Abstract
We designed five degenerate primers for detection of novel cry genes from Bacillus thuringiensis strains. An efficient strategy was developed based on a two-step PCR approach with these primers in five pair combinations. In the first step, only one of the primer pairs is used in the PCR, which allows amplification of DNA fragments encoding protein regions that include consensus domains of representative proteins belonging to different Cry groups. A second PCR is performed by using the first-step amplification products as DNA templates and the set of five primer combinations. Cloning and sequencing of the last-step amplicons allow both the identification of known cry genes encoding Cry proteins covering a wide phylogenetic distance and the detection and characterization of cry-related sequences from novel B. thuringiensis isolates.
Application of chemical control agents against insect pests, although efficacious in most cases, has severe drawbacks related to the appearance of insect resistance, the emergence of secondary pests, the impact on nontarget organisms, environmental pollution, and residues on the agriculture products and animals (29). There is an urgent need for environmentally friendly pesticides to reduce contamination and the likelihood of insect resistance (34).
Bacillus thuringiensis is a gram-positive bacterium that is characterized by the production of insecticidal crystal proteins (Cry proteins or δ-endotoxins that are encoded by cry genes). B. thuringiensis has been used as a successful biological insecticide for more than 40 years and is a uniquely specific, safe, and effective tool for the control of a wide variety of insect pests (29). Cry toxins constitute a family of related proteins that can kill insects belonging to the Lepidoptera, Coleoptera, Diptera, Hymenoptera, Homoptera, and Mallophaga, as well as other invertebrates (17, 32). These toxins can be grouped according to the degree of amino acid homology. Cry proteins with the same primary rank in a phylogenetic tree proposed by Crickmore et al. (13) often affect the same order of insects. The incorporation of cry genes into major crops, providing insect-resistant plants, is the most recent application of B. thuringiensis in agriculture (29).
Intensive screening programs leading to important collections of isolates have been conducted in the last few decades (10, 11, 15, 19, 27, 35, 37). The need for novel Cry proteins with toxic potential against different organisms with specificity for a much broader range of pests or to provide alternatives after the appearance of insect resistance has resulted in a continuous search for new experimental approaches in order to expand the host ranges of the strains available (12).
Several PCR-based methodologies with universal primers and sets of primers directed against specific regions of type-specific cry genes have been proposed, and these approaches allow the detection of cry genes and prediction of their insecticidal activities (2, 4, 7, 9, 20, 23). However, these approaches (mostly multiplex PCR) are mainly focused on screening most currently known cry genes or gene families and do not ensure isolation of novel genes belonging to undescribed groups (3, 5, 6, 8, 25).
We report here a two-step PCR-based approach which allows amplification of unknown cry-related sequences that can be characterized after DNA sequencing. To prove the efficiency of the strategy, we showed that it was possible to identify coding sequences for known proteins, as well as for unknown proteins, that are members of different Cry families that are widespread in a phylogenetic tree.
(This work is part of C.M.B.'s Ph.D. thesis at FCEyN, Universidad Nacional de Mar del Plata.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Known strains that served as references were kindly supplied by D. Zeigler (Bacillus Genetic Stock Centre, Columbus, Ohio) and J. Ibarra (CINVESTAV, Irapuato, Mexico). Argentinean strains were obtained from the FIBA Culture Collection (Fundación para Investigaciones Biológicas Aplicadas, Mar del Plata, Argentina). Cultures of B. thuringiensis strains were grown at 28°C in nutrient broth (Difco) with vigorous shaking or on nutrient broth agar. Cells of transformed Escherichia coli DH5α were grown at 37°C in Luria-Bertani medium supplemented with 50 μg of carbenicillin per ml (31).
Primer design and PCR conditions.
