Abstract
Replication by many positive-strand RNA viruses includes genomic RNA amplification and subgenomic mRNA (sgRNA) transcription. For brome mosaic virus (BMV), both processes occur in virus-induced, membrane-associated compartments, require BMV replication factors 1a and 2a, and use negative-strand RNA3 as a template for genomic RNA3 and sgRNA syntheses. To begin elucidating their relations, we examined the interaction of RNA3 replication and sgRNA transcription in Saccharomyces cerevisiae expressing 1a and 2a, which support the full RNA3 replication cycle. Blocking sgRNA transcription stimulated RNA3 replication by up to 350%, implying that sgRNA transcription inhibits RNA3 replication. Such inhibition was independent of the sgRNA-encoded coat protein and operated in cis. We further found that sgRNA transcription inhibited RNA3 replication at a step or steps after negative-strand RNA3 synthesis, implying competition with positive-strand RNA3 synthesis for negative-strand RNA3 templates, viral replication factors, or common host components. Consistent with this, sgRNA transcription was stimulated by up to 400% when mutations inhibiting positive-strand RNA3 synthesis were introduced into the RNA3 5′-untranslated region. Thus, BMV subgenomic and genomic RNA syntheses mutually interfered with each other, apparently by competition for one or more common factors. In plant protoplasts replicating all three BMV genomic RNAs, mutations blocking sgRNA transcription often had lesser effects on RNA3 accumulation, possibly because RNA3 also competed with RNA1 and RNA2 replication templates and because any increase in RNA3 replication at the expense of RNA1 and RNA2 would be self-limited by decreased 1a and 2a expression from RNA1 and RNA2.
Subgenomic mRNA (sgRNA) transcription is a mechanism used by many eukaryotic RNA viruses to multiply the number of separate genes expressed and to regulate the timing and magnitude of their expression. Numerous positive-strand RNA viruses produce sgRNAs, using diverse transcription mechanisms distinguished by internal initiation or termination, continuous or discontinuous RNA synthesis, etc., to link internal open reading frames (ORFs) to a translatable 5′ end, producing sgRNAs that are 3′-coterminal with the viral genomic RNA (40). Many studies have revealed cis-signals and mechanistic features of sgRNA transcription and genomic RNA replication in different positive-strand RNA viruses, but relatively little is known about how these two processes, which depend on common viral replication factors and RNA templates, are coordinated.
Relevant infectious processes, such as RNA replication, sgRNA transcription, and host factor involvement, have been studied for a number of positive-strand RNA viruses, including brome mosaic virus (BMV), a member of the alphavirus superfamily. The BMV genome consists of three genomic RNAs. RNA1 and RNA2 encode viral RNA synthesis factors 1a and 2a, which are required for genome replication and sgRNA synthesis. 2a is the viral RNA polymerase, while 1a is a multifunctional RNA replication factor that directs assembly of the membrane-associated RNA replication complex, recruits viral RNA templates and 2a polymerase, and contributes RNA capping and potentially helicase functions to RNA synthesis (4, 18, 30, 34). RNA3 encodes movement protein 3a and coat protein (CP), which are dispensable for RNA replication but required for systemic spread in plants. RNA3 is a template for RNA replication and sgRNA transcription (Fig. 1), which initiates internally on negative-strand RNA3 templates (39).
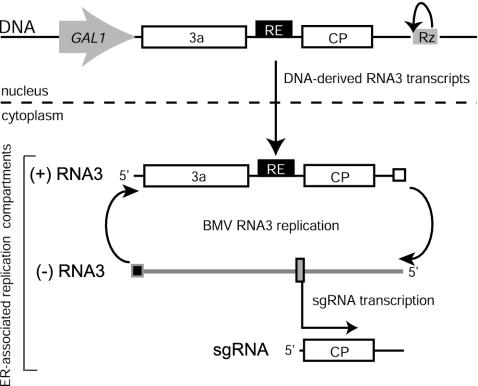
FIG. 1.
Schematic illustration of DNA-based expression, genomic RNA replication, and sgRNA transcription of BMV RNA3 in yeast. “DNA” indicates a plasmid with RNA3 sequences (5′-UTR, 3a ORF, intergenic UTR with RE [black box], CP ORF, and 3′-UTR) flanked by the yeast GAL1 promoter and hepatitis δ virus ribozyme (Rz). Upon galactose induction, yeast RNA polymerase II synthesizes positive-strand RNA3 transcripts that, upon transport to the cytoplasm, serve as the templates for 1a- and 2a-dependent RNA3 replication and sgRNA transcription, which occur in 1a-induced ER membrane-associated replication compartments. Small white, black, and gray boxes indicate negative-strand, positive-strand, and sgRNA promoters, respectively.
Whether and how RNA3 replication and sgRNA transcription are coordinated in BMV are poorly understood. Syntheses of negative-strand RNA3, positive-strand RNA3, and sgRNA all utilize GTP as a priming nucleotide and require stem-loop RNA secondary structures for recognition of corresponding promoters (17, 29). Nevertheless, the differing nucleotide sequences of promoters for replication and sgRNA transcription (29, 45) suggest that recognition of these varied promoters may involve distinct domains of viral and/or host factors. Alternatively or in addition, an “induced fit” mechanism may adjust the viral replicase to different promoters (59, 63). Despite their shared features, it has not been established whether BMV genomic and sgRNA syntheses are dependent, mutually interfering, or independent. The possibility that these processes might be largely or completely independent is supported by multiple findings. Among these is that alphaviruses and many other positive-strand RNA viruses produce partially double-stranded replicative intermediate RNAs bearing multiple nascent strands, showing that multiple elongating polymerases can simultaneously traverse a single RNA (8). Thus, genomic and sgRNA syntheses might occur simultaneously on a single negative-strand RNA template without interference. Moreover, under normal conditions of BMV infection, factors other than the viral RNA-dependent RNA polymerase 2a are limiting (13), suggesting that ample polymerase may be available for both genomic and subgenomic RNA syntheses.
Whether and how RNA3 replication and sgRNA transcription might interact also is intriguing in light of recent findings that BMV replication and sgRNA transcription occur in constrained endoplasmic reticulum (ER) membrane-associated compartments (50). As a first step to elucidate the mechanistic relationships between genomic RNA replication and sgRNA transcription, we examined how these two processes interact with each other in vivo. These experiments used a yeast (Saccharomyces cerevisiae) system that duplicates all known features of BMV replication and gene expression in its natural plant host cells, up to and including formation of progeny virions (31, 46, 47). Here, we report that BMV sgRNA transcription and genomic RNA3 replication strongly and mutually interfere with each other. Our data indicate that such interference occurs only in cis and is a result of competition between positive-strand genomic RNA3 synthesis and sgRNA transcription. The implications of these findings for RNA replication complex function and for defining limiting steps and components in BMV replication are discussed.
MATERIALS AND METHODS
Yeast methods.
Yeast strain YPH500 (MAT ura3-52 lys2-801 ade2-101 trp1-63 his3-200 leu2-1) was used in all experiments. Yeast cultures were grown at 30°C in defined synthetic medium containing 2% galactose, 2% glucose, or 2% raffinose (6) and lacking histidine, leucine, tryptophan, uracil, or their combinations to maintain plasmids. The lithium acetate-polyethylene glycol method was used to transform plasmids into yeast cells (24).
Plasmids.
Where applicable, laboratory designations for plasmids are given in parentheses. BMV replication factors 1a and 2a were expressed from yeast plasmids pB1YT3H and pB2Ag, respectively. pB1YT3H is a centromeric plasmid containing the HIS3 selectable marker and the 1a ORF flanked by the galactose-inducible GAL1 promoter and the ADH1 polyadenylation sequence (5). pB2Ag was derived from pB2CT15, a 2μm plasmid containing the LEU2 selectable marker and the 2a ORF flanked by the constitutive ADH1 promoter and the ADH1 polyadenylation sequence (26), by replacing the ADH1 mRNA 5′-untranslated region (5′-UTR) with 40 nucleotides (nt) from the 5′ end of GAL1 mRNA. In some experiments, pRS313 (HIS3 marker), pRS315 (LEU2 marker), pRS314 (TRP1 marker), or pRS316 (URA3 marker) “empty” plasmids (53) were transformed into yeast strains lacking BMV expression plasmids with the corresponding marker genes to allow growing all tested yeast strains in the same selective media. pB3WT (pB3VG46) contained BMV RNA3 expressed from the GAL1 promoter and terminated by a hepatitis δ virus ribozyme. pB3WT was based on pB3RQ39 (23) but had PCR-introduced XhoI and MluI sites immediately flanking the 3a ORF. These changes had no effect on RNA3 replication. pB3CPfs (pB3VG103) was based on pB3WT but had the CP ORF inactivated by a double mutation as in pB3MS82 (5). pB3ΔSG (pB3VG84) was constructed by digesting pB3WT at unique BglII and SalI sites, filling in the ends using T4 DNA polymerase, and religating the vector, resulting in deletion of the 28-nt “core” sgRNA promoter (RNA3 nt 1226 to 1253) (14). pB3SG[+1,-2] (pB3VG85) and pB3SG[-13] (pB3VG109) were obtained by introducing (G+1C,G-2U) and (G-13C) point mutations (relative to the +1 sgRNA transcription start site, positive-strand RNA3 polarity), correspondingly, into pB3WT by PCR amplification with pairs of overlapping primers bearing the desired point mutations. To obtain pB3ΔCP (pB3VG54), pB3WT was digested with SalI and StuI, filled with T4 DNA polymerase, and religated. pB3ΔSGΔCP (pB3VG66) was constructed by digesting pB3ΔCP with BglII and SalI, filling the ends with T4 DNA polymerase, and religating. pB3(CPfs-BoxB*) (pB3VG104), pB3(ΔSG-BoxB*) (pB3VG105), and pB3(SG[-13]-BoxB*) (pB3VG106) were based on pB3CPfs, pB3ΔSG, and pB3SG[-13], respectively, but had a GGG-to-CCC substitution at RNA3 nt 1099 to 1101 (7, 60), which was introduced using PCR and a pair of overlapping mutant primers. pB3GAL1-CPfs (pB3VG39), pB3GAL1-ΔSG (pB3VG110), and pB3GAL1-SG[-13] (pB3VG112) were based on pB3CPfs, pB3ΔSG, and pB3SG[-13], respectively, but had identical replacements of the RNA3 5′-UTR with the GAL1 5′-UTR. pCP-trans was pMK-CP3, which contains the URA3 selectable marker and the BMV CP gene under control of the GAL1 promoter (31). pB4[sgRNA] was pB4MK1, which contains the URA3 selectable marker and the BMV sgRNA sequence expressed from the GAL1 promoter and terminated by a hepatitis δ virus ribozyme (41). pB4[sgRNA-CPfs] (pB4VG94) was based on pB4MK1 but with the CP ORF inactivated by frameshift mutation as in pB3MS82 (5).
