Abstract
The SAGA complex is a multisubunit protein complex involved in transcriptional regulation in Saccharomyces cerevisiae. SAGA combines proteins involved in interactions with DNA-bound activators and TATA-binding protein (TBP), as well as enzymes for histone acetylation (Gcn5) and histone deubiquitylation (Ubp8). We recently showed that H2B ubiquitylation and Ubp8-mediated deubiquitylation are both required for transcriptional activation. For this study, we investigated the interaction of Ubp8 with SAGA. Using mutagenesis, we identified a putative zinc (Zn) binding domain within Ubp8 as being critical for the association with SAGA. The Zn binding domain is required for H2B deubiquitylation and for growth on media requiring Ubp8's function in gene activation. Furthermore, we identified an 11-kDa subunit of SAGA, Sgf11, and showed that it is required for the Ubp8 association with SAGA and for H2B deubiquitylation. Different approaches indicated that the functions of Ubp8 and Sgf11 are related and separable from those of other components of SAGA. In particular, the profiles of Ubp8 and Sgf11 deletions were remarkably similar in microarray analyses and synthetic genetic interactions and were distinct from those of the Spt3 and Spt8 subunits of SAGA, which are involved in TBP regulation. These data indicate that Ubp8 and Sgf11 likely represent a new functional module within SAGA that is involved in gene regulation through H2B deubiquitylation.
Transcriptional regulation involves covalent chemical modifications of the core histone proteins that comprise the nucleosome. Each histone is modified, and modifications occur on the amino- and carboxyl-terminal extensions as well as the globular domains. The modifications can be either dynamic or static. Dynamic modifications include acetylation, phosphorylation, and ubiquitylation (60). Many of the enzymes that add and remove these modifications have been identified. Methylation is a more static modification, and lysine demethylating enzymes have not been reported (4).
Certain large multicomponent protein complexes possess more than one enzyme to regulate histone modifications. The 1.8-MDa SAGA complex in Saccharomyces cerevisiae contains enzymes that act to acetylate primarily histones H3 and H2B and to deubiquitylate histone H2B (22, 24). Acetylation is carried out by Gcn5, and this reaction has been intensively studied. The Gcn5-mediated acetylation of histones is required for the full activation of many genes in organisms ranging from yeast to humans (12, 25) and is reversed by deacetylation through a number of histone deacetylase (HDAC) enzymes (47). Gcn5 is essential for viability in mice (68a, 70). The biochemistry and atomic structure of Gcn5 have been characterized, revealing the basis of the substrate specificity for Lys-14 of the histone H3 amino terminus (13, 51, 64). H3 acetylation by Gcn5 requires prior H3 phosphorylation for the full activation of certain genes in yeast (34). An interrelationship between phosphorylation and acetylation is conserved from yeast to humans (2, 69).
The SAGA complex also harbors Ubp8, a histone H2B-deubiquitylating enzyme (14, 24, 52). Lys-123 of the carboxyl terminus of H2B is ubiquitylated by an E2 conjugase-E3 ligase pair (Rad6/Bre1) (28, 47, 66). Ubiquitylation is transient during gene activation, and ubiquitin is removed by Ubp8 acting within the SAGA complex (14, 24). Both ubiquitylation and deubiquitylation are required for optimal gene activation. The ubiquitylation of H2B is linked to histone methylation (24, 61). Specifically, ubiquitylated H2B (ubH2B) is required for Set1-mediated histone H3 Lys-4 methylation and Dot1-mediated Lys-79 methylation (10, 62). Although the mechanism is still unknown, this trans-tail modification pattern requires a portion of the proteasome, which is present at the promoter (17, 21). In contrast, Set2-mediated histone H3 Lys-36 methylation does not require ubiquitylation (61). SAGA-dependent H2B deubiquitylation by Ubp8 is required for the correct levels of Lys-4 vis-à-vis Lys-36 methylation (24). Specifically, the absence of deubiquitylation leads to high levels of Lys-4 methylation, whereas the absence of ubiquitylation leads to high levels of Lys-36 methylation (24).
Little is known about the biochemistry of deubiquitylating enzymes, including how they are targeted to substrates for specificity. One way to target enzymes to their substrates is to integrate them into multicomponent protein complexes. For example, SAGA delivers Gcn5 to appropriate promoters in response to certain inducing conditions via associations of the Tra1 subunit with DNA-bound activators (23). In this case, the presence of Gcn5 within SAGA is a key aspect of delivery to the substrate. How Ubp8 is integrated into SAGA or whether its association within SAGA is an important feature of substrate targeting is not known.
SAGA is modular in structure and function, incorporating multiple components for each of several functions, including acetylation, TATA-binding protein (TBP) regulation, and activator interactions. The histone acetyltransferase module consists of Gcn5, Ada2, and Ada3 (3, 11). TBP is regulated both positively and negatively by a module consisting of Spt3, Spt7, and Spt8 (7, 16, 71). The complex may bind to DNA through a Taf module that may form a nucleosome-like structure through histone fold domains (19). Tra1 provides key interactions with acidic activators (23), along with contributions by Ada2 (5). It has not previously been understood whether the Ubp8 function is correlated with a known module within SAGA or whether it constitutes a distinct module.
Here we describe a structure-function study of the association of Ubp8 with SAGA, using biochemical, genetic, microarray, and proteomic analyses. We provide evidence that a putative zinc finger domain of Ubp8 is required for the association of Ubp8 with the SAGA complex. Furthermore, our results indicate that Ubp8 constitutes a functional module within SAGA through its association with a newly identified component of SAGA, called Sgf11.
MATERIALS AND METHODS
Yeast strains and plasmids.
