Abstract
One impediment to effective cancer-specific gene therapy is the rarity of regulatory sequences targeting gene expression selectively in tumor cells. Although many tissue-specific promoters are recognized, few cancer-selective gene promoters are available. Progression-elevated gene-3 (PEG-3) is a rodent gene identified by subtraction hybridization that displays elevated expression as a function of transformation by diversely acting oncogenes, DNA damage, and cancer cell progression. The promoter of PEG-3, PEG-Prom, displays robust expression in a broad spectrum of human cancer cell lines with marginal expression in normal cellular counterparts. Whereas GFP expression, when under the control of a CMV promoter, is detected in both normal and cancer cells, when GFP is expressed under the control of the PEG-Prom, cancer-selective expression is evident. Mutational analysis identifies the AP-1 and PEA-3 transcription factors as primary mediators of selective, cancer-specific expression of the PEG-Prom. Synthesis of apoptosis-inducing genes, under the control of the CMV promoter, inhibits the growth of both normal and cancer cells, whereas PEG-Prom-mediated expression of these genes kills only cancer cells and spares normal cells. The efficacy of the PEG-Prom as part of a cancer gene therapeutic regimen is further documented by in vivo experiments in which PEG-Prom-controlled expression of an apoptosis-inducing gene completely inhibited prostate cancer xenograft growth in nude mice. These compelling observations indicate that the PEG-Prom, with its cancer-specific expression, provides a means of selectively delivering genes to cancer cells, thereby providing a crucial component in developing effective cancer gene therapies.
Keywords: gene therapy, PEG-3, mda-7/IL-24, p53
Eradication of cancer, especially when tumor cells acquire metastatic potential, is an extremely difficult and frequently unsuccessful endeavor even when combinatorial therapeutic approaches, i.e., surgery, radiotherapy, and chemotherapy, are used (1). Moreover, even with such aggressive therapeutic approaches, the relapse rate of patients with disseminated cancers is significantly high, and disease-free survival time becomes progressively shortened. These observations underscore the need for additional innovative treatment modalities, and gene therapy is one approach that, with appropriate improvements, may provide such an option (2). Cancer gene therapy applications have used a number of strategies, including replacement and overexpression of tumor-suppressor and suicide genes, targeted inhibition of oncogenic changes in cancer cells, and selective viral replication and cytolysis in tumor cells (3, 4). In many cases, using a gene promoter to drive appropriate gene expression selectively in a cancer cell context provides a means of avoiding toxicity to normal cells and tissues (2, 4). Additionally, a gene delivery vector that will transduce its payload into a majority of the cancer cell population with the potential to be administered to patients multiple times without eliciting an immune response represents an essential requirement for successful cancer gene therapy (2, 4). Unfortunately, the combination of all these elements to generate an efficacious gene therapy approach has not been achieved.
Progression-elevated gene-3 (PEG-3) was cloned from a tumor-progression model based on rat embryo cells E-11 and E11-NMT (5–7). E11 is a mutant adenovirus type 5 (H5ts125)-transformed rat embryo cell clone that forms small, slow-growing, compact tumors. E11-NMT is a clone of E11 that has been selected for aggressiveness by passage through a nude mouse and that forms rapidly growing, highly aggressive tumors (5). Subtraction hybridization of an E11 cDNA library from an E11-NMT cDNA library identified PEG-3 (7), which has recently been determined to be a C-terminal truncated mutant form of the rat-growth-arrest and DNA-damage-inducible gene-34 (GADD-34) (8, 9). Elevated PEG-3 expression has been documented in E11-NMT cells in comparison with E11 cells and also in normal cloned rat embryo fibroblast (CREF) cells displaying a transformed/tumorigenic phenotype as a consequence of the expression of diversely acting oncogenes, including Ha-ras, v-src, human papilloma virus type-18-transforming genes, and a specific mutant of adenovirus (Ad) 5 (H5hr1), relative to parental CREF cells (7). Ectopic expression of PEG-3 in E11 cells markedly augments in vitro anchorage-independent growth and increases their oncogenic potential in nude mice as reflected by a shorter tumor-latency time and the production of larger tumors with increased vascularization (7, 10). As a corollary, E11-NMT cells stably expressing antisense PEG-3 lose their progressed-cancer phenotype (10). Overexpression of PEG-3 induces genomic instability, modulates the expression of important genes involved in centrosomal duplication, and augments invasive capability by increasing matrix metalloproteinase activity, indicating that PEG-3 facilitates tumor progression by multiple pathways (11, 12).
The promoter region of the PEG-3 gene (PEG-Prom) was cloned to investigate the mechanism of induction of PEG-3 expression as a consequence of oncogenic transformation (13, 14). It was observed that the PEG-Prom was ≈8- to 10-fold more active in CREF cells transformed with either Ha-ras or v-raf than were the parental CREF cells, and a minimum region of the promoter that extends from –118 to +194 (when the transcription initiation site is regarded as +1) was shown to be sufficient for the increased activity associated with transformation and cancer progression (13, 14). This region contains a binding site for PEA-3 at –104 and for AP-1 at +8, and sequence-specific mutational analysis revealed that both of these transcription factors are important for regulating the basal and oncogene-induced activity of the PEG-Prom (13, 14). Interestingly, PEA-3 expression or DNA binding could be detected only in CREF-ras cells but not in CREF cells, indicating that oncogenic transformation alters the transcription-factor expression pattern leading to increased PEG-Prom activity (14).
