ABSTRACT
The nitrogen phosphotransferase system (PTSNtr) is a regulatory cascade present in many bacteria, where it controls different functions. This system is usually composed of three basic components: enzyme INtr (EINtr), NPr, and EIIANtr (encoded by the ptsP, ptsO, and ptsN genes, respectively). In Legionella pneumophila, as well as in many other Legionella species, the EIIANtr component is missing. However, we found that deletion mutations in both ptsP and ptsO are partially attenuated for intracellular growth. Furthermore, these two PTSNtr components were found to be required for maximal expression of effector-encoding genes regulated by the transcriptional activator PmrA. Genetic analyses which include the construction of single and double deletion mutants and overexpression of wild-type and mutated forms of EINtr, NPr, and PmrA indicated that the PTSNtr components affect the expression of PmrA-regulated genes via PmrA and independently from PmrB and that EINtr and NPr are part of the same cascade and require their conserved histidine residues in order to function. Furthermore, expression of the Legionella micdadei EIINtr component in L. pneumophila resulted in a reduction in the levels of expression of PmrA-regulated genes which was completely dependent on the L. pneumophila PTS components and the L. micdadei EIINtr conserved histidine residue. Moreover, reconstruction of the L. pneumophila PTS in vitro indicated that EINtr is phosphorylated by phosphoenolpyruvate (PEP) and transfers its phosphate to NPr. Our results demonstrate that the L. pneumophila incomplete PTSNtr is functional and involved in the expression of effector-encoding genes regulated by PmrA.
KEYWORDS: Legionella, effector gene expression, phosphotransferase system (PTS), PmrA two-component system
INTRODUCTION
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular pathogen which utilizes the Icm/Dot type IV secretion system for pathogenesis (1, 2). The Icm/Dot secretion system was shown to translocate a cohort of approximately 300 effector proteins into host cells during infection (3). The levels of expression of many of the genes encoding these effectors were found to be regulated by three two-component systems (TCSs): (i) the PmrAB TCS, which consists of the PmrA response regulator (RR) and the PmrB sensor histidine kinase (SHK), was shown to directly activate the expression of 43 effector-encoding genes (4, 5); (ii) the CpxRA TCS, which consists of the CpxR RR and the CpxA SHK, was shown to directly activate or repress the expression of 27 effector-encoding genes and four Icm/Dot components (6–8); (iii) the LetAS TCS, which consists of the LetA RR and the LetS SHK (9, 10), was shown to regulate the transcription of two small regulatory RNAs, RsmY and RsmZ, which act in a redundant fashion to jointly antagonize CsrA, an RNA-binding protein that negatively regulates the expression of 26 effector-encoding genes at the translational level (11–15). In addition, the L. pneumophila Lqs system and its signaling molecule LAI-1 positively regulate the transcription of these small regulatory RNAs and negatively regulate the RNA-binding protein CsrA (16). Such TCSs are usually activated by phosphorylation of a conserved histidine residue located in their SHKs, from which the phosphate group is then transferred to a conserved aspartic acid residue located in the receiver domain of the cognate RR, which in turn directly activates or represses gene expression (17).
In many bacteria, global regulatory systems such as the phosphoenolpyruvate (PEP) phosphotransferase system (PTS) are involved in various regulatory functions. The sugar-related PTS consists of two cytoplasmic proteins, enzyme I (EI), which is phosphorylated by PEP, and histidine phosphocarrier protein (HPr), which is phosphorylated by the EI component, both of which lack sugar specificity, and sugar-specific enzyme II (EII) components (18). Many Gram-negative bacteria also contain the nitrogen PTS (PTSNtr). The PTSNtr constitutes another phosphorylation cascade which proceeds sequentially from PEP to EINtr encoded by ptsP, to NPr encoded by ptsO, and to EIIANtr encoded by ptsN, proteins which are homologous to the sugar PTS components EI, HPr, and EIIA, respectively (19, 20). In different bacterial species the PTSNtr regulates diverse processes implicated in metabolism of nitrogen and carbon (21), it plays a role in potassium homeostasis (22) and biofilm formation (23), and is essential for virulence in some bacteria such as Salmonella enterica (24) and Brucella melitensis (25), as well as in L. pneumophila (26).
In this report, we demonstrate by in vitro and in vivo experiments that the incomplete PTSNtr (lacking an EIIANtr) found in L. pneumophila functions as a phosphorelay. We also demonstrate a link between the PTSNtr and the RR PmrA which controls the expression of numerous effector-encoding genes. We show that ptsP and ptsO deletion mutants are defective for intracellular growth and that the levels of expression of PmrA-regulated genes are reduced in these mutants, thus for the first time connecting the PTSNtr to effector gene expression in L. pneumophila.
RESULTS
L. pneumophila contains an incomplete PTSNtr lacking an EIIANtr component.
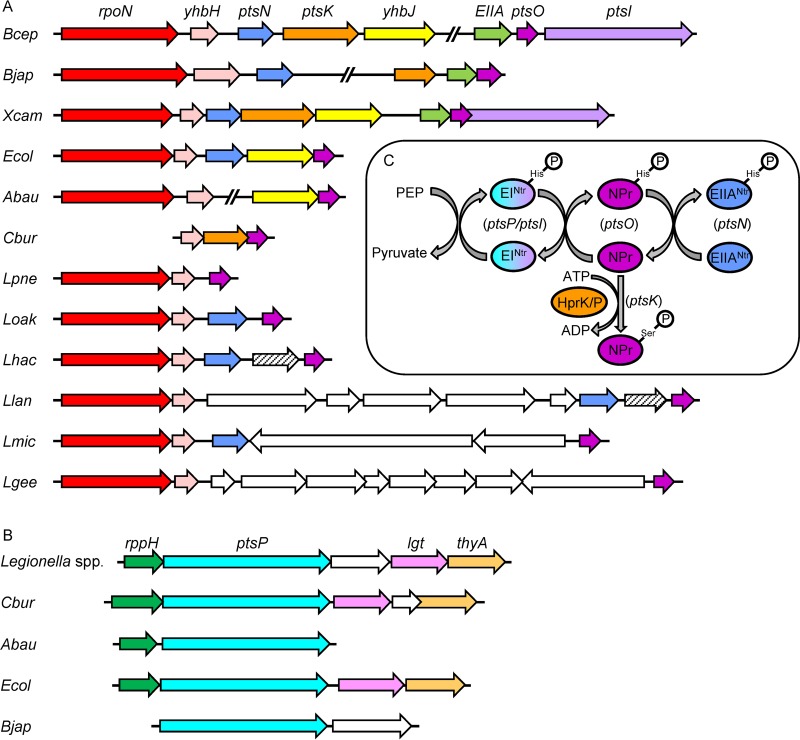
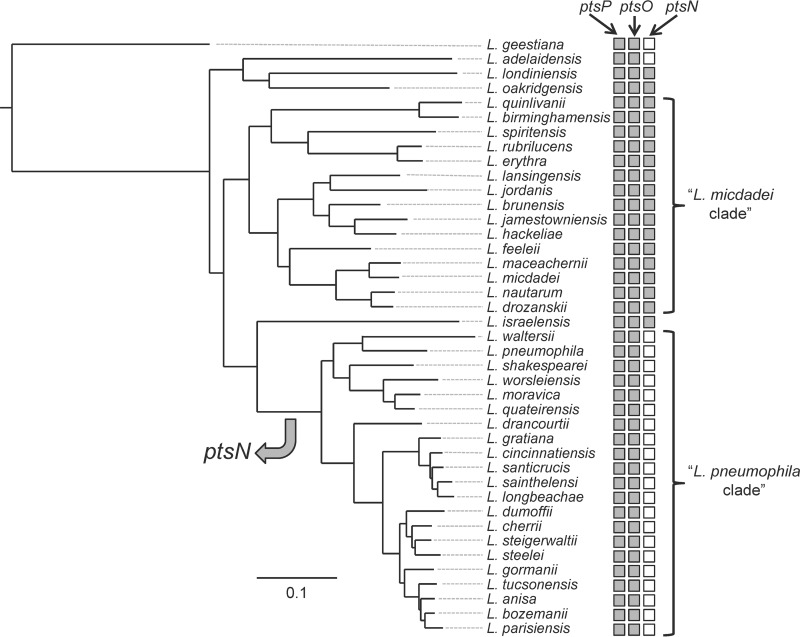
A genomic search performed at the NCBI database revealed that L. pneumophila contains only the first two components of the PTSNtr, the ptsP gene encoding EINtr and the ptsO gene encoding NPr (Fig. 1A to C). The overall operon organization of the PTSNtr genes is known to be conserved in bacteria (20). In most cases the rpoN gene encoding the nitrogen sigma factor (RpoN) is located first in an operon which also includes ptsN (encoding EIIANtr) and ptsO (encoding NPr) (Fig. 1A). This operon was also shown to contain additional genes, some of which are conserved in most bacterial species (such as the yhbH gene, encoding a ribosome binding protein), and in several bacteria additional genes belonging to the PTSNtr are also part of this operon (such as ptsK encoding HPr kinase/phosphorylase and ptsI encoding an EI-like protein). Examination of this operon in many bacteria indicated that the ptsN gene, when present, is always located between the rpoN and the ptsO genes (Fig. 1A). However, this was found not to be the case in L. pneumophila (as well as in Acinetobacter baumannii), which does not contain a ptsN gene as part of the rpoN operon. In addition, a BLAST search indicated that there is no EIIANtr homologue in L. pneumophila (an E score of 0.1 was used as a cutoff to determine similarity). Importantly, the absence of the ptsN gene encoding EIIANtr is not unique to L. pneumophila. This was also found to be the case in all the Legionella species belonging to the L. pneumophila clade (27), which includes 21 species, while it is present in most of the other Legionella species and in all of the members of the Legionella micdadei clade (Fig. 2). Even though the EIIANtr is absent in the L. pneumophila clade, the EINtr and NPr proteins are conserved in the entire Legionella genus (see Fig. S1 in the supplemental material and data not shown), suggesting that they are functional.
