Abstract
Adenylate kinases (AKs) play important roles in nucleotide metabolism in all organisms and in cellular energetics by means of phosphotransfer networks in eukaryotes. The crystal structure of a human AK named AK6 was determined by in-house sulfur single-wavelength anomalous dispersion phasing methods and refined to 2.0-Å resolution with a free R factor of 21.8%. Sequence analyses revealed that human AK6 belongs to a distinct subfamily of AKs present in all eukaryotic organisms sequenced so far. Enzymatic assays show that human AK6 has properties similar with other AKs, particularly with AK5. Fluorescence microscopy showed that human AK6 is localized predominantly to the nucleus of HeLa cells. The identification of a nuclear-localized AK sheds light on nucleotide metabolism in the nucleus and the energetic communication between mitochondria and nucleus by means of phosphotransfer networks.
Keywords: x-ray crystallography, nuclear localization, nucleotide metabolism, phosphotransfer networks
Nucleoside monophosphate kinases (NMPKs), which phosphorylate nucleoside monophosphates or deoxynucleoside monophosphates, which produce nucleoside diphosphates or deoxynucleoside diphosphates, play important roles in the maintenance of intracellular nucleotide pools in all organisms. Adenylate kinases (AKs) (ATP:AMP phophotransferases, EC 2.7.4.3) catalyze the reversible transfer of the γ-phosphate group from a phosphate donor (normally ATP) to AMP, releasing two molecules of ADP (1). Besides crucial roles in homeostasis of adenine nucleotide metabolism, AKs are involved in cellular energetics through complex phosphotransfer networks regulating intracellular ATP-producing processes (2, 3).
At present, five AK isoforms with different subcellular localization and substrate specificity have been characterized in mammalian tissues (4). In this work, we have identified a sixth AK isoform through a characterization of the adrenal gland protein AD-004 gene as part of a structural genomics project involving human genes (5).
The AD-004 gene was first identified in a gene expression profiling study of the human hypothalamus-pituitary-adrenal axis (6) and has also appeared in analyses of genomic and cDNA sequences (7, 8). The putative protein sequence for AD-004 contains 172 residues with an expected molecular mass of 20 kDa. AD-004 shows low overall sequence identity with proteins of known structures; however, in its N-terminal regions are sequence motifs [most notably a Walker motif (9)] that are characteristic of NMPKs. Through the work presented here, including crystal structure determination, enzymatic assays, and subcellular localization experiments, we show that AD-004 constitutes a AKs isoform that has been named human AK6, with many characteristics that distinguish it from the other eukaryotic AKs.
Materials and Methods
Protein Purification and Crystallization. The preparation of protein used for both enzymatic assays and crystallization trials of human AD-004 (also referred to as AK6) has been described in ref. 10. Crystals of AK6 were obtained at room temperature within 2 weeks from conditions containing 1.44 M Li2SO4 in 0.1M Hepes, pH 7.5, by using the hanging-drop vapor diffusion method. The crystals belong to the space group P61 with unit cell parameters a = b = 99.56 Å and c = 57.19 Å.
Data Collection, Structure Determination, and Refinement. Diffraction data sets were collected on a Bruker Smart 6000 charge-coupled device detector mounted with a Nonius FR591 x-ray generator and Cu rotating anode. The 2.0- and 2.6-Å resolution data sets were collected from the same crystal, which was flash cooled to 100 K. An oscillation scan of 0.15° was used for both data sets. All diffraction data were indexed and integrated by using the Bruker on-line program suite proteum. Scaling and merging of the data were performed with proscale of the on-line programs.
The sulfur sites were determined by the program solve (11) with data from 20 to ≈3.0 Å. The initial phases and their figures of merit (FOM) were then input into resolve for solvent flattening and protein autotracing (12). resolve automatically traced 113 residues with correct side chains, and the remainder of the structure was built manually by using the program xtalview (13).
Refinement of the structure with cns (14) involved steps of energy minimization, simulated annealing, and individual B factor refinement. Reference and model rebuilding with 3Fo - 2Fc and Fo - Fc electron density maps were made by using the program o (15). The program procheck (16) was used to check the quality of the model.