These provocative observations in the rodent system prompted us to test the hypothesis that the PEG-Prom might be an effective reagent to facilitate transgene expression in a transformed, cell-selective manner in human tumor cells. Considering this possibility, the activity of the PEG-Prom was evaluated in a battery of diverse human cancer cells, including glioblastoma multiforme and carcinomas of the breast and prostate, and their normal counterparts, astrocytes and epithelial cells, confirming that the PEG-Prom functions selectively in human cancer cells. As observed in a rodent transformation context, the importance of both PEA-3 (or E1AF, the human homologue) and AP-1 transcription factors in regulating PEG-Prom activity in human tumor cells has been verified. Of direct relevance in the context of enhancing cancer gene therapy applications, targeting expression of tumor-suppressing/apoptosis-inducing genes, including wild-type p53 and melanoma differentiation-associated gene-7/IL-24 (mda-7/IL-24), in tumor cells results in growth suppression and cell death without inducing similar effects in normal tissue. In contrast, when these genes are regulated by a nonspecific CMV, minimum promoter growth is suppressed in both normal and cancer cells, indicating a lack of targeting specificity. These results provide support for the use of the PEG-Prom as a means of selectively targeting gene expression in human tumor cells for directed cancer gene therapy applications.
Materials and Methods
Cell Lines and Culture Conditions. Normal human prostate epithelial cells (HuPEC) and Du-145, PC-3, and LNCaP prostate carcinoma cells were cultured as described in ref. 15. P69, M2182, and M12 cells were cultured as described in ref. 16. Normal human mammary epithelial cells (HMEC), normal human immortal mammary epithelial cells HBL-100 and MCF-7, T47D, MDA-MB-231, MDA-MB-157, and MDA-MB-453 human breast cancer cells were cultured as described in ref. 17. Primary human fetal astrocytes (PHFA) and T98G, U87MG, U251MG, U373, and H4 human malignant glioma cell lines were cultured as described in ref. 18. All cultures were maintained at 37°C in a humidified 5% CO2/95% air incubator.
Construction of Plasmids, Transient Transfection, and Luciferase Assay. The minimum region of PEG-3 promoter (–118 to +194) was cloned into pGL3-basic Vector (Promega) to generate pPEG-Luc. Mutant pPEG-Luc constructs containing a mutation in either PEA-3 or AP-1 or in both sequences were generated by using the Altered Sites II in vitro Mutagenesis System (Promega) as described in ref. 13. Transient transfection was performed by using Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's instructions and as described in ref. 13. Luciferase and β-galactosidase assays were performed by using commercial kits (Promega and Tropix, respectively) 48 h after transfection as described in ref. 13.
Construction of Ads and Infection Protocol. The recombinant replication-incompetent Ads, Ad.CMV-GFP, Ad.PEG-GFP, Ad.CMV-p53 (CMV promoter driving wild-type p53 expression), Ad.PEG-p53 (PEG-Prom driving wild-type p53 expression), Ad.CMV-mda-7 (CMV promoter driving mda-7/IL-24 expression), and Ad.PEG-mda-7 (PEG-Prom driving mda-7/IL-24 expression) were created in two steps as described previously and plaque purified by standard procedures (17, 19). As a control, empty replication-incompetent Ad.vec was used. Cells were infected with a multiplicity of infection (moi) of 100 plaque-forming units (pfu) per cell of different Ads as described in ref. 19.
RNA Extraction and Northern Blot Analysis. Total RNA was extracted, and Northern blotting was performed as described in ref. 14. The cDNA probes used were full-length human PEA-3, c-jun, and GAPDH.
Western Blotting. Whole-cell lysates were prepared, and Western blotting was performed as described in ref. 17. The primary antibodies used were anti-PEA-3, anti-c-JUN, and anti-actin (Santa Cruz Biotechnology).
EMSA. Nuclear extracts were prepared, and EMSA was performed as described in ref. 13. The sequences of the probes were as follows: wild-type AP-1, 5′-CGCAGATTGACTCAGTTCGC-3′; mutant AP-1, 5′-CGCAGATAAACTACGTTCGC-3′; wild-type PEA-3, 5′-GTGTTGTTTTCCTCTCTCCA-3′; and mutant PEA-3, 5′-GTGTTGTTCCCATCTCTCCA-3′.
DNA Fragmentation Analysis. DNA fragmentation was analyzed as described in ref. 20.
Animal Studies. Tumorigenicity studies were performed as described in ref. 17. Briefly, Du-145 cells were either uninfected or infected with Ad.vec, Ad.CMV-p53, Ad.PEG-p53, Ad.CMV-mda-7, or Ad.PEG-mda-7 at a moi of 100 pfu per cell, and 1 × 106 cells were s.c. injected into athymic nude mice 48 h later. Animals were monitored for tumor formation, and tumor volume was determined (17).
Statistical Analysis. All of the experiments were performed at least three times. The results are expressed as mean ± SD. Statistical comparisons were made by using an unpaired two-tailed Student t test. P < 0.05 was considered as significant.
