Abstract
Members of the kinesin superfamily are microtubule-based motor proteins that transport molecules/organelles along microtubules. We have identified similar internal motor kinesins, Kinesin-13A, from the cotton Gossypium hirsutum and Arabidopsis thaliana. Their motor domains share high degree of similarity with those of internal motor kinesins of animals and protists in the MCAK/Kinesin13 subfamily. However, no significant sequence similarities were detected in sequences outside the motor domain. In Arabidopsis plants carrying the T-DNA knockout kinesin-13a-1 and kinesin-13a-2 mutations at the Kinesin-13A locus, >70% leaf trichomes had four branches, whereas wild-type trichomes had three. Immunofluorescent results showed that AtKinesin-13A and GhKinesin-13A localized to entire Golgi stacks. In both wild-type and kinesin-13a mutant cells, the Golgi stacks were frequently associated with microtubules and with actin microfilaments. Aggregation/clustering of Golgi stacks was often observed in the kinesin-13a mutant trichomes and other epidermal cells. This suggested that the distribution of the Golgi apparatus in cell cortex might require microtubules and Kinesin-13A, and the organization of Golgi stacks could play a regulatory role in trichome morphogenesis. Our results also indicate that plant kinesins in the MCAK/Kinesin-13 subfamily have evolved to take on different tasks than their animal counterparts.
INTRODUCTION
Cytoskeletal networks often serve as tracks along which molecules and organelles are transported by cytoskeletal motor proteins toward different intracellular destinations (Vale, 2003). Members of the kinesin superfamily, or kinesins, are microtubule-based motor proteins found in all eukaryotic organisms that contain a conserved motor domain responsible for ATP hydrolysis and microtubule binding. In the model plant Arabidopsis thaliana, there are at least 61 genes encoding divergent kinesins (Reddy and Day, 2001). Although they have a high degree of similarity in their motor domains, the nonmotor sequences of these Arabidopsis kinesins do not have common features. Moreover, the motor domains are located either at the N terminus or the C terminus or in the middle of these 61 polypeptides (Reddy and Day, 2001). Some of the Arabidopsis kinesins are believed to have diverse functions as motors to transport their cargoes of organelles and molecules. Others may contribute to the organization of microtubules, possibly by affecting the stability of microtubules or by rearranging individual microtubules. However, the roles of most Arabidopsis kinesins have not been defined. The specific cargoes that they transport and the cellular activities in which they participate are also unknown.
Plant cell enlargement is typically the result of organized secretion of Golgi vesicles, organized deposition of cell wall materials (mainly cellulose microfibrils), and the modification of the existing cell wall. In interphase plant cells, microtubules are often organized in a parallel cortical array. Although the cortical microtubule array has been considered to serve as a template for the deposition of cellulose microfibrils, this hypothesis has been challenged by a number of recent studies (Baskin, 2001; Sugimoto et al., 2003). The cortical array may provide tracks along which molecules and vesicles are transported. Growing evidence suggests that microtubule-based motility, driven by kinesins, plays active roles in plant cell enlargement (Smith, 2003). For example, mutations in the FRA1 kinesin gene cause aberrant cellulose microfibril organization in the secondary cell wall without affecting cortical microtubule organization in fiber cells of Arabidopsis (Zhong et al., 2002). Consequently, mutant cells fail to elongate properly, and mutant plants have a retarded growth phenotype. Because cortical microtubules have mixed polarities (Shaw et al., 2003; Tian et al., 2004), how a unidirectional transport powered by the FRA1 kinesin contributes to cell enlargement is still a mystery.
The leaf trichome is an extreme example of anisotropic cell enlargement. In Arabidopsis, a mature trichome is a single polyploid cell with three sharp branches (Schnittger and Hülskamp, 2002). Pharmacological studies have indicated that trichome morphogenesis requires microtubules (Mathur et al., 1999; Szymanski et al., 1999; Mathur and Chua, 2000; Qiu et al., 2002). Genetic studies have provided further evidence for the roles of microtubules and their associated factors in trichome branching. For example, the absence of the KCBP/ZWI calmodulin-binding kinesin results in shortened trichome stalk and reduced branches (Oppenheimer et al., 1997). A mutation in the AtKTN1 gene encoding a microtubule-severing protein causes trichomes to have only one branching point (Burk et al., 2001). Although many tubulin/microtubule-interacting proteins are critical for trichome morphogenesis (Schnittger and Hülskamp, 2002; Szymanski, 2002), the molecular mechanisms by which microtubules affect anisotropic cell enlargement are largely unknown.
Cotton fibers, which also are referred as trichomes, are single cells that elongate from the epidermis of the ovule. The unidirectional elongation process of cotton fibers can last for up to 30 d to render cells several centimeters in length. Immediately after the elongation process is the thickening of the fiber cell wall by cellulose microfibrils. In the process of fiber development, cortical microtubules undergo drastic reorganization from a transverse orientation to a steeply pitched array (Seagull, 1992). Studies using microtubule drugs have indicated that cortical microtubules are required for the organized deposition of cellulose microfibrils in cotton fibers (Seagull, 1990). However, it remains unclear whether different arrays of cortical microtubules imply directional changes of active transports by kinesins in the cotton fiber.
Some plant kinesins may be exclusively required for microtubule-based motility for cell enlargement during interphase. Because cotton fibers can be easily separated from the rest of the ovule, they are a homogeneous source for identification of kinesin genes robustly expressed during fiber elongation. Such kinesins could play important roles in general cell enlargement during interphase cell growth. We previously reported that the cotton Gossypium hirsutum homolog of the kinesin KCBP decorates cortical microtubules during cotton fiber development (Preuss et al., 2003). In this article, we report the initial characterization of a new cotton kinesin, GhKinesin-13A, and its Arabidopsis homolog, which are divergent from previously identified plant kinesins. Our results demonstrate that the similar Kinesin-13A proteins are associated with the Golgi apparatus. In addition, we found that in Arabidopsis Kinesin-13A plays a role in trichome morphogenesis. The results suggest that microtubules, kinesin(s), and the Golgi apparatus work in concert to regulate cell morphogenesis in flowering plants.
MATERIALS AND METHODS
Plant Materials
Cotton bolls were collected from cotton Gossypium hirsutum`Coker 130′ plants grown under greenhouse conditions.
The A. thaliana plants used in this study are the ecotype Columbia. The kinesin-13a-1 mutant plant was generated at Salk Institute as the SALK_047048 line (La Jolla, CA) and obtained through the Arabidopsis Biological Resource Center (Columbus, OH). The kinesin13a-2 mutant plant was obtained from the Syngenta Arabidopsis Insertion Library (La Jolla, CA) as the SAIL761 B04 line. The STtmd-GFP line was made available by the Hawes laboratory (Oxford Brooks University, Oxford, United Kingdom).
Cloning of the GhKinesin-13A cDNA
A cotton fiber EST sequence showed a high degree of sequence homology to the A. thaliana kinesin gene At3g16630 in the on-line Clemson cotton database (http://www.genome.clemson.edu/projects/cotton/est/). PCR primers Gh-KIM1-F (5′-CTACGAATTCAAATGAGGA(A/G)TCTTCAAGG-3′) and Gh-KIM1-R (5′-GATGTCTAGAAAGCATC(C/T)(G/T)TATTACTTGG-3′), were designed according to the expressed sequence tag (EST) sequence. A 510-base pair fragment was amplified by polymerase chain reaction (PCR) from a 21-DPA cotton (`Acala SJ-2') fiber-specific Lambda Uni-Zap (Stratagene, LaJolla, CA) cDNA library (Pear et al., 1996). The PCR fragment was gel purified and used as a probe to screen the cDNA library by using a digoxigenin-labeling method according to manufacturer's procedure (Roche Diagnostics, Indianapolis, IN). Recovered cDNA clones were sequenced at the DBS sequencing facility (Davis, CA). Sequence assembly and analysis were performed with the GCG software package (GCG, Madison, WI).
A full-length cDNA clone of the At3g16630 was obtained from the Kazusa DNA Research Institute (Chiba, Japan) according to the EST sequence in the GenBank (accession no. AV529483). The cDNA sequence matches perfectly with the one deposited by the Riken Arabidopsis Full-Length cDNA project (GenBank accession no. AY056129).
Phylogenetic Analysis
The phylogenetic analysis was carried out by comparing the amino acid sequences of the catalytic core and the neck motifs only. The sequences were from the following kinesins with the GenBank accession numbers in parentheses: AtKinesin-13A, AtNACK1/AtHIK (AB081599), AtNACK2/AtTES (AB088121), AtPAKRP1/AtKinesin-12A (AF193767), AtKATA/AtKinesin-14A (D11371), AtKATD (AF080249), AtKCBP (L40358), AtFRA1/AtKinesin-4 (AY158083) from A. thaliana; GhKinesin-13A from G. hirsutum; NtTKRP125/NtKinesin-5A (D83711) from Nicotiana tabacum; CgMCAK/CgKinesin-13 (U11790) from Cricetulus griseus; DmKHC/DmKinesin-1 (M24441) and DmNCD/DmKinesin-14 (X52814) from Drosophila melanogaster; SpKRP95/SpKinesin-2A (U00996) from Strongylocentrotus purpuratus; XKLP2/XlKinesin-12 (X94082) from Xenopus laevis; GgChrkin/GgKinesin-4 (U18309) from Gallus gallus; HsMKLP1/HsKinesin-6 (X67155) from Homo sapiens; and An-BIMC/AnKinesin-5 (M69021) from Aspergillus nidulans. Phylogenetic trees were built in PAUP by using maximum parsimony. The phylogenetic tree shown in Figure 1B was obtained by using a heuristic search method with random stepwise addition of sequences. Boostrap support values were obtained from 500 replicates, and the numerical values referred to the percentage of trees containing a specific branch. Only values >50% were shown in the tree.