For the cloning of UBP8-3HA into pCR2.1-TOPO, genomic DNA was prepared from the YKH013 strain (24), which has a chromosomal copy of UBP8 C-terminally tagged with three copies of the hemagglutinin (HA) epitope and a his5+ marker. PCRs were then performed on the genomic DNA by the use of primer pairs spanning the UBP8-3×HA-his5+ cassette and the Expand high-fidelity PCR system (Roche). Agarose gel-purified PCR products were cloned into the pCR2.1-TOPO vector according to the manufacturer's recommendations (Invitrogen). A recombinant plasmid was sequenced to confirm proper integration of the insert. This plasmid served as a template for the creation of UBP8 point mutations and deletion constructs by use of a QuickChange site-directed mutagenesis kit (Stratagene). The resulting plasmids were digested with EcoRI and XhoI, which cut on either side of the UBP8 sequence (including upstream and downstream sequences flanking the open reading frame [ORF], the tag, and the selectable marker), and these fragments were used for integration into the endogenous UBP8 locus in ubp8 deletion strains. Correct integration was confirmed by PCR amplification of genomic DNAs.
Ubp8 episomal constructs were generated by amplifying UBP8 from pCR2.1-TOPO constructs by the use of primers containing SacI and NotI restriction enzyme sites and then ligating those fragments into the pRS416 vector. Correct clones were confirmed by sequencing, cotransformed into a gcn5Δ ubp8Δ strain (YKH068) (24) along with an empty TRP1-containing vector (pRS314), and maintained in synthetic medium lacking Ura and Trp.
Gene deletions and tagging were performed either as described previously (35) or by transforming PCR amplification products of genomic DNAs from previously established strains. All S. cerevisiae strains used for this study are listed in Table 1.
TABLE 1.
S. cerevisiae strains used for this study
| Strain | Genotype | Reference |
|---|---|---|
| JR5-2A | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 YCp50 (HTB1-CEN-URA3) | 48 |
| JR5-2A FLAG-HTB1 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) | 48 |
| JR5-2A FLAG-htb1 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-htb1-K123R-CEN-TRP1) | 48 |
| SB301 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG | 57 |
| SB301 Ada2-TAP | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 | This study |
| SB303 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 | 57 |
| YKI011 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8-ZnFΔ-3HA::his5+ | This study |
| YKI012 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8-H77A-3HA::his5+ | This study |
| YKI013 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8-H419A-3HA::his5+ | This study |
| YKI014 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8-C46/49A-3HA:his5+ | This study |
| YKI015 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) UBP8-3HA:his5+ | This study |
| YKI016 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8ZnFΔ-3HA:his5+ | This study |
| YKI017 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-H77A-3HA:his5+ | This study |
| YKI019 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-C46/49A-3HA:his5+ | This study |
| YKI020 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 UBP8-3HA:his5+ | This study |
| YKI041 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (UBP8-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI042 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (ubp8-ZnFΔ-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI043 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (ubp8-C46/49A-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI044 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (ubp8-H77A-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI045 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (ubp8-C146S-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI046 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (ubp8-H419A-CEN-URA3) pRS314 (CEN-TRP1) | This study |
| YKI047 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG UBP8-3FLAG:KanMX SGF11-3HA:his5+ | This study |
| YKI048 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 UBP8-2FLAG:KanMX sgf11::his5+ | This study |
| YKI049 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade 2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) sgf11::his5+ | This study |
| YKI050 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-C46/49A-3HA:his5+ | This study |
| YKI051 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-H77A-3HA:his5+ SGF11-3FLAG:KanMX | This study |
| YKI052 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-H419A-3HA:his5+ SGF11-3FLAG:KanMX | This study |
| T64 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8-C146S-3HA:his5+ | This study |
| T67 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8-C146S-3HA:his5+ | This study |
| YKH002 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 UBP8-2FLAG:KanMX | 24 |
| YKH007 | MATaura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (FLAG-HTB1-CEN-TRP1) ubp8::KanMX | 24 |
| YKH039 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG ADA2-TAP:URA3 ubp8::KanMX | 24 |
| YKH068 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX | 24 |
| YKH083 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG gcn5::HIS3 ubp8::KanMX pRS416 (CEN-URA3) pRS314 (CEN-TRP1) | 24 |
| YKH094 | MATahis3Δ200 leu2Δ1 ura3-52 trp1 ΔhisG UBP8-3FLAG:KanMX | This study |
Immunoprecipitations.
Coimmunoprecipitations of Ubp8 and other SAGA subunits with tandem affinity purification (TAP)-tagged Ada2 were performed with immunoglobulin G (IgG)-Sepharose (Amersham). Overnight immunoprecipitation reactions were washed twice in extraction buffer (40 mM HEPES [pH 7.5], 250 mM NaCl, 0.1% Tween 20, 10% glycerol, protease inhibitors) and twice in extraction buffer containing 350 mM NaCl. Immunoprecipitates were eluted by boiling, and input and eluate samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis with the anti-HA 12CA5 monoclonal antibody (Roche).
The same conditions were used for coimmunoprecipitations of Ubp8 and Sgf11. Using a strain expressing FLAG-tagged Ubp8 and HA-tagged Sgf11, we performed immunoprecipitations with either anti-HA affinity matrix (Covance) or anti-FLAG M2 agarose (Sigma). For Western blot analysis, either the anti-HA 12CA5 monoclonal antibody (Roche) or the anti-FLAG M2 antibody (Sigma) was used. For coimmunoprecipitations of Sgf11 and mutant forms of Ubp8, strains containing Sgf11-FLAG and various point mutants of Ubp8-HA were used. Immunoprecipitations and Western blot analysis were performed as described above.
Global levels of ubH2B were assessed by the use of JR5-2A strains expressing FLAG-tagged H2B (48). Whole-cell extracts were made in 20% trichloroacetic acid to precipitate histones, and the resuspended precipitates were either used directly for Western blot analysis or used for immunoprecipitation with anti-FLAG M2 agarose (Sigma). The immunoprecipitates were washed twice in IP buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% NP-40, 0.5% bovine serum albumin, protease inhibitors) and three times in IP buffer without bovine serum albumin. Elution was performed with 300 ng of 3× FLAG peptide (Sigma)/μl, and input and eluate samples were subjected to SDS-PAGE and Western blotting with the anti-FLAG M2 antibody or an anti-FLAG-horseradish peroxidase conjugate antibody (Sigma).
