Abstract
We have employed whole genome sequencing to define and evaluate a core genome multilocus sequence typing (cgMLST) scheme for Acinetobacter baumannii. To define a core genome we downloaded a total of 1,573 putative A. baumannii genomes from NCBI as well as representative isolates belonging to the eight previously described international A. baumannii clonal lineages. The core genome was then employed against a total of fifty-three carbapenem-resistant A. baumannii isolates that were previously typed by PFGE and linked to hospital outbreaks in eight German cities. We defined a core genome of 2,390 genes of which an average 98.4% were called successfully from 1,339 A. baumannii genomes, while Acinetobacter nosocomialis, Acinetobacter pittii, and Acinetobacter calcoaceticus resulted in 71.2%, 33.3%, and 23.2% good targets, respectively. When tested against the previously identified outbreak strains, we found good correlation between PFGE and cgMLST clustering, with 0–8 allelic differences within a pulsotype, and 40–2,166 differences between pulsotypes. The highest number of allelic differences was between the isolates representing the international clones. This typing scheme was highly discriminatory and identified separate A. baumannii outbreaks. Moreover, because a standardised cgMLST nomenclature is used, the system will allow inter-laboratory exchange of data.
Introduction
Acinetobacter baumannii is now a recognised serious nosocomial pathogen and is isolated frequently particularly in intensive care unit settings where it is a cause of serious infections such as ventilator-associated pneumonia, wound and bloodstream infections [1]. It affects mainly severely debilitated patients and is typically selected by prior antimicrobial therapy [2]. A. baumannii shares several characteristics with methicillin-resistant Staphylococcus aureus (MRSA) such as multidrug resistance and long-term survival on inanimate surfaces such as computer keyboards, pillows, curtains and other dry surfaces. [2–4]. This longevity contributes to hospital outbreaks and the clonal spread of isolates, and it facilitates person-to-person transmission and environmental contamination. Strict adherence to infection control measures and sometimes even the closure of wards is required for the control of outbreaks [5].
Many typing methods have been used to investigate outbreaks of A. baumannii, and its clonal population structure is now well-established. Molecular typing of isolates obtained from various locations in the EU has shown the existence of three distinct clonal lineages that were termed pan-European clonal complexes I, II and III [6,7]. Rep-PCR (DiversiLab) has shown that these lineages are no longer restricted to Europe and that there exist at least eight distinct international clonal lineages that are particularly associated with carbapenem-resistance [8]. Furthermore, multilocus sequence typing (MLST) has corroborated these data [9–11].
Pulsed-field gel electrophoresis (PFGE) is still considered the gold standard for outbreak analysis and although the method has been standardized, there is still the problem of reproducibility and portability of these data outside of reference laboratories [12–14]. Similarly, with rep-PCR we demonstrated that overlaying data generated from different centres is not reliable [15]. Perhaps the only truly portable established method is MLST, but this lacks the resolution for outbreak investigations [16].
With the advent of relatively cheap whole genome sequencing (WGS), there is now the possibility of comparing whole genomes and not relying on a few loci for typing purposes. There have been several studies that have addressed this by either comparing isolates on single nucleotide variants (SNVs) or genome-wide gene-by-gene allelic profiling, which is now termed core genome MLST (cgMLST) [17–20]. One barrier to the ready adoption of WGS for routine typing is the data analysis, which can be difficult for the non-bioinformatician. Therefore the development of user-friendly software is expected to greatly enhance the adoption of WGS [21,22].
The objective of this study was to establish a cgMLST scheme for A. baumannii that can form the basis of a standardised nomenclature for typing this organism. To enable this, we first defined a core genome gene set that represented the genetic diversity of A. baumannii based on well-characterised reference strains from our collection and those available online. We then challenged the scheme against data available from NCBI. Well-defined outbreaks were sequenced and analysed to determine the scheme’s suitability for outbreak investigations.
Material and methods
Bacterial isolates and DNA extraction
For scheme calibration, a total of 53 carbapenem-susceptible and -resistant A. baumannii isolates from well-described hospital outbreaks occurring throughout Germany were used, some of which have been the subject of a previous report (Table 1) [23]. The isolates had previously been assigned to international clones (IC) 1, 2, 4, 7, and 8 using rep-PCR, and their carbapenem resistance mechanisms determined by PCR [8,11,24]. Two isolates were not considered as clustering with the ICs. For the purpose of this study, an outbreak was defined as two or more isolates from a given hospital and from patients that were linked in time and space that had highly similar or identical PFGE patterns as had been determined previously. PFGE subtypes were defined as having 1–3 band differences. In some cases, there was more than one circulating clone in the same hospital. Isolates were routinely grown on blood agar and after overnight growth in LB liquid media, DNA was extracted using the MagAttract HMW DNA isolation kit following the manufacturer’s instructions (Qiagen, Hilden, Germany) and quantified using the Qubit dsDNA BR assay (Fisher Scientific GmbH, Schwerte, Germany).