Figure 1.
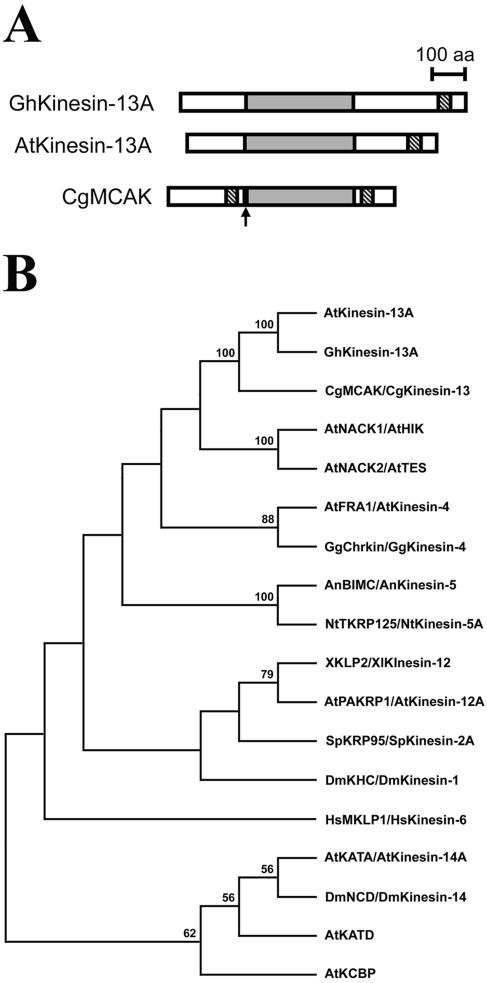
Structural analysis of the internal motor kinesins GhKinesin-13A and AtKinesin-13A. (A) Schematic structures of GhKinesin-13A, AtKinesin-13A, and CgMCAK. The motor domain is shaded, and the coiled-coil domain is shown in hatched lines. The position of the MCAK neck was pointed by an arrow. (B) Phylogenetic analysis of the motor domains of GhKinesin-13A/AtKinesin-13A and other representative kinesins. Numerical values referred to the percentage of trees containing a specific branch in 500 replications. Only values >50% were shown.
Isolation of T-DNA Insertional Mutants
Homozygous and heterozygous T-DNA insertional mutant plants were identified according to a PCR-based method as described previously (Krysan et al., 1999; McElver et al., 2001). The Salk_047048 line containing the kinesin-13a-1 mutation was detected by the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress). The gene-specific primer KIM1F1 (5′-GTGGAGGCTCAAAGACATCTTC-3′) and the T-DNA border-specific primer LBa1 (5′ TGGTTCACGTAGTGGGCCATCG-3′) were used to detect plants that carried the T-DNA insertion. The gene-specific primers KIM1F1′ (5′-GCTGTTCGATCTTCTCAGTG-3′) and KIM1R1 (5′-TGCCCACAAGTCCAACAGAG-3′) were used for the homozygosity test of the mutant allele. The SAIL761 B04 line carrying the kinesin-13a-2 mutation was created by the Syngenta Terrey Mesa Research Institute (La Jolla, CA). The T-DNA insertion was detected by the primer KIM1F2 (5′-ACCTCGATGCACTGCTTGAG-3′) and the T-DNA border-specific primer LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The gene specific primers KIM1F2 and KIM1R2 (5′-GCCCAAATTCACAAACATGAAGG-3′) were used for the homozygosity test.
Standard genetic crosses were carried out for the dominant/recessive test and for the allelic test. Segregations of mutations were analyzed in progeny of crosses.
Genetic Suppression (Complementation) of the kinesin-13a-1 Mutation
The full-length AtKinesin-13A–coding sequence was amplified from the cDNA clone mentioned above by using two primers: KIM1R4 (5′-GACAAGCTTCTACCGAGGAACTCTCTTACG-3′) and KIM1F4 (5′-CTAAGGTACCATGGGCGGCCAAATGCAGCAAAAC-3′). The PCR product was digested with KpnI and HindIII, and subsequently cloned into the pART7 vector (Gleave, 1992) at the corresponding sites. The expression cassette was then digested with NotI, and then cloned into the binary vector pMLBART at the NotI site. This construct was transformed into a homozygous kinesin-13a-1 plant by the standard flower dipping method. Transgenic lines were selected by resistance to the herbicide BASTA.
Production of Fusion Proteins
An AtKinesin-13A cDNA fragment encoding amino acids 523–794 was released from the EST clone 150N20 obtained from the Arabidopsis Biological Resource Center, by SalI and NotI. This fragment was ligated into the glutathione S-transferase (GST) vector pGEX4T1 (Amersham Biosciences, Piscataway, NJ) digested with SalI and NotI. The resulting GST-AtKinesin-13A fusion protein was expressed in BL21 (DE3) pLysS cells (Novagen, Madison, WI) and purified with glutathione agarose matrix (Pierce Endogen, Rockford, IL) according to manufacturer's instructions.
The cDNA fragment encoding amino acids 1–204 of GhKinesin-13A was amplified using the primers KIM1F3 (5′-GGCCGAATTCAGCAAAGCAATGCTG-3′) and KIM1R3 (5′-TCCCAAGCTTTCATCTCTTTTTCATTGG-3′) with the Vent DNA polymerase (New England Biolabs, Beverly, MA). After digested with EcoRI and HindIII, the fragment was cloned into the pGEX-KG vector (Guan and Dixon, 1991) at the EcoRI and HindIII sites. The GST-GhKinesin-13A fusion protein was expressed and purified as described above. A construct for a 6 × His-tagged GhKinesin-13A fusion protein was made by subcloning the BamHI/HindIII fragment from the pGEX-KG-GhKinesin-13A construct, and ligating it into the pQE-30 vector at the BamHI and HindIII sites (QIAGEN, Valencia, CA). The 6 × His-GhKinesin-13A fusion protein was expressed in M15pREP4 cells and purified using the Talon resin (BD Biosciences Clontech, Palo Alto, CA).
Antibody Production and Purification
The GST-AtKinesin-13A fusion protein was used as the antigen for raising rabbit anti-AtKinesin-13A antibodies at a commercial facility (Antibodies, Davis, CA). Rat anti-GhKinesin-13A antibodies were raised at the Comparative Pathology Laboratory on the University of California, Davis, campus.
Anti-AtKinesin-13A antibodies were purified by affinity chromatography. Bacterially purified GST, GST-AtKinesin-13A, and 6 × His-GhKinesin-13A proteins were separately conjugated to agarose matrix by using the Amino-Link Plus coupling gel (Pierce Endogen) according to manufacturer's instructions. To purify AtKinesin-13A–specific antibodies, anti-AtKinesin-13A serum was first passed through the GST column to remove anti-GST antibodies. The flow-through was then loaded onto the GST-AtKinesin-13A column. Anti-GhKinesin-13A serum was subject to purification using the 6 × His-GhKinesin-13A-agarose column. In both cases, specific antibodies were eluted using 100 mM glycine (pH 2.5), and neutralized immediately with 1 M Tris-HCl (pH 8.0). Qualities of purified antibodies were first monitored by immunoblotting against the corresponding fusion proteins, and then tested with the immunofluorescent localization.
Protein Extraction and Immunoblotting
Arabidopsis seeds were germinated at room temperature in 1/2 Murashige and Skoog (MS) liquid medium (Invitrogen, Carlsbad, CA) supplemented with 1% sucrose by shaking at 200 rpm for 3 d in dark. Approximately 1-g seedlings were ground in liquid nitrogen with a mortar and a pestle, and the powder was mixed with 1 ml of sample buffer containing 6% (wt/vol) SDS, 0.12 M dithiothreitol, 20%(vol/vol) glycerol, and 4 M urea in 100 mM Tris-HCl, pH 6.8.
Five days postanthesis, cotton bolls were harvested from the cotton plants, and fibers were excised using a razor blade and frozen in liquid nitrogen immediately. After grinding with a mortar and pestle in liquid nitrogen, powders of cotton fibers were mixed with the above-mentioned sample buffer. Protein extracts were separated in a 7.5% (wt/vol) SDS-PAGE and processed for immunoblotting analysis as described previously (Lee et al., 2001).
