Abstract
Reactive oxygen species (ROS), such as O2− and H2O2, play a key role in plant metabolism, cellular signaling, and defense. In leaf cells, the chloroplast is considered to be a focal point of ROS metabolism. It is a major producer of O2− and H2O2 during photosynthesis, and it contains a large array of ROS-scavenging mechanisms that have been extensively studied. By contrast, the function of the cytosolic ROS-scavenging mechanisms of leaf cells is largely unknown. In this study, we demonstrate that in the absence of the cytosolic H2O2-scavenging enzyme ascorbate peroxidase 1 (APX1), the entire chloroplastic H2O2-scavenging system of Arabidopsis thaliana collapses, H2O2 levels increase, and protein oxidation occurs. We further identify specific proteins oxidized in APX1-deficient plants and characterize the signaling events that ensue in knockout-Apx1 plants in response to a moderate level of light stress. Using a dominant-negative approach, we demonstrate that heat shock transcription factors play a central role in the early sensing of H2O2 stress in plants. Using knockout plants for the NADPH oxidase D protein (knockout-RbohD), we demonstrate that RbohD might be required for ROS signal amplification during light stress. Our study points to a key role for the cytosol in protecting the chloroplast during light stress and provides evidence for cross-compartment protection of thylakoid and stromal/mitochondrial APXs by cytosolic APX1.
INTRODUCTION
Reactive oxygen species (ROS) are partially reduced or excited forms of atmospheric oxygen (O2) continuously produced in cells during aerobic metabolism (Halliwell and Gutteridge, 1989). They can cause extensive cell injury or death, but they play a central role in many signaling pathways in plants involved in stress perception, photosynthesis regulation, pathogen response, programmed cell death, hormonal action, and plant growth and development (Dat et al., 2000; Mittler, 2002; Mullineaux and Karpinski, 2002; Neill et al., 2002; Apel and Hirt, 2004).
In Arabidopsis thaliana, a network of at least 152 genes controls ROS metabolism (Mittler et al., 2004). The network is thought to regulate the rates of ROS production and ROS scavenging in the different cellular compartments and to modulate the steady state level of ROS for signaling as well as defense purposes. In leaf cells, an intricate balance exists between H2O2 and O2− production in the chloroplast and peroxisome during photosynthesis and the activities of the ROS-scavenging enzymes superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (Asada, 1999; Mittler, 2002; Apel and Hirt, 2004). The chloroplast contains at least three different isozymes of APX: a thylakoid-bound APX (At1g77490), a lumen APX (At4g09010), and a stromal APX (At4g08390; stromal APX was recently shown to be dually targeted to the stroma and mitochondria in Arabidopsis and would be referred to as stromal/mitochondrial APX; Chew et al., 2003). The chloroplast also contains four different isozymes of SOD: a CuZnSOD (CSD2), three FeSODs, and enzymes of the ascorbate-glutathione cycle capable of reducing oxidized ascorbic acid and glutathione (Asada and Takahashi, 1987). By contrast, the cytosol contains one cytosolic APX (APX1, At1g07890), with an additional APX (APX2, At3g09640) that is inducible mainly under extreme light or heat stress conditions (Karpinski et al., 1999; Panchuk et al., 2002), and different enzymes of the ascorbate-glutathione cycle (Mittler et al., 2004). Leaf peroxisomes contain three isozymes of catalase, one isozyme of APX (At4g35000) that is bound to the outer layer of the peroxisome, and enzymes of the ascorbate-glutathione cycle (Corpas et al., 2001; Shigeoka et al., 2002). Compared with other cellular compartments, chloroplasts contain high levels of the antioxidants ascorbic acid and glutathione (up to 25 and 5 mM, respectively; Noctor and Foyer, 1998).
Mathematical calculations, as well as computer model simulations, based on enzyme concentration and reaction rate, the rates of O2− and H2O2 production, and the concentrations of the antioxidants ascorbic acid and glutathione in chloroplasts, conclude that the chloroplast is well equipped to scavenge the O2− and H2O2 produced during photosynthesis in plants grown under controlled conditions or subjected to light stress (Asada and Takahashi, 1987; Asada, 1999; Polle, 2001). In addition, the expression of transcripts encoding the stromal/mitochondrial and thylakoid-bound APXs was not enhanced during light stress, whereas the expression of the cytosolic APXs was enhanced (Karpinski et al., 1997; Shigeoka et al., 2002). This finding was mainly interpreted as supportive of the hypothesis that within leaf cells the chloroplastic scavenging systems are sufficient to handle ROS production even under stressful conditions (Shigeoka et al., 2002).
The signal transduction pathway leading to the enhanced expression of cytosolic APXs during light stress was proposed to involve a chloroplast-to-nuclei signal that depends on the pool of reduced plastoquinone in the chloroplast and may involve H2O2 (Karpinski et al., 1997; Rodermel, 2001; Mullineaux and Karpinski, 2002). Moreover, the signal that enhanced APX2 expression in response to high light stress was shown to be a systemic signal (Karpinski et al., 1999). However, in view of the supposition that the chloroplast is well equipped to scavenge ROS produced during photosynthesis, the significance of the enhanced cytosolic APX expression during light stress is unknown. Recent analysis of knockout plants deficient in cytosolic APX1 (KO-Apx1) revealed that in the absence of APX1 photosynthetic activity was suppressed, suggesting that cytosolic APXs might be essential for chloroplast protection during light stress (Pnueli et al., 2003; Mittler et al., 2004).
In this study, we demonstrate that in the absence of cytosolic APX1, the entire chloroplastic H2O2-scavenging system of Arabidopsis collapses, H2O2 levels increase, and protein oxidation occurs. Using Affymetrix GeneChip and matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) technology, we identify specific proteins oxidized in KO-Apx1 plants and characterize the signaling events that ensue in KO-Apx1 plants in response to a moderate level of light stress. Using a dominant-negative approach, we demonstrate that heat shock transcription factors (HSFs) play a key role in the early sensing of H2O2 stress in KO-Apx1 plants, and using knockout plants for the NADPH oxidase D protein (KO-RbohD), we demonstrate that RbohD might be required for ROS signal amplification during light stress in Arabidopsis. Our study points to a key role for the cytosol in protecting the chloroplast during light stress and provides evidence for cross-compartment protection of thylakoid and stromal/mitochondrial APXs by cytosolic APX1.
RESULTS
H2O2 Accumulation and Protein Oxidation in KO-Apx1 Plants
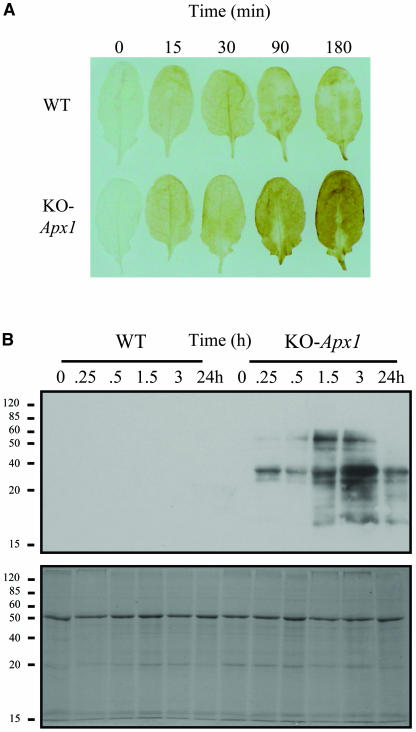
As shown in Figure 1A, KO-Apx1 plants grown under low light conditions (25 μmol m−2 s−1) and transferred to a moderate light level of 250 μmol m−2 s−1 accumulated H2O2 in leaves. By contrast, wild-type plants subjected to the same treatment did not accumulate H2O2 to the same level. The accumulation of H2O2 appeared to occur in a homogeneous manner in most leaf cells and was not confined to a particular tissue, such as the vascular tissue.
Figure 1.
Deficiency in APX1 Results in H2O2 Accumulation and Protein Oxidation in Arabidopsis Leaves Subjected to a Moderate Level of Light Stress.
(A) Accumulation of H2O2 in wild-type and knockout-Apx1 (KO-Apx1) plants in response to a moderate level of light stress. Compared with wild-type plants, KO-Apx1 plants are shown to accumulate higher levels of H2O2 (evident by darker staining of leaves at 90 and 180 min after the application of light stress).
(B) Detection of proteins containing carbonyl groups (indicative of protein oxidation) in leaf extracts obtained from wild-type and KO-Apx1 plants by a protein blot assay (top). A protein gel stained with Coomassie blue is used to demonstrate equal loading of proteins (bottom). Compared with wild-type plants, KO-Apx1 plants are shown to accumulate a high level of oxidized proteins. Detection of H2O2 and protein oxidation was performed as described in Methods. All experiments were repeated at least three times. Representative results are shown.
