mRNAs coding for ribosomal proteins are preferentially stored during heat shock and released during recovery phase to enhance ribosome production in an HSP101-dependent manner.
Abstract
Heat shock (HS) is known to have a profound impact on gene expression at different levels, such as inhibition of protein synthesis, in which HS blocks translation initiation and induces the sequestration of mRNAs into stress granules (SGs) or P-bodies for storage and/or decay. SGs prevent the degradation of the stored mRNAs, which can be reengaged into translation in the recovery period. However, little is known on the mRNAs stored during the stress, how these mRNAs are released from SGs afterward, and what the functional importance is of this process. In this work, we report that Arabidopsis HEAT SHOCK PROTEIN101 (HSP101) knockout mutant (hsp101) presented a defect in translation recovery and SG dissociation after HS. Using RNA sequencing and RNA immunoprecipitation approaches, we show that mRNAs encoding ribosomal proteins (RPs) were preferentially stored during HS and that these mRNAs were released and translated in an HSP101-dependent manner during recovery. By 15N incorporation and polysome profile analyses, we observed that these released mRNAs contributed to the production of new ribosomes to enhance translation. We propose that, after HS, HSP101 is required for the efficient release of RP mRNAs from SGs resulting in a rapid restoration of the translation machinery by producing new RPs.
Plants are often exposed to elevated temperature and have developed different layers of regulation to counteract adverse effects induced by heat stress. A well-defined plant response is the HS response, resulting in the production of molecular chaperones, including heat shock proteins (HSPs; Kotak et al., 2007). The typical roles of these HSPs are to maintain the cellular proteostasis in limiting the production and accumulation of protein aggregates induced by HS and to confer thermotolerance (Kotak et al., 2007). Recently, the functions of HSPs in eukaryotes are extended to a wider scope. It appears that some of these proteins are also involved in the translational control of mRNAs under normal and stress conditions or during recovery after stress (Walters and Parker, 2015).
Especially, certain HSPs have been demonstrated to be involved in translation recovery after stress. Recently in Arabidopsis (Arabidopsis thaliana), small HSPs and HSP101 were shown to protect protein translation factors during HS (McLoughlin et al., 2016). Small HSPs and HSP101 are involved in the resolubilization of translation factors like eEF1B and eIF4A during the recovery phase. A similar mechanism was reported for human cells (Cuesta et al., 2000). During HS, HSP27 binds eIF4G and traps it in aggregates to prevent assembly of the cap-initiation complex and to rapidly enhance translation recovery. During the recovery phase, these aggregates, called stress granules (SGs), have to be dissociated, a process which also implicates HSPs. In yeast, it was reported that HSP104 (the homolog of Arabidopsis HSP101), together with HSP70, is involved in disaggregation of SGs after HS (Cherkasov et al., 2013). Yeast hsp104 knockout mutant presents some defect in SG dissociation, resulting in a slower translation recovery. In fact, the authors proposed that a slower dissociation of SGs results in a longer release of translation factors trapped in SG, thus limiting translation initiation recovery.
The assembly of SG, in the same time with P-bodies, is generated by a massive inhibition of translation induced by stress. This repression was observed under many stresses in yeast (Kuhn et al., 2001; Melamed et al., 2008), mammals (Hamilton et al., 2006; Liu et al., 2013), as well as in plants like Arabidopsis (Juntawong and Bailey-Serres, 2012), Medicago truncatula (Puckette et al., 2012), and rice (Oryza sativa; Park et al., 2012). HS is well known to repress translation, inducing massive translation repression (Ueda et al., 2012; Merret et al., 2013). According to protein composition, SG was attributed to mRNA storage, while P-bodies are involved in mRNA decay. Although proteomic composition and assembly of these aggregates are increasingly documented (Buchan, 2014; Protter and Parker, 2016), little is known about the mRNAs stored in SGs, what the functional role is of the stored mRNAs during recovery, and how these mRNAs are released. Nonetheless, it is now well accepted that the dynamics of SGs arise by an ATP-dependent mechanism and play a crucial role in mRNA metabolism (Jain et al., 2016).
Recently, in Arabidopsis, formation and dissociation of SG were reported under hypoxia and reoxygenation (Sorenson and Bailey-Serres, 2014). The authors demonstrated the role of UBP1 in sequestration of poorly translated mRNAs during stress. They also showed that UBP1 granules are dissociated during reoxygenation. The authors proposed that UBP1 orchestrates the translational control of mRNAs during and after stress. As in other organisms, SGs play an important role in plant stress tolerance and development, as mutants of SG components present some deficiency in acclimation to stress and/or development (Xu et al., 2006; Xu and Chua, 2009; Merret et al., 2013; Sorenson and Bailey-Serres, 2014; Perea-Resa et al., 2016).
Nonetheless, few analyses address the involvement of the stored mRNAs in translation recovery. Here, we show that the Arabidopsis hsp101 knockout mutant presents defects in SG dissociation and polysome recovery. We took advantage of this defect to identify mRNAs stored during HS and released at recovery. By RNA sequencing and RNA coimmunoprecipitation (RIP) validation, we found that ribosomal protein (RP) mRNAs are preferentially stored during stress and released during recovery phase, the latter of which requires the chaperone function of HSP101. By 15N incorporation, we have further demonstrated that new ribosomes can be produced independently of transcription, thus contributing to revive translation during the recovery period. We postulate that this mechanism could be a mean to rapidly adjust translational levels following successive stresses.
