This study |
| PFG63 | FC526 groEL100 purA::Tn10 | 2 | This study |
| PFG65 | FC40 groES30 purA::Tn10 | 2 | This study and reference 33 |
| PFG67 | FC526 groES30 purA::Tn10 | 2 | This study |
| PFG250 | FC40 groE+purA::Tn10 ΔdinB::Zeo on chromosome and episome | None | This study and reference 5 |
| PFG251 | FC40 groEL100 purA::Tn10 ΔdinB::Zeo on chromosome and episome | None | This study |
| PFG305 | FC526 groE+purA::Tn10 ΔdinB::Zeo on chromosome and episome | None | This study |
| PFG307 | FC526 groEL100 purA::Tn10 ΔdinB::Zeo on chromosome and episome | None | This study |
| PFG308 | FC36 sulA-lacZ groEL+purA::Tn10 | 1 | This study |
| PFG309 | FC36 sulA-lacZ groEL100 purA::Tn10 | 1 | This study |
| PFG332 | PFG308 recG263::Kn | 1 | This study |
| PFG333 | PFG309 recG263::Kn | 1 | This study |
| PFG324 | FC36 sulA11 groEL+purA::Tn10 lexA71::Kn | 1 | This study and reference 34 |
| PFG325 | FC36 sulA11 groEL100 purA::Tn10 lexA71::Kn | 1 | This study and reference 34 |
| PFG326 | FC36 sulA11 groEL+purA::Tn10 | 1 | This study and reference 34 |
| PFG327 | FC36 sulA11 groEL100 purA::Tn10 | 1 | This study and reference 34 |
| PFG328 | PFG324/F′ Φ(lacI33-lacZ) Pro+ | 2 | This study |
| PFG329 | PFG325/F′ Φ(lac/33-lacZ) Pro+ | 2 | This study |
| PFG330 | PFG326/F′ Φ(lacI33-lacZ) Pro+ | 2 | This study |
| PFG331 | PFG327/F′ Φ(lacI33-lacZ) Pro+ | 2 | This study |
| PFG334 | GJ2770egroEL+purA::Tn10 | 1 | This study |
| PFG335 | GJ2770egroEL100 purA::Tn10 | 1 | This study |
| PFG336 | PFG334 rpoS::Cm | 1 | This study |
| PFG340 | PF2244 groEL+purA::Tn10 | 1 | This study |
| PFG341 | PF2244 groEL100 purA::Tn10 | 1 | This study |
| PFG342 | PF2245 groEL+purA::Tn10 | 1 | This study |
| PFG343 | PF2245 groEL100 purA::Tn10 | 1 | This study |
| PFG321 | FC40 groES::Cm | 2 | This study |
| PFG351 | FC526 groES::Cm | 2 | This study |
TE8197 is MG1655 Δ(lacIZ. argA::Tn10 trpDC700::putPA1303::Knr-rpoS-lac (protein fusion) (26). MG1655 is considered to be a wild-type strain (4).
TE8222 is MG1655 Δ(lacIZ. argA::Tn10 trpDC700::putPA1303::Knr-rpoS-lac (operon fusion) (26).
WAM106 is F− araD139 Δ(argF-lac)U169 Δ(his-gnd. thi rpsL150 flbB5301 relA1 deoC1 rbsR (57).
Both strains were constructed by transduction; the donor for the first was GW7503 (=AB1157 groEL100 purA::Tn10), and the donor for the second was GW7504 (=AB1157 groES30 purA::Tn10) (9). Tetr Ade− λvir-sensitive transductants were isolated.
GJ2770 is MC4100 λ [katE::lac(Kan)] (47). MC4100 is Δ(argF-lac)U169 rpsL150 relA1 araD139 flbB5301 deoC1 ptsF25.
Mutation, growth, and viability assays.
Media and experimental protocols were as previously described (6, 15, 20). As required, minimal medium was supplemented with 100 μg of adenine or histidine per ml, 50 μg of tryptophan per ml, 10 μg of tetracycline per ml, 15 μg of kanamycin per ml, or 10 μg of chloramphenicol per ml. Rich medium was supplemented with 100 μg of adenine per ml, 20 μg of tetracycline per ml, 30 μg of kanamycin per ml, 10 μg of chloramphenicol per ml, or 25 μg of zeomycin per ml.
For adaptive-mutation experiments, cells were grown to saturation at 32°C in 1% glycerol-M9 minimal medium (plus adenine when required) (43). Approximately 107 cells from each culture were spread on each quadrant of a 1% lactose-M9 minimum medium plate (plus adenine when required), or an appropriate number of cells (106 to 108) was added to approximately 109 FC29 scavenger cells and spread on a 1% lactose-M9 minimum medium plate. Plates were incubated for 5 days at 37°C, and newly arising Lac+ colonies were counted each day. To measure viability, plugs were removed from between the Lac+ colonies and the number of viable cells was determined each day by plating appropriate dilutions on Luria-Bertani plates plus chloramphenicol or tetracycline plus adenine, on which FC29 cannot grow (6). Mutation to Lac+ is given either as the mean number of Lac+ colonies appearing each day from days 3 to 5 or as the mean number of Lac+ colonies accumulating each day divided by the number of Lac− cells present 2 days earlier. Statistical calculations were as given in references 49 and 65.