To examine whether H2O2 accumulation in KO-Apx1 resulted in oxidative damage to cells, we tested protein extracts from leaf cells using a protein gel blot assay that detects protein oxidation (detects the presence of carbonyl groups on proteins by first reacting proteins with 2,4-dintrophenylhydrazine (DNP) and then detecting the DNP-bound proteins on protein blots using a DNP-specific antibody; Johansson et al., 2004; Rizhsky et al., 2004). Leaf cells grown under controlled conditions contain a low baseline level of oxidized proteins (data not shown; Johansson et al., 2004). As shown in Figure 1B, KO-Apx1 plants subjected to the light treatment described above (25 to 250 μmol m−2 s−1) accumulated high levels of oxidized proteins. By contrast, wild-type plants subjected to the same treatment did not accumulate a similar level of oxidized proteins. This result suggests that the absence of APX1 results not only in the accumulation of H2O2 (Figure 1A) but also in damage to specific proteins in leaf cells (Figure 1B).
To identify specific proteins oxidized in KO-Apx1 plants in response to the light stress treatment described above, we performed proteomic analysis of protein extracts and determined the identity of specific proteins detected by the DNP-antibody using MALDI-TOF (see Methods for details). As shown in Table 1, the lack of APX1 resulted in the oxidation of different proteins belonging to different metabolic pathways. Interestingly, three of the proteins identified by our study as oxidized in knockout cytosolic Apx1 plants were chloroplastic proteins (Cys synthase, Asp kinase, and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit; Table 1). Other proteins oxidized in KO-Apx1 plants are possibly localized to the nuclei, cytosol, or the secretory pathway. At least one protein was predicted to be a membrane protein (Table 1). Low levels of oxidized Rubisco large subunit were also detected in wild-type plants subjected to the light stress treatment (data not shown; see also Rizhsky et al., 2004). The accumulation of H2O2 and the oxidation of chloroplastic proteins in KO-Apx1 plants during the light shift treatment (Figure 1, Table 1) suggest that, in the absence of cytosolic APX1, the chloroplastic or peroxisomal H2O2-scavenging system(s) of Arabidopsis are unable to scavenge the H2O2 produced during photosynthesis or photorespiration. The accumulation of H2O2 and protein oxidation (Figure 1, Table 1) did not result in the activation of cell death in leaves of wild-type or KO-Apx1 plants, and KO-Apx1 plants were able to make a complete recovery from the light stress treatment (data not shown; see also Pnueli et al., 2003).
Table 1.
Proteins Oxidized in KO-Apx1 Plants in Response to a Moderate Level of Light Stress
| Protein | Locus Number | No. of Fragments Matched | Coverage | Predicted Location |
|---|---|---|---|---|
| Cys synthase [O-acetylserine (thiol)lyase] | At3g22460 | 5 | 28% | Plastid |
| Asp kinase/homoserine dehydrogenase | At1g31230 | 18 | 27% | Plastid |
| Putative transposase | At2g14140 | 20 | 28% | Cytosol/nuclear |
| Rubisco large subunit | ArthCp030 | 14 | 26% | Plastid |
| Zinc-finger, C3HC4 (RING finger) | At5g60710 | 10 | 22% | Nuclear |
| Cytochrome P450 | At4g15396 | 22 | 39% | Secretory |
| Putative cation/H+ exchanger | At5g58460 | 15 | 23% | Membrane |
| Transcription activator | At3g52910 | 4 | 22% | Cytosol/nuclear |
Proteomic analysis of protein oxidation was determined as described in Methods. Predicted subcellular localization was performed as described in Mittler et al. (2004).
In the Absence of APX1, the Chloroplastic H2O2-Scavenging System Collapses
The lack of cytosolic APX1 resulted in the oxidation of chloroplastic proteins (Table 1), suggesting that APX1 activity might be important for chloroplast protection. Previous studies have shown that chloroplasts are extremely sensitive to external application of H2O2 (Asada, 2000) and that chloroplastic APXs are inactivated by H2O2 (Mano et al., 2001). The lack of cytosolic APX1 may therefore affect H2O2 scavenging systems in the chloroplast. We therefore studied the relationship between cytosolic APX1 and the chloroplastic APXs, thylakoid and stromal/mitochondrial APX.
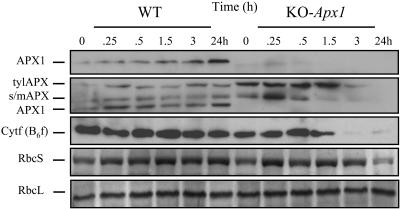
As shown in Figure 2, compared with KO-Apx1 plants, the steady state protein level of thylakoid and stromal/mitochondrial APX did not appear to change during this treatment in wild-type plants. The level of cytosolic APX1 did however increase in wild-type plants during the light stress treatment (Figure 2). By contrast, in the absence of APX1 (KO-Apx1), the steady state level of thylakoid and stromal/mitochondrial APX was elevated, at least during early time points of light stress. However, the lack of APX1 resulted in the apparent degradation of thylakoid and stromal/mitochondrial APXs, a b6f complex subunit protein (Cytf) and the small subunit protein of Rubisco (Figure 2). These findings suggest that although APX1 is localized to the cytosol, it is critical for protecting the H2O2 scavenging system in the chloroplast during light stress. The inactivation of chloroplastic APXs by H2O2 (Mano et al., 2001) may serve as the initiation point for their degradation by chloroplastic proteases.
Figure 2.
Deficiency in APX1 Results in a Decrease in the Steady State Level of Chloroplastic Proteins, Including Chloroplastic APXs, during a Moderate Level of Light Stress.
Detection of thylakoid APX (tylAPX), stromal/mitochondrial APX (s/mAPX), APX1, Cytf (b6f), and Rubisco (RbcS and RbcL) steady state protein levels by protein blots in leaf extracts from wild-type and knockout-Apx1 (KO-Apx1) plants subjected to a moderate level of light stress. Deficiency in APX1 is shown to result in a decrease in the steady state protein level of the chloroplastic H2O2-scavenging enzymes tylAPX and s/mAPX as well as in a decrease in the steady state protein level of the chloroplastic proteins Cytf (b6f) and RbcS. Light stress experiments and protein blots were performed as described in Methods. All experiments were repeated at least three times. Representative results are shown. APX1 was detected with two different antibodies: one against purified APX1 (top panel), and one against a conserved domain from tylAPX (second panel from top).
Cytosolic APX1 Might Be Sufficient to Protect the Chloroplast in the Absence of Stromal/Mitochondrial APX
To further characterize the relationship between cytosolic APX1 and chloroplastic APXs, we studied knockout plants deficient in stromal/mitochondrial APX (KO-s/mApx). Because chloroplasts are sensitive to the external application of H2O2 (Asada, 2000) and the lack of APX1 might allow H2O2 to diffuse into the chloroplast from the cytosol, we focused our study on stromal/mitochondrial APX. This enzyme might serve as the first chloroplastic line of defense against diffusion of H2O2 from the cytosol into the chloroplast. However, if cytosolic APX1 is sufficient to prevent this process, then stromal/mitochondrial APX might not be required for chloroplast protection under the experimental conditions shown in Figures 1 and 2.
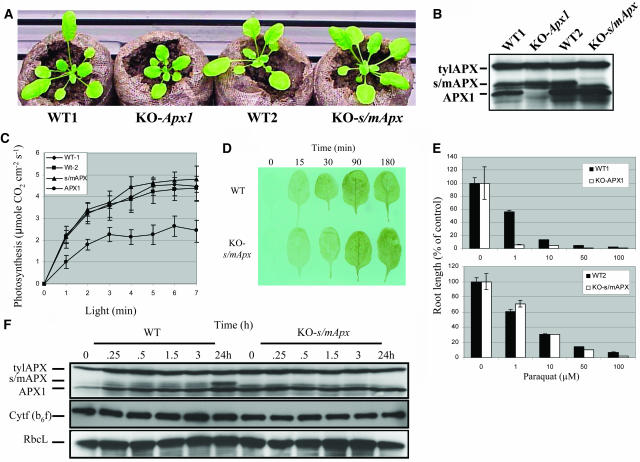
As shown in Figure 3A, KO-s/mApx plants did not have a visible phenotype when grown under controlled conditions (25 to 75 μmol m−2 s−1, 21°C). Protein blot analysis of KO-s/mApx plants confirmed that the stromal/mitochondrial protein was not detected in KO-s/mApx plants grown under controlled conditions (Figure 3B). Measurements of photosynthetic activity in KO-s/mApx and KO-Apx1 revealed that the lack of stromal/mitochondrial APX did not significantly affect photosynthesis (Figure 3C). By contrast, under the same conditions, the photosynthetic activity of KO-Apx1 plants was suppressed (Figure 3C; Pnueli et al., 2003). However, it should be noted that in certain plants, stromal APX could be generated by the alternative splicing of thylakoid APX (Yoshimura et al., 2002). If this process occurs in Arabidopsis as well, a low level of stromal/mitochondrial APX protein, that is not detected by our protein blots (Figure 3B), might be present in KO-s/mApx plants. A search of the Arabidopsis EST database (http://www.arabidopsis.org/), however, did not provide evidence for alternative splicing of thylakoid APX (data not shown).