RESULTS
Polysome Recovery Is Affected in hsp101 after HS
To check translational activity in wild type compared to hsp101 (hot1-3; Lee et al., 2005), we analyzed polysome profiles on 7-d-old seedlings under the normal condition at 22°C (control condition), after an HS treatment at 37°C for 1 h, and after 2 h of recovery at 22°C following the HS (referred to as 2 h recovery hereafter; Fig. 1). Under the normal condition and during HS, no obvious differences were observed between wild type and hsp101 (Fig. 1, A and D). After 1 h at 37°C, a massive polysome dissociation occurred that induces accumulation of monosomes (80S) and free 40S/60S ribosomal subunits (Fig. 1, B and E). In wild type, after 2 h recovery, polysomes massively reappeared, suggesting a translational restart (Fig. 1C). Nonetheless, polysome recovery in hsp101 is affected compared to wild type (Fig. 1, C and F). In fact, polysome increase is slower in hsp101, resulting in a higher amount of monosomes (compare the 80S peaks). To confirm this result, polysome area was quantified for each condition using three biological replicates (Fig. 1G). A significant difference was only observed between wild type and hsp101 after 2 h recovery. HSP101 is a molecular chaperone protein; some missense mutants (dlt1-1 and dlt1-2) were shown to differentially affect its chaperone activity (Wu et al., 2013). dlt1-1 contains a single mutation C-to-T in HSP101 at position 599 near the N terminus of Nucleotide-Binding Domain2, causing the replacement of Thr by Ile. In dlt1-2, a single mutation C-to-T appears at position 33 in the N-terminal domain, resulting in a His-to-Tyr substitution (Wu et al., 2013). Both maintained a normal level of HSP101 protein, but dlt1-2 results in substantial loss of the chaperone function of HSP101, as dlt1-1 mutation has little effect on HSP101 chaperone activity. Moreover dlt1-2 mutation induces thermotolerance defect, while dlt1-1 does not. Like hsp101, dlt1-2 seedlings are not able to survive under short-acquired thermotolerance (Wu et al., 2013). To check if HSP101 chaperone activity is involved in translation recovery, the polysome profiles of dlt1-1 and dlt1-2 were analyzed after 2 h recovery (Supplemental Fig. S1). Only dlt1-2 presented defect in polysome recovery, suggesting that HSP101 chaperone activity is involved in translation recovery. We tried to correlate this defect with a defect in growth rate after HS. To this end, the extension of root length was measured, but no differences were observed between the wild type and hsp101 during recovery (data not shown).
Figure 1.
hsp101 knockout mutant is affected in polysome recovery. A to F, Polysome profiles on 7-d-old seedlings in the wild type (A–C) or hsp101 (D–F). A and D, Control 22°C. B and E, HS for 1 h at 37°C. C and F, Two hours of recovery at 22°C after HS. G, Percentage of polysomes for each condition. n = 3 biological repeats, mean ± sd. A t test was performed between wild type and hsp101 for control (P > 0.1), at 1 h 37°C (P > 0.1), and at 2 h recovery (P < 0.05).
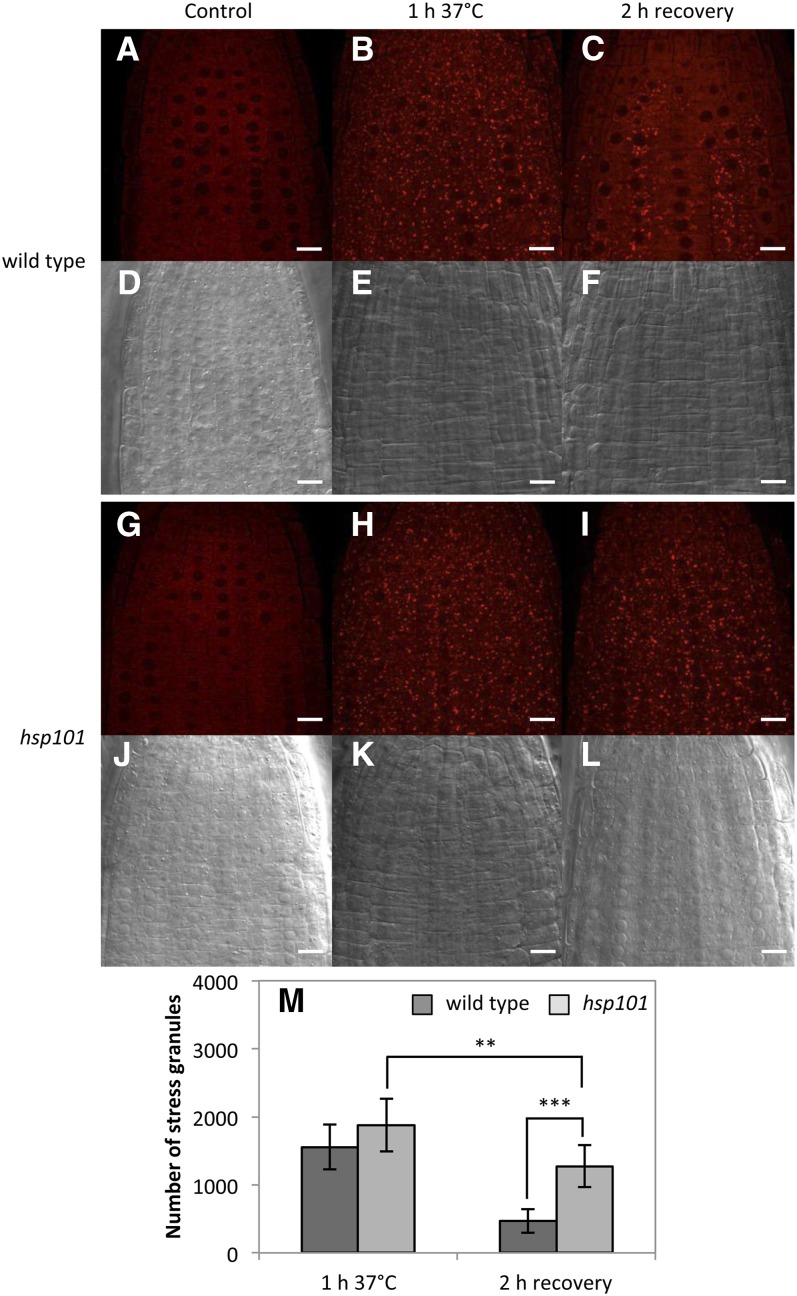
To check if this process is also correlated with a defect in SG dissociation, appearance and disappearance of SGs were monitored (Fig. 2; Supplemental Fig. S2) with the Arabidopsis transgenic lines expressing the poly(A) binding protein2 fused to the red fluorescent protein (PABP2-RFP) under the PABP2 promoter in the wild type or hsp101 background. The levels of PABP2-RFP and HSP101 were checked by western blot (Supplemental Fig. S2A). SG accumulation and dissociation were analyzed on different roots by confocal microscope (Fig. 2; Supplemental Fig. S2B). A defect in SG dissociation was observed in hsp101 compared to wild type after 2 h recovery (Fig. 2C versus Fig. 2I; Supplemental Fig. S2B), whereas no difference can be observed under normal and HS conditions (Fig. 2A versus Fig. 2G, and Fig. 2B versus Fig. 2H). A SG quantification was performed in wild type and hsp101 to confirm this observation (Fig. 2M). According to these data, we proposed that HSP101 could be involved in translation recovery by regulating the dissociation of SG.