AK Activity Assays. Radio-labeled nucleotides [3H]AMP [24 mCi/mmol (1 Ci = 37 GBq)] and [γ-32P]ATP (3,000 Ci/mmol) were purchased from Amersham Pharmacia Biosciences. Other nucleosides and nucleotides were purchased from Sigma. The AK activity of human AK6 was measured by radiochemical methods either with [3H]AMP as substrate or by using a phosphoryl transfer assay with [γ-32P]ATP as substrate (17), with modifications as described below. Standard reaction mixture contained 50 mM Tris·HCl (pH 8.0), 0.5 mg/ml BSA, 5 mM DTT, 5 mM MgCl2,[3H]AMP, and 50 ng of purified AK6 protein in a total volume of 50 μl. The reaction was started by addition of the protein, and 10-μl aliquots were withdrawn at 0-, 10-, 20-, and 30-min intervals and spotted on DE-81 filter paper and dried. The filters were washed three times with 50 mM formic acid/5 mM ammonium formate and rinsed once with water; then the products were eluted with 0.1 M HCl/0.2 M KCl, and the radioactivity was determined in a liquid scintillation counter. Enzymatic activity was calculated based on the formation of [3H]ADP in the reaction.
Phosphoryl transfer assays were performed with 0.1 mM [γ-32P]ATP as the phosphate donor and nonradioactive nucleoside monophosphate as the substrate in the buffer described above. The reaction proceeded for 20 min at 37°C and was stopped by heating at 70°C for 2 min. After a brief centrifugation, 1 μl of the supernatant was spotted on a polyethyleneimine-cellulose plate (Merck). The plates were developed in 0.2 M NaH2PO4 and autoradiographed. Nonradioactive nucleoside monophosphates, diphosphates, and triphosphates were used as standards.
Immunofluorescence Assays. An antiserum against AK6 was prepared by immunizing a rabbit with purified AK6 protein as follows. A New Zealand rabbit was immunized s.c. with 0.5 mg of recombinant protein in Freud complete adjuvant (1:1 vol/vol) at five sites. Booster injections were given at 15 and 25 days in Freund's incomplete adjuvant (1:1 vol/vol). Blood samples were taken right before the immunization as negative control and test bleeds were performed 7 days after the third immunization, and the sera were checked for its potency against AK6 by ELISA assays. The antiserum was obtained 11 days after the third booster immunization.
HeLa cells were incubated in a medium containing 10% FCS, 1% penicillin/streptomycin, and 89% DMEM on a cover glass in 35-mm dishes for 24 h before an immunofluorescence experiment. Cells were fixed with 3.7% paraformaldehyde for 30 min at room temperature, then washed with PBS and permeablized with 0.2% Triton X-100 for 5 min on ice. After being washed with PBS, the cells were incubated with the rabbit antiserum raised against AK6 (1:100 in PBS) for 1 h at 37°C. The cells were rinsed six times with PBS and incubated for 1 h more with a fluorescent-labeled secondary antibody (1:50 in PBS) (goat anti-rabbit IgG-FITC, Santa Cruz Biotechnology) at room temperature. After being mounted in Mowiol (Sigma-Aldrich), the cells were examined by using a 40× or 100× objective, and images were captured on a Zeiss Axioskop fluorescence microscope (Axiovert 2000M) coupled to a cooled charge-coupled device camera (AxioCam MRM, Zeiss) and processed by using the spot advanced program. The fluorescent DNA-intercalating dye, DAPI (1 μg/ml in Mowiol mounting medium), was used to stain nuclear DNA.
Construction of the Expression Vector pEGFP-AK6 and Transfection Assays. The cDNA of AK6 was amplified by PCR, with pET21DEST-AK6 as a template and gfp-5′ (ccggaattcatgttgcttccgaacatcc) and gfp-3′ (ccgggatccagagttatgatctttgatcc) as primers, to introduce an EcoRI site at the 5′ end and a BamHI site at the 3′ end of the AK6 gene. The resulting PCR product was digested with EcoRI and BamHI and ligated into vector pEGFP-C2 (Clontech), which had been cut with the same restriction enzymes, to generate an N-terminal GFP fusion AK6, pEGFP-AK6. A vector to produce C-terminal GFP-AK fusion protein was constructed similarly.
HeLa cells were incubated without antibiotics in 35-mm dishes 1 day before transfection. pEGFP-AK6 plasmid (6 μl) was diluted in 250 μl of DMEM and mixed gently. Meanwhile, 8 μl of Lipofectamine 2000 (Invitrogen) was diluted in 250 μl of DMEM. The diluted Lipofectamine 2000 was mixed with the plasmid and incubated at room temperature for 20 min. The medium was discarded, and the cells were incubated at 37°C in 5% CO2 for 4-6 h; the final volume was set to 2 ml with medium. Cells were then fixed with 3.7% paraformaldehyde for 30 min at room temperature and mounted in Mowiol, and GFP fluorescence from the fusion proteins was visualized with a fluorescent microscope as described above 24 h after the transfection. DAPI stain (1 μg/ml in Mowiol) was used to visualize the nucleus.