FIG 1.
The PTSNtr components in different bacteria. The operon organization of genes encoding the PTSNtr components NPr (A) and EINtr (B) in various bacteria is shown. Homologous genes are shown with the same color; open reading frames that have no homologues in the other regions presented are represented by white arrows. rpoN encodes the nitrogen sigma factor, yhbH encodes a ribosome binding protein, ptsN encodes EIIANtr, ptsK encodes NPr kinase/phosphorylase, ptsO encodes NPr, ptsI encodes an EI-like protein, rppH encodes RNA pyrophosphohydrolase, ptsP encodes EINtr, lgt encodes diacylglyceryl transferase, and thyA encodes thymidylate synthetase. Bcep, Burkholderia cepacia; Bjap, Bradyrhizobium japonicum; Xcam, Xanthomonas campestris; Ecol, Escherichia coli; Abau, Acinetobacter baumannii; Cbur, Coxiella burnetii; Lpne, L. pneumophila; Loak, Legionella oakridgensis; Lhac, Legionella hackeliae; Llan, Legionella lansingensis; Lmic, L. micdadei; and Lgee, Legionella geestiana. The ptsO operon organization found in L. pneumophila is the same in 21 other Legionella species (Fig. 2). (C) Schematic representation of the PTSNtr phosphorylation cascade. Phosphoryl groups are sequentially transferred on histidine residues from PEP to EINtr, NPr, and subsequently to EIINtr. Phosphorylation of NPr by HPrK/P on serine is also indicated.
FIG 2.
Presence and absence of PTSNtr components in the Legionella genus. A maximum-likelihood tree of 41 sequenced Legionella species was reconstructed on the basis of concatenated amino acid alignment of 78 orthologous open reading frames (27). For each species, the presence (gray) or absence (white) of the ptsP, ptsO, and ptsN genes of the PTSNtr is indicated.
The absence of the ptsN gene from L. pneumophila, together with the conservation of the NPr protein in all members of the Legionella genus, might indicate that the PTSNtr of L. pneumophila participates in other functions which probably involve the transfer of phosphate from EINtr to NPr and then to a component which is different from EIIANtr.
The L. pneumophila ptsP and ptsO genes are required for optimal intracellular growth.
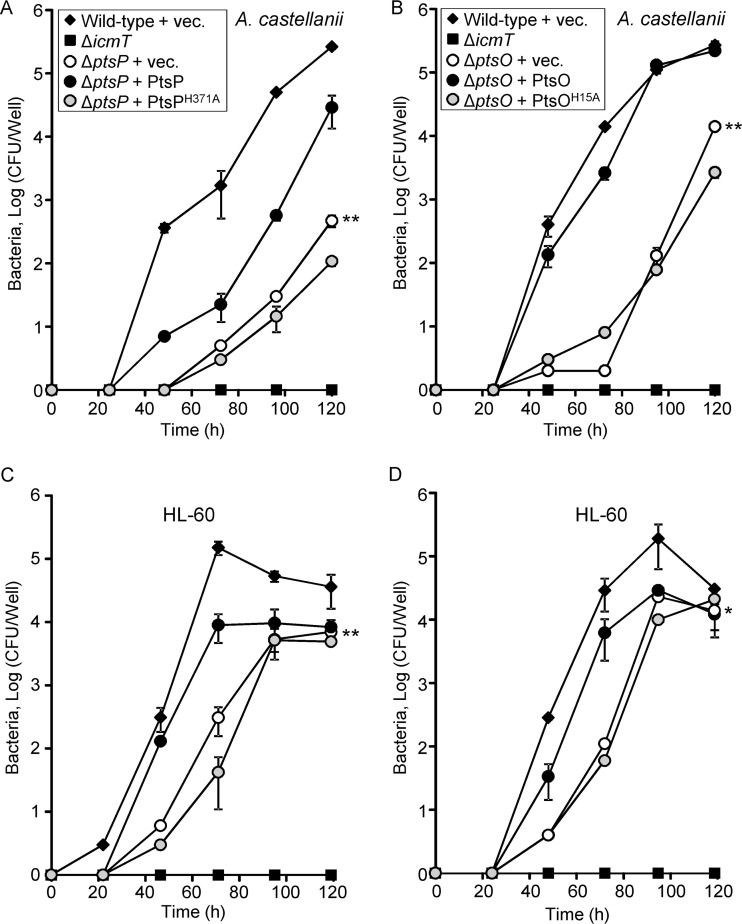
Since only two components of the PTSNtr are present in L. pneumophila, we constructed deletion mutations in each of the genes encoding these components and examined them for intracellular growth in the amoeba host Acanthamoeba castellanii (28). Since ptsP is located in the middle of an operon (29), we generated a nonpolar in-frame deletion mutation in this gene and a kanamycin deletion substitution mutant in ptsO. Examination of these mutants revealed that both the ptsP and ptsO deletion mutants are partially defective for intracellular growth in A. castellanii (Fig. 3A and B). The intracellular growth phenotype of these mutants was complemented by introducing plasmids containing the ptsP and ptsO genes, respectively, cloned under the control of the Ptac promoter (induced by isopropyl-β-d-thiogalactopyranoside [IPTG]) (Fig. 3A and B). These two deletion mutants were also examined for intracellular growth in HL-60-derived human macrophages (Fig. 3C and D) (28). In these cells, both the ptsP and ptsO deletion mutants showed a mild intracellular growth phenotype which was also complemented using the plasmids described above (Fig. 3C and D). Examination of a double deletion mutant of both ptsP and ptsO (ptsP-ptsO) did not result in an additive effect on intracellular growth in comparison to the single deletion mutants (data not shown). The growth rates of ptsP and ptsO single deletion mutants, as well as the growth rate of the double ptsP-ptsO deletion mutant, are similar to the growth rate of the wild-type strain when the mutants are examined in vitro in regular growth medium (Fig. S2).
FIG 3.
The PTSNtr is required for L. pneumophila optimal intracellular multiplication. The ability of ptsP (A and C) and ptsO (B and D) deletion mutants to grow intracellularly was examined in A. castellanii (A and B) and HL-60-derived human macrophages (C and D). Symbols: diamonds, L. pneumophila wild-type strain JR32; squares, the icmT mutant strain GS3011; open circles, ptsP and ptsO deletion mutants SY-ΔptsP and SY-ptsO-Km, respectively, containing the vector (pMMB207); black circles, ptsP and ptsO deletion mutants SY-ΔptsP and SY-ptsO-Km, respectively, containing the complementing plasmids expressing the wild-type EINtr and NPr (pSY-207-Ptac-ptsP in panels A and C and pSY-207-Ptac-ptsO in panels B and D); and gray circles, ptsP and ptsO deletion mutants SY-ΔptsP and SY-ptsO-Km, respectively, containing plasmids expressing the mutated EINtr-H371A and NPrH15A (pSY-207-Ptac-ptsP-H371A in panels A and C and pSY-207-Ptac-ptsO-H15A in panels B and D). The experiments were performed as described in Materials and Methods. The experiments were done three times, and similar results were obtained; error bars indicate standard deviations. The intracellular growth rates were found to be significantly different (*, P < 0.01; **, P < 0.001, two-way repeated analysis of variance) in comparisons between results for the pstP and ptsO deletion mutants and those of the wild-type strain.
The results described above agree with previous results showing that the L. pneumophila ptsP gene is required for full virulence in guinea pigs (26) and clearly indicate that the PTSNtr is functional in L. pneumophila even though it lacks the EIIANtr component.
Identification of genes whose levels of expression are reduced in the ptsP deletion mutant.