β-Galactosidase assays.
Saturated cultures were diluted 1:1,000 into 1% glycerol-M9 minimal medium supplemented with the additive required for each strain and allowed to reach saturation at 37°C. Appropriate dilutions of the cultures were assayed for optical density at 600 nm and β-galactosidase production as described previously (43).
Molecular techniques.
Standard molecular biology techniques were used (1). For immunoblots, cells were grown in 1% glycerol-M9 minimal medium plus appropriate additives at 37°C to saturation, and then incubation was continued for 16 to 24 h; this procedure was previously found to result in maximum levels of Pol IV (34). Cells were harvested and boiled in sample loading buffer, and the total protein was measured by Bradford assays (Bio-Rad Laboratories). Samples containing 40 μg of total protein were subjected to electrophoresis in a sodium dodecyl sulfate-12% polyacrylamide gel and then electro-transferred to Immobilon-P membranes (pore size, 0.45 μm; Millipore Co.). Pol IV was visualized with rabbit anti-Pol IV polyclonal antiserum (obtained from H. Ohmori) clarified with acetone powder made from a Δ(dinB) strain, followed by reaction with alkaline phosphatase-conjugated goat anti-rabbit secondary antibody, and developed using the Western-light chemiluminescence reagent (Applied Biosystems). Bands were quantitated using ImageJ software (W. Rasband, National Institutes of Health). Coimmunoprecipitations were done with monoclonal antibody to GroEL (Stressgen Biotechnologies Corp.) and protein A or protein G immunoprecipitation kits (Roche Diagnostics Corp). For metal affinity purification, Talon cobalt-resin columns (BD Biosciences Clontech) were used. Yeast two-hybrid analysis was performed with Matchmaker T System 3 (BD Biosciences Clontech).
RESULTS
An insertion in the groES/groEL operon reduces adaptive mutation to Lac+.
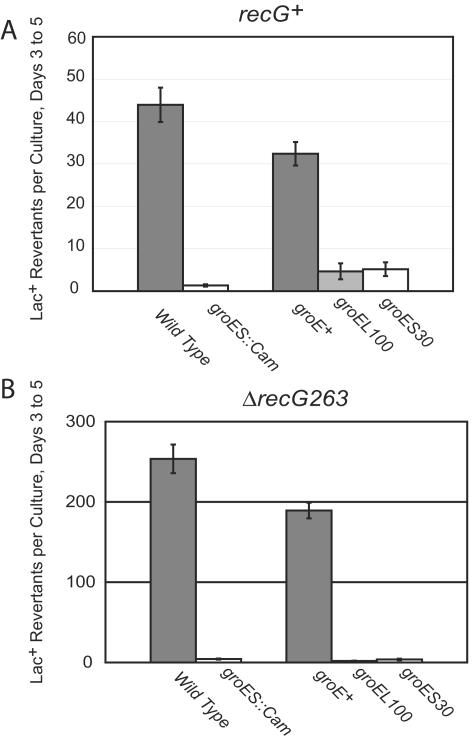
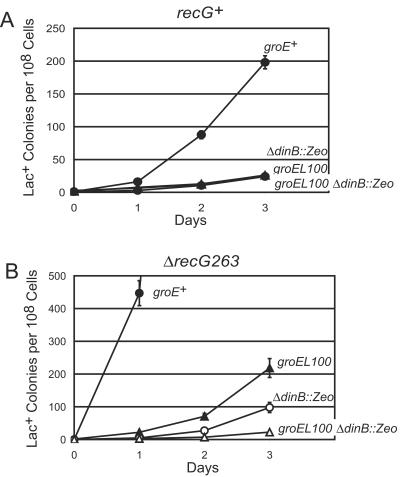
The rate of adaptive mutation to Lac+ in E. coli strain FC40 is elevated 10- to 100-fold in recG mutant strains (20, 24). This increase is substantially, or entirely, due to elevated levels of Pol IV in recG mutant strains (34). To find mutations in genes that regulate the amount or activity of Pol IV, we mutated a recG mutant strain with a transposon conveying chloramphenicol resistance, mini-Tn10d Cm (31), and screened Cmr isolates for reduction in adaptive mutation to Lac+ (34). One such mutant was an insertion in the groES/groEL operon. groES and groEL are essential genes that encode the two subunits of the heat shock-inducible molecular chaperone GroE. The mini-Tn10d Cm inserted between the −10 and −35 regions of the heat shock (σ32-dependent) promoter, leaving intact the proposed vegetative (σ70-dependent) promoter (see reference 67 for a description of the promoters). Like E. coli strains with other mutations in groES or groEL (58), groES::Cm mutant strains are temperature sensitive for viability (at 43°C) and resistant to bacteriophage lambda (data not shown). As shown in Fig. 1, the groES::Cm allele reduced Lac+ adaptive mutation in both recG+ and recG mutant strains. The effect of the groES::Cm allele was stronger in the recG mutant strain than in the recG+ strain.