Results
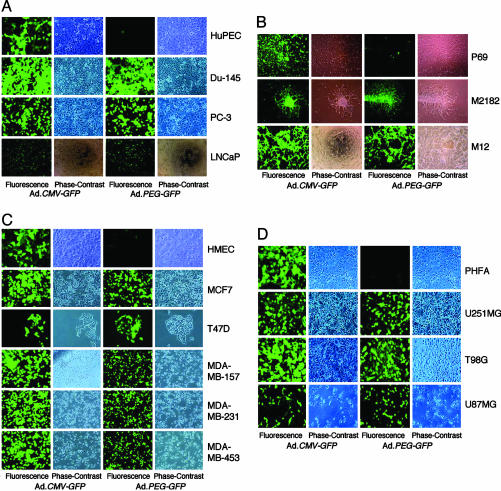
PEG-Prom Functions Selectively in Cancer Cells. Previous studies document significantly higher activity of the PEG-Prom in transformed rodent cell lines that are highly aggressive and metastatic in comparison with their less aggressive and nonmetastatic variants (13, 14). To determine whether this phenomenon is also evident in human tumors, experiments were performed by using a battery of human cancer cell lines of diverse origins and their normal cellular counterparts. Two replication-incompetent adenoviral vectors, Ad.CMV-GFP, in which the expression of the GFP was driven by the CMV minimum promoter that is routinely used in expression vectors, and Ad.PEG-GFP, in which GFP expression was driven by the minimal region of the PEG-3 promoter (–118 to +194 when the transcription start site is regarded as +1), were constructed. Different cell lines and their normal counterparts were infected with the Ad at a moi of 100 pfu per cell because, at this moi, >90% of the cells were infected. GFP expression was analyzed by using an immunofluorescence microscope at 2 d postinfection. After infection with Ad.CMV-GFP, high GFP expression could be detected in normal HuPEC and in prostate carcinoma cell lines Du-145, PC-3, and LNCaP (Fig. 1A). In contrast, with Ad.PEG-GFP infection, none-to-barely detectable levels of GFP could be observed in HuPEC, whereas in the prostate carcinoma cell lines, especially in Du-145 and PC-3, the GFP-expression level was robust and was comparable to that observed after Ad.CMV-GFP infection. These findings were extended further in a set of HuPEC lines displaying different stages of tumor progression, i.e., SV40-TAg-immortalized (P69), tumorigenic but not metastatic P69 variant (M2182), and a tumorigenic and metastatic variant of P69 (M12) (16). As shown in Fig. 1B, in P69, M2182, and M12 cells, >90% of the cells showed high GFP expression upon Ad.CMV-GFP infection. However, with Ad.PEG-GFP infection, very few P69 cells displayed only low GFP expression, whereas >90% of M2182 and M12 cells showed strong GFP expression.
Fig. 1.
PEG-Prom drives the expression of GFP only in cancer cells but not in normal cells. The indicated cells were infected with either Ad.CMV-GFP or Ad.PEG-GFP at a moi of 100 pfu per cell, and GFP expression was analyzed by an immunofluorescence microscope at 2 d postinfection.
To determine whether the tumor-suppressor status of a cancer cell line might affect PEG-Prom activity, we infected normal HMEC and a series of breast cancer cell lines that have different p53 status with Ad.CMV-GFP and Ad.PEG-GFP. Similar to prostate cancer cell lines, high-level GFP expression could be detected in HMEC and MCF-7 (WT-p53), T47D, MDA-MB-231, MDA-MB-453 (Mut-p53), and MDA-MB-157 (p53-null) breast cancer cell lines after Ad.CMV-GFP infection (Fig. 1C). Whereas all of the breast cancer cell lines showed significant GFP expression, with Ad.PEG-GFP infection, no-to-barely detectable levels of GFP could be observed in HMEC, indicating that PEG-Prom functions selectively in cancer cells and independently of their tumor-suppressor status. These findings were reinforced by similar results in PHFA and human malignant glioma cell lines, U87MG (WT-p53), T98G, and U251MG (Mut-p53). Whereas Ad.CMV-GFP infection resulted in high GFP expression in all four cell types, with Ad.PEG-GFP infection, high GFP expression could be detected only in malignant glioma cell lines but not in PHFA (Fig. 1D). Similarly, by using Ad.PEG-GFP, no or minimal GFP expression was evident in normal human melanocytes, normal human mesothelial cells, normal human colonic and pancreatic epithelial cells, normal human hepatocytes, and normal human fibroblasts, whereas high GFP expression was observed in malignant melanoma cells and carcinomas of the colon, liver, ovary, and pancreas (data not shown), indicating the highly selective activity of the PEG-Prom in cancer cells of all origins and genetic backgrounds tested so far with almost no activity in their normal counterparts.
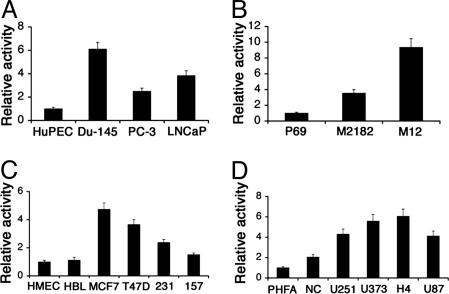
To quantify the activity of the PEG-Prom, a luciferase-assay-based approach was used. We constructed pPEG-Luc in which the expression of the luciferase reporter gene was under the control of the PEG-Prom and transfected it into different cancer cell lines and primary or immortal normal cells obtained from corresponding organs. Cotransfection of a β-galactosidase expression plasmid was performed to normalize for transfection efficiency. As shown in Fig. 2A, the relative activity of pPEG-Luc was 6.1-, 2.5-, and 3.9-fold higher in Du-145, PC-3, and LNCaP cells, respectively, in comparison with that in HuPEC. Similarly, the activity of pPEG-Luc was 3.6- and 9.4-fold higher in the M2182 and M12 cell lines, respectively, in comparison with that in P69 cells (Fig. 2B). MCF-7, T47D, MDA-MB-231, and MDA-MB-157 cells showed 4.8-, 3.7-, 2.4-, and 1.5-fold activity of pPEG-Luc, respectively, over that in HMEC and HBL100, which is an immortal but nontumorigenic human breast epithelial cell line (Fig. 2C). The activity of pPEG-Luc in NC (immortal normal cerebellum cell line), U251MG, U373, H4, and U87MG cells was 1.5-, 4.3-, 5.6-, 6-, and 4.1-fold higher, respectively, than that in PHFA (Fig. 2D). Similar findings were observed in human malignant melanoma, ovarian, colon, pancreatic, and hepatic carcinoma cell lines (data not shown). These findings indicate that the activity of the PEG-Prom is significantly higher in all of the cancer cell lines tested than in their normal counterparts. It should be noted that in some cell lines (such as MDA-MB-157) the high GFP-expression level did not correlate with a high luciferase-expression level, most likely due to the highly efficient Ad-mediated gene-delivery method versus the conventional transient-transfection approach.