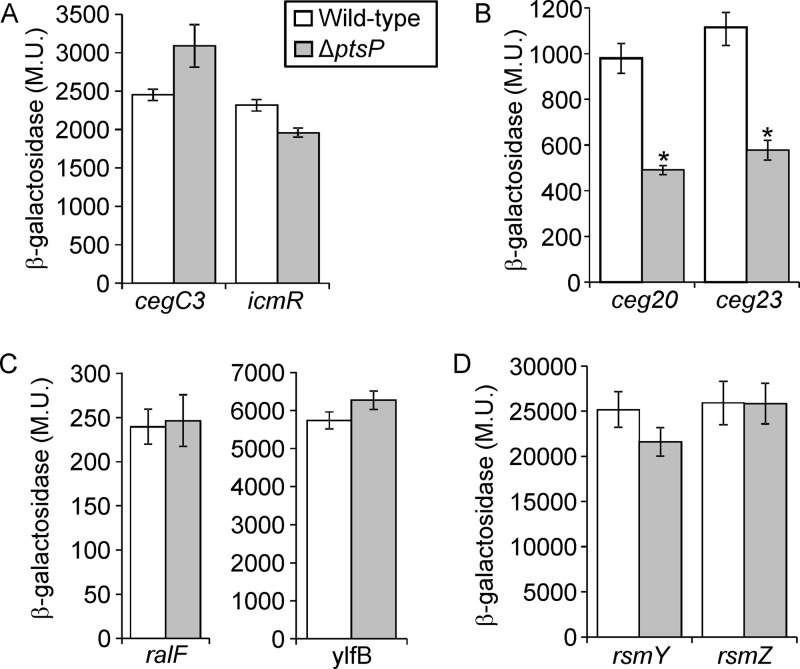
Thus far, three regulatory systems (CpxRA, PmrAB, and LetAS) (see introduction) which are activated by phosphorylation were shown to be involved in the expression of L. pneumophila virulence-related genes (30). Therefore, we decided to examine the possibility that one of these systems is regulated by the PTSNtr and leads to the intracellular growth phenotype observed. To this end, genes known to be regulated by these two-component regulatory systems were examined: (i) an effector-encoding gene (cegC3) and an Icm/Dot-encoding gene (icmR) activated by the CpxRA TCS (7), (ii) two effector-encoding genes (ceg20 and ceg23) activated by the PmrAB TCS (4), (iii) two effector-encoding genes (ralF and ylfB) regulated by the LetAS-RsmYZ-CsrA regulatory cascade (12, 13), and (iv) two genes encoding small RNAs (sRNAs) (rsmY and rsmZ) which are directly activated by the LetAS TCS (13, 14). The examination of these eight genes revealed that only the expression levels of the PmrA-regulated genes were reduced in the ptsP deletion mutant (Fig. 4), indicating that the PTSNtr specifically affects the levels of expression of PmrA-regulated genes.
FIG 4.
The absence of PtsP affects the expression levels of PmrA-regulated genes. The expression of effector translational lacZ fusions regulated by CpxR (A), PmrA (B), and CsrA (C) as well as sRNA transcriptional lacZ fusions regulated by LetA (D) (the genes examined are indicated below the bars) were examined in the wild-type strain (JR32) and in the ptsP deletion mutant (SY-ΔptsP) at the stationary phase. β-Galactosidase activity was measured as described in Materials and Methods. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, Student's t test) in comparisons between results in the wild-type strain and those in the pmrA deletion mutant. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
The two PTSNtr components affect the levels of expression of PmrA-regulated genes.
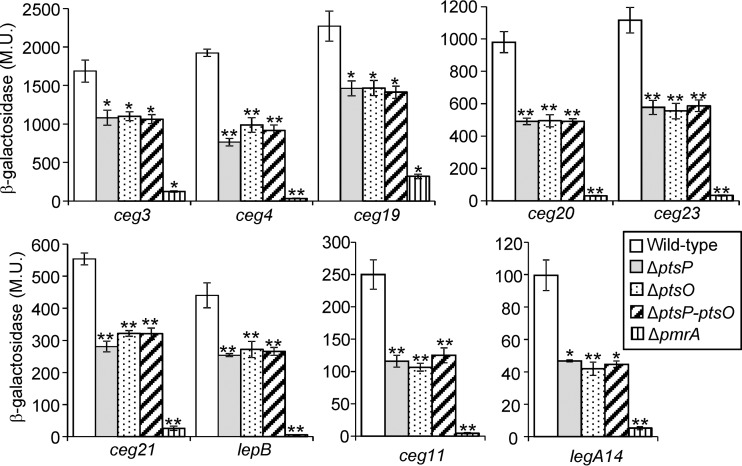
To further substantiate the result described above, the levels of expression of lacZ fusions of nine PmrA-regulated genes (ceg3, ceg4, ceg11, ceg19, ceg20, ceg21, ceg23, lepB, and legA14) (4, 5) were examined in the wild-type strain and in the pmrA, ptsP, and ptsO deletion mutants. The expression levels of all these genes were found to be completely dependent on PmrA, and their levels of expression were very low in the absence of PmrA (Fig. 5). Importantly, all the examined genes had reduced levels of expression in the ptsP and ptsO deletion mutants (Fig. 5). The reduction in the levels of expression of the PmrA-regulated effector-encoding genes in these two mutants was about 2-fold in comparison to their expression levels in the wild-type strain (for complementation analysis of the levels of expression using ptsP and ptsO, see below). When the levels of expression of the PmrA regulated genes were further examined in a ptsP-ptsO double deletion mutant, no additional reduction in expression levels were observed in comparison to the levels in the ptsP and ptsO single deletion mutants (Fig. 5). These results indicate that the two PTSNtr components present in L. pneumophila affect the levels of expression of PmrA-regulated genes similarly and are most likely part of a single regulatory cascade.
FIG 5.
The levels of expression of PmrA-regulated effector-encoding genes are reduced in mutants of the PTSNtr. The expression of effector translational lacZ fusions (the effectors examined are indicated below the bars) was examined in the wild-type strain (JR32), the ptsP deletion mutant (SY-ΔptsP), the ptsO deletion mutant (SY-ptsO-Km), the ptsP-ptsO double deletion mutant (SY-ΔptsP-ptsO-Km), and the pmrA deletion mutant (HK-PQ1) at the stationary phase. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 0.001; **, P < 10−5, Student's t test) in comparisons between results in the wild-type strain and those in the different deletion mutants. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments. The effector-encoding genes were divided according to their levels of expression.
The PTSNtr components do not affect the expression of the pmrA gene.
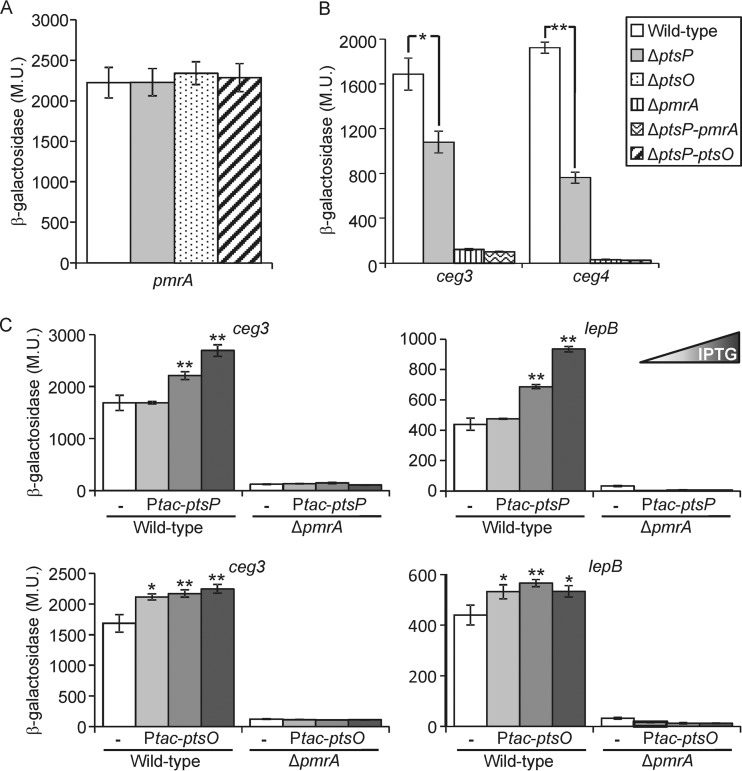
The similar effect of the ptsP, ptsO, and the double ptsP-ptsO deletion mutants on the levels of expression of PmrA-regulated genes (Fig. 5) might result from the PTSNtr affecting (i) the level of expression of the pmrA gene itself or (ii) the function of the PmrAB TCS. To distinguish between these possibilities, we examined the level of expression of the pmrA gene in the ptsP and ptsO single deletion mutants and in the ptsP-ptsO double deletion mutant and found that the pmrA gene is similarly expressed in these three mutants and in the wild-type strain (Fig. 6A). The lack of effect on the level of expression of the pmrA gene in the deletion mutant examined indicates that the PTSNtr most likely affects the functionality of the PmrA regulator since it had no effect on its level of expression.
FIG 6.
The PTSNtr affects the levels of expression of PmrA-regulated genes via PmrA. (A) The expression of the PmrA translational lacZ fusion was examined in the wild-type strain (JR32), in the ptsP deletion mutant (SY-ΔptsP), in the ptsO deletion mutant (SY-ptsO-Km), and in the ptsP-ptsO double deletion mutant (SY-ΔptsP-ptsO-Km) at the stationary phase. (B) The expression of effector translational lacZ fusions (the effectors examined are indicated below the bars) was examined in the wild-type strain (JR32), in the ptsP deletion mutant (SY-ΔptsP), in the pmrA deletion mutant (HK-PQ1), and in the ptsP-pmrA double-deletion mutant (SY-ΔptsP-pmrA-Km) at the stationary phase. The levels of expression of the marked lacZ fusions were found to be significantly different (*, P < 0.01; **, P < 0.001, Student's t test). For panels A and B, strains are identified according to the legend on the figure. (C) The levels of expression of two PmrA-regulated effector-encoding genes (ceg3 and lepB) were examined in the L. pneumophila wild-type strain (JR32) and the pmrA deletion mutant (HK-PQ1). The bacteria contained a plasmid with the ptsP gene (two upper graphs) or the ptsO gene (two lower graphs) cloned under the control of the IPTG-inducible Ptac promoter. The plasmids containing the corresponding lacZ fusion of the examined gene without the ptsP or ptsO gene were used as controls (white bars). The IPTG concentrations used to express the wild-type and mutated ptsP were 0.005, 0.01, and 0.05 mM, and they were 0, 0.005, and 0.01 mM for ptsO. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 0.01; **, P < 0.001, Student's t test), in comparisons between results with the fusions with the ptsP or ptsO gene expressed from the Ptac promoter and those with the corresponding lacZ fusions without the ptsP and ptsO genes. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
The PTSNtr components affect the levels of expression of the PmrA-regulated effector-encoding genes via PmrA.
