Abstract
Background: The plants in the Erigeron genus of the Compositae (Asteraceae) family are commonly called fleabanes, possibly due to the belief that certain chemicals in these plants repel fleas. In the traditional Chinese medicine, Erigeron breviscapus, which is native to China, was widely used in the treatment of cerebrovascular disease. A handful of bioactive compounds, including scutellarin, 3,5-dicaffeoylquinic acid, and 3,4-dicaffeoylquinic acid, have been isolated from the plant. With the purpose of finding novel medicinal compounds and understanding their biosynthetic pathways, we propose to sequence the genome of E. breviscapus. Findings: We assembled the highly heterozygous E. breviscapus genome using a combination of PacBio single-molecular real-time sequencing and next-generation sequencing methods on the Illumina HiSeq platform. The final draft genome is approximately 1.2 Gb, with contig and scaffold N50 sizes of 18.8 kb and 31.5 kb, respectively. Further analyses predicted 37 504 protein-coding genes in the E. breviscapus genome and 8172 shared gene families among Compositae species. Conclusions: The E. breviscapus genome provides a valuable resource for the investigation of novel bioactive compounds in this Chinese herb.
Keywords: Erigeron breviscapus, Illumina sequencing, PacBio sequencing
Background
Erigeron breviscapus (also known as dengzhanhua in Chinese) is a perennial flower in the Erigeron genus of the Compositae (Asteraceae) family. Its flower head is comprised of yellow disk florets and multiple surrounding blue to purple ray florets (Fig. 1). This species is endemic to Southwestern China, and it grows in mid-altitude mountains, subalpine open slopes, grasslands, and forest margins from 1000 m to 3500 m [1, 2]. In traditional Chinese medicine, E. breviscapus is believed to improve blood circulation and ameliorate platelet coagulation [3, 4]. Since the 1980s, herbal extracts and bioactive compounds from E. breviscapus have been widely used for the treatment of cerebral embolism and its complications, cerebral thrombosis, coronary heart disease, angina pectoris, acute renal failure, and nephritic syndrome [5]. At present, more than 1000 tons of dry E. breviscapus are collected and used in the pharmaceutical industry each year, greatly exhausting the wild resources of this species [6, 7]. In this study, we report the draft genome assembly of E. breviscapus. Because of the high heterozygosity of the E. breviscapus genome, we adopted both Illumina sequencing and PacBio single-molecular real-time sequencing in the assembly procedure.
Figure 1:
Example of the E. breviscapus (image from Shengchao Yang).
Data Description
Whole-genome shotgun sequencing of E. breviscapus on Illumina platform
E. breviscapus seedlings were provided by Longjin Pharmaceutical Co., Ltd., and maintained in a greenhouse at the Yunnan Agricultural University. Genomic DNA was extracted from the leaf tissues of a single E. breviscapus plant using the GenEluteTM Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, USA). Paired-end libraries with insert sizes ranging from 150 bp to 800 bp were constructed using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, USA), and mate pair libraries with insert sizes from 2 kb to 20 kb were constructed using Illumina Nextera Mate Pair Library Preparation Kit (Illumina, USA). All constructed libraries were sequenced on a HiSeq 2500 platform (Illumina, USA) using either a PE-100 or PE-90 module (Additional File 1: Table S1). In total, about ∼413.4 Gb of raw data were generated on the Illumina platform. All reads were preprocessed for quality control and filtered using our in-house Perl script. The raw data were initially filtered by removing reads with more than 10% N or more than 40 bp low-quality bases. Next, redundant reads resulting in duplicate base calls were filtered at a threshold of Euclidean distance ≤3 and mismatch rate of ≤0.1. Only one copy of any duplicated paired-end reads was retained. Finally, both read 1 and read 2 were removed if they contained an adapter ≥10 bp with a mismatch rate ≤0.1. This process yielded ∼275.1 Gb of clean data for the de novo assembly of the E. breviscapus genome (Additional File 1: Table S1).