The consensus amino acid sequences of Cry proteins were selected by using the Protein Domain Database, ProDom (http://prodes.toulouse.inra.fr/prodom/doc/prodom.html), and Cry1Aa as the query (33). After analysis of all available sequences of Cry-related proteins, we retrieved Cry subgroup clusters and selected nine representative sequences of most members of Cry families. We identified the PD000873 domain in 115 proteins and the PD14498 domain in 102 proteins. Both of these domains contain conserved blocks that have been reported previously (32). By using multiple-sequence alignment and taking into account Cry1Aa structural determinants (21), we obtained four consensus oligopeptides (Fig. 1). The nucleotides encoding sequences of oligopeptides I to IV present in some Cry proteins, which were selected as being widespread in the Cry phylogeny (13), were aligned by using the CLUSTAL X method of the MegAlign program of the DNAStar software package (22). The consensus of these DNA sequence alignments gave rise to five degenerate oligonucleotides (OL1 to OL5), which are listed in Table 1. The genomic DNA of B. thuringiensis used for PCR amplification was isolated as previously described (14, 31). The PCRs were carried out by using 0.2 μg of DNA template in a reaction mixture (total volume, 25 μl) containing each deoxynucleoside triphosphate at a concentration of 400 μM, each primer at a concentration of 2 μM, and 0.5 U of Taq DNA polymerase dissolved in the corresponding reaction buffer (Promega). Amplifications were performed with a PTC-100 thermal cycler (model-96V; MJ Research, Inc., Watertown, Mass.) under the following conditions: 2 min of denaturation at 94°C, followed by 35 cycles of denaturation for 45 s at 94°C, annealing for 1 min at 35 or 40°C depending on the DNA template, and extension for 1 min at 72°C. An extra extension step consisting of 3 min at 72°C was added after completion of the 35 cycles. Primers OL1 and OL5 (Table 1) were used in a first PCR amplification. After this, 1 μl of a 1:50 dilution of each PCR product was used as a template in a second-step PCR amplification with the five primer pairs to increase the probability for successful amplification of sequences corresponding to cry fragments. PCR products were analyzed by electrophoresis in 0.8 to 1% (wt/vol) agarose gels in Tris-acetate buffer and ethidium bromide staining (31).
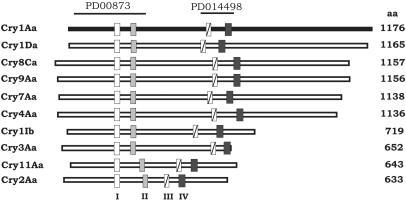
FIG. 1.
Alignment of representative Cry proteins. Consensus oligopeptides were obtained from a ProDom analysis by using Cry1Aa (accession no. AAA22353) as the query (solid bar), and these oligopeptides corresponded to amino acids 153 to 183 (oligopeptide I; YEVPLLVYAQAANLHLLLRDVFGQWG), 229 to 251 (oligopeptide II; YNQFRREMTLVLDVALFPYD), 520 to 530 (oligopeptide III; QRYRVRIRYAS), and 595 to 605 (oligopeptide IV; EVYIDRIEFIP) of Cry1Aa, respectively. The two ProDom domains are indicated (PD000873 and PD14498). Five degenerate oligonucleotides were designed from the nucleotides encoding the underlined amino acid sequences indicated above. The number of amino acid (aa) residues in each Cry protein is indicated on the right.
TABLE 1.