Table 1. A. baumannii strains used for A. baumannii cgMLST cluster calibration.
| Cluster | Strain | Hospital | City of Isolation | Year of Isolation | PFGE Typea | ST (‘Oxford’/ ‘Pasteur’) | Clonal Lineageb | BAPS Partition | Carbapenem-resistance Determinant | Coverage (Assembled) | Contig Count (Assembled) | % good cgMLST Genes | ENA Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | UKK_0008 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 108 | 112 | 99.8 | ERS1042999 |
| I | UKK_0012 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 98 | 107 | 99.8 | ERS1043003 |
| I | UKK_0016 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 99 | 160 | 99.7 | ERS1043004 |
| I | UKK_0024 | 3 | Berlin | 2006 | not done | 436/2 | IC2 | 2 | OXA-23 | 114 | 130 | 99.7 | ERS1043005 |
| I | UKK_0031 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 116 | 102 | 99.8 | ERS1043006 |
| I | UKK_0038 | 3 | Berlin | 2006 | 2-A2 | 436/2 | IC2 | 2 | OXA-23 | 109 | 99 | 99.8 | ERS1043007 |
| I | UKK_0047 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 94 | 145 | 99.8 | ERS1043008 |
| I | UKK_0052 | 3 | Berlin | 2006 | 2-A1 | 436/2 | IC2 | 2 | OXA-23 | 74 | 123 | 99.8 | ERS1043009 |
| II | UKK_0252 | 8 | Leverkusen | 2011 | 4-A | 218/2 | IC2 | 2 | OXA-23 | 56 | 71 | 99.8 | ERS1043096 |
| II | UKK_0254 | 8 | Leverkusen | 2011 | 4-A | 218/2 | IC2 | 2 | OXA-23 | 104 | 121 | 99.7 | ERS1043128 |
| II | UKK_0260 | 8 | Leverkusen | 2011 | 4-A | 218/2 | IC2 | 2 | OXA-23 | 116 | 178 | 99.8 | ERS1043151 |
| II | UKK_0263 | 8 | Leverkusen | 2011 | 4-A | 218/2 | IC2 | 2 | OXA-23 | 37 | 75 | 99.7 | ERS1043152 |
| II | UKK_0266 | 8 | Leverkusen | 2012 | 4-A | 218/2 | IC2 | 2 | OXA-23 | 92 | 67 | 99.9 | ERS1043178 |
| III | UKK_0253 | 8 | Leverkusen | 2011 | 4-B | 195/2 | IC2 | 2 | OXA-23 | 118 | 157 | 99.7 | ERS1043117 |
| III | UKK_0255 | 8 | Leverkusen | 2011 | 4-B | 195/2 | IC2 | 2 | OXA-23 | 119 | 108 | 99.9 | ERS1043139 |
| III | UKK_0256 | 8 | Leverkusen | 2011 | 4-B | 195/2 | IC2 | 2 | OXA-23 | 119 | 183 | 99.7 | ERS1043150 |
| III | UKK_0264 | 8 | Leverkusen | 2011 | 4-B | 195/2 | IC2 | 2 | OXA-23 | 73 | 140 | 99.7 | ERS1043177 |
| IV | UKK_0173 | 5 | Cologne | 2011 | 6A | 558/2 | IC2 | 2 | no | 119 | 120 | 99.6 | ERS1043040 |
| IV | UKK_0174 | 5 | Cologne | 2011 | 6A | 558/2 | IC2 | 2 | no | 113 | 104 | 99.6 | ERS1043042 |
| IV | UKK_0177 | 5 | Cologne | 2011 | 6A | 558/2 | IC2 | 2 | no | 117 | 159 | 99.6 | ERS1043043 |
| V | UKK_0178 | 5 | Cologne | 2011 | 6-B | 448/2 | IC2 | admixture | OXA-23 | 102 | 178 | 99.3 | ERS1043044 |
| V | UKK_0179 | 5 | Cologne | 2011 | 6-B | 448/2 | IC2 | admixture | OXA-23 | 79 | 159 | 99.2 | ERS1043045 |
| V | UKK_0180 | 5 | Cologne | 2011 | 6-B | 448/2 | IC2 | admixture | OXA-23 | 116 | 276 | 99.2 | ERS1043057 |
| VI | UKK_0182 | 6 | Cologne | 2011 | 7-A1 | 448/2 | IC2 | admixture | OXA-23 | 123 | 147 | 99.3 | ERS1043059 |
| VI | UKK_0183 | 6 | Cologne | 2011 | 7-A2 | 448/2 | IC2 | admixture | OXA-23 | 124 | 156 | 99.0 | ERS1043089 |
| VI | UKK_0185 | 6 | Cologne | 2011 | 7-A1 | 448/2 | IC2 | admixture | OXA-23 | 124 | 142 | 99.2 | ERS1043090 |
| VI | UKK_0193 | 6 | Cologne | 2011 | 7-A2 | 448/2 | IC2 | admixture | OXA-23 | 122 | 123 | 99.2 | ERS1043092 |