Single-molecule real-time sequencing of long reads on PacBio platform
Single-molecule real-time (SMRT) sequencing of long reads on a PacBio RS II platform (Pacific Biosciences, USA) was used to assist the subsequent de novo genome assembly process [8]. In brief, 40 μg of sheared DNA was used to construct 26 SMRT Cell libraries with an insert size of 17 kb. These libraries were sequenced in 105 SMRT DNA sequencing cells using the P6 polymerase/C4 chemistry combination and a data collection time of 240 minutes per cell. The sequencing produced about 62.4 Gb of clean data, consisting of 6 802 553 reads with an average read length of 9175 bp (Additional File 1: Table S1).
Estimation of the E. breviscapus genome size
The genome size of E. breviscapus was estimated by flow cytometry using Oryza sativa Nipponbare as internal standard and propidium iodide as the stain. The result showed that the genome size of E. breviscapus was approximately 1.52 Gb (Additional File 1: Figure S1).
Estimation of the E. breviscapus genome heterozygosity using k-mer analysis
Quality-filtered reads from the Illumina platform were subjected to 23-mer frequency distribution analysis with Jellyfish (v. 2.2.5) [9]. Analysis parameters were set at -k 23, and the final result was plotted as a frequency graph (Additional File 1: Figure S2). Two distinctive modes were observed from the distribution curve: the lower peak at a depth of 57 reflected the high heterozygosity of the E. breviscapus genome.
Hybrid de novo genome assembly of E. breviscapus
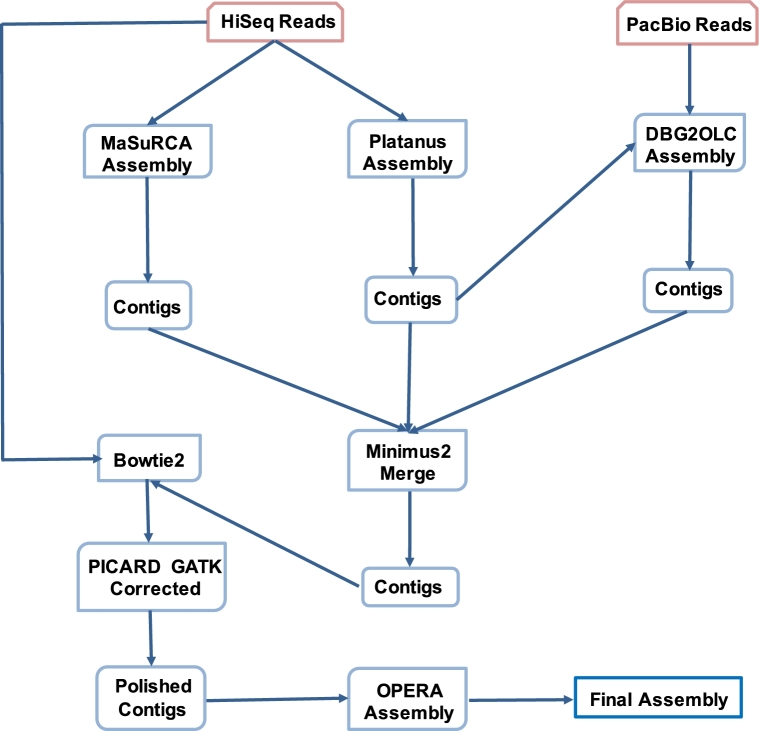
A hybrid genome assembly pipeline was used to overcome challenges posed by the heterozygous E. breviscapus genome (Fig. 2). HiSeq reads were first assembled using MaSuRCA (v. 3.1.3) [10] with default parameters, and also using Platanus (v. 1.2.1) [11] with parameters “-m 500 -k 43 -s 5 -d 0.3 -u 0.15 -c 3,” resulting in two contig assemblies. The Platanus-generated contigs, together with PacBio reads, were used to generate a third contig assembly by DBG2OLC with default parameters [12]. The three different contig assemblies were merged together by Minimus2 Amos (v. 3.1.0) using default parameters [13]. To eliminate possible errors of the merged contig assembly: (i) Bowtie2 (v. 2.1.0) [14] was used to align Hiseq reads back to this contig assembly. The resultant SAM file was changed into a BAM file by SAMtools (v. 0.1.19-44428cd) [15] with the command “samtools view -bS.” (ii) Redundant sequences resulting from polymerase chain reaction amplification were removed with PICARD (v. 1.134; http://picard.sourceforge.net) using the command “MarkDuplicates.” (iii) The single nucleotide polymorphisms and indels were called from short-read alignments and used to correct the contigs by GATK (v. 3.4-0-g7e26428) [16, 17] with the command “HaplotypeCaller” and “FastaAlternateReferenceMaker,” respectively. The final polished contig number was 464 088 with N50 of 18.8 kb. Polished contigs were then used to build scaffolds using OPERA (v. 2.0.1) [18] with a k-mer of 39. This process yielded a final draft E. breviscapus genome of 1.2 Gb, with a contig N50 size of 18.8 kb and a scaffold N50 size of 31.5 kb (Additional File 1: Table S2).