FIG. 1.
Mutant alleles of groE decrease adaptive mutation in E. coli. The accumulation of Lac+ revertants during incubation on lactose-minimal medium. Four to six independent cultures were plated for each strain; data are the mean numbers of Lac+ colonies appearing each day from days 3 to 5 ± standard errors of the means (SEM). The results of four experiments are shown. (A) Wild type = FC40; groES::Cm = PFG361; groE+ = PFG60 and PFG64; groEL100 = PFG61; groES30 = PFG65. (B) Wild type = FC526; groES::Cm = PFG351; groE+ = PFG62 and PFG66; groEL100 = G63; groES30 = PFG67. PFG60 through PFG67 are also purA::Tn10.
Point mutations in groES and groEL also reduce adaptive mutation to Lac+.
Because our groES::Cm mutation is unique, we sought to confirm its effects with previously isolated and well-characterized groE mutant alleles. The groES30 allele has two mutations, a G-to-A change in the Shine-Dalgarno sequence of the gene and an A31V mutation in the coding sequence (33). The groEL100 allele has an S201F mutation that affects the ability of GroEL to bind to GroES and to bind and release substrate proteins (3, 66). Both mutant alleles make cells resistant to bacteriophage, but only the groEL100 allele makes cells temperature sensitive (58). As shown in Fig. 1, both of these mutant alleles reduced the level of adaptive mutation. As with the groES::Cm allele, the reduction in adaptive mutation was greater in the recG mutant strain.
The reduction in adaptive mutation due to GroE deficiency is not due to loss of viability.
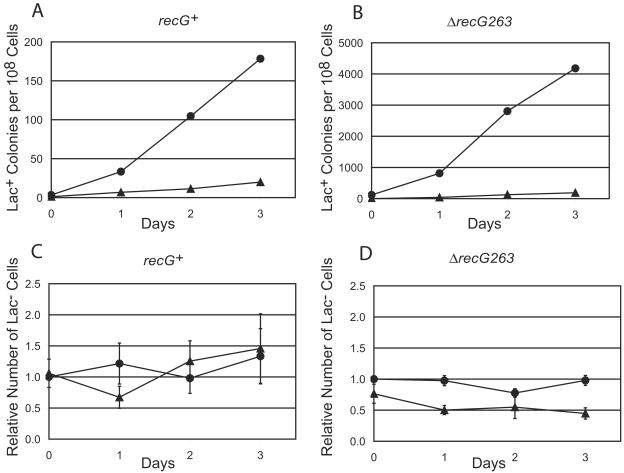
The results in Fig. 1 were obtained with our semiquantitative assay (20). To obtain quantitative results and to correct for any loss of viability during the experiments, we did large-scale experiments monitoring the number of viable Lac− cells on the lactose plates every day (6). These results are shown in Fig. 2, which compares groEL100 and groE+ in both recG+ and recG mutant backgrounds. The large-scale experiments confirmed the previous results: after correction for viable-cell number, GroE deficiency reduced the rate of adaptive mutation to Lac+ about 10-fold in recG+ cells and about 20-fold in recG mutant cells.
FIG. 2.
The reduction in the level of adaptive mutation due to deficiency of GroE is not due to loss of viability. (A and B) Accumulation of Lac+ revertants of groE+ and groEL100 mutant strains during incubation on lactose-minimal medium. Data are the cumulative number of Lac+ colonies divided by the number of viable Lac− cells on the plate 2 days earlier; each point is the mean ± SEM of results for 20 independent cultures (some error bars are smaller than the symbols). Because it takes 2 days for a Lac+ revertant to make a visible colony, the values are displaced 2 days earlier to correspond to the points in the survival curves in panels C and D. (A) Circles, PFG60 (groE+); triangles, PFG61 (groEL100). (B) Circles, PFG62 (ΔrecG263 groE+); triangles, PFG63 (ΔrecG263 groEL100). All strains are also purA::Tn10. (C and D) Survival of Lac− cells during incubation on lactose-minimal medium. Each point is the mean ± SEM of results for three independent cultures. (C) Circles, PFG60 (groE+); triangles, PFG61 (groEL100). Numbers have been normalized to the value for PFG60 on day 0. (D) Circles, PFG62 (ΔrecG263 groE+); triangles, PFG63 (ΔrecG263 groEL100). Because about 100-fold-more PFG63 cells than PFG62 cells were plated, the numbers of PFG62 cells were multiplied by 100; then the results for both strains were normalized to this value for PFG62 on day 0.
Deficiency of GroE reduces the cellular amount of DNA Pol IV.