Characteristics of the general primers designed and predicted sizes of DNA PCR amplification fragments of known cry genes
| Primer paira | Primer sequenceb | Positionc | Product sizes (bp)d
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cry1Aa | cry3Aa | cry4Aa | cry7Aa | cry8Aa | cry9Aa | cry2Aa | cry11Aa | cry14Aa | |||
| OL1 (d) | 5′ TAHCANYATAYGCACARGCHGCMAAYTTHCAT 3′ | 510-1788 | 1,308 | 1,311 | 1,374 | 1,317 | 1,362 | 1,359 | 735 | 816 | 1377 |
| OL5 (r) | 5′ GGAATAAATTCRATTYTRTCTATATAAA 3′ | ||||||||||
| OL1 (d) | 5′ TAHCANYATAYGCACARGCHGCMAAYTTHCAT 3′ | ||||||||||
| OL4 (r) | 5′ AGCATADCGRAHNCYHRYDYVATA 3′ | 510-1566 | 1,089 | 1,122 | 1,125 | 1,080 | 1,137 | 1,134 | 696 | 771 | 1113 |
| OL2 (d) | 5′ AGAGAHRTGAHWDTDAHRGTATTRGAT 3′ | ||||||||||
| OL5 (r) | 5′ GGAATAAATTCRATTYTRTCTATATAAA 3′ | 699-1788 | 1,119 | 1,122 | 1,143 | 1,128 | 1,167 | 1,158 | 558 | 636 | 1152 |
| OL2 (d) | 5′ AGAGAHRTGAHWDTDAHRGTATTRGAT 3′ | ||||||||||
| OL4 (r) | 5′ AGCATADCGRAHNCYHRYDYVATA 3′ | 699-1566 | 891 | 900 | 894 | 891 | 942 | 933 | 519 | 591 | 888 |
| OL3 (d) | 5′ TATBRHRYDRGNDTYCGHTATGCT 3′ | ||||||||||
| OL5 (r) | 5′ GGAATAAATTCRATTYTRTCTATATAAA 3′ | 1566-1788 | 252 | 246 | 273 | 261 | 249 | 249 | 57 | 54 | 288 |
d, direct primer; r, reverse primer.
The primers are degenerate primers, and the sequences are indicated according to the degenerate DNA genetic code as follows: B = C, G, or T; D = A, G, or T; H = A, C, or T; M = A or C; R = A or G; Y = T or C; W = A or T; N = A, C, G, or T; and V = A, C, or G.
Position starting from the first base of the start codon sequence of the cry1Aa gene (accession number M11250) according to sequences obtained from the National Center for Biotechnology Information database.
Cloning, sequencing, and database analysis of the PCR fragments.
PCR-amplified products were ligated to the cloning vector pGEM-T Easy (Promega, Madison, Wis.), which was used for transforming E. coli DH5α by standard protocols (31). DNA sequencing was carried out by Medigenomix GmbH, Inc. and Macrogen Services. BLAST X (version 2.2.6) was used for DNA sequence analysis (1). Known Cry sequences were obtained from the nonredundant protein database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The sequences of the following proteins were used: Cry1Ac (accession no. AAA22331), Cry1Aa (AAA22353), Cry1Ca (CAA30396), CryEa (CAA37933), Cry1Ha (CAA80236), Cry1Da (CAA38099), Cry1Fa (AAA22348), Cry1Ga (CAA80233), Cry1Ja (AAA22341), Cry1Ba (CAA29898), Cry1Bf (CAC50778), Cry1Bb (AAA22344), Cry1Ka (AAB00376), Cry1Ic (AAC62933), Cry1Ib (AAA82114), Cry1Ia (CAA44633), Cry1Id (AAD44366), Cry7Aa (M64478), Cry26Aa (AF122897), Cry8Bb (CAD57542), Cry8Aa (U04364), Cry8Ca (U04366), Cry9Da (D85560), Cry9Ea (AB011496), Cry9Aa (X58120), Cry3Bb (M89794), Cry3Aa (M22472), Cry32Aa (AY008143), Cry32Da (BAB78603), Cry4Aa (Y00423), Cry10Aa (M12662), Cry30Aa (AJ251978), Cry19Aa (Y07603), Cry44Aa (BAD08532), Cry27Aa (AB023293), Cry16Aa (X94146), Cry24Aa (U88188), Cry40Ba (BAC77648), Cry20Aa (U82518), Cry25Aa (U88189), Cry5Aa (L07025), Cry5Ba (U19725), Cry12Aa (L07027), Cry21Ba (BAC06484), Cry14Aa (U13955), Cry13Aa (L07023), Cry2Ab (M23724), Cry2Aa (M31738), Cry2Ac (X57252), Cry18Aa (X99049), Cry18Ba (AF169250), Cry11Ba (X86902), Cry11Aa (M31737), and Cry31Aa (AB031065). Sequence alignments were generated with the CLUSTAL X software program (version 1.8) (22). Dendrograms were compiled from 1,000 independent trials of CLUSTAL X. The graphic representation of the tree was generated by using the TREEVIEW 16 program.