Figure 2:
Assembly pipeline for the E. breviscapus genome.
Evaluation of the completeness of the E. breviscapus genome assembly
We evaluated the completeness of the final assembly using the Core Eukaryotic Genes Mapping Approach (CEGMA; v. 2.5) [19] with a set of 248 ultra-conserved core eukaryotic genes and Benchmarking Universal Single-Copy Orthologs (BUSCO; v. 2.0) [20] with the Embryophyta gene set. CEGMA assessment showed that our assembly captured 240 (96.9%) of the 248 ultra-conserved core eukaryotic genes, of which 217 (87.5%) were complete (Table 1). BUSCO analysis showed that 80.6% and 6.3% of the 1440 expected embryophytic genes were identified as complete and fragmented, respectively (Table 2).
Table 1:
Statistics of the completeness of the hybrid de novo assembly genome of E. breviscapus by CEGMA.
| Group | Protein Numa | Completeness (%)b | Total Numc | Average Numd | Ortholog (%)e |
|---|---|---|---|---|---|
| Complete | 217 | 87.50 | 633 | 2.92 | 82.95 |
| Group 1 | 58 | 87.88 | 158 | 2.72 | 77.59 |
| Group 2 | 49 | 87.50 | 126 | 2.57 | 77.55 |
|
Group 3 |
53 | 86.89 | 171 | 3.23 | 96.23 |
| Group 4 | 57 | 87.69 | 178 | 3.12 | 80.70 |
| Partial | 240 | 96.77 | 856 | 3.57 | 89.58 |
| Group 1 | 63 | 95.45 | 206 | 3.27 | 85.71 |
| Group 2 | 55 | 98.21 | 185 | 3.36 | 83.64 |
| Group 3 | 59 | 96.72 | 232 | 3.93 | 98.31 |
| Group 4 | 63 | 96.92 | 233 | 3.70 | 90.48 |
aProtein Num.: Number of 248 ultra-conserved core eukaryotic genes (CEGs) present in the E. breviscapus genome.
bCompleteness (%): Percentage of 248 ultra-conserved CEGs present in the E. breviscapus genome.
cTotal Num.: Total number of CEGs including putative orthologs present in the E. breviscapus genome.
dAverage Num: Average number of orthologs per CEG.
eOrtholog (%): Percentage of detected CEGs that have more than one ortholog.
Table 2:
Statistics of the completeness of the hybrid de novo assembly genome of E. breviscapus by BUSCO.
| BUSCO benchmark | Number | Percentage (%) |
|---|---|---|
| Total BUSCO groups searched | 1440 | – |
| Complete BUSCOs | 1161 | 80.63 |
| Complete and single-copy BUSCOs | 635 | 44.10 |
| Complete and duplicated BUSCOs | 526 | 36.53 |
| Fragmented BUSCOs | 90 | 6.25 |
| Missing BUSCOs | 189 | 13.13 |
Transcriptome sequencing
Total RNA was extracted from the leaf, root, stem, and flower tissues of a cultivated E. breviscapus individual using Qiagen RNeasy Plant Mini Kits. Additional RNA samples of the leaf tissues were acquired from six more cultivated individuals and five wild individuals (Additional File 1: Table S3). All cultivated samples were acquired from the greenhouse, and all wild samples were collected from Dali, Yunnan Province. Total RNA-seq libraries were prepared using TruSeq RNA Library Preparation Kit, v. 2 (Illumina, CA, USA), according to the manufacturer's instructions and subsequently sequenced on the HiSeq 2500 platform. In total, about 1.1 billion RNA-seq reads were obtained, representing ∼117.6 Gb of raw data. We aligned all the RNA-seq reads back to the E. breviscapus genome assembly using TopHat (v. 2.0.10) [21] with default parameters (Additional File 1: Table S3). The percentage of aligned reads ranged from 60.6% for the root to 80.9% for the leaf. We also calculated that 177 886 122 RNA-seq reads were mapped outside of the annotated regions using HTSeq (v. 0.6.1p1) [22] with the command “htseq-count –a 0.” The FPKM value was calculated for each protein-coding gene by Cufflinks (v. 2.1.1) using default parameters. FPKM >0.05 was used as the cutoff value to identify expressed genes.