Immunolocalization
Immunolocalizations in Arabidopsis root tip cells of wild-type and kinesin-13a mutants were carried out as described previously (Lee et al., 2001). Affinity purified anti-AtKinesin-13A antibodies and the DM1A anti-α-tubulin antibodies were used followed by the secondary antibodies of fluorescein isothiocyanate-conjugated goat anti-rabbit and Texas Red-conjugated goat antimouse antibodies (Sigma-Aldrich, St. Louis, MO). A monoclonal anti-SABP antibody (Chen et al., 1993) was used to detect peroxisomes.
Test of colocalization of AtKinesin-13A and the Golgi apparatus was carried out in mesophyll protoplasts of the STtmd-GFP plants. Briefly, leaves of the STtmd-GFP plants were floated on the cell wall-digesting solution containing 1.0% (wt/vol) Cellulase-RS (Yakult Pharmaceutical, Tokyo, Japan) and 0.1% (wt/vol) Macerozyme R-10 (Yakult Pharmaceutical) in 0.6 M mannitol, pH 5.3. Protoplast were collected and processed for immunolocalization as described previously (Preuss et al., 2003). The primary antibodies were rabbit anti-AtKinesin-13A and monoclonal mouse anti-green fluorescent protein (GFP) antibodies (Covance Research, Berkeley, CA).
Immunolocalization in leaf segments was carried out according to a published protocol (Cleary and Smith, 1998). Microtubules were labeled with DM1A, and Golgi stacks were detected by an antibody against α-mannosidase (Preuss et al., 2004). Immunolocalization of GhKinesin-13A and α-mannosidase in cotton cytoplasts was carried out as described previously (Preuss et al., 2003).
Establishing GFP-tagged Transgenic Lines
To create a construct expressing the GFP-tubulin fusion, GFP-coding sequence was first amplified by PCR with a forward primer: 5′-CCAGCTCGAGCATGAGTAAAGGAGAAG-3′, and a reverse primer: 5′-ATATGGTACCTTCGCCCTCTCCGCCATGTGTAATCCCAGC-3′. The amplified fragment was digested with XhoI and KpnI, and cloned into the pART7 vector to give rise to the pART7-GFP plasmid. Full-length cDNA sequence of the Arabidopsis α-6 tubulin (Kopczak et al., 1992) was amplified from the cDNA clone 209P19 available from the ABRC using the forward primer: 5′-TGCAGGTACCATGAGAGAGTGCATTTCG-3′, and the reverse primer: 5′-GCGCTCTAGATTAGTATTCCTCTCCTTC-3′. The amplified fragment was digested with KpnI and XbaI, and ligated into pART7-GFP at the corresponding sites. The GFP-tubulin expression cassette was then digested by NotI, and ligated into pMLBart at the NotI site.
A N-acetylglucosaminyl transferase I (Nag)-GFP construct was made to label Golgi stacks. Briefly, the DNA fragment encoding the transmembrane-stem region of Arabidopsis Nag was amplified from a Nag-RFP construct (Dixit and Cyr, 2002) by PCR with the forward primer 5′-AGTCGACATGGCGAGGATCTCGTGTG-3′ and the reverse primer 5′-CTGGATCTCGGAGATAAGTTCTTCGTCCTGG-3′. The GFP-coding sequence also was amplified by PCR with the forward primer 5′-ATCTCCGAGATCCAGAGTAAAGGAGAAGAAC-3′ and the reverse primer: 5′-GGATCCTATTTGTATAGTTCATCCATG-3′. The Nag-GFP fusion was generated using two-step recombinant PCR, digested with SalI and BamHI, and cloned into pART7 at the XhoI and BamHI sites. The Nag-GFP expression cassette was then digested by NotI and ligated into pMLBart at the NotI site.
GFP fusions were transformed into wild-type and kinesin-13a mutant plants by standard flower dipping methods. Transformants were selected by applying the herbicide BASTA.
Transient Expression by Particle Bombardment
The plasmid pBI221-GFP-mTalin (Fu et al., 2002) was used to label actin microfilaments in live cells. The procedures for the particle bombardment-mediated transient expression in Arabidopsis leaves were carried out according to a published procedure (Fu et al., 2002).
Electron Microscopy
For scanning electron microscopy, leaves were dissected away from wild-type and mutant A. thaliana plants and placed on sample holder via the Tissue-Tek adhesive (Electron Microscopy Sciences, Fort Washington, PA). After the samples were frozen in liquid nitrogen, they were coated with Au and scanned with a S3500N scanning electron microscope equipped with a cryo stage (Hitachi, Tokyo, Japan).
For transmission microscopy, roots of seedlings germinated on 1/2 MS agar medium were processed by a published freeze substitution method using the HPM 010 HPF high-pressure freezer (Balzers, Liechtenstein, Austria) (Ding et al., 1991). OsO4, uranyl acetate, and lead citrate were used to enhance the contrast of subcellular structures. Sections were obtained by using the Ultra-T ultramicrotome (Leica, Bannockburn, IL). Sections were observed under a Philips 410 LS electron microscope (Philips, Hillsboro, OR).
Fluorescence Microscopy and Image Processing
Samples were observed on an Eclipse E600 microscope equipped with epi-fluorescence optics (Nikon, Melville, NY). Images were acquired with a charge-couple device camera (Hamamatsu Photonics, Tokyo, Japan) by using the ImageProPlus 4.0 software package (Media Cybernetics, Silver Spring, MD). Confocal images were collected with a TCS-SP laser scanning confocal microscope (Leica) by using argon and krypton lasers. All images presented in this article were assembled in Photoshop 7.0 (Adobe Systems, San Jose, CA).
The distribution pattern of anti-Golgi immunofluorescence intensity was analyzed quantitatively by measuring 100 randomly chosen 6.4 × 6.4-μm areas in Figure 11, A and D. Analysis of pixel count variance was performed using R 2.0 (http://www.R-project.org) (R Development Core Team, 2004). By using mean-centered data, the variance of Figure 11A (wild-type) pixel counts was 254.7, whereas the variance of Figure 11D (kinesin-13a-1) pixel counts was 755.0; the variance ratio Fs was 2.96. This suggested that the mutant count data were further dispersed from its mean than wild type. Because the data did not follow a normal, Poisson, or binomial distribution, a bootstrapping approach to determine whether the observed difference in variances was significant. Using a null hypothesis that the mutant and wild-type count data came from the same population, we pooled the observations. A null distribution for the variance ratio F was created by taking 100,000 pairs of n = 100 random samples, with replacement, from the pooled observations and calculating F for each pair. For each pair, F was calculated using the smaller variance in the denominator. The observed Fs of 2.96 was larger than all but 1.523% of the randomly generated F values.
Figure 11.
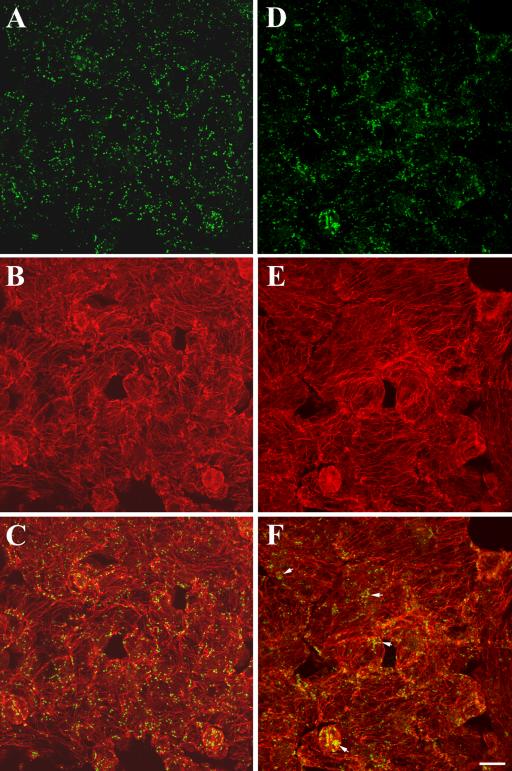
Distribution of Golgi stacks in leaf epidermal cells of wild-type and the kinesin-13a-1 mutant. (A–C) Wild-type cells. (A) Golgi stacks revealed by anti-α-mannosidase were evenly distributed in epidermal cells. (B) Cortical microtubules stained by anti-α-tubulin. (C) Merged image showing both Golgi stacks and microtubules. (D–F) kinesin-13a-1 mutant cells. (D) Golgi stacks revealed by anti-α-mannosidase. Although many Golgi stacks were still dispersed, in a number of places, Golgi stacks seemed to be more aggregated than in wild-type cells. (E) Similar cortical microtubule network was observed as in wild-type cells. (F) Merged image. Note aggregation of Golgi stacks was obvious in guard cells and other epidermal cells (arrows). Bar, 10 μm.