Figure 3.
Characterization of Knockout Plants Deficient in Stromal/Mitochondrial APX.
(A) Photograph of wild-type (WT1 and WT2 for Columbia and Wassilewskija [Ws], respectively), knockout-Apx1 (KO-Apx1, Ws), and knockout stromal/mitochondrial APX (KO-s/mApx, Columbia) plants grown under controlled conditions. No visible phenotype is shown to be associated with the lack of s/mAPX under these conditions.
(B) Protein blot analysis showing the lack of the s/mAPX proteins in KO-s/mApx plants.
(C) Measurements of photosynthetic activity (CO2 gas exchange) in WT1, WT2, KO-Apx1, and KO-s/mApx plants. In contrast with KO-Apx1 (APX1) plants, the photosynthetic activity of KO-s/mApx (s/mAPX) plants is shown not to be suppressed. Conditions for photosynthetic measurements were as follows: CO2, 400ppm; light intensity, 1000 μmol m−2 s−1; temperature, 21°C.
(D) Accumulation of H2O2 in wild-type and knockout-s/mApx (KO-s/mApx) plants in response to a moderate level of light stress. Compared with wild-type or KO-Apx1 plants (Figure 1A), KO-s/mApx are shown not to accumulate high levels of H2O2 in response to a moderate level of light stress.
(E) Inhibition of root growth in 5-d-old seedlings grown on agar plates in the presence of different concentrations of the superoxide-generating compound paraquat. In contrast with seedlings of KO-Apx1 plants that show high sensitivity to paraquat (top panel), the root growth of KO-s/mApx seedlings is shown to be less sensitive to the paraquat treatment.
(F) Detection of thylakoid APX (tylAPX), stromal/mitochondrial APX (s/mAPX), APX1, Cytf (b6f), and Rubisco (RbcL) steady state protein levels by protein blots in leaf extracts from wild-type and KO-s/mApx plants subjected to a moderate level of light stress. Compared with wild-type or KO-Apx1 plants (Figure 2), the steady state protein levels of tylAPX and Cytf (b6f) are shown not to be suppressed in response to a moderate level of light stress. Detection of H2O2 in leaves, protein blots, and measurements of photosynthetic activity were performed as described in Methods. No differences were found between the sensitivity of Ws and Columbia cultivars to the treatments shown in Figures 1 to 3 (data not shown). All experiments were repeated at least three times. Representative results are shown.
As shown in Figures 3D and 3F, KO-s/mApx plants subjected to the same light stress treatment that results in the accumulation of H2O2, protein oxidation, and damage to the chloroplastic H2O2-scavenging system in KO-Apx1 plants (Figures 1 and 2) did not accumulate H2O2 or displayed enhanced protein turnover. In addition, KO-s/mApx plants did not accumulate products of protein oxidation in response to the light stress treatment (data not shown). In roots, the stromal/mitochondrial APX is expected to play a key role in protecting the mitochondria. To test whether the lack of stromal/mitochondrial APX (KO-s/mApx) or cytosolic APX (KO-Apx1) results in a heightened sensitivity of roots to oxidative stress, we tested the effect of the superoxide-generating agent paraquat on root growth in the two knockout lines (paraquat is expected to enhance the production of superoxide radicals in the mitochondria and cytosol of root cells, leading to the accumulation of hydrogen peroxide). As shown in Figure 3E, compared with KO-s/mApx, KO-Apx1 displayed high sensitivity to paraquat stress. Sensitivity to paraquat was displayed by KO-s/mApx plants at concentrations higher than 50 μM (Figure 3E).
The apparent lack of damage in KO-s/mApx plants in response to the moderate light stress treatment described above suggests that in contrast with cytosolic APX1, stromal/mitochondrial APX might not be essential for cellular protection under these conditions. It is also possible that cytosolic APX1 could compensate for the lack of stromal/mitochondrial APX during a moderate level of light stress.
Microarray Analysis of Transcript Expression in KO-Apx1 Plants Subjected to Light Stress
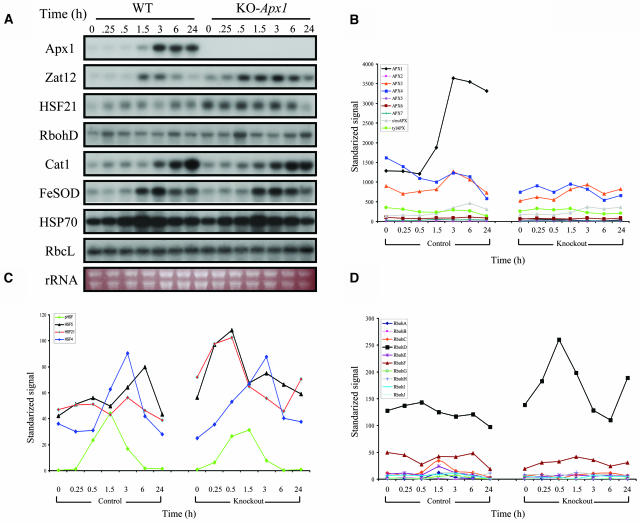
The central role that APX1 appears to play in protecting the cell during light stress (Figures 1 to 3; Pnueli et al., 2003) prompted us to study the signal transduction pathway that controls Apx1 expression during this treatment. For this purpose, we performed a time-course microarray analysis of wild-type and KO-Apx1 plants subjected to the same light stress treatment described above (Figures 1 to 3; Affymetrix ATH1 chips). As shown in Figure 4A, the steady state level of transcripts encoding APX1, the H2O2-related zinc-finger protein Zat12 (also known as RHL41, Iida et al., 2000; Rizhsky et al., 2004), catalase (Cat1), iron-SOD (FeSOD), and heat shock protein 70 (HSP70) is elevated in wild-type plants in response to the moderate light stress treatment. The steady state transcript level of Cat1, FeSOD, and HSP70 is similarly elevated in KO-Apx1 plants; however, compared with wild-type plants, the elevation in Zat12 transcript level is more pronounced in KO-Apx1 plants.
Figure 4.
Microarray Analysis of Knockout-Apx1 Plants Subjected to a Moderate Level of Light Stress.
(A) RNA gel blots showing an increase in the steady state level of transcripts encoding APX1 (Apx1), the zinc-finger protein Zat12 (Zat12), HSF21, NADPH oxidase D (RbohD), catalase (Cat1), iron superoxide dismutase (FeSOD), and heat shock protein 70 (Hsp70) in plants subjected to a moderate level of light stress. RNA gel blots showing the transcript level of transcripts encoding the large subunit of Rubisco (RbcL) as well as a photograph of total RNA are shown to demonstrate equal loading of RNA. Experiments were repeated at least six times. Representative results are shown.
(B) Changes in steady state transcript level of transcripts encoding all members of the APX gene family (APX1-7, tylAPX, and s/mAPX) in wild-type (control) and knockout-Apx1 (knockout) plants in response to a moderate level of light stress.
(C) Changes in steady state transcript level of transcripts encoding four HSFs (HSF21, HSF5, HSF4, and a putative HSF [pHSF]) all with an increase in expression of more than twofold during the light stress treatment.
(D) Changes in steady state level of transcripts encoding all members of the NADPH oxidase gene family (RbohA to J) in control and knockout-Apx1 (knockout) plants in response to a moderate level of light stress.
Time-course microarray analsysis ([B] to [D]) was repeated twice with similar results, and representative results are shown. RNA gel blots and microarray analysis were performed as described in Methods. Visualization of transcript levels in (B), (C), and (D) was performed with ArrayAssist. Statistical analysis of microarray results was performed as described in Methods. Transcripts that were significantly elevated in KO-Apx1 compared with wild-type plants are shown in Table 2 and in the supplemental data online.
