Abstract
FoF1-ATP synthase (FoF1) is a motor enzyme that couples ATP synthesis/hydrolysis with a transmembrane proton translocation. F1, a water-soluble ATPase portion of FoF1, rotates by repeating ATP-waiting dwell, 80° substep rotation, catalytic dwell, and 40°-substep rotation. Compared with F1, rotation of FoF1 has yet been poorly understood, and, here, we analyzed ATP-driven rotations of FoF1. Rotation was probed with an 80-nm bead attached to the ring of c subunits in the immobilized FoF1 and recorded with a submillisecond fast camera. The rotation rates at various ATP concentrations obeyed the curve defined by a Km of ≈30 μM and a Vmax of ≈350 revolutions per second (at 37°C). At low ATP, ATP-waiting dwell was seen and the kon-ATP was estimated to be 3.6 × 107 M-1·s-1. At high ATP, fast, poorly defined stepwise motions were observed that probably reflect the catalytic dwells. When a slowly hydrolyzable substrate, adenosine 5′-[γ-thio]triphosphate, was used, the catalytic dwells consisting of two events were seen more clearly at the angular position of ≈80°. The rotational behavior of FoF1 resembles that of F1. This finding indicates that “friction” in Fo motor is negligible during the ATP-driven rotation. Tributyltin chloride, a specific inhibitor of proton translocation, slowed the rotation rate by 96%. However, dwells at clearly defined angular positions were not observed under these conditions, indicating that inhibition by tributyltin chloride is complex.
Keywords: ATP hydrolysis, binding change mechanism, membrane protein, single-molecule imaging
FoF1-ATPase/synthase (FoF1) is a large protein complex (≈500 kDa) that catalyzes ATP synthesis/hydrolysis coupled with a transmembrane H+ (proton)-translocation in bacteria, chloroplasts, and mitochondria (1-5). The enzyme is easily and reversibly separated into two portions, termed F1 and Fo. In its simplest prototype bacterial enzyme, a water-soluble F1 portion consists of five different subunits, α3β3γ1δ1ε1, and catalyzes ATP hydrolysis (hence, often called F1-ATPase). Three α-subunits and three β-subunits are arranged alternately, forming a hexagonal cylinder around the coiled-coil structure of the γ-subunit (6). Membrane-integrated Fo portion has three different subunits, a1b2cn (n; variable among species) and mediates proton transport across the membrane. The c-subunits forms a ring structure, and ab2 associates with the c-subunit ring peripherally (7-10). FoF1 is a motor enzyme. When the magnitude of electrochemical potential of protons is large enough, downhill proton flow through Fo causes rotation of the rotor subunits (cn-γε) relative to the stator subunits (ab2-α3β3δ), and rotation of the γ-subunit forces the β-subunits of F1 to change conformations sequentially that result in ATP synthesis. In the reverse reaction, ATP hydrolysis at F1 causes the reverse rotation of the rotor subunits that drives Fo to pump protons (Fig. 1) (11).
Fig. 1.
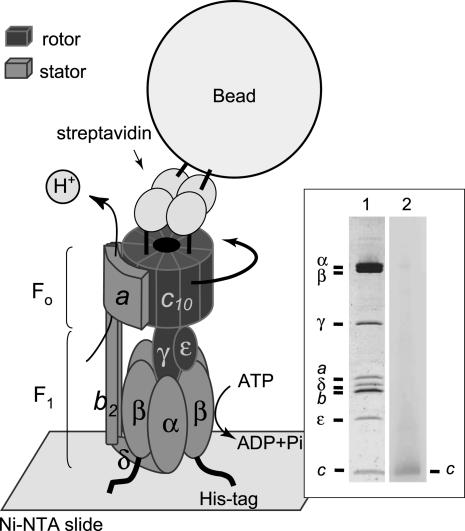
Experimental system. ATP-driven rotation of the bead attached to the c-subunit ring in FoF1 was observed under dark-field microscopy and recorded with a fast camera. (Inset)FoF1 from thermophilic Bacillus PS3 used for observation of rotation. Lane 1, PAGE in the presence of sodium dodecyl sulfate of FoF1 purified in LPC and stained by Coomassie brilliant Blue; lane 2, immunoblots stained by alkaline phosphatase-streptavidin conjugates that show specific biotin labeling of the c-subunit.
We have been studying rotation of thermophilic Bacillus F1 since the first direct visualization of ATP-driven rotation of F1 immobilized on the glass surface (12). A rotation probe attached on the γ-subunit rotates unidirectionally counterclockwise when viewed from membrane side. It repeats a pause and a 120° step rotation when medium ATP concentration ([ATP]) is low (13). The duration of the pause becomes shorter as [ATP] increases and is finally invisible beyond the limit of the observation system. This [ATP]-dependent pause corresponds to the period during which the enzyme waits for medium ATP to come into the empty catalytic site, and, hence, is called the ATP-waiting dwell. With high-speed imaging, it was found that the 120° step of rotation is further split into 90° and 30° substeps (14). A pause between two substeps, ≈2 ms at 23°C, is not influenced by [ATP]. This [ATP]-independent nature of the pause means that catalytic events after substrate binding should occur during this pause, and we call this pause the catalytic dwell. The histogram of durations of the catalytic dwells did not obey a single exponential but did obey double exponentials, indicating that two catalytic reactions of ≈1 ms occur in the catalytic dwell (14). More recent experiments using a slowly hydrolyzable ATP analog, adenosine 5′-[γ-thio]triphosphate (ATPγS) and a slow mutant in ATP hydrolysis, clarified that one of the two 1-ms events is cleavage of a bound ATP at a catalytic site (15). Also, previous 90° and 30° substeps were recently revised to be 80° and 40° substeps (15, 16). Thus, F1 rotates by repeating four stages; ATP-waiting dwell, rapid 80° substep rotation upon ATP binding, catalytic dwell in which ATP hydrolysis occurs, and rapid 40° substep rotation, probably upon the release of the last product.