RESULTS
Determination of cDNA Sequences of Kinesin-13A from Cotton and Arabidopsis
In an analysis of kinesins that were abundantly expressed in cotton fiber growth, a 3650-base pair cDNA (GenBank accession no. AY 283056) was isolated that encoded a kinesin of 909 amino acids, with a calculated molecular mass of 100,818 Da and an isoelectric point of 3.59. A similar kinesin is encoded by the At3g16630 gene in Arabidopsis. A full-length cDNA sequence of At3g16630 was determined with an EST clone from the Kazusa DNA Research Institute (GenBank accession no. AV528755). This cDNA sequence encodes a polypeptide of 794 amino acids, with a calculated molecular mass of 89,084 Da and an isoelectric point of 5.07. Because these two kinesins have their motor domains located in the middle of the polypeptides, they were named as GhKinesin-13A/AtKinesin-13A for G. hirsutum/A. thaliana Kinesin-13A (Figure 1A). This name is adopted based on a recent agreement on the kinesin nomenclature (Lawrence etal., 2004). GhKinesin-13A and AtKinesin-13A share 76% sequence identity across the entire polypeptides (Figure 2A). The motor domains of amino acids 207–549 and amino acids 185–533 for GhKinesin-13A and AtKinesin-13A, respectively, have 88% identity.
Figure 2.
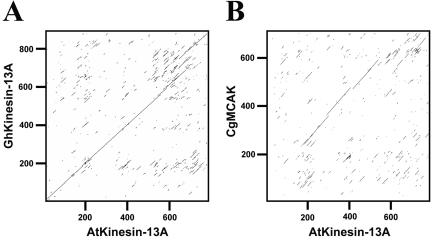
Sequence comparison of AtKinesin-13A, GhKinesin-13A, and CgMCAK. (A) Comparison of amino acid sequences of GhKinesin-13A and AtKinesin-13A was presented in dot matrix plot. Note that significant homology was seen from the N terminus to the C terminus. (B) Comparison of amino acid sequences of AtKinesin-13A and CgMCAK was presented in dot matrix plot. Note that the only significant homology was seen in the central region.
Analysis of Kinesin-13A Sequences
To reveal how these polypeptides relate to other kinesins, we first analyzed their highly conserved motor domains. A phylogenetic analysis of representative members of the kinesin superfamily showed that GhKinesin-13A and AtKinesin-13A were placed into the MCAK/Kinesin-13 subfamily (Figure 1B). To date, all nonplant kinesins in the MCAK/Kinesin-13 subfamily have internal motor domains, and outside the motor domain they are closely related to each other (Ovechkina and Wordeman, 2003). However, sequence similarity between the plant Kinesin-13A and animal MCAK/Kinesin-13 was only limited to their motor domains (Figure 2B).
According to the Lupas algorithm (Lupas et al., 1991), GhKinesin-13A and AtKinesin-13A have predicted coiled-coil regions of amino acid 820–859 and 703–742, respectively, toward their C termini (Figure 1A). We did not detect any other structural or functional motif/domain in Kinesin-13A, nor did we detect the signature neck sequence, which is critical for the microtubule depolymerization activities for animal MCAK/Kinesin-13 (Ovechkina et al., 2002).
AtKinesin-13A Is Required for Normal Trichome Development
To understand the function of Kinesin-13A, we isolated T-DNA insertional mutations at the Kinesin-13A locus in Arabidopsis. Two independent T-DNA insertional mutations, kinesin-13a-1 and kinesin-13a-2, have been confirmed at intron 10 and exon 14 (Figure 3). Compared with wild-type plants, homozygous kinesin-13a-1 and kinesin-13a-2 plants did not exhibit obvious growth phenotype in seed germination, root and shoot formation, and flowering (our unpublished data).
Figure 3.
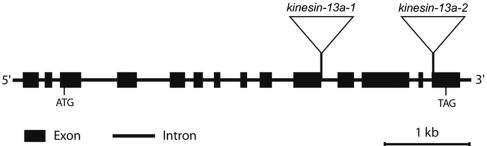
kinesin-13a-1 and kinesin-13a-2 mutations. The bar graph showed the organization of introns and exons of the AtKinesin-13A gene. kinesin-13a-1 had the T-DNA inserted at the intron 10, and kinesin-13a-2 had the insertion at the exon 14.
The mutant leaves, however, had abnormal trichomes (Figure 4). In wild-type plants (ecotype Columbia), trichomes normally had two branching points with three branches (Figure 4, A and C). In the kinesin-13a-1 homozygous mutant, trichomes predominantly had four to five branches (Figure 4, B and D). Similar trichome phenotypes were observed in the kinesin-13a-2 homozygous mutant (our unpublished data). To determine how such abnormal trichomes were formed in the mutants, we analyzed trichomes at different developmental stages. After initial cell elongation (Figure 5, A and B), typical wild-type trichomes form three branches via the primary and secondary branching events (Figure 5, C and D) (Hülskamp et al., 1994; Folkers et al., 1997). In kinesin-13a-1 mutants, both these branching events were clearly present (Figure 5, E–G). However, at least one and occasionally two additional branches were formed after the second branching event (Figure 5H).
Figure 4.
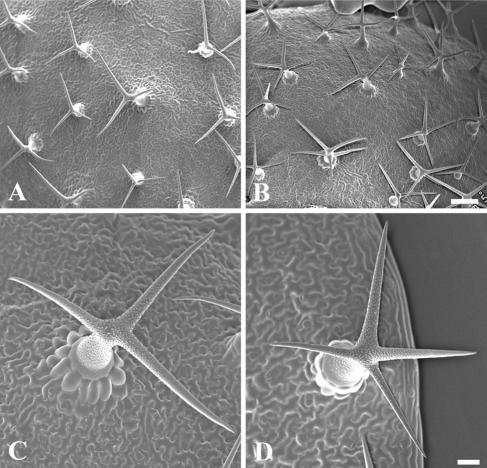
Trichome morphology in wild-type and the kinesin-13a-1 mutant. Scanning electron microscopic images of wild type (A and C) and kinesin-13a-1 mutant (B and D) trichomes are shown. (A and B) Most if not all wild-type trichomes bear three branches with two branching points, and most kinesin-13a-1 trichomes bear four branches, and occasionally five-branch trichomes can be seen. Bar, 200 μm. (C and D) High-magnification views of a wild-type trichome (C) and a kinesin-13a-1 one (D). Bar, 20 μm.
Figure 5.
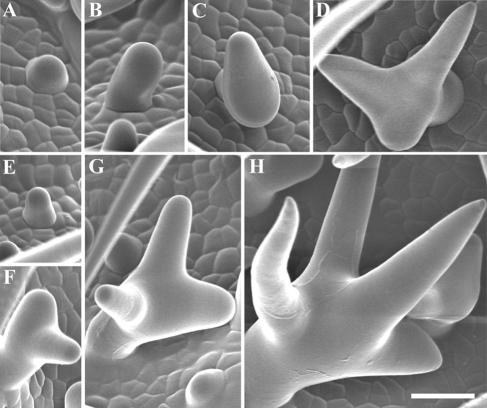
Developmental stages of trichomes in wild-type and kinesin-13a-1 mutant. (A–D) In wild type, trichomes were initiated on the epidermis (A), followed by elongation (B) and the first branching event (C), and then with an additional branching event at the secondary branching point (D). (E–(H) In kinesin-13a-1 mutant, the first three stages of trichome development were all observed (E–G). In addition, a new branch usually occurred much later when the trichome cells were much larger (H). Bar, 40 μm.
We quantified branching patterns in wild-type and kinesin-13a mutants (Table 1). Five rosette leaves were taken from each plant. The wild-type trichomes predominantly had three branches (93.05%). Although occasionally four branches were found, the number was minimal. In both kinesin-13a-1 and kinesin-13a-2 mutants, 74.01 and 81.21% trichomes had four branches, respectively. Some of the mutant trichomes even had five branches (Table 1). Therefore, we concluded that the morphology of mutant trichomes was significantly different from that of the wild type.
Table 1.
Percentages of trichomes with different branches from mutant and wild-type leaves
| No. of branches on mutant and wild-type trichome
|
||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Totala | |
| kinesin-13a-1 | 0 | 0.3 | 24.51 | 74.01 | 1.2 | 608 |
| kinesin-13a-2 | 0 | 0 | 16.30 | 81.21 | 2.5 | 583 |
| Wild type | 0 | 2.89 | 93.05 | 4.05 | 0 | 518 |
| kinesin-13a-1 + KIM1 cDNA | 0 | 2.09 | 94.38 | 3.53 | 0 | 671 |
Data represent percentage of trichomes having the indicated number of branches.
Total number of trichome counted on fifth or sixth rosette leaves of five plants
Segregation analysis of progeny from a cross between homozygous kinesin-13a-1 mutant and a wild-type Columbia plant also was performed. Our results indicated that the kinesin-13a-1 mutation was recessive, and the trichome branching phenotype was mapped to a single locus (n = 57, p > 0.9). Furthermore, genetic complementation analysis of kinesin-13a-1 and kinesin-13a-2 showed that these two mutations are allelic. The results collectively suggested that defects in trichome morphology were caused by the inactivation of the Kinesin-13A gene.
To further confirm that loss of AtKinesin-13A caused the trichome defects, we carried out genetic suppression (complementation) experiments with the AtKinesin-13A cDNA. Homozygous kinesin-13a-1 plants were transformed with AtKinesin-13A cDNA under the control of the 35S promoter. T1 transformants had majority of trichomes with restored three-branch morphology, and the number of such trichome matched that in wild type (Table 1). Together, we concluded that mutations at the Kinesin-13A locus caused defects in trichome morphology.