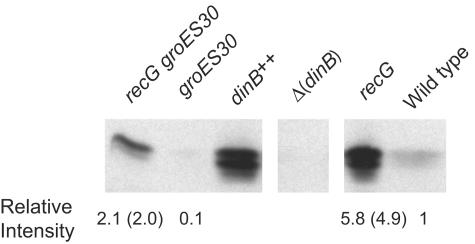
GroE is required for normal levels of SOS-dependent mutagenesis after DNA damage because the polymerase subunit of DNA Pol V, the product of the umuC gene, is destabilized in groE mutants (9). The dinB gene, which encodes Pol IV, is homologous to the umuC gene (45). Therefore, we checked whether the amount of Pol IV was also reduced in groE mutant cells. Because Pol IV is induced late in stationary phase (34), we grew cells in minimal medium to saturation and then incubated the cultures for an additional 16 to 24 h before preparing samples for Western blots. As shown in Fig. 3, a groES30 mutant strain had about 1/10 the amount of Pol IV as a groE+ strain. Pol IV levels are elevated in recG mutant cells (34) (Fig. 3), and the groES30 mutant allele reduced the amount of Pol IV in these cells about threefold. Similar results were obtained with the groEL100 and the groES::Cm mutant strains (data not shown). (The faster-running band in Fig. 3 is probably a degradation product; see reference 34).
FIG. 3.
A mutant allele of groE reduces the cellular amount of Pol IV protein. Shown is a Western blot indicating the amounts of Pol IV in (from left to right) PFG67 (recG groES30), PFG65 (recG+ groES30), FC1230 (dinB++), FC1240 (ΔdinB), FC526 (recG), and FC40 (wild type). Only one blot is shown, but lanes with irrelevant samples have been removed; the rightmost three lanes have been previously published (34). Samples consisting of 40 μg of total protein were loaded in each lane. The intensities of the bands for each strain relative to those of the wild-type strain are given below each lane; the intensity of only the upper band relative to that of the wild-type strain is in parentheses.
GroE may affect the levels of Pol IV indirectly.
We were unable to detect a direct interaction between GroE and Pol IV using coimmunoprecipitation with anti-GroEL antibody, a technique that has previously been used to detect an interaction between GroE and Pol V (10). We also failed to detect GroE copurifying on a metal resin with His-tagged Pol IV from a crude cell lysate. We used Pol IV and the peptide-binding domain of GroEL (14) in the yeast two-hybrid analysis and again failed to detect an interaction, although this technique has been used to reveal other substrates of GroE (12). In addition, the yeast two-hybrid system detects an interaction between DnaN and Pol IV (35), which we confirmed and used as a positive control in our experiments (data not shown).
GroE affects the amount of Pol IV independently of LexA and when Pol IV is expressed only from the chromosome.
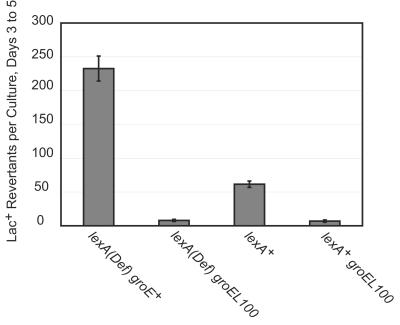
Pol IV is induced after DNA damage and in LexA-deficient [LexA(Def)] strains in which all SOS genes are derepressed (8, 28, 34). As shown in Fig. 4, the groEL100 allele reduced the amount Pol IV in a LexA(Def) strain about sixfold, demonstrating that GroE affects the levels of Pol IV independently of LexA.
FIG. 4.
A mutant allele of groE reduces the cellular amount of Pol IV protein in the absence of LexA. Shown is a Western blot indicating the levels of Pol IV in (from left to right): PFG324 [sulA lexA(Def)], PFG325 [sulA lexA(Def) groEL100], PFG326 (sulA), FC526 (dinB++), and FC1240 [Δ(dinB)]. PFG324, PFG325, and PFG326 are F− strains, so their only dinB alleles are chromosomal; these strains are also sulA11 [which prevents lethal filamentation due to the lexA(Def) allele]. Only one blot is shown, but lanes with irrelevant samples have been removed. Samples consisting of 40 μg of total protein were loaded. The intensities of the bands for each strain relative to those of the wild-type strain are given below each lane.
There are two copies of dinB in most of our strains, one on the chromosome and one on the episome. However, the three experimental strains shown in Fig. 4 are all F−, so dinB is expressed only from their chromosomes. Thus, the effect of GroE on Pol IV is not a peculiarity of the episome or the episomal allele but is also exerted when Pol IV is expressed solely from the chromosome.
GroE affects adaptive mutation independently of LexA.
Although dinB is derepressed in LexA(Def) mutant strains (34), these strains normally have adaptive mutation rates lower than those of LexA+ strains because another gene, psiB, whose product interferes with the expression of the SOS response, is also derepressed in LexA(Def) cells (41). However, in the purA::Tn10 background, the LexA(Def) mutant strain showed a fourfold increase in adaptive mutation relative to that of the LexA+ control (Fig. 5). Apparently, the psiB+ phenotype is suppressed by the purA::Tn10 allele or by a mutation that maps close to it. This fortuitous result allowed observation of the 30-fold reduction in the level of adaptive mutation of the LexA(Def) strain due to the groEL100 allele (Fig. 5).