Repeat annotation of the E. breviscapus genome assembly
The E. breviscapus genome was searched for tandem repeats using the Tandem Repeat Finder (v. 4.07b) [23]. RepeatMasker (v. 3.3.0) and RepeatProteinMasker [24] were used against Repbase library (v. 18.07) [25] to identify known transposable element repeats. De novo evolved transposable element annotation was performed using RepeatModeler (v. 1.0.8) [24] and LTR FINDER (v. 1.0.5) [26]. The combined results show that the total length of repeated sequences is about 664.2 Mb, accounting for ∼54.58% of the E. breviscapus genome assembly (Additional File 1: Tables S4 and S5).
Gene prediction
We used multiple methods to annotate protein-coding genes in the E. breviscapus genome, including homology-based predictions, de novo predictions, and transcriptome-based predictions. For homology-based predictions, protein sequences of Arabidopsis thaliana, Fragaria vesca, Malus domestica, Oryza sativa, Prunus persica, and Vitis vinifera were obtained from Phytozome, v. 9.1 (http://www.phytozome.net/), Pyrus communis from Genome Database for Rosaceae (https://www.rosaceae.org), and Prunus mume from the National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/genomes/Prunus_mume). First, query sequences were subjected to TBLASTN analysis with a cutoff E-value of 1e−5. BLAST hits corresponding to reference proteins were concatenated by Solar (v. 0.9.6; The Beijing Genomics Institute [BGI] development) [27] after low-quality records were removed. The genomic sequence of each reference protein was extended upstream and downstream by 2000 bp to represent a protein-coding region. GeneWise (v. 2.2.0) [28] was used to predict the gene structure contained in each protein region. For de novo predictions, AUGUSTUS (v. 2.5.5) [29], GENSCAN (v. 1.0) [30], SNAP (released 29 November 2013) [31], and glimmerHMM (v. 3.0.2) [32] analyses were performed on the repeat-masked genome, with parameters trained from A. thaliana. For transcriptome-based predictions, RNA-seq data from the leaves of six cultivated individuals were used for gene annotation, processed by Tophat and Cufflinks. The homology, de novo–, and transcriptomic-based predicted gene sets were merged to form a comprehensive and non-redundant reference gene set using Evidence Modeler (released 25 June 2012) [33]. We filtered gene models using our in-house Perl script in by the following criteria: (i) genes with incomplete open reading frames, (ii) small genes with a protein-coding region <150 bp, (iii) stop codons present in the middle of the gene, (iv) genes containing only one exon, and not supported by transcriptome-based evidence. Our analysis indicates that the E. breviscapus genome contains 37 504 protein-coding genes with an average coding DNA sequence length of 1034 bp (Additional File 1: Table S6).
Non-coding RNA annotation
tRNAscan-SE (v. 1.3.1) [34] with default parameters for eukaryotes was used for tRNA annotation. Homology-based rRNA annotation was performed by mapping plant rRNAs to the E. breviscapus genome using BLASTN with parameters of “E-value = 1e−5.” miRNA and snRNA genes were predicted by INFERNAL (v. 1.1) [35] using the Rfam database (release 11.0) [36]. The final results include 504 miRNAs, 751 tRNAs, 159 rRNAs, and 385 snRNAs (Additional File 1: Table S7).