Kinesin-13A Localization in Arabidopsis and Cotton Cells
To understand how Kinesin-13A affects cell growth, we determined its intracellular localization. Affinity-purified anti-AtKinesin-13A antibodies were used to detect AtKinesin-13A in Arabidopsis. The antibodies detected a band with ∼110-kDa by immunoblotting with wild-type protein extracts (Figure 6). When protein extracts of homozygous kinesin-13a-1 and kinesin-13a-2 were probed with identical antibodies under identical conditions, no band was detected in the region of the similar molecular mass (Figure 6). Two weaker bands at the 90–100-kDa region also were detected in wild-type extract, but not in either mutant extract (Figure 6). Thus, they were most likely the degradation products of AtKinesin-13A. An anti-α-tubulin antibody was used in a positive control experiment, and the α-tubulin band was revealed in all samples (Figure 6). The results suggested that the antibodies specifically recognized AtKinesin-13A as a 110-kDa band in the wild-type extract.
Figure 6.
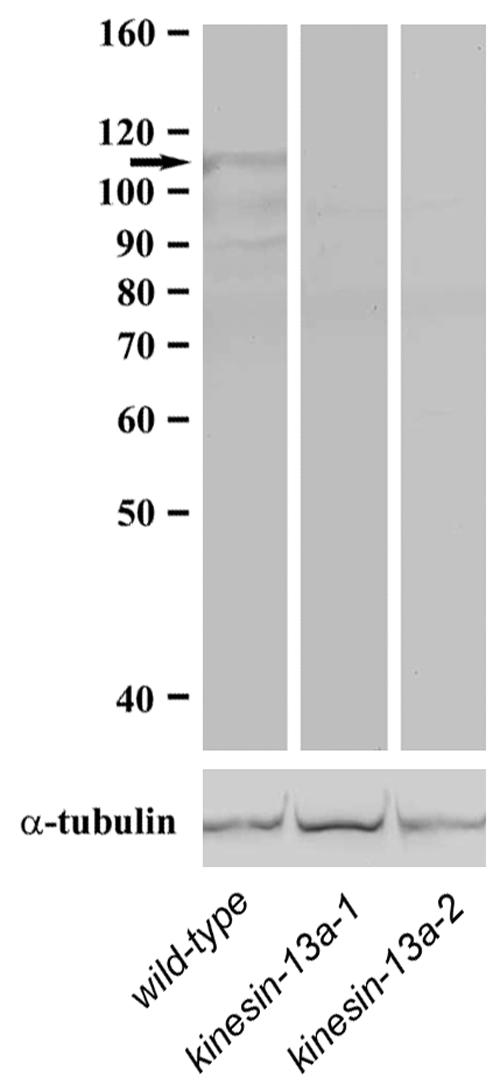
Immunoblotting with Anti-AtKinesin-13A antibodies. Protein extracts from the wild-type seedlings, and kinesin-13a-1 and kinesin-13a-2 mutant seedlings were probed with affinity purified anti-AtKinesin-13A antibodies (top), and the DM1A anti-α-tubulin antibody (bottom). Note the 110-kDa AtKinesin-13A band (arrow) shown with the wild-type extract was missing in the mutants, whereas the tubulin band persisted. Molecular weight markers were shown on the left.
Based on more than 60 ESTs, AtKinesin-13A RNA seems to be present in young and mature seedlings, and leaves and roots. Therefore, we initially localized AtKinesin-13A in root meristematic cells by immunofluorescence. Figure 7, A and B, show that anti-AtKinesin-13A antibodies detected particles smaller than 1 μm in the cytoplasm. The same antibodies did not react with particles in the cytoplasm of kinesin-13a-1 cells (Figure 7, C and D), demonstrating the specificity of the antibodies. AtKinesin-13A might be specifically associated with an organelle.
Figure 7.
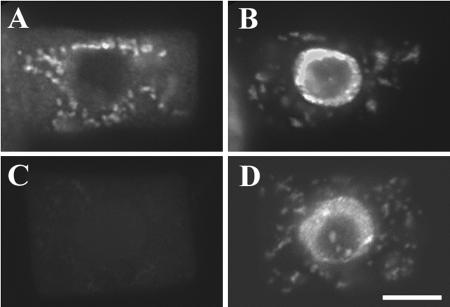
AtKinesin-13A localization in Arabidopsis root cells. (A and B) A wild-type root cell was probed with affinity-purified anti-AtKinesin-13A antibodies. Abundant organelles were labeled (A). The same cell was labeled with DAPI to show its nucleus (B). (C) and (D) An kinesin-13a-1 mutant cell did not reveal noticeable signal with AtKinesin-13A antibodies (C), whereas its nucleus was shown by 4,6-diamidino-2-phenylindole (D). Bar, 5 μm.
Antibodies also were prepared against GhKinesin-13A. Affinity-purified anti-GhKinesin-13A antibodies recognized a band of ∼133-kDa (our unpublished data). When the antibodies were used to detect GhKinesin-13A in cotton fibers, organelles similar to those in Arabidopsis cells were detected (Figure 8D). Therefore, it is likely that Kinesin-13A proteins from these two different plants are associated with similar organelles.
Figure 8.
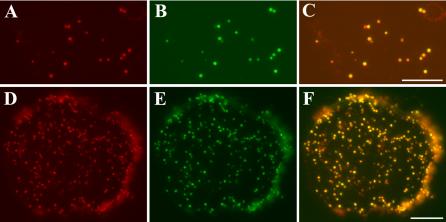
Colocalization of AtKinesin-13A and the Golgi apparatus. (A–C) Leaf mesophyll protoplast isolated from a STtmd-GFP plant was labeled with Anti-AtKinesin-13A antibodies. (A) Anti-AtKinesin-13A immunofluorescence. (B) STtmd-GFP signal showed the distribution of Golgi stacks. (C) A merged image had AtKinesin-13A signal pseudocolored in red and ST-tmd-GFP in green. Appearance of yellow indicated strong colocalization. Bar, 10 μm. (D and E) Cotton cytoplast isolated from a cotton fiber was labeled with anti-GhKinesin-13A and anti-α-mannosidase. (D) Anti-GhKinesin-13A immunofluorescence. (E) Anti-α-mannosidase signal showed the distribution of Golgi stacks. (F) Merged image had GhKinesin-13A signal pseudocolored in red and α-mannosidase in green. Appearance of yellow indicated strong colocalization. Bar, 10 μm.
Kinesin-13A Is Associated with Golgi Stacks
The anti-Kinesin-13A antibody staining pattern most closely reflected the size and distribution of the Golgi apparatus (Boevink et al., 1998; Nebenführ et al., 1999; Dixit and Cyr, 2002). Therefore, we asked whether Kinesin-13A was indeed associated with the Golgi apparatus. It has been reported that a fusion protein of the transmembrane domain of a rat sialyl transferase (STtmd) and GFP decorates Golgi stacks in plant cells (Boevink et al., 1998). To visualize the Golgi apparatus, we used an Arabidopsis line expressing this fusion protein (generous gift from Dr. C. Hawes and F. Brandizzi, Oxford Brooks University). Protoplasts isolated from leaf mesophyll cells were used to discern the relationship between AtKinesin-13A localization and individual Golgi stacks marked by STtmd-GFP. Figure 8, A and B, respectively, showed immunofluorescence with anti-AtKinesin-13A antibodies and the anti-GFP antibody. As shown in Figure 8C, merging of the two images suggest that AtKinesin-13A is localized to the Golgi stacks. The intensity of anti-AtKinesin-13A staining and the STtmd-GFP signal varied among some Golgi stacks. The significance of such variation is not clear.
Because an association of kinesins with entire Golgi stacks has not been reported, we used rabbit antibodies against α-mannosidase to obtain additional evidence. α-Mannosidase has been previously reported as an endogenous Golgi marker (Nebenführ et al., 1999). Because anti-GhKinesin-13A antibodies were raised in rats, dual localization experiments were carried out in cytoplasts isolated from cotton fibers to clearly distinguish individual Golgi stacks. The cytoplasm of cotton fibers had numerous Golgi stacks as detected by the anti-α-mannosidase antibodies (Figure 8E). Similar anti-GhKinesin-13A staining pattern also was observed (Figure 8D). As shown in a pseudocolored merged image in Figure 8F, Golgi stacks labeled with anti-α-mannosidase also were decorated by anti-GhKinesin-13A (Figure 8, D–F). Therefore, we concluded that GhKinesin-13A decorated the Golgi stacks in cotton fibers as seen in Arabidopsis cells.
To rule out that Kinesin-13A may be associated with other abundant organelles, we used an anti-salicylic acid-binding protein antibody (SABP) that specifically detects peroxisomes (Chen et al., 1993). Peroxisomes labeled by anti-SABP were much smaller and had different distribution pattern than that of Kinesin-13A in Arabidopsis root cells (our unpublished data). Therefore, we concluded that AtKinesin-13A and GhKinesin-13A represented a new class of plant kinesins that were associated specifically with the Golgi apparatus in both Arabidopsis and cotton cells.