ATP-driven rotation of FoF1 was demonstrated for Propionigenium modestum FoF1 with single-fluorophore polarization (17), and for Escherichia coli FoF1 with disulfide cross-linking (18, 19), direct visualization (20), and fluorescence resonance energy transfer (21). In general, ATP-driven rotation of FoF1 can differ from that of F1 because proton transport through Fo and interaction between the c-subunit ring (rotor) and ab2 (stator) during rotation may modify the rotation. It was reported that the central rotor rotates counterclockwise when viewed from membrane side, and there are three pauses (each 19-30 ms) in one revolution, likely corresponding to the catalytic dwell (21, 22). The rotation of FoF1 driven by proton flow was also observed, and the direction of the rotation is opposite of that of the ATP-driven rotation (22). However, in general, knowledge on rotation of FoF1 has yet been very limited. For example, even the following basic motor natures of FoF1 remain unknown: (i) whether ATP-binding dwell appears at low [ATP], (ii) how the rotation depends on [ATP], (iii) whether the observed rotation consists of the sequence of 80° and 40° substep rotation or other new substep(s) exists at different angular position(s), (iv) whether catalytic dwell is composed of two events, as observed for F1, and (v) how rotation changes when a reversible Fo inhibitor is present. To address these questions, we isolated thermophilic FoF1, which kept structural integrity in a detergent, immobilized it on a glass surface through the β-subunits, attached a small (80 nm) bead to the c-subunit ring as a rotation probe whose viscous friction was low enough to allow full-speed rotation, and observed ATP-driven rotation with a fast camera. The results reveal that basic natures of ATP-driven rotation of FoF1 are almost unaltered from those of F1, except for the response to an Fo-specific inhibitor.
Materials and Methods
Isolation of FoF1. A plasmid pTR19-ASDS-CNCR3 for mutant FoF1 (cSer2Cys/β-His10 tags) was made by the Mega-primer method (23) using a plasmid, pTR19-ASDS (24), an expression vector for FoF1 complex of thermophilic Bacillus PS3. The mutant FoF1 was expressed constitutively in E. coli strain DK8 [Δ(uncB-uncC), ilv::Tn10] that lacks whole FoF1 genes, and the inverted membrane vesicles were prepared by the procedures described (24). After washing a 1-ml suspension of the vesicles (20 mg of protein per ml) with 5-fold volume of buffer PA3 (10 mM Hepes/KOH, pH 7.5/5 mM MgCl2/10%glycerol) containing 2% sodium cholate by centrifugation, the vesicles were dissolved in 2 ml of buffer PA3 containing 2% (wt/vol) octaethylene glycol monododecyl ether and 100 μM Tris (2-carboxyethyl) phosphine, and incubated for 30 min at room temperature. After adjusting the pH of the solution to 7.0 by phosphate buffer, 6-N′-[2-(N-maleimido)ethyl]-N-piperazinylamidohexyl-d-biotinamide (Dojindo) was added (50 μM), and incubated for 30 min at room temperature. Biotinylation was quenched with 100 μM DTT, and the suspension was diluted 6-fold with buffer M (20 mM potassium phosphate buffer, pH 7.5/100 mM KCl) containing 0.1% lysophosphatidylcholine (LPC) and 20 mM imidazole. This suspension was applied to a Ni2+-nitrilotriacetic acid (Ni2+-NTA) column (Qiagen, Valencia, CA) equilibrated with the same buffer. After washing with 10 volumes of the equilibration buffer, the protein was eluted with buffer M containing 0.1% LPC and 200 mM imidazole. The eluate containing FoF1 was applied to a Soft-Link avidin column (Promega) equilibrated with buffer M containing 0.05% LPC. The column was washed with 10 volumes of the same buffer, and the protein was eluted with buffer M containing 0.05% LPC and 10 mM d-biotin. The purification was finished within 4 h, and the purified sample was used within 1 day. The purified FoF1 contained 0.9 mol of ADP and 0.8 mol of ATP per mol of FoF1 as endogenously bound nucleotides. Specific biotinylation of the Foc-subunit was confirmed by immunoblotting using streptavidin-alkaline phosphatase conjugates (Promega).
Beads. Colloidal gold (diameter of 40 or 80 nm; British BioCell International) was incubated in 100 mM borate buffer, pH 8.2, containing 20 mg/ml BSA for 16 h at 37°C. BSA-coated colloidal gold was precipitated by centrifugation, and resuspended in 100 mM borate buffer, pH 8.2, with 10 mg/ml BSA. The suspension was biotinylated with 0.4 mg/ml 15-([biotinoyl]amino)-4,7,10,13-tetraoxapentadecanoic acid, N-hydroxysuccinimidylester (Pierce) for 3 h at room temperature. After removing unreacted biotin by centrifugation, 1 mg/ml streptavidin was added. After the excess streptavidin was removed by centrifugation, the pellet was resuspended in 2 mM potassium phosphate, pH 7.0, containing 0.05% polyethylene glycol and stored at 4°C. In the experiments to test the inhibition of rotation by N,N′-dicyclohexylcarbodiimide (DCCD), larger beads (streptavidin-coated microspheres, 0.56 μm, Bangs Laboratories, Carmel, IN) were used.