Additional Programs Used for Data Analysis and Presentation. Multiple sequence alignments and calculation of amino acid sequence identities were performed by using clustal x 1.81 (18) and visualized by alscript 2.03 (19). Phylogenetic analysis was performed by clustal x with a neighbor-joining method, and the unrooted radial tree was drawn with the treeview32 software (20). Surface electrostatics potentials were calculated, and the images were made by the program grasp (21).
Results and Discussion
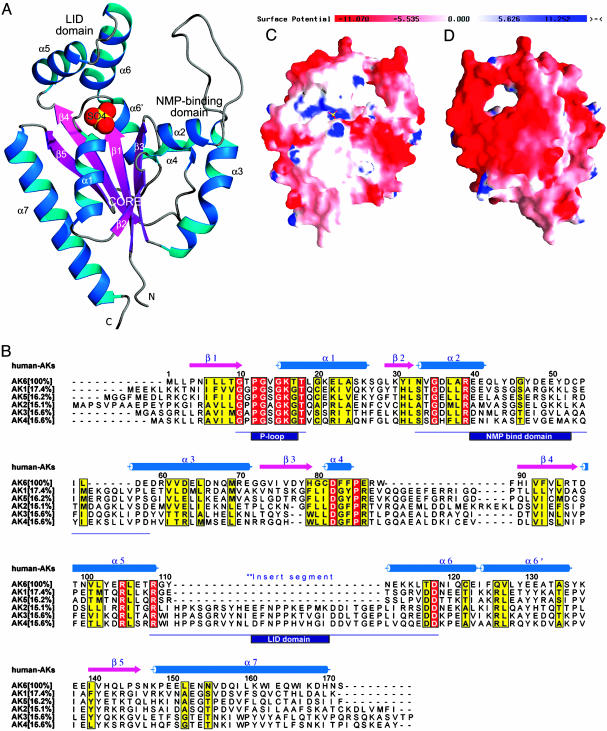
Structure Determination by In-House Sulfur Single-Wavelength Anomalous Dispersion (SAD) Methods. To solve the crystal structure of human AK6 (the name AD-004 will also be used as appropriate during the discussion of enzymatic activity), the contribution of the SAD from sulfur atoms to the observed intensities was measured to obtain initial phase information. Data were collected in-house on one single crystal, first to 2.0 Å, which were used for structure refinement, and then to 2.6 Å, which were used for SAD phasing (Table 1). To increase the accuracy of the intensity measurements, a high redundancy of observations and the use of the fine-slicing oscillations (0.15°) proved to be effective; the overall Rmerge of the SAD data set was 3.0%. In total, six sulfur sites could be identified from the Bijvoet pair differences with the program solve (11). These sites were later confirmed by the final structure to correspond to Sδ atoms from two Met residues and Sγ atoms from three Cys residues and one sulfate ion bound to the protein in the P loop (Fig. 1A).
Table 1. Data collection, phase determination, and refinement statistics.
| SAD phasing (2.6 Å) Refinement (2.0 Å) | ||
|---|---|---|
| Data collection statistics | ||
| Resolution range, Å | 57–2.6 (2.70–2.60) | 57–2.0 (2.04–2.0) |
| Space group/unit cell parameters, Å | P61/a = b = 99.56; c = 57.23 | |
| Unique reflections, n | 9,954 (999) | 21,969 (1,125) |
| Rmerge, % | 3.04 (7.48) | 3.37 (20.65) |
| Completeness, % | 98.34 (89.84) | 99.7 (99.4) |
| Redundancy | 33.48 (11.16) | 5.15 (2.37) |
| I/σ(I)* | 29.44 (5.41) | 15.15 (1.73) |
| Vmax, Å3/Da | 3.93 | 3.93 |
| Molecules per a.u. | 1 | 1 |
| Refinement statistics | ||
| Test set for Rfree | 1,071 (174) | |
| Rfactor,† %/Rfree,‡ % | 20.3/21.8 | |
| Deviation from ideality | ||
| Bonds, Å | 0.006 | |
| Angles, ° | 1.2 | |
| Average B factors, Å2 | 30 | |
| No. of nonhydrogen atoms | ||
| Protein | 173 | |
| Water | 161 | |
| Other | 5 SO42-, 3 Li+ | |
| Ramachandran plot statistics | ||
| Most favored regions, % | 92.9 | |
| Allowed regions, % | 7.1 | |
| Generously allowed regions | 0 | |
| Disallowed regions | 0 | |
Values in parentheses correspond to the highest resolution data shell. For phase statistics, the mean figure of merit for phases calculated to 3.0 Å were 0.333 (before solvent flattening) and 0.642 (after solvent flattening).