The Golgi Apparatus Is Associated with Microtubules and Microfilaments
Other studies suggest that the motility of plant Golgi only relies on actin microfilaments (Boevink et al., 1998; Nebenführ et al., 1999). Because we showed that a microtubule-based motor was clearly associated with the Golgi apparatus, we also wanted to test whether the plant Golgi apparatus was associated with microtubules. Because kinesin-13A mutations caused defects in leaves, we carried out simultaneous observation of microtubules and Golgi stacks in live cells of Arabidopsis leaves. Cortical microtubules were observed in leaf epidermal cells stably expressing GFP-α-tubulin (Figure 9A). Because both STtmd-GFP and Nag-GFP indistinguishably decorate Golgi stacks (Boevink et al., 1998; Dixit and Cyr, 2002), we chose Nag-GFP to label the Golgi apparatus as the Nag sequence was from Arabidopsis. When Nag-GFP was transiently expressed by particle bombardment, Golgi stacks were labeled as particles throughout the cytoplasm (Figure 9A). Some Golgi stacks were found to be associated along the cortical microtubule network (Figure 9A, arrows), although not all Golgi stacks were attached to microtubules. Similar association of Golgi stacks labeled by anti-GhKinesin-13A with cortical microtubules was also observed in cytoplasts isolated from cotton fibers (our unpublished data). To further confirm that the Golgi-microtubule association was not an artifact caused by chemical fixation or other means, we examined cells preserved by rapid freeze and freeze substitution under an electron microscope. A close association between a Golgi stack and a microtubule was observed (Figure 9B). Thus, our observation of the possible association of Golgi stacks with cytoplasmic microtubules in vivo is consistent with the notion that a novel kinesin mediates the functional association of the two components.
Figure 9.
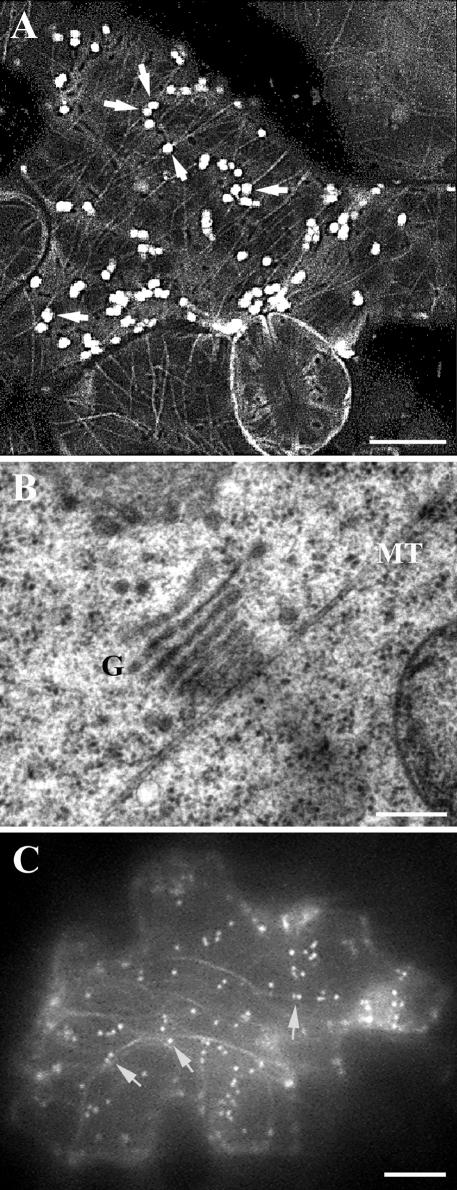
Association of Golgi stacks with microtubules and actin microfilaments. (A) Confocal image showed association of Golgi stacks labeled with Nag-GFP with cortical microtubules labeled with GFP-α6-tubulin (arrows). Note cortical microtubules were present in all cells. Bar, 10 μm. (B) Transmission electron microscopic image showed that a Golgi stack (G) was associated with a microtubule (MT). Bar, 0.2 μm. (C) Wide-field image showed association of Golgi stacks labeled with Nag-GFP and GFP-talin (arrows). Nag-GFP labeled Golgi stacks as bright particles, and GFP-talin labeled actin microfilaments as dimmer filamentous structures. Bar, 10 μm.
We also determined whether the Golgi apparatus was associated with microfilaments. A GFP-talin fusion protein had been shown to label actin microfilaments in live cells (Fu et al., 2002; Jedd and Chua, 2002). Among leaf epidermal cells expressing both GFP-talin and Nag-GFP fusions, a number of fluorescent Golgi stacks were associated with the fluorescent microfilaments (Figure 9C). However, some Golgi stacks did not show a close association with microfilaments (Figure 9C). Therefore, we conclude that in plant cells, the Golgi apparatus is capable of interacting with both microtubules and microfilaments.
AtKinesin-13A and Golgi Distribution
Because kinesin-13a mutations affect the branching pattern of trichomes in Arabidopsis, we wondered whether the distribution of Golgi stacks was different in mutant trichomes compared with those of wild type. Nag-GFP was stably expressed in the wild-type and kinesin-13a mutation backgrounds, which revealed Golgi stacks in leaf cells including trichomes. Although we were able to image Golgi stacks in trichomes by confocal microscopy, it was impossible to superimpose the optical sections of whole trichomes as Golgi stacks moved rapidly inside the trichomes so that different sections reflected Golgi distribution patterns at different time points. Thus, only single optical sections are presented here. In wild-type trichomes, Golgi stacks distributed more or less loosely in the cortex in the trichome stalk as well as branches (Figure 10A). The kinesin-13a trichomes also contained numerous Golgi stacks in the cortical cytoplasm (Figure 10B). The mutant trichomes consistently showed Golgi aggregation phenomenon (Figure 10B, arrows). This result suggested to us that this additional aggregation event might lead to the formation of the fourth branch in mutant trichomes.
Figure 10.
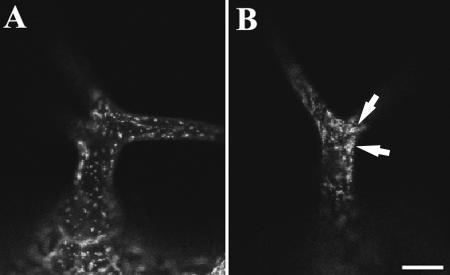
Distribution of Golgi stacks labeled with Nag-GFP in wild-type and kinesin-13a-1 trichomes. (A) Golgi stacks occurred loosely in the wild-type trichome cytoplasm. (B) Golgi stacks often form aggregates (arrows) in the kinesin-13a-1 mutant trichomes. Bar, 20 μm.
We also carried out immunolocalization of the Golgi apparatus and microtubules in other leaf epidermal cells in both wild-type and the kinesin-13a-1 mutant. Anti-α-mannosidase immunofluorescence revealed discrete particles of Golgi stacks in both wild-type and kinesin-13a-1 mutant cells (Figure 11, A and D). Networks of cortical microtubules were revealed by anti-tubulin immunofluorescence, and no obvious difference was detected between wild-type and mutant cells (Figure 11, B and E). Merged images were shown in Figure 11, C and F. In wild-type cells, the Golgi stacks were evenly distributed throughout the cortical cytoplasm where cortical microtubules were present (Figure 11C). In kinesin-13a-1 cells, Golgi stacks also were largely dispersed across the cytoplasm (Figure 11, D and F). However, in mutant cells, one or more aggregates of Golgi stacks were found in mutant epidermal cells (Figure 11F, arrows). The distribution pattern also was analyzed quantitatively by measuring anti-Golgi immunofluorescence intensity in 100 randomly chosen 6.4 × 6.4-μm areas in Figure 11, A and D. The result of the measurement is presented in Table 2. More areas in the kinesin-13a-1 mutant were distributed toward highest intensities and lowest intensities than those in the wild-type cells (Table 2). Consistent with the observation that Golgi stacks seemed more clumped in the mutant, and the observed distribution of pixel counts (Table 2), the pixel count variance was 3 times higher in the kinesin-13a-1 mutant than in wild-type (p = 0.01523). Therefore, this result suggested that a significant difference in the distribution of Golgi stacks occurred between the two samples. Similar phenomenon was also observed with independent samples. We also examined whether Golgi-microtubule association was dependent on the AtKinesin-13A protein. The Golgi apparatus was still found in frequent association with cortical microtubules in the kinesin-13a-1 mutant (Figure 11F).
Table 2.
Percentages of randomly chosen 6.4 × 6.4-μ m areas with different intensities of the Golgi immunofluorescent signal
| Intensity of Golgi immunofluorescent signal
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0–10 | 11–20 | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | 71–80 | >81 | Total | |
| Wild type | 38 | 24 | 17 | 9 | 7 | 3 | 1 | 1 | 0 | 100 |
| kinesin-13a–1 | 60 | 14 | 5 | 8 | 2 | 1 | 3 | 4 | 3 | 100 |
Data represent precntage of areas having the indicated value of fluorescent intensity. The relative fluorescent intensity was given arbitrarily by the ImagePro software package.