Rotation Assay. A flow cell was made of a Ni2+-NTA-coated cover glass and a slide glass separated by two spacers with 50-μm thickness. At first, buffer R (50 mM Hepes/KOH, pH 7.5/100 mM KCl/10 mg/ml BSA/0.05% LPC/20 mM imidazole) was infused into the flow cell and incubated for 5 min to block nonspecific binding of the enzyme. Biotinylated His10-tagged FoF1 (1-5 nM) in buffer R was infused. After 10 min, unbound FoF1 was washed with buffer R. Then, streptavidin-coated beads (2 × 109 particles per milliliter) in buffer R1 (50 mM Hepes/KOH, pH 7.5/100 mM KCl/10 mg/ml BSA/0.05% LPC) were infused, and incubated for 10 min. Unbound beads were removed with buffer R1 containing 5 mM MgCl2, an ATP-regeneration system, and ATP at indicated concentrations, and observation of rotation was started. When indicated, ATPγS was added instead of ATP, and an ATP-regeneration system was omitted in this case. The number of the beads at the glass surface depended on the concentrations of biotinylated FoF1 infused to the flow cell. When nonbiotinylated enzyme was infused, it was almost the same as the case when no protein was infused, and we could not find any rotating beads in this case. Inhibitory effect of DCCD was assessed by observing the rotation of FoF1 incubated with 50 μM DCCD for 30 min at room temperature before infusion into the flow cell. Beads were observed with a dark-field microscopy (IX-70, Olympus) with a ×100 objective lens (numerical aperture of 1.35, Olympus) and a dark-field condenser (numerical aperture of 1.2-1.4, Olympus). Both the objective lens and the condenser were warmed by lens (condenser) heater (Tokai Hit) to maintain temperature of the flow cell at 37°C. Bead images were recorded as an eight-bit AVI file with a fast-framing charge-coupled device camera (Hi-Dcam, NAC Image Technology) at the indicated frame rate. To analyze the acquired image data, custom software (created by R. Yasuda; Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) was used. Rotation rate was obtained from the average of 20 continuous revolutions without unnatural interruption.
Other Assays. ATPase activity was measured at 37°C with an ATP regeneration system (25). The assay solution was composed of buffer R1 containing 5 mM MgCl2, 1 mM ATP-Na, 2 mM phosphoenolpyruvate, 100 μg/ml lactate dehydrogenase, 100 μg/ml pyruvate kinase, and 0.2 mM NADH. DCCD inhibition of ATP hydrolyzing activity was measured as described for the inverted membrane vesicles (24), or after a 30-min preincubation with DCCD (50 μM) for the purified enzymes. To measure inhibitory effect of tributyltin chloride (TBT-Cl) on ATP hydrolyzing activity, indicated concentrations of TBT-Cl was added to the assay solution before the start of the reaction. Because the inhibitory effect of TBT-Cl on ATPase activity, as well as on rotation, tended to be relieved gradually as time passed (for an unknown reason), we collected rotation data quickly after the initiation of measurements. Reconstitution of FoF1 into liposomes was performed by the method described (24). Protein concentrations were determined by the BCA protein assay kit (Pierce) with BSA as a standard.
Results
Intact FoF1 in Detergent. In this study, we used FoF1 of thermophilic Bacillus PS3 expressed in the plasma membranes of an FoF1-deficient E. coli strain, DK8. A detergent octaethylene glycol monododecyl ether was used to solubilize FoF1 from the membrane vesicles and it was substituted with LPC at the next step of purification. We have tested various detergents, but this combination of detergents gave the most efficient solubilization, and the FoF1 preparation had the most stable, intact coupling properties. The FoF1 has His10 tags at the N terminus of the β-subunits to immobilize onto a glass surface and has a cysteine residue at the second position from N terminus of the c-subunit for biotinylation to attach the beads. The enzyme was purified with a Ni2+-NTA column and a Soft-Link avidin column (Fig. 1 Inset, lane 1). These procedures should help to remove free F1 and Fo, if any. Specific biotinylation of the c-subunit was confirmed by immunoblotting (Fig. 1 Inset, lane 2). The purified FoF1 in LPC comprised eight kinds of subunits and exhibited DCCD-sensitive ATPase activity; ≈85% of the activity was inhibited by DCCD in the solution used for observation of rotation (Tables 1 and 2). This degree of inhibition is similar to that of the intact FoF1 embedded in membranes, either in the membrane vesicles, or in the reconstituted vesicles. It has been known that DCCD covalently labels an essential carboxyl residue of the c-subunit, blocks proton transport, and consequently, if FoF1 is intact, prevents ATP hydrolysis/synthesis. Unlike FoF1, ATPase activity of the isolated F1 was not affected by DCCD at pH values of >7.5. (data not shown). Therefore, high sensitivity to DCCD inhibition is a good indication of the intactness of our FoF1 preparation. The reconstituted vesicles containing purified FoF1 showed substantial ATP-driven proton-pumping activity, comparable with that of the authentic wild-type thermophilic FoF1 (24). Based on these observations, we concluded that the purified FoF1 in LPC had intact ATPase activity that coupled proton transport. This conclusion was further confirmed by TBT-Cl sensitivity of FoF1 as described later.