Rmerge = Σ|I - 〈I〉|/Σ 〈I〉
Rfactor = Σ||Fobs| - |Fcalc||/Σ|Fobs|
Rfree is as for Rfactor but calculated by using a 4.9% test set of reflections excluded from the refinement
Fig. 1.
Characterization of AK6 by 3D structure analysis and sequence alignment. (A) Ribbon diagram of human AK6 produced by using the program prepi v0.9; the protein contains seven α-helices (blue ribbons), five β-sheets (magenta arrows), and a long NMP-binding loop (silver gray coil) in three domains: lid, core, and NMP-binding. The sulfate ion located in the P loop is shown as a Corey-Pauling-Koltun sphere with the default atomic color scheme (S, yellow; O, red). (B) Structure-based sequence alignment of AK6 (AD-004) with human AK1, AK2, AK3, AK4, and AK5. Secondary structure elements are depicted with the color scheme used in A. The percentage of amino acid sequence identity shared between AK6 and other AKs is shown in square brackets. Twenty-seven residues of a conserved insert segment for AK2, AK3, and AK4 is the ancient divergence of long isoforms (AK2, AK3, and AK4) and short isoforms (AK1, AK5, and AK6) (26). All sequences contain a highly conserved P loop, a NMP-binding domain, and a lid domain, all with different lengths (boxed in black). Conserved residues are labeled in red, and similar residues are labeled in yellow. (C) Charge distribution on AK6 structure surfaces. Negative charges are represented by red, and positive charges are represented by blue (see color key bar). The sulfate ion located in the P loop is shown as a ball-and-stick model with the default atomic color scheme (S, yellow; O, red). The front side is defined as facing the P loop. (D) The back side has striking patches of negative charges, which is a unique feature of AK6.
The current model for AK6 has been refined to 2.0 Å with final crystallographic Rfactor and free Rfactor values of 20.3% and 21.8%, respectively (Table 1). All residues of the protein could be well located in electron density maps, except for a few residues (48-52) in the NMP-binding region (Fig. 1 A and B) and one additional fusion residue (numbered as Leu-0) at the N-terminus could be located. Nonprotein atoms that were also identified included a sulfate ion bound to the P loop, 161 water molecules, 3 lithium ions, and 4 more sulfate ions.
Overall Structure of AK6. The overall structure of AK6 is shown in Fig. 1 A and B. Compared with other known NMPK structures (22), AK6 can be best described as a single α/β domain containing three functional subdomains (23): (i) the core domain with an ATP-binding site containing a central five-stranded, parallel open-twisted β-sheet surrounded by seven α-helices; (ii) the NMP-binding domain composed of helices α2, α3 and α4 and a long loop (residues 33-58) that, to our knowledge, has not been observed in other AKs before; and (iii) the so-called lid domain containing helices α5 and α6 (residues 109-118).
During their enzymatic cycles, NMPKs undergo considerable conformational changes, most notable are the movements of the two arms of NMPKs: the NMP-binding domain and the lid domain (24). In the human AK6 crystal structure, the enzyme is in the “open” conformation, i.e., waiting for ATP and AMP to bind, a conformation that has been observed before for AK1 and other AKs (reviewed in ref. 25). From this crystal structure, we can only make assessments of AK6 in the open conformation. It remains to be determined whether the conformational states of AK6 and other NMPKs are homologous.
From a sequence alignment among all human AKs (Fig. 1B), we can identify a number of local regions where certain sequence conservation does exist among all AK isoforms. For example, the helices α2 and α5 are sites involved in AMP and ATP binding in known AK structures, respectively. A short form of the lid domain exists in AK6, which has been found in AK1 and AK5 isoforms as well (26). However, unlike the other AK structures where the lid and NMP binding domains do not interact, in our current crystal structure of human AK6 with the open conformation, the short lid domain interacts with the tip of the NMP-binding domain (Fig. 1 A) by means of salt bridges, as if two “hands” are being held together, and presumably serves to stabilize AK6 in the open state.