Figure 2.
hsp101 knockout mutant is affected in SG dissociation after HS. A to L, Monitoring of SG formation using pPABP2-RFP-PABP2 in wild type (A–F) or the hsp101 background (G–L). A, D, G, and J, Control at 22°C. B, E, H, and K, HS for 1 h at 37°C. C, F, I, and L, Two hours recovery at 22°C after HS. Pictures are representative of at least three independent analyses. White lines correspond to 10 μm. M, SG quantification in wild type and hsp101 after 1 h at 37°C and after 2 h recovery. n = 5 biological repeats, mean ± sd. A t test was performed between wild type and hsp101 at 1 h 37°C (P > 0.1), at 2 h recovery (P < 0.001), and for hsp101 between 1 h 37°C and 2 h recovery (P < 0.01). Quantification was performed on the same volume of stacks for each condition.
Identification of Stored mRNAs Released in an HSP101-Dependent Manner
Although the involvement of the yeast HSP104 in SG dissociation is known (Cherkasov et al., 2013), the nature of mRNA stored and released was not identified. As stored mRNAs could play a role in translation recovery, we designed an RNA sequencing strategy to identify them. Because SG purification could be difficult and quite challenging, we decided to take advantage of the hsp101 translation defect to identity these mRNAs with the idea that stored mRNAs will come back slower in translation in hsp101 compared to wild type. To do that, we performed RNA sequencing on total and polysome-associated mRNAs purified from wild type and hsp101 seedlings under the normal condition at 22°C, after 1 h at 37°C, and after 2 h recovery (Supplemental Table S1). Each condition was performed on two biological repeats.
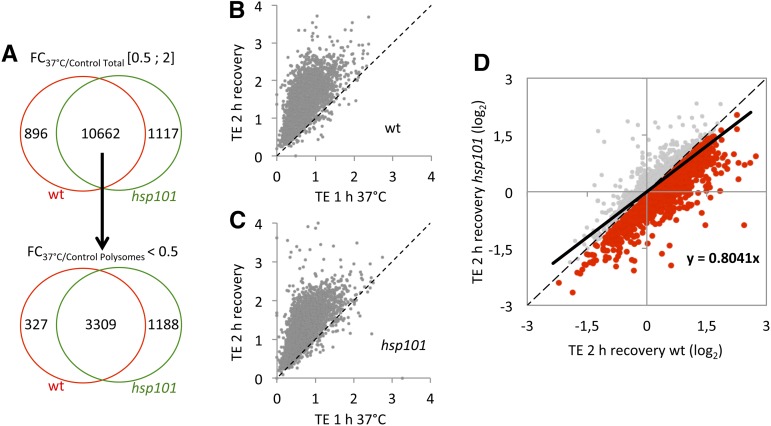
Prior to analysis, for each condition, reads per kilobase per million mapped reads (RPKM) values obtained between replicates were compared. Each set of experiment was found to be reproducible as demonstrated by plotting read counts pairwise between replicates (with R2 ∼0.99; Supplemental Table S1). Next, only transcripts with at least 1 RPKM in one library were kept, resulting in 19,599 transcripts. The aim of the RNA sequencing is to identify mRNAs first stored during HS and next released in an HSP101-dependent manner. For stored mRNAs, we reasoned as follows: A stored mRNA must be stable and released from translation during HS, i.e. down-regulated at polysome level. To identify those mRNAs, a fold-change (FC) analysis was performed at total RNA level between HS and control condition. The same analysis was also applied for polysomal RNA. A transcript having an FC below 0.5 was considered as down-regulated and was considered as stable with an FC between 0.5 and 2. To identify transcripts that were stored during HS (and not degraded as in Merret et al., 2013, 2015), we selected from the total RNA population heat “stable” mRNAs (having an FC between 0.5 and 2) that were clearly down-represented (having an FC below 0.5) in the polysomal fraction after HS. According to the criteria, 11,558 and 11,779 transcripts are stable during HS in wild type and hsp101, respectively, with an overlap of 90% between both genotypes (Fig. 3A). Then, only common transcripts between the wild type and hsp101 were kept, and their translation repression was analyzed. We found 3309 transcripts to be down-regulated at the polysomal level in both genotypes (Fig. 3A; Supplemental Table S2).
Figure 3.
Translation efficiency of mRNAs released from translation during HS is affected in the hsp101 mutant during recovery phase. A, Workflow used to identify transcripts stored in SG and released from translation during recovery. Only mRNAs with an FC between 0.5 and 2 on total RNA population and FC below 0.5 on polysomal RNA population in wild type (wt) and hsp101 were kept. B and C, TE of the 3309 mRNAs during HS versus recovery period in wt (B) and hsp101 (C). D, The log2 value of TE of the 3309 mRNAs during recovery period in wt versus hsp101. Red dots correspond to mRNAs affected in translation recovery in hsp101 (2103). The black line corresponds to a linear regression of the 3309 dots.
To identify among the 3309 transcripts those released during recovery in an HSP101-dependent manner, we measured their translation efficiency (TE; their ratio in polysomes versus total RNA), during and after HS, in wild type and hsp101. As shown in Figure 3, B and C, TEs of these mRNAs were generally higher during recovery compared to HS, suggesting that most of the stored mRNAs during HS can come back rapidly in translation after stress. As hsp101 presents a defect in polysome recovery, we decided to compare the TEs of these 3309 mRNAs during recovery in wild type and hsp101. To do that, a scatter plot was performed between wild type and hsp101 for the three conditions (Fig. 3D; Supplemental Fig. S3). Next, a linear regression was calculated for each condition with the 3309 dots. Although their TEs are identical under control and HS conditions (Supplemental Fig. S3; linear regression close to 1), during recovery, TEs in hsp101 appeared to be affected compared to wild type as the linear regression decrease close to 0.8. Out of 3309 transcripts, 2103 showed TEs lower in hsp101 compared to wild type (red dots on Fig. 3D; Supplemental Table S3). Since SG dissociation was affected in hsp101, we propose that these 2103 mRNAs are stored in SG during HS and released in an HSP101-dependent manner.