Table 1. ATPase activity sensitivity of FoF1 to DCCD inhibition.
| Samples | Residual activity +DCCD, % |
|---|---|
| Membrane vesicles | 25 |
| Reconstituted vesicles | 20 |
| FoF1 | 15 |
The samples were pretreated with 50 μM DCCD for 30 min and subjected to the assays. Membrane vesicles were prepared from Escherichia coli cells expressing FoF1 from thermophilic Bacillus PS3. Purified FoF1 was incorporated into reconstituted vesicles. ATPase activity was measured in the same solution used for rotation observation. Other experimental details are described in Materials and Methods.
Table 2. Effect of DCCD on rotation of FoF1.
| No. of rotating beads
|
||
|---|---|---|
| Trial | +DCCD | –DCCD |
| 1 | 2 | 10 |
| 2 | 1 | 10 |
| 3 | 0 | 9 |
| 4 | 2 | 12 |
| 5 | 1 | 11 |
| 6 | 1 | 8 |
| Total | 7 | 60 |
Rotating beads were looked for in 120 optical fields of a unit area (∼45 × 55 μm2) in 10 min in one trial. Other experimental details are described in Materials and Methods.
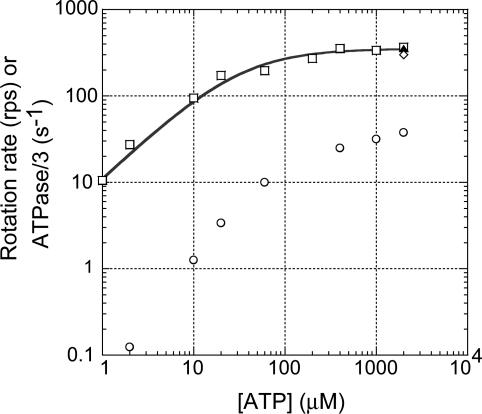
Vmax Rotation of FoF1. FoF1 was immobilized on a glass surface through β-subunits, and an 80-nm bead was attached to the c-subunit ring. Because the size of the bead was much larger than the c-subunit ring (≈5 nm) (26), a single bead could be tethered to several c-subunits in the same ring, although two beads could not attach to the same c-subunit ring. ATP-driven rotation of the bead attached obliquely to the c-subunit ring was observed at 37°C under dark-field microscopy equipped with a fast camera (Fig. 1). Functional integrity of immobilized FoF1 was confirmed by the observations that the number of rotating beads was drastically decreased to 12% (7:60) by DCCD-pretreatment of FoF1 (Table 2). In the case of rotation of F1, on the contrary, pretreatment of F1 with DCCD did not cause decrease in the number of the rotating molecules (data not shown). The direction of rotation was exclusively viewed counter clockwise from the membrane side. We observed the rotation at various [ATP] and found that rotation rates obeyed a simple Michaelis-Menten kinetics with a Vmax of 352 ± 16 revolutions per second (rps) and a Km of 31 ± 7 μM (Fig. 2, □). We used an 80-nm single bead as a rotation probe, but the rotation rate observed with a 40-nm single bead at 2 mM ATP (≈300 rps, Fig. 2, ⋄) was almost the same as that of an 80-nm bead, indicating that the viscous friction in the rotating 80-nm bead did not slow down the rotation rate of FoF1. It is noteworthy that the rotation rate of FoF1 under Vmax conditions was very similar to the rotation rate of F1 (≈320 rps, Fig. 2, ▴) under the same conditions. Therefore, any events occurring at the Fo portion during rotation, such as the interaction between rotor (c-subunit ring) and stator (ab2-subunits) and proton transport through Fo, does not limit rotation rate of FoF1, at least in the absence of the electrochemical gradient of protons.
Fig. 2.
Dependency of rotation rate and bulk-phase ATPase of FoF1 on [ATP]. □, rotation observed with an 80 nm-bead; ⋄, rotation observed with a 40 nm-bead; ▴ rotation of F1 observed with an 80 nm-bead; ○, bulk-phase ATPase activities measured in the rotation buffer. The line shows fit with Michaelis-Menten kinetics, V = Vmax[ATP]/(Km+[ATP]), where Vmax = 352 ± 16 rps, and Km = 31 ± 7 μM. Values are means ± SE.
Rotation Rate Versus ATPase Activity. The bulk-phase steady-state ATPase activities of the purified FoF1 in the same solution used for observation of rotation were only ≈10% at 2 mM ATP, and even <1% at 2 μM ATP of the expected values from the rotation rates (Fig. 2, ○). This result indicates that >90% population of FoF1 molecules are not working at a given moment, or, in other words, a single molecule spends ≈90% of time in inactive state(s). Indeed, we noticed that rotating molecules usually stopped rotation after several seconds of continuous rotation. Sometimes, the same molecule resumed rotation after a while. We also often saw, under the microscopic field, that a previously nonrotating bead started rotating. The real reason why such high fractions of FoF1 molecules are in inactive state(s) is not known, but it should be noted that F1 (α3β3γ subcomplex) also shows similar large discrepancy of rates between rotation and bulk-phase steady-state ATPase activity. In the case of F1, the responsibility for the discrepancy is thought to be the ADP-Mg inhibition, which is caused from the nonturnover retention of ADP-Mg at a catalytic site (27-29). The steady-state ATPase activity of F1 is largely suppressed by the ADP-Mg inhibition, but, uninhibited activity that is almost consistent with the rotation rate, is estimated from the initial burst activity that appears upon initiation of ATPase assays (14). We speculate that ADP-Mg inhibition occurring in FoF1 can explain, at least partly, the discrepancy between rotation and bulk-phase ATPase activity, although we cannot estimate its quantitative contribution because FoF1 does not show the uninhibited initial-burst activity. Pronounced inhibition of ATPase activity of FoF1 at low [ATP] in Fig. 2 might be due to the inhibition by the endogenous inhibitor, ε-subunit, that exhibits inhibition at low [ATP] (30).