Nucleotide sequence accession number.
The novel cry-related sequence reported in this paper (FCC4) has been deposited in the EMBL database under accession number AJ640131.
RESULTS
Primer design.
The need to characterize novel entomopathogenic proteins led us to search for novel native B. thuringiensis strains isolated from Argentinean soil samples. In order to identify the majority of possible of cry genes present in the isolates, consensus predicted amino acid sequences were determined after compilation of homologous domains present in most Cry protein sequences by using the ProDom database with Cry1Aa as the query (33). A structural comparison of the four consensus oligopeptides identified (Fig. 1) revealed that oligopeptides I and II fold like the Cry1Aa structurally conserved α6 and α8 helixes, respectively; oligopeptide III corresponds to an amino acid stretch between the conserved β19 and β20 sheets, and oligopeptide IV folds like the conserved β27 sheet (21). After alignment of the DNA sequences encoding the four conserved regions, a set of five degenerate primers was designed to be used in PCR amplification in five pair combinations (Table 1).
Identification of cry genes from B. thuringiensis strains.
To test the efficiency of the primers that were designed for cry sequence detection, PCRs were carried out with total DNA isolated from the following 13 representatives of B. thuringiensis: HD1 (B. thuringiensis subsp. kurstaki), HD525 (B. thuringiensis subsp. wuhanensis), serotype 8a8b (B. thuringiensis subsp. morrisoni), BTC007 (36), BP465 (B. thuringiensis subsp. entomocidus), PGSI245 (B. thuringiensis subsp. galleriae), HD511 (B. thuringiensis subsp. dakota), B. thuringiensis subsp. san diego, H-14 (B. thuringiensis subsp. israeliensis), T03C001 (B. thuringiensis subsp. fukuokaensis), PS17 (B. thuringiensis subsp. darmstadiensis), P580JJ1 (B. thuringiensis subsp. sotto), and HD12 (B. thuringiensis subsp. kenyae), which produce (among other Cry proteins) the Cry1Aa, Cry1Bd, Cry1Bc, Cry1Ie, Cry1Ib, Cry7Aa, Cry7Ab, Cry3Aa, Cry4Aa, Cry4Ab, Cry11Aa, Cry20Aa, Cry5Aa, Cry14Aa, and Cry2Aa toxins (13). In order to characterize novel cry sequences, the technique described here was performed by using 40 native B. thuringiensis isolates collected from Argentinean soil samples. In most cases, the DNA product of the first PCR amplification with primers OL1 and OL5 was not visible. Amplicons of the second PCR step that were the expected sizes (Table 1) were obtained with DNA of the 53 strains analyzed (Fig. 2 and data not shown). The five possible expected amplicons were not visible in all cases; however, we always obtained PCR amplification products with at least two primer combinations. To illustrate this, while for B. thuringiensis subsp. kurstaki HD1 (Fig. 2A, lanes 1 to 5) and B. thuringiensis subsp. san diego (data not shown) the five combinations of oligonucleotides gave PCR amplification products, DNA from B. thuringiensis subsp. sotto P580JJ1 could be amplified only with two of the primer pairs (Fig. 2A, lanes 8 and 9). Similarly, the set of primer combinations was used to analyze 40 native B. thuringiensis isolates. DNA fragments whose sizes were in the expected ranges were obtained in all cases with at least two primer combinations, as shown for strain FCC4 in Fig. 2B. When the OL2-OL4 pair was used, DNA fragments that were about 900 bp long were amplified with more than 90% of the total DNA samples isolated from the native B. thuringiensis strains (Fig. 2C and data not shown).
FIG. 2.