Fig. 2.
PEG-Prom drives high luciferase expression in cancer cells but not in normal cells. The indicated cells were transfected with pPEG-Luc, and luciferase activity was measured at 48 h posttransfection. The luciferase activity was normalized by β-galactosidase activity. The data represent the mean ± SD of three independent experiments, each performed in triplicate. HBL, HBL100; 231, MDA-MB-231; 157, MDA-MB-157; NC, normal cerebellum.
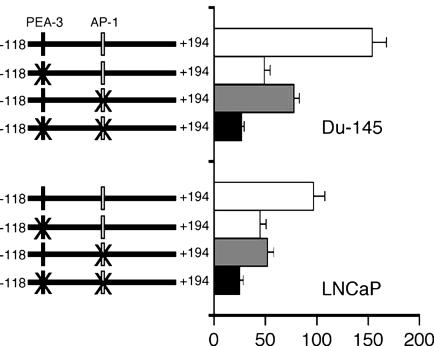
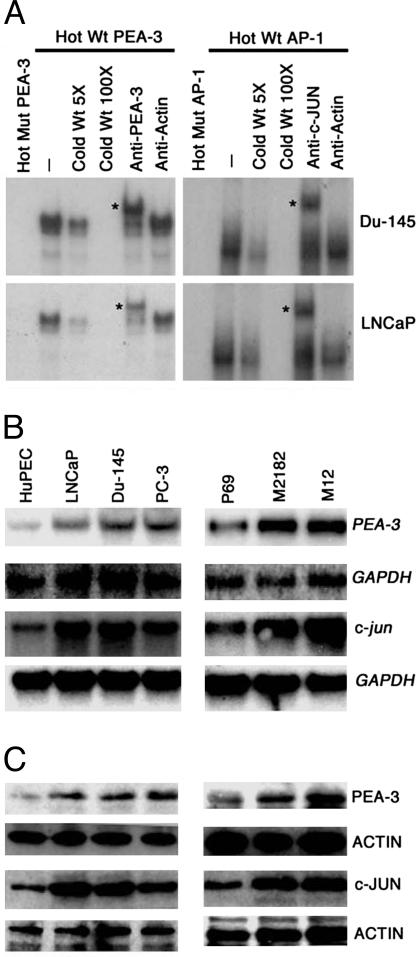
PEA-3 and AP-1 Transcription Factors Are Central for PEG-Prom Activity. In the minimum effective element of PEG-Prom (–118 to +194) there are two important transcription factor binding sites, one for PEA-3 at –104 and another at +8 for AP-1. In the rodent cell system, these two transcription factors play an essential role in regulating PEG-Prom activity. Based on this consideration, the potential involvement of PEA-3 and AP-1 in regulating PEG-Prom activity was evaluated in human cell systems. Data are presented from prostate carcinoma cell lines, although similar findings were obtained from other types of cells such as breast carcinoma and malignant glioma (data not shown). As shown in Fig. 3, mutation of the PEA-3 site reduced pPEG-Luc activity by 68% and 54% in Du-145 and LNCaP cells, respectively. Mutation of the AP-1 site reduced the pPEG-Luc activity by 50% and 47% in Du-145 and LNCaP cells, respectively, whereas mutations of both sites reduced pPEG-Luc activity by 82% and 75% in Du-145 and LNCaP cells, respectively. EMSAs with nuclear extracts from Du-145 and LNCaP cells revealed that PEA-3 and AP-1 transcription factors bind specifically to the corresponding sites (Fig. 4A). With both Du-145 and LNCaP nuclear extracts, whereas no shifted band could be detected with a labeled mutated PEA-3 probe, a single shifted band could be observed with a labeled wild-type PEA-3 probe, which could be competed by a cold, wild-type PEA-3 probe and supershifted by anti-PEA-3 antibody but not by anti-actin antibody (Fig. 4A Left). The labeled mutated AP-1 probe did not give rise to any shifted band with Du-145 and LNCaP nuclear extracts, whereas a shifted band was observed with a labeled wild-type AP-1 probe, which was competed by a cold, wild-type AP-1 probe and supershifted by antibody against c-JUN, a component of AP-1, but not with anti-actin antibody (Fig. 4A Right).
Fig. 3.
PEA-3 and AP-1 are essential for PEG-Prom activity. Du-145 and LNCaP cells were transfected with the indicated plasmids, and luciferase activity was measured at 48 h posttransfection. The luciferase activity was normalized by β-galactosidase activity. The data represent the mean ± SD of three independent experiments, each performed in triplicate.
Fig. 4.
PEA-3 and AP-1 bind to PEG-Prom and are overexpressed in prostate cancer cells versus HuPEC or P69 cells. (A) PEA-3 and AP-1 DNA binding was analyzed by EMSA by using nuclear extracts from Du-145 and LNCaP cells. The asterisks represent supershifted bands. (B) The expressions of PEA-3, GAPDH, and c-jun mRNAs were analyzed in the indicated cells by Northern blot analysis. (C) The expressions of PEA-3, ACTIN, and c-JUN proteins were analyzed by Western blot analysis in the same cells as shown in B.