Analysis of the steady state transcript level of all nine members of the Apx gene family in Arabidopsis during the light stress treatment revealed that the absence of APX1 did not result in the enhanced expression of transcripts encoding other members of this gene family (Figure 4B). We were also unable to detect a significant difference between the changes in the steady state level of other transcripts encoding H2O2-scavenging enzymes, such as catalase (three genes in Arabidopsis; Vandenabeele et al., 2004), peroxiredoxin (11 genes in Arabidopsis; Dietz, 2003), or glutathione peroxidase (eight genes in Arabidopsis; Rodriguez-Milla et al., 2003), in wild-type and KO-Apx1 plants during the light stress treatment (data not shown; Figure 4A for Cat1). We have previously shown that the HSF21 is constitutively elevated in KO-Apx1 plants grown at a light intensity of 100 μmol m−2 s−1 (Pnueli et al., 2003; see also Figure 4 for plants grown at 25 μmol m−2 s−1 and shifted to 250 μmol m−2 s−1). As shown in Figure 4C, a gene-specific analysis of the steady state transcript level of HSFs performed with the DNA chips (ATH1-Affymetrix) revealed that the steady state level of HSF21 and HSF5 is elevated in KO-Apx1 plants during early stages of response to light stress (only HSFs with a change of more than twofold in expression are shown in Figure 4C).
To study signal transduction events associated with H2O2 accumulation in KO-Apx1 plants, we identified all transcripts that were significantly elevated in KO-Apx1 plants compared with wild-type plants during the light stress treatment (see Methods). Table 2 presents 158 signal transduction, ROS-related, defense, transcription factor, and zinc-finger protein transcripts significantly elevated in KO-Apx1 plants compared with wild-type plants during the light stress treatment (more than twofold). Supplemental Table 1 (general metabolism and cell function), Supplemental Table 2 (putative disease resistance genes), Supplemental Table 3 (P450s), and Supplemental Table 4 (unknowns) online present all other transcripts significantly elevated in KO-Apx1 (more than twofold) compared with wild-type plants during the light stress treatment.
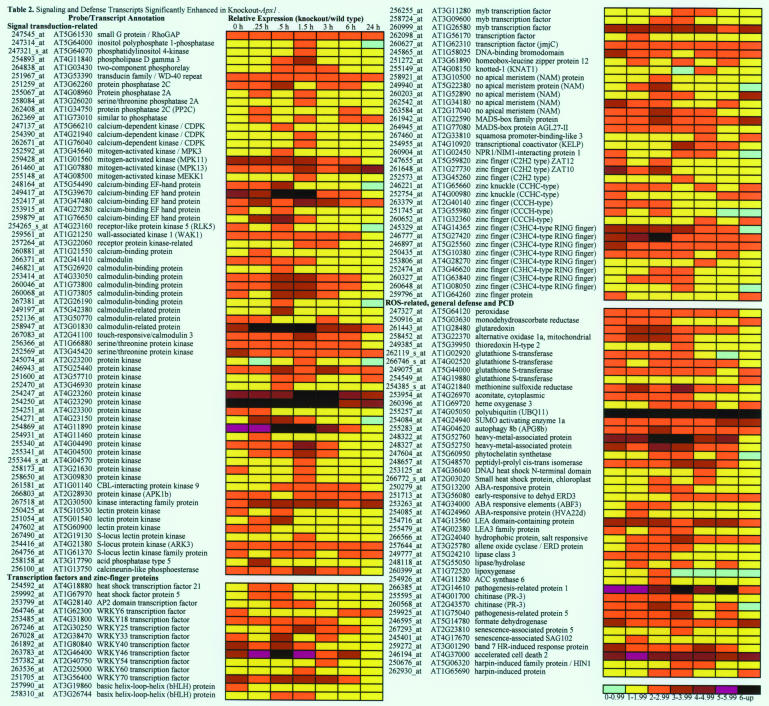
Table 2.
Signaling and Defense Transcripts Significantly Enhanced in Knockout-Apx1.
A heat map showing changes in the steady state level of transcripts encoding putative signaling, zinc-finger, transcription factors, and defense and programmed cell death (PCD) related proteins in response to a moderate level of light stress is shown on the right (relative expression is shown as knockout/wild type). Each probe is identified by its Affymetrix accession number (ATH1 chip) as well as by a locus identifier number on the left. All transcripts included in the table, as well as in Supplemental Tables 1 to 6 online, are significantly elevated in knockout-Apx1 plants compared with wild-type plants. Conditions for microarray analysis and statistical and data analysis are described in Methods.
As shown in Table 2, the level of transcripts encoding protein phosphatase 2C and calcium and calmodulin binding proteins is elevated in KO-Apx1 plants. This finding is in accordance with the proposed involvement of Ca2+ in H2O2 signaling (Rentel and Knight, 2004) and the possible function of protein phosphates 2C in sensing H2O2 (Desikan et al., 2004). The steady state level of a transcript encoding mitogen-activated protein kinase 3 (MAPK3) and other MAPK enzymes, several zinc-finger proteins, HSF5 and HSF21, and the WRKY transcription factors 6, 18, 25, 33, 40, 46, 54, 60, and 70 is also elevated in KO-Apx1 plants during light stress. Some of the WRKY transcription factors, as well as MAPK3, elevated in KO-Apx1 plants might be involved in ROS signaling and the enhanced expression of the pathogenesis-related (PR) proteins PR-1, -3, and -5 during light stress in KO-Apx1 plants (Table 2; Zhang and Klessig, 2001; Apel and Hirt, 2004; Ulker and Somssich, 2004). The enhanced expression of these pathogen defense mechanisms in KO-Apx1 plants suggest a high degree of cross talk between biotic and abiotic stresses mediated by H2O2 (Bowler and Fluhr, 2000; Mittler, 2002; Dat et al., 2003; see also Supplemental Table 2 online for disease resistance genes).
ROS-related transcripts enhanced in KO-Apx1 plants compared with wild-type plants included putative metal binding proteins, mitochondrial alternative oxidase (Aox1A), a class III peroxidase, thioredoxin, cytosolic glutaredoxin, and cytosolic monodehydroascorbate reductase (MDAR). Cytosolic glutaredoxin and MDAR are components of the ascorbate-glutathione cycle and might be elevated to compensate for the lack of cytosolic APX1 (Rizhsky et al., 2002; Pnueli et al., 2003; Mittler et al., 2004). The enhanced level of transcripts encoding mitochondrial Aox1A in KO-Apx1 plants support our findings that in the absence of cytosolic APX1, proteins from other compartments are subjected to oxidative stress (Table 1). The expression of at least two different transcripts associated with protein oxidation was also elevated in KO-Apx1 plants compared with wild-type plants. These encode for Met sulfoxide reductase that reduces oxidized Met residues in proteins (Bechtold et al., 2004) and aconitase that is highly sensitive to ROS damage (Yoo and Regnier, 2004). This finding supports our direct measurements of protein oxidation in KO-Apx1 plants (Table 1).
Additional transcripts with a possible involvement in cellular detoxification elevated in KO-Apx1 plants compared with wild-type plants included glutathione S-transferase (Table 2) and cytochrome P-450 (see Supplemental Table 3 online). The steady state level of transcripts encoding accelerated cell death 2 (Acd2), a lesion mimic gene, is also elevated in KO-Apx1 plants compared with wild-type plants. ACD2 might be linked to pathogen responses activated in KO-Apx1, or it might be required to suppress programmed cell death in KO-Apx1. It is also possible that the function of ACD2 (chlorophyll catabolite reductase; Mach et al., 2001) is related to oxidative stress induced in the chloroplast in KO-Apx1 plants.
Interestingly, the steady state level of most signaling and defense transcripts shown in Table 2 decreased at the 6- and 24-h time points, suggesting that the stress that was imposed on KO-Apx1 plants by the light shift treatment might have been transient. It is possible that KO-Apx1 plants were able to acclimate and decrease the level of stress by activating alternative H2O2 scavenging mechanisms, by activating photoprotective mechanisms, or by decreasing the rate of H2O2 production (Mittler, 2002; Muller-Moule et al., 2004). This finding might explain why KO-Apx1 plants were able to recover from the light stress treatment (data not shown).
Because NADPH oxidases were recently proposed to function as mediators of positive amplification loops during ROS signaling (Mittler et al., 2004), we tested the steady state transcript level of all members of the NADPH oxidase gene family during light stress. As shown in Figure 4D, the steady state level of transcripts encoding the NADPH oxidase D protein (RbohD) was transiently elevated in KO-Apx1 plants compared with wild-type plants (see also Figure 4A). The steady state level of transcripts encoding all other members of this gene family was, however, unchanged during the moderate light stress treatment in wild-type and KO-Apx1 plants (Figure 4D).