PCR amplification products from different B. thuringiensis strains separated by agarose electrophoresis. (A and B) Amplified DNA fragments obtained with the five primer pairs in the second PCR amplification step by using DNA from two known B. thuringiensis (Bt) strains (A) or DNA of FCC4, a native strain (B), as the template. The cry1Aa and cry14Aa genes are present in B. thuringiensis subsp. kurstaki HD1 and B. thuringiensis subsp. sotto PS80JJI, respectively. Lanes 1 and 6, primers OL1 and OL5; lanes 2 and 7, primers OL1 and OL4; lanes 3 and 8, primers OL2 and OL5; lanes 4 and 9, primers OL2 and OL4; lanes 5 and 10, primers OL3 and OL5. (C) PCR products obtained after amplification from DNA of 19 native strains by using primers OL2 and OL4 in the second PCR amplification step. The positions of molecular weight markers (1-kb DNA ladder; GIBCO BRL) are indicated. The arrows indicate the positions of the predicted amplified DNA fragments.
For known B. thuringiensis strains, we selected one or two of the amplicons for cloning, and for novel strains FCC4, FCC7, and FCC41 we cloned the DNA fragments amplified with the OL2-OL4 and OL2-OL5 primer pairs. The cloned fragments were sequenced, and they were identified as partial cry genes after BLAST X and multiple-sequence alignment analysis. We identified partial cry sequences of the cry1Aa, cry1Ea, cry1Ga, cry1Bc, cry1Ib, cry1Ie, cry7Aa, cry7Ab, cry3Aa, cry4Aa, cry20Aa, cry5Aa, cry14Aa, cry2Aa, and cry11Aa genes. Also, we characterized as cry-related sequences DNA fragments amplified from native strains FCC4, FCC7, and FCC41.
The deduced amino acid sequences of known toxins and of the three partial polypeptide sequences (FCC7, FCC41, and FCC4) corresponding to DNA fragments of the undescribed B. thuringiensis strains were analyzed by phylogenetic methods (Fig. 3). FCC7 and FCC41 partial deduced amino acid sequences clustered together with Cry8 and Cry24, respectively, and remarkably, the FCC4 sequence was one of the most divergent Cry sequences in our phylogenetic analysis.
FIG. 3.
Phylogenetic analysis of Cry proteins. Neighbor-joining phylograms were constructed after sequence alignment of the deduced amino acid sequences of selected known toxins and protein fragments corresponding to partial DNA sequenced fragments from FCC4, FCC7, and FCC41 by using the CLUSTAL X program and a BLOSSUM matrix. The tree was generated by using TREEVIEW. Sequences were obtained from the databases of the National Center for Biotechnology Information. The arrows indicate the positions of three novel Cry sequences. The positions of amino acid sequences deduced from known cry genes used in the analysis are indicated by asterisks. Numbers at nodes represent the percentages of bootstrap resamplings based on 1,000 replicates.
DISCUSSION
PCR is a tool that has been widely used for characterization of genes coding for Cry proteins and for analysis of B. thuringiensis collections (30). This technique was first introduced by Carozzi et al. (7) to identify cry genes in order to predict insecticidal activity. Over the last decade, PCR methods for screening cry genes present in B. thuringiensis collections, including multiplex PCR methods, have been used to identify strains that harbor genes coding for known Cry types (2-6, 8, 9, 18, 23, 24, 37). Undoubtedly, the use of PCR has greatly improved cry gene detection; however, this method is mostly limited to members of previously described gene families and requires a large number of primers (2-4, 8, 9). Also, universal degenerate primers were designed to amplify all the members of different subfamilies of cry genes. Although the use of these degenerate oligonucleotides increases the probability of amplifying novel genes, the efficiency is restricted to detection of closely related genes in the same group (23, 24, 28). The relatively modest success of other PCR-based methods (PCR-restriction fragment length polymorphism analysis and exclusive PCR) in the identification of novel cry genes was associated with technical problems related to the complexity of multigenic cry strains or the adaptation of the techniques in each laboratory (30).