mRNAs Coding for RPs Are Stored during HS and Released during Recovery
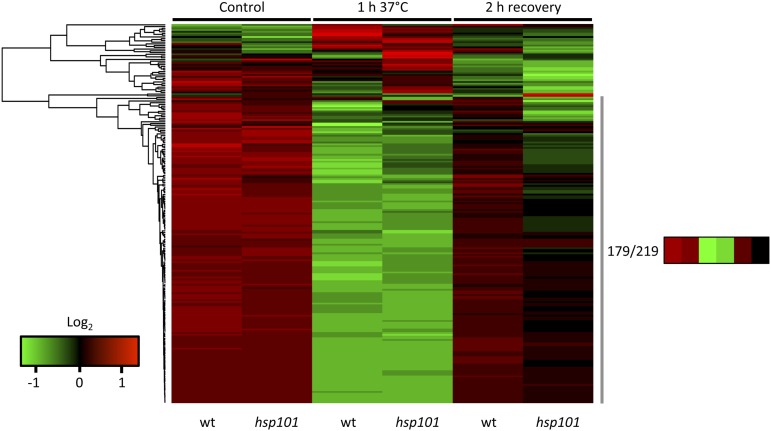
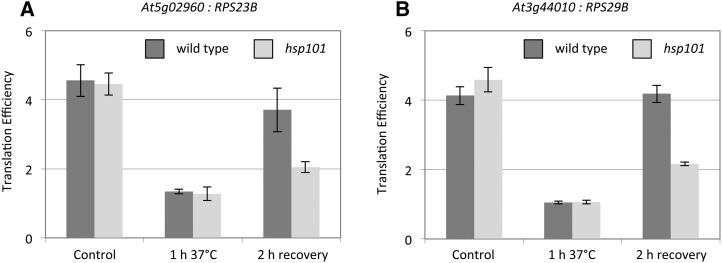
To go further in the functional role of the stored mRNAs during recovery, we performed a Gene Ontology (GO) analysis on the stored mRNAs released during recovery in an HSP101-dependent manner. Two molecular functions were enriched in the 2103 mRNA population: “protein binding” (GO:0005515, P-value 0.000202) and “structural constituent of ribosome” (GO:0003735, P-value 6.81e−54; Supplemental Fig. S4). The second GO term is highly enriched, and most of the mRNAs present in this group encode large or small subunit ribosomal proteins (RPL or RPS). According to our RNA-seq database (Supplemental Table S1), 276 nuclear genes coding for RPS or RPL are expressed at seedlings stage. Two hundred nineteen correspond to cytoplasmic RPs, 50 to chloroplastic RPs, and seven to mitochondrial RPs. Thus, to have a better overview of translation activity of mRNAs coding for RPs, TE was determined for all of them in the three conditions (control, HS, and recovery) in wild type and hsp101 (Supplemental Table S4). TEs were clustered according to their behavior on three heat maps (Fig. 4; Supplemental Fig. S5). According to the clustering, a major group can be distinguished in the three class of RPs. This group is highly present for cytoplasmic RPs with 179 members (81.7%) compared to chloroplastic and mitochondrial RPs with 27 (54%) and four (57%) members, respectively. This group has members with low TE during HS compared to the control, and their translation recovery seemed to be HSP101 dependent (Fig. 4; Supplemental Fig. S5). We propose that members of this group are stored during HS and released directly or indirectly by HSP101 during the recovery phase. To confirm this hypothesis, we performed quantitative polymerase chain reaction (qPCR) validation on two candidate transcripts of RPS23B (At5g02960) and RPS29B (At3g44010) (Fig. 5, A and B). As expected, their TEs were significantly and identically reduced during HS in wild type and hsp101, whereas translation recovery was affected in hsp101 for both transcripts.
Figure 4.
Translation efficiency of cytoplasmic RP mRNAs in wild type and hsp101 under normal condition, after 1 h at 37°C, and after 2 h recovery. Heat map of cytoplasmic RP mRNA translation efficiency in log2 value. The heat map was performed on the 219 cytoplasmic RP mRNAs identified in RNA-seq database. One hundred seventy-nine of them are grouped in the same cluster. The numbers of transcripts and the mean value of TE of this cluster are marked. Red values correspond to a high translation efficiency and green values to a low translation efficiency. wt, Wild type.
Figure 5.
Determination of RP mRNA translation efficiency by qPCR. A and B, Translation efficiency of the At5g02960 and At3g44010 genes determined by qPCR. Translation efficiency was determined as the ratio of mRNA quantity between polysomal and total RNA values. Values are normalized to ACTIN7 level. n = 3 biological repeats, mean ± sd.
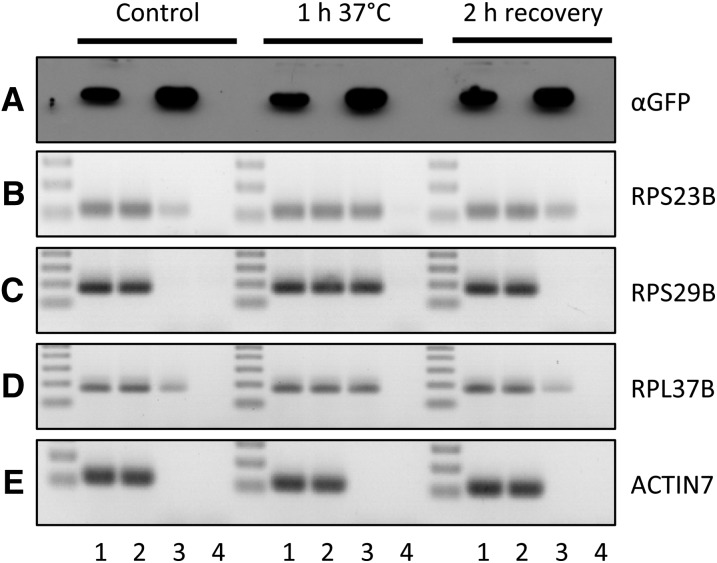
To confirm that these mRNAs are stored in SGs during HS, a RIP approach coupled to RT-PCR was performed using a transgenic line expressing UBP1a fused to the GFP. UBP1a was previously reported to localize in SG during stress (Weber et al., 2008; Sorenson and Bailey-Serres, 2014). The RIP experiment was performed under the normal condition at 22°C, after 1 h at 37°C, and after 2 h recovery. After checking immunoprecipitation efficiency (Fig. 6A), the association of RPS23B and RPS29B mRNAs with UBP1a was analyzed. RPL37B mRNA was used as a positive control, as this transcript was previously shown as an mRNA partner of UBP1 under hypoxia (Sorenson and Bailey-Serres, 2014). According to our data, this transcript seems to be stored during HS and released in an HSP101-dependent manner. For all transcripts, association with UBP1a was highly induced during HS (Fig. 6, B–D) and reduced or abolished during the recovery phase, coincident with the model where RP mRNAs are stored during HS and released during the recovery phase. Thus, we propose that HS induces a massive storage of mRNAs coding RPs and these mRNAs are released during recovery to enhance translation.
Figure 6.