For barley protoplast experiments, plasmids pB1TP3, pB2TP5, and pT7-B3WT(pB3TP8) were used, containing complete cDNA copies of wild-type (wt) BMV RNA1, RNA2, and RNA3, respectively, and T7 RNA polymerase promoters allowing in vitro synthesis of infectious BMV transcripts (3, 27). To obtain pT7-B3CPfs, pT7-B3ΔSG, pT7-B3SG[+1,-2], and pT7-B3SG[-13], the BglII-AflII fragment from yeast plasmid pB3CPfs, pB3SG[+1,-2], or pB3SG[-13], respectively, was inserted into BglII-AflII-digested pB3TP8. pT7-ΔCP was obtained by digesting pB3TP8 with SalI and StuI, filling the ends with T4 DNA polymerase, and religating. pT7-ΔSGΔCP was obtained by digesting pB3TP8 with BglII and StuI, filling the ends with T4 DNA polymerase, and religating.
RNA and protein analyses.
Single colonies from selective plates containing glucose were used to inoculate 5 ml of selective liquid medium containing 2% galactose. All yeast transformations were repeated at least twice, and RNA analysis was performed for at least three independent yeast colonies from each transformation. Representative results are shown in the figures. Yeast cultures were grown for about 48 h and harvested in mid-log phase (optical density at 600 nm of 0.5 ± 0.1 [mean ± standard deviation]). For time course experiments, yeast cells were first grown in 2% raffinose medium and then switched to galactose medium to prevent the delay in GAL1 promoter induction associated with prior growth in glucose medium (23). To induce the GAL1 promoter, yeast cells were washed twice with water, resuspended (zero hours), and grown in medium containing 2% galactose. Total yeast RNA was isolated using acidic hot phenol and ethanol precipitation, as described elsewhere (35). For Northern blotting, 1.5 μg of total yeast RNA was analyzed to detect positive- and negative-strand RNA3 as described in reference 23. To detect negative-strand RNA3 for replicons B3ΔSGΔCP and B3ΔCP, which lacked the CP ORF, an RNA probe complementary to the RNA3 intergenic region was used. To produce this probe, plasmid pProbe(IGR) was constructed by inserting the MluI-SalI fragment from pB3WT (the MluI end was filled using T4 DNA polymerase) into pBluescript II KS(+) (Stratagene) digested at the SacI and SalI sites. pProbe(IGR) was digested with SalI and transcribed in vitro using T7 RNA polymerase. Radioactive signals were measured using a Molecular Dynamics PhosphorImager 425 and ImageQuant 1.2 software (Molecular Dynamics). Primer extension analysis was performed as described in reference 23, using 5 μg of total RNA and oligonucleotide primers complementary to RNA3 nt 33 to 40 or nt 112 to 132 for RNA3 derivatives with the viral 5′-UTR or GAL1 5′-UTR, respectively. For cell fractionation, yeast cells were grown to mid-log phase, converted to spheroplasts, and fractionated as described previously (10, 11). Western blotting was performed as described elsewhere (4), using polyclonal rabbit BMV CP antiserum (1:25,000; American Type Culture Collection).
Barley protoplasts.
In vitro transcription with T7 DNA polymerase, barley protoplast isolation, and inoculation were performed as described by Kroner et al. (32, 33). A total of 1.0 × 106 protoplasts were inoculated with 5 μg of BMV transcripts and incubated for 20 h. Total RNA was isolated as described in reference 32, and 10 μg of total barley RNA was analyzed to detect positive- or negative-strand RNA3 by Northern blotting, as described in reference 23.
RESULTS
Inactivating the BMV sgRNA promoter stimulates RNA3 replication.
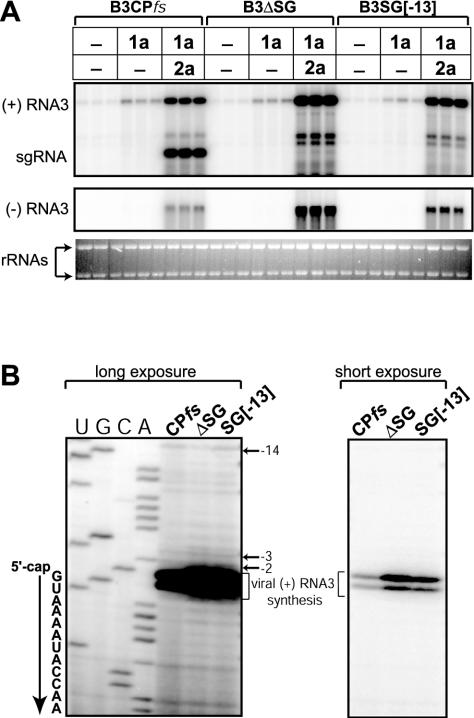
To test for possible effects of sgRNA transcription on genomic RNA3 replication, we constructed RNA3 derivative B3ΔSG by deleting the 28-nt core sgRNA promoter region (RNA3 nt 1226 to 1253) (14). This mutation was expected to block sgRNA synthesis and, therefore, CP expression. To focus on the effect of sgRNA transcription on RNA3 replication while avoiding the contribution of CP to RNA3 stability due to viral RNA encapsidation (31), replication of B3ΔSG (and other sgRNA promoter mutants) was compared to that of B3CPfs, which is wt BMV RNA3 with an intact sgRNA promoter but with CP expression blocked by an early frameshift mutation (5). Yeast expressing BMV RNA replication factors 1a and 2a were transformed with plasmids expressing B3CPfs or B3ΔSG from the galactose-inducible yeast GAL1 promoter, grown in galactose-containing liquid medium, and collected for RNA analysis 48 h postinduction (p.i.) with galactose. As shown in Fig. 2B, deletion of the core sgRNA promoter in B3ΔSG reduced sgRNA accumulation to background levels while increasing positive-strand and negative-strand RNA3 accumulation to 350 and 275% of that of B3CPfs, respectively (Fig. 2C). As expected, expression of BMV replication factors 1a and 2a from their independent plasmids was unaffected by these and later manipulations of the RNA3 expression plasmids.
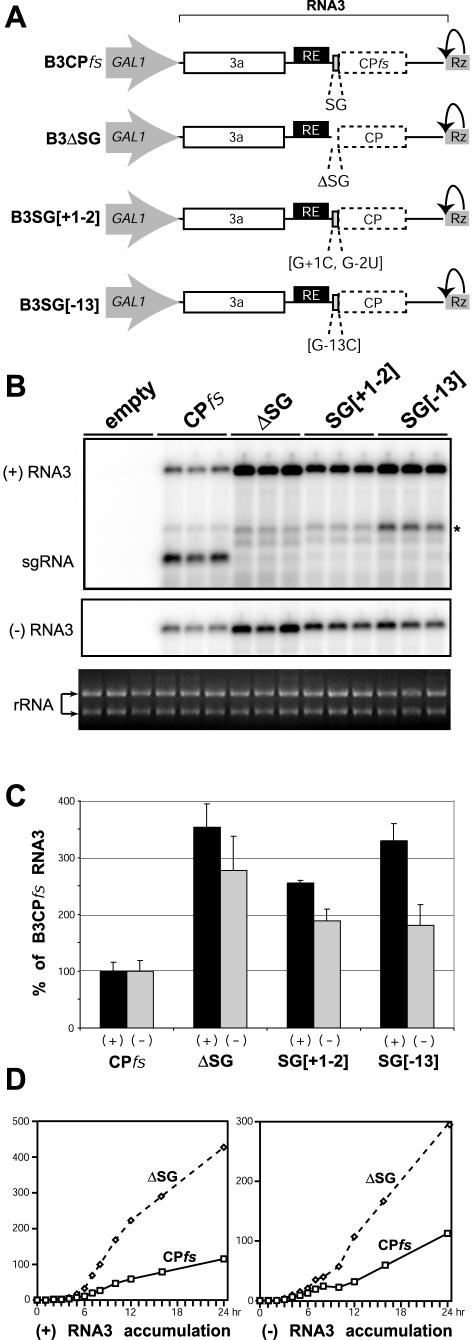
FIG. 2.