To obtain better insights into the role of PEA-3 and AP-1 in regulating PEG-Prom activity, the relative abundance of PEA-3 and AP-1 mRNA and proteins in HuPEC, P69, and prostate carcinoma cell lines was analyzed by Northern and Western blot analysis (Fig. 4 B and C). Low-level PEA-3 mRNA and protein were detected in HuPEC and P69 cells. However, both the mRNA and protein levels were significantly higher in Du-145, PC-3, LNCaP, M2182, and M12 cells. Similar findings were also observed for c-jun mRNA and protein, indicating that the relative abundance of PEA-3 and AP-1 in prostate carcinoma cells, in comparison with their normal counterparts, might be the critical factor for the higher PEG-Prom activity in these cells. Similarly elevated levels of PEA-3 and AP-1 were found in a broad spectrum of additional human cancers, including carcinomas of the breast, colon, liver, and lung versus their normal counterparts, and this elevated expression correlated with increased PEG-Prom activity in a cancer-cell-specific context (data not shown).
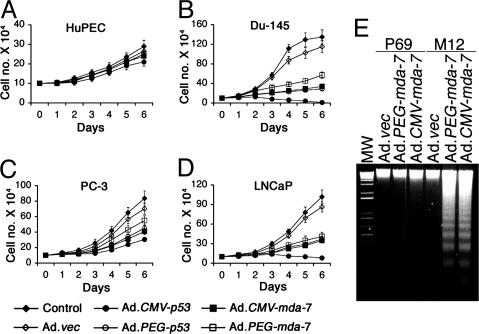
PEG-Prom Is a Useful Tool for Cancer Gene Therapy. An important question was whether the PEG-Prom could drive transgene expression for actual cancer gene therapy purposes. We chose wild-type p53 and mda-7/IL-24 as the transgenes to target growth inhibition in cancer cells. It should be noted that a unique property of mda-7/IL-24 is that, when administered via an adenoviral vector, mda-7/IL-24 induces apoptosis selectively in cancer cells without affecting the normal cells (21, 22). We constructed replication-incompetent Ads, namely, Ad.CMV-p53, Ad.PEG-p53, Ad.CMV-mda-7, and Ad.PEG-mda-7. These Ads were infected into HuPEC and prostate carcinoma cell lines at a moi of 100 pfu per cell, and cell survival was analyzed by the trypan-blue-exclusion test. Among the Ads, only Ad.CMV-p53 inhibited the growth of HuPEC (Fig. 5A). The growth inhibition was small but statistically significant. The observation that Ad.PEG-p53, Ad.CMV-mda-7, and Ad.PEG-mda-7 did not inhibit the growth of HuPEC further strengthens the cancer-cell-specific activity of the PEG-Prom and cancer-cell-specific killing effect of mda-7/IL-24 (Fig. 5A). This cancer-specific expression of wild-type p53 and mda-7/IL-24 was verified on both mRNA and protein levels, where expression controlled by the PEG-Prom occurred in the prostate tumor cells but not in normal HuPEC cells (data not shown). In contrast, the CMV-Prom resulted in transgene expression in both normal and cancer cells (data not shown). In the case of Du-145, PC-3, and LNCaP prostate cancer cells, except for Ad.vec, all of the Ads markedly inhibited cell growth (Fig. 5 B–D). The growth inhibition was profound in Du-145 and LNCaP cells whereas, in PC-3 cells, growth was moderate. Ad.CMV-p53 was the most potent virus in inhibiting growth, whereas Ad.PEG-p53, Ad.CMV-mda-7, and Ad.PEG-mda-7 showed similar efficacy. However, the growth-inhibitory effect of Ad.CMV-p53 on HuPEC precludes its selective employment for gene therapy purposes and highlights the enhanced selectivity of Ad.PEG-p53, Ad.CMV-mda-7, or Ad.PEG-mda-7 viruses for applications in patients. Similar results were obtained with HMEC and breast cancer cells and PHFA and malignant glioma cells (data not shown). mda-7/IL-24 is an apoptosis-inducing gene, and the mechanism of growth inhibition was determined by DNA fragmentation assay, an indicator of apoptosis. Infection with Ad.vec, Ad.CMV-mda-7, or Ad.PEG-mda-7 did not induce apoptosis in P69 cells, whereas infection with Ad.CMV-mda-7 or Ad.PEG-mda-7 induced significant apoptosis in M12 cells (Fig. 5E), indicating the cancer-selective function of both PEG-Prom and mda-7/IL-24.
Fig. 5.
PEG-Prom driving expression of apoptosis-inducing genes inhibits growth of prostate cancer cells but not normal prostate epithelial cells. (A–D) The indicated cells were either uninfected or infected with the indicated Ad, and cell survival was analyzed by the trypan-blue-exclusion assay. The data represent the mean ± SD of three independent experiments, each performed in triplicate. (E) P69 and M12 cells were infected with the indicated viruses, and DNA fragmentation was analyzed.
The in vitro findings were further extended into in vivo studies. Du-145 cells were either uninfected or infected with Ad.vec, Ad.CMV-p53, Ad.PEG-p53, Ad.CMV-mda-7, or Ad.PEG-mda-7 at a moi of 100 pfu per cell ex vivo, and 48 h later, the cells were s.c. injected into athymic nude mice. The growth of the tumor xenografts was monitored every week. Except for Ad.vec, infection with all of the other Ads resulted in complete eradication of the tumor xenografts (data not shown), indicating that these Ads might be highly efficacious tools for the treatment of cancers.