RP mRNAs are associated with UBP1 during HS and released after stress. A, Western-blot analysis of UBP1a-GFP immunoprecipitation efficiency using GFP antibody. B to E, PCR amplification (30 cycles) of the At3g44010 (RPS29B), At5g02960 (RPS23B), At1g52300 (RPL37B), and At5g09810 (ACTIN7) genes after RNA immunoprecipitation with the 35S-UBP1a-GFP line. Wild-type line was used as a negative control. 1, Input UBP1a; 2, input wild type; 3, eluate UBP1a; 4, eluate wild type.
Polysome Recovery Can Be Independent of Transcription Restart
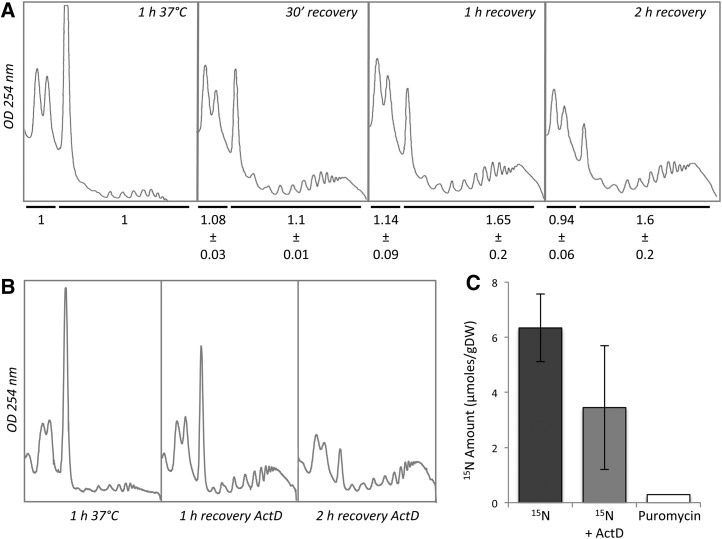
The storage of mRNA coding for RPs during HS suggests that the production of new ribosomes is a critical step to resume translation at recovery, meaning the new subunits have to be produced. To test this hypothesis, polysome profiles were done at different time points during recovery. We hypothesized that if preexisting free ribosomal subunits (40S and 60S) can be reused during recovery, their amount will be decreased with the concomitant increase of 80S/polysomes. To test this hypothesis, the area of 40S/60S and 80S/polysomes were determined for each condition and normalized according to the respective area for HS condition. After 30 min recovery, monosomes (the 80S peak) decreased and polysomes increased as compared to the HS situation, suggesting a translation restart (Fig. 7A). However, curiously, the amount of free ribosomal subunits did not change as compared to HS condition (ratio = 1.08 ± 0.03). After 1 h recovery, polysomes continued to increase and reached a maximum level while the amount of free ribosomal subunits remained, again, constant (ratio = 1.14 ± 0.09). After 2 h recovery, the amount of free ribosomal subunits decreased, but the polysomes stopped to increase. These observations strongly suggest that, up to 1 h recovery, preexisting free ribosomal subunits could not fully account for the translation restart and that new ribosomal subunits must come into play with the synthesis of RPs.
Figure 7.
New ribosomes can be produced during recovery independently of 40S and 60S reassembly. A, Polysome profiles analysis in wild type after HS 1 h at 37°C, and after 30 min, 1 h, and 2 h recovery at 22°C. Polysome profile analysis was performed three times on three biological repeats. The area of 40/60S and 80S/polysomes were determined for each condition and normalized according to the respective area for HS condition. Data are presented below each profile. n = 3 biological repeats, mean ± sd. B, Polysome recovery after HS on liquid medium supplemented with actinomycin D. C, 15N incorporation in polysomal proteins after 2 h recovery with or without actinomycin D treatment; n = 3 biological repeats, mean ± sd. Puromycin treatment was used as a negative control. Values were normalized by 15N natural abundance.
The production of new RPs could result from the translation of either de novo transcribed mRNAs or those released from SGs. To discriminate between these two possibilities, seedlings in the recovery phase were incubated on liquid medium supplemented or not with the transcription inhibitor, actinomycin D, to block both polymerase I and II activities (Bensaude, 2011). After 2 h recovery, polysome profiles were performed. As shown in Figure 7B, although transcription is blocked, polysome recovery still took place, suggesting that a least a portion of new polysomes can be formed without transcription. To prove that new ribosomes can be formed in the time frame of the recovery period without necessarily mobilizing preexisting, heat-dissociated ribosomal subunits, a 15N incorporation experiment was performed after stress. After 15N incorporation and polysome purification, the 15N amount was determined for proteins present in the polysomal fractions (essentially composed of ribosomes). These data were compared to a negative control (puromycin treatment) to be sure that incorporated 15N signal only comes from proteins associated with polysomes. As shown in Figure 7C, 15N was detected in polysomes after 2 h recovery, suggesting the formation of new ribosomes in this frame time. The same incorporation was performed with actinomycin D treatment (Fig. 7C). Although transcription is blocked, 15N incorporation was still detected in polysomes after 2 h recovery.
As ribosome biogenesis needs mature rRNAs produced by RNA polymerase I, we investigated the effect of HS on nucleoli architecture. Nucleolus architecture is highly dependent on the production of a proper amount of rRNAs. One of the major components of the nucleolus is the dense fibrillar component that consists of newly transcribed rRNA.
Fibrillarin is a nucleolar protein known to localize to the dense fibrillar component, forming ring-like structures in the nucleolus (Picart and Pontvianne, 2016) and can be used as a marker of nucleolus integrity (Pontvianne et al., 2013). Using FIB2-YFP stable line, the nucleolus status was checked under normal and HS conditions. As shown in Supplemental Figure S6, A and B, FIB2 formed ring-like structures both in normal and HS conditions, whereas actinomycin D treatment (that inhibits polymerase I activity) disrupts FIB2 localization (Supplemental Fig. S6C). Thus, HS did not affect nucleolus architecture, suggesting that RNA polymerase I activity was not significantly affected under this condition. Altogether, these data suggest that new ribosomes can be formed shortly after HS by mobilizing stored RP-coding mRNAs.