Gene family clustering analysis
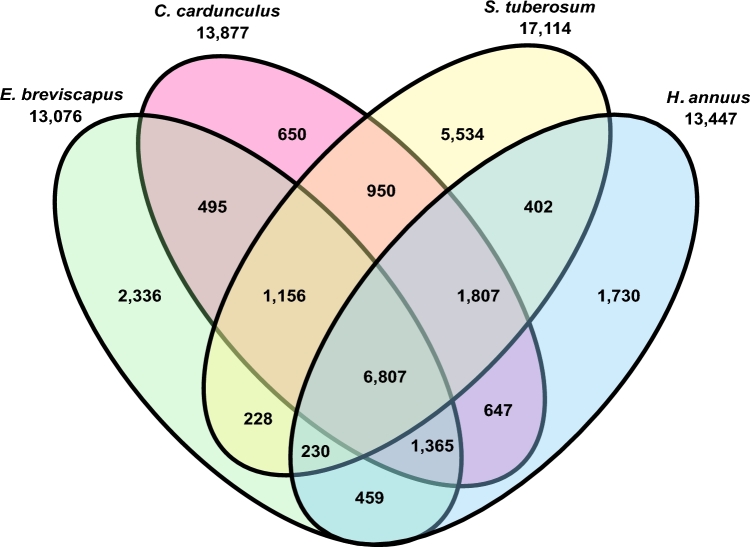
To identify and estimate the number of potential orthologous gene families between E. breviscapus, Helianthus annuus, Cynara cardunculus, Solanum tuberosum, Solanum lycopersicum, V. vinifera, and O. sativa, we applied the OrthoMCL (v. 2.0.9) pipeline [37] using standard settings (BLASTP E-value < 1e−5) to compute the all-against-all similarities. Gene sequences from S. tuberosum, S. lycopersicum, V. vinifera, and O. sativa were downloaded from Phytozome, v. 11.0. Gene sequences from H. annuus and C. cardunculus were downloaded from the Sunflower Genome Database (http://www.sunflowergenome.org) and Globe artichoke GBrowse (http://gviewer.gc.ucdavis.edu/cgi-bin/gbrowse/Artichoke_v1_1), respectively. Among the total 13 076 E. breviscapus gene families, 2336 (17.9%) appear to be lineage specific. There are 8172 (41.8%) gene families shared among Compositae species including E. breviscapus, H. annuus, and C. cardunculus. In addition, E. breviscapus shared 8421 (64.4%) gene families with S. tuberosum (Fig. 3).
Figure 3:
Venn diagram showing unique and shared gene families among four sequenced dicotyledonous species.
Phylogenetic tree construction and divergence time estimation
All 389 single-copy orthologous genes identified in the gene family clustering analysis from the S. lycopersicum, V. vinifera, O. sativa, E. breviscapus, H. annuus, C. cardunculus, and S. tuberosum were used to construct a phylogenetic tree. Orthologous genes from the seven species were aligned using MUSCLE (v3.8.31) with default settings [38] for each gene. Four-fold degenerate sites were extracted from each gene and concatenated into a ‘‘super gene’’ for each species. PhyML (v. 3.0) [39] was used to reconstruct phylogenetic trees between species. We implemented a Monte Carlo Markov chain (MCMC) algorithm for the estimation of divergence times using the program MCMCtree from the PAML package [40]. The result showed that E. breviscapus shared a closer phylogenetic relationship with H. annuus than C. cardunculus in the Compositae family (Additional File 1: Figure S3). The estimated divergence time was 29.7 million years ago between E. breviscapus and H. annuus (Additional File 1: Figure S4).
Expansion and contraction of gene families
CAFE (v. 2.1) [41] is a tool for analyzing the evolution of gene family size based on the stochastic birth and death model. With the calculated phylogeny and the divergence time, this software was applied to identify gene families that had undergone expansion and/or contraction in S. lycopersicum, V. vinifera, O. sativa, E. breviscapus, H. annuus, C. cardunculus, and S. tuberosum with the parameters “P = 0.05, number of threads = 10, number of random = 1000, and search for lambda.” We identified 5730 expanded gene families in the E. breviscapus genome, which is more than that in two other species, C. cardunculus (1336) and H. annuus (3897) in Compositae (Additional File 1: Figure S5).
In summary, we reported the genome sequencing, assembly, annotation, and evolution analysis of the E. breviscapus. This genome assembly will provide a valuable resource for studying the biosynthetic pathways of the medicinal components in E. breviscapus. This information will also help find novel bioactive compounds and improve the molecular breeding of this medicinal herb.