FIG. 5.
A groE mutant allele reduces adaptive mutation in the absence of LexA. The accumulation of Lac+ revertants during incubation on lactose-minimal medium is shown. Three independent cultures were plated for each strain; data are the mean numbers of Lac+ colonies appearing each day from days 3 to 5 ± SEM. lexA(Def) groE+ = PFG328: lexA(Def) groEL100 = PFG329; lexA+ = PFG340; lexA+ groEL100 = PFG341. All strains are sulA11 and purA::Tn10.
Mutations in groE and dinB are epistatic in recG+ cells but not in recG mutant cells.
We wished to determine whether the reduced amount of Pol IV in groE mutant strains was sufficient to explain their reduction in adaptive mutation. Epistasis is the classical genetic test to determine if two genes are in the same or different pathways. As shown in Fig. 6A, in recG+ cells the groEL100 ΔdinB::Zeo double mutant strain had the same low rate of adaptive mutation to Lac+ as did strains carrying either single mutant. However, as shown in Fig. 6B, in recG mutant cells the double mutant had a significantly lower rate of adaptive mutation than either single mutant. These results support the hypotheses that, in otherwise wild-type cells, GroE affects adaptive mutation solely by regulating the amount of Pol IV, but in recG mutant cells, GroE has an additional effect.
FIG. 6.
The groEL100 and ΔdinB::Zeo mutant alleles are epistatic in recG+ but not in recG mutant strains. Shown is the accumulation of Lac+ revertants of groE+, ΔdinB::Zeo, groEL100, and groEL100 ΔdinB::Zeo strains during incubation on lactose-minimal medium. Data are the cumulative number of Lac+ colonies divided by the number of viable Lac− cells on the plate 2 days earlier. Because it takes 2 days for a Lac+ revertant to make a visible colony, the values shown are displaced 2 days earlier. Each point is the mean ± 95% confidence limits of results for six cultures (some error bars are smaller than the symbols). (A) Closed circles, PFG60 (groE+); open circles, PFG250 (ΔdinB::Zeo); closed triangles, PFG61 (groEL100); open triangles, PFG251 (ΔdinB::Zeo groEL100). (B) Closed circles, PFG62 (groE+); open circles, PFG305 (ΔdinB::Zeo); closed triangles, PFG63 (groEL100); open triangles, PFG307 (ΔdinB::Zeo groEL100). For clarity, the results for PF62 after day 1 are not shown; see Fig. 2 for comparable data for this strain. All strains are also purA::Tn10.
GroE is unlikely to act via RpoS but does influence SOS induction.
In our previous paper on the regulation of Pol IV, we reported that the general stress response sigma factor, RpoS (σ38), is required for the induction and maintenance of Pol IV in stationary-phase cells. So, an obvious hypothesis is that GroE and RpoS act together in a single pathway affecting Pol IV. However, we were unable to construct stable rpoS groES or rpoS groEL double mutants to test this hypothesis directly by using epistasis. Therefore, we used ectopic lacZ fusions to determine if mutations in each gene influence the expression of the other gene. For these experiments we grew the cells under the same conditions that were used to determine the amount of Pol IV protein.
Since Pol IV levels are more severely reduced by a GroE defect than by loss of RpoS (34), it is unlikely that our results can be explained by a positive effect of GroE on RpoS. In confirmation, the groEL100 allele did not decrease, and in fact slightly increased, the expression of lacZ transcriptional and translational fusions to rpoS (Table 2). However, when RpoS activity was measured by using a lacZ fusion to katE, a gene whose expression is rpoS dependent (53), the groEL100 allele resulted in a 30% decrease in β-galactosidase activity (Table 2). Although statistically significant (t = 3.9; P = 0.002), this reduction is far less than the 97% decrease in katE expression caused by total loss of RpoS (Table 2) and seems of insufficient magnitude to account for the 10-fold reduction in the levels of Pol IV observed in groE mutant strains (Fig. 3).
TABLE 2.
Expression of lacZ under the control of various promoters in different genetic backgrounds
| Strain | Fusion | Relevant genotype | No. of cultures | Mean β-galactosidase activity (Miller units) ± 95% CLa |
|---|---|---|---|---|
| PFG340 | rpoS-lacZb | groE+ | 3 | 2,053 ± 191 |
| PFG341 | rpoS-lacZb | groEL100 | 3 | 2,286 ± 44 |
| PFG342 | PrpoS-lacZc | groE+ | 3 | 7,426 ± 873 |
| PFG343 | PrpoS-lacZc | groEL100 | 3 | 8,031 ± 1,186 |
| PFG334 | katE-lacZd | groE+ | 7 | 662 ± 76 |
| PFG335 | katE-lacZd | groEL100 | 7 | 461 ± 99 |
| PFG336 | katE-lacZd | groE+rpoS::Cm | 3 | 23 ± 3 |
| PF2253 | PgroE-lacZe | rpoS+ | 7 | 585 ± 164 |
| PF2259 | PgroE-lacZe | rpoS::Cm | 7 | 361 ± 41 |
| PF2252 | PL-lacZf | rpoS+ | 7 | 6,112 ± 261 |
| PF2258 | PL-lacZf | rpoS::Cm | 7 | 5,525 ± 1,194 |
| PFG308 | sulA-lacZg | groE+ | 7 | 224 ± 59 |
| PFG309 | sulA-lacZg | groEL100 | 7 | 168 ± 28 |
| PFG332 | sulA-lacZg | recG groE+ | 6 | 610 ± 339 |
| PFG333 | sulA-lacZg | recG groEL100 | 7 | 256 ± 38 |
Values are the means of results with the indicated number of independent cultures ± the 95% confidence limits (CL). Results from several experiments have been combined.