To examine if the PTSNtr functions via PmrA, a double deletion mutant of ptsP and pmrA was constructed. Because the deletion of the pmrA gene leads to very low levels of expression of most of its target genes (Fig. 5), we chose the ceg3 and ceg4 lacZ fusions for this analysis since their levels of expression, in the absence of the PmrA activator, would still make it possible to observe an additional reduction in their levels of expression in the ptsP-pmrA double deletion mutant. Examination of the ceg3 and ceg4 lacZ fusions in the wild-type strain, in the ptsP and pmrA single deletion mutants, and in the ptsP-pmrA double deletion mutant resulted in no additive reduction in the levels of expression of these two genes in the double deletion mutant (Fig. 6B), suggesting that the PTSNtr affects the level of expression of PmrA-regulated genes only in the presence of PmrA.
To further substantiate the requirement of PmrA in order to observe the effect of the PTSNtr on the levels of expression of PmrA-regulated effector-encoding genes, we constructed a system in which the EINtr and NPr were expressed under the Ptac promoter (activated by IPTG), and their ability to affect the levels of expression of PmrA-regulated genes was examined. Overexpression of EINtr and NPr from a Ptac promoter resulted in an increase in the levels of expression of ceg3 and lepB lacZ fusions as the concentrations of IPTG increased (Fig. 6C), indicating that when the components of the PTSNtr are overexpressed, they can increase the levels of expression of the PmrA-regulated genes above the level found in the wild-type strain. However, in the absence of PmrA, overexpression of EINtr and NPr did not result in an increase in the levels of expression of ceg3 and lepB (Fig. 6C).
Collectively, these results clearly show that the PTSNtr requires the presence of PmrA in order to exert its effect on the levels of expression of PmrA-regulated effector-encoding genes.
The PTSNtr components affect the levels of expression of PmrA-regulated genes in the absence of PmrB.
The requirement of the PmrA activator in order for the PTSNtr components to affect the expression levels of the PmrA-regulated effector-encoding genes suggests that this effect might be mediated also via the PmrA cognate SHK PmrB. To this end, a pmrB single deletion mutant and a pmrB-ptsP double deletion mutant were constructed. If the PTSNtr components function via PmrB, the expected result would be that in the absence of PmrB the ptsP deletion would not affect the expression levels of the PmrA-regulated genes. In contrast, if the PTSNtr components function independently from PmrB, the expected result would be that in the absence of PmrB, the ptsP deletion would continue to affect the levels of expression of the PmrA-regulated genes in a way similar to that of the wild-type strain.
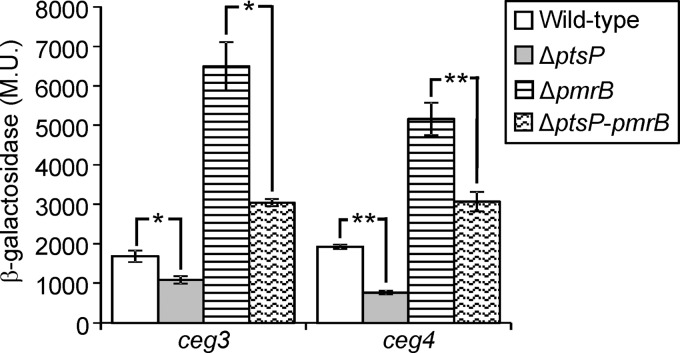
To examine this, we determined the levels of expression of two PmrA-regulated genes (ceg3 and ceg4) in the pmrB deletion mutant and in the ptsP-pmrB double deletion mutant. Examination of the levels of expression of PmrA-regulated genes in a deletion mutant of pmrB revealed that these genes are expressed at higher levels in the absence of PmrB than in the wild-type strain (Fig. 7 and S3). Previous studies have found that removal of the PmrB SHK results in a higher level of expression due to the absence of the phosphatase activity usually elicited by PmrB (31). Importantly, examination of the levels of expression of the ceg3 and ceg4 lacZ fusions in the ptsP-pmrB double deletion mutant showed that their expression levels were reduced in comparison to their levels in the pmrB deletion mutant (Fig. 7), indicating that the effect of PtsP is independent of the presence of PmrB in the bacterial cell. In addition, similar reductions were observed between the pmrB deletion mutant and the pmrB-ptsP double deletion mutant and between the wild-type strain and the ptsP deletion mutant (Fig. 7).
FIG 7.
The PTSNtr affects the levels of expression of PmrA-regulated genes and is independent of PmrB. The expression of effector translational lacZ fusions (the effectors examined are indicated below the bars) was examined in the wild-type strain (JR32), in the ptsP deletion mutant (SY-ΔptsP), in the pmrB deletion mutant (EA-pmrB), and in the ptsP-pmrB double deletion mutant (SY-ΔptsP-pmrB-Km). The levels of expression of the marked lacZ fusions were found to be significantly different (*, P < 0.001; **, P < 10−5, Student's t test). β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
The similar reductions in the levels of expression of PmrA-regulated genes mediated by the deletion of ptsP in the presence or absence of PmrB indicate that the PTSNtr affects the levels of expression of these genes independently from PmrB.
Both EINtr and NPr require their conserved phosphorylation sites in order to function.
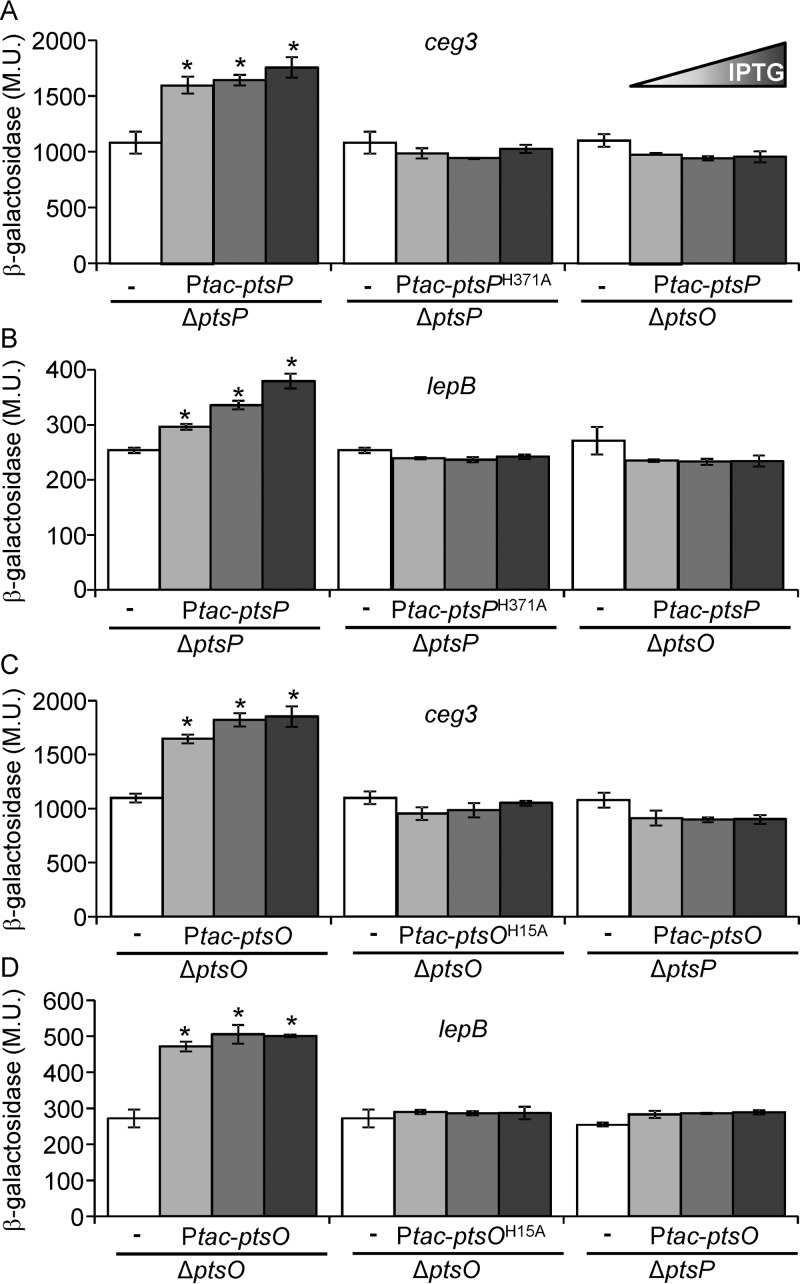
Components of the PTSNtr are highly conserved throughout bacterial species, and the histidine residues used for phosphorylation are universally conserved in all of them (19). We therefore constructed mutant versions of the L. pneumophila EINtr and NPr in which the highly conserved histidine residues of both proteins were replaced with alanines (EINtr-H371A and NPrH15A, respectively). These mutants were examined for their ability to activate the expression of the PmrA-regulated genes. Overexpression of the wild-type EINtr (encoded by ptsP) and NPr (encoded by ptsO) from a Ptac promoter in the ptsP and ptsO deletion mutants, respectively, resulted in an increase in the levels of expression of ceg3 and lepB lacZ fusions as the concentrations of IPTG increased (Fig. 8). However, when the mutated EINtr-H371A or NPrH15A was expressed in the same mutants, under the same conditions, no increase in the expression levels of these fusions was observed. Moreover, when the mutated EINtr-H371A and NPrH15A were used for complementation of the ptsP and ptsO deletion mutants for intracellular growth in amoeba, no complementation was observed (Fig. 3).