Fig. 1 C (front view) and D (back view) show the surface rendering of charge distribution of the AK6 structure. Perhaps the most striking features of human AK6 are the large regions of negative charges on its surface on the back view (Fig. 1D). From the sequence alignment of AK6, it can be seen that the acidic residues that contribute to these charges are well conserved (indicated by open arrows in Fig. 4, which is published as supporting information on the PNAS web site). It is also noticeable that the loop belonging to the NMP-binding domain, residues 40-59 of human AK6, contains 10 acidic residues, without a single basic residue that is a conserved feature of the AK6 proteins (Fig. 4). By contrast, the comparable region of the AK1 to AK5 proteins is longer and contains basic residues.
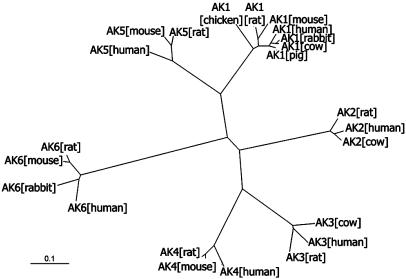
Sequence Analysis Defines a New Family of AKs. The human AK6 gene is localized on chromosome 12p11, spanning 1,119 bp composed of four exons and three introns (National Center for Biotechnology Information human genome). A blast search of databases revealed that this gene exists in all eukaryotic genomes sequenced so far; e.g., mammals, plants, insects, and yeasts. An alignment of sequence homologous to human AK6 derived from many eukaryotic organisms (Fig. 4) reveals the basis for proposing that they belong to a distinct AK subfamily. According to the alignment, there is >86% sequence identity of AK6 amino acid sequences among human, mouse, and rat, and even the most distant member, yeast Fap7p, shares ≈40% sequence identity with human AK6, whereas AK6 and other AKs, AK1 to AK5, share <18% sequence identity (Fig. 1B). We also constructed an unrooted radial phylogenetic tree (Fig. 2) to show the evolutionary relationship of AK6 with the other vertebrate AKs. Six subgroups corresponding to the six isoforms of AK can be discerned. From these analyses, it can be seen that AK6 is indeed a most distinct AK isoform in eukaryotes.
Fig. 2.
An unrooted radial phylogenetic tree was constructed on the basis of sequence alignments of AK isoforms found in vertebrates. Six subgroups corresponding to six isoforms of AKs can be discerned. The following is a list of the organisms, with corresponding accession numbers in parentheses: AK1, including human (NP_000467), pig (P00571), cow (P00570), rabbit (P00569), mouse (NP_067490), rat (NP_077325), and chicken (P05081); AK2, including human (AAH09405), rat (JQ1944), and cow (JS0422); AK3, including human (BAA87913), rat (NP_037350), and cow (NP_776662); AK4, including human (P27144), mouse (NP_033777), and rat (NP_058831); AK5, including human (Q9Y6K8), mouse (NP_694706), and rat (XP_345320); AK6, including human, mouse, rat, and rabbit. (Scale bar, evolutionary distance value of 0.1 substitutions per site.)
Human AD-004 Identified as AK6 by Enzymatic Assays. Although the structure of AD-004 (AK6 has been named only after the enzymatic characterization) revealed a fold very similar to nucleotide kinases after ab initio structure determination, this fold is also commonly seen in kinases that phosphorylate substrates other than nucleotides. A search by the dali Server (www.ebi.ac.uk/dali) (27) with the initial AD-004 structure identified numerous close homologues, including the closest hit Bacillus stearothermophilus AK and bacterial shikimate kinases. It was therefore necessary to evaluate the catalytic activity of human AD-004 before definitively assigning it as an AK.
Natural ribonucleosides and deoxyribonucleosides, as well as their corresponding monophosphates, were tested in a phosphoryltransfer assay with [γ-32P]ATP as a phosphate donor. The results showed that AD-004 was an NMPK. AMP and dAMP were the preferred substrates of all tested NMPs, but CMP and dCMP were also good substrates. IMP could be phosphorylated, but to a much lesser extent (Fig. 5, which is published as supporting information on the PNAS web site).
Because AD-004 shows sequence and structural similarity to bacterial shikimate kinase, we examined whether the enzyme can phosphorylate nonnucleoside substrates such as d-galactose, l-homoserine, and mevalonate, which are substrates for the bacterial shikimate kinase. No formation of phosphorylated products was found in a phosphoryl transfer assay with 10 mM concentrations of these three compounds.
The nucleoside triphosphates ATP, GTP, UTP, CTP, dATP, dCTP, dGTP, and TTP were then evaluated as phosphate donors at 5 mM concentrations. All of the triphosphates were able to serve as substrates for AK6. CTP was the best phosphate donor, with 5-fold higher activity than that of ATP. UTP was also a good phosphate donor that gave 3-fold higher activity than ATP. GTP and dCTP showed activities very similar to ATP. The nucleotides dGTP, TTP, and dATP were, however, 3- to 5-fold less active compared with ATP (Table 2, which is published as supporting information on the PNAS web site).