Inactivation of sgRNA transcription stimulates RNA3 replication. (A) Schematic diagram of expression cassettes for RNA3 derivatives with wt or mutated sgRNA promoter. Dashed boxes represent a translationally inactive CP ORF due to a frameshift mutation (in B3CPfs) or sgRNA transcription block. SG indicates wt RNA3 sequence complementary to the core sgRNA promoter. +1, -2, and -13 indicate mutated nucleotide positions within the sgRNA promoter relative to the +1 sgRNA transcription start site. (B) Northern blot analysis of BMV-specific RNA products for RNA3 derivatives with wt or mutated sgRNA promoters. Total RNA was extracted from yeast expressing BMV 1a, 2a, and RNA3 derivatives (indicated at the top; “empty” is the yeast plasmid without RNA3 expression cassette), and 1.5 μg of total RNA was analyzed by Northern blotting using specific probes to detect RNA3 positive or negative strands as indicated on the left. Triplicate samples represent three independent transformants for each plasmid combination. The asterisk indicates a minor RNA band, discussed in the text. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (C) Relative accumulation of positive-strand RNA3, sgRNA, and negative-strand RNA3 from the RNA3 derivatives in panel B. The histogram compares positive-strand (black bars) and negative-strand (gray bars) accumulation levels for tested RNA3 derivatives, measured using a Molecular Dynamics PhosphorImager. (D) Time course analysis of positive-strand RNA3 (left panel) and negative-strand RNA3 (right panel) accumulation for RNA3 derivatives B3CPfs and B3ΔSG. Yeast cells with 1a, 2a, and RNA3 expression plasmids were first grown in raffinose medium to prevent GAL1 promoter induction. To induce the GAL1 promoter, yeast cells were transferred into galactose-containing medium (zero hours p.i.), and cell aliquots were collected at the indicated time points. Northern blot analysis was performed as for panel B, and the relative accumulation of positive- and negative-strand RNA3 was measured.
To confirm that stimulation of RNA3 replication in B3ΔSG resulted from blocking sgRNA transcription, additional RNA3 derivatives with point mutations in the core sgRNA promoter were constructed (Fig. 2A). As demonstrated previously in vivo (37) and in vitro (2, 16, 51), detectable sgRNA transcription was blocked in the RNA3 derivative B3SG[+1,-2] with substitutions at sgRNA initiation site +1 and at −2, and in B3SG[-13] with a G-to-C substitution at position −13 (Fig. 2B). Moreover, as for B3ΔSG, these mutants similarly stimulated positive- and negative-strand RNA3 accumulation (Fig. 2B and C). Thus, all tested sgRNA promoter mutations stimulated accumulation of RNA3 of both polarities, implying that sgRNA transcription or its products inhibited RNA3 replication. Particularly for B3SG[-13], these mutations and their associated increased RNA3 replication were accompanied by increased accumulation of a positive-strand RNA band intermediate in size between genomic RNA3 and wt sgRNA (Fig. 2B). Unlike sgRNA or genomic RNA3, this intermediate band was not accompanied by a negative-strand RNA of corresponding size (result not shown), suggesting that this RNA was not found in the RNA replication complex and may be a cytoplasmic degradation product of RNA3.
To rule out the possibility that the effect of sgRNA transcription on RNA3 accumulation was specific to the late stage of RNA3 replication tested in Fig. 2B (48 h p.i.), we compared the levels of RNA replication products for B3CPfs and the sgRNA promoter mutants at multiple time points in the first 24 h p.i. of the GAL1 promoter. Figure 2D shows representative results for B3CPfs and B3ΔSG, and similar results were found for B3SG[+1,-2] and B3SG[-13]. For B3CPfs, sgRNA was detectable by 2 h p.i., while inhibition of positive-strand and negative-strand RNA3 accumulation relative to that of B3ΔSG was evident from 3 h p.i. (Fig. 2D and data not shown). Thus, sgRNA transcription interfered with RNA3 replication throughout the entire RNA3 replication cycle.
sgRNA transcription-mediated inhibition of RNA3 replication is independent of CP expression.
Since blocking sgRNA synthesis also blocks production of CP from wt RNA3 (Fig. 1), the Fig. 2 experiments compared the sgRNA promoter mutants to B3CPfs, an RNA3 derivative with normal sgRNA synthesis but no CP expression, to best compare the interaction of RNA synthesis pathways in the absence of any secondary effects from CP. Nevertheless, since CP selectively packages both BMV genomic and sgRNAs in yeast and plant cells (31) and can alter the balance of positive- and negative-strand RNA accumulation (see below), it was of interest to see whether and, if so, how CP production might perturb the observed results. To test whether the effect of sgRNA transcription on RNA3 accumulation was affected by the presence or absence of CP, we expressed CP in cis from B3WT or in trans from a separate plasmid (Fig. 3A and B). As shown in Fig. 3C, the presence or absence of CP did not significantly alter the inhibition of positive-strand RNA3 accumulation by sgRNA transcription (Fig. 3C), indicating that the effect of sgRNA transcription on RNA3 replication was CP independent.
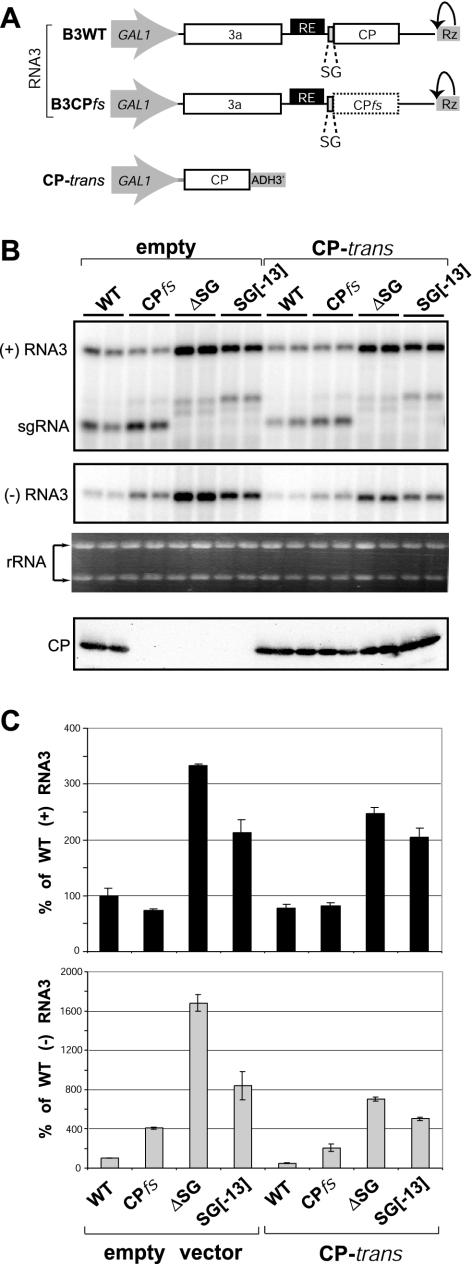
FIG. 3.
Effect of CP on sgRNA transcription-mediated inhibition of RNA3 replication. (A) Schematic diagram of expression cassettesfor RNA3 derivatives and CP mRNA used in this experiment and not introduced in Fig. 2. A solid box used for the CP ORF indicates a translationally active wt CP ORF; a dashed box represents a translationally inactive CP ORF due to a frameshift mutation. CP-trans mRNA has the BMV CP ORF and yeast 5′- and 3′-UTRs and is expressed under control of the GAL1 promoter and ADH1 polyadenylation sequences. (B) Northern blot and Western blot analyses of BMV RNA3-specific products and CP, respectively. Yeast expressing BMV 1a, 2a, and RNA3 derivatives plus empty plasmid or CP-trans plasmid were grown in galactose medium, collected, and divided for RNA (as for Fig. 2) or protein analysis. CP expression was analyzed by Western blotting using total cell lysate and polyclonal anti-CP serum. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (C) Effect of CP expression on the relative accumulation of positive- and negative-strand RNA3 derivatives. The histogram compares accumulation of the positive strand (black bars) and negative strand (gray bars) for the RNA3 derivatives shown in panel B.
While CP had at most modest effects on positive-strand RNA3 accumulation, CP expression significantly reduced negative-strand RNA3 accumulation (Fig. 3B and C). This is in keeping with prior results from yeast and plant cells that selective packaging by BMV CP (31) removes positive-strand RNA from the pool of templates available for replicase copying, reducing the accumulation of negative-strand RNA (20, 31, 37, 42, 48).
sgRNA transcription does not interfere with RNA3 replication in trans.