rpoS-lacZ is a gene fusion of lacZ to rpoS under the control of the rpoS promoter (26).
PropS-lacZ is an operon fusion that puts lacZ under the control of the rpoS promoter (26).
katE-lacZ is λ(katE::lac Kn) that puts lacZ under the control of the σ38-dependent katE promoter (53).
PgroE-lacZ is an operon fusion that puts lacZ under the control of the σ32-dependent groE promoter; the fusion does not retain the σ70-dependent groE promoter (61).
PL-lacZ is an operon fusion that puts lacZ under the control of the σ70-dependent lambda promoter (61).
sulA-lacZ is an operon fusion of Mud(Ap lac B::Tn9) to the sulA gene (D. Mount, personal communication).
Our Pol IV results also could be explained if RpoS were a positive effector of GroE, although there are no reports of this in the literature. As shown in Table 2, the rpoS::Cm allele caused about a 40% reduction in the expression of a lacZ fusion to the groE heat shock (σ32-dependent) promoter (Table 2), and this reduction is statistically significant (t = 3.2; P = 0.01). Again, the magnitude of this effect seems insufficient to account for the 10-fold reduction in the levels of Pol IV caused by GroE deficiency (Fig. 3).
Using a lacZ fusion to the SOS-induced gene, sulA, we also tested whether GroE was required for expression of the SOS response. As shown in Table 2, in wild-type cells the groEL100 allele reduced sulA expression by about 25%, but this reduction was significant, with a probability of only 8% (t = 1.9; P = 0.08). Interestingly, in recG mutant cells in which the SOS response is partially induced (38) (Table 2), GroE deficiency resulted in a larger decrease (58%), and this decrease was statistically significant despite the variability of results with the sulA-lacZ fusion (t = 3.16; P = 0.03). Thus, the 20-fold reduction in the level of adaptive mutation of recG mutant strains caused by a deficiency in GroE (Fig. 2) can be attributed to both a reduction in the amount of Pol IV and a decrease in the degree of SOS induction.
DISCUSSION
The heat shock response in E. coli is induced by temperature shift and other conditions that result in unfolded proteins. Approximately 30 genes are positively regulated by the heat shock sigma factor RpoH (σ32). GroE, E. coli's Hps60 chaperone, is part of this regulon and is required at all temperatures to aid essential proteins to fold and maintain their proper conformation (40). GroE consists of two subunits that in E. coli are encoded by the groES/groEL operon; both genes are essential, and mutation of each results in the same phenotypes (13, 58, 66). In addition to its RpoH-dependent promoter, the groES/groEL operon has a promoter recognized by the vegetative sigma factor, RpoD (σ70), allowing for expression of GroE at all temperatures (67). GroE and certain other components of the heat shock regulon are also induced by DNA damage, oxidative stress, antibiotics and heavy metals, phage infection, and carbon source or amino acid starvation (reviewed in reference 64). Thus, the RpoH regulon can be considered a general stress response. The discovery, reported here, that cellular levels of DNA Pol IV are dependent on GroE provides a new link between this error-prone polymerase, adaptive mutation, and global stress responses.
GroE is required for the recovery of mutations after DNA-damaging treatments that induce the SOS response (9, 37). There is strong evidence that GroE interacts with the polymerase subunit of Pol V and protects it from degradation (10). Because Pol IV is a homolog of Pol V, it seemed likely that GroE would also interact with Pol IV, but we failed to detect an interaction between GroE and Pol IV using three methods: coimmunoprecipitation, copurification on a metal resin, and yeast two-hybrid analysis. Possibly, our techniques were not robust enough to detect an interaction; alternatively, the effect of GroE on Pol IV levels may be indirect. For example, some substrate of GroE may be required for Pol IV stability.
GroE appears to affect expression of the SOS response, particularly in recG mutant cells (Table 2). Mass coimmunoprecipitation experiments identified RecA as a GroE substrate (27). Since RecA promotes the proteolytic inactivation of LexA (36), it is possible that when GroE is deficient, levels of RecA are low, levels of active LexA are relatively high, and SOS genes, including dinB, are repressed. However, this sequence of events cannot account for the entire effect of GroE because even in the absence of LexA, a deficiency in GroE reduced both the cellular amount of Pol IV and the rate of adaptive mutation (Fig. 4 and 5).