Discussion
Strict cancer-cell-specific expression of cell-death-promoting genes is mandatory for developing effective cancer gene therapies because inappropriate transgene expression can result in nonspecific toxicity. Currently, this problem is addressed by using tissue-specific promoters that have a high level of activity in transformed cells versus normal cells (23, 24). However, the spectrum of regulatory sequences available for this approach is limited, and many promoters are leaky, resulting in the gradual/temporal production of toxic products. The only verified promoter that functions selectively in a diverse cancer-cell-specific manner without having significant activity in normal cells, is the telomerase promoter, which is being extensively used to drive transgene expression to eradicate cancer cells (25–27). Our demonstration that the PEG-Prom displays similar robust cancer-specific expression now provides an additional reagent for developing cancer-specific gene therapy vectors with clear cancer therapeutic potential.
A significant structural aspect of the PEG-Prom is that the minimum promoter element is regulated predominantly by two transcription factors, PEA-3 and AP-1, which regulate the expression of a spectrum of genes associated with transformation, tumor progression, and invasion (28, 29). PEA-3 and AP-1 cooperate to regulate the expression of diverse genes involved in carcinogenesis, such as cyclooxegenase-2, osteopontin, and IL-8 (30–32). The expression of PEA-3 is very weak in normal human tissues. However, PEA-3 is overexpressed in multiple types of cancers including breast, ovarian, colon, lung, and oral cancers and PEA-3 expression correlated with poor overall survival for breast, ovarian, and oral cancers (33–37). One preliminary immunohistochemical study failed to detect PEA-3 expression in high-grade prostate cancers (38). However, we detected high PEA-3 mRNA and protein expression in multiple prostate cancer cell lines, and our in vivo studies confirmed the effective activity of the PEG-Prom in tumor xenografts generated from Du-145 prostate cancer cells. Moreover, it is possible that in prostate cancers in patients, up-regulation of AP-1 might compensate for reduced PEA-3 expression, thereby permitting robust PEG-Prom transgene expression (39). The components of AP-1, such as c-fos, fosB, and c-jun, are potent oncogenes, the overexpression of which results in cell transformation (29). PEA-3 and AP-1 transactivation and PEG-Prom activity are positively regulated by the Ras-dependent signaling cascade (28, 29, 40), and, because activation of the Ras pathway is a frequent event in diverse cancers including pancreatic and colorectal cancers (41), the efficacy of PEG-Prom to drive transgene expression in these cancers would be specific and robust. The fact that the activity of both PEA-3 and AP-1 is low in normal cells and enhanced in the contexts of cell transformation and tumor progression ensures the tight control of the PEG-Prom in a transformed cell-specific manner. It should be noted that in addition to AP-1 and PEA-3, other as yet uncharacterized transactivating factor(s) might be involved in regulating the cancer-cell-specific expression of the PEG-Prom.
PEG-3 is a C-terminally truncated mutant form of the rat GADD-34 (8, 9). We have recently documented that mutation in the GADD34 gene resulting in PEG-3-like C-terminally truncated molecules is a frequent event during rodent tumorigenesis in multiple tissues, and PEG-3 functions as a dominant negative inhibitor of the apoptosis-inducing and growth-suppressing properties of both human and rat GADD34 (9). We could not identify any PEG-3-like molecule in human contexts, indicating that this mutational event is most likely very specific for rodents. We cloned the human GADD34 promoter and compared its sequence and activity to that of the PEG-Prom (unpublished data). The sequence similarity between these two promoters was very low, and there were no PEA-3 or AP-1 binding sites in the proximal promoter of the human GADD34 gene. More importantly, the human GADD34 promoter showed equal activity in both normal and cancer cells (data not shown), thus bolstering the uniqueness of functional activity of the rodent PEG-Prom.
The cancer-cell-specific, apoptosis-inducing properties of mda-7/IL-24 have been documented by numerous studies (21, 22). We demonstrate that, whereas Ad.CMV-p53 inhibited the growth of both HuPEC and prostate cancer cells, Ad.PEG-p53, Ad.CMV-mda-7, and Ad.PEG-mda-7 had no adverse effect on HuPEC and killed only prostate cancer cells. The combination of the PEG-Prom and mda-7/IL-24 in Ad.PEG-mda-7 ensures minimum-to-no toxicity to normal cells and provides effective, selective killing of cancer cells. This cancer-specific activity of Ad.PEG-mda-7 was observed not only in prostate cancer models but also in breast cancer cells, malignant glioma cells, malignant melanoma cells, and ovarian carcinoma cells, versus their respective normal counterparts, HMEC, PHFA, melanocytes, and mesothelial cells. Because Ad.CMV-mda-7 has been shown to be highly effective in a phase I clinical trial for advanced carcinomas and melanomas and is now being evaluated in phase II clinical trials (22), the profound observations relative to the selective activity of the PEG-Prom indicates that Ad.PEG-mda-7 could be a very effective tool for the treatment of diverse cancers without harming normal tissue.
Although many innovative therapies are now coming to the forefront and may eventually become mainstream approaches for cancer therapy, much work still remains to optimize these tactics. This is highlighted in the context of cancer gene therapy, in which appropriate vectors that display high tumor-specific transduction and expression profiles with minimal immunogenicity remain to be developed. One impediment to effective gene-based therapies for cancer relates to the scarceness of promoters that display wide-ranging cancer-selective activity. In this report, we describe a promoter, the PEG-Prom, that displays cancer-specific expression in a spectrum of histologically distinct human tumors with the capacity to deliver growth-suppressing and apoptosis-inducing genes uniquely to the cancer cell. This initial prototype-targeting vector promotes the delivery of transgenes by means of a nonreplicating Ad. However, in principle, the PEG-Prom could also be used to develop Ad in which this promoter drives replication resulting in conditionally replication-competent Ad (CRAD). Additionally, bipartite CRAD could be generated in which the PEG-Prom drives the expression of both replication-controlling genes and mda-7/IL-24 or an immunomodulatory gene, thus generating a scenario in which a complete cancer “cure” might actually be an achievable endpoint. In this context, the PEG-Prom has moved us closer to achieving the objective of developing an effective cancer gene therapy.