Fig. 2 and 3 showed that RNA3 replication was inhibited by sgRNA transcription in cis. Next, we decided to test whether sgRNA molecules or sgRNA transcription interfere with RNA3 replication in trans. Previously, it was demonstrated that BMV replication can be dramatically inhibited by 200-nt transcripts corresponding to the 3′-UTR of positive-strand RNA3, which contains the promoter for negative-strand RNA3 (21). In part, such inhibition was attributed to the template activity of such transcripts because 200-nt negative-strand RNA products were detected (21). As BMV sgRNA is 3′-coterminal with genomic positive-strand RNA3 and serves, though inefficiently, as a template for negative-strand sgRNA synthesis in vivo (23, 34), we tested whether trans-expressed BMV sgRNA was able to inhibit RNA3 replication. Accordingly, artificial sgRNAs with an intact or frameshifted CP ORF were expressed from a separate plasmid and tested for their influence on replication of B3ΔSG (Fig. 4A). As shown in Fig. 4A, no inhibition of B3ΔSG replication was observed, showing that DNA-transcribed sgRNA supplied in trans was unable to interfere with RNA3 replication.
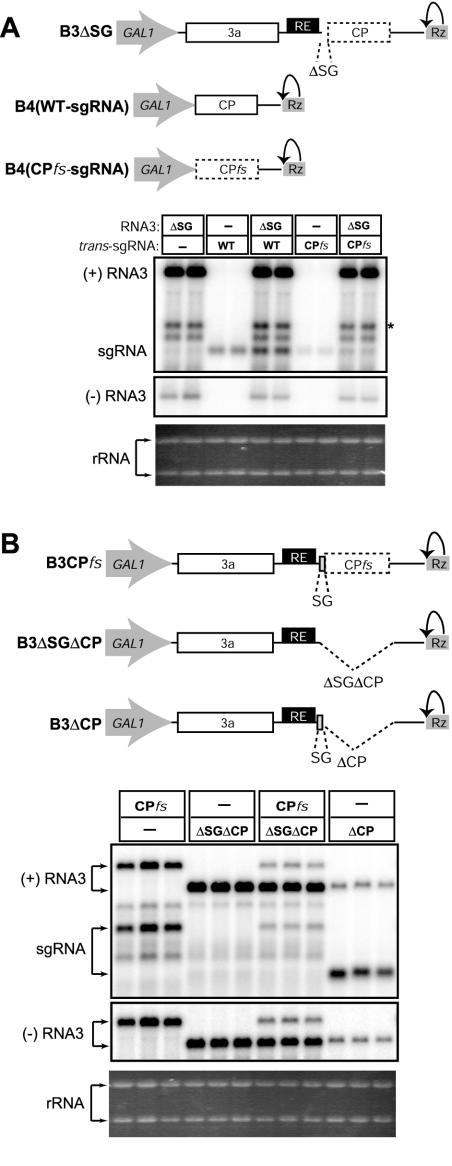
FIG. 4.
Neither sgRNA transcription nor sgRNA itself inhibits replication of exogenous RNA3 replicons. (A) Effect of trans-expressed sgRNA on the replication of an RNA3-derived replicon. The upper diagram represents expression cassettes for B3ΔSG and DNA-derived BMV sgRNAs with wt [B4(WT-sgRNA)] and frameshifted CP ORF [B4(CPfs)]. The lower panel shows a Northern blot analysis of total RNA from yeast expressing B3ΔSG and sgRNA (with or without translationally active CP ORF) alone or in combination with BMV 1a and 2a replication factors. The asterisk indicates a positive-strandRNA whose appearance is associated with the inactivation of sgRNA transcription. (B) Effect of sgRNA transcription on the replication of an exogenous RNA3-derived replicon. The upper diagram depicts the RNA3 derivatives used in this experiment. The lower panel shows Northern blot analyses of total RNA from yeast expressing 1a, 2a, and the indicated RNA3 cassettes to compare the replication level of B3ΔSGΔCP to that of B3ΔCP. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control.
Next, we tested whether RNA3-directed sgRNA transcription could inhibit RNA3 replication in trans. Previously, we have observed interference between replication of some RNA3 replicons when present in the same cells (V. Grdzelishvili and P. Ahlquist, unpublished results), suggesting competition between replicons for limiting replication factors. Similarly, sgRNA transcription might competitively inhibit replication of its cognate, parental genomic RNA or replication of independent RNA replicons. To test this possibility, we analyzed the replication levels of RNA3 derivative B3CPfs alone or together with B3ΔSGΔCP, whose CP ORF deletion allowed us to distinguish its size on Northern blotting. As shown in Fig. 4B, when B3ΔSGΔCP and B3CPfs were present together, sgRNA transcription by B3CPfs did not inhibit replication of B3ΔSGΔCP. Rather, B3ΔSGΔCP replication inhibited B3CPfs replication. Nevertheless, when the core sgRNA promoter was restored in B3ΔCP, a more-than-10-fold decrease in RNA3 replication level was observed (Fig. 4B, right three lanes), confirming the dramatic ability of sgRNA transcription to interfere in cis despite the lack of a demonstrable effect in trans.
Taken together, the results showed that neither DNA-derived sgRNA transcripts nor BMV-directed, RNA-dependent sgRNA transcription inhibited replication of independent RNA3 replicons in trans, in contrast to strong interference with replication in cis.
sgRNA transcription does not inhibit RNA3 template recruitment into replication compartments.
RNA3 replication and sgRNA transcription occur in ER membrane-associated compartments induced by BMV 1a protein, which also recruits 2a polymerase and genomic positive-strand RNA3 templates (50). Viral RNA recruitment into replication compartments, which dramatically increases RNA3 stability, is a crucial step of BMV replication, and its efficiency is strongly linked to the fitness of a BMV RNA derivative as a replicon (11, 12, 15, 50, 60). The cis-signal for RNA3 recruitment into the replication compartments, the replication enhancer (RE), is located within RNA3 intergenic region immediately upstream of the sgRNA promoter (15, 60). To confirm that deletion or mutation of the core sgRNA promoter region did not increase the efficiency of 1a-mediated RNA3 stabilization, we compared the accumulation of B3CPfs, B3ΔSG, and B3SG[-13] in the presence of 1a and thus of recruitment to the replication compartment, but in the absence of 2a polymerase and therefore of viral RNA synthesis. As shown in Fig. 5A, in the absence of 2a, 1a stimulated the accumulation of all three RNA3 derivatives to a closely similar level, while in the presence of 1a and 2a, RNA3 accumulation was again stimulated by mutations blocking sgRNA transcription. Thus, sgRNA promoter mutations did not affect the efficiency of 1a-mediated RNA3 recruitment and stabilization.
FIG. 5.
Mutations in the sgRNA promoter do not alter the 1a-dependent accumulation of DNA-derived RNA3 transcripts. (A) Effects of core sgRNA promoter mutations (B3ΔSG and B3SG[-13]) on 1a-dependent RNA3 accumulation. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (B) Primer extension analysis of the 5′ ends of RNA3 species in yeast expressing BMV replication factors 1a and 2a and either B3CPfs, B3ΔSG, or B3SG[-13]. Total RNA from yeast was used in primer extension reactions containing a 5′ 32P-labeled oligonucleotide primer complementary to RNA3 bases 30 to 44. A sequencing ladder, shown on the left, was prepared by extending the same labeled primer using pB3CPfs as template, and the sequence corresponding to the 5′-end of the positive-strand RNA3 is shown. As demonstrated previously, two major primer extension bands were detected due to the cap-dependent incorporation of an additional nucleotide by reverse transcriptase (22, 24). Longer (shown next to the sequencing ladder) and shorter (right panel) exposures of the same gel are presented to better visualize the relative accumulation of the indicated RNA3 species.
These results showed that, in the absence of 2a, the wt core sgRNA promoter region did not inhibit DNA-derived RNA3 recruitment into 1a-induced RNA replication compartments. It remained possible that, in the presence of 2a, sgRNA products made in BMV RNA replication complexes might inhibit further recruitment of DNA-derived RNA3 templates. To examine this possibility, we used primer extension analysis to differentiate DNA-derived RNA3 and RNA3 produced by 1a- and 2a-dependent viral replication based on their 5′ ends. Specifically, GAL1-promoted DNA transcription produces RNA3 species with multiple 5′ ends, including bands at positions −2, −3, and −14 relative to the 5′ end of natural RNA3 (23, 25). In contrast, 1a- and 2a-dependent RNA replication in yeast specifically amplifies RNA3 with the natural 5′ end, which produces a doublet primer extension product whose upper band is derived from the 5′ cap (23, 25). As shown in Fig. 5B, primer extension analysis of total RNA from yeast with replicating B3CPfs, B3ΔSG, and B3SG[-13] confirmed the Northern blotting results that RNA3 replication, as evidenced by the major virus-specific primer extension doublet, was stimulated by mutations blocking sgRNA production. Nevertheless, the same primer extension reactions showed equal levels of accumulation of DNA-derived RNA3 transcripts (Fig. 5B, upper bands). As total accumulation of DNA-derived RNA3 in yeast harboring 1a is directly dependent upon the efficiency of 1a-mediated RNA3 recruitment and consequent stabilization in the replication compartment (25, 50, 60), sgRNA transcription did not interfere with recruitment of DNA-derived RNA3.