| VI | UKK_0196 | 6 | Cologne | 2011 | 7-A2 | 448/2 | IC2 | admixture | OXA-23 | 98 | 153 | 99.2 | ERS1043093 |
| VII | UKK_0056 | 2 | Berlin | 2007 | 1-B | 350/2 | IC2 | 2 | OXA-58 | 69 | 70 | 99.9 | ERS1043010 |
| VII | UKK_0057 | 2 | Berlin | 2007 | 1-B | 350/2 | IC2 | 2 | OXA-58 | 67 | 78 | 99.9 | ERS1043029 |
| VII | UKK_0058 | 2 | Berlin | 2007 | 1-B | 350/2 | IC2 | 2 | OXA-58 | 64 | 75 | 99.9 | ERS1043030 |
| VIII | UKK_0004 | 3 | Berlin | 2005 | 2-B | 438/15 | IC4 | 4 | OXA-58 | 116 | 95 | 99.4 | ERS1042932 |
| VIII | UKK_0005 | 3 | Berlin | 2005 | 2-B | 438/15 | IC4 | 4 | OXA-58 | 111 | 118 | 99.3 | ERS1042933 |
| VIII | UKK_0006 | 3 | Berlin | 2005 | 2-B | 438/15 | IC4 | 4 | OXA-58 | 114 | 142 | 99.2 | ERS1042934 |
| VIII | UKK_0007 | 3 | Berlin | 2005 | 2-B | 438/15 | IC4 | 4 | OXA-58 | 116 | 124 | 99.1 | ERS1042935 |
| IX | UKK_0412 | 9 | Ludwigshafen | 2013 | 8-A | 933/94 | IC1 | not doned | OXA-23 | 117 | 104 | 99.1 | ERS1043424 |
| IX | UKK_0413 | 9 | Ludwigshafen | 2013 | 8-A | 933/94 | IC1 | not done | OXA-23 | 57 | 150 | 99.2 | ERS1043425 |
| IX | UKK_0414 | 9 | Ludwigshafen | 2013 | 8-A | 933/94 | IC1 | not done | OXA-23 | 104 | 155 | 99.1 | ERS1043426 |
| IX | UKK_0415 | 9 | Ludwigshafen | 2014 | 8-A | 933/94 | IC1 | 1C | OXA-23 | 101 | 173 | 99.1 | ERS1043427 |
| X | UKK_0283 | 1 | Aachen | 2010 | 9-A1 | 229/25 | IC7 | 1A | OXA-23 | 59 | 141 | 99.2 | ERS1043179 |
| X | UKK_0284 | 1 | Aachen | 2010 | 9-A1 | 229/25 | IC7 | 1A | OXA-23 | 113 | 80 | 99.2 | ERS1043215 |
| X | UKK_0285 | 1 | Aachen | 2011 | 9-A2 | 229/25 | IC7 | 1A | OXA-23 | 104 | 96 | 99.2 | ERS1043216 |
| X | UKK_0286 | 1 | Aachen | 2011 | 9-A2 | 229/25 | IC7 | 1A | OXA-23 | 58 | 186 | 99.1 | ERS1043217 |
| X | UKK_0290 | 1 | Aachen | 2011 | not done | 229/25 | IC7 | 1A | OXA-23 | 115 | 100 | 98.2 | ERS1043218 |
| X | UKK_0295 | 1 | Aachen | 2011 | not done | 229/25 | IC7 | 1A | OXA-23 | 98 | 91 | 99.2 | ERS1043219 |
| XI | UKK_0386 | 4 | Bonn | 2013 | 10-A | 229/25 | IC7 | 1A | OXA-23 | 110 | 125 | 99.0 | ERS1043422 |
| XI | UKK_0409 | 4 | Bonn | 2013 | 10-A | 229/25 | IC7 | 1A | OXA-23 | 93 | 79 | 99.1 | ERS1043423 |
| XII | UKK_0306 | 9 | Ludwigshafen | 2012 | 8-B1 | 1118/136 | uncc | not done | no | 58 | 90 | 99.0 | ERS1043220 |
| XII | UKK_0318 | 9 | Ludwigshafen | 2012 | 8-B2 | 1118/136 | unc | not done | no | 95 | 333 | 99.0 | ERS1043221 |
| XIII | UKK_0333 | 7 | Cologne | 2013 | 11-A | 391/157 | IC8 | 1C | OXA-23 | 68 | 194 | 98.8 | ERS1043342 |
| XIII | UKK_0334 | 7 | Cologne | 2012 | 11-A | 391/157 | IC8 | 1C | OXA-23 | 59 | 93 | 98.8 | ERS1043375 |
| XIII | UKK_0368 | 7 | Cologne | 2013 | 11-A | 391/157 | IC8 | 1C | OXA-23 | 53 | 103 | 98.7 | ERS1043384 |
| XIII | UKK_0369 | 7 | Cologne | 2013 | 11-A | 391/157 | IC8 | 1C | OXA-23 | 87 | 100 | 98.7 | ERS1043385 |
aPFGE types: The first numeral indicates the hospital, the letter is for pulsotype which is specific to that hospital, and in some cases there is a second numeral for subtype.
bClonal lineage based on rep-PCR clustering.
cunc; unclassified as it did not cluster with one of the ICs.
dSTs above ST920 were not available when BAPS analysis was performed, therefore not included in BAPS partitioning.