When radiolabeled AMP ([3H]AMP) was used as substrate, we observed that the reaction product included a substantial amount of [3H]ATP and [3H]ADP (data not shown), indicating that the reaction catalyzed by this enzyme is reversible, as has been observed with the other NMPKs (4). The kinetic parameters for AMP and ATP were determined by using [3H]AMP as a substrate under conditions in which only the forward reaction occurred. The Km and Vmax values for AMP were 192 μM and 955 nmol·min-1·mg-1, respectively, and the Km and Vmax values for ATP were 44 μM and 982 nmol·min-1·mg-1, respectively (Table 3, which is published as supporting information on the PNAS web site).
From the above results, we concluded that human AD-004 is an AK and named it AK6 after the five known AKs. Human AK6 is found to be broadly similar to other AKs, particularly to AK5, in many of its enzymatic properties. However, the other human AK isoforms have more restricted phosphate donor and/or acceptor specificity (4). For human AK6, all NTPs and dNTPs can serve as phosphate donors, and adenylate and cytidylate can serve as phosphate acceptors, with CTP and UTP as more preferred donors than ATP and GTP at these assay conditions (Table 2).
The preference of AK6 for CTP and UTP as phosphate donors compared with ATP is apparently a unique feature of AK6 and indicates a different phosphoryl transfer reaction. The broad activity of AK6 with all NTPs and dNTPs (Table 2) as phosphate donors may reflect a functional role in maintenance of nuclear homeostasis of nucleotide and deoxynucleotide pools because AK6 has been identified as a nuclear enzyme, as shown below. Furthermore, when coupled with nucleoside diphosphate kinase or creatine kinase, AK6, like the other AKs, may form phosphotransfer networks to control and regulate cellular (nuclear) energetics (3).
Structural Comparison of AK6 with Other AKs. Previously investigated NMPKs possess the conserved motif GXPGXGKGT (residues 10-18 in AK6) at the P loop (25). This feature is conserved perfectly in the five known human AKs but not in AK6 (Fig. 1B); instead, the last Gly in the motif is changed to Thr-17 in the whole AK6 family (Fig. 2). Structural comparison shows that the rms deviations between the overall structure Cα atoms (≈140 atoms in the calculation) of human AK6 and those of AK1 [Protein Data Bank (PDB) ID code 3ADK, porcine], AK2 (PDB ID code 1AK2, bovine), and AK3 (PDB ID code 2AK3, bovine) are 2.083, 1.977, and 2.162 Å, respectively, and the superposition of the four structures shows that the overall structures in the core regions are all very similar (Fig. 6A, which is published as supporting information on the PNAS web site). However, the lid- and NMP-binding domains greatly differ between AK6 and the other AKs. The corresponding Cα atoms (≈20 atoms in the calculation) rms deviations around the active site P loop are 0.613, 0.618, and 0.989 Å.
Most of the catalytically important residues, such as Arg-39, Arg-105, and Arg-109, are conserved in the AK6 structure and are supposed to play similar roles in AK6 during catalysis. However, besides Thr-17, another feature of AK6 distinct from the other AKs is that His-79 inserts into the activity site and is positioned close to Arg-39. Quite interestingly, the positioning of Thr-17 and His-79 in the active site of AK6 is similar to that of an archaeal AK, AKsa (PDB ID code 1NKS) (Fig. 6B). Because the other catalytically important residues are conserved in AKsa as well, we can anticipate that the phosphoryl transfer mechanism of AK6 would be more similar to that AKsa than to that of the other human AKs.
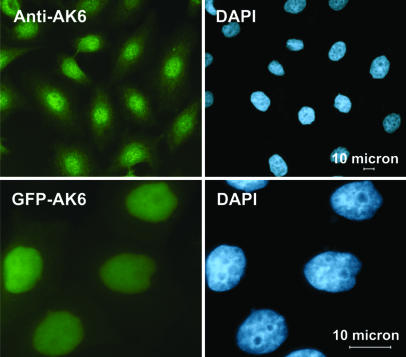
AK6 Localizes to the Nucleus of HeLa Cells. Inspired by the observations that Fap7p, the baker's yeast AK6 homologue, localizes to the cell nucleus (28), the intracellular localization of human AK6 was determined by using immunofluorescent microscopy. HeLa cells were stained with a rabbit antiserum raised against human AK6. Comparison between the AK6 antiserum staining and the staining of the nucleus with a DNA dye (DAPI) demonstrates that human AK6 is primarily localized to the nuclei of HeLa cells (Fig. 3). Similar results were obtained in A549 [a human lung carcinoma (II) cell line] and A375 cells (a human melanoma cell line) (data not shown).