It is unlikely that GroE affects Pol IV via RpoS for the following reasons. First, we could find no reports in the literature that GroE is a positive effector of the amount or activity of RpoS. Second, a defect in GroE reduces the amount of Pol IV to a greater extent than does the loss of RpoS (34); if GroE were a positive effector of RpoS, the loss of RpoS should have the greater impact. Third, the groEL100 mutation had only a modest effect on the expression and activity of RpoS (Table 1). On the other hand, the results in Table 2 suggest that RpoS may increase the transcription of the groE operon from its heat shock promoter by about 40%, suggesting that RpoS may be a modest positive effector of GroE. However, these small effects may not be biologically significant and are unlikely to account for the large effects that GroE has on levels of Pol IV.
Although it appears that RpoS and GroE do not positively affect each other, it is still possible that they are in the same pathway that regulates Pol IV. For example, each may be a positive effector of yet a third component that is itself a positive effector of Pol IV. Further experiments are necessary to test this hypothesis.
Deficiency in GroE had a greater relative effect on Pol IV in recG mutant strains than in wild-type strains (Fig. 1 and 2). The results of epistasis tests (Fig. 6) indicate that in recG+ cells, the reduction in Lac+ adaptive mutations when GroE is deficient is due solely to reduction in the amount of Pol IV, but this is not the case in recG mutant cells. Interestingly, the adaptive mutation rate of the recG groEL100 mutant strain was nearly the same as that of the recG+ groE+ strain (for example, see Fig. 6), yet from the Western results shown in Fig. 3, the double mutant has twice the amount of Pol IV. Thus, the large decrease in adaptive mutation caused by deficiency of GroE in recG mutant strains can be attributed to a reduced amount of Pol IV (Fig. 3) plus a decrease in the degree of SOS induction (Table 1). The LexA-repressed genes that are known to be required for adaptive mutation are recA and ruvAB (6, 17, 20, 23, 24); in addition there may be yet-unknown SOS genes involved. To determine how GroE affects expression of SOS-induced genes requires further experimentation.
Overproduction of Pol IV is a powerful mutator (29, 30, 60), but deletion of Pol IV has little effect on growth-dependent spontaneous mutation rates (42, 51, 62; P. L. Foster, unpublished results), although it does contribute to spontaneous mutation on extrachromosomal elements, such as the episome (32; Foster, unpublished). These observations suggest that Pol IV levels and activity are normally tightly regulated and targeted. Previously we reported that Pol IV is induced late in stationary phase and that RpoS, the general stress response sigma factor, is required for this induction and for the continued maintenance of high levels of Pol IV during starvation (34). The results presented here show that levels of Pol IV are sustained, directly or indirectly, by GroE. Since GroE is part of the RpoH regulon, which is induced by a variety of stresses that perturb or prevent growth, our results suggest that it is important to survival to have adequate levels of this error-prone polymerase. Indeed, all three of E. coli's inducible DNA polymerases, Pol II, IV, and V, confer a competitive advantage during prolonged stationary phase (63). The links between Pol IV and global responses to DNA damage, protein damage, and starvation provide evidence that the ability to increase genetic diversity has evolved to promote survival during stress.
Acknowledgments
We thank the following people for bacterial strains, plasmids, and reagents: T. Elliot, A. R. Fernández de Henestrosa, J. Krueger, J. H. Miller, R. Lloyd, S. T. Lovett, R. Maurer, T. Nohmi, H. Ohmori, G. C. Walker, G. Wegrzyn, and R. Woodgate. We are grateful to the other members of our laboratory for useful suggestions and comments.