Whole genome sequencing and assembly
Sequencing libraries were prepared using the Nextera XT library prep kit (Illumina GmbH, Munich, Germany) for a 250bp paired-end sequencing run on an Illumina MiSeq sequencer. Samples were sequenced to aim for a minimum 100-fold coverage using Illumina’s recommended standard protocols with dual-index barcoding and rotation of barcodes over time. Sequencing run quality (Q30 and output) had to fulfill the manufacturer’s minimum specifications. The resulting FASTQ files were quality trimmed and assembled de novo using the Velvet assembler that is integrated in Ridom SeqSphere+ v.3.0 software (Ridom GmbH, Münster, Germany) [25]. Here, reads were trimmed at their 5'- and 3'-ends until an average base quality of 30 was reached in a window of 20 bases, and the assembly was performed with Velvet version 1.1.04 [26] using optimized k-mer size and coverage cutoff values based on the average length of contigs with > 1000 bp.
BAPS
To determine the overall A. baumannii species variation, we applied a Bayesian analysis of population structure (BAPS) [27] with the more discriminatory MLST scheme described by Bartual et al [28]. All MLST profiles available as of March 25th 2015 (913 sequence types [ST]) were downloaded from the PubMLST web site (http://pubmlst.org/abaumannii/), all allelic gene sequences per locus were multiple aligned using MUSCLE [29] and finally concatenated for each ST. The BAPS analysis was carried out using the clustering of linked molecular data functionality. Ten runs were performed setting an upper limit of 20 partitions. Admixture analysis was performed using the following parameters: minimum population size considered 5, iterations 50, number of reference individuals simulated from each population 50, number of iterations for each reference individual 10. Those STs that had significant (P< 0.05) admixture were not assigned to a partition. The aligned and concatenated ST sequences were used to produce a maximum likelihood (ML) tree using FastTree 2 [30]. Finally, the assignment of sequence types to BAPS partitions was visualized by coloring the nodes (representing the individual STs) of the radial phylogram calculated with FastTree 2 that was drawn by Dendroscope 3 [31]. The largest resulting partition was further subdivided by visual inspection of the phylogram.
cgMLST target gene definition
To determine the cgMLST gene set, a genome-wide gene-by-gene comparison was performed using the cgMLST target definer (version 1.1) function of the SeqSphere+ (Ridom GmbH, Münster, Germany) software with default parameters as described previously [17]. The ACICU strain served as reference genome (NC_010611.1, dated 12 August 2015) [32]. BLAST version 2.2.12 was used for pairwise comparison with the A. baumannii query genomes (Table 2).
Table 2. List of A. baumannii strains and genomes used for cgMLST A. baumannii target definition.
| Strain | Lineage | BAPS Partition | NCBI Genome Status | ST ‘Oxford’ | ST ‘Pasteur’ | Avg. Coverage (Assembled) | NCBI/ENA Accession |
|---|---|---|---|---|---|---|---|
| ACICU (Reference) | IC2 | 2 | Complete Genome | 437 | 2 | n/a | NC_010611.1 |
| AYE | IC1 | 4 | Complete Genome | 231 | 1 | n/a | NC_010410.1 |
| AC30 | IC2 | 2 | Complete Genome | 195 | 2 | 245.0 | CP007577.1 |
| NIPH 1669 | IC3 | 4 | Scaffold | 106 | 3 | 139.0 | APOQ01 |
| UKK_0004 | IC4 | 4 | This study | 438 | 15 | 113.0 | ERS1042932 |
| TG02011 | IC5 | 1C | Contig | 205 | 79 | 79.0 | ASES01 |
| BMBF-448 | IC6 | not donea | This study | 944 | 78 | 76.0 | ERS1047685 |
| 1429530 | IC7 | 1A | Contig | 229 | 25 | 113.3 | JEWM01 |
| LAC-4 | IC8 | 4 | Complete Genome | 447 | 10 | 116.0 | NZ_CP007712.1 |
| NIPH 601 | n/a | 1B | Scaffold | 373 | 40 | 86.0 | APQZ01 |
| AA-014 | n/a | 1D | Contig | 499 | 158 | 63.9 | AMGA01 |
| 6013150 | n/a | 1E | Scaffold | 498 | 81 | 59.8 | ACYQ02 |
| 268680 | n/a | 6 | Contig | 355 | 16 | 95.2 | JEYN01 |
n/a: not available
aSTs above ST920 were not available when BAPS analysis was performed, therefore not included in BAPS partitioning
Evaluation and calibration of the cgMLST target gene set
To evaluate the newly defined cgMLST scheme, all available A. baumannii NCBI genome datasets (as of 2016-08-29) were downloaded, analyzed with the cgMLST and the ‘Oxford’ and ‘Pasteur’ MLST schemes, and filtered by ST. Also NCBI data that were used as reference or query genomes for scheme definition were removed. It was assumed that a suitable cgMLST scheme should reach on average at least 97.5% cgMLST called targets for all of those quality-filtered genomes. In addition, genome data for the three closely related species from the ACB complex, i.e. A. nosocomialis, A. pittii, and A. calcoaceticus were added for demonstration of the applicability of the defined cgMLST scheme for A. baumannii sensu stricto only.