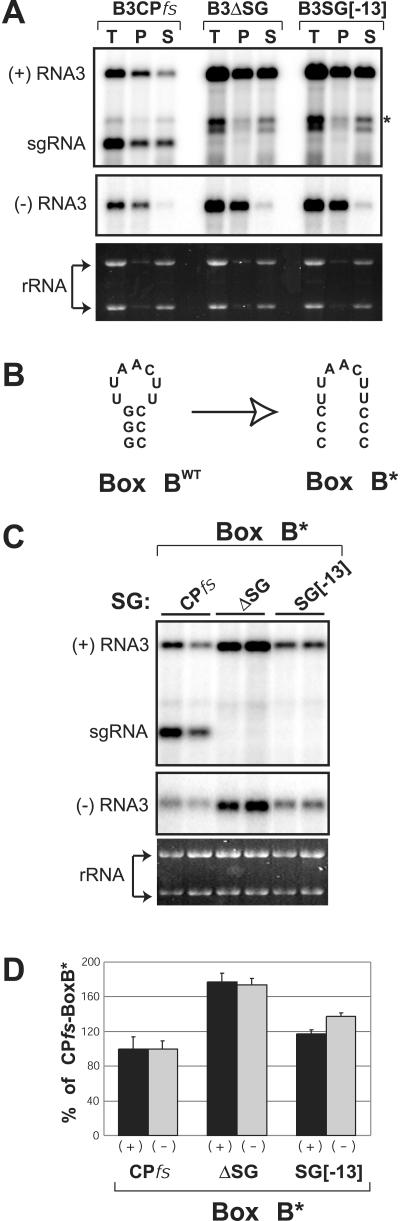
Cell fractionation showed that, in cells replicating B3CPfs, about 40% of sgRNA accumulated in the stable, membrane-associated, replication complex pool rather than in the rapidly degraded cytoplasmic pool (Fig. 6A). This accumulation of sgRNA in the limited space of new or existing replication compartments might interfere with recruitment of additional RNA3 templates, thus contributing to the observed inhibition of RNA3 replication. While sgRNAs lack the RE recruitment signal required for efficient 1a recruitment of general cytoplasmic or polysomal RNAs (50, 60), such competition might occur because sgRNAs newly exported from one replication complex might have an enhanced ability to be captured due to their immediate proximity to high concentrations of membrane-bound 1a and other replication complexes (46, 47, 50). To test whether newly synthesized sgRNA interfered with 1a-mediated RNA3 recruitment, a 3-nt substitution disrupting functionally important base-pairing in the secondary structure of an essential tRNA-related motif, box B (7, 12), was introduced into the RE sequence of B3CPfs, B3ΔSG, and B3SG[-13] replicons in order to inhibit 1a-mediated recruitment of these RNA3 derivatives into replication compartments (Fig. 6B). As this mutation was positioned upstream of the sgRNA transcription start site, B3CPfs and B3(CPfs-BoxB*) produced identical sgRNA transcripts. If sgRNA transcription interfered with RNA3 recruitment into replication compartments, this mutation would be expected to greatly exacerbate the negative effect of sgRNA transcription by making RNA3 transcripts less competitive for 1a-mediated recruitment. However, as shown in Fig. 6C and D, while this box B mutation lowered the absolute replication level of all three recipient RNAs by nearly 10-fold as expected (58, 60), it did not reduce the relative fitness of B3CPfs relative to that of B3ΔSG and B3SG[-13] (compare Fig. 6D and 2C), implying that sgRNA produced in cis did not significantly compete with RNA3 for recruitment into replication compartments.
FIG. 6.
A box B mutation inactivating the template recruitment element does not exacerbate the negative effect of sgRNA transcription on RNA3 replication. (A) Distribution of RNA3 and sgRNA in yeast cells. Yeast cells expressing replication factors 1a and 2a and either B3CPfs, B3ΔSG, or B3SG[-13] were spheroplasted and lysed osmotically to yield a total RNA fraction (T). A portion of the lysate then was centrifuged at 20,000 × g to yield a membrane-enrichedpellet (P) fraction and a supernatant (S) fraction. RNA was isolated from each fraction and analyzed by Northern blotting to detect positive-strand and negative-strand products of viral replication. Substantial amounts of sgRNA produced by B3CPfs were detected in the pellet fraction. The asterisk indicates an RNA band, discussed in the text. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (B) Schematic diagram of wt box B (Box BWT) and mutated box B (Box B*). A 3-nt replacement in the box B motif of the RE template recruitment element was designed to destabilize functionally important base-pairing at the top of the RE stem-loop structure. (C) Effect of the box B mutation on the interaction of RNA3 replication and sgRNA transcription. Northern blot analysis of total RNA was performed for yeast expressing 1a, 2a, and RNA3-derived replicons containing the Box B* mutation and either a wt or mutated sgRNA promoter. (D) The histogram compares accumulation of positive-strand (black bars) and negative-strand (gray bars) RNAs for RNA3 derivatives with the Box B* mutation.
sgRNA transcription inhibits an RNA3 replication step after negative-strand RNA3 synthesis.
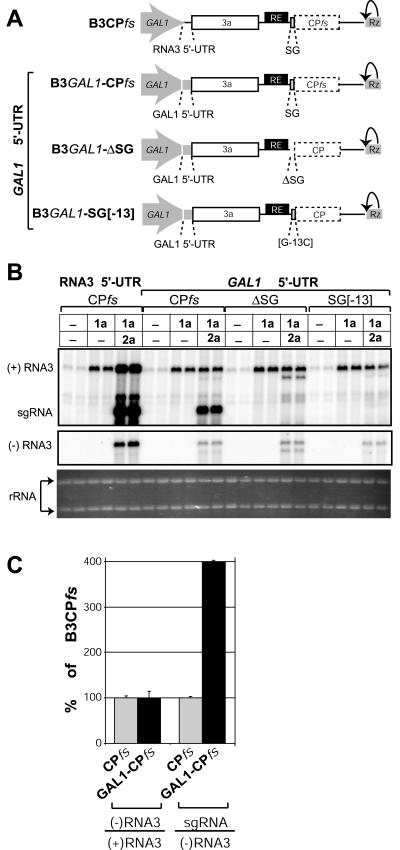
Fig. 2C showed that sgRNA transcription inhibited both positive-strand and negative-strand RNA3 accumulation. As positive-strand and negative-strand RNA3 syntheses are codependent in the BMV replication cycle, specific inhibition of either step would decrease the accumulation of RNA3 of both polarities (Fig. 1). To bypass this problem and test directly whether sgRNA transcription inhibited negative-strand RNA3 synthesis, viral positive-strand RNA3 synthesis was artificially blocked by replacing the natural RNA3 5′-UTR with the 5′-UTR of the yeast GAL1 mRNA in RNA3 derivatives with or without active sgRNA promoter (Fig. 7A). As found previously for such RNA3 derivatives with the viral 5′-UTR deleted or replaced (23, 44), this replacement failed to direct viral positive-strand RNA3 synthesis, but the original DNA-derived RNA3 transcripts served as templates for the synthesis of negative-strand RNA3, which further directed sgRNA transcription. If interference of sgRNA transcription with RNA3 replication included inhibition of negative-strand RNA3 synthesis, we would expect to see a similar inhibition for RNA3 derivatives with the GAL1 5′-UTR. As shown in Fig. 7, RNA3 derivative B3GAL1-CPfs had normal 1a responsiveness, produced negative-strand RNA3 with an efficiency equal to that of B3CPfs, as measured by the negative-strand/positive-strand RNA3 ratio, and synthesized sgRNA, indicating that the 5′-UTR had no detectable effects beyond blocking positive-strand RNA3 synthesis. Most importantly, GAL1 5′-UTR RNA3 derivatives lacking an active sgRNA promoter produced negative-strand RNA3 at levels equal to those of B3GAL1-CPfs (Fig. 7B). Thus, sgRNA transcription did not inhibit negative-strand RNA3 synthesis, and sgRNA-mediated inhibition of RNA3 replication must occur at a step or steps after negative-strand RNA3 synthesis.
FIG. 7.
Subgenomic RNA transcription does not inhibit negative-strand RNA3 accumulation. (A) Schematic diagram of expression cassettes for B3CPfs and RNA3 derivatives in which the viral 5′-UTR was replaced with the GAL1 mRNA 5′-UTR. (B) Northern blot analysis of BMV-specific RNA products. Total RNA was extracted from yeast expressing RNA3 alone or in combination with 1a or both 1a and 2a (indicated at the top). Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (C) GAL1 5′-UTR replacement strongly stimulates sgRNA transcription efficiency. The histogram compares negative-strand RNA3 synthesis efficiency (measured as the ratio of negative-strand RNA3 to positive-strand RNA3) and sgRNA transcription efficiency (measured as the ratio of sgRNA to negative-strand RNA3) for B3CPfs (gray boxes) and B3GAL1-CPfs (black boxes).
Mutations blocking positive-strand RNA3 synthesis stimulate sgRNA transcription efficiency.
