Abstract
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus (KSHV) binds to two sites within the 801-bp-long terminal repeat (TR) and is the only viral protein required for episomal maintenance. While two or more copies of TR are required for long-term maintenance, a single TR confers LANA-dependent origin activity on plasmid DNA. Deletion mapping revealed a 71-bp-long minimal replicator containing two distinctive sequence elements: LANA binding sites (LBS1/2) and an adjacent 29- to 32-bp-long GC-rich sequence which we termed the replication element. Furthermore, the transcription factor Sp1 can bind to TR outside the minimal replicator and contributes to TR's previously reported enhancer activity.
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with Kaposi's sarcoma (KS) and two lymphoproliferative diseases: primary effusion lymphoma and multicentric Castleman's disease (5, 6, 30). The 140-kbp KSHV long unique region encodes approximately 90 genes, which are flanked on both sides with 20 to 40 copies of 801-bp-long GC-rich terminal repeats (TR) (20, 24, 29). During latency, only a small subset of viral genes, including the latency-associated nuclear antigen (LANA), is expressed and the viral genome is maintained as multiple copies of episomal DNA (10, 11, 28, 32). LANA is a functional homologue of origin binding proteins EBNA-1 from Epstein-Barr virus (EBV) and E2 from human papillomavirus (15, 22). LANA is necessary and sufficient to support stable long-term episomal maintenance in dividing cells by (i) tethering viral episomes to cellular chromosomes (1, 2, 7, 8, 12, 13, 19, 26) and (ii) supporting the initiation of DNA replication of TR-containing plasmids (15, 17, 23), presumably by bridging the viral origin of replication (ori) and the host cellular replication machinery. Using short-term replication assays, we have previously demonstrated that all cis-regulatory sequences required for LANA-dependent ori activity are located within a single copy of TR (17). By performing a detailed deletion analysis, we determined the minimal replicator within TR and showed that the transcription factor Sp1 can bind to at least two sites within TR and contributes to its previously described enhancer activity (13).
Mapping the minimal replicator within TR.
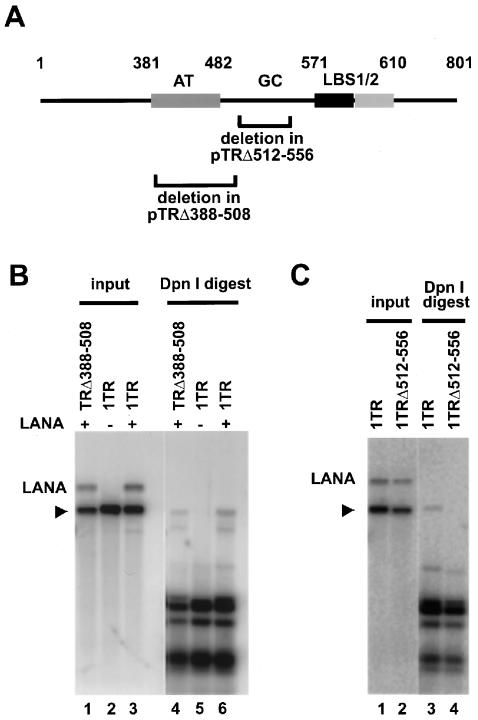
The TR unit is 801 bp long and 89% GC rich (20, 29). LANA binds in a cooperative fashion to two sites (LBS1/2) in TR located between nucleotides (nt) 571 and 610, and both binding sites contribute to ori activity as determined by short-term replication assays (12, 17). Located directly upstream of LBS1/2 is an 89-bp highly GC-rich element adjacent to a 101-bp AT-rich region (Fig. 1A). AT-rich stretches are often found in origins of replication and are believed to function in DNA unwinding (4). We have previously reported that deletion of TR sequences from nt 1 to 482, including the AT-rich region, and nt 610 to 801 downstream of LBS1/2 does not decrease replication efficiency. In contrast, a deletion of the GC-rich region (up to nt 551) completely abolished replication activity. These data suggested that, in addition to LBS1/2, sequences of this GC-rich element are required for ori function while the AT-rich element is dispensable (17).
FIG. 1.