Plate assays.
Ubp8 episomal constructs in pRS416 and an empty TRP1-containing vector (pRS314) were cotransformed into YKH068 (ubp8Δ gcn5Δ), grown to exponential phase, and spotted in 5× dilutions on SC-Ura-Trp plates containing different carbon sources (i.e., glucose or galactose). Plates were incubated at 30°C for 48 h.
Ubp8-2×FLAG purification.
Ubp8-containing complexes were purified from the YKH002 strain, in which Ubp8 bears a C-terminal 2× FLAG epitope tag. Lysates were prepared from 15 liters of culture, followed by immunoprecipitation with anti-FLAG M2 affinity resin (Sigma) according to the manufacturer's protocol. Bound proteins were washed and then eluted with a 3× FLAG peptide (Sigma) at a concentration of 300 ng/μl. The eluate was then subjected to MonoQ ion-exchange chromatography followed by anti-FLAG Western blotting to identify the fractions in which Ubp8 eluted. Ubp8-containing fractions were then pooled and loaded onto a Superose 6 column, from which Ubp8 eluted in a single peak. Fractions containing Ubp8-2×FLAG, as determined by Western blotting, were pooled, concentrated, and then loaded into a 4 to 20% SDS-polyacrylamide gradient gel. Proteins were then visualized by silver staining and anti-FLAG Western blotting (Sigma) to determine the migration of the tagged Ubp8.
TAP purification.
TAP-tagged components of SAGA were purified on IgG and calmodulin columns from extracts of yeast cells (3 liters) grown in YPD medium to an optical density at 600 nm of 1.0 to 1.5. The complexes were analyzed by SDS-PAGE, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF), and tandem mass spectrometry as previously described (33).
SGA and microarray analyses.
Synthetic genetic array (SGA) analysis was performed as previously described (63). Automated analyses of the results were performed by procedures that will be described elsewhere (H. Ding and C. Boone, unpublished data). RNA preparation and microarray analysis were performed as previously described (42) after isogenic strains were grown in parallel in SC medium at 30°C.
RESULTS
The catalytic domain and a putative Zn finger domain within Ubp8 are both required for in vivo function.
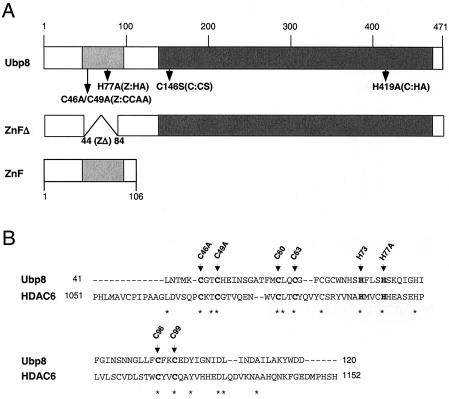
We used a mutagenesis approach to analyze the function of Ubp8. In addition to the putative catalytic domain (Fig. 1A), there is another notable structural feature within the predicted protein sequence of Ubp8. This is a cysteine- and histidine-rich region within the amino-terminal quarter of the protein, which is likely a zinc finger domain (6, 54) (Fig. 1A). This region of Ubp8 is similar to a region within HDAC6 (Fig. 1B), as previously proposed for a related Ubp (26, 55). To test whether the Zn finger is required for the function of Ubp8 within SAGA, we prepared deletion (ZnFΔ; amino acid residues 46 to 83) and substitution mutations of predicted critical residues for the structure of the Zn finger domain (Fig. 1). The substitution mutations at predicted critical Zn finger residues were cysteines 46 and 49 to alanine (C46/49A mutant, called Z:CCAA in this paper) and histidine 77 to alanine (Z:HA). As controls for Ubp8 function, we generated substitution mutations within the catalytic domain based on homology to residues that were experimentally identified to be required for catalysis (27). These substitutions were cysteine 146 to serine (C146S mutant, or C:CS) and histidine 419 to alanine (C:HA) (Fig. 1A).
FIG. 1.
Schematic structure of Ubp8. (A) The Zn finger (ZnF) and catalytic domains are indicated, as well as the mutation and deletion constructs generated for this study. (B) The Ubp8 Zn finger shares homology with murine HDAC6. The Cys and His residues believed to be involved in Zn binding are shown in bold and indicated by arrows.
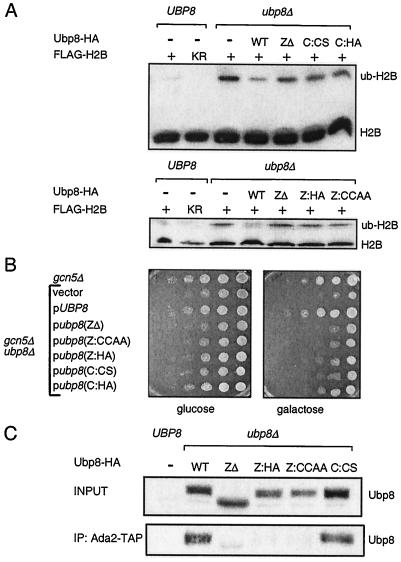
We tested the importance of these regions for function in vivo by using two assays. First, we monitored the effect on ubiquitylation of H2B compared to that of a mutant with a complete deletion of UBP8 (24). We employed an H2B-FLAG epitope pull-down assay followed by gel electrophoresis and Western blotting of FLAG (48). The larger size of ubH2B-FLAG than of H2B-FLAG causes retardation in its gel mobility. As previously shown (24), the level of ubH2B-FLAG increased in cells bearing a deletion of Ubp8 (Fig. 2A). Just as substitutions within the putative catalytic domain of Ubp8 increased the level of ubH2B (Fig. 2A, C:CS and C:HA), so did a deletion (Fig. 2A, ZΔ) and substitutions within the Zn finger (Fig. 2A, Z:CCAA and Z:HA).