To further calibrate the A. baumannii cgMLST scheme to investigate outbreaks, 53 sequenced carbapenem-resistant isolates were analysed using Ridom SeqSphere+ software to determine the presence of the target genes. Again we assumed that a well-defined core genome would cover at least 97.5% of the cgMLST genes used in this scheme. The target genes were extracted as previously described with “required identity to reference sequence of 90%”, “required aligned to reference sequence with 100%” [17] and the process included an assessment of the quality of the target genes, i.e. the absence of frame shifts and ambiguous nucleotides. A core genome gene was considered a “good target” only if all of the above criteria were met, in which case the complete sequence was analyzed in comparison to the ACICU reference. Alleles for each gene were called and assigned automatically by the SeqSphere+ software to ensure unique nomenclature. The combination of all alleles in each strain formed an allelic profile that was used to generate minimum spanning trees (MST) using the parameter “pairwise ignore missing values” during distance calculation. The MST was used to determine if outbreak isolates could be attributed to the same cluster and clearly separated from other clusters. To maintain backwards compatibility, classical MLST alleles (Oxford and Pasteur schemes) were extracted from the assembled genomes with SeqSphere+ using the PubMLST nomenclature (http://pubmlst.org/).
Results
Species variability was checked by BAPS analysis based on 913 ‘Oxford‘ MLST ST which resulted in 8 partitions with ST 262 being the only member of BAPS partition 8. From the remaining 912 analysed STs, 312 showed significant admixture (P> 0.05) and were removed from the analysis, and 600 STs could be grouped into 7 partitions with a probability of ≥ 0.95 (S1 Table). Members of the largest partition 1 were located at five distinct branches of the ML tree, therefore this partition was further subdivided manually into five subgroups according to the branching of the tree (Partition 1A – 1E) (S1 Fig). For BAPS partitions 5 and 7 no strains, genomic data, or taxonomic information were available neither from NCBI or PubMLST. Strain data of BAPS partitions 3 and 8 were not considered for core genome genes definition because all known isolates of these groups were taxonomically identified as A. nosocomialis. Members from the remaining BAPS partitions and eight international clone lineages were included as query genomes for core genome definition (Table 2).
Based on these data the cgMLST Target Definer created a cgMLST scheme comprising 2,390 targets of the ACICU reference genome (58.6% of the complete genome) (S2 Table). A total of 1,573 A. baumannii datasets could be downloaded from NCBI Genomes and used. Genome data for which no MLST ST (‘Oxford’ and/or ‘Pasteur’) could be extracted were removed. Furthermore, the ST information was used to remove wrongly as A. baumannii identified genomes (e.g., A. nosocomialis). Thereby, in total 1,339 A. baumannii genomes were used to challenge the newly defined cgMLST scheme. On average 98.4% cgMLST genes were called successfully for those genomes. Analysis of datasets from the closely related species A. nosocomialis, A. pittii, and A. calcoaceticus resulted in 71.2%, 33.3%, and 23.2% good targets, respectively, thus demonstrating the applicability of the defined cgMLST scheme for A. baumannii sensu stricto only (S3 Table).
Fifty-three A. baumannii genomes were sequenced from isolates which were collected from patients hospitalized in nine hospitals located in eight German cities between 2005–2013. A summary of the genome assembly is shown in Table 1. Average coverage ranged from 37- to 124-fold, with a median of 102-fold. The number of contigs ranged from 67–333, with a median of 123. The percentage of good targets based on the core genome ranged from 98.7%−99.9% with a median of 99.3%.