The GC-rich fragment upstream of LBS1/2 but not the AT-rich sequence is required for LANA-dependent replication of TR-containing plasmids. Replication assays were performed as previously described (17). Briefly, 10 μg of each mutant TR construct was cotransfected with 10 μg of pcDNA3/orf73 or carrier DNA into 293 cells. Seventy-two hours after transfection, extrachromosomal DNA was recovered by Hirt extraction. Ten percent of the episomal DNA was digested with 20 U of HindIII (input), while 90% was double digested with 20 U of HindIII and 200 U of DpnI for 16 h (DpnI digest). After electrophoresis, DNA was immobilized on nylon membranes and hybridized with a radiolabeled probe of the vector backbone. (A) Schematic representation of TR and two deletion mutants. Deleted regions in the context of full-length TR are indicated by brackets. (B) Deletion of AT-rich sequence nt 388 to 508 does not eliminate replication. (C) TR sequences within nt 512 to 556 are essential for LANA-dependent DNA replication. Positions of linearized test plasmids are indicated by arrowheads; also indicated is the position of the cotransfected LANA expression vector. Nucleotide numbers within TR are based on the work of Lagunoff and Ganem (20).
To confirm these observations, which were based on large truncations, we generated two constructs in which the AT-rich or the GC-rich element was specifically deleted from a full-length TR. pTRΔnt388-508 contains a 120-bp-long deletion encompassing the AT-rich stretch. In pTRΔ512-556, 48 bp was deleted from the GC-rich element (Fig. 1A). Constructs were cotransfected with pcDNA3/LANA into 293 cells, and replication ability was scored by Southern blot analysis of DpnI-resistant DNA as previously described (17). Plasmid replication was LANA dependent, since pTR did not replicate in the absence of LANA (Fig. 1B, lane 5). In the presence of LANA, pTRΔnt388-508 replicated with efficiency similar to that of pTR, further confirming that the AT-rich region does not contribute to replication of small plasmids (Fig. 1B, lanes 4 and 6). We cannot rule out a role for this element in the context of the viral episome. In contrast, pTRΔ512-556 did not replicate to any detectable level (Fig. 1C, lane 4), confirming that sequences near LBS1/2 are required for ori function and that this requirement cannot be compensated for by GC-rich sequences located elsewhere in TR.
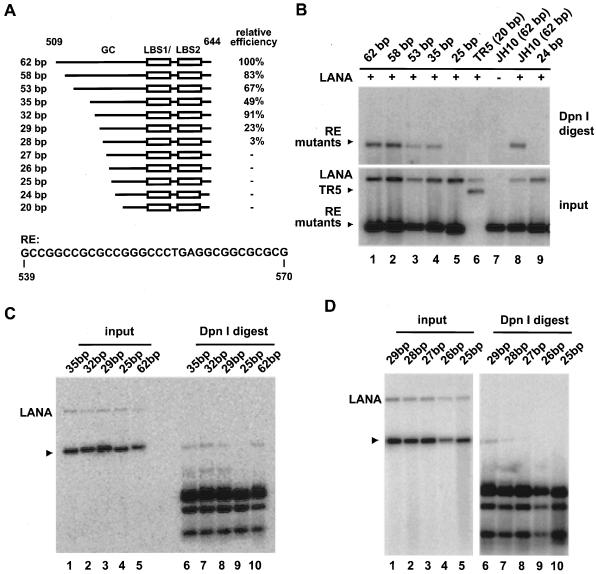
To map which nucleotides within this GC-rich region are vital for replication, we created 5′ truncation mutants containing various lengths of the GC-rich element adjacent to LBS1/2 by enzyme digestion or by ligating synthetic oligonucleotides (Fig. 2A). Due to the lack of restriction enzyme sites, the first set of mutants with lengths between 62 and 20 bp was generated by digestion of pJH10 (TR nt 509 to 644) with KpnI/XhoI, followed by exonuclease III nuclease treatment at 4°C. As shown in Fig. 2B, plasmids containing 58, 53, or 35 bp of the GC-rich fragment replicated in the presence of LANA, while mutants containing 25 and 24 bp did not. To map the 5′ border of this element, additional mutants covering the region between residues 35 and 25 were constructed. A plasmid containing 32 bp of the fragment replicated with activity comparable (91%) to that of pJH10 containing 62 bp (compare Fig. 2C, lane 7, to Fig. 2B, lane 8). A 29-bp-long GC-rich element still replicated at 23% of wild-type activity (Fig. 2C, lane 8, and Fig. 2D, lane 6), while removing one more nucleotide dramatically decreased replication to only 3% of wild-type levels (Fig. 2D, lane 7); a mutant containing 27 bp showed no detectable replication (Fig. 2D, compare lanes 6, 7, and 8). These data show that the minimal GC-rich sequence that confers LANA-dependent DNA replication is between 29 and 32 bp long. Furthermore, the functional minimal replicator is located between nt 539 and 610 and consists of two functional elements, LBS1/2 and a 29- to 32-bp-long GC-rich sequence, which we termed the replication element (RE) (Fig. 2A).