Fig. 3.
Subcellular localization of AK6 in HeLa cells by means of fluorescence microscopy. (Upper Left) AK6 was detected in vivo by rabbit anti-AK6 and goat anti rabbit IgG-FITC. (Upper Right) DAPI staining on the same cells confirmed a nuclear localization of AK6. (Lower Left) HeLa cells were transfected by pEGFP-AK6. (Lower Right) After transfection (24 h), cells were visualized with a fluorescent microscope, and strong nuclear fluorescence was observed. (Lower Right) DAPI staining on the same cells again proved the nuclear localization of AK6.
The localization of AK6 in the nucleus was further confirmed by determining the location of a transfected AK6-GFP fusion protein (Fig. 3). A vector encoding GFP fused to the N terminus of human AK6 was transfected into HeLa cells, and the cells were visualized by fluorescence microscopy after DAPI staining. GFP fluorescence from the N-terminal GFP fusion protein (Fig. 3) and from a C-terminal GFP fusion protein (data not shown) were found to colocalize with the DAPI staining, confirming that human AK6 is predominantly localized in the nucleus.
From the sequence analysis of AK6s (Fig. 4), mammalian AK6s appear to lack a nuclear localization sequence, whereas Fap7p and the AK6 homologues of rice, Arabidopsis and C. elegans contain putative nuclear localization sequence at their N termini. From an examination of the AK6 crystal structure, we propose that the acidic domains on its surface (Fig. 1D) could be important for its nuclear localization. For example, AK6 may be cotransported to the nucleus with a basic protein. The mechanism for how human AK6 is transported into the nucleus would be an interesting task for further studies.
The broad activity of AK6 with all types of NTPs and dNTPs as phosphate donors may be correlated to its nuclear localization. AK6 can use all of the (deoxy)nucleoside triphosphates there to produce ADP and CDP. These ADP and CDP can then be used by NDPKs for nucleotide metabolism in the nucleus or for the energetic communication between mitochondria and nucleus by means of phosphotransfer networks (3).
Furthermore, because ATP/ADP and GTP/GDP ratios are important in nuclear transport and energetics, specific inhibition of AKs can disrupt nuclear transport processes (2). AK6 may thus be an important player with the ability to regulate ATP/ADP and GTP/GDP ratios and their concentrations simultaneously.
Adenine nucleotide homoeostasis is essential for multiple cellular functions, such as cellular energetics, but it also plays an important role in providing nucleotides for RNA and DNA synthesis. Each of the five known human AK isoforms show specific tissue distribution and subcellular localization. AK1 is a cytosolic enzyme mainly expressed in skeletal muscle, brain, and erythrocytes, and AK2 is located both in the cytosol and mitochondrial intermembrane space and is expressed mainly in liver, kidney, and, to a lower extent, spleen and heart. AK3 and AK4 are mitochondrial matrix enzymes with AK3 ubiquitously expressed in all tissues, whereas AK4 is mainly expressed in kidney, heart, and liver (29). AK5 was found in brain tissue and is a cytosolic enzyme (30). Now we have determined that human AK6 is a nuclear enzyme. This result has confirmed the previous evidence for a nuclear form of AKs (31) and provided solid basis for AK as an important component in nuclear phosphotransfer networks (3). The discovery of AK6 is important considering the many roles such an enzyme could have. The AK6 gene (AD-004) was initially cloned from a human hypothalamus-pituitary-adrenal cDNA library as an adrenal gland-specific gene (6). The exact tissue distribution of AK6 remains to be determined.
Together, the results presented here reveal that human AK6, although having much in common with other AK isoforms in terms of general structure, because of its sequence, nuclear localization, and enzymatic properties, is a unique AK and offers exciting avenues for future research.
Supplementary Material
Acknowledgments
This work was supported by from Grants 2001AA233011 and 2002BA711A13 from National High Technology and Development Program 863 of China, Peking University Grants 985 and 211, and grants from the Swedish Research Council and the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (L.W. and S.E.). X.D.S. is a recipient of Fund 30325012 from the National Science Fund for Distinguished Young Scholars of China. C.Z. was supported by National Natural Science Foundation of China Grants 30225016 and 30330200 and State Key Basic Research and Development Plan Grant 2004CB720003.