FIG. 2.
Effects of Ubp8 mutations on global H2B ubiquitylation, plate growth, and association with SAGA. (A) H2B ubiquitylation assay. Top panel, Western blot analysis of FLAG-H2B immunoprecipitates in strains containing the Ubp8 Zn finger deletion and catalytic point mutants. The faster migrating band represents H2B and the slower migrating band represents ubiquitylated H2B. Bottom panel, Western blot analysis of extracts containing FLAG-H2B and Ubp8 Zn finger deletion and point mutants. The catalytic domain substitution mutants were C:CS (C146S) and C:HA (H419A). The Zn finger domain substitution mutants were Z:HA (H77A) and Z:CCAA (CC46/49AA). KR, K123R mutation in the H2B ubiquitylation site. (B) Plate phenotype assay. Left panel, phenotype assay of Ubp8 mutant strains on selective plates containing 2% glucose as carbon source. Exponentially growing yeast strains were used to make fivefold serial dilutions, which were spotted onto plates and incubated at 30°C for 48 h. Right panel, phenotype assay on selective plates containing 2% galactose. Substitution mutants were the same as those described for panel A. (C) Association of Ubp8 mutants with Ada2. Strains containing TAP-tagged Ada2 and various HA-tagged Ubp8 mutants were subjected to IgG immunoprecipitation and Western blot analysis to determine the amounts of Ubp8 associated with SAGA subunits. Whole-cell extracts made from the Ubp8-ZΔ strain contained less Ubp8 protein than extracts from the other strains, as judged by Western blotting of HA. To include equal amounts of Ubp8 in each reaction, we used five times the amount of Ubp8-ZΔ whole-cell extract for this experiment as that used for other strains.
We then tested the role of the Zn finger in Ubp8 functions during growth. A galactose indicator plate assay was used to test the role of Ubp8 in activating a pathway of gene expression that includes the GAL1 gene (24). We previously found that a loss of Ubp8 combined with a loss of Gcn5 causes very poor growth on these plates (24), and we therefore tested the effect of the mutations within Ubp8 in the absence of Gcn5. Substitution mutations within both the catalytic domain and the Zn finger caused poor growth in the absence of Gcn5, to a similar extent as the complete deletion of UBP8 (Fig. 2B). These assays indicated that the Zn finger of Ubp8 is required for its function, likely through Ubp8's role in deubiquitylating H2B.
The putative Zn binding domain of Ubp8 is required for its association with SAGA.
Zn binding domains often serve to promote protein-protein or protein-DNA interactions. Therefore, we further tested whether the Ubp8 Zn finger provides a surface for an association with SAGA. We used the set of deletion and substitution mutants of the Zn finger and examined their ability to associate with SAGA compared to that of mutants with substitutions within the catalytic domain. Wild-type or mutant Ubp8 was integrated into a strain bearing a deletion of UBP8 and a TAP-tagged version of Ada2. We previously showed that the immunoprecipitation of Ada2-TAP with IgG coprecipitates components of SAGA, including Ubp8 (24). Deletion of the entire Zn finger caused a loss of Ubp8 from the immunoprecipitate, as did substitution mutants within this domain (Z:HA and Z:CCAA) (Fig. 2C). In contrast, substitution within the catalytic domain (C:CS), which lowered the function of Ubp8 (Fig. 2A and B), did not reduce the level of Ubp8 within SAGA (Fig. 2C).
We then examined whether the Zn finger is sufficient for SAGA interaction by expressing only the amino-terminal portion of Ubp8 (amino acid residues 1 to 106) (Fig. 1A) in the Ada2-TAP strain. The Zn finger domain was able to interact with SAGA, although at greatly reduced levels relative to the full-length Ubp8 protein (data not shown). We concluded that the putative Zn finger of Ubp8 is necessary for Ubp8 interaction with SAGA but that other surfaces within Ubp8 likely contribute to the interaction.
Purification of SAGA reveals a new component, Sgf11.
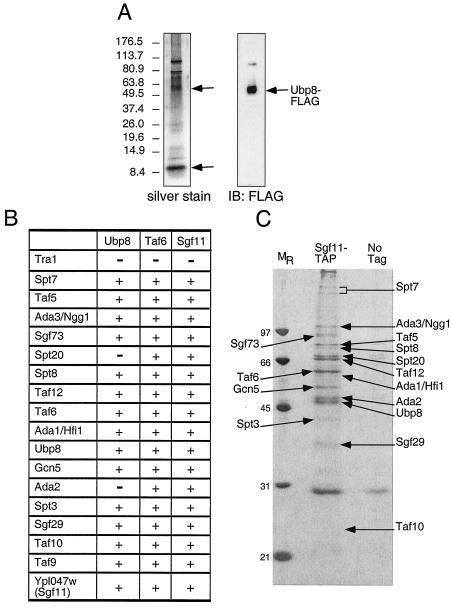
Having determined the region within Ubp8 that is required for its interaction with SAGA, we then investigated how SAGA interacts with Ubp8, that is, which component of SAGA associates with Ubp8. We tagged Ubp8 with the FLAG epitope and purified Ubp8-FLAG to near homogeneity, using a series of chromatography steps consisting of FLAG affinity purification and MonoQ ion-exchange chromatography followed by anti-FLAG Western blotting to identify fractions containing Ubp8. These were pooled and separated by size in a Superose 6 column, and a single peak of Ubp8-FLAG was found by Western blotting. This peak fraction was then electrophoresed in a gradient gel. Silver staining revealed several prominent peptides of approximately 60 to 110 kDa, and Western blotting of FLAG identified the smallest of these as Ubp8 (Fig. 3A). One notable, possibly stoichiometric polypeptide was approximately 10 kDa.
FIG. 3.