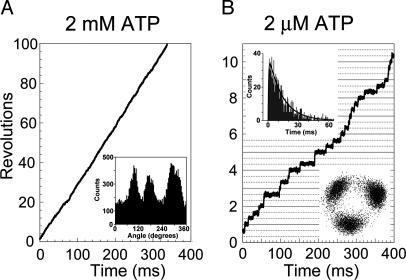
Stepwise Rotation. We analyzed the time course of rotation at 2 mM ATP (Fig. 3A). Under Vmax conditions, it is expected that ATP binds to FoF1 very quickly, and the catalytic events occurring in the enzyme determine the rates of ATP hydrolysis and rotation. Therefore, the rotation would show only the catalytic dwell. Indeed, the histogram of angular distribution of centroid of bead (Fig. 3A Inset) indicated the presence of three favorable positions for a bead to make a brief pause. However, the expanded time course of rotation did not always show clear steps (see Fig. 6, which is published as supporting information on the PNAS web site), and we could not define each dwell time with certainty.
Fig. 3.
Rotation of FoF1 driven by ATP. Bead images were recorded at 8,000 frames per second. (A) Time course of rotation at 2 mM ATP. (Inset) Histogram of angular distribution of the centroid of the bead image. (B) Time course of rotation at 2 μM ATP. (Upper Inset) Histogram of the total dwell times of rotation at 2 μM ATP. The total dwell time is defined as the time period from the start of the short dwell to the end of the ATP-waiting dwell. The black line is fit with the two rate constants, constant × [exp(-k1t)-exp(-k2t)], k1 = 0.073 ± 0.002 ms-1 (time constant = 13.8 ± 0.4 ms) and k2 = 1.72 ± 0.18 ms-1 (time constant = 0.58 ± 0.06 ms). k1 is [ATP]-dependent and corresponds to the ATP-waiting dwell. k2 is [ATP]-independent and corresponds to the catalytic dwell. Values are means ± SE. Total counts of dwells are 1,079. Bin width is 0.5 ms. (Lower Inset) Trace of the centroid of the bead image.
At 2 μM ATP, FoF1 rotated with discrete 120° steps (Fig. 3B), and the dwell time of the pauses between adjacent step-rotations was apparently dependent on [ATP], indicating that the observed pauses were the ATP-waiting dwells. In addition, very short dwells were often seen in the intermediate position in a 120° rotation. They are not obvious in the bead-centroid distribution plots (Fig. 3B Lower Inset) because these dwells would be buried behind spreading distribution of the long ATP-waiting dwells. Usually, the starting time points of the short dwells could be recognized in the rotation time course as an interruption of 120° rotation, but the end points were mostly unclear; the dwells transited to the next ATP-waiting dwell without showing discrete step rotation. Therefore, the total dwell time from the start of the short dwell to the end of the ATP-waiting dwell was analyzed. The histogram of the total dwells at 2 μM ATP showed a distinct peak, and was fitted by the sum of two exponential components that assumed two rate-limiting reactions (Fig. 3B Upper Inset). One of the time constants was [ATP]-dependent and corresponds to the ATP-waiting dwell. From the time constant, the ATP-binding rate (kon-ATP) of FoF1 was estimated to be 3.6 ± 0.1 × 107 M-1·s-1. This value is close to that of F1 at 23°C (3.0 ± 0.1 × 107 M-1·s-1) (14). The other time constant (≈0.58 ms) was [ATP]-independent and corresponds to the catalytic dwell. This value is consistent with that (≈0.95 ms) obtained from the Vmax rotation.
From these results, we learned that FoF1 rotates by repeating ATP-waiting dwell and catalytic dwell. However, the very short lifetime of the catalytic dwell did not allow for its further analysis, and the angular position of the catalytic dwell and the number of events occurring in the catalytic dwell were not determined. To learn these answers, we extended the duration of the catalytic dwell by adopting a slowly hydrolyzable ATP analog, ATPγS, as a substrate (15).
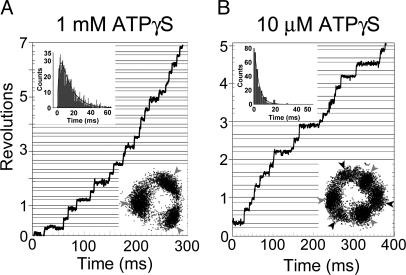
Rotation Driven by ATPγS. FoF1 rotated in 1 mM ATPγS at ≈20 rps, >10 times slower rate compared with the rate in 1 mM ATP (≈335 rps), and the discrete 120 ° steps were observed (Fig. 4A Lower Inset). The rotation in 100 μM ATPγS was apparently very similar to that observed in 1 mM ATPγS; rotation rate of ≈18 rps and 120° step rotation (data not shown). The dwell time between steps was not changed in two ATPγS concentrations and should be the catalytic dwell. The histogram of the catalytic dwell of the rotation in 1 mM ATPγS showed a peak, and was fitted with the sum of two exponential components that provided two time constants, ≈12.4 and ≈2.3 ms (Fig. 4A Upper Inset). Taking the F1 study on hydrolysis of ATPγS as a reference (15), the long time constant is likely the one for cleavage of ATPγS, and the short time constant for the release of the last product. The total of these time constants (≈14.7 ms) agrees well with that (≈16.7 ms) obtained from Vmax (≈20 rps). In 10 μMATPγS, the 120° step was further split into two substeps (Fig. 4B Lower Inset). The angles of the two substep rotations were roughly 80° and 40° that are the same as observed for F1. The pauses between adjacent 40° and 80° substep rotations depended on ATPγS concentrations, and, hence, the ATPγS-waiting dwell. The histogram of the ATPγS-waiting dwell was fitted well with a single-exponential component (Fig. 4B Upper Inset), and the ATPγS-binding rate (kon-ATPγS) of FoF1 was estimated to be 2.4 ± 0.1 × 107 M-1·s-1. This value is in the same order of kon-ATP (3.6 ± 0.1 × 107 M-1·s-1) of FoF1, and kon-ATPγS (2.6 ± 0.1 × 107 M-1·s-1) of F1 (15). The pauses between adjacent 80° and 40° substep rotations corresponded to the catalytic dwell, and the analysis of the dwell-time histogram (not shown) gave two time constants, ≈17.3 and ≈2.4 ms, which is consistent with the values obtained from the rotation in 1 mM ATPγS.