FIG 8.
The conserved histidine residues of EINtr and NPr are required for their function. The levels of expression of two PmrA-regulated effector-encoding genes (ceg3 and lepB) were examined in the L. pneumophila ptsP deletion mutant (SY-ΔptsP) and the ptsO deletion mutant (SY-ΔptsO-Km). The bacteria contained a plasmid carrying the wild-type ptsP gene, the wild-type ptsO gene, the mutated ptsPH371A gene, or the mutated ptsOH15A gene cloned under the control of the IPTG inducible Ptac promoter. The plasmids containing the corresponding lacZ fusion of the examined gene without the ptsP or ptsO gene were used as controls (white bars). The IPTG concentrations used to express the wild-type and mutated ptsP genes were 0.005, 0.01, and 0.05 mM, and they were 0, 0.005, and 0.01 mM for ptsO. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 0.001, Student's t test) in comparisons between results for the lacZ fusions containing the ptsP or ptsO gene expressed from the Ptac promoter and those for the fusions without the ptsP and ptsO genes. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
Collectively, the observation that the conserved histidine residues of the L. pneumophila EINtr and NPr are required for their function suggests that their effect on PmrA-regulated genes is mediated by phosphorylation.
Both EINtr and NPr require the presence of each other in order to function.
To determine if EINtr and NPr require one another in order to affect the levels of expression of PmrA regulated effector-encoding genes, we examined whether the wild-type EINtr protein can increase the expression levels of PmrA-regulated genes in the absence of NPr and vice versa. Expression of the wild-type EINtr (encoded by ptsP) and NPr (encoded by ptsO) proteins from a Ptac promoter in the ptsO and ptsP deletion mutants, respectively, resulted in no increase in the expression levels of ceg3 and lepB lacZ fusions as opposed to the increase in the levels of expression that was obtained when each of them was expressed in its corresponding deletion mutant (Fig. 8, compare the left and right bar groups). These results indicate that EINtr and NPr can affect the expression levels of PmrA-regulated genes only when both components are present in the bacterial cell.
The expression of the L. micdadei ptsN in L. pneumophila abolishes the effect of the PTSNtr on PmrA-regulated genes.
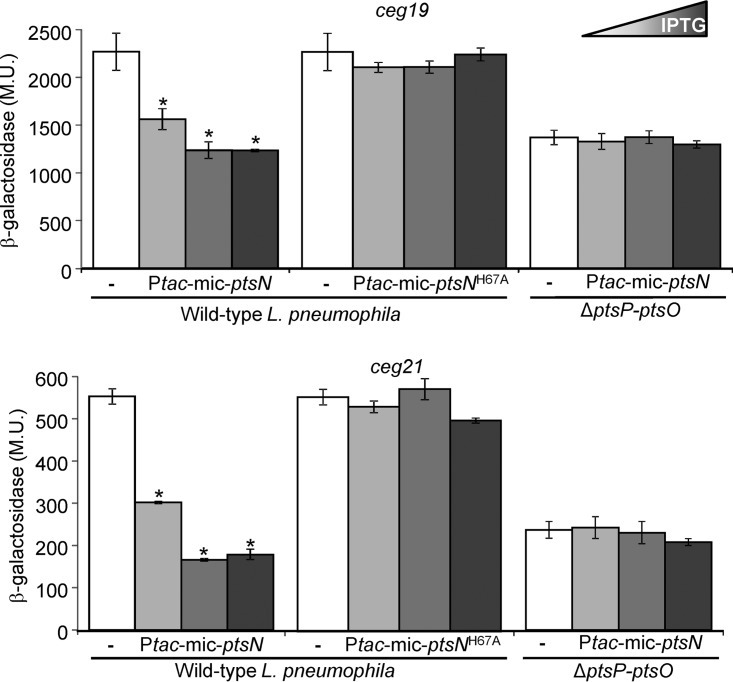
As described above (Fig. 2), about 20 Legionella species belonging to the L. micdadei clade (27) contain a ptsN gene encoding the EIIANtr component, which is missing in the L. pneumophila clade. To further explore the function of the L. pneumophila incomplete PTSNtr, we examined the effect of expressing the L. micdadei ptsN gene on the expression levels of PmrA-regulated genes in L. pneumophila. To this end, we cloned and expressed the L. micdadei EIIANtr (encoded by ptsN) under the control of the Ptac promoter (activated by IPTG) in L. pneumophila and examined its effect on the levels of expression of two PmrA-regulated effector-encoding genes, ceg19 and ceg21. The expression of the L. micdadei ptsN gene in L. pneumophila resulted in a reduction in the levels of expression of both the ceg19 and ceg21 lacZ fusions, and the reduction strengthened as the concentration of the added IPTG increased (Fig. 9). At the highest IPTG concentration (1 mM), the expression levels of both genes were reduced to about half of their levels of expression in the absence of the L. micdadei ptsN. The extent of this reduction in the expression is similar to the reduction obtained when these two genes were examined in the ptsP and ptsO single deletion mutants or the ptsP-ptsO double deletion mutant (compare Fig. 5 and 9). This result suggests that the overexpression of L. micdadei ptsN completely abolished the effect of the L. pneumophila PTSNtr on the PmrA-regulated effector-encoding genes. To further support the conclusion that the effect of the L. micdadei ptsN resulted from its natural function encoding a protein that is phosphorylated by NPr, we constructed a mutated form of the L. micdadei ptsN in which the conserved histidine residue, which is phosphorylated by NPr, was mutated to alanine (EIIANtr-H67A). Overexpression of this mutated form of ptsN had no effect on the levels of expression of the PmrA-regulated effector-encoding genes ceg19 and ceg21 (Fig. 9). Moreover, when the L. micdadei ptsN was expressed in the ptsP-ptsO double deletion mutant, no reduction in the levels of expression of the ceg19 and ceg21 lacZ fusions was observed, indicating that the L. pneumophila PTSNtr components are required in order for the L. micdadei ptsN to affect the expression levels of PmrA-regulated genes in L. pneumophila.
FIG 9.
Overexpression of the L. micdadei ptsN abolishes the effect of PTSNtr on PmrA-regulated genes in L. pneumophila. The levels of expression of two PmrA-regulated effector-encoding genes (ceg19 and ceg21) were examined in the L. pneumophila wild-type strain (JR32) or the ptsP-ptsO double deletion mutant (SY-ΔptsP-ptsO-Km). The bacteria contained a plasmid with the L. micdadei ptsN gene (Ptac-mic-ptsN) or the mutated L. micdadei ptsNH67A gene (Ptac-mic-ptsNH67A) cloned under the control of the IPTG-inducible Ptac promoter. The plasmids containing the corresponding lacZ fusions of the examined genes without the ptsN gene were used as controls (white bars). The IPTG concentrations used to express the wild-type and mutated L. micdadei ptsN genes were 0, 0.1, and 1 mM. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, Student's t test) in comparisons between results for the lacZ fusions containing the wild-type L. micdadei ptsN gene expressed from the Ptac promoter and those for the fusions without the ptsN gene. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results of at least three different experiments.
Collectively, the effect of the wild-type L. micdadei ptsN and the lack of effect of the mutated L. micdadei ptsN on the levels of expression of the PmrA-regulated genes in L. pneumophila indicate that the L. micdadei EIIANtr functions as a phosphate sink for the PTSNtr in L. pneumophila. This result further supports the possibility that the L. pneumophila NPr transfers its phosphate directly or indirectly to PmrA.
EINtr and NPr are phosphorylated in vitro by PEP.
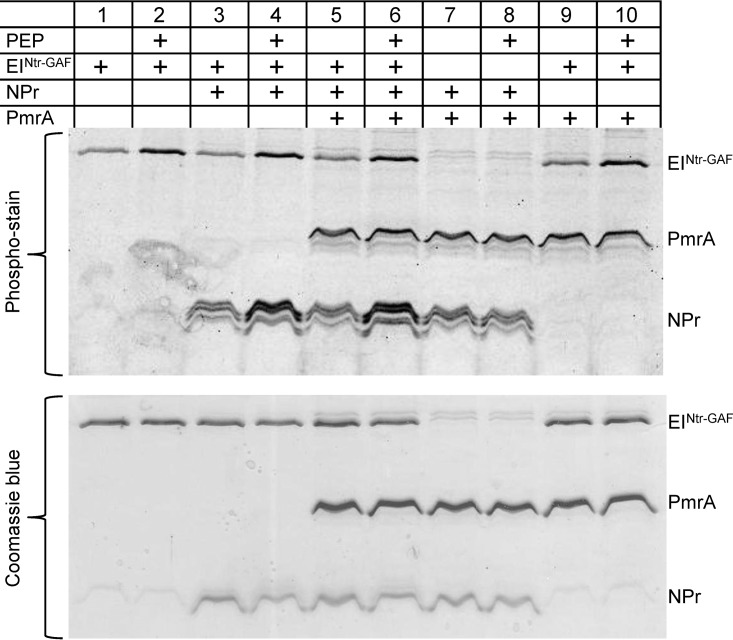
To determine if the PTSNtr phosphorylation cascade can be recapitulated in vitro, the EINtr, NPr, and PmrA proteins were His tagged, overexpressed, purified, and used for an in vitro phosphorylation assay using PEP as a phosphate donor (Fig. 10). The results obtained show that the phosphorylation level of EINtr was increased 3- to 4-fold following incubation with PEP (Fig. 10, compare lanes 1 and 2, lanes 3 and 4, lanes 5 and 6, and lanes 9 and 10), and the phosphorylation level of NPr was increased similarly following incubation with PEP and EINtr (Fig. 10, compare lanes 3 and 4 and lanes 5 and 6). However, no increase in the phosphorylation level of NPr was observed in the absence of EINtr with or without PEP (compare lanes 7 and 8). When EINtr and NPr were incubated together with PmrA, a very slight increase (1.2-fold) in the PmrA levels of phosphorylation was observed in the presence of PEP (Fig. 10, compare lanes 5 and 6); this slight increase in phosphorylation was not observed when the same reactions were performed without NPr (compare lanes 9 and 10). The slight increase in the phosphorylation of PmrA might occur since there is another component present in vivo which functions between NPr and PmrA, or the slight increase may arise because an unknown component is required in order to stabilize a direct phosphotransfer between NPr and PmrA or because the conditions in vitro do not allow the phosphotransfer from NPr to PmrA to take place in an efficient manner. Attempts to change the concentrations of the proteins as well as the PEP concentration did not result in phosphorylation of PmrA. In addition, PmrA was phosphorylated in vitro by acetyl-phosphate, but no reverse phosphorylation from PmrA to NPr or EINtr was observed (data not shown).