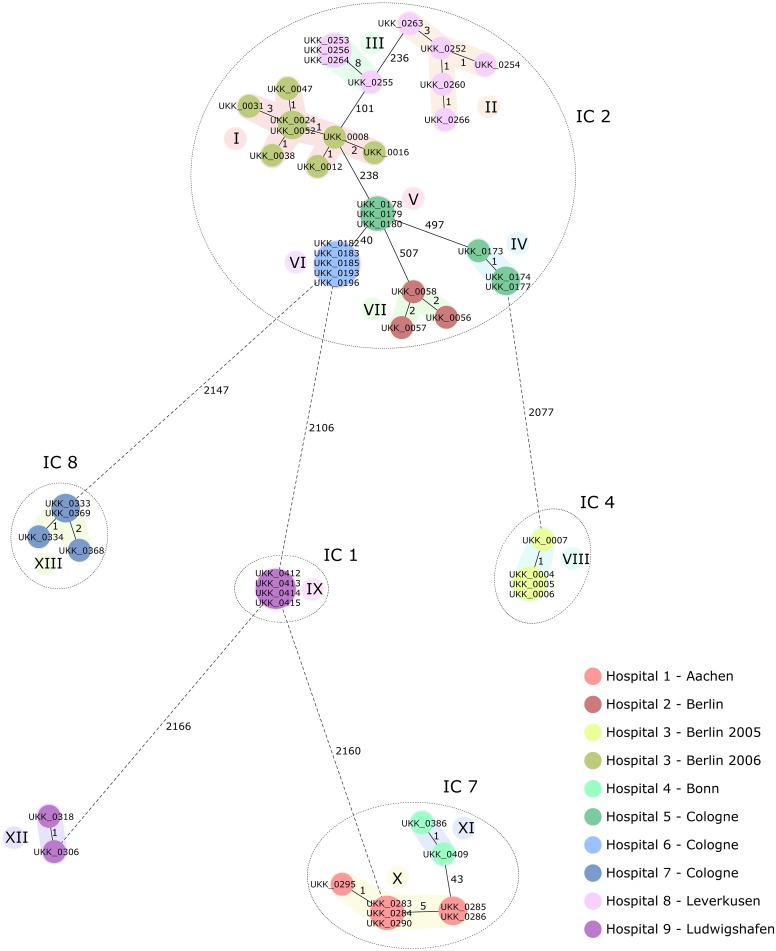
There was a good correlation between PFGE types and cgMLST clusters. Based on their cgMLST profiles (S4 Table), a minimum spanning tree was generated (Fig 1). Fifty-three isolates were grouped into 13 distinct clusters (I to XIII, Table 1). Within a cluster, there were several examples where isolates were identical within the same hospital outbreak (clusters V, VI and IX), ≤ 5 differences (clusters I, II, IV, VII, VIII, XI, XII, XIII) and ≥ 5 differences (III andX,). Clusters IV and V contained isolates from the same hospital that were isolated within a 5-week period. However, although they shared the same Pasteur sequence type (ST2) and were double locus variants (DLV) using the Oxford scheme, they differed in 497 alleles. The number of allelic differences did not correlate with length of outbreak. For example isolate UKK_0255 from cluster III was collected on the same day as UKK_0253 and UKK_0256 but differed by 8 alleles, while cluster VI isolates were collected over a 5-week period and were identical.
Fig 1. Minimum spanning tree based on cgMLST allelic profiles of 53 A. baumannii isolates.
Each circle represents an allelic profile based on sequence analysis of 2,390 cgMLST target genes. The numbers on the connecting lines illustrate the numbers of target genes with different alleles. Colours of the circles represent the different isolation sources. Closely related genotypes (≤10 alleles difference) are shaded and clusters are numbered consecutively (I to XIII). Isolates belonging to the same IC lineage are surrounded by dotted circles.
The cgMLST clustering also matched the classification of international clones as previously determined by DiversiLab typing or MLST. The investigated outbreaks included strains that we determined to belong to IC1, IC2, IC4, IC7, IC8, and one set of strains that were not part of these clonal lineages (Fig 1). Clusters I-VII had up to 507 differences within IC2, but had >2000 differences to clusters belonging to other lineages.
Discussion
For the thorough investigation of hospital outbreaks of bacterial infections, especially in the globalised society we now live in, simple, accurate and portable typing methods are essential. Thus, the ability to determine clonality among bacterial strains and to share this information in a centralised database has many advantages. MLST can be considered the proof of principle of a sequence-based method with a curated and standardised nomenclature [33]. However classical MLST does not have the resolution to determine person-person spread [16]. More discriminatory DNA fingerprinting methods such as PFGE and rep-PCR (DiversiLab) are not always comparable between different laboratories [15]. The use of WGS and user-friendly software means that whole genomes can be sequenced and compared within a few working days [17]. In our present study, we demonstrated that our A. baumannii cgMLST scheme was able to distinguish different outbreaks from the same hospital which shared the same or similar MLST profiles. An additional result was that we were clearly able to distinguish between different international A. baumannii clones.