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA35675, CA97318, and CA98712, the Samuel Waxman Cancer Research Foundation (SWCRF), and the Chernow Endowment. P.B.F. is the Michael and Stella Chernow Urological Cancer Research Scientist and a SWCRF Investigator.
Author contributions: Z.-Z.S., D.S., and P.B.F. designed research; Z.-Z.S., D.S., L.E., and P.B.F. performed research; Z.-Z.S., D.S., G.J.D., C.S.H.Y., J.W., A.R., K.V., and P.B.F. contributed new reagents/analytic tools; Z.-Z.S., D.S., L.E., and P.B.F. analyzed data; and Z.-Z.S., D.S., and P.B.F. wrote the paper.
Abbreviations: PEG-3, progression-elevated gene-3; GADD, growth-arrest and DNA-damage-inducible gene; CREF, cloned rat embryo fibroblast; mda-7/IL-24, melanoma differentiation-associated gene-7/IL-24; PEG-Prom, promoter region of the PEG-3 gene; HuPEC, human prostate epithelial cells; HMEC, normal human mammary epithelial cells; PHFA, primary human fetal astrocytes; Ad, adenoviruses; moi, multiplicity of infection; pfu, plaque-forming units; CMV-p53, CMV-promoter driving wild-type p53 expression; PEG-p53, PEG-Prom driving wild-type p53 expression; CMV-mda-7, CMV-promoter driving mda-7/IL-24 expression; PEG-mda-7, PEG-Prom driving mda-7/IL-24 expression.
References
- 1.Bartelink, H., Schellens, J. H. & Verheij, M. (2002) Eur. J. Cancer 38, 216–222. [DOI] [PubMed] [Google Scholar]
- 2.Buchsbaum, D. J. & Curiel, D. T. (2001) Cancer Biother. Radiopharm. 16, 275–288. [DOI] [PubMed] [Google Scholar]
- 3.Lebedeva, I. V., Su, Z.-Z., Sarkar, D. & Fisher, P. B. (2003) Semin. Cancer Biol. 13, 169–178. [DOI] [PubMed] [Google Scholar]
- 4.Scanlon, K. J. (2004) Anticancer Res. 24, 501–504. [PubMed] [Google Scholar]
- 5.Babiss, L. E., Zimmer, S. G. & Fisher, P. B. (1985) Science 228, 1099–1101. [DOI] [PubMed] [Google Scholar]
- 6.Fisher, P. B., Weinstein, I. B., Eisenberg, D. & Ginsberg, H. S. (1978) Proc. Natl. Acad. Sci. USA 75, 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su, Z.-Z., Shi, Y. & Fisher, P. B. (1997) Proc. Natl. Acad. Sci. USA 94, 9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollander, M. C., Poola-Kella, S. & Fornace, A. J., Jr. (2003) Oncogene 22, 3827–3832. [DOI] [PubMed] [Google Scholar]
- 9.Su, Z.-Z., Emdad, L., Sarkar, D., Randolph, A., Valerie, K., Yacoub, A., Dent, P. & Fisher, P. B. (2005) Oncogene, in press. [DOI] [PubMed]
- 10.Su, Z.-Z., Goldstein, N. I., Jiang, H., Wang, M. N., Duigou, G. J., Young, C. S. & Fisher, P. B. (1999) Proc. Natl. Acad. Sci. USA 96, 15115–15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su, Z.-Z., Gopalkrishnan, R. V., Narayan, G., Dent, P. & Fisher, P. B. (2002) J. Cell Physiol. 192, 34–44. [DOI] [PubMed] [Google Scholar]
- 12.Emdad, L., Sarkar, D., Su, Z.-Z., Boukerche, H., Bar-Eli, M. & Fisher, P. B. (2005) J. Cell. Physiol. 202, 135–146. [DOI] [PubMed] [Google Scholar]
- 13.Su, Z.-Z., Shi, Y. & Fisher, P. B. (2000) Oncogene 19, 3411–3421. [DOI] [PubMed] [Google Scholar]
- 14.Su, Z.-Z., Shi, Y., Friedman, R., Qiao, L., McKinstry, R., Hinman, D., Dent, P. & Fisher, P. B. (2001) Nucleic Acids Res. 29, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebedeva, I. V., Sarkar, D., Su, Z.-Z., Kitada, S., Dent, P., Stein, C. A., Reed, J. C. & Fisher, P. B. (2003) Oncogene 22, 8758–8773. [DOI] [PubMed] [Google Scholar]
- 16.Bae, V. L., Jackson-Cook, C. K., Maygarden, S. J., Plymate, S. R., Chen, J. & Ware, J. L. (1998) Prostate 34, 275–282. [DOI] [PubMed] [Google Scholar]
- 17.Su, Z.-Z., Madireddi, M. T., Lin, J. J., Young, C. S., Kitada, S., Reed, J. C., Goldstein, N. I. & Fisher, P. B. (1998) Proc. Natl. Acad. Sci. USA 95, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su, Z.-Z., Lebedeva, I. V., Sarkar, D., Gopalkrishnan, R. V., Sauane, M., Sigmon, C., Yacoub, A., Valerie, K., Dent, P. & Fisher, P. B. (2003) Oncogene 22, 1164–1180. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, M., Rosenberg, E. & Valerie, K. (2003) Methods Mol. Biol. 234, 1–16. [DOI] [PubMed] [Google Scholar]