DISCUSSION
Translation Repression and Storage of RP mRNAs during Stress
Although conditions leading to SG formation are now well documented, how specific mRNAs are stored and released from these aggregates and the functional consequences of this process remain unclear. Here, using the hsp101 translation recovery defect, we observed that 210 of 276 nuclear mRNAs coding for RPs are preferentially stored under HS and released in the recovery period in an HSP101-dependent manner. Translation inhibition of RP mRNAs was already observed for different stresses like hypoxia (Branco-Price et al., 2008), Suc deprivation (Nicolaï et al., 2006), or water deficit (Kawaguchi et al., 2004), suggesting that inhibition of ribosome biogenesis is a conserved stress response. This dampening of translation during stress was attributed to energy conservation (Branco-Price et al., 2008), as ribosome biogenesis is highly ATP consuming. Recently, it was reported that the association of RP mRNAs with UBP1 is enhanced under hypoxia (Sorenson and Bailey-Serres, 2014). Our data suggest that the same phenomenon occurs under HS (Fig. 6). Thus, it appears that RP mRNAs are translationally repressed and stored during stress. In yeast, the same observation was reported under Glc deprivation, while adding back Glc led to RP mRNAs translation recovery (Arribere et al., 2011). Interestingly, in this case, few genes can be reactivated for translation upon Glc replenishing, and most of them were attributed to ribosomal protein genes (Arribere et al., 2011). We observed the same mechanism in Arabidopsis under HS, suggesting a common signaling pathway in regulating RP mRNAs translation across eukaryotes. In yeast, it was suggested that hsp104 translational defect is due to a defect in the release of preinitiation complexes stored in SG (Cherkasov et al., 2013). Our data go further in this process and show that the nature of stored mRNAs could also influence the translation recovery.
How to Coordinate de Novo Ribosome Synthesis after Stress
We show in this work that new ribosomes can be formed shortly after HS by mobilizing stored RP-coding mRNAs. Ribosome biogenesis needs a fine-tuning between rRNA production mediated by RNA polymerase I and RP production mediated by RNA polymerase II. Therefore, an important question is how to coordinate the timely production of rRNAs and RPs to assemble new ribosomes shortly after a stress event. Previously it was reported that, in tomato, the production of rRNA primary transcripts is maintained under HS, while the maturation process is abolished, leading to the accumulation of primary transcripts in the nucleolus (Nover et al., 1986). The same observation was recently proposed also for yeast (Kos-braun et al., 2017). The authors showed that pre-rRNA continues to be synthesized upon stress and suggested that continued transcription of rDNA allows maintenance of nucleolar integrity to rapidly respond to environmental changes. This result agrees with our observation that the nucleolus architecture (a proxy of polymerase I activity) is maintained during HS in Arabidopsis (Supplemental Fig. S6, A and B). The coordinated control during the stress of essential molecules needed to generate new ribosomes (primary rRNA production in the nucleolus and storage of RP-coding mRNAs in SGs) could be critical to synchronize ribosome synthesis in the recovery period. This hypothesis is in agreement with our data showing that RNA polymerase I and II activities are not needed to synthesize new ribosomes early in the recovery period. The rapid restart of translation, only minutes after stress, could therefore benefit from the HSP101-dependent release of RP-coding mRNAs from SGs, which is to be synchronized with the maturation of primary rRNAs in the nucleolus, leading to new ribosome formation.
Ribosomal Subunits under Stress and during Recovery
The dynamics of ribosomal subunits we observed during and after the stress support our major conclusion that RP synthesis with stored mRNAs is needed to resume translation in the recovery period. As shown in our polysome profiles (Fig. 7A), HS induces a massive dissociation of polysomes and the accumulation of monosomes and free 40S and 60S subunits. This is expected, as the ribosomal subunits released following translation termination cannot easily reengage in new rounds of protein synthesis in a stress situation. Early in the recovery period, one would expect that the reactivation of translation will increase the level of polysomes with a corresponding decrease in monosomes and free subunits. However, this is not what we observed, as the level of free subunits was stable even 1 h in the recovery period, when the polysome level is strongly increased. This phenomenon suggests that newly synthesized ribosomal subunits (and not only the preexisting ones), are contributing to the build up of polysome recovery.
Why new ribosomes were produced instead of salvaging the preexisting ones? One possibility is that part of the preexisting subunits have been damaged or irreversibly modified by the stress and cannot be easily recycled. Indeed, under stress conditions that affect energy status, alteration of ribosome subunits is one strategy that can preserve cellular energy homeostasis. It was reported that under stress conditions or after translation inhibition, modification of ribosomal subunits could occur through ubiquitination both for 40S and 60S subunits (Kraft et al., 2008; Higgins et al., 2015). It was proposed that this modification not only controls translational status under stress but also induces the ribosome degradation pathway, called ribophagy (Kraft et al., 2008). To evade HS-inducing modifications, the protein aggregation process was proposed to be a specific and reversible pathway to maintain protein activity for recovery phase (Wallace et al., 2015). This study suggests that free ribosome subunits may not be able to efficiently aggregate under HS, making them more sensitive to modification and/or degradation. This could be physiologically relevant, as the reduction in ribosome abundance was proposed to be a rapid and adaptive stress response during proteotoxic stress in yeast (Guerra-Moreno et al., 2015). Ribosomal subunit ubiquitination can occur rapidly upon stress (after 1 h dithiothreitol [DTT] for human cells; Higgins et al., 2015), whereas ribosomal subunit cleavage increases only significantly after 4 h (Kraft et al., 2008). Thus, we postulate that the same mechanisms could occur during and after HS in Arabidopsis, and one way to recover from these effects would be to store RP mRNAs in SG to enhance ribosome production during recovery phase.
Another challenging question is how RP mRNAs could be specifically targeted for this massive sequestration and release process. A common cis-element “TAGGGTTT” was previously identify on Arabidopsis RP mRNAs and was shown to be important in the context of translation efficiency during photomorphogenesis (Liu et al., 2012). However, this cis-element is also present on RP mRNAs that are apparently not stored and released following HS. Therefore, it is unclear at the moment if this cis-element is involved or not, and further effort will be needed to determine the regulators, both cis and trans, required for sequestrate and release of most RP mRNAs in a heat-stress situation.
In this study, we identified SG-associated mRNAs using RNA sequencing approach. According to our criteria, 210 of 276 nuclear RP mRNAs are stored during HS and released in the recovery period in an HSP101-dependent manner (Supplemental Table S4). This allows us to identify the functional role of stored RP mRNAs for recovery period. Moreover, another GO term was enriched in our analysis (“protein binding” GO:0005515). As this term encompasses a large group of proteins with distinct functions, it is difficult to draw a model around this term. Nonetheless, we cannot exclude that other mRNAs with specific functions are stored during HS and also play a role during recovery.