Purification of SAGA through the use of different tagged subunits. (A) Ubp8-FLAG purification. Ubp8 was subjected to affinity chromatography purification, followed by ion-exchange chromatography and gel filtration. Ubp8-containing fractions were pooled, concentrated, and analyzed by SDS-PAGE followed by silver staining and Western blotting. (B) Summary of tandem affinity purifications of SAGA that were performed with strains containing either no tagged protein or a TAP-tagged version of Ubp8, Taf6, or Sgf11. The protein complexes were purified in the presence of 100 mM NaCl and were then analyzed by SDS-PAGE. The subunits of SAGA were identified by trypsin digestion and MALDI-TOF mass spectrometry or by tandem mass spectrometry after subjecting an aliquot of the eluate from the final column directly to trypsin digestion. Proteins that were present in at least two of the three purifications are represented. (C) Silver staining of Sgf11-TAP purification. The proteins identified by MALDI-TOF mass spectrometry are labeled. Taf9 and Sgf11-TAP were not identified by MALDI-TOF mass spectrometry since they are small proteins and ran off the gel, but we were able to identify them by liquid chromatography-mass spectrometry.
To further characterize components of the SAGA complex and, in particular, to determine the identity of the 10-kDa putative subunit, we used a strain bearing a TAP-tagged version of Ubp8. Ubp8-TAP was purified sequentially on IgG and calmodulin columns and analyzed by SDS-PAGE followed by silver staining. Protein bands corresponding to the tagged protein and any associated proteins that were absent from a control preparation were then identified by MALDI-TOF and tandem mass spectrometry. Ubp8-TAP copurified with almost every known component of SAGA, as well as with a previously uncharacterized, 11-kDa protein encoded by the ORF YPL047W (Fig. 3B and data not shown). Tandem affinity purification of another subunit of SAGA, Taf6, which is also a component of TFIID, resulted in the copurification of not only SAGA and TFIID (Fig. 3B and data not shown), but also the 11-kDa ORF, YPL047W. To confirm that this previously uncharacterized protein was a bona fide member of SAGA, we purified Ypl047w-TAP and demonstrated that it copurified with the known components of SAGA (Fig. 3B and C). We and others have named this novel subunit of SAGA Sgf11 (44).
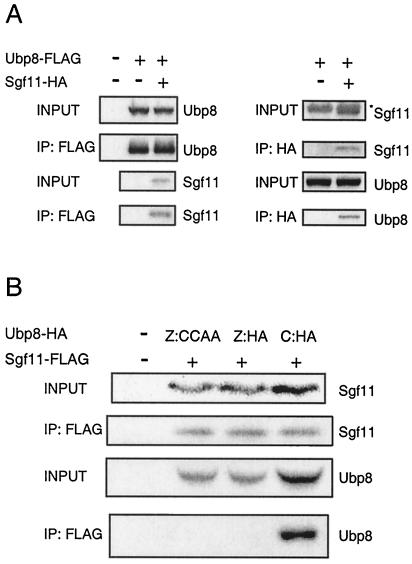
To confirm the association of Sgf11 with SAGA, we determined whether Ubp8 and Sgf11 interact. The SGF11 and UBP8 genes were tagged in the genome with HA and FLAG epitopes, respectively. The immunoprecipitation of Ubp8 with FLAG followed by Western blotting of HA showed that Sgf11 interacted with Ubp8 (Fig. 4A). The reverse test demonstrated that the immunoprecipitation of Sgf11-HA coprecipitated Ubp8-FLAG (Fig. 4A).
FIG. 4.
Interaction between Ubp8 and Sgf11. (A) Coimmunoprecipitation of Ubp8-FLAG and Sgf11-HA. Left panel, FLAG-immunoprecipitation analyzed by Western blotting of FLAG and HA. Right panel, HA immunoprecipitation followed by Western blotting of HA and FLAG. The asterisk in the right panel marks a nonspecific band migrating right above Sgf11-HA. (B) Coimmunoprecipitation of Sgf11-FLAG and Ubp8-HA containing Zn finger domain and catalytic domain substitution mutations. The images are for FLAG immunoprecipitation followed by Western blotting of FLAG and HA. The first lane represents a strain without tags on either Ubp8 or Sgf11. The following three lanes all contained FLAG-tagged Sgf11 and various mutants of Ubp8-HA.
To determine whether the interaction between Ubp8 and Sgf11 is mediated by the Zn finger domain of Ubp8, we performed a coimmunoprecipitation assay with strains containing Ubp8-HA bearing either Zn finger domain or catalytic domain substitution mutations and Sgf11-FLAG. We predicted that Zn finger mutations in Ubp8, but not catalytic mutations, would lower the amount of interaction with Sgf11. Indeed, the precipitation of Sgf11 pulled down the catalytic mutant of Ubp8 (Fig. 4B), but it did not pull down the two Zn finger mutants (Fig. 4B). These results mirrored the results of the SAGA (Ada2-TAP) association experiment (Fig. 2C), suggesting that the Ubp8 association with SAGA is dependent on its Zn finger domain through binding to Sgf11.
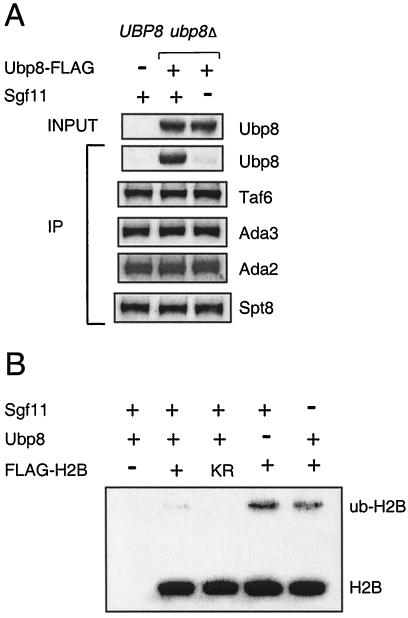
Sgf11 is required for Ubp8 association with SAGA and for H2B deubiquitylation.