Although there are now a plethora of sequenced A. baumannii genomes, few outbreaks have been investigated by WGS, and where applied to outbreak analysis, all the studies have performed SNV analysis, whereas our approach was allele-based [34–37]. In all of these cases, the SNV approach was compared to PFGE and was found to be generally in accordance with a few exceptions. For example, Salipante et al reported discrepancies between PFGE and WGS [36]. Halachev et al. applied SNV analysis during a prolonged A. baumannii outbreak and was able to differentiate between outbreak and non-outbreak strains and when combined with epidemiological data was able to reconstruct potential transmissions [37]. The ability to differentiate between different A. baumannii clonal lineages was also investigated and a threshold of 2,500 core SNPs was determined to accurately distinguish isolates from different clonal lineages [35]. In our study, we found that there were in the order of approximately 2000 allelic differences between different clonal lineages.
However, SNV analysis has to be carefully analysed and a common nomenclature may be difficult to adopt as there are multiple variations at the genome level both in ORFs and intergenic regions. Furthermore, during infection, several studies have shown that SNVs can develop rapidly and are often associated with antibiotic resistance determinants [38,39]. Indeed, Halachev et al. found several SNVs in pmrB which was associated with reduced susceptibility to colistin. Our cgMLST scheme does not follow the SNV approach, instead we have opted for an allele-based method. This has the advantage that it can “soften” the conflicting signals of horizontal-gene transfer such as a bias introduced by a single homologous recombination in a gene that typically results in multiple SNVs but only in single allele change. A further advantage of our allele-based typing is its relatively easy storage and curation in a centralised database, similar that seen with classical MLST [16,40]. To date, cgMLST schemes using the Ridom SeqSphere+ software have been established with Listeria monocytogenes, Staphylococcus aureus, Legionella pneumophila, Francisella tularensis, and Mycobacterium tuberculosis [17,41–44].
Bacterial genomes show high diversity and this makes the definition of a core-genome difficult. Previous work has shown that there are eight international A. baumannii clonal lineages. Of these, IC2 is the most prevalent clone worldwide and indeed is responsible for the majority of outbreaks [8,11]. Therefore, we took the approach that the core genome should be built around an IC2 isolate, which was why ACICU strain was chosen. From a genome of 3972 ORFs (ACICU genome), we determined that the core genome was made up of 2,390 genes. Based on the strains that we investigated, the number of missing targets was very small, the maximum was always below <2.5% of total targets. Some of these missing targets are caused by the presence of IS elements that insert into genes. These IS elements can be present in multiple copies and are one of the reasons for genome misassembly and multiple contigs [45].
In conclusion, we present a highly representative and discriminatory cgMLST scheme for WGS-based typing of A. baumannii. We were able to differentiate between strains obtained from patients at the same hospital and to link isolates from patients hospitalized in different cities. Easy to use software and public nomenclature will be the key for wide-spread adoption and contribution to outbreak analysis and general epidemiological surveillance.
Supporting information
(A) Overview and (B) Zoom into the central part of the tree. BAPS partition 1 was further subdivided manually into five subgroups (1A-1E) according to the branching of the tree. STs that have significant admixture are colored in black.
(PDF)
(XLSX)
(XLS)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files. All raw reads generated were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) of EMBL European Bioinformatics Institute (EBI) under the study accession number PRJEB12082.
Funding Statement
PGH was supported by grant FOR-2251 from the German Research Council (DFG) (www.acinetobacter.de). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Peleg AY, Seifert H, Paterson DL (2008) Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev 21: 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nature Rev Microbiol 5: 939–951. [DOI] [PubMed] [Google Scholar]
- 3.Wagenvoort JHT, De Brauwer EIGB, Toenbreker HMJ, van der Linden CJ (2002) Epidemic Acinetobacter baumannii strain with MRSA-like behaviour carried by healthcare staff. Eur J Clin Microbiol Infect Dis 21: 326–327. 10.1007/s10096-002-0716-2 [DOI] [PubMed] [Google Scholar]
- 4.Husni RN, Goldstein LS, Arroliga AC, Hall GS, Fatica C, Stoller JK, et al. (1999) Risk factors for an outbreak of multi-drug-resistant Acinetobacter nosocomial pneumonia among intubated patients. Chest 115: 1378–1382. [DOI] [PubMed] [Google Scholar]
- 5.Chmielarczyk A, Higgins PG, Wojkowska-Mach J, Synowiec E, Zander E, Romaniszyn D, et al. (2012) Control of an outbreak of Acinetobacter baumannii infections using vaporized hydrogen peroxide. J Hosp Infect 81: 239–245. 10.1016/j.jhin.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 6.Dijkshoorn L, Aucken H, Gerner Smidt P, Janssen P, Kaufmann ME, Garaizar J, et al. (1996) Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J Clin Microbiol 34: 1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, et al. (2004) Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol 155: 105–112. 10.1016/j.resmic.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 8.Higgins PG, Dammhayn C, Hackel M, Seifert H (2010) Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65: 233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 9.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PloS ONE 5: e10034 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins PG, Janßen K, Fresen MM, Wisplinghoff H, Seifert H (2012) Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol 50: 3493–3500. 10.1128/JCM.01759-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomaschek F, Higgins PG, Stefanik D, Wisplinghoff H, Seifert H (2016) Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PloS ONE 11: e0153014 10.1371/journal.pone.0153014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. (2006) Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3: 59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 13.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. (2003) Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol 41: 1574–1585. 10.1128/JCM.41.4.1574-1585.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, et al. (2005) Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43: 4328–4335. 10.1128/JCM.43.9.4328-4335.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H (2012) Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacter baumannii isolates. J Med Microbiol 61: 137–141. 10.1099/jmm.0.036046-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Losada M, Cabezas P, Castro-Nallar E, Crandall KA (2013) Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infecta Genet Evol 16: 38–53. [DOI] [PubMed] [Google Scholar]