Functional Analysis of the Role of HSF21 and RbohD in Apx1 Transcript Expression during Light Stress in Wild-Type Plants
The steady state level of transcripts encoding the transcription factors HSF21 and HSF5 is rapidly elevated in KO-Apx1 plants in response to the light stress treatment (Figure 4C). At least in Drosophila melanogaster and mammalian cells the oligomerization and DNA binding of HSFs were shown to be induced in vivo and in vitro by H2O2, suggesting that HSFs act as direct sensors of H2O2 in cells (Zhong et al., 1998; Ahn and Thiele, 2003). The promoter of Apx1, as well as the promoters of many defense genes and transcription factors involved in H2O2 signaling and defense, contains an HSF binding motif (Mittler and Zilinskas, 1992; Rizhsky et al., 2004). Promoter analyses, as well as overexpression studies of HSF3 in Arabidopsis, suggest that the HSF binding site at the Apx1 promoter is functional (Storozhenko et al., 1998; Panchuk et al., 2002). However, no genetic evidence was presented for the role of HSFs in H2O2 signaling in plants.
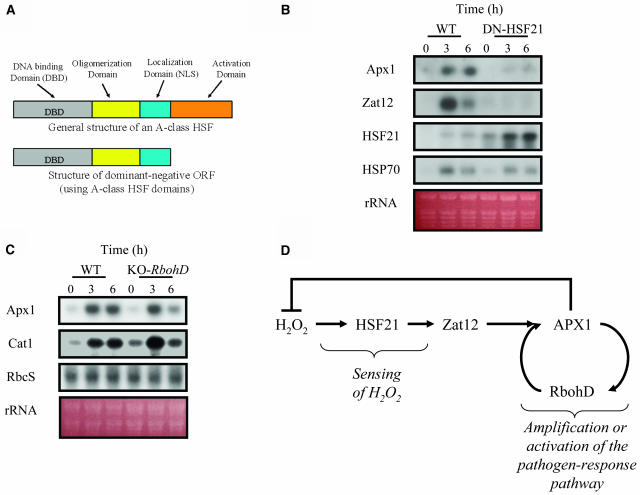
To study the importance of HSF21 (AtHSFA4a, an A-class HSF; Nover et al., 2001) to Apx1 expression during the light stress treatment described above, we generated transgenic plants that constitutively express a dominant-negative construct for HSF21. The expressed construct contained the entire HSF21 open reading frame, but without the HSF activation domain (Figure 5A). As shown in Figure 5B, constitutive expression of the dominant-negative construct for HSF21 prevented the accumulation of transcripts encoding APX1 and the zinc-finger protein Zat12 in response to light stress. By contrast, the accumulation of transcripts encoding the heat shock protein HSP70 (Figure 5B) or catalase (data not shown) was not inhibited in the dominant-negative expressing plants. Interestingly, the expression of transcripts encoding HSF21 was strongly elevated in the dominant-negative plants in response to light stress (tested with a probe for the activation domain of HSF21 that is missing from the dominant-negative construct; data not shown). Our findings that Apx1 and Zat12 transcript accumulation is inhibited in plants expressing the dominant-negative HSF21 construct suggest that HSF21 function is required for Zat12 and Apx1 expression during light stress (Figure 5B).
Figure 5.
Functional Analysis of HSF21 and RbohD in Transgenic and Knockout Arabidopsis Plants.
(A) A scheme describing the structure of the HSF21 dominant-negative construct expressed in transgenic plants.
(B) RNA gel blots showing the steady state transcript level of Apx1, Zat12, HSF21, and HSP70 in wild-type plants and transgenic plants expressing the dominant-negative construct for HSF21 (DN-HSF21) plants in response to a moderate level of light stress.
(C) RNA gel blots showing the steady state transcript level of Apx1 and Cat1 in wild-type plants and knockout plants lacking RbohD in response to a moderate level of light stress.
(D) A model showing the putative signal transduction pathway that controls the expression of Apx1 during a moderate level of light stress in Arabidopsis. Construction of transgenic plants, light stress experiments, and RNA gel blots were performed as described in Methods.
To study the relative importance of RbohD to Apx1 expression during the light stress treatment described above, we used knockout plants deficient in RbohD (KO-RbohD; Torres et al., 2002). As shown in Figure 5C, the steady state level of transcripts encoding APX1 was enhanced in response to light stress in both wild-type and KO-RbohD plants at 3 h. However, in contrast with wild-type plants, the steady state level of transcripts encoding APX1 declined at the 6-h time point in KO-RbohD plants. Similar results were found with transcripts encoding Cat1 (Figure 5C). Because the steady state level of transcripts encoding RbohD was elevated in KO-Apx1 plants at an early time point (0.5 h; Figure 5D), it is possible that RbohD is required for Apx1 and Cat1 expression during late stages of response to light stress, acting to amplify the signal that controls Apx1 and Cat1 expression (see Discussion; Figure 5D).
DISCUSSION
Cross-Compartment Protection by APX1
Several recent studies have demonstrated that ROS function as important signaling molecules involved in the control of processes such as pathogen defense, hormonal signaling, stress response, and plant growth and development (Torres et al., 2002; Foreman et al., 2003; Kwak et al., 2003). However, ROS are also toxic molecules capable of injuring or even killing plant cells, and their level in cells needs to be tightly regulated (Neill et al., 2002; Mittler et al., 2004). To prevent ROS, produced in the chloroplast, from damaging cells and possibly interfering with ROS signaling in other compartments, the chloroplast contains multiple ROS scavenging systems, including at least two complete pathways for H2O2 removal at the thylakoid (water–water cycle; Asada, 1999) and the stroma (ascorbate-glutathione cycle; Asada and Takahashi, 1987). Although these pathways are thought to be sufficient for proper H2O2 scavenging during photosynthesis (Asada and Takahashi, 1987; Asada, 1999; Polle, 2001), our findings suggest that the cytosolic ascorbate-glutathione cycle is required for H2O2 removal during photosynthesis (Figures 1 and 2, Table 1). Moreover, our findings suggest that the cytosolic pathway could even compensate or protect the cell when the stromal pathway is absent (Figure 3). This type of cross-compartment protection might also be found with other organelles, such as peroxisomes or mitochondria, because H2O2 is transported across biological membranes (Willekens et al., 1997; Henzler and Steudle, 2000). The extent of cross-compartment protection, however, might be limited because at least in tobacco (Nicotiana tabacum) and wheat (Triticum aestivum) a deficiency in thylakoid-bound APX was shown to have adverse effects on plant growth and photosynthetic performance (Yabuta et al., 2002; Danna et al., 2003). These findings suggest that cytosolic APX1 might not be able to protect the chloroplast against H2O2 produced at the surface of the thylakoid membrane (see also Rizhsky et al., 2003 for the effect of disrupting the water–water cycle in Arabidopsis).
Interestingly, at least at the steady state transcript level, none of the other Apx genes, including the cytosolic Apx2 gene, responded to the deficiency in APX1 (Figure 4B; Apx2 expression was however elevated in KO-Apx1 plants in response to heat shock; data not shown; Pnueli et al., 2003). KO-Apx1 plants grown under controlled growth conditions are stunted and have a late flowering phenotype; however, they are viable and productive (Figure 3A; see also Pnueli et al., 2003). Although the steady state level of transcripts encoding several ROS-scavenging enzymes, including MDAR and glutaredoxin, is elevated in KO-Apx1 plants, it is not entirely clear how they compensate for the lack of APX1 (Table 2; Mittler et al., 2004). The exposure of KO-Apx1 plants to a moderate level of light stress does, however, reveal that APX1 is important during early stages of plant acclimation to this treatment (Figures 1 and 2, Tables 1 and 2). There may be two different explanations for the function of APX1 under these conditions: (1) APX1 might be directly involved in scavenging of H2O2 that leaks from the chloroplast or peroxisomes, and/or (2) APX1 might be required for the signal transduction pathway that is activated in plants during the light stress treatment, and in its absence the activation of certain defense mechanisms is prevented. We were unable to find significant differences in transcript expression between the wild type and KO-Apx1 that could account for the later explanation. Asada (2000) demonstrated that intact chloroplasts are sensitive to the external application of H2O2. In view of this report, the enhanced production of H2O2 in KO-Apx1 plants and the accumulation of oxidized proteins from different cellular compartments in KO-Apx1, we favor the first explanation suggesting that APX1 is directly involved in H2O2 scavenging during light stress. As a key cytosolic enzyme responsible for H2O2 scavenging in Arabidopsis, it is possible that APX1 functions as a defense barrier between the three major ROS-producing organelles of plant cells (i.e., the chloroplast, mitochondria, and peroxisomes) and/or the redox- and oxidative stress–sensitive regulatory mechanisms of the nuclei. The damage to chloroplastic APXs in KO-Apx1 plants (Figure 2) suggests that a key role of cytosolic APX1 is to protect the chloroplast from H2O2 that may diffuse from the mitochondria, cytosol, or peroxisome during photosynthesis.