- 20.Lebedeva, I. V., Su, Z.-Z., Chang, Y., Kitada, S., Reed, J. C. & Fisher, P. B. (2002) Oncogene 21, 708–718. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar, D., Su, Z.-Z., Lebedeva, I. V., Sauane, M., Gopalkrishnan, R. V., Dent, P. & Fisher, P. B. (2002) BioTechniques 33, 30–39. [PubMed] [Google Scholar]
- 22.Fisher, P. B., Gopalkrishnan, R. V., Chada, S., Ramesh, R., Grimm, E. A., Rosengeld, M. R., Curiel, D. T. & Dent, P. (2003) Cancer Biol. Ther. 2, S23–S37. [PubMed] [Google Scholar]
- 23.Shirakawa, T., Hamada, K., Zhang, Z., Okada, H., Tagawa, M., Kamidono, S., Kawabata, M. & Gotoh, A. (2004) Clin. Cancer Res. 10, 4342–4348. [DOI] [PubMed] [Google Scholar]
- 24.Greenberger, S., Shaish, A., Varda-Bloom, N., Levanon, K., Breitbart, E., Goldberg, I., Barshack, I., Hodish, I., Yaacov, N., Bangio, L., et al. (2004) J. Clin. Invest. 113, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu, J., Kagawa, S., Takakura, M., Kyo, S., Inoue, M., Roth, J. A. & Fang, B. (2000) Cancer Res. 60, 5359–5364. [PubMed] [Google Scholar]
- 26.Irving, J., Wang, Z., Powell, S., O'Sullivan, C., Mok, M., Murphy, B., Cardoza, L., Lebkowski, J. S. & Majumdar, A. S. (2004) Cancer Gene Ther. 11, 174–185. [DOI] [PubMed] [Google Scholar]
- 27.Shay, J. W. & Bacchetti, S. (1997) Eur. J. Cancer 33, 787–791. [DOI] [PubMed] [Google Scholar]
- 28.de Launoit, Y., Baert, J. L., Chotteau, A., Monte, D., Defossez, P. A., Coutte, L., Pelczar, H. & Leenders, F. (1997) Biochem. Mol. Med. 61, 127–135. [DOI] [PubMed] [Google Scholar]
- 29.Eferl, R. & Wagner, E. F. (2003) Nat. Rev. Cancer 3, 859–868. [DOI] [PubMed] [Google Scholar]
- 30.Subbaramaiah, K., Norton, L., Gerald, W. & Dannenberg, A. J. (2002) J. Biol. Chem. 277, 18649–18657. [DOI] [PubMed] [Google Scholar]
- 31.Iguchi, A., Kitajima, I., Yamakuchi, M., Ueno, S., Aikou, T., Kubo, T., Matsushima, K., Mukaida, N. & Maruyama, I. (2000) Biochem. Biophys. Res. Commun. 279, 166–171. [DOI] [PubMed] [Google Scholar]
- 32.El-Tanani, M., Platt-Higgins, A., Rudland, P. S. & Campbell, F. C. (2004) J. Biol. Chem. 279, 20794–20806. [DOI] [PubMed] [Google Scholar]
- 33.Shepherd, T. G., Kockeritz, L., Szrajber, M. R., Muller, W. J. & Hassell, J. A. (2001) Curr. Biol. 11, 1739–1748. [DOI] [PubMed] [Google Scholar]
- 34.Davidson, B., Goldberg, I., Gotlieb, W. H., Kopolovic, J., Ben-Baruch, G. & Reich, R. (2003) Clin. Cancer Res. 9, 1412–1419. [PubMed] [Google Scholar]
- 35.Crawford, H. C., Fingleton, B., Gustavson, M. D., Kurpios, N., Wagenaar, R. A., Hassell, J. A. & Matrisian, L. M. (2001) Mol. Cell. Biol. 21, 1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiroumi, H., Dosaka-Akita, H., Yoshida, K., Shindoh, M., Ohbuchi, T., Fujinaga, K. & Nishimura, M. (2001) Int. J. Cancer 93, 786–791. [DOI] [PubMed] [Google Scholar]
- 37.Hida, K., Shindoh, M., Yoshida, K., Kudoh, A., Furaoka, K., Kohgo, T., Fujinaga, K. & Totsuka, Y. (1997) Oral Oncol. 33, 426–430. [DOI] [PubMed] [Google Scholar]
- 38.Gavrilov, D., Kenzior, O., Evans, M., Calaluce, R. & Folk, W. R. (2001) Eur. J. Cancer 37, 1033–1040. [DOI] [PubMed] [Google Scholar]
- 39.Payson, R. A., Chotani, M. A. & Chiu, I. M. (1998) J. Steroid Biochem. Mol. Biol. 66, 93–103. [DOI] [PubMed] [Google Scholar]
- 40.Park, J. S., Qiao, L., Su, Z.-Z., Hinman, D., Willoughby, K., McKinstry, R., Yacoub, A., Duigou, G. J., Young, C. S., Grant, S., et al. (2001) Oncogene. 20, 3266–3280. [DOI] [PubMed] [Google Scholar]
- 41.Adjei, A. A. (2001) J. Natl. Cancer Inst. 93, 1062–1074. [DOI] [PubMed] [Google Scholar]