FIG 10.
EINtr and NPr are phosphorylated by PEP. Results of an in vitro phosphorylation assay of EINtr, NPr, and PmrA are shown. Wild-type His6-tagged EINtr without its GAF domain (His6-EINtrΔGAF), NPr-His6, and His6-PmrA were overexpressed and purified. Different combinations of the purified proteins were incubated with or without PEP and separated on 15% SDS-polyacrylamide gels, as described in Materials and Methods. Phosphorylated proteins were specifically detected by the Pro-Q Diamond phosphoprotein gel stain technique, while total protein was visualized by Coomassie blue.
The results obtained demonstrate that the L. pneumophila PTSNtr is functional. EINtr is phosphorylated by PEP and can transfer its phosphate group to NPr, similarly to what was previously shown in other bacterial PTSs. It is currently not possible to draw a definite conclusion about whether NPr can transfer its phosphate directly to PmrA or to another component which functions between NPr and PmrA in order to mediate the effect on the PmrA-regulated genes.
DISCUSSION
The term “phosphotransferase system” is used for a group of proteins that transfer phosphate derived from PEP from one member of the system to the next in a given order. Two general types of PTSs are known (19): the sugar PTS, which is responsible for phosphorylation and transport of sugars into the cell, and the nitrogen PTS (PTSNtr), which does not transport sugars but exerts regulatory functions. In both PTSs, the active phosphate moiety is derived from PEP and then transferred through two general phosphotransferase proteins: enzyme I (EI or EINtr in PTSNtr) and histidine protein (HPr or NPr in PTSNtr). In the sugar PTS, HPr subsequently phosphorylates the sugar-specific components enzyme IIA (EIIA) and enzyme IIB (EIIB) allowing uptake of the sugar (18). The sugar PTSs do not exclusively catalyze carbohydrate uptake but also regulate the activities of a huge number of genes and proteins in response to available carbon sources (18, 32). In the PTSNtr, NPr phosphorylates EIIANtr; however, the EIIANtr is not active in transport. The PTSNtr regulates diverse processes implicated in the metabolism of nitrogen and carbon (21), it plays a role in potassium homeostasis (22) and biofilm formation (23), and is essential for virulence in several bacteria such as S. enterica (24), B. melitensis (25), and L. pneumophila (26). In many bacteria the PTSNtr constitutes a phosphorylation cascade that works in parallel with the sugar PTS, while in other bacteria only one of the two systems exists. In addition to the three basic PTS components (EI, HPr, and EIIA), some gammaproteobacteria also contain an HPr kinase/phosphorylase (HPrK/P). HPrK/P controls the phosphorylation state of HPr at a serine residue, whereas EI phosphorylates HPr at a histidine. When phosphorylated on a serine residue, HPr mediates different functions involved in gene regulation (33).
All the components of the sugar PTS are absent in L. pneumophila; no gene encoding HPrK/P was found in this bacterium, and only the first two components of the PTSNtr are present in it (Fig. 2). Even though L. pneumophila possesses an incomplete PTSNtr, the remaining phosphorylation cascade (which includes only EINtr and NPr) is functional. We could demonstrate that EINtr becomes phosphorylated by PEP, that NPr becomes phosphorylated by EINtr (Fig. 11), and that the conserved histidine residues of both components are essential for their function (Fig. 3 and 8). The observation that inactivation of both EINtr and NPr, as well as mutations in their conserved histidine residues, similarly affected L. pneumophila intracellular growth suggests that NPr might phosphorylate or interact with a protein or an unknown regulatory factor involved in intracellular growth. Along these lines, our results demonstrate that all the PmrA-regulated genes examined are downregulated in the absence of EINtr and NPr (Fig. 5). The effect of the PTSNtr components on PmrA-regulated genes was completely dependent on the presence of PmrA (Fig. 6) and independent of the presence of PmrB (Fig. 7). In addition, the ability of EINtr and NPr to activate the expression of the PmrA-regulated effector-encoding genes was dependent on the presence of their conserved histidine residues and on the presence of both components (Fig. 8). Moreover, adding the L. micdadei EIIANtr reduced the levels of expression of PmrA-regulated effector-encoding genes to levels similar to those of the ptsP and ptsO deletion mutants (Fig. 9). Together, our results indicate that the L. pneumophila incomplete PTSNtr functions in a manner similar to that of bacteria containing a complete PTSNtr, but in L. pneumophila it was rewired to affect the activity of the PmrA response regulator which regulates the expression of the largest regulon of effector-encoding genes in L. pneumophila (more than 40 effectors).
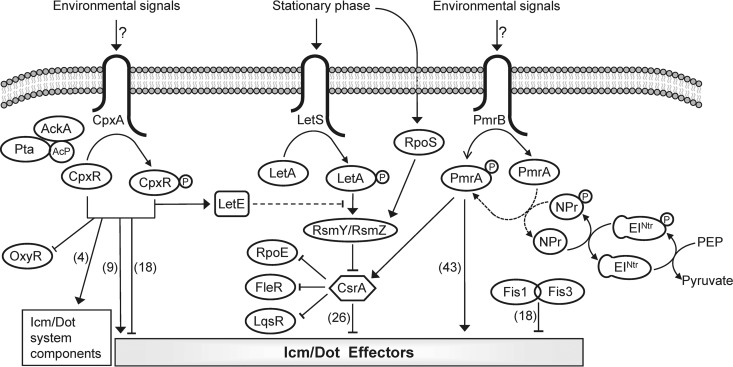
FIG 11.
Model of the regulatory systems that control the expression of the L. pneumophila pathogenesis-related genes. The three TCSs (CpxRA, PmrAB, and LetAS), the components of the LetAS-RsmYZ-CsrA regulatory cascade, and the two Fis regulators (Fis1 and Fis3) are schematically illustrated. The environmental signals sensed by CpxA and PmrB are currently not known, and the phosphorylation of these components is expected to be activated by transfer of the phosphate group to their cognate response regulators CpxR and PmrA, respectively, which then directly activate or repress the transcription of their target effector-encoding genes. Acetyl-phosphate (Ac-P) produced by AckA and Pta was previously shown to transfer a phosphate group to CpxR. The PTSNtr was shown in this study to be functional in L. pneumophila; EINtr is phosphorylated by PEP and then transfers the phosphate group to NPr. In addition, the PTSNtr affects the levels of expression of PmrA-regulated effector-encoding genes via PmrA in a direct or indirect manner. The numbers of effector-encoding genes which were shown to be regulated by each of the regulatory systems are indicated in parentheses. Solid arrows and T-shaped symbols indicate activation and repression, respectively; the dotted line indicates a possible indirect effect.
Even though we could demonstrate in vitro phosphorylation of EINtr by PEP and phosphorylation of NPr by EINtr, we did not observe significant phosphorylation of PmrA by NPr. However, all our results suggest that the phosphorylated NPr can activate the expression of PmrA-regulated genes via PmrA. There are several possible explanations of how this activation might occur: (i) PmrA is directly phosphorylated by the phosphorylated NPr (which we were unable to observe in vitro); (ii) an unknown regulatory factor is phosphorylated by the phosphorylated NPr, which in turn transfers its phosphate to PmrA; (iii) an unknown regulatory factor stabilizes the direct phosphotransfer between the phosphorylated NPr and PmrA; or (iv) a metabolite or second messenger affects the interaction/phosphotransfer of phosphorylated NPr with PmrA.
Direct involvement of the PTS and PTSNtr in regulation of gene expression was described before. For example, the SsrB RR (which is part of the SsrAB TCS) directly promotes the transcription of multiple genes within Salmonella pathogenicity island 2 (SPI-2). It was found that EIIANtr controls SPI-2 genes by acting on the SsrB protein at the posttranscriptional level. EIIANtr was found to directly interact with SsrB, preventing the SsrB protein from binding to its target promoters (24). However, since there is no EIIANtr in L. pneumophila, this is not the mechanism by which the L. pneumophila PTSNtr functions. In firmicutes, PTS regulation domains (PRDs) are found fused to transcription regulators. Transcription activators such as MltR, LicR, and LevR contain different numbers of PRDs, and they were found to become phosphorylated by EI and HPr. However, no transcription regulators containing a PRD were found in L. pneumophila, and they are very rarely found in gammaproteobacteria (18).