MATERIALS AND METHODS
Growth Conditions
Analyses were carried out with 5- or 7-d-old whole plantlets grown on synthetic Murashige and Skoog medium (Duchefa) containing 1% Suc and 0.8% plant agar at 22°C under a 16-h-light/8-h-dark regime. For HS treatment, plates were immerged in a water bath at 37°C. For recovery, plates were transferred at 22°C under previous conditions.
Generation of Transgenic Plants
A vector containing pPABP2-tRFP-PABP2 was transformed into wild type (Col0) by floral dip transformation. Next, a cross between this line and the hsp101 mutant (hot1-3; Lee et al., 2005) was realized to obtain pPABP2-tRFP-PABP2 in the hsp101 background. The expression level of RFP-PAB2 and HSP101 was confirmed by western-blot analysis. Transgenic lines with levels similar to wild type were kept for confocal analysis.
Polysome Profile Analysis and Quantification
Polysome profiles were performed as described previously (Merret et al., 2013). In brief, 400 mg of tissue powder were homogenized with 1.2 mL of lysis buffer (200 mm Tris-HCl, pH 9.0, 200 mm KCl, 25 mm EGTA, 35 mm MgCl2, 1% detergent mix [1% Tween 20, 1% Triton, 1% Brij35, and 1% Igepal], 1% sodium deoxycholate, 1% polyoxyethylene tridecyl ether, 5 mm DTT, 50 μg mL−1 cycloheximide, 50 μg mL−1 chloramphenicol, and 1% protease inhibitor cocktail [Sigma-Aldrich]). Crude extract was incubated 10 min on ice. After centrifugation, the supernatant was clarified with a 0.2-μm filter. Three hundred sixty microliters (=120 mg) of crude extract was loaded on a 15% to 60% Suc gradient (9 mL). Ultracentrifugation was performed with an SW41 rotor at 38,000 rpm (∼180,000g) for 3 h. Polysome profile analyses were performed with an ISCO absorbance detector at 254 nm. For normalization and direct comparison of profiles, the same volume of crude extract was loaded on the Suc gradient for each condition. For quantification, the percentage of polysomes for each condition was determined using Image J as the ratio between polysomes area and total area (including 40S, 60S, monosome, and polysome areas).
Confocal Microscopy Analysis and SG Quantification
Confocal microscopy was performed on 5-d-old whole seedlings as described in Merret et al. (2015). For SG monitoring, at least five different roots for each condition were analyzed. For nucleoli analysis, FIB2-GFP line was used (Pontvianne et al., 2013). Twenty-five nucleoli were analyzed for each condition on five different seedlings. SGs were quantified using plug-ins of Fidji (http://fiji.sc/; Schindelin et al., 2012) on five seedling roots per condition. First, 20 iterations of deconvolution were applied using Iterative Deconvolve 3D on 15-slide (overlapping) stacks of 5.46-μm thick. Centre of granules were then counted using 3D Objects Counter with a threshold of 1000, excluding signal from structures smaller than 50 voxels.
RNA Sequencing
Total RNA extraction was performed by using RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. For polysomal RNA extraction, fractions corresponding to polysomes were pooled. Two volumes of 8 m guanidium and three volumes of absolute ethanol were added. After overnight precipitation, centrifugation (16,000g, 45 min, 4°C) was performed. Pellets were resuspended in RLT buffer (Qiagen). Next, RNA was extracted with RNeasy Micro Kit. RNA quality was assessed with Agilent 2100 bioanalyzer prior to RNA sequencing. Libraries preparation was performed with TrueSeq Illumina kit. Libraries were multiplexed and sequenced (PE 2×150) on an Illumina HiSeq 2500. For each library, at least 40 million reads were obtained. Reads filtering and mapping were performed as described in Merret et al. (2015). For each condition, the mean value of RPKM was determined between replicates. Transcripts, which did not have at least 1 RPKM in one condition, were removed from the analysis. FC was determined as the ratio between transcript quantity during HS and normal condition or during recovery and normal condition. TE was determined as the ratio between transcript quantity in polysomal RNA and total RNA. Raw data are reported in Supplemental Table S1.
GO and Heat Map Analysis
The GO analyses were performed with method available at the AgriGO website (http://bioinfo.cau.edu.cn/agriGO/; Du et al., 2010), with the following parameters: hypergeometric statistical test method, Yekutieli (FDR under dependency) multitest adjustment method, and a 0.05 significance level. The heat map was obtained using heatmap.2 function of gplots R package (https://cran.r-project.org/web/packages/gplots/index.html) using Pearson distance and an average method for the hierarchical clustering.
qPCR Validation
For qPCR validation, 500 ng of total or polysomal RNA was reverse-transcribed with SuperScript IV Kit (Life Technology) using oligo dT18. Real-time PCR was performed in an LC 480 384-well thermocycler. The following program was used: 5 min 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. The PCR mix contained Takyon qPCR master mix (Eurogentec), 500 nm gene-specific primers, and 1.8 μL cDNA in a total volume of 9 μL. Primer efficiencies were determined on standard curves. Primers used in this study are presented in Supplemental Table S5.
RNA Coimmunoprecipitation
RNA coimmunoprecipitation was performed as described previously using the UBP1a-GFP line or Col0 line as a negative control (Sorenson and Bailey-Serres, 2014). In brief, 400 mg of tissue powder were incubated in 3 mL of lysis buffer (200 mm Tris, pH 9.0, 110 mm potassium acetate, 0.5% Triton X-100, 0.1% Tween 20, 5 mm DTT, and 1.5% protease inhibitor). Lysate was clarified using Miracloth (Millipore) layer and centrifuged at 1,500g during 2 min at 4°C, and 1.5 mL of crude extract was incubated with 15 μL of GFP-trap beads (ChromoTek) during 1.5 h at 4°C under rotation. Crude extract (0.75 mL) was incubated with 7.5 μL of GFP-trap beads for western-blot analysis. Beads were washed five times with 0.75 mL of lysis buffer. For RIP experiments, elution was performed with 200 μL of 8 m guanidium for 5 min on ice and precipitated overnight with 300 μL of 100% ethanol before RNA extraction as described above. For western blot, elution was performed by adding 20 μL of Laemmli 2× to beads and incubated 5 min at 95°C. Reverse transcription was performed on 10 μL of eluate or 500 ng of input as described above. One microliter of cDNA was used for PCR amplification with specific primers (Supplemental Table S5).