FIG. 2.
Fine mapping of the minimal sequence requirement for RE. (A) Panel of RE mutants. Plasmids contain different lengths of the GC-rich element as indicated upstream of LBS1/2 and 34 bp downstream. Mutants were generated by exonuclease III nuclease digestion of pJH10 (TR nt 509 to 644) or by ligating synthetic oligonucleotides into pBluescript II SK(+). The lower panel shows the RE sequence of 32 bp in length (nt 539 to 570). All replication assays were performed as described for Fig. 1. Replication efficiency was calculated by determining the ratio of input and replicated DNA for each construct in comparison to pJH10, which was set to 100%. Values were derived from two or three independent experiments for each construct. (B) The plasmid containing 35 bp, but not that containing 25 bp, of the GC-rich fragment replicates in the presence of LANA. (C and D) The plasmid containing 29 bp of the GC-rich sequence replicates, the plasmid containing 28 bp has reduced activity, and the plasmid containing 27 bp is not active. Test plasmids are indicated by arrowheads in panels B to D.
Sp1 binds to TR and contributes to TR enhancer activity but not to ori activity.
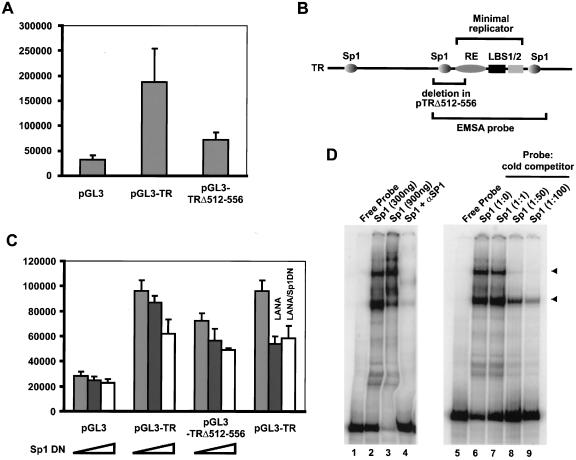
TR can function as a transcriptional enhancer, suggesting that cellular transcription factors bind TR (13, 34). We wanted to determine whether sequences within the GC-rich element contribute to the observed enhancer activity. A luciferase reporter was prepared by cloning TRΔnt512-556 into the pGL3 promoter upstream of a simian virus 40 (SV40) promoter and used in transient-transfection assays in CV-1 cells. pGL3-TR augmented transcription four- to fivefold more than pGL3 did; however, the deletion mutant TRΔnt512-556 showed about a 50% decrease in luciferase activity (Fig. 3A). Analysis of the TR sequence for the presence of transcription factor binding sites revealed three GC-box consensus sites for Sp1 (GGGGCGGGG) (18). Surprisingly, one of these sites is located at nt positions 523 to 531 within the deleted sequence of TRΔnt512-556, while two more are located at nt 242 to 259 and nt 623 to 631 (Fig. 3B). Sp1 binding sites have been identified in a variety of origins of replication, including the ori of SV40; however, it is not clear how transcription factors contribute to replication (for a review see references 4 and 14). All three Sp1 sites are located outside of the minimal replicator (nt 539 to 610) and therefore are not required for LANA-dependent replication of plasmid DNA.
FIG. 3.