Based on these results, we next tested whether the deletion of Sgf11 would alter the Ubp8 association with SAGA. Ubp8 was tagged with FLAG in a strain bearing Ada2-TAP and in an equivalent strain that also had a deletion of SGF11. IgG immunoprecipitation of Ada2 revealed the coprecipitation of Ubp8-FLAG (Fig. 5A). The deletion of SGF11 eliminated the coprecipitation of Ubp8 with Ada2-TAP, whereas Ada3, Spt8, and Taf6 coprecipitation was maintained, indicating that the SAGA complex was largely intact.
FIG. 5.
Role of Sgf11 in Ubp8 interaction with SAGA and in deubiquitylation of H2B. (A) Effect of sgf11Δ mutation on Ubp8 interaction with SAGA subunits. Strains containing TAP-tagged Ada2 were immunoprecipitated with IgG and analyzed by Western blotting of SAGA subunits. (B) Comparison of ubp8Δ and sgf11Δ effects on global H2B ubiquitylation. Strains containing FLAG-H2B were subjected to FLAG immunoprecipitation and Western blot analysis to study the levels of ubiquitylated H2B. KR, K123R mutation in the H2B ubiquitylation site.
Thus, Ubp8 requires Sgf11 for its association with SAGA. These findings suggested that, like Ubp8, Sgf11 is required for H2B deubiquitylation. This was examined by assaying the level of ubH2B in an sgfllΔ strain compared to that in a ubp8Δ strain. The amount of ubH2B was increased to a comparable level in either deletion strain (Fig. 5B), suggesting a role for Sgf11 in Ubp8-mediated histone H2B deubiquitylation.
Loss of Ubp8 or Sgf11 results in similar synthetic genetic interactions and microarray expression profiles, which are distinct from those of other components of SAGA.
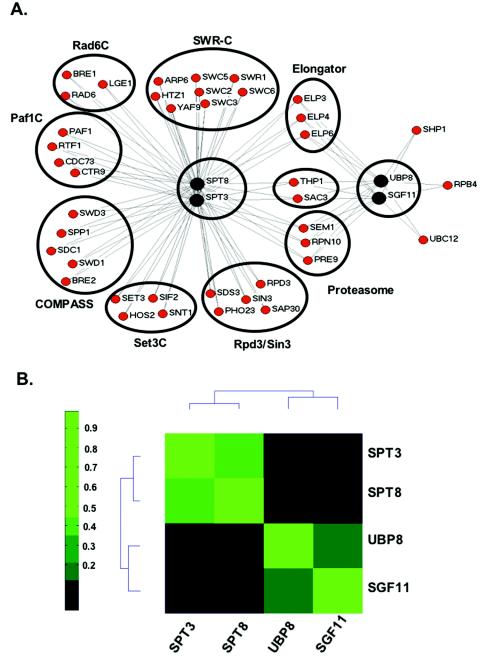
Previous analyses of SAGA subunits have demonstrated their modularity in structure and function. For example, certain subunits form subcomplexes and have similar biochemical or genetic functions, such as Gcn5/Ada2/Ada3 (3, 11, 46, 58), Spt3/Spt7/Spt8 (7, 58), and Ada1/Spt20 (Ada5) (22, 58). Genes encoding proteins in the same functionally distinct subcomplex should, in principle, have similar effects on gene expression and similar sets of synthetic genetic interactions. To identify the functional module of SAGA to which Sgf11 belongs, we first used automated SGA analysis (63). Natr strains harboring individual gene deletions of nonessential components of SAGA (SPT3, SPT8, UBP8, SGF11, GCN5, ADA2, ADA3, SPT20, and ADA1) were generated and crossed to a set of viable deletion strains selected for their involvement in gene expression, and resulting double mutant strains were analyzed (data not shown). Interestingly, strains harboring deletions of genes encoding proteins that are in two of the functional subgroups (Gcn5/Ada2/Ada3 and Ada1/Spt20) were unable to be analyzed by this technology. In every case, the selection of haploid cells from the heterozygous double mutant diploid strains was not successful, presumably because these factors are needed for the expression of genes involved in sporulation. The fact that the deletion of genes from these two subcomplexes resulted in a sporulation-defective phenotype is consistent with the existence of similar functions between them.
We were, however, able to generate double mutants from starting strains that contained deletions of SPT3, SPT8, UBP8, or SGF11. Similar sets of genetic interactions were obtained with spt3Δ and spt8Δ mutants, a result consistent with both being involved in the same functional module. These included genetic interactions with components of the Set1 histone methyltransferase-containing complex, COMPASS (SPP1, SDC1, SWD3, BRE2, and SPP1) (9, 29, 36, 39, 50); the PAF complex (RTF1, PAF1, CTR9, and CDC73) (30, 38, 56), which is required for the recruitment of COMPASS to chromatin (30, 40); and the Rad6 complex (RAD6, BRE1, and LGE1), which ubiquitylates Lys-123 of histone H2B (28, 48, 66), an event required for the methylation of H3 Lys-4 by COMPASS (Fig. 6A) (15, 62). Genetic interactions were also detected with components of the SWR-C remodeling complex (SWC2, SWR1, SWC3, SWC5, SWC6, ARP6, and YAF9) and the histone variant HTZ1, which is incorporated into chromatin via SWR-C (32, 37). Interactions were also seen with components of two HDAC complexes, the Rpd3-containing complex (RPD3, SIN3, SAP30, SDS3, and PHO23) (20) and the SET3 complex (SET3, HOS2, SNT1, and SIF2), which contains two histone deacetylases, Hst1 and Hos2 (43).
FIG. 6.