Fig. 4.
Rotation of FoF1 driven by ATPγS. Bead images were recorded at 2,000 frames per second. Gray horizontal lines are placed 40° below black lines. (A) Time course of the rotation of FoF1 at 1 mM ATPγS. (Upper Inset) Histogram of the pauses before 40° substeps (catalytic dwell) in rotation at 1 mM ATPγS. The line is fit with the two rate constants, constant × [exp(-k1t)-exp(-k2t)], k1 = 80.8 ± 3.5 s-1 (time constant = 12.4 ± 0.5 ms) and k2 = 445 ± 36 s-1 (time constant = 2.25 ± 0.18 ms). Values are means ± SE. Total counts of dwells are 1,028. Bin width is 0.5 ms. (Lower Inset) Trace of the centroid of the bead image. Gray arrows indicate the positions of catalytic dwell. (B) Time course of the rotation of FoF1 at 10 μM ATPγS. (Upper Inset) Histogram of the pauses before 80° substep rotations (ATPγS-binding dwell) in rotation at 10 μM ATPγS. The line is a single-exponential fit, constant × exp(-kt) where k = 238.3 ± 5.3 s-1 (time constant = 4.2 ± 0.1 ms). Values are means ± SE. Total counts of dwells are 202. Bin width is 2 ms. (Lower Inset) Trace of the centroid of the bead image. Black arrows indicate the positions of ATPγS-binding dwell.
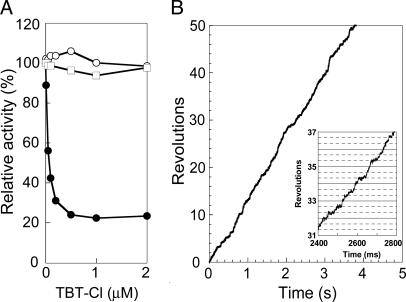
Effect of TBT-Cl on FoF1 Rotation. TBT-Cl has been known as an inhibitor of FoF1 (31, 32). It reacts noncovalently with the Fo portion, probably with the Foa-subunit, and prevents proton translocation-coupled ATP hydrolysis/synthesis (33). ATPase activity of our purified FoF1 measured in the same solution used for observation of rotation was efficiently (≈80%) inhibited by 1 μM TBT-Cl (Fig. 5A, •). No inhibition was observed in the presence of 0.3% lauryldodecylamine oxide (Fig. 5A, ○), a detergent known to disrupt stator/rotor interface of Fo. ATPase activity of the isolated F1 was also totally insensitive to TBT-Cl (Fig. 5A, □). Rotation of FoF1 in 1 mM ATP was slowed down significantly by TBT-Cl (Fig. 5B); the average rotation rate decreased to 4% (≈15 rps). Rotation of F1 under the same condition was not affected by TBT-Cl (data not shown). We had expected to identify the angular position(s) where rotation would have paused in TBT-Cl, and 15 rps was slow enough to show any prolonged dwells. However, the rotating bead did not show any obvious pause at certain angular position (Fig. 5 Inset). We speculate that TBT-Cl interferes with the rotation of the Fo portion at many angular positions, and makes the step-wise rotation obscure.
Fig. 5.
Effect of TBT-Cl on ATPase activity of FoF1 (A) and rotation of FoF1 (B). (A) The ATPase activities of FoF1 were measured in the presence of indicated concentrations of TBT-Cl. Samples are FoF1 (•), FoF1 plus 0.3% lauryldodecylamine oxide (○), and F1 (□). (B) Time course of the rotation of FoF1 in 1 mM ATP in the presence of 1 μM TBT-Cl. (Inset) Expanded time course of rotation.
Discussion
In this study, we directly observed ATP-driven rotation of single-molecule FoF1 immobilized on a glass surface. To do so, FoF1 must be solubilized and purified without losing original structural integrity. It should be noted that the previous demonstrations of rotation of purified E. coli FoF1 immobilized on a glass surface were carried out by using the enzyme preparation, of which ATPase activity in the rotation buffer and the rotation itself were totally insensitive to DCCD (34, 35), and might have been the rotation of incomplete FoF1, as stated in the subsequent paper (36). We have spent considerable effort to establish the procedures to isolate intact thermophilic FoF1 from the recombinant E. coli cells, and found that extraction from membranes by octaethylene glycol monododecyl ether and isolation in LPC gave the best yield and preparation. Both bulk-phase ATPase activity in the rotation buffer and the observed rotations under microscopy were efficiently inhibited by DCCD and TBT-Cl. Combination of this intact FoF1 and the submillisecond fast camera enabled us to learn several features of ATP-driven rotation of FoF1.