Abbreviations
BUSCO: Benchmarking Universal Single-Copy Orthologs; CEGMA: Core Eukaryotic Genes Mapping Approach.
Supplementary Material
Table S1: Raw sequencing statistics from the Illumina platform and PacBio platform.
Table S2: Summary of genome assembly.
Table S3: Summary of transcriptomes.
Table S4: Statistics of repeats in the E. breviscapus genome.
Table S5: Repeat annotation of the E. breviscapus genome assembly.
Table S6: Gene annotation statistics for the E. breviscapus genome.
Table S7: Summary of non-protein-coding gene annotation in the E. breviscapus genome assembly.
Figure S1: The estimated genome size of E. breviscapus with flow cytometry.
Figure S2: Frequency distribution of the 23-mer graph.
Figure S3: Phylogenetic reconstruction of the E. breviscapus and six other plant species.
Figure S4: Divergence time estimation of the E. breviscapus and six other plant species.
Figure S5: Gene family expansions and contractions in the E. breviscapus.
Additional File
Table S1: Raw sequencing statistics from the Illumina platform and PacBio platform.
Table S2: Summary of genome assembly.
Table S3: Summary of transcriptomes.
Table S4: Statistics of repeats in the E. breviscapus genome.
Table S5: Repeat annotation of the E. breviscapus genome assembly.
Table S6: Gene annotation statistics for the E. breviscapus genome.
Table S7: Summary of non-protein-coding gene annotation in the E. breviscapus genome assembly.
Figure S1: The estimated genome size of E. breviscapus with flow cytometry.
Figure S2: Frequency distribution of the 23-mer graph.
Figure S3: Phylogenetic reconstruction of the E. breviscapus and six other plant species.
Figure S4: Divergence time estimation of the E. breviscapus and six other plant species.
Figure S5: Gene family expansions and contractions in the E. breviscapus.
Funding
This work was supported by the National Natural Science Foundation of China (81260614) and the pilot project for establishing a new socialized service system through a combination of agricultural science and education in Yunnan Province (Medical Plant Unit; 2014NG003).
Availability of supporting data
Sequencing reads of each sequencing library and RNA-seq data have been deposited at the National Center for Biotechnology Information with the project ID PRJNA352312. Supporting data, including alignments, annotations, and custom scripts, are available in the GigaScience database (GigaDB) [42]. All supplementary figures and tables are provided in Additional File 1.
Conflicts of interest
The authors declare that they have no competing interests.
Authors’ contributions
W.C., Y.D., G.Z., and S.Y. designed the study. H.L. assembled the genome. J.Y., X.W., Y.L., and J.Z. analyzed the data. J.Y., W.C., and Y.D. wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Longjin Pharmaceutical Co., Ltd., for providing samples of the E. breviscapus plant.
References
- 1. Lin R, Chen Y, Shi Z. Erigeron breviscapus in Flora Reipublicae Popularis Sinicae. Vol. 74 Beijing: Science Press;1985:308–9. [Google Scholar]
- 2. Li X, Zhang S, Yang Z et al. . Conservation genetics and population diversity of Erigeron breviscapus (Asteraceae), an important Chinese herb. Biochem Syst Ecol 2013;49(2):156–66. [Google Scholar]
- 3. Sheng J, Zhao P, Huang Z. Influence of deng zhan xi xin (Erigeron breviscapus) on thrombolytic treatment during acute coronary thrombosis by affecting function of blood platelet and coagulation. Chin J Cardiol 1999;27(2):115–7. [Google Scholar]
- 4. Liu H, Tang X, Wang Y et al. . Effects of scutellarin on rat cerebral blood flow determined by laser speckle imagine system. Chin Hosp Pharm J 2010;30(9):719–22. [Google Scholar]
- 5. Sun H. A drug for treating cardio-cerebrovascular diseases-phenolic compounds of Erigeron breviscapus. Prog Chem 2009;21(1):77–83. [Google Scholar]
- 6. Yu H, Chen Z. Study on artificial culture of Erigeron breviscapus. Acta Bot Yunnanica 2002;24:115–20. [Google Scholar]
- 7. Li X, Song K, Yang J, et al. . Isolation and characterization of 11 new microsatellite loci in Erigeron breviscapus (Asteraceae), an important chinese traditional herb. Int J Mol Sci 2011;12(10):7265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eid J, Fehr A, Gray J et al. . Real-time DNA sequencing from single polymerase molecules. Science 2009;323:133–8. [DOI] [PubMed] [Google Scholar]