Sp1 binds to TR and contributes to TR enhancer activity. In transient-transfection assays luciferase reporter plasmids were transfected into CV-1 cells (African green monkey kidney cells), and luciferase activity was determined 48 h posttransfection as previously described (13). (A) Transient-transfection assays indicate that TRΔ512-556 has decreased enhancer activity compared to full-length TR. (B) Diagram of Sp1 binding sites in TR. EMSA, electrophoretic mobility shift assay. (C) DN Sp1 suppressed TR enhancer activity in a dose-dependent manner. Reporter plasmid (100 ng) was cotransfected with an increasing amount of DN Sp1 expression vector (0 and 500 ng and 1 μg). The last group of bars shows that cotransfection of a LANA expression vector (pcDNA3/orf73) and DN Sp1 does not further repress transcription of pGL3-TR. Data shown are derived from two independent experiments performed in triplicate. (D) Sp1 binds to two sites within TR. Electrophoretic mobility shift assay analysis with a 32P-labeled probe spanning LBS1/2, the RE, and the two flanking Sp1 binding sites together with recombinant Sp1 protein reveals two specific complexes (lanes 2 and 3). Sp1-specific antibody (lane 4) and unlabeled competitor DNA (lanes 6 to 9) inhibit Sp1 binding. Positions of DNA-Sp1 complexes are indicated by arrowheads.
To determine whether Sp1 binding is responsible for the TR enhancer activity, we cotransfected CV-1 cells with either pGL3-TR or pGL3-TRΔnt512-556 and increasing amounts of pEBGN-Sp1 (25) encoding a dominant-negative (DN) Sp1. Luciferase expression of both constructs was inhibited in a dose-dependent manner, suggesting that Sp1 augments transcription from TR (Fig. 3C). The observed inhibition was similar to that observed when LANA is bound to LBS1/2; cotransfection of both LANA and DN Sp1 did not further suppress transcription, suggesting that the TR enhancer activity is suppressed under conditions where LANA is bound to TR (Fig. 3C). To directly prove Sp1 binding to TR, we performed an electrophoretic mobility shift assay with recombinant Sp1 and a radiolabeled probe containing LBS1/2, the RE, and two flanking Sp1 binding sites (Fig. 3D). In the presence of Sp1, two complexes formed (Fig. 3D, lane 2), consistent with the presence of two Sp1 binding sites. Specificity was further confirmed by addition of a monoclonal Sp1 antibody (Fig. 3D, lane 4) and by adding increasing amounts of cold competitor (Fig. 3D, lanes 6 to 9), both of which prevented complex formation. These data, together with the reporter assays, demonstrate that Sp1 binds to TR and contributes to its enhancer activity.
The role of Sp1 binding to TR in the viral life cycle is currently unclear. The fact that LANA suppresses transcription when bound to TR seems to negate a model by which Sp1 directly contributes to DNA replication. This notion is further strengthened by the fact that all three Sp1 sites within TR are located outside the minimal replicator (nt 539 to 610); hence, Sp1 binding does not contribute to LANA-dependent DNA replication. The ability of TR to function as a transcriptional enhancer might contribute to the regulation of viral gene expression in a LANA-dependent manner. For instance, expression of K1, the gene at the left end of the KSHV genome, is suppressed during latency but significantly induced during lytic growth (20, 21). Whether this suppression is caused by LANA binding to TR needs to be further investigated in latently infected cells. Interestingly, the TR of EBV also contain Sp1 and Sp3 binding sites, and it has been suggested that these factors do not regulate transcription but might play a role in genome circularization following de novo infection (31, 33).
Our mapping data demonstrate the presence of a second essential cis-regulatory element, termed RE. Deletion analysis mapped the length of RE between 29 and 32 bp, which, together with LBS1/2, constitutes the minimal replicator of 71 bp in length. The RE may function as a loading pad for cellular proteins involved in DNA replication; however, computer-assisted sequence analysis did not reveal any homologies to known cis-regulatory elements. In vitro, LANA has been shown to interact with origin recognition complex proteins (23), and studies to address whether cellular proteins bind directly to RE (in the presence and absence of LANA) are ongoing. While most origins of replication contain AT-rich sequences close to the initiation site, it was shown for the origin of SV40 that melting occurs at a GC-rich element (3).