Since negative-strand RNA3 serves as a template for genomic positive-strand RNA3 synthesis and sgRNA transcription, these two processes might interfere with each other by competing for negative-strand RNA3 or other replication factors. Accordingly, when the ratios of sgRNA to negative-strand RNA3 were calculated for B3CPfs and B3GAL1-CPfs as a measure of sgRNA transcription efficiency, we found that blocking positive-strand RNA3 synthesis stimulated sgRNA transcription efficiency in B3GAL1-CPfs by 400% (Fig. 7C). To confirm that positive-strand RNA3 synthesis inhibited sgRNA transcription and to rule out the possibility that the stimulation of sgRNA transcription in Fig. 7 was specifically associated with the GAL1 mRNA 5′-UTR, selected single and multiple point mutations were introduced into the 5′-UTR of B3CPfs to downregulate positive-strand RNA3 synthesis. Positions for mutagenesis (Fig. 8A) were chosen based on in vitro studies with BMV RNA-dependent RNA polymerase extracts and short templates corresponding to the core positive-strand RNA3 promoter (56, 57) and on secondary structure predictions for the positive-strand RNA3 promoter (29). All tested mutations decreased positive-strand RNA3 accumulation below that of the wt 5′-UTR (Fig. 8B) and increased sgRNA transcription efficiency (Fig. 8B and C). Moreover, the degree of stimulation was correlated with how severely each mutation inhibited positive-strand RNA3 accumulation (Fig. 8B and C). Thus, the data indicate that positive-strand RNA3 synthesis interferes with sgRNA transcription.
FIG. 8.
Inhibition of positive-strand RNA3 synthesis stimulates sgRNA transcription. (A) Point mutations (bold letters in MUT1 to MUT5) were introduced into the putative positive-strand RNA3 promoter of BMV expression cassettes. Shown in the table are the sequences of the wt and mutant cDNAs in the region corresponding to the 5′-end of wt RNA3. (B) Northern blot analysis of BMV-specific RNA products. Total RNA was extracted from yeast expressing BMV 1a, 2a, and RNA3 derivatives (indicated at the top) and analyzed by Northern blotting using specific probes to detect positive-strand RNA3 and sgRNA or negative-strand RNA3. Ethidium bromide-stained rRNA is shown below the Northern blots as a loading control. (C) The histogram compares the relative sgRNA transcription efficiency (measured as a ratio of the sgRNA to negative-strand RNA3) for B3CPfs and the 5′-UTR mutants.
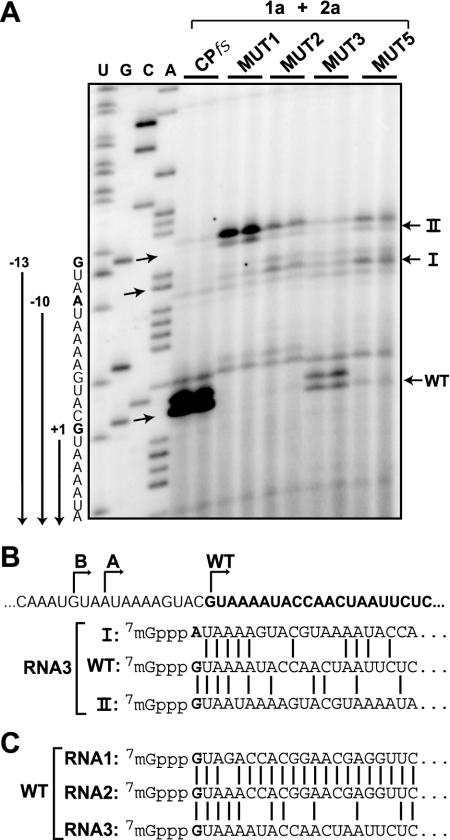
The highest level of positive-strand RNA3 accumulation among the 5′-UTR mutants was produced by B3MUT1 (Fig. 8B), even though prior in vitro results indicated that its G-to-C substitution at the wt initiation site should dramatically inhibit positive-strand RNA3 initiation (56). Primer extension analysis showed that for B3MUT1, B3MUT2, B3MUT5, and to a lesser extent the other 5′-UTR mutants, doublet bands characteristic of 5′-capped BMV RNA replication products appeared at upstream positions where flanking GAL1-derived transcript sequences shared similarity with the wt RNA3 initiation site (Fig. 9A and B, sites I and II; see also Fig. 5B and associated comments). These doublet bands were selectively amplified above the level of starting DNA-derived RNA3 transcripts and were dependent on coexpression of 1a and 2a, confirming that they represented BMV RNA replication products. Thus, in several cases, mutating the natural initiation site led to synthesis from nearby sites satisfying the apparently limited local sequence requirements for positive-strand initiation.
FIG. 9.
Generation of RNA3 derivatives with additional, nonviral 5′ nucleotides. (A) Primer extension analysis of the 5′ ends of RNA3 species from yeast expressing BMV replication factors 1a and 2a and either B3CPfs or its derivatives containing mutations in the 5′-UTR as described in the legend for Fig. 8B. Primer extension was performed on total yeast RNA as for Fig. 5B. Arrows indicate the natural site of initiation for positive-strand RNA3 (WT) and novel positive-strand RNA3 initiation sites at position −10 (I) or −13 (II). Two major primer extension bands for each initiation position were detected due to cap-dependent incorporation of an additional nucleotide by reverse transcriptase (22, 24). (B) Alignment of the sequences of wt RNA3 and derivatives A and B produced as a result of upstream initiation at the positions indicated in panel A. (C) Alignment of the wt sequences of wt RNA1, wt RNA2, and wt RNA3.
Analysis of RNA3 sgRNA promoter mutants in barley protoplasts.
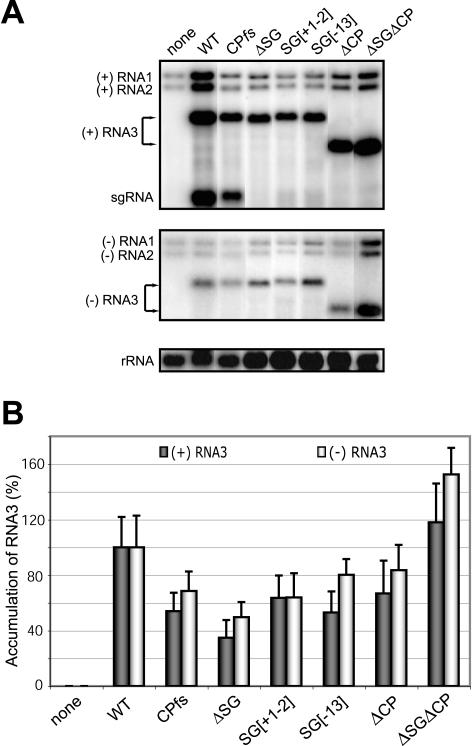
Recent studies of the effects of different mutations in RNA3 genomic and sgRNA promoters on replication and sgRNA transcription in barley protoplasts also observed increased sgRNA accumulation when positive-strand RNA3 synthesis was inhibited by 5′-UTR mutations but failed to observe increased RNA3 accumulation when sgRNA synthesis was inhibited by sgRNA promoter mutations (19, 55). To further explore such effects, we tested the effects of sgRNA promoter mutations from this study on RNA3 accumulation in barley protoplasts. Protoplasts were transfected with in vitro transcripts of wt BMV genomic RNA1 and RNA2 plus wt RNA3 or RNA3 derivatives B3CPfs, B3ΔSG, B3SG[+1,-2], B3SG[-13], B3ΔCP, or B3ΔSGΔCP (Fig. 2A, 6B, and 10), and positive- and negative-strand RNA3 accumulation were analyzed by Northern blotting. Representative Northern blots are shown in Fig. 10A, and the histogram in Fig. 10B summarizes the results of three independent experiments, including nine independent transformations for each RNA3 derivative.
FIG. 10.
Effect of inactivating sgRNA transcription on RNA3 accumulation in barley protoplasts inoculated with all three BMV genomic RNAs. (A) Northern blot analysis of BMV-specific RNA products in barley protoplasts. Total RNA was extracted from barley protoplasts 20 h p.i. with BMV wt RNA1, wt RNA2, and one of the indicated RNA3 derivatives. Ten micrograms of total RNA was analyzed by Northern blotting using specific probes to detect positive- or negative-strand BMV RNA or 18S rRNA as indicated on the left. Representative results are shown. (B) Relative accumulation of positive-strand (dark bars) and negative-strand (light bars) RNA3 derivatives. The histogram summarizes results from nine independent transfections for each RNA3 derivative.
Relative to wt RNA3 expressing CP from sgRNA (T7-B3WT), the sgRNA mutants displayed a decrease in RNA3 positive- and negative-strand RNA3 accumulation, as reported in reference 19. However, CP expression modulates BMV RNA accumulation due to encapsidation, as shown previously (20, 31, 37, 42, 48) and in this study (Fig. 3). Therefore, as in the yeast experiments, we compared the replication products of such RNA3 mutants to more relevant controls retaining sgRNA synthesis but with CP expression blocked by CP ORF mutations (B3CPfs) or deletion (B3ΔCP).
Consistent with the yeast results and prior barley protoplast studies (15), deletion of sgRNA promoter in B3ΔSGΔCP increased positive- and negative-strand RNA3 accumulation by 80% relative to that of its parental RNA, B3ΔCP (Fig. 10). However, RNA3 derivatives with inactivated subgenomic promoters but retaining the full CP ORF produced positive- and negative-strand RNA3 accumulation that was generally similar to that of B3CPfs, with B3SG[+1,-2] and B3SG[-13] modestly increased and B3ΔSG modestly decreased by increments near the standard deviations of the experiments. Thus, while the data show that sgRNA promoter deletion can stimulate RNA3 accumulation in barley cells, these findings and the independent results (19, 55) show that sgRNA promoter inactivation generally has a lesser effect on RNA3 accumulation in barley protoplasts replicating all three BMV genomic RNAs than in 1a- and 2a-expressing yeast replicating RNA3 alone. Possible reasons for such a difference are considered in the Discussion.