This work was supported by USPHS grant NIH-NIGMS G651575.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bagg, A., C. J. Kenyon, and G. C. Walker. 1981. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baneyx, F., and A. A. Gatenby. 1992. A mutation in GroEL interferes with protein folding by reducing the rate of discharge of sequestered polypeptides. J. Biol. Chem. 267:11637-11644. [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1479. [DOI] [PubMed] [Google Scholar]
- 5.Borden, A., P. I. O'Grady, D. Vandewiele, A. R. Fernandez de Henestrosa, C. W. Lawrence, and R. Woodgate. 2002. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J. Bacteriol. 184:2674-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, J., J. Overbaugh, and S. Miller. 1988. The origin of mutants. Nature 335:142-145. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly, C. E., and G. C. Walker. 1989. groE mutants of Escherichia coli are defective in umuDC-dependent UV mutagenesis. J. Bacteriol. 171:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly, C. E., and G. C. Walker. 1992. Coexpression of UmuD′ with UmuC suppresses the UV mutagenesis deficiency of groE mutants. J. Bacteriol. 174:3133-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dri, A.-M., and P. L. Moreau. 1994. Control of the LexA regulon by pH: evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Mol. Microbiol. 12:621-629. [DOI] [PubMed] [Google Scholar]
- 12.Edenhofer, F., R. Rieger, M. Famulok, W. Wendler, S. Weiss, and E.-L. Winnacker. 1996. Prion protein PrPc interacts with molecular chaperones of the Hsp60 family. J. Virol. 70:4724-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171:1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenton, W. A., Y. Kashi, K. Furtak, and A. L. Horwich. 1994. Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371:614-619. [DOI] [PubMed] [Google Scholar]
- 15.Foster, P. L. 1994. Population dynamics of a Lac− strain of Escherichia coli during selection for lactose utilization. Genetics 138:253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, P. L. 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33:57-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, P. L. 1993. Adaptive mutation: the uses of adversity. Annu. Rev. Microbiol. 47:467-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, P. L., and J. M. Trimarchi. 1995. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92:5487-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galitski, T., and J. R. Roth. 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268:421-423. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 23.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 24.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch, M., and T. Elliott. 2002. Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli. J. Bacteriol. 184:5077-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houry, W. A., D. Frishman, C. Eckerskorn, F. Lottspeich, and F. U. Hartl. 1999. Identification of in vivo substrates of the chaperonin GroEL. Nature 402:147-154. [DOI] [PubMed] [Google Scholar]
- 28.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S.-R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA Pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 31.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 32.Kuban, W., P. Jonczyk, D. Gawel, K. Malanowska, R. M. Schaaper, and I. J. Fijalkowska. 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 186:4802-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry, S. J., J. Zeilstra-Ryalls, O. Fayet, C. Georgopoulos, and L. M. Gierasch. 1993. Characterization of a functionally important mobile domain of GroES. Nature 362:255-258. [DOI] [PubMed] [Google Scholar]
- 34.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenne-Samuel, N., J. Wagner, H. Etienne, and R. P. Fuchs. 2002. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 3:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 37.Liu, S.-K., and I. Tessman. 1990. groE genes affect SOS repair in Escherichia coli. J. Bacteriol. 172:6135-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 173:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardo, M.-J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 41.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenzie, G. J., P. L. Lee, M.-J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohmori, H., E. Hatada, Y. Qiao, M. Tsuji, and R. Fukuda. 1995. dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutat. Res. 347:1-7. [DOI] [PubMed] [Google Scholar]
- 46.Radicella, J. P., P. U. Park, and M. S. Fox. 1995. Adaptive mutation in Escherichia coli: a role for conjugation. Science 268:418-420. [DOI] [PubMed] [Google Scholar]
- 47.Rajkumari, K., and J. Gowrishankar. 2002. An N-terminally truncated RpoS σS protein in Escherichia coli is active in vivo and exhibits normal environmental regulation even in the absence of rpoS transcriptional and translational control signals. J. Bacteriol. 184:3167-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuven, N. B., G. Arad, A. Maor-Shoshani, and Z. Livneh. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763-31766. [DOI] [PubMed] [Google Scholar]
- 49.Rice, J. A. 1995. Mathematical statistics and data analysis. Wadsworth Publishing Company, Belmont, Calif.
- 50.Rodriguez, C., J. Tompkin, J. Hazel, and P. L. Foster. 2002. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J. Bacteriol. 184:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka, K., K. Handel, P. C. Loewen, and H. Takahashi. 1997. Identification and analysis of the rpoS-dependent promoter of katE, encoding catalase HPII in Escherichia coli. Biochim. Biophys. Acta 1352:161-166. [DOI] [PubMed] [Google Scholar]
- 54.Tang, M., I. Bruck, R. Eritja, J. Turner, E. G. Frank, R. Woodgate, M. O'Donnell, and M. F. Goodman. 1998. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc. Natl. Acad. Sci. USA 95:9755-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang, M., P. Pham, X. Shen, J.-S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 56.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli Pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, M. S., and R. E. Glass. 1991. Escherichia coli rpoA mutation which impairs transcription of positively regulated systems. Mol. Microbiol. 5:2719-2725. [DOI] [PubMed] [Google Scholar]
- 58.Tilly, K., H. Murialdo, and C. Georgopoulos. 1981. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc. Natl. Acad. Sci. USA 78:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA Pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 60.Wagner, J., and T. Nohmi. 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182:4587-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wegrzyn, A., A. Szalewska-Palasz, A. Blaszczak, K. Liberek, and G. Wegrzyn. 1998. Differential inhibition of transcription from σ70- and σ32-dependent promoters by rifampicin. FEBS Lett. 440:172-174. [DOI] [PubMed] [Google Scholar]
- 62.Wolff, E., M. Kim, K. Hu, H. Yang, and J. H. Miller. 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186:2900-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 65.Zar, J. H. 1984. Biostatistical analysis. Prentice Hall, Englewood Cliffs, N.J.
- 66.Zeilstra-Ryalls, J., O. Fayet, L. Baird, and C. Georgopoulos. 1993. Sequence analysis and phenotypic characterization of groEL mutations that block lambda and T4 bacteriophage growth. J. Bacteriol. 175:1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou, Y. N., N. Kusukawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J. Bacteriol. 170:3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]