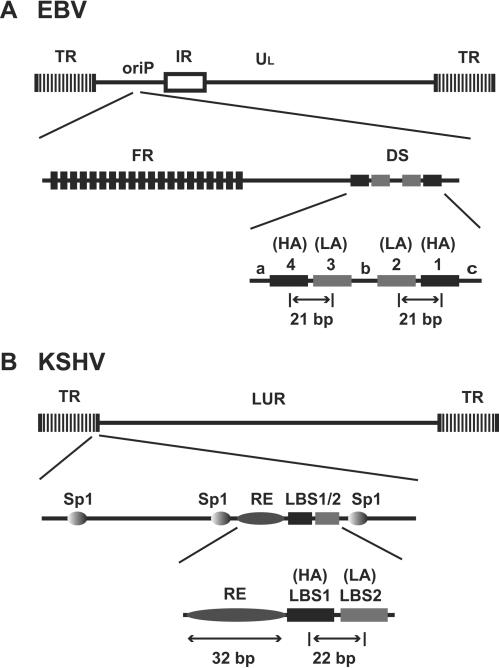
These data together with our previously reported characterization of LANA binding to TR (12, 13) define an ori structure, which is strikingly different from oriP of EBV (Fig. 4). The KSHV ori is positioned in TR, while EBV oriP is located within the coding region of the viral genome. As a consequence, KSHV has multiple origins, which may provide a function for segregation similar to that of the family of repeats in oriP. This hypothesis is supported by the fact that a single TR is sufficient for DNA replication while long-term episome maintenance requires multiple copies of TR (1, 2, 12, 13). The dyad symmetry (DS) element of EBV contains four EBNA-1 binding sites that are organized as two pairs, each containing a high-affinity site and a low-affinity site spaced by 21 bp (22, 27, 35, 37). Similarly, LANA has a high- and a low-affinity binding site spaced by 22 bp (12). Although the sequences differ, the organization of LBS1/2 resembles half of a DS element, which is sufficient for replication in plasmid-based assays (16, 36). Flanking sequences of the DS element modulate ori activity but are not required for function (9, 36). This is different for KSHV, where LBS1/2 alone does not have ori activity and in addition requires the RE. In summary, the origins of replication of rhadinoviruses and lymphocryptoviruses have significantly diverged. Therefore, a detailed analysis of the KSHV ori structure may not only increase our knowledge of gammaherpesvirus latent replication but also provide an attractive model to study the interaction of host cellular factors and origins of replication in metazoan cells.
FIG. 4.
Comparison of the latent origins between EBV and KSHV. (A) Genome structure and oriP of EBV. (B) Genome structure and latent origin of KSHV. IR, internal repeat; DS, dyad symmetry; FR, family of repeat; HA, high-affinity binding site; LA, low-affinity binding site; LBS1 and LBS2, LANA binding sites 1 and 2, respectively.
Acknowledgments
We thank Gerald Thiel, University of Saarland Medical Center, Germany, for providing pEBGSp1. We thank Rebecca Johnson, Mark Samols, and Feng-Qi An for fruitful discussions and critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health (RO1 CA 88763 and R21 CA97939) to R.R.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec, J. A., and J. Hurwitz. 1988. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 7:3149-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulikas, T. 1996. Common structural features of replication origins in all life forms. J. Cell. Biochem. 60:297-316. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 9.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 13.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadonaga, J. T., A. J. Courey, J. Ladika, and R. Tjian. 1988. Promoter-selective activation of transcription by Sp1, p. 239-250. In B. R. Franza, Jr., B. R. Cullen, and F. Wong-Staal (ed.), The control of human retrovirus gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 21.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 23.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersohn, D., and G. Thiel. 1996. Role of zinc-finger proteins Sp1 and zif268/egr-1 in transcriptional regulation of the human synaptobrevin II gene. Eur. J. Biochem. 239:827-834. [DOI] [PubMed] [Google Scholar]
- 26.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 31.Spain, T. A., R. Sun, and G. Miller. 1997. The locus of Epstein-Barr virus terminal repeat processing is bound with enhanced affinity by Sp1 and Sp3. Virology 237:137-147. [DOI] [PubMed] [Google Scholar]
- 32.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, R., T. A. Spain, S. F. Lin, and G. Miller. 1997. Sp1 binds to the precise locus of end processing within the terminal repeats of Epstein-Barr virus DNA. J. Virol. 71:6136-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 77:7093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]