Author contributions: M.L. and X.-D.S. designed research; H.R., L.W., M.B., Y.L., X.Z., F.L., L.L., and X.-D.S. performed research; H.R., M.B., Y.L., X.Z., J.N., S.E., C.Z., and X.-D.S. analyzed data; and M.B. and X.-D.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AK, adenylate kinase; NMPK, nucleoside monophosphate kinase; SAD, single-wavelength anomalous dispersion.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1RKB).
References
- 1.Noda, L. (1973) in The Enzymes, ed. Boyer, P.D. (Academic, New York), Vol. 8, pp. 279-305. [Google Scholar]
- 2.Dzeja, P. P., Bortolon, R., Perez-Terzic, C., Holmuhamedov, E. L. & Terzic, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10156-10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dzeja, P. P. & Terzic, A. (2003) J. Exp. Biol. 206, 2039-2047. [DOI] [PubMed] [Google Scholar]
- 4.Van Rompay, A. R., Johansson, M. & Karlsson, A. (2000) Pharmacol. Ther. 87, 189-198. [DOI] [PubMed] [Google Scholar]
- 5.Ding, H. T., Ren, H., Chen, Q., Fang, G., Li, L. F., Li, R., Wang, Z., Jia, X. Y., Liang, Y. H., Hu, M. H., et al. (2002) Acta. Crystallogr. D 58, 2102-2108. [DOI] [PubMed] [Google Scholar]
- 6.Hu, R. M., Han, Z. G., Song, H. D., Peng, Y. D., Huang, Q. H., Ren, S. X., Gu, Y. J., Huang, C. H., Li, Y. B., Jiang, C. L., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 9543-9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai, C. H., Chou, C. Y., Chang, L. Y., Liu, C. S. & Lin, W. (2000) Genome Res. 10, 703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. (1982) EMBO J. 1, 945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren, H., Liang, Y. H., Li, R., Ding, H., Qiu, S., Lu, S., An, J., Li, L., Luo, M., Zheng, X. & Su, X. D. (2004) Acta. Crystallogr. D 60, 1292-1294. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger, T. C. & Berendzen, J. (1996) Acta. Crystallogr. D 52, 749-757. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger, T. C. (2003) Methods Enzymol. 374, 22-37. [DOI] [PubMed] [Google Scholar]
- 13.McRee, D. E. (1992) J. Mol. Graphics 10, 44-46. [Google Scholar]
- 14.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta. Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta. Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 16.Morris, A. L., MacArthur, M. W., Hutchinson, E. G. & Thornton, J. M. (1992) Proteins 12, 345-364. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsson, B., Britton, S., Tornevik, Y. & Eriksson, S. (1998) Biochem. Pharmacol. 56, 389-395. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barton, G. J. (1993) Protein Eng. 6, 37-40. [DOI] [PubMed] [Google Scholar]
- 20.Page, R. D. (1996) Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- 21.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 22.Vonrhein, C., Schlauderer, G. J. & Schulz, G. E. (1995) Structure (Cambridge, MA, U.S.) 3, 483-490. [DOI] [PubMed] [Google Scholar]
- 23.Schulz, G. E., Muller, C. W. & Diederichs, K. (1990) J. Mol. Biol. 213, 627-630. [DOI] [PubMed] [Google Scholar]
- 24.Dreusicke, D. & Schulz, G. E. (1988) J. Mol. Biol. 203, 1021-1028. [DOI] [PubMed] [Google Scholar]
- 25.Yan, H. & Tsai, M. D. (1999) Adv. Enzymol. Relat. Areas Mol. Biol. 73, 103-134. [DOI] [PubMed] [Google Scholar]
- 26.Fukami-Kobayashi, K., Nosaka, M., Nakazawa, A. & Go, M. (1996) FEBS Lett. 385, 214-220. [DOI] [PubMed] [Google Scholar]
- 27.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 28.Juhnke, H., Charizanis, C., Latifi, F., Krems, B. & Entian, K. D. (2000) Mol. Microbiol. 35, 936-948. [DOI] [PubMed] [Google Scholar]
- 29.Noma, T., Fujisawa, K., Yamashiro, Y., Shinohara, M., Nakazawa, A., Gondo, T., Ishihara, T. & Yoshinobu, K. (2001) Biochem. J. 358, 225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Rompay, A. R., Johansson, M. & Karlsson, A. (1999) Eur. J. Biochem. 261, 509-517. [DOI] [PubMed] [Google Scholar]
- 31.Criss, W. E. (1970) J. Biol. Chem. 245, 6352-6356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.