In this study we designed five degenerate primers that were used in a two-step PCR approach that was followed by DNA sequencing. We demonstrated that this strategy is an efficient strategy for identifying cry-related sequences present in novel B. thuringiensis isolates. In particular, the second PCR step was shown to be important for amplifying DNA fragments of the expected size that were to be cloned and sequenced for identification. Primer design was a key factor in the proposed strategy. The five degenerate primers have advantages over previous primers used in PCR-based methodologies, which did not amplify novel genes encoding proteins belonging to undescribed Cry groups. The possibility of detecting and characterizing unknown cry sequences, which was proved by characterization of novel cry-related sequences from native B. thuringiensis strains (FCC4, FCC7, and FCC41), was a clear demonstration of the potential of the set of primers and the experimental methodology described in this report (Fig. 3).
The discovery of novel B. thuringiensis toxins is likely to continue at least into the near future (12). Instead of alternative approaches, based on the use of libraries, on polypeptide sequencing, or on PCR-restriction fragment length polymorphism analysis (16, 25, 26), we propose an efficient strategy that can be routinely used for starting the cloning of cry genes of novel B. thuringiensis strains.
Acknowledgments
This investigation was supported by Fundación para Investigaciones Biológicas Aplicadas and by the Universidad Nacional de Mar del Plata (grant 15/E142). G.L.S. is a career investigator and C.M.B. is a fellow of Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Argentina.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Dov, E., A. Zaritsky, E. Dahan, Z. Barak, R. Sinai, R. Manasherob, A. Khameaev, E. Troitskaya, A. Dubitsky, N. Berezina, and Y. Margalith. 1997. Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 63:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Dov, E., Q. Wang, A. Zaritsky, R. Manasherob, Z. Barak, B. Schneider, A. Khamraev, M. Baizhanov, V. Glupov, and Y. Margalith. 1999. Multiplex PCR screening to detect cry9 genes in Bacillus thuringiensis strains. Appl. Environ. Microbiol. 65:3714-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Dov, E., R. Manasherob, A. Zaritsky, Z. Barak, and Y. Margalith. 2001. PCR analysis of cry7 genes in Bacillus thuringiensis by the five conserved blocks of toxins. Curr. Microbiol. 42:96-99. [DOI] [PubMed] [Google Scholar]
- 5.Bourque, S. N., J. R. Valéro, J. Mercier, M. C. Lavoie, and R. C. Levesque. 1993. Multiplex polymerase chain reaction for detection and differentiation of the microbial insecticide Bacillus thuringiensis. Appl. Environ. Microbiol. 59:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo, A., S. Sarabia, L. Lopez, H. Ontiveros, C. Abarca, A. Ortíz, M. Ortíz, L. Lina, F. J. Villalobos, G. Peña, M. E. Nuñez-Valdez, M. Soberón, and R. Quintero. 1998. Characterization of cry genes in a Mexican Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 64:4965-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carozzi, N. B., V. C. Kramer, G. W. Warren, S. Evola, and M. G Koziel. 1991. Prediction of insecticidal activity of Bacillus thuringiensis strains by polymerase chain reaction product profiles. Appl. Environ. Microbiol. 57:3057-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerón, J. L., A. Ortíz, R. Quintero, L. Güereca, and A. Bravo. 1995. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 61:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerón, J., L. Covarrubias, R. Quintero, A. Ortíz, M. Ortíz, E. Aranda, L. Lina, and A. Bravo. 1994. PCR analysis of the cryI insecticidal family genes from Bacillus thuringiensis. Appl. Environ. Microbiol. 60:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaufaux, J., M. Marchal, N. Gilois, I. Jehanno, and C. Buisson. 1997. Investigation of natural strains of Bacillus thuringiensis in different biotopes throughout the world. Can. J. Microbiol. 43:337-343. [Google Scholar]
- 11.Chilcot, C. N., and P. J. Wigley. 1993. Isolation and toxicity of Bacillus thuringiensis from soil and insect habitats in New Zealand. J. Invertebr. Pathol. 61:244-247. [Google Scholar]
- 12.Crickmore, N. 2002. The diverse armoury of the Bt crystal, p. 147-152. In The Society for Invertebrate Pathology (ed.), Embrapa Soja, Londrina, Brazil. Proceedings of the 6th International Conference on Bacillus thuringiensis.