Because our experimental design involved acclimating plants to a low level of light intensity before the application of light stress, it is possible that some photoprotective mechanisms were not activated in our pretreated plants, exacerbating the effect of the light stress treatment and making the dependence on H2O2-scavenging systems central to the acclimation of plants to this treatment. At later time points, these mechanisms might be activated to protect the cell, and the dependence on APXs might be reduced (see above for Table 2; Muller-Moule et al., 2004).
Apx1 Signal Transduction: A Key Role for HSFs and Cross Talk between Biotic and Abiotic Stresses
At least three different types of sensor molecules might be involved in H2O2 perception in plants. Hydrogen peroxide might be sensed by a two-component kinase system or other receptor-type molecules, by direct inhibition of phosphatase activity, or by redox-sensitive transcription factors such as HSF (Desikan et al., 2004; Mittler et al., 2004). Transcripts encoding some of these putative sensing mechanisms might have been identified by our microarray analysis (Table 2; see supplemental data online). The finding that expression of a dominant-negative construct for HSF21 in transgenic Arabidopsis plants prevented the accumulation of transcripts encoding APX1 during light stress (Figure 5) strongly suggests that HSFs are involved in the signal transduction pathway controlling Apx1 expression under the conditions tested. Expression of the dominant-negative construct also prevented the accumulation of transcripts encoding Zat12, an H2O2-responsive zinc-finger protein required for Apx1 expression during oxidative stress (Figure 5; Rizhsky et al., 2004). This finding suggests that HSF21 is required at a relatively early stage of the oxidative stress acclimation response. It should be noted that the promoter of Zat12 contains at least two copies of the HSF binding element (Rizhsky et al., 2004), suggesting that HSF21 might function by directly binding to the promoter of Zat12.
Although the dominant-negative construct was designed using the HSF21 open reading frame (Figure 5A), there are 21 different HSF genes in Arabidopsis, and oligomers (three subunits per DNA binding domain) of HSFs could contain multiple copies of a particular factor or monomers encoded by different HSF genes (Nover et al., 2001). Thus, it is not possible to determine without further experimental work the specificity of the effect of the dominant-negative construct to the HSF21 protein. Nonetheless, our findings with the dominant-negative construct for HSF21 could be viewed as the first genetic evidence that HSFs are important sensors for H2O2. HSF21 might be a good candidate for a sensor because it is constitutively expressed in cells in the absence of stress and its expression is elevated in KO-Apx1 plants (Figure 4C).
The putative order of events that follows the accumulation of H2O2 in Arabidopsis leaf cells in response to a moderate level of light stress is shown in Figure 5D. NADPH oxidases were recently proposed to function as generators of ROS for different signal transduction purposes (Torres et al., 2002; Foreman et al., 2003; Kwak et al., 2003). Our microarray analysis (Figure 4D), as well as our analysis of KO-RbohD plants (Figure 5C), suggest that the NADPH oxidase protein RbohD might be involved in the signal transduction pathway of Apx1 as a positive amplification factor that maintains the expression of the Apx1 transcript at a high steady state level (Figure 5D). Thus, in the absence of RbohD, the steady state level of transcripts encoding APX1 is elevated at 3 h but declines at 6 h. It is not clear, however, whether RbohD functions as part of the oxidative stress-response pathway of Arabidopsis or whether it is activated in KO-Apx1 plants as part of the pathogen response pathway, similar to transcripts encoding PR proteins elevated in KO-Apx1 plants (Table 2). Perhaps these two responses (i.e., the oxidative stress response and the pathogen response) could not be separated in KO-Apx1 plants because the APX1 protein is involved in both (Mittler et al., 1998, 1999). The large number of pathogen response transcripts, including disease resistance genes (see Supplemental Table 2 online), elevated in KO-Apx1 plants in response to the light stress treatment, demonstrate the high degree of overlap that exists between biotic and abiotic stresses. It is likely that H2O2 is the mediator of this cross talk (Bowler and Fluhr, 2000; Mittler, 2002; Dat et al., 2003). Because the steady state level of Apx1 is enhanced in response to oxidative stress, as well as pathogen attack (Mittler and Zilinskas, 1992; Mittler et al., 1998), it is possible that the elevation in Apx1 steady state transcript level at the 3-h time point is a result of activating the oxidative stress response pathway, whereas the elevation of Apx1 transcripts at the 6-h time point is a result of activating the pathogen response pathway (Figure 5C). In KO-RbohD plants, the later pathway might be suppressed, whereas in the dominant-negative HSF21 plants, both are suppressed (Figure 5). Our findings might suggest that the expression of transcripts encoding other ROS response and general defense proteins, in response to abiotic stress, should be reevaluated in view of the cross talk between biotic and abiotic stresses that might result from the accumulation of H2O2 in cells. Thus, a biphasic mechanism might control the expression of these transcripts. In the first phase, their expression might be elevated because of direct sensing of abiotic stress–generated H2O2 in cells, and in the second phase their expression might be elevated as an integral part of the pathogen response pathway activated during abiotic stress (this pathway might be suppressed in KO-RbohD plants; Figure 5C). A similar biphasic process might be activated in response to pathogen attack; however, during this process, the pathogen response pathway is activated first (this pathway will generate H2O2 in cells through the action of NADPH oxidases), and the abiotic stress, H2O2 response pathway is second.
Proteomics of Protein Oxidation
Protein carbonylation is an irreversible oxidative process leading to a loss of function and often proteolysis of the modified protein (Stadtman, 1992). Proteomic analysis of protein oxidation is emerging as a new tool to study oxidative stress and damage caused to cells during disease or different environmental stresses (Akenov et al., 2001; Rizhsky et al., 2004). Johansson et al. (2004) have identified different proteins oxidized in Arabidopsis plants grown under controlled conditions. They have identified seven chloroplastic and five mitochondrial proteins oxidized in cells at the end of the vegetative phase of development (i.e., before bolting). Interestingly, only one of the proteins Johansson et al. (2004) have identified in their study (Rubisco large subunit; RbcL) was identified by our proteomic analysis of proteins oxidized in KO-Apx1 plants during the moderate light stress treatment (Figure 1B, Table 1). The RbcL protein was initially identified by Rizhsky et al. (2004) as the major protein carbonylated in knockout-Zat12 plants in response to oxidative stress. Prior analysis of Rubisco has demonstrated that this protein is prone to different types of oxidative damage (Mehta et al., 1992). However, because of the high abundance of the RbcL protein in chloroplasts, it is not entirely clear whether it is oxidized as a general byproduct of oxidative stress in plants or whether its oxidation is a specific process associated with oxidative damage to cells and inhibition of photosynthesis.
Proteomic analysis of protein oxidation during stress, especially in different genetic backgrounds lacking specific ROS-defense enzymes, is a particularly powerful approach because it can pinpoint the specific proteins damaged in specific mutants and link between enzymatic activities in particular compartments and oxidative damage at the cellular level. Thus, our findings that chloroplastic, mitochondrial, and membrane-bound proteins are oxidized in mutant plants lacking a cytosolic H2O2-scavenging enzyme (KO-Apx1; Table 1) provides direct evidence that this enzyme is key to the protection of adjacent compartments (see also Figures 1 to 3). The notion that each individual compartment is protected by its own set of ROS-scavenging enzymes should therefore be reevaluated, and a new view of the ROS network should be adopted as a highly interlinked network that requires the coordinated function of ROS-scavenging pathways from different cellular compartments to modulate the level of ROS in cells, prevent cellular damage, and control ROS signaling.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (cv Columbia and Ws) plants were grown in growth chambers (Percival E-30, AR-66; Percival Scientific, Perry, IA) under controlled conditions: 21 to 22°C, constant light, 25 μmol m−2 s−1, and a relative humidity of 70%. Knockout Arabidopsis lines containing a T-DNA insert in the stromal/mitochondrial APX gene (s/mApx, At4g08390; obtained through the SIGnAL project, http://signal.salk.edu/tabout.html) were out-crossed and selfed to check for segregation as previously described (Pnueli et al., 2003; Rizhsky et al., 2004). Screening for expression of s/mAPX was performed by protein blots using an antibody raised against a conserved domain of cytosolic and chloroplastic APXs. Knockout plants deficient in cytosolic Apx1 (KO-Apx1) were obtained as described by Pnueli et al. (2003). Light stress treatments were performed by changing the light intensity from 25 to 250 μmol m−2 s−1, and all other parameters were maintained constant. At different times, leaves were excised, flash frozen in liquid nitrogen, and stored for further analysis at −80°C. All experiments were performed in triplicate and repeated at least three times (with the exception of DNA arrays that were performed as described below). Representative results are shown in the different figures. Analysis of seedling tolerance to paraquat was performed on agar plates as described by Rizhsky et al. (2004). Vector construction, PCR cloning and sequencing, and transformation of Arabidopsis plants was performed as described by Rizhsky et al. (2004). Transgenic plants were screened by RNA and protein blots (Pnueli et al., 2003; Rizhsky et al., 2004).