Motility of Golgi Stacks
We also examined the motility of Golgi stacks in both wild-type and mutant cells. When Nag-GFP was transiently and stably expressed in leaf epidermal cells and trichomes, Golgi stacks were labeled across the cytoplasm in both wild-type and mutant cells. In both wild-type and mutant cells, certain Golgi stacks were found to undergo rapid movement at a rate of several micrometers per second (our unpublished data). Other Golgi stacks, however, remained stationary. This is consistent with the report of a maximum velocity of 4.1 μm/s for some Golgi stacks in tobacco cells (Nebenführ et al., 1999). We were not able to detect any difference in the velocity of Golgi motility between wild-type and mutant cells. Thus, we conclude that Kinesin-13A does not contribute to the primary movement of the Golgi apparatus in plant cells.
DISCUSSION
In higher plants, the specific functions of most kinesins have not been determined. We used cotton fibers as starting materials to isolate candidate kinesins that are required for anisotropic cell enlargement. The results presented here describe the identification of a novel internal motor kinesin, Kinesin-13A from both cotton and Arabidopsis. Our data indicate that Kinesin-13A is associated with intact Golgi stacks. Although AtKinesin-13A is not essential for cell growth in Arabidopsis, it is required for normal trichome development in leaves.
Potential Roles of Kinesin-13A at the Golgi Apparatus
Activities of organelles like the Golgi apparatus require the cytoskeleton. In animal cells, the Golgi apparatus is positioned near the microtubule-organizing center, the centrosome. The dynamics of the Golgi apparatus involves constant addition of endoplasmic reticulum-derived compartments, in addition to deploying vesicles to various cellular destinations (Thyberg and Moskalewski, 1999). Studies in animal cells indicate that both microtubule-based and microfilament-based motor proteins play critical roles in Golgi dynamics (Allan et al., 2002). In higher plants cells, however, the Golgi apparatus is present as a large number of stacks throughout the cytoplasm (Dupree and Sherrier, 1998). In contrast to the movement of endoplasmic reticulum-to-Golgi transport compartment seen in animal cells, plant cells have the intact Golgi stacks move around the cytoplasm (Boevink et al., 1998; Nebenführ et al., 1999). Pharmacological evidence indicates that the rapid movement of Golgi stacks is dependent on actin microfilaments instead of microtubules (Boevink et al., 1998; Nebenführ et al., 1999). Therefore, the primary movement of the Golgi apparatus is likely driven by myosin(s), although the Golgi-associated myosin(s) are yet to be identified in plants.
Our data demonstrate that Kinesin-13A decorates the whole Golgi stacks, and functions in trichome morphogenesis. We postulate two possible roles for Kinesin-13A that may reconcile those two observations. First, Kinesin-13A may act as a motor that functions in the distribution of the Golgi apparatus in cell cortex. Although myosin(s) play a dominant role in Golgi distribution, Kinesin-13A's role could be less critical. For example, unidentified myosin(s) may play a role in driving long-distance transport of Golgi stacks, and Kinesin-13A plays a role in local dispersal of the stacks (Figure 12). The observation of Golgi aggregates in kinesin-13a mutant leaf epidermal cells and trichomes supports the hypothesis that Kinesin-13A plays a role in the dispersal of Golgi stacks. Second, Kinesin-13A may help distribute vesicles released from the Golgi apparatus. A prediction of this interpretation is that more Kinesin-13A would be expected to accumulate at one side of the Golgi stack than the opposite side. Because Kinesin-13A decorates evenly across the Golgi stacks, it is less likely that Kinesin-13A acts as a transporter for vesicles coming off from the Golgi apparatus.
Figure 12.
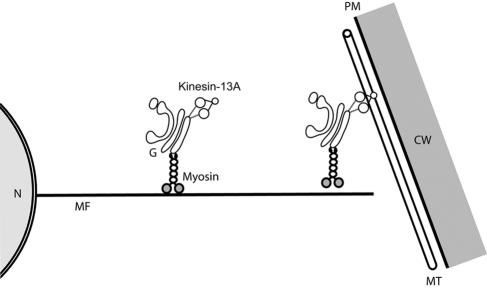
Proposed model for Kinesin-13A functions in plant cells. The plant Golgi apparatus may be associated with unknown myosin(s) and Kinesin-13A. From the perinuclear region to the cell cortex, the motility of Golgi stacks depends on myosin(s). In the cell cortex, Kinesin-13A plays a role in either the dispersal of the Golgi apparatus along cortical microtubules. CW, cell wall; G, Golgi apparatus; MF, actin microfilaments; MT, microtubules; N, nucleus.
To our knowledge, it is not understood how positioning of Golgi stacks affects plant cell morphogenesis. Obviously, the Golgi apparatus is responsible for secretory activities. It has been determined that hypersecretory activities accompany the appearance of large numbers of Golgi stacks (Dupree and Sherrier, 1998). Our data suggest that during trichome development, a branching event may follow the aggregation of Golgi stacks toward a site of branch emergence to provide robust secretory activities. To prevent a branch from emerging, a developing trichome cell may need to prevent Golgi stacks from accumulating at a particular site. AtKinesin-13A may play a role in the dispersal of the Golgi apparatus. Therefore, when AtKinesin-13A is not present, an additional clustering event of the Golgi stacks is not broken, so that a new branch emerges.
In kinesin-13a mutants, the only visible mutant phenotype was in trichome branching. The reason that no other defects were observed in other cell types might be because function(s) of Kinesin-13A could be shared by other kinesins in these cells. Trichome cells are highly differentiated in which only AtKinesin-13A might have been expressed to carry out designated function(s). Therefore, when AtKinesin-13A was lost, a phenotype was shown.
Control of the Number of Trichome Branches
Trichome branching is a sophisticated cellular process which is regulated by a number of factors. In Arabidopsis, trichome cell growth is preceded by approximately four rounds of DNA replication (endoreduplication) because trichome nuclei are normally 32C (Hülskamp et al., 1994). Ectopic expression of a cyclin D protein promotes DNA replication and cell division, so that more branches are formed (Schnittger et al., 2002). Conversely, when a cyclin-dependent kinase inhibitor is misexpressed in trichomes, a reduction of trichome branch number is seen (Schnittger et al., 2003). Trichomes with additional branches are present in mutants with increased trichome DNA content and in tetraploid plants (Perazza et al., 1999; Szymanski and Marks, 1998). We compared trichome DNA contents in wild-type, kinesin-13a mutants, and the try mutant that had higher DNA content than in wild type (Szymanski and Marks, 1998). It was found that kinesin-13a-1 trichomes showed similar nuclear DNA content as the wild-type trichomes, which was consistently lower than that in the try mutant. Therefore, it is unlikely that kinesin-13a mutations alter DNA replication/cell cycle events in the trichome.
After endoreduplication, trichome morphogenesis initiates with a microtubule-dependent polarization event, which is followed by an actin microfilament-dependent morphogenesis process (Mathur et al., 1999; Szymanski et al., 1999; Mathur and Chua, 2000; Szymanski, 2001). Loss of function mutations in genes encoding a microtubule-severing protein AtKTN or the kinesin KCBP/ZWI leads to the formation of two-branched trichomes (Burk et al., 2001; Oppenheimer et al., 1997). It has been hypothesized that the branching event requires localized microtubule stabilization activity (Mathur and Chua, 2000). Because kinesin-driven transport activities require microtubule tracks, microtubule organization can be considered as an early event relative to vesicle transport by kinesins. A Golgi-dependent process, regulated by AtKinesin-13A, may act later to specify branching events.
By using forward genetic approaches, mutations have been isolated that increase the branch number in trichomes. For example, extragenic suz suppressors of the zwi-3 mutation have been isolated in Arabidopsis (Krishnakumar and Oppenheimer, 1999). Among the SUZ genes whose mutations increase the branch numbers in the background of zwi-3, SUZ2 acts as a negative regulator for trichome branching (Krishnakumar and Oppenheimer, 1999). Because the SUZ2 gene is located on chromosome IV, and AtKinesin-13A is on chromosome III, SUZ2 cannot be allelic to AtKinesin-13A. Experiments need to be done to test whether the kinesin-13a mutations suppress the zwi-3 mutation as suz2 does.
Plant Kinesin-13A Is Functionally Divergent from Animal MCAK/Kinesin-13 Proteins
Because our phylogenetic analysis of the motor domain showed that plant Kinesin-13A is part of the MCAK/Kinesin-13 subfamily, it was important to determine whether members of this subfamily have conserved functions. Animal MCAK/Kinesin13 typically plays critical roles in spindle assembly and chromosome segregation (Walczak, 2003). Because no cell division phenotype has been observed in kinesin-13a mutants, unlike MCAK/Kinesin-13 members from animals, plant Kinesin-13A does not seem to be involved in cell division directly. Three lines of evidence further suggest to us that plant Kinesin-13A functions in a different manner than the animal members of the MCAK/Kinesin-13 subfamily.