Firstly, FoF1 rotates as fast as 350 rps (37°C). We observed rotations at 25°C and 45°C and obtained the Vmax rotation rates of ≈230 and ≈650 rps, respectively. Therefore, extrapolated rotation rate at 60°C, an optimum growth temperature of Bacillus PS3, can reach ≈1,600 rps. Although reservation is needed concerning whether these enormous numbers are really the case, it is certain that the rotation rate of thermophilic FoF1 at 25°C is much faster than the rotation rate (19 ms/120°, that is, 17 rps, at 23°C) reported for E. coli FoF1 reconstituted into liposomes measured with fluorescence resonance energy transfer (22). However, in these measurements, uphill proton gradient should be immediately established by proton-pumping ATP hydrolysis to reach an equilibrium, and the rate limiting can be the rate of passive proton leak through membranes. It is interesting to discover whether the rotation rate will increase by addition of an uncoupler. One might argue whether fast rotation of >200 rps at Vmax is common in FoF1 (and F1) from various sources. Some reported values of very high ATPase activities would predict rapid rotations such as bovine mitochondrial F1 (≈310 rps) (37), yeast mitochondrial F1 (≈280 rps) (38), and E. coli FoF1 (≈300 rps) (39). Much lower ATPase activities corresponding to 10 ≈ 100 rps were also reported in many papers. However, if a significant fraction of molecules in the bulk solution are in the ADP-Mg-inhibited state or other inactive states, as in the case of thermophilic FoF1, real ATPase activity specific for the working enzymes should be higher, and the rotation rates can be much faster. It is intriguing to learn whether these rapid rotations are really occurring in living cells.
Second, FoF1 rotates in the almost same manner as F1. Vmax rotation rate, [ATP] dependency, ATP-waiting dwell, and catalytic dwell are all very similar to those observed for F1. Apparently, friction in Fo motor is negligible, that is, Fo does not impose significant drag during ATP-driven rotation of FoF1. This finding means that Fo keeps the interaction between the c-subunit ring and ab2 neither too strong nor too weak. The c-subunit ring seems to be able to pause rotation at the angular positions dictated by F1. We recently proposed that b2 in Fo act as a strong brake of ATP-driven rotation when the a-subunit is removed (40). The association of the a-subunit makes b2 to be a proper anchor rail that allows the c-subunit ring to slide without breaking association during rotation.
Finally, an Fo-specific inhibitor, TBT-Cl, obscures the steps in ATP-driven rotation of FoF1. Obviously, now the Fo portion resists the rotary torque generated in F1 portion. Because the rotor of Fo in thermophilic FoF1 is a decamer c-subunit ring (41), it is likely that resistance occurs at every 36°. As discussed in the recent paper (41), mismatch of the unit-rotation angle in F1 (120°) and in Fo (36°) assumes a torsion-spring-like motion of the central rotor shaft (and/or the peripheral stalk) that makes the dwelling position of the c-subunit ring in ATP-driven rotation obscure. We don't know whether this explanation is really the case, but we expect that further study on TBT-Cl inhibition will provide the answer.
Supplementary Material
Acknowledgments
We thank J. Suzuki for technical assistance; K. Shimabukuro, E. Muneyuki, R. Iino, T. Masaike, T. Ariga, and M. Takeda for valuable discussion and technical advice; and H. Noji, R. Yasuda, and K. Adachi for creating and developing the single-molecule observation system. This work was supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to H.U.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ATPγS, adenosine 5′-[γ-thio]triphosphate; DCCD, N,N′-dicyclohexylcarbodiimide; LPC, lysophosphatidylcholine; Ni2+-NTA, Ni2+-nitrilotriacetic acid; rps, revolutions per second; TBT-Cl, tributyltin chloride.