Molecular, Physiological, and Biochemical Analysis
RNA and protein were isolated and analyzed by RNA and protein blots as previously described (Pnueli et al., 2003; Rizhsky et al., 2004). RNA staining or a ribosomal 18S rRNA probe were used to control for RNA loading. Coomassie Brilliant Blue staining of protein gels was used to control for protein loading. Photosynthetic activity was measured with a LI-6400 apparatus (LI-COR Biosciences, Lincoln, NE) as described by Rizhsky et al. (2003). Detection of H2O2 was performed by infiltrating excised leaves with a solution of 1 mg/mL diaminobenzidine prepared in water. Leaves were placed on two layers of 3MM Whatman paper (Clifton, NJ) soaked with water and exposed to light for different times. Leaves were then fixed with a solution of 3:1:1 (v:v:v) ethanol:lactic acid:glycerol, washed with 75, 50, and 25% ethanol, equilibrated with water, and photographed. Detection of protein oxidation by protein blots was performed with the OxyBlot protein oxidation kit (Chemicon International, Temecula, CA) as recommended by the manufacturer (see also Rizhsky et al., 2004). Antibodies against APX1 were obtained as described by Mittler and Zilinskas (1991). Polyclonal antibodies that react with tylAPX, s/mAPX, and cAPX (APX1) were prepared in rabbits against a fragment of thyAPX (from Lys100 to Ile341) cloned into pCAL-n, expressed in bacteria and purified by affinity chromatography. Antibodies against RbcL, RbcS, and Cytf (a component of the b6f complex) were a gift of R. Nechushtai (Hebrew University, Jerusalem, Israel).
Proteomic Analysis of Oxidized Proteins
Oxidized proteins were extracted by grinding 200 mg of tissue in liquid nitrogen. The powder was dissolved in 1 mL of extraction buffer (25 mM Tris-Cl, pH 8, 0.1% Triton X-100, 50 mM DTT, and the protease inhibitors leupeptin [0.5 μg/mL], trypsin inhibitor [0.5 μg/mL], and PMSF [40 μg/mL]). After centrifugation, the supernatant was stored at −80°C until analysis. The protein (250 μL containing ∼500 μg protein) was derivatized by adding 1 mL of 10 mM 2,4-dinitrophenylhydrazine in 2 M HCl and incubating for 30 min. The protein was precipitated with 25% trichloroacetic acid, and the pellet, collected by centrifugation, was washed with cold ethanol/ethyl acetate (1:1; v:v), dried in a vacuum, and redissolved in 100 μL of rehydration buffer (7 M urea, 2 M thiourea, 2 mM tributyl phosphine, 4% CHAPS, and 40 mM Tris). After centrifugation, the resulting solution was diluted with 2 mL of coupling buffer (0.1 M NaHCO3 and 0.5 M NaCl, pH 8.3 to 8.5) and mixed with 15 mg of anti-DNP affinity column matrix. The column matrix was produced using cyanogen bromide–activated beads (Sigma-Aldrich, St. Louis, MO) and anti- dinitrophenylhydrazone (DNP) antibody (Bethyl Lab, Montgomery, TX) using the methods included with the beads. After incubating for 1 h, the matrix was loaded into a column made from a 200-μL pipette tip, washed with 4 mL of coupling buffer, and eluted with 200 μL of 5% SDS. Samples were resolved on one- or two-dimensional PAGE. The oxidized proteins were then detected by protein gel blotting.
For MALDI-TOF analysis, proteins identified on the protein gel blots were punched out of a Coomassie Brilliant Blue–stained gel and processed as described by Porubleva et al. (2001). Peptide masses were analyzed with MS-Fit (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm). Matches were deemed significant when at least four fragments were matched, covering more than 20% of the protein.
GeneChip Microarray Experiments
In six independent experiments, RNA was isolated from wild-type and KO-Apx1 plants grown under controlled conditions and subjected to a moderate level of light stress as described above (samples were obtained at 0, 0.25, 0.5, 1.5, 3, 6, and 24 h after light stress application, 15 plants per time point, and RNA was isolated using Trizol; Pnueli et al., 2003; Rizhsky et al., 2004). For each time point, RNA from three independent experiments was pooled to generate 28 RNA pools (two RNA pools for each time point for a total of 14 wild-type and 14 KO-Apx1 pools). These RNA samples were used to perform chip hybridization analyses (Arabidopsis ATH1 chips; Affymetrix, Santa Clara, CA) at the Virginia Bioinformatics Institute Core Laboratory Gene Expression Facility (https://www.vbi.vt.edu/). Conditions for RNA isolation, labeling, hybridization, and data analysis are described by Pnueli et al. (2003) and Rizhsky et al. (2004). Data visualization and analysis were performed with the GeneChip mining tool version 5.0 (Silicon Genetics, Redwood City, CA), GeneSpring version 5.1, and ArraAassit (IobionLab, La Jolla, CA). Some of the results were confirmed by RNA gel blots. See details for data analysis below.
GeneChip Data Processing and Analysis
All GeneChip arrays were processed first by robust multiarray average (RMA) (Irizarry et al., 2003) using the R package affy (Gautier et al., 2004). Specifically, expression values were computed from raw CEL files by first applying the RMA model of probe-specific correction of perfect match probes. These corrected probe values were then normalized via quantile normalization, and a median polish was applied to compute one expression measure from all probe values. Resulting RMA expression values were log2-transformed. (Please see the affy manual at www.bioconductor.org/repository/devel/vignette/affy.pdf for details.) Density plots and box plots of RMA expression value distributions of all arrays were very similar with no apparent outlying arrays (see Supplemental Figure 1 onlinw). Digestion curves describing trends in RNA degradation between the 5′ end and the 3′ end in each probe set were generated, and all 28 proved very similar, with a downward trend at the 5′ end (see Supplemental Figure 2 online).
Pearson correlation coefficients and Spearman rank coefficients were computed on the RMA expression values (log base 2) for each set of biological replicates. Spearman coefficients ranged from 0.977 to 0.994; Pearson coefficients ranged between 0.982 and 0.995 (see Supplemental Table 5 online). A scatter plot of RMA expression values between the two wild-type biological replicates at 6 h is presented in Supplemental Figure 3 online.
To determine whether genes were differentially expressed between genotypes across the temporal states, analysis of variance (ANOVA) was performed on the RMA expression values. (For an overview on the application of ANOVA to microarray data, please see Kerr et al., 2000.) The following model was used for this analysis: yijk = Vi + Tj + (VT)ij + ɛijk, where yijk denotes the log2 signal measured for variety i, time j, and biological replicate k, with 1 ≤ i ≤ 2, 1 ≤ j ≤ 7, and 1 ≤ k ≤ 2. The terms Vi and Tj measure the effect of the variety and time point, respectively, and the interaction term (VT)ij accounts for the interaction between variety and time. An ANOVA was performed on each gene using the linear model above, and six contrasts based on differences of genotypes with respect to the last six time points. (Differences between genotypes at time 0 were not of interest in this study.) The R package limma was used for ANOVA methods (www.bioconductor.org/repository/devel/vignette/affy.pdf) A multiple testing correction (Benjamini and Hochberg, 1995) was applied to the P values of the F-statistics to adjust the false discovery rate. Genes with adjusted F-statistic P values < 0.05 were extracted for further analysis. This resulted in 3915 genes with significant F-statistics based on the ANOVA above (see Supplementary Table 6 online). Expression values of this selection were then inverse log transformed, and genes with differential expression between wild-type and knockout expression values of more than twofold during at least one time point were selected. This selection included 843 genes. Similarly, genes with differential expression between wild-type and knockout expression values of <0.5-fold during at least one time point were selected as suppressed (see Supplemental Table 7 online). Interestingly, no gene was both upregulated and downregulated by more than twofold in knockout across the time series, enabling a clear separation between upregulated and downregulated genes. Storey multiple testing adjustments (Storey and Tibshirani, 2003) were also performed on the P values of the F-statistics. All P values found significant by the Benjamini-Hochberg correction were also found to be significant in the Storey correction. The Storey correction contained 691 additional genes that were not included in the analysis.