First, there is no significant sequence homology outside the motor domain between Kinesin-13A and previously reported MCAK/Kinesin-13 proteins. Animal kinesins in the MCAK/Kinesin-13 subfamily typically have a unique neck sequence upstream of the catalytic core (Ovechkina and Wordeman, 2003). This signature K (lysine)-loop neck region is essential for their depolymerase activity (Ovechkina et al., 2002). Such a neck sequence is absent in plant Kinesin-13A. MCAK/Kinesin-13 proteins also have predicted coiled-coils after the catalytic core (Ovechkina and Wordeman, 2003). Plant Kinesin-13A, however, has predicted coiled-coils toward the C terminus.
Second, plant Kinesin-13A and animal MCAK/Kinesin-13 proteins have different localization patterns. Most animal MCAK/Kinesin-13 proteins localize to microtubule ends at kinetochores/centromeres and centrosomes (Wordeman and Mitchison, 1995; Walczak et al., 1996; Rogers et al., 2004). Kinesin-13A presence in Golgi stacks provides strong evidence that it does not perform cellular activities similar to animal MCAK/Kinesin-13 proteins.
Third, a distinct characteristic of animal MCAK/Kinesin-13 proteins is their microtubule end-stimulated ATPase (Walczak, 2003). This activity is required for establishing mitotic spindles bearing shorter microtubules than ones in interphase (Walczak et al., 1998). Our preliminary results indicate that plant Kinesin-13A is unlikely to possess the depolymerization activity. When the AtKinesin-13A protein was ectopically expressed in the filamentous fungus Aspergillus nidulans, we did not detect an obvious microtubule depolymerization phenotype (Lu and Liu, unpublished data). Moreover, the depolymerase activity is probably not associated with Kinesin-13A because of the absence of the signature neck sequence of animal MCAK/Kinesin-13, which is essential for their microtubule depolymerase activity.
In summary, the Kinesin-13A protein from flowering plants is a novel member of the MCAK/Kinesin-13 subfamily. Members of this subfamily may have evolved from a common ancestor but have later emerged as structurally and functionally divergent proteins.
Acknowledgments
We thank Dr. Debby Delmer and laboratory for generous help throughout the course of this project. We thank Drs. Robert Roberson and Fengli Guo for assistance in Figure 9B, and Samantha Barling-Silva for Figure 11. We thank Drs. Sebastian Bednarek, Federica Brandizzi, Richard Cyr, Ram Dixit, Chris Hawes, Eliot Herman, Dan Szymanski, Richard Tralease, and Zhenbiao Yang for providing plasmids, antibodies, and seeds for this study. We are also grateful to the Arabidopsis Biological Resources Center, the Kazusa DNA Research Institute, the Salk Institute, and the Syngenta Terra Mesa Research Institute for services. We especially thank Professor John Harada for critical comments on the manuscript. This work was supported by a grant from the U.S. Department of Energy. Our studies on plant kinesins also were supported by a grant from the U.S. Department of Agriculture.
Article published online ahead of print in MBC in Press on December 1, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-05-0400).
References
- Allan, V., Thompson, H., and McNiven, M. (2002). Motoring around the Golgi. Nat. Cell Biol. 4, E236-E242. [DOI] [PubMed] [Google Scholar]
- Baskin, T. I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215, 150-171. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Cruz, S. S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441-447. [DOI] [PubMed] [Google Scholar]
- Burk, D. H., Liu, B., Zhong, R., Morrison, W. H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807-827. [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Ricigliano, J., and Klessig, D. (1993). Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc. Natl. Acad. Sci. USA 90, 9533-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, A. L., and Smith, L. G. (1998). The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10, 1875-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Turgeon, R., and Parthasarathy, M. V. (1991). Microfilaments in the preprophase band of freeze substituted tobacco root cells. Protoplasma 165, 209-211. [Google Scholar]
- Dixit, R., and Cyr, R. J. (2002). Golgi secretion is not required for marking the preprophase band site in cultured tobacco cells. Plant J. 29, 99-108. [DOI] [PubMed] [Google Scholar]
- Dupree, P., and Sherrier, D. (1998). The plant Golgi apparatus. Biochim. Biophys. Acta 1404, 259-270. [DOI] [PubMed] [Google Scholar]
- Folkers, U., Berger, J., and Hülskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis - differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779-3786. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203-1207. [DOI] [PubMed] [Google Scholar]
- Guan, K. L., and Dixon, J. E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262-267. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Misera, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555-566. [DOI] [PubMed] [Google Scholar]
- Jedd, G., and Chua, N. (2002). Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43, 384-392. [DOI] [PubMed] [Google Scholar]
- Kopczak, S. D., Haas, N. A., Hussey, P. J., Silflow, C. D., and Snustad, D. P. (1992). The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell 4, 539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, S., and Oppenheimer, D. (1999). Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation sand pollen tube growth. Development 126, 3079-3088. [DOI] [PubMed] [Google Scholar]
- Krysan, P. J., Young, J. C., and Sussman, M. R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standarized kinesin nomenclature. J. Cell Biol. 167, 19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.-R.J., Giang, H. M., and Liu, B. (2001). A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13, 2427-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Spielhofer, P., Kost, B., and Chua, N. (1999). The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126, 5559-5568. [DOI] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Gallagher, L. A., Dunahay, T. G., Frohlick, J. A., Mazurkiewicz, A. M., Meehl, J. B., and Staehelin, L. A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D. G., Pollock, M. A., Vacik, J., Szymanski, D. B., Ericson, B., Feldmann, K., and Marks, M. D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261-6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina, Y., Wagenbach, M., and Wordeman, L. (2002). K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J. Cell Biol. 159, 557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina, Y., and Wordeman, L. (2003). Unconventional motoring: an overview of the Kin C and Kin I kinesins. Traffic 4, 367-375. [DOI] [PubMed] [Google Scholar]
- Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., and Stalker, D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93, 12637-12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza, D., Herzog, M., Hülskamp, M., Brown, S., Dorne, A., and Bonneville, J. (1999). Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, 461-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, M. L., Delmer, D. P., and Liu, B. (2003). The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol. 132, 154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, M. L., Serna, J., Falbel, T. G., Bednarek, S. Y., and Nielsen, E. (2004). The Arabidopsis Rab GTPase, RabA4b, localizes to the tips of growing root hair cells. Plant Cell 16, 1589-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., Jilk, R., Marks, M., and Szymanski, D. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14, 101-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2004). R: a language and environment for statistical computing. Vienna, R Foundation for Statistical Computing. ISBN 3–900051-07-0, URL http://www.R-project.org.
- Reddy, A.S.N., and Day, I. S. (2001). Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G., Rogers, S., Schwimmer, T., Ems-McClung, S., Walczak, C., Vale, R., Scholey, J., and Sharp, D. (2004). Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364-370. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., and Hülskamp, M. (2002). Trichome morphogenesis: a cell-cycle perspective. Phil. Trans. R. Soc. Lond. B Biol. Sci. 357, 823-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y., and Hülskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99, 6410-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hülskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15, 303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagull, R. W. (1990). The effects of microtubule and microfilament disrupting agents on cytoskeletal arrays and wall deposition in developing cotton fibers. Protoplasma 159, 44-59. [Google Scholar]
- Seagull, R. W. (1992). A quantitative electron microscopic study of changes in microtubule arrays and wall microfibril orientation during in-vitro cotton fiber development. J. Cell Sci. 101, 561-577. [Google Scholar]
- Shaw, S. L., Kamyar, R., and Ehrhardt, D. W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715-1718. [DOI] [PubMed] [Google Scholar]
- Smith, L. G. (2003). Cytoskeletal control of plant cell shape: getting the fine points. Curr. Opin. Plant Biol. 6, 63-73. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Himmelspach, R., Williamson, R. E., and Wasteneys, G. O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15, 1414-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D. (2001). Arabidopsis trichome morphogenesis: a genetic approach to studying cytoskeletal function. J. Plant Growth Reg. 20, 131-140. [Google Scholar]
- Szymanski, D. (2002). Tubulin folding cofactors: half a dozen for a dimer. Curr. Biol. 12, R767-R769. [DOI] [PubMed] [Google Scholar]
- Szymanski, D., and Marks, M. (1998). GLABROUS1 overexpression and TRIP-TYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D., Marks, M., and Wick, S. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg, J., and Moskalewski, S. (1999). Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 246, 263-279. [DOI] [PubMed] [Google Scholar]
- Tian, G. W., Smith, D., Gluck, S., and Baskin, T. I. (2004). Higher plant cortical microtubule array analyzed in vitro in the presence of the cell wall. Cell Motil. Cytoskeleton 57, 26-36. [DOI] [PubMed] [Google Scholar]
- Vale, R. D. (2003). The molecular motor toolbox for intracellular transport. Cell 112, 467-480. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E. (2003). The Kin I kinesins are microtubule end-stimulated ATPases. Mol. Cell 11, 286-288. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Mitchison, T. J., and Desai, A. (1996). XKCM 1, A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37-47. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Vernos, I., Mitchison, T. J., Karsenti, E., and Heald, R. (1998). (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 13. [DOI] [PubMed] [Google Scholar]
- Wordeman, L., and Mitchison, T. (1995). Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128, 95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D. H., Morrison W Herbert, III and Ye, Z.-H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]