DISCUSSION
In this study, we analyzed the functional interactions between BMV genomic RNA3 replication and sgRNA transcription in vivo and found that these two processes mutually interfere. The results led to three principle conclusions. First, sgRNA synthesis, rather than its RNA or protein products, inhibits genomic RNA3 replication in cis, strictly targeting amplification of its parental RNA3 but not separate RNA3 derivatives replicating in the same cell. Second, sgRNA transcription interferes with RNA3 replication at a step or steps after negative-strand RNA3 synthesis, implying specific inhibition of positive-strand RNA3 synthesis. Third, consistent with interference in the usage of their common negative-strand RNA3 template, positive-strand RNA3 synthesis inhibits sgRNA transcription efficiency. Overall, the results suggest that sgRNA transcription and positive-strand RNA3 synthesis compete for negative-strand RNA3 templates, viral replication factors, or limiting host components. As discussed below, these findings have implications for the mechanisms of RNA replication and the nature of limiting steps and components in the RNA replication complexes.
sgRNA transcription inhibits RNA3 replication.
Previous studies demonstrated that BMV sgRNAs are initiated internally on negative-strand RNA3 templates (39), and they explored promoter sequence (1, 14, 36, 51, 52, 58) and secondary structure requirements for sgRNA initiation (16, 17). To test for interactions between sgRNA transcription and RNA3 replication, selected mutations were introduced into the sgRNA promoter region and the effects on sgRNA synthesis and genomic RNA3 replication were analyzed. All tested mutations markedly inhibited sgRNA synthesis while increasing RNA3 replication (Fig. 2). Thus, in wt RNA3, the presence of an active sgRNA promoter inhibited RNA3 replication. Previously, an inhibiting effect of an active sgRNA promoter on genomic RNA replication was documented for a defective interfering RNA of the coronavirus mouse hepatitis virus (28). Nevertheless, since coronaviruses employ unusually complex mechanisms of discontinuous sgRNA synthesis (40), it remains unclear whether common mechanisms underlie such interference in mouse hepatitis virus and BMV.
Further experiments above, including trans-expression studies, showed that the inhibition of RNA3 replication associated with sgRNA transcription was independent of the products of sgRNA synthesis, CP (Fig. 3) and sgRNA (Fig. 4A). Moreover, sgRNA synthesis only inhibited RNA3 replication in cis (Fig. 4A and B). As deletion of the core sgRNA promoter blocked inhibition of RNA3 replication, it was possible that such inhibition was not due to sgRNA synthesis per se, but to removing a hypothetical, overlapping negative regulator of RNA3 replication, similar to a “replication silencer” recently described for tomato bushy stunt virus (43). However, since RNA3 replication was similarly stimulated by this 28-nt deletion or alternate, separated point mutations in the sgRNA promoter (Fig. 2), the results suggest that stimulation of RNA3 replication was due to the well-documented, shared effects of these mutations on sgRNA synthesis, rather than to potential effects on a hypothetical replication silencer. Thus, the data imply that sgRNA synthesis itself, rather than sgRNA promoter sequences or products of sgRNA synthesis, inhibited RNA3 replication.
Competition between RNA3 replication and sgRNA transcription occurs after negative-strand RNA synthesis.
RNA3 replication has been resolved into multiple, experimentally separable steps, involving distinct RNA3 cis-signals and, in some cases, distinguishable replication factor requirements (Fig. 1). Recruitment of positive-strand RNA3 into replication complexes requires BMV 1a protein and the cis-acting, intergenic RE element (7, 50, 60). Negative-strand synthesis from these RNA3 templates requires 1a and 2a and is driven by a promoter in the 3′ tRNA-like sequence of RNA3 (9, 22, 38). In turn, negative-strand RNA3 is a template for positive-strand RNA3 synthesis and sgRNA transcription, driven by promoters adjacent to their respective initiation sites (56, 57).
Examination of these RNA replication steps showed that sgRNA transcription does not interfere either with 1a-mediated recruitment of RNA3 into replication compartments (Fig. 5 and 6) or with negative-strand RNA3 synthesis (Fig. 7). Therefore, sgRNA transcription must inhibit RNA3 replication at a step or steps downstream of negative-strand RNA3 synthesis, and thus at or immediately before positive-strand RNA3 synthesis.
As discussed further below, these findings suggested that sgRNA synthesis may inhibit RNA3 replication by competing with positive-strand RNA3 synthesis for one or more common factors, such as negative-strand RNA3 templates, 1a and 2a, and possibly host factors. Such direct competition would imply that positive-strand RNA3 synthesis should reciprocally interfere with sgRNA synthesis. To test this possibility, we made a series of RNA3 derivatives in which the RNA3 5′-UTR bore mutations designed to inhibit positive-strand RNA3 synthesis, based on prior studies (56, 57). These mutants inhibited RNA3 replication as intended and simultaneously increased the ratio of sgRNA to negative-strand RNA3 template in proportion to the severity of the block to positive-strand RNA3 synthesis (Fig. 7 and 8). Thus, synthesis of positive-strand genomic RNA3 inhibited sgRNA synthesis from their common negative-strand RNA3 template.
Limiting components for RNA3 replication and sgRNA synthesis.
The observed mutual interference between sgRNA transcription and positive-strand RNA3 synthesis suggests that these two processes may compete for one or more common, limiting components of the BMV RNA synthesis machinery. Such competition is easily envisioned, since sgRNA and positive-strand RNA3 synthesis both occur in ER membrane-bound RNA replication compartments (50), use the same negative-strand RNA3 templates (29), require BMV replication factors 1a and 2a, and have similar kinetics (23, 34). Such direct competition within the confines of individual, 50- to 70-nm-diameter, membrane-bound RNA replication complexes also would explain our finding that sgRNA transcription strictly inhibited replication of the parental RNA3 in cis but had no effect in trans on other RNA3-derived replicons in the same cells (Fig. 4).
Although more than one polymerase can simultaneously copy a single viral RNA template (8), possible competition between genomic RNA replication and sgRNA synthesis for negative-strand RNA templates is supported by prior findings with other members of the alphavirus superfamily. Cells infected with Sindbis virus (54) or Semliki Forest virus (49) were found to contain two types of replicative intermediates, one associated with positive-strand genomic RNA and another with sgRNA and a partial positive-strand RNA product 5′-coterminal with genomic RNA. Studies with Sindbis virus showed that termination of the partial, 5′-coterminal, genomic positive-strand RNA was due to sgRNA transcription and occurred 4 nt upstream of the sgRNA transcription start site, possibly due to relatively long-lived protein-RNA interactions at the sgRNA promoter (62). Thus, initiation at the sgRNA promoter may at least temporarily block genomic RNA synthesis from the same negative-strand RNA template.
Alternatively or in addition to competition for negative-strand RNA3 templates, the observed interference between positive-strand genomic RNA and sgRNA synthesis may involve competition for virus- or host-encoded RNA replication factors. As noted above, both processes require BMV RNA replication factors 1a and 2a. Prior experiments showed that, in BMV-infected cells, 1a was more limiting than 2a polymerase for RNA synthesis (13), suggesting that competition for 1a or the replication compartments that it induces might be the limiting factor in this competition. Moreover, while 2a polymerase may not be the limiting factor for overall RNA synthesis, the number of 2a polymerases within individual replication complexes is limited to approximately 10 to 15 copies (50), so that the genomic and subgenomic promoters may compete locally for 2a within individual complexes.
Alternate genomic RNA initiation sites.
In addition to their effects on sgRNA transcription efficiency (see above), the mutations that we introduced into the RNA3 5′ sequences to inhibit genomic RNA3 synthesis (Fig. 8A) provided unexpected insights into positive-strand RNA3 initiation. For some of these mutants, inhibition of positive-strand RNA3 synthesis from the wt initiation site was accompanied by the selective amplification of elongated RNA3 derivatives initiated from novel upstream sites in the flanking GAL1 sequences (Fig. 9A). Previously we showed that DNA transcription from the GAL1 promoter initiates at multiple sites at and before the start of BMV RNA3 sequences (23, 25), providing the initial RNA templates for these novel, elongated RNA replicons (Fig. 9B). The novel 5′ ends of these elongated replicons resembled the wt 5′ ends of BMV RNAs 1, 2, and 3 over the first 4 to 6 nt (Fig. 9C), suggesting that these 5′-proximal positions contribute to successful BMV RNA replication in vivo. Nevertheless, replication of these elongated RNA3 derivatives was not observed in the presence of the wt RNA3 5′ end (Fig. 9A, B3CPfs), presumably due to the greater replicative fitness of wt RNA3. Moreover, these elongated replicons also failed to be amplified when the natural RNA3 5′-UTR was replaced with the yeast GAL1 5′-UTR (Fig. 7 and results not shown), suggesting that additional cis-requirements for positive-strand RNA synthesis may be positioned in the RNA3 5′-UTR. The observed flexibility in initiation site usage is consistent with in vitro data on positive-strand initiation from templates complementary to 5′-proximal sequences of BMV RNA2, where artificial repetition of the initiating trinucleotide stimulated initiation from multiple sites (56). Similarly, deleting 22 5′ nucleotides or adding nonviral sequences to the 5′ end of RNA3 from another bromovirus, alfalfa mosaic virus, gave rise in vivo to novel RNA3 replicons lacking the 5′ 79 nt of wt RNA3 (61). Further studies are in progress to characterize the detailed requirements for BMV positive-strand RNA synthesis.