Supplementary Material
Acknowledgments
We thank Jeffery Dangl and Miguel Angel Torres for the gift of the RbohD probe and KO-RbohD seeds and Rachel Nechushtai for the gift of antisera to RbcL, RbcS, and Cytf. This work was supported by funding from the National Science Foundation (NSF-0431327), the Plant Sciences Institute at Iowa State University, and the Biotechnology Council of Iowa State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Ron Mittler (ronm@unr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026971.
References
- Ahn, S.G., and Thiele, D.J. (2003). Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akenov, M.Y., Aksenova, M.V., Butterfield, D.A., Geddes, J.W., and Markesbery, W.R. (2001). Protein oxidation in the brain in Alzheimer's disease. Neuroscience 103, 373–383. [DOI] [PubMed] [Google Scholar]
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygen and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. [DOI] [PubMed] [Google Scholar]
- Asada, K. (2000). The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, K., and Takahashi, M. (1987). Production and scavenging of active oxygen in photosynthesis. In Photoinhibition, D.J. Kyle, C.B. Osmond, and C.J. Arntzen, eds (Amsterdam: Elsevier), pp. 227–287.
- Bechtold, U., Murphy, D.J., and Mullineaux, P.M. (2004). Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights. Plant Cell 16, 908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bowler, C., and Fluhr, R. (2000). The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 5, 241–246. [DOI] [PubMed] [Google Scholar]
- Chew, O., Whelan, J., and Millar, A.H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. [DOI] [PubMed] [Google Scholar]
- Corpas, F.J., Barroso, J.B., and del Rio, L.A. (2001). Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 6, 145–150. [DOI] [PubMed] [Google Scholar]
- Danna, C.H., Bartoli, C.G., Sacco, F., Ingala, L.R., Santa-Maria, G.E., Guiamet, J.J., and Ugalde, R.A. (2003). Thylakoid-bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiol. 132, 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat, J., Vandenabeele, S., Vranova, E., Van Montagu, M., Inze, D., and Van Breusegem, F. (2000). Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 57, 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat, J.F., Pellinen, R., Beeckman, T., Van De Cotte, B., Langebartels, C., Kangasjarvi, J., Inze, D., and Van Breusegem, F. (2003). Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 33, 621–632. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Cheung, M.K., Bright, J., Henson, D., Hancock, J.T., and Neill, S.J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55, 205–212. [DOI] [PubMed] [Google Scholar]
- Dietz, K.J. (2003). Plant peroxiredoxins. Annu. Rev. Plant Biol. 54, 93–107. [DOI] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Gautier, L., Cope, L., Bolstad, B., and Irizarry, R.A. (2004). Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J.M.C. (1989). Free Radicals in Biology and Medicine. (Oxford: Clarendon Press).
- Henzler, T., and Steudle, E. (2000). Transport and metabolic degradation of hydrogen peroxide in Chara corallina: Model calculations and measurements with the pressure probe suggest transport of H(2)O(2) across water channels. J. Exp. Bot. 51, 2053–2066. [DOI] [PubMed] [Google Scholar]
- Iida, A., Kazuoka, T., Torikai, S., Kikuchi, H., and Oeda, K. (2000). A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 24, 191–203. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, R., Collin, R., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Johansson, E., Olsson, O., and Nystrom, T. (2004). Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 279, 22204–22208. [DOI] [PubMed] [Google Scholar]
- Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, S., Reynolds, H., Karpinska, B., Wingsle, G., Creissen, G., and Mullineaux, P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284, 654–657. [DOI] [PubMed] [Google Scholar]
- Kerr, M.K., Martin, M., and Churchill, G.A. (2000). Analysis of variance for gene expression microarray data. J. Comput. Biol. 7, 819–837. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, J., Ohno, C., Domae, Y., and Asada, K. (2001). Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: Its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim. Biophys. Acta 1504, 275–287. [DOI] [PubMed] [Google Scholar]
- Mehta, R.A., Fawcett, T.W., Porath, D., and Mattoo, A.K. (1992). Oxidative stress causes rapid membrane translocation and in vivo degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 267, 2810–2816. [PubMed] [Google Scholar]
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler, R., Feng, X., and Cohen, M. (1998). Post-transcriptional suppression of cytosolic ascorbate peroxidase expression during pathogen-induced programmed cell death in tobacco. Plant Cell 10, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Hallak-Herr, E., Orvar, B.L., Van Camp, W., Willekens, H., Inze, D., and Ellis, B. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyper-responsive to pathogen infection. Proc. Natl. Acad. Sci. USA 96, 14165–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). The reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Mittler, R., and Zilinskas, B. (1991). Purification and characterization of pea cytosolic ascorbate peroxidase. Plant Physiol. 97, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., and Zilinskas, B. (1992). Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J. Biol. Chem. 267, 21802–21807. [PubMed] [Google Scholar]
- Muller-Moule, P., Golan, T., and Niyogi, K.K. (2004). Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 134, 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux, P., and Karpinski, S. (2002). Signal transduction in response to excess light: getting out of the chloroplast. Curr. Opin. Plant Biol. 5, 43–48. [DOI] [PubMed] [Google Scholar]
- Neill, S., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5, 388–395. [DOI] [PubMed] [Google Scholar]
- Noctor, G., and Foyer, C. (1998). Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. [DOI] [PubMed] [Google Scholar]
- Nover, L., Bharti, K., Doring, P., Mishra, S.K., Ganguli, A., and Scharf, K.D. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk, I.I., Volkov, R.A., and Schoffl, F. (2002). Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129, 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli, L., Hongjian, L., and Mittler, R. (2003). Growth suppression, abnormal guard cell response, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J. 34, 187–203. [DOI] [PubMed] [Google Scholar]
- Polle, A. (2001). Dissecting the superoxide dismutase-ascorbate peroxidase-glutathione pathway in chloroplasts by metabolic modeling. Computer simulations as a step towards flux analysis. Plant Physiol. 126, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubleva, L., Vander Velden, K., Kothari, S., Oliver, D.J., and Chitnis, P.R. (2001). The proteome of maize leaves: Use of gene sequences and EST data for identification of proteins with peptide mass fingerprints. Electrophoresis 22, 1724–1783. [DOI] [PubMed] [Google Scholar]
- Rentel, M.C., and Knight, M.R. (2004). Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol. 135, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky, L., Davletova, S., Liang, H., and Mittler, R. (2004). The zinc-finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279, 11736–11743. [DOI] [PubMed] [Google Scholar]
- Rizhsky, L., Hallak-Herr, E., Van Breusegem, F., Rachmilevitch, S., Rodermel, S., Inze, D., and Mittler, R. (2002). Double antisense plants with suppressed expression of ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants with suppressed expression of ascorbate peroxidase or catalase. Plant J. 32, 329–342. [DOI] [PubMed] [Google Scholar]
- Rizhsky, L., Liang, H., and Mittler, R. (2003). The water-water cycle is essential for chloroplast protection in the absence of stress. J. Biol. Chem. 278, 38921–38925. [DOI] [PubMed] [Google Scholar]
- Rodermel, S. (2001). Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 6, 471–478. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Milla, M.A., Maurer, A., Rodriguez-Huete, A., and Gustafson, J.P. (2003). Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 36, 602–615. [DOI] [PubMed] [Google Scholar]
- Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., and Yoshimura, K. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. [PubMed] [Google Scholar]
- Stadtman, E.R. (1992). Protein oxidation and aging. Science 257, 1220–1224. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko, S., De Pauw, P., Van Montagu, M., Inze, D., and Kushnir, S. (1998). The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 118, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker, B., and Somssich, I.E. (2004). WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. [DOI] [PubMed] [Google Scholar]
- Vandenabeele, S., Vanderauwera, S., Vuylsteke, M., Rombauts, S., Langebartels, C., Seidlitz, H.K., Zabeau, M., Van Montagu, M., Inze, D., and Van Breusegem, F. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58. [DOI] [PubMed] [Google Scholar]
- Willekens, H., Chamnongpol, S., Davey, M., Schraudner, M., Langebartels, C., Van Montagu, M., Inze, D., and Van Camp, W. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C-3 plants. EMBO J. 16, 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta, Y., Motoki, T., Yoshimura, K., Takeda, T., Ishikawa, T., and Shigeoka, S. (2002). Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J. 32, 915–925. [DOI] [PubMed] [Google Scholar]
- Yoo, B.S., and Regnier, F.E. (2004). Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis 25, 1334–1341. [DOI] [PubMed] [Google Scholar]
- Yoshimura, K., Yabuta, Y., Ishikawa, T., and Shigeoka, S. (2002). Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 277, 40623–40632. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhong, M., Orosz, A., and Wu, C. (1998). Direct sensing of heat shock and oxidation by Drosophila heat shock transcription factor. Mol. Cell 2, 101–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.