Synthetic genetic interactions and microarray analysis of gene expression of UBP8 and SGF11 deletions compared to SPT3 and SPT8 deletions. (A) Synthetic genetic interactions of SPT3, SPT8, UBP8, and SGF11. SGA technology was used to cross Natr strains harboring individual deletions of genes encoding Spt3, Spt8, Ubp8, and Sgf11 with a transcription-targeted array of deletion strains to create sets of Natr Kanr haploid double mutants. Growth rates were assessed by automated image analyses of colony size. Lines connect genes with synthetic genetic interactions. The lengths of lines and proximity of boxes in this diagram are unrelated to the strengths of the indicated synthetic genetic interactions. (B) Microarray analysis of gene expression was performed for the indicated deletion strains. Pearson correlation coefficients were then calculated for each pair of deletions, and the deletions were organized by two-dimensional hierarchical clustering.
In contrast, strains containing a deletion of either UBP8 or SGF11 did not result in synthetic growth defects when combined with deletions in these genes, arguing that they are functionally distinct from the module that contains Spt3/Spt8. Deletions in either of these two genes did result in synthetic sick phenotypes in combination with a deletion of RPB4, which encodes a component of RNA polymerase II, Shp1, which functions as an adaptor for Cdc48-mediated protein degradation via the ubiquitin-proteasome pathway (53), and Ubc12, an E2 ubiquitin-conjugating enzyme. Thus, UBP8 and SGF11 genetically interact with two genes that have been implicated in ubiquitylation, further strengthening the idea that Sgf11 may function together with Ubp8 during histone deubiquitylation. The introduction of spt3Δ and spt8Δ mutations into strains already containing deletions of RPB4, SHP1, and UBC12 did not generate a slow growth phenotype. However, we did find a set of genes that genetically interacted with all four starting genes, including components of the 19S (RPN10 and SEM1) and 20S (PRE9) proteasome (65), the histone acetyltransferase complex elongator (ELP3, ELP4, and ELP6) (31, 41), and an mRNA transport complex (THP1 and SAC3) (18), which interestingly shares a component, Sus1, with SAGA (49).
To further characterize these functionally distinct sets of proteins contained within SAGA, we performed microarray analyses of gene expression in strains containing deletions of SPT3, SPT8, UBP8, and SGF11 (raw microarray data are available at http://www.utoronto.ca/greenblattlab/SAGA.xls). Pearson correlation coefficients were calculated for each pair of deletions, and the strains were organized by two-dimensional hierarchical clustering according to the similarities of their effects on gene expression (Fig. 6B). As observed with our genetic analyses, the gene expression profiles of strains containing deletions of UBP8 and SGF11 were similar to each other and different from those of strains containing deletions of SPT3 and SPT8. In contrast, SPT3 and SPT8 clustered next to each other and were distinct from UBP8 and SGF11 in this gene expression analysis, behaving just as they had in our genetic analysis. Microarray experiments have also been conducted with over 300 other strains containing individual deletions of genes that have been implicated in some aspect of transcription or chromatin metabolism (N. J. Krogan, T. R. Hughes, and J. F. Greenblatt, unpublished data). The clustering of this larger data set (not shown) reflects the data represented in Fig. 6B, in which SPT3 and SPT8 clustered next to each other and away from SGF11 and UBP8, which also clustered next to each other. Therefore, both the genetic data and the gene expression data reported here suggest that the Spt3 and Spt8 subunits of SAGA are functionally similar and predict a role for Sgf11 in Ubp8 function, consistent with the biochemical results described above.
DISCUSSION
SAGA is a large (1.8 MDa) multicomponent protein complex. We and others recently showed that Ubp8 is a subunit of SAGA and is a histone H2B-deubiquitylating enzyme. In this report, we provide structural and functional data showing that Ubp8 is associated with SAGA via a Cys-His-rich region (a probable zinc finger domain) within Ubp8 and that the association with SAGA is through an 11-kDa subunit of SAGA, which we and others (44) have named Sgf11. Sgf11 and Ubp8 both function to promote H2B deubiquitylation, and genetic and gene expression analyses demonstrated that they have a discrete function compared to other components of the SAGA complex.
Our results showed that the Zn finger within the amino terminus of Ubp8 is necessary for the interaction of Ubp8 with SAGA. Furthermore, deletion or substitution mutations within this region eliminated Upb8's interaction with Sgf11 and its association with SAGA. These mutations also eliminated in vivo Ubp8 function in assays for the deubiquitylation of H2B and for growth on galactose medium (which requires Ubp8 for full activation of the GAL1 gene) (24). However, it appears that there is at least one other Ubp8 surface for interaction with SAGA, as the Zn binding domain by itself interacted only weakly with SAGA.
It is interesting that the Zn binding domain of Ubp8 has striking homology to a region within the mammalian histone deacetylase HDAC6. This region in HDAC6 interacts with ubiquitin (26, 55). We tested whether the Zn finger or Ubp8 itself interacts with ubiquitin conjugated to agarose beads, but we were unable to detect an interaction (K. Ingvarsdottir and S. L. Berger, unpublished observations). It is possible that Sgf11 is ubiquitylated and that this promotes Ubp8 binding in vivo, but we have not examined this possibility.
The Zn finger may be critical for substrate targeting at the right location in the genome and under appropriate conditions via this association with SAGA. Indeed, there are 17 Ubp proteins in S. cerevisiae (1), and others may also target ubH2B for Lys-123 deubiquitylation (N. C. Tolga Emre, S. L. Berger, M. Osley, and C.-F. Kao, unpublished observations). Therefore, it is possible that the association into distinct protein complexes targets similar catalytic deubiquitylation domains to the common H2B substrate.