- 17.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. (2015) Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53: 2869–2876. 10.1128/JCM.01193-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eyre DW, Golubchik T, Gordon N, Bowden R, Piazza P, Batty EM, et al. (2012) A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2: e001124 10.1136/bmjopen-2012-001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, et al. (2011) Prospective genomic characterization of the German Enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PloS ONE 6: e22751 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, et al. (2013) Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J Clin Microbiol 51: 3950–3954. 10.1128/JCM.01930-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyres KL, Conway TC, Garg S, Queiroz C, Reumann M, Holt K,et al. (2014) WGS analysis and interpretation in clinical and public health microbiology laboratories: what are the requirements and how do existing tools compare? Pathogens 3: 437–458. 10.3390/pathogens3020437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fricke W, Rasko DA (2014) Bacterial genome sequencing in the clinic: bioinformatic challenges and solutions. Nat Rev Genet 15: 49–55. 10.1038/nrg3624 [DOI] [PubMed] [Google Scholar]
- 23.Kohlenberg A, Brummer S, Higgins PG, Sohr D, Piening BC, de Grahl C, et al. (2009) Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in a German University Medical Centre. J Med Microbiol 58: 1499–1507. 10.1099/jmm.0.012302-0 [DOI] [PubMed] [Google Scholar]
- 24.Molter G, Seifert H, Mandraka F, Kasper G, Weidmann B, Hornei B, et al. (2016) Outbreak of carbapenem-resistant Acinetobacter baumannii in the intensive care unit: a multi-level strategic management approach. J Hospital Infect 92: 194–198. [DOI] [PubMed] [Google Scholar]
- 25.Juenemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, et al. (2013) Updating benchtop sequencing performance comparison. Nat Biotech 31: 294–296. [DOI] [PubMed] [Google Scholar]
- 26.Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corander J, Marttinen P, Siren J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9: 539 10.1186/1471-2105-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, Rodriguez-Valera F (2005) Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43: 4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price MN, Dehal PS, Arkin AP (2010) FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PloS ONE 5: e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R (2007) Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJP, et al. (2008) Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52: 2616–2625. 10.1128/AAC.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. (1998) Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 95: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, Holm A, et al. (2015) Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother 70: 1965–1968. 10.1093/jac/dkv072 [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick MA, Ozer EA, Hauser AR (2016) Utility of whole-genome sequencing in characterizing Acinetobacter epidemiology and analyzing hospital outbreaks. J Clin Microbiol 54: 593–612. 10.1128/JCM.01818-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT (2015) Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53: 1072–1079. 10.1128/JCM.03385-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halachev M, Chan J, Constantinidou C, Cumley N, Bradley C, Smith-Banks M, et al. (2014) Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham, England. Genome Med 6: 70 10.1186/s13073-014-0070-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, et al. (2011) Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J Antimicrob Chemother 66: 1499–1503. 10.1093/jac/dkr168 [DOI] [PubMed] [Google Scholar]
- 39.Higgins PG, Schneiders T, Hamprecht A, Seifert H (2010) In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolated from a hospitalized patient. Antimicrob Agents Chemother 54: 5021–5027. 10.1128/AAC.00598-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiden MC, van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA,et al. (2013) MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11: 728–736. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohl TA, Diel R, Harmsen D, Rothgaenger J, Walter KM, Merker M, et al. (2014) Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52: 2479–2486. 10.1128/JCM.00567-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran-Gilad J, Prior K, Yakunin E, Harrison TG, Underwood A, Lazarovitch T, et al. (2015) Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires' disease incidents. Eurosurveillance 20: 21087. [DOI] [PubMed] [Google Scholar]
- 43.Antwerpen MH, Prior K, Mellmann A, Hoppner S, Splettstoesser WD, Harmsen D (2015) Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes ("MLST+"). PloS ONE 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A (2014) Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52: 2365–2370. 10.1128/JCM.00262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, et al. (2008) Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190: 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Overview and (B) Zoom into the central part of the tree. BAPS partition 1 was further subdivided manually into five subgroups (1A-1E) according to the branching of the tree. STs that have significant admixture are colored in black.
(PDF)
(XLSX)
(XLS)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All raw reads generated were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) of EMBL European Bioinformatics Institute (EBI) under the study accession number PRJEB12082.