15N Incorporation and Elemental Analysis
After heat treatment (as described above), seedlings were transferred in a liquid medium containing 1 mm KH2PO4, 50 μm FeNaEDTA, 0.5 mm MgCl2, 0.5 mm CaCl2 2H2O, and 10 mm KNO3. For 15N incorporation, KNO3 was replaced by 99% K15NO3. Incorporation was performed during 2 h recovery. For transcription inhibition, actinomycin D was added to the medium at 0.2 mg mL−1 during recovery. After incorporation, polysomes were extracted as described previously. Polysomal proteins were precipitated 6 h at 4°C by adding two volumes of 100% ethanol. After centrifugation, pellets were dried at 70°C for 48 h and analyzed for total N content and atom % 15N abundance by continuous-flow mass spectrometry, using a Euro-EA Elemental Analyzer (Eurovector SPA) coupled with an IsoPrime mass spectrometer (Isoprime).
Accession Numbers
The accession number for the RNA-seq data reported in this article is NCBI Bioproject PRJNA345276.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Polysome profiles after 2 h of recovery in wild type, dlt1-1, hsp101, and dlt1-2.
Supplemental Figure S2. Western-blot analysis of RFP-PABP2 lines.
Supplemental Figure S3. Log2 value of the 3309 mRNAs in control and after 1 h at 37°C in wild type versus hsp101.
Supplemental Figure S4. GO analysis of mRNAs poorly translated during HS and affected in translation recovery in hsp101 (2103/3309).
Supplemental Figure S5. Translation efficiency of chloroplastic and mitochondrial RP mRNAs in wild type and hsp101 under normal condition, after 1 h at 37°C, and after 2 h recovery.
Supplemental Figure S6. Heat stress did not affect nucleoli architecture.
Supplemental Table S1. RNA sequencing raw data.
Supplemental Table S2. Transcripts stable and released from translation during HS in wild type and hsp101 (3309).
Supplemental Table S3. Transcripts affected in translation recovery in hsp101 (2103).
Supplemental Table S4. Translation efficiency of ribosomal protein mRNAs.
Supplemental Table S5. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Academia Sinica NGS Core Facility for library preparation and RNA sequencing. We thank Dr. Frédéric Pontvianne and Prof. Julia Bailey-Serres for kindly giving us FIB2-YFP and UBP1a-GFP lines, respectively.
Glossary
- HS
heat shock
- HSP
heat shock protein
- SG
stress granule
- RIP
RNA coimmunoprecipitation
- RP
ribosomal protein
- FC
fold change
- TE
translation efficiency
Footnotes
This article was supported Ministry of Science and Technology, Taiwan, ROC (MOST 103-2311-B-001-011-MY3 and 104-2923-B-001-001-MY3 to Y.-y.C.), Agence Nationale de la Recherche (ANR-2010 BLANC1707-01 and ANR-14-CE10-0015 to J.-M.D.). R.M. was the recipient of an Academia Sinica Postdoctoral Research Fellowship. This work was supported by CNRS and UVPD through utilization of confocal microscope and qPCR devices at the Technoviv platform.
Articles can be viewed without a subscription.
References
- Arribere JA, Doudna JA, Gilbert WV (2011) Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol Cell 44: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. (2011) Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2: 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Buchan JR. (2014) mRNP granules. Assembly, function, and connections with disease. RNA Biol 11: 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–2462 [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ (2000) Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev 14: 1460–1470 [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Moreno A, Isasa M, Bhanu MK, Waterman DP, Eapen VV, Gygi SP, Hanna J (2015) Proteomic analysis identifies ribosome reduction as an effective proteotoxic stress response. J Biol Chem 290: 29695–29706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TL, Stoneley M, Spriggs KA, Bushell M (2006) TOPs and their regulation. Biochem Soc Trans 34: 12–16 [DOI] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. (2015) The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol Cell 59: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Bailey-Serres J (2012) Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kos-braun IC, Jung I, Kos M (2017) Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. Plos Biol 15: e2000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kuhn KM, DeRisi JL, Brown PO, Sarnow P (2001) Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol 21: 916–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Escobar M, Williams B, Hong SW, Vierling E (2005) Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell 17: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB (2013) Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell 49: 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Chen H-M, Wu S-H (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F, Basha E, Fowler ME, Kim M, Bordowitz J, Katiyar-Agarwal S, Vierling E (2016) Class I and II small heat-shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiol 172: 1221–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Pnueli L, Arava Y (2008) Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. RNA 14: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Reports 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Merret R, Nagarajan VK, Carpentier MC, Park S, Favory JJ, Descombin J, Picart C, Charng YY, Green PJ, Deragon JM, et al. (2015) Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res 43: 4121–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï M, Roncato MA, Canoy AS, Rouquié D, Sarda X, Freyssinet G, Robaglia C (2006) Large-scale analysis of mRNA translation states during sucrose starvation in arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol 141: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Munsche D, Neumann D, Ohme K, Scharf KD (1986) Control of ribosome biosynthesis in plant cell cultures under heat-shock conditions. Ribosomal RNA. Eur J Biochem 160: 297–304 [DOI] [PubMed] [Google Scholar]
- Park S-H, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim Y-K, Do Choi Y, Kim J-K (2012) Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol 159: 1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C, Carrasco-López C, Catalá R, Turečková V, Novak O, Zhang W, Sieburth L, Jiménez-Gómez JM, Salinas J (2016) The LSM1-7 complex differentially regulates Arabidopsis tolerance to abiotic stress conditions by promoting selective mRNA decapping. Plant Cell 28: 505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart C, Pontvianne F (2016) Plant nucleolar DNA: Green light shed on the role of Nucleolin in genome organization. Nucleus 8: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgová I, Hassel C, Pontes OMF, Tucker S, Mokroš P, Muchová V, Fajkus J, et al. (2013) Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 27: 1545–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckette M, Iyer NJ, Tang Y, Dai XB, Zhao P, Mahalingam R (2012) Differential mRNA translation in Medicago truncatula accessions with contrasting responses to ozone-induced oxidative stress. Mol Plant 5: 187–204 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 111: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Matsuura H, Yamaguchi M, Demura T, Kato K (2012) Genome-wide analyses of changes in translation state caused by elevated temperature in Oryza sativa. Plant Cell Physiol 53: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Parker R (2015) Coupling of ribostasis and proteostasis: Hsp70 proteins in mRNA metabolism. Trends Biochem Sci 40: 552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Wu T-Y, Juan Y-T, Hsu Y-H, Wu S-H, Liao H-T, Fung RWM, Charng Y-Y (2013) Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol 161: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua N-H (2009) Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang J-Y, Niu Q-W, Chua N-H (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.