In addition to the two modes of involvement of PTS and PTSNtr in transcription regulation described above, direct involvement of HPr/NPr in regulation of gene expression was also described previously. In Bacillus subtilis it was found that the AraC type transcriptional activator YesS, which regulates the expression of pectin/rhamnogalacturonan utilization genes, interacts with HPr. YesS was found to interact with HPr and His-phosphorylated HPr but not with Ser-phosphorylated HPr (34). In Neisseria meningitidis HPr and Ser-phosphorylated HPr were found to interact with the LysR-type transcriptional repressor CrgA involved in capsule production in response to host cell contact (35). Our attempts to find interaction between EINtr or NPr and PmrA as well as between their mutated forms did not reveal any interaction (data not shown).
The PmrAB TCS controls the level of expression of the largest regulon of effector-encoding genes (more than 40 effectors) in L. pneumophila (4, 5) (Fig. 11). The results described in the manuscript suggest that the levels of expression of these PmrA-regulated effector-encoding genes are affected by at least three factors: (i) the level of expression of the pmrAB operon itself, which was found to be relatively high (Fig. 6A) even though no regulators which control the expression of this operon were described; (ii) the function of PmrB SHK as a kinase or phosphatase of PmrA that can activate or inactivate PmrA, respectively; and (iii) the activity of the PTS components EINtr and NPr that can directly or indirectly affect the activity of PmrA. The PmrA protein must be phosphorylated in order to activate gene expression (see Fig. S4 in the supplemental material), and PmrB seems to function as a phosphatase of PmrA under the growth conditions examined (Fig. 7); but it is highly likely that PmrB also functions as a kinase of PmrA under specific conditions. In S. enterica the PmrB SHK was also shown to function as a phosphatase of PmrA (31), but under conditions of mildly acidic pH (36) or the presence of Fe3+ (37); in vivo during infection of macrophages (38), PmrB was shown to function as a kinase of PmrA. The conditions under which the L. pneumophila PmrB SHK phosphorylates and thus activates PmrA are currently not known. The finding that the L. pneumophila PTSNtr also contributes to the activity of PmrA suggests that cytoplasmic signals activate the expression of the PmrA-regulated effector-encoding genes as well (Fig. 11). Moreover, the deletion of the L. pneumophila PTSNtr resulted in a reduction of only 2-fold in the expression of PmrA-regulated genes in comparison to a very strong reduction in their levels of expression in the pmrA deletion mutant (Fig. 5), indicating that additional components probably phosphorylate PmrA. The most notifiable difference between the EI of the sugar PTS and the EINtr is the presence of a GAF (cyclic GMP phosphodiesterase, Anabaena adenylate cyclase, and Escherichia coli FhlA) domain at the N terminus of EINtr (39). This domain was also found to be present in the N terminus of the L. pneumophila EINtr. N-terminal GAF domains are often responsible for detection of small-molecule signals, such as cyclic GMP (cGMP), formate, glutamine, and α-ketoglutarate (tricarboxylic acid [TCA] cycle intermediate) (40–42), and they frequently exert regulatory effects on adjacent catalytic domains. The putative ligands of many of the GAF domains remain unidentified, and such is the case with the L. pneumophila EINtr GAF domain. Nonetheless, the cytoplasmic/metabolic signal recognized by the L. pneumophila EINtr GAF domain together with the environmental signal sensed by the L. pneumophila PmrB SHK is expected to result in activation of the expression levels of the effector-encoding genes regulated by PmrA.
To conclude, the way by which the L. pneumophila PTSNtr activates the levels of expression of PmrA-regulated genes via PmrA is currently not known. However, our results demonstrate that the L. pneumophila incomplete PTSNtr is functional, that it is required for optimal intracellular growth, and that it mediates its effect on intracellular growth by controlling the expression levels of more than 40 effector-encoding genes regulated by PmrA.
MATERIALS AND METHODS
Bacterial strains and media.
The L. pneumophila parental strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which behaves as a wild-type strain in terms of intracellular growth (43). In addition, mutant strains derived from JR32 which contain a kanamycin (Km) cassette instead of the icmT gene (GS3011) (44), the pmrA gene (HK-PQ1) (4), the pmrB gene (EA-pmrB) (this study), and the ptsO gene (SY-ptsO-Km) (this study) and a clean deletion mutation in the ptsP gene (SY-ΔptsP) (this study) were used. In addition, three double deletion mutants were constructed, all of them containing the clean deletion in the ptsP gene together with a deletion in the pmrA gene (SY-ΔptsP-pmrA-Km), the pmrB gene (SY-ΔptsP-pmrB-Km), or the ptsO gene (SY-ΔptsP-ptsO-Km). The L. micdadei strain used in this work was ATCC 33218 (45). The E. coli strains used in this work were MC1022, MC1061 (46), BL21(DE3) (47), and SY327 λpir (48). Bacterial media, plates, and antibiotics were as previously described (49).
Plasmid construction.
To construct lacZ translational fusions, the 300-bp regulatory regions of four effector-encoding genes (ceg3, ceg4, ceg11, and legA14) and 900 bp of one regulator (pmrA) (see Data Set S1 in the supplemental material) were amplified by PCR using the primers listed in Data Set S2. The PCR products were then digested with BamHI and EcoRI, cloned into pGS-lac-02, and sequenced. These five new lacZ fusions, as well as 11 lacZ fusions that were previously constructed and used in this study, are listed in Data Set S1.
To construct IPTG-inducible ptsP and ptsO genes, the L. pneumophila ptsP and ptsO genes were amplified by PCR using the primers listed in Data Set S2. The PCR products were then digested with NdeI and BamHI for ptsP and EcoRI and BamHI for ptsO and cloned into pMMB207 downstream of the Ptac promoter to generate pSY-207-Ptac-ptsP and pSY-207-Ptac-ptsO, respectively. In addition, the ptsP and ptsO genes were mutated by PCR to encode substitutions of alanines for the conserved histidines at position 371 (ptsPH371A) for EINtr (EINtr-H371A) and at position 15 (ptsOH15A) for NPr (NPrH15A) using the primers listed in Data Set S2 and cloned into pMMB207 downstream of the Ptac promoter to generate pSY-207-Ptac-ptsP-H371A and pSY-207-Ptac-ptsO-H15A, respectively (Data Set S1). The resulting four plasmids were then digested with PstI and EheI, and the resulting fragment, containing Ptac-ptsP, Ptac-ptsO, Ptac-ptsPH371A, or Ptac-ptsOH15A together with the lacI gene, was cloned into plasmids containing the lacZ fusions of the ceg3 and lepB genes digested with PstI and XmnI, resulting in the plasmids listed in Data Set S1.
To construct IPTG-inducible wild-type and mutated L. micdadei ptsN genes, the ptsN gene was amplified by PCR, using the primers listed in Data Set S2, and cloned into pMMB207 downstream of the Ptac promoter to generate pZT-207-mic-ptsN (Data Set S1). In addition, the ptsN gene was mutated by PCR to encode a substitution of alanine for the conserved histidine residue at position 67 (ptsNH67A), using the primers listed in Data Set S2, and cloned into pMMB207 downstream of the Ptac promoter to generate pZT-207-mic-ptsNmut (Data Set S1). These two plasmids were then digested with PstI and EheI, and the resulting fragment, containing either Ptac-ptsN together with the lacI gene or Ptac-ptsNH67A together with the lacI gene, was cloned into plasmids containing the lacZ fusions of the ceg19 and ceg21 genes digested with PstI and XmnI, resulting in the plasmids listed in Data Set S1.
To construct a deletion substitution in the L. pneumophila ptsO and pmrB genes, a 1-kb DNA fragment located on each side of the planned deletions was amplified by PCR using the primers listed in Data Set S2. The primers were designed to contain an SalI site at the place of the deletion. The four fragments that were amplified were cloned into pUC-18 digested with suitable enzymes, and the inserts were sequenced to generate the plasmids listed in Data Set S1. The resulting plasmids were digested with suitable enzymes, and the inserts were used for a four-way ligation containing the Km resistance cassette (Pharmacia) digested with SalI and the pUC-18 vector digested with EcoRI and BamHI. The correct plasmids were identified by restriction digests. The generated plasmids (Data Set S1) were digested with PvuII, which cuts on both sides of the pUC-18 polylinker, and the resulting fragments were cloned into the pLAW344 allelic exchange vector digested with EcoRV to generate the plasmids pEA-pmrB-Km-GR and pSY-ptsO-Km-GR that were used for allelic exchange, as previously described (49).
To construct a nonpolar in-frame deletion mutation in the L. pneumophila ptsP gene, a 1-kb DNA fragment located on each side of the planned deletion was amplified by PCR using the primers listed in Data Set S2. The primers were designed to contain an SalI site at the place of the deletion. The two fragments that were amplified were cloned into pUC-18 digested with suitable enzymes, and the inserts were sequenced to generate the plasmids listed in Data Set S1. The resulting plasmids were digested with suitable enzymes, and the inserts were used for a four-way ligation containing the Km resistance cassette (Pharmacia) digested with SalI and the pUC-18 vector digested with EcoRI and BamHI. The generated plasmid (Data Set S1) was digested with PvuII, and the resulting fragment was cloned into the pSY100 allelic exchange vector digested with EcoRV to generate the plasmid pSY100-ptsP-Km, which was then digested with SalI and self-ligated to generate pSY100-ptsP-GR, which was used for allelic exchange, as previously described (44). To construct double deletion ptsP-ptsO, ptsP-pmrA, and ptsP-pmrB mutants, the ptsP clean deletion mutant was used instead of the wild-type strain.