- 9. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27:764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zimin AV, Marçais G, Puiu D et al. . The MaSuRCA genome assembler. Bioinformatics 2013;29(21):2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kajitani R, Toshimoto K, Noguchi H et al. . Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 2014;24:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye C, Hill C, Wu S et al. . DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep 2016;6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Treangen TJ, Sommer DD, Angly FE et al. . Next generation sequence assembly with AMOS. Curr Protoc Bioinformatics 2011;Chapter 11: Unit 11.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Handsaker B, Wysoker A et al. . The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKenna A, Hanna M, Banks E et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DePristo MA, Banks E, Poplin R et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao S, Nagarajan N, Sung WK. Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol 2011;18(11):1681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007;23:1061–7. [DOI] [PubMed] [Google Scholar]
- 20. Simão FA, Waterhouse RM, Ioannidis P et al. . BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 21. Trapnell C, Roberts A, Goff L et al. . Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Prot 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999;27:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009;3:4–14. [DOI] [PubMed] [Google Scholar]
- 25. Jurka J, Kapitonov VV, Pavlicek A et al. . Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 2005;110(1–4):462–7. [DOI] [PubMed] [Google Scholar]
- 26. Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 2007;35:W265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X, Kui L, Zhang J et al. . Improved hybrid de novo genome assembly of domesticated apple (malus x domestica). Gigascience 2016;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birney E, Durbin R. Using genewise in the drosophila annotation experiment. Genome Res 2000;10:547–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stanke M, Keller O, Gunduz I et al. . AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 2006;34(suppl 2):W435–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai Y, Gonzalez JV, Liu Z, et al. . Computational systems biology methods in molecular biology, chemistry biology, molecular biomedicine, and biopharmacy. Biomed Res Int 2014;2014:746814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korf I. Gene finding in novel genomes. BMC Bioinformatics 2004;5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–9. [DOI] [PubMed] [Google Scholar]
- 33. Haas BJ, Salzberg SL, Zhu W et al. . Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 2008;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997;25:955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics 2009;25:1335–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardner PP, Daub J, Tate J et al. . Rfam: Wikipedia, clans and the ‘‘decimal’’ release. Nucleic Acids Res 2011;39(suppl 1):D141–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13:2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guindon S, Dufayard JF, Lefort V et al. . New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 2010;59(3):307–21. [DOI] [PubMed] [Google Scholar]
- 40. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007;24(8):1586–91. [DOI] [PubMed] [Google Scholar]
- 41. De Bie T, Cristianini N, Demuth JP, et al. . CAFE: a computational tool for the study of gene family evolution. Bioinformatics 2006;22(10):1269–71. [DOI] [PubMed] [Google Scholar]
- 42. Yang J, Zhang G, Zhang J, Liu H, Chen W, Li Y, Wang X, Dong Y, Yang S (2017). Supporting data for “Hybrid de novo genome assembly of the Chinese herbal fleabane Erigeron breviscapus” GigaScience Database. http://dx.doi.org/10.5524/100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Raw sequencing statistics from the Illumina platform and PacBio platform.
Table S2: Summary of genome assembly.
Table S3: Summary of transcriptomes.
Table S4: Statistics of repeats in the E. breviscapus genome.
Table S5: Repeat annotation of the E. breviscapus genome assembly.
Table S6: Gene annotation statistics for the E. breviscapus genome.
Table S7: Summary of non-protein-coding gene annotation in the E. breviscapus genome assembly.
Figure S1: The estimated genome size of E. breviscapus with flow cytometry.
Figure S2: Frequency distribution of the 23-mer graph.
Figure S3: Phylogenetic reconstruction of the E. breviscapus and six other plant species.
Figure S4: Divergence time estimation of the E. breviscapus and six other plant species.
Figure S5: Gene family expansions and contractions in the E. breviscapus.