Interaction of sgRNA and genomic RNA3 synthesis in barley protoplasts.
While this work was in progress, other reports appeared on the effects of mutations in RNA3 genomic and sgRNA promoters on RNA3 replication and sgRNA transcription in barley protoplasts transfected with BMV RNAs 1 to 3 (19, 55). In agreement with the results presented above in yeast, several mutations in the 5′-UTR-encoded promoter for positive-strand RNA3 substantially increased sgRNA accumulation (up to 358%), indicating interference of positive-strand RNA3 synthesis with sgRNA transcription (19).
This interference between genomic RNA and sgRNA syntheses suggested that sgRNA promoter mutations should conversely stimulate RNA3 accumulation in barley cells (19). However, in contrast to this expectation and our findings in yeast replicating RNA3 (Fig. 7 and 8), various mutations inhibiting sgRNA transcription were not found to stimulate RNA3 accumulation in protoplasts replicating all three BMV genomic RNAs, 1 to 3 (19, 55). Rather, in many cases, inhibiting sgRNA transcription reduced RNA3 accumulation relative to that of wt. Our results in barley protoplasts (Fig. 10) showed that this reduction in RNA3 accumulation relative to that of wt RNA3 largely was due to blocking CP expression and thus viral RNA encapsidation. Consistent with this, inactivating sgRNA synthesis also inhibited RNA1 and RNA2 accumulation, particularly of positive-strand polarity (19, 55) (Fig. 10).
When using CP ORF deletion to eliminate encapsidation effects, the results of sgRNA promoter deletion correlated well with those in yeast, with B3ΔSGΔCP stimulating positive- and negative-strand RNA accumulation approximately twofold relative to that with B3ΔCP (Fig. 4B and 10). Similar stimulation of CP-negative RNA3 accumulation by 2.5- to >4-fold in barley protoplasts upon sgRNA promoter inactivation was reported previously for RNA3 derivatives B3BHI and B3BX1 relative to that for B3SHI (15).
However, even after accounting for loss of CP expression, sgRNA promoter mutations tested in the context of the full CP ORF showed little or no change in RNA3 accumulation in barley protoplasts relative to B3CPfs (B3ΔSG, B3SG[+1,-2], and B3SG[-13] in Fig. 10B). Further experiments are needed to determine how the presence or absence of the CP ORF, or length of the sgRNA produced, modifies these interference effects. Nevertheless, these and independent results (19, 55) show that blocking sgRNA synthesis often has less effect on RNA3 accumulation in the barley protoplast system used than in yeast.
Several significant differences exist in the design of the yeast and protoplast experiments that may modulate the effects of sgRNA synthesis on RNA3 replication. First, in yeast, RNA3 was the only RNA replication template, while in barley protoplasts all three BMV genomic RNAs replicated simultaneously. Thus, while in yeast RNA3 synthesis competed only with RNA4, in protoplasts RNA3 synthesis competed with RNAs 1, 2, and 4, and blocking RNA4 synthesis only removed a fraction of the competition with RNA3 synthesis. Second, in yeast, BMV replication factors 1a and 2a were expressed at stable levels from DNA plasmids, while in protoplasts, 1a and 2a expression was from RNA1 and RNA2 and thus was directly dependent on RNA1 and RNA2 replication levels. In barley protoplasts any increase in RNA3 competitiveness for genomic RNA replication thus may tend to be self-limiting, since it would be at the expense of RNA1 and RNA2 and reduce expression of the very proteins that drive replication. Third, while barley protoplasts are essentially quiescent and plateau in negative-strand accumulation by 6 to 7 h p.i. (34), yeast cells divide every several hours and are constantly producing new RNA replication complexes. This dynamic expansion, which mimics the expansion of natural infections through the tissues of plant hosts, may amplify effects associated with competition for replication factors. As with in vitro and in vivo systems, such differences reflect the synergistic benefits of different experimental designs for revealing various important aspects of infection. Future experiments will continue to benefit from integrating the insights revealed from all such approaches.
Acknowledgments
We thank all members of our laboratory for stimulating discussions throughout the course of this work, Billy Dye for helpful comments on the manuscript, and Michael Janda for assistance with barley protoplast experiments.
This research was supported by National Institutes of Health grant GM35072. T.W. was partially supported by the Uehara Memorial Foundation, Japan. P.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, S., R. W. Siegel, J. H. Sun, and C. C. Kao. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634-647. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist, P., R. French, M. Janda, and L. S. Loesch-Fries. 1984. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA 81:7066-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 7.Baumstark, T., and P. Ahlquist. 2001. Brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TΨC stemloop and is modified in vivo. RNA 7:1652-1670. [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, M. R., and C. C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., A. Noueiry, and P. Ahlquist. 2003. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 77:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′-proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinant, S., M. Janda, P. A. Kroner, and P. Ahlquist. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 67:7181-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haasnoot, P. C., F. T. Brederode, R. C. Olsthoorn, and J. F. Bol. 2000. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haasnoot, P. C., R. C. Olsthoorn, and J. F. Bol. 2002. The brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8:110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haseloff, J., P. Goelet, D. Zimmern, P. Ahlquist, R. Dasgupta, and P. Kaesberg. 1984. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. USA 81:4358-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hema, M., and C. C. Kao. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikoshi, M., M. Nakayama, N. Yamaoka, I. Furusawa, and J. Shishiyama. 1987. Brome mosaic virus coat protein inhibits viral RNA synthesis in vivo. Virology 158:15-19. [DOI] [PubMed] [Google Scholar]
- 21.Huntley, C. C., and T. C. Hall. 1993. Interference with brome mosaic virus replication by targeting the minus strand promoter. J. Gen. Virol. 74:2445-2452. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa, M., J. Diez, M. Restrepo-Hartwig, and P. Ahlquist. 1997. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. USA 94:13810-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 71:7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 27.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 28.Jeong, Y. S., and S. Makino. 1992. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J. Virol. 66:3339-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, C. C. 2002. Lessons learned from the core RNA promoters of brome mosaic virus and cucumber mosaic virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 30.Kong, F., K. Sivakumaran, and C. Kao. 1999. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200-210. [DOI] [PubMed] [Google Scholar]
- 31.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroner, P., and P. Ahlquist. 1992. RNA-based viruses, p. 23-34. In S. J. Gurr, M. J. McPherson, and D. Bowles (ed.), Molecular plant pathology-a practical approach, vol. I. IRL/Oxford University Press, Oxford, England. [Google Scholar]
- 33.Kroner, P., D. Richards, P. Traynor, and P. Ahlquist. 1989. Defined mutations in a small region of the brome mosaic virus 2 gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 63:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroner, P. A., B. M. Young, and P. Ahlquist. 1990. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J. Virol. 64:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 36.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsh, L. E., C. C. Huntley, G. P. Pogue, J. P. Connell, and T. C. Hall. 1991. Regulation of (+):(−)-strand asymmetry in replication of brome mosaic virus RNA. Virology 182:76-83. [DOI] [PubMed] [Google Scholar]
- 38.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537-546. [DOI] [PubMed] [Google Scholar]
- 39.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 40.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Noueiry, A. O., J. Diez, S. P. Falk, J. Chen, and P. Ahlquist. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pacha, R. F., and P. Ahlquist. 1991. Use of bromovirus RNA3 hybrids to study template specificity in viral RNA amplification. J. Virol. 65:3693-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranjith-Kumar, C. T., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sacher, R., and P. Ahlquist. 1989. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 63:4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawicki, D. L., L. Kaariainen, C. Lambek, and P. J. Gomatos. 1978. Mechanism for control of synthesis of Semliki Forest virus 26S and 42S RNA. J. Virol. 25:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 51.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel, R. W., L. Bellon, L. Beigelman, and C. C. Kao. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 95:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons, D. T., and J. H. Strauss. 1972. Replication of Sindbis virus. II. Multiple forms of double-stranded RNA isolated from infected cells. J. Mol. Biol. 71:615-631. [DOI] [PubMed] [Google Scholar]
- 55.Sivakumaran, K., S. K. Choi, M. Hema, and C. C. Kao. 2004. Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 78:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sivakumaran, K., and C. C. Kao. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 73:6415-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivakumaran, K., C. H. Kim, R. Tayon, and C. C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smirnyagina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198:427-436. [DOI] [PubMed] [Google Scholar]
- 59.Stawicki, S. S., and C. C. Kao. 1999. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRp) reveal that RdRp can adjust its promoter binding sites. J. Virol. 73:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan, M., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Vossen, E. A., L. Neeleman, and J. F. Bol. 1996. The 5′ terminal sequence of alfalfa mosaic virus RNA 3 is dispensable for replication and contains a determinant for symptom formation. Virology 221:271-280. [DOI] [PubMed] [Google Scholar]
- 62.Wielgosz, M. M., and H. V. Huang. 1997. A novel viral RNA species in Sindbis virus-infected cells. J. Virol. 71:9108-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson, J. R. 2000. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7:834-837. [DOI] [PubMed] [Google Scholar]