To overexpress the NPr, EINtr, and the EINtr protein without its GAF domain (EINtrΔGAF) for in vitro phosphorylation assays, a fragment containing the L. pneumophila ptsO or ptsP gene or the ptsP gene encoding a deletion of the GAF domain (ptsPΔGAF) was amplified by PCR using the primers listed in Data Set S2. The resulting fragments were digested with NdeI and BamHI and cloned into the pET-21a vector for ptsO and into pET-15b for ptsP and ptsPΔGAF digested with the same enzymes to generate the plasmids listed in Data Set S1. The resulting plasmids express the full-length NPr fused to a His6 tag on the C terminus, the full-length EINtr fused to a His6 tag on the N terminus, and the EINtrΔGAF protein fused to a His6 tag on the N terminus. The plasmid pZT-His-lpn-pmrA (Data Set S1) was used to overexpress the His-tagged PmrA protein.
Growth analysis.
Intracellular growth assays of L. pneumophila mutants were performed in Acanthamoeba castellanii (ATCC 30234) and HL-60-derived human macrophages (ATCC CCL-240), as previously described (28). In vitro growth assays were performed in AYE growth medium [N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract]. Bacteria grown on plates were scraped and resuspended to an optical density at 600 nm (OD600) of 0.10 for time zero measurement, and OD600 measurements were conducted in a Synergy HT2011 plate reader in triplicate.
β-Galactosidase assay.
The levels of expression of the individual lacZ fusions were examined in the L. pneumophila wild-type and mutant strains at stationary phase, as previously described (4).
In vitro phosphorylation assay.
L. pneumophila His6-PmrA, His6-EINtr, His6-EINtrΔGAF, and NPr-His6 were purified from E. coli BL21(DE3) containing the plasmid pZT-His-lpn-pmrA (4), pZT-His-ptsP, pZT-His-ptsP-GAF, or pZT-His-ptsO, respectively. The four proteins were purified by nickel-affinity chromatography using Ni-nitrilotriacetic acid (Ni-NTA) resin (Qiagen). Protein purification was performed as previously described (4, 8). In vitro phosphorylation was carried out by incubating 10 mM PEP (Sigma) with different combinations of purified 4 μM His6-PmrA, 1 μM His6-EINtrΔGAF, and 10 μM NPr-His6 at 37°C for 20 min in a buffer containing 100 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM EDTA, 5 mM KCl, and 0.5 mM dithiothreitol (DTT). The results presented were obtained with His6-EINtrΔGAF, and similar results were obtained also with the full-length His6-EINtr as well as after treatment of the PmrA protein with a phosphatase (data not shown). Samples were separated on 15% SDS-polyacrylamide gels, and phosphorylated proteins were detected by a Pro-Q Diamond phosphoprotein gel stain (Invitrogen), according to the manufacturer's instructions. Bands were quantified by densitometry of the autoradiograms using ImageQuant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Efrat Altman for plasmids and strain construction and Tal Pupko and David Burstein for their help with statistical analysis.
This research was supported by Israeli Science Foundation grant 877/15 (to G.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00121-17.
REFERENCES
- 1.Segal G, Feldman M, Zusman T. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev 29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 4.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 5.Al-Khodor S, Kalachikov S, Morozova I, Price CT, Abu Kwaik Y. 2009. The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect Immun 77:374–786. doi: 10.1128/IAI.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman E, Segal G. 2008. The response regulator CpxR directly regulates the expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol 190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldheim YS, Zusman T, Speiser Y, Segal G. 2016. The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR's function as a dual regulator and its connection to the effectors regulatory network. Mol Microbiol 99:1059–1079. doi: 10.1111/mmi.13290. [DOI] [PubMed] [Google Scholar]
- 8.Gal-Mor O, Segal G. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol 185:4908–4919. doi: 10.1128/JB.185.16.4908-4919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gal-Mor O, Segal G. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb Pathog 34:187–194. doi: 10.1016/S0882-4010(03)00027-5. [DOI] [PubMed] [Google Scholar]
- 10.Hammer BK, Tateda ES, Swanson MS. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol 44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 11.Hovel-Miner G, Pampou S, Faucher SP, Clarke M, Morozova I, Morozov P, Russo JJ, Shuman HA, Kalachikov S. 2009. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol 191:2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol 196:681–692. doi: 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol 72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 14.Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi C, Forsbach-Birk V, Marre R, McNealy TL. 2006. The Legionella pneumophila global regulatory protein LetA affects DotA and Mip. Int J Med Microbiol 296:15–24. doi: 10.1016/j.ijmm.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Schell U, Simon S, Sahr T, Hager D, Albers MF, Kessler A, Fahrnbauer F, Trauner D, Hedberg C, Buchrieser C, Hilbi H. 2016. The alpha-hydroxyketone LAI-1 regulates motility, Lqs-dependent phosphorylation signalling and gene expression of Legionella pneumophila. Mol Microbiol 99:778–793. doi: 10.1111/mmi.13265. [DOI] [PubMed] [Google Scholar]
- 17.Gao R, Stock AM. 2010. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol 13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deutscher J, Ake FM, Derkaoui M, Zebre AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterkofsky A, Wang G, Seok YJ. 2006. Parallel PTS systems. Arch Biochem Biophys 453:101–107. doi: 10.1016/j.abb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Pfluger-Grau K, Gorke B. 2010. Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol 18:205–214. doi: 10.1016/j.tim.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Powell BS, Court DL, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier MH Jr, Reizer J. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J Biol Chem 270:4822–4839. [DOI] [PubMed] [Google Scholar]
- 22.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci U S A 104:4124–4129. doi: 10.1073/pnas.0609897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabeen MT, Leiman SA, Losick R. 2016. Colony-morphology screening uncovers a role for the Pseudomonas aeruginosa nitrogen-related phosphotransferase system in biofilm formation. Mol Microbiol 99:557–570. doi: 10.1111/mmi.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J, Shin D, Yoon H, Kim J, Lee CR, Kim M, Seok YJ, Ryu S. 2010. Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc Natl Acad Sci U S A 107:20506–20511. doi: 10.1073/pnas.1000759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dozot M, Poncet S, Nicolas C, Copin R, Bouraoui H, Maze A, Deutscher J, De Bolle X, Letesson JJ. 2010. Functional characterization of the incomplete phosphotransferase system (PTS) of the intracellular pathogen Brucella melitensis. PLoS One 5:e12679. doi: 10.1371/journal.pone.0012679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higa F, Edelstein PH. 2001. Potential virulence role of the Legionella pneumophila ptsP ortholog. Infect Immun 69:4782–4789. doi: 10.1128/IAI.69.8.4782-4789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA, Pupko T, Shuman HA, Segal G. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal G, Shuman HA. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67:2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindre T, Bruggemann H, Buchrieser C, Hechard Y. 2008. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154:30–41. doi: 10.1099/mic.0.2007/008698-0. [DOI] [PubMed] [Google Scholar]
- 30.Segal G. 2013. The Legionella pneumophila two-component regulatory systems that participate in the regulation of Icm/Dot effectors. Curr Top Microbiol Immunol 376:35–52. doi: 10.1007/82_2013_346. [DOI] [PubMed] [Google Scholar]
- 31.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nessler S. 2005. The bacterial HPr kinase/phosphorylase: a new type of Ser/Thr kinase as antimicrobial target. Biochim Biophys Acta 1754:126–131. doi: 10.1016/j.bbapap.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 34.Poncet S, Soret M, Mervelet P, Deutscher J, Noirot P. 2009. Transcriptional activator YesS is stimulated by histidine-phosphorylated HPr of the Bacillus subtilis phosphotransferase system. J Biol Chem 284:28188–28197. doi: 10.1074/jbc.M109.046334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derkaoui M, Antunes A, Poncet S, Nait Abdallah J, Joyet P, Maze A, Henry C, Taha MK, Deutscher J, Deghmane AE. 2016. The phosphocarrier protein HPr of Neisseria meningitidis interacts with the transcription regulator CrgA and its deletion affects capsule production, cell adhesion, and virulence. Mol Microbiol 100:788–807. doi: 10.1111/mmi.13349. [DOI] [PubMed] [Google Scholar]
- 36.Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol 63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125. doi: 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 38.Merighi M, Ellermeier CD, Slauch JM, Gunn JS. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J Bacteriol 187:7407–7416. doi: 10.1128/JB.187.21.7407-7416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anantharaman V, Koonin EV, Aravind L. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol 307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 40.Lee CR, Park YH, Kim M, Kim YR, Park S, Peterkofsky A, Seok YJ. 2013. Reciprocal regulation of the autophosphorylation of enzyme INtr by glutamine and α-ketoglutarate in Escherichia coli. Mol Microbiol 88:473–485. doi: 10.1111/mmi.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Argudo I, Little R, Dixon R. 2004. Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Mol Microbiol 52:1731–1744. doi: 10.1111/j.1365-2958.2004.04089.x. [DOI] [PubMed] [Google Scholar]
- 42.Ho YS, Burden LM, Hurley JH. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J 19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zusman T, Yerushalmi G, Segal G. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 71:3714–3723. doi: 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers RM, Brown A, Garrity GM. 1981. Dye-containing buffered charcoal-yeast extract medium for differentiation of members of the family Legionellaceae. J Clin Microbiol 13:380–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol 138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 47.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 48.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59:4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segal G, Shuman HA. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun 65:5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.