References
- 1.Boyer, P. D. (1997) Annu. Rev. Biochem. 66, 717-749. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, M., Muneyuki, E. & Hisabori, T. (2001) Nat. Rev. Mol. Cell Biol. 2, 669-677. [DOI] [PubMed] [Google Scholar]
- 3.Capaldi, R. A. & Aggeler, R. (2002) Trends Biochem. Sci. 27, 154-160. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen, P. L. (2002) J. Bioenerg. Biomembr. 34, 327-332. [DOI] [PubMed] [Google Scholar]
- 5.Senior, A. E., Nadanaciva, S. & Weber, J. (2002) Biochim. Biophys. Acta 1553, 188-211. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. (1994) Nature 370, 621-628. [DOI] [PubMed] [Google Scholar]
- 7.Seelert, H., Poetsch, A., Dencher, N. A., Engel, A., Stahlberg, H. & Müller, D. J. (2000) Nature 405, 418-419. [DOI] [PubMed] [Google Scholar]
- 8.Birkenhäger, R., Hoppert, M., Deckers-Hebestreit, G., Mayer, F. & Altendorf, K. (1995) Eur. J. Biochem. 230, 58-67. [PubMed] [Google Scholar]
- 9.Singh, S., Turina, P., Bustamante, C. J., Keller, D. J. & Capaldi, R. (1996) FEBS Lett. 397, 30-34. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, W. & Fillingame, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 6607-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer, P. D. (1993) Biochim. Biophys. Acta 1140, 215-250. [DOI] [PubMed] [Google Scholar]
- 12.Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K., Jr. (1997) Nature 386, 299-302. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda, R., Noji, H., Kinosita, K., Jr., & Yoshida, M. (1998) Cell 93, 1117-1124. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., Jr., & Itoh, H. (2001) Nature 410, 898-904. [DOI] [PubMed] [Google Scholar]
- 15.Shimabukuro, K., Yasuda, R., Muneyuki, E., Hara, K. Y., Kinosita, K., Jr., & Yoshida, M. (2003) Proc. Natl. Acad. Sci. USA 100, 14731-14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishizaka, T., Oiwa, K., Noji, H., Kimura, S., Muneyuki, E., Yoshida, M. & Kinosita, K., Jr. (2004) Nat. Struct. Mol. Biol. 11, 142-148. [DOI] [PubMed] [Google Scholar]
- 17.Kaim, G., Prummer, M., Sick, B., Zumofen, G., Renn, A., Wild, U. P. & Dimroth, P. (2002) FEBS Lett. 525, 156-163. [DOI] [PubMed] [Google Scholar]
- 18.Hutcheon, M. L., Duncan, T. M., Ngai, H. & Cross, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8519-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsunoda, S. P., Aggeler, R., Yoshida, M. & Capaldi, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishio, K., Iwamoto-Kihara, A., Yamamoto, A., Wada, Y. & Futai, M. (2002) Proc. Natl. Acad. Sci. USA 99, 13448-13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Börsch, M., Diez, M., Zimmermann, B., Reuter, R. & Gräber, P. (2002) FEBS Lett. 527, 147-152. [DOI] [PubMed] [Google Scholar]
- 22.Diez, M., Zimmermann, B., Börsch, M., König, M., Schweinberger, E., Steigmiller, S., Reuter, R., Felekyan, S., Kudryavtsev, V., Seidel, C. A. & Gräber, P. (2004) Nat. Struct. Mol. Biol. 11, 135-141. [DOI] [PubMed] [Google Scholar]
- 23.Landt, O., Grunert, H. P. & Hahn, U. (1990) Gene 96, 125-128. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, T., Ueno, H., Mitome, N., Suzuki, J. & Yoshida, M. (2002) J. Biol. Chem. 277, 13281-13285. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, T., Suzuki, J., Mitome, N., Ueno, H. & Yoshida, M. (2000) J. Biol. Chem. 275, 37902-37906. [DOI] [PubMed] [Google Scholar]
- 26.Stock, D., Leslie, A. G. & Walker, J. E. (1999) Science 286, 1700-1705. [DOI] [PubMed] [Google Scholar]
- 27.Hirono-Hara, Y., Noji, H., Nishiura, M., Muneyuki, E., Hara, K. Y., Yasuda, R., Kinosita, K., Jr., & Yoshida, M. (2001) Proc. Natl. Acad. Sci. USA 98, 13649-13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui, T., Muneyuki, E., Honda, M., Allison, W. S., Dou, C. & Yoshida, M. (1997) J. Biol. Chem. 272, 8215-8221. [DOI] [PubMed] [Google Scholar]
- 29.Jault, J. M., Matsui, T., Jault, F. M., Kaibara, C., Muneyuki, E., Yoshida, M., Kagawa, Y. & Allison, W. S. (1995) Biochemistry 34, 16412-16418. [DOI] [PubMed] [Google Scholar]
- 30.Kato-Yamada, Y., Bald, D., Koike, M., Motohashi, K., Hisabori, T. & Yoshida, M. (1999) J. Biol. Chem. 274, 33991-33994. [DOI] [PubMed] [Google Scholar]
- 31.Cain, K. & Griffiths, D. E. (1977) Biochem. J. 162, 575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuno-Yagi, A. & Hatefi, Y. (1993) J. Biol. Chem. 268, 6168-6173. [PubMed] [Google Scholar]
- 33.von Ballmoos, C., Brunner, J. & Dimroth, P. (2004) Proc. Natl. Acad. Sci. USA 101, 11239-11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambongi, Y., Iko, Y., Tanabe, M., Omote, H., Iwamoto-Kihara, A., Ueda, I., Yanagida, T., Wada, Y. & Futai, M. (1999) Science 286, 1722-1724. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe, M., Nishio, K., Iko, Y., Sambongi, Y., Iwamoto-Kihara, A., Wada, Y. & Futai, M. (2001) J. Biol. Chem. 276, 15269-15274. [DOI] [PubMed] [Google Scholar]
- 36.Pänke, O., Gumbiowski, K., Junge, W. & Engelbrecht, S. (2000) FEBS Lett. 472, 34-38. [DOI] [PubMed] [Google Scholar]
- 37.Feinstein, D. L. & Moudrianakis, E. N. (1984) J. Biol. Chem. 259, 4230-4236. [PubMed] [Google Scholar]
- 38.Ichikawa, N. & Mizuno, M. (2004) Protein Expression Purif. 37, 97-101. [DOI] [PubMed] [Google Scholar]
- 39.Moriyama, Y., Iwamoto, A., Hanada, H., Maeda, M. & Futai, M. (1991) J. Biol. Chem. 266, 22141-22146. [PubMed] [Google Scholar]
- 40.Ono, S., Sone, N., Yoshida, M. & Suzuki, T. (2004) J. Biol. Chem. 279, 33409-33412. [DOI] [PubMed] [Google Scholar]
- 41.Mitome, N., Suzuki, T., Hayashi, S. & Yoshida, M. (2004) Proc. Natl. Acad. Sci. USA 101, 12159-12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.