Our analyses further showed that Ubp8 and Sgf11 may function as a unit within SAGA. These data add to previous models arguing for the modularity of SAGA in its structure and function. Gcn5 is a histone acetyltransferase that associates with Ada2 and Ada3 to form a unit (11), and the last two subunits are required for acetylation activity (3, 59). Spt3, Spt7, and Spt8 are physically associated and regulate TBP's association with the TATA box (45, 57, 58, 68). These groups of proteins have been distinguished in genetic and biochemical assays (7, 16, 58). For example, mutations in the acetylation and TBP regulation subunits cause distinct growth patterns on indicator media that assay for certain gene expression pathways and for DNA damage (58). Further evidence for modularity is the independence of enzyme activities. First, the deletion of Ubp8 does not lower the histone acetyltransferase activity of the purified SAGA complex (24), and second, the deletion of Gcn5 or catalytic inactivation of Gcn5 does not reduce the H2B-deubiquitylating activity of purified SAGA (K. K. Lee and J. L. Workman, personal communication). Our new data suggest that Ubp8 and Sgf11 may comprise a deubiquitylation module for which ubH2B is one substrate. We have provided biochemical evidence that the proteins are closely associated and that Sgf11 is required specifically for Ubp8 interaction with SAGA, since other tested subunits were not lost from SAGA upon the deletion of SGF11. We also observed similar functions of Sgf11 and Ubp8 in limiting intracellular ubH2B levels.
Additional functional analyses demonstrated that Ubp8 and Sgf11 have similar roles and, moreover, that these are distinct from those of other subunits of SAGA that have different biochemical activities. First, the deletion of Ubp8 and Sgf11 caused very similar patterns of altered gene expression in whole-genome microarray analyses. These patterns were strikingly different from those for the loss of SAGA subunits involved in TBP regulation, Spt3 and Spt8. Second, synthetic genetic analyses that combined the deletion of UBP8 and SGF11 with the deletion of a large number of other transcription-linked factors showed a consistent pattern of synthetic growth interactions. Again, these patterns were strikingly different from those displayed by the deletion of subunits involved in TBP regulation (Spt3 and Spt8). These data indicate that Ubp8 and Sgf11 form a unit within SAGA that coregulates gene expression in a similar fashion. It is not yet clear whether Sgf11 plays a regulatory role for Ubp8 in addition to its function in promoting Ubp8 interaction with SAGA.
The picture that emerges from these biochemical and genetic observations is that SAGA possesses distinct structural and functional modules. Previous low-resolution structural analyses by cryo-electron microscopy of human TFTC/STAGA and yeast SAGA complexes revealed spatially separated lobes (8, 67), within which specific biochemical features are localized (67). Our results predict that Sgf11-Ubp8 comprises a distinct and mutually overlapping structural module in SAGA. Further three-dimensional analysis of SAGA will help us to unravel how specific histone substrates within nucleosomes are targeted by the deubiquitylation and acetylation enzymes of SAGA. In summary, our study indicates that SAGA possesses multiple functional modules that use different mechanisms to regulate transcription.
Acknowledgments
We gratefully acknowledge S. Kochbin for directing our attention to the similar Zn finger sequences in HDAC6 and Ubp8 and thank L. J. Duggan for the generation of the Ada2-TAP strain used for the immunoprecipitation of SAGA subunits.
This work was supported by research grants from the National Institutes of Health (GM 55360 to S.L.B) and the National Science Foundation (MCB-0078940 to S.L.B). An NIH training grant supported A.W. (T32 GM008216). A doctoral fellowship from the University of Pennsylvania—Biomedical Graduate Studies supported K.I. N.J.K. was supported by a doctoral fellowship from the Canadian Institutes of Health Research. This research was supported by grants to J.F.G. from the Canadian Institutes of Health Research, the Ontario Genomics Institute, and the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
REFERENCES
- 1.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 2.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 423:659-663. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 5.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TBP function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAF(II)-containing complexes TFIID and TFTC. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 9.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 11.Candau, R., and S. L. Berger. 1996. Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J. Biol. Chem. 271:5237-5245. [DOI] [PubMed] [Google Scholar]
- 12.Candau, R., et al. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements, A., A. N. Poux, W. S. Lo, L. Pillus, S. L. Berger, and R. Marmorstein. 2003. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12:461-473. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 15.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 16.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, T., K. Strasser, A. Racz, S. Rodriguez-Navarro, M. Oppizzi, P. Ihrig, J. Lechner, and E. Hurt. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez, F., A. Delahodde, T. Kodadek, and S. A. Johnston. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548-550. [DOI] [PubMed] [Google Scholar]
- 22.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 23.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 24.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 26.Hook, S. S., A. Orian, S. M. Cowley, and R. N. Eisenman. 2002. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 99:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, M., P. Li, M. Li, W. Li, T. Yao, J. W. Wu, W. Gu, R. E. Cohen, and Y. Shi. 2002. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041-1054. [DOI] [PubMed] [Google Scholar]
- 28.Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone, and H. D. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 29.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 30.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 31.Krogan, N. J., and J. F. Greenblatt. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:8203-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 33.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, W.-S., R. C. Trievel, J. R. Rojas, L. Duggan, J.-Y. Hsu, C. D. Allis, R. Marmorstein, and L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 35.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 36.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 38.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 41.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 42.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 43.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22:8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 48.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Navarro, S., T. Fischer, M. J. Luo, O. Antunez, S. Brettschneider, J. Lechner, J. E. Perez-Ortin, R. Reed, and E. Hurt. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116:75-86. [DOI] [PubMed] [Google Scholar]
- 50.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojas, J. R., R. C. Trievel, J. Zhou, Y. Mo, X. Li, S. L. Berger, C. D. Allis, and R. Marmorstein. 1999. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401:93-98. [DOI] [PubMed] [Google Scholar]
- 52.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuberth, C., H. Richly, S. Rumpf, and A. Buchberger. 2004. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sterner, D. E., R. Belotserkovskaya, and S. L. Berger. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99:11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation and TBP binding. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 60.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 61.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 63.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 64.Trievel, R. C., J. R. Rojas, D. E. Sterner, R. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, and R. Marmorstein. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96:8931-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 66.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 67.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 68.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22:5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68a.Xu, W., D. G. Edmondson, Y. A. Errard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn512 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed]
- 69.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423:655-659. [DOI] [PubMed] [Google Scholar]
- 70.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu, Y., P. Eriksson, L. T. Bhoite, and D. J. Stillman. 2003. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 23:1910-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]