- 13.Crickmore, N., D. R. Zeigler, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, A. Bravo, and D. H. Dean. 19 April 2004, revision date. Bacillus thuringiensis toxin nomenclature. http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html.
- 14.Delécluse, A., J. F., Charles, A. Klier, and G. Rapoport. 1991. Deletion by in vivo recombination shows that the 28-kilodalton cytolytic polypeptide from Bacillus thuringiensis subsp. israelensis is not essential for mosquitocidal activity. J. Bacteriol. 173:3374-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delucca, A. J., II, J. Simonson, and A. D. Larson. 1981. Bacillus thuringiensis distribution in soils of the United States. Can. J. Microbiol. 27:865-870. [DOI] [PubMed] [Google Scholar]
- 16.Ellis, R. T., B. A. Stockhoff, L. Stamp, H. E. Schnepf, G. E. Schwab, M. Knuth, J. Russell, G. A. Cardineau, and K. E. Narva. 2002. Novel Bacillus thuringiensis binary insecticidal crystal proteins active on western corn rootworm, Diabrotica virgifera virgifera LeConte. Appl. Environ. Microbiol. 68:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feitelson, J. S., J. Payne, and L. Kim. 1999. Bacillus thuringiensis: insects and beyond. Bio/Technology 10:271-275. [Google Scholar]
- 18.Ferrandis, M. D., V. M. Juárez-Pérez, R. Frutos, Y. Bel, and J. Ferré. 1999. Distribution of cryI, cryII and cryV genes within Bacillus thuringiensis isolates from Spain. Syst. Appl. Microbiol. 22:179-185. [Google Scholar]
- 19.Forsyth, G., and N. A. Logan. 2000. Isolation of Bacillus thuringiensis from Northern Victoria Land, Antarctica. Lett. Appl. Microbiol. 30:263-266. [DOI] [PubMed] [Google Scholar]
- 20.Gleave, A. P., R. Williams, and R. J. Hedges. 1993. Screening by polymerase chain reaction of Bacillus thuringiensis serotypes for the presence of cryV-like insecticidal protein genes and characterization of a cryV gene cloned from B. thuringiensis subsp. kurstaki. Appl. Environ. Microbiol. 59:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grochulski, P., L. Masson, S. Borisova, M. Pusztai-Carey, J. L. Schwartz, R. Brousseau, and M. Cygler. 1995. Bacillus thuringiensis CryIA(a) insecticidal toxin: structure and channel formation. J. Mol. Biol. 254:447-464. [DOI] [PubMed] [Google Scholar]
- 22.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 23.Juárez-Pérez, V. M., M. D. Ferrandis, and R. Frutos. 1997. PCR-based approach for detection of novel Bacillus thuringiensis cry genes. Appl. Environ. Microbiol. 63:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, W. S., J. H. Lin, C. C. Tzeng, S. S. Kao, and K. F. Chak. 2000. Cloning of two new cry genes from Bacillus thuringiensis subsp. wuhanensis strain. Curr. Microbiol. 40:227-232. [DOI] [PubMed] [Google Scholar]
- 25.Kuo, W. S., and K. F.Chak. 1996. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl. Environ. Microbiol. 62:1369-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, H.-K., and S. S. Gill. 1997. Molecular cloning and characterization of a novel mosquitocidal protein gene from Bacillus thuringiensis subsp. fukuokaensis. Appl. Environ. Microbiol. 63:4664-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, P. A. W., and R. S. Travers. 1989. World wide abundance and distribution of Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 55:2437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson, L., M. Erlandson, M. Puzstai-Carey, R. Brousseau, V. Juárez-Pérez, and R. Frutos. 1998. A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl. Environ. Microbiol. 64:4782-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nester, E. W., L. S. Thomashow, M. Metz, and M. Gordon. 2002. 100 Years of Bacillus thuringiensis: a critical scientific assessment. [Online.] American Society for Microbiology, Washington, D.C. http://www.asmusa.org. [PubMed]
- 30.Porcar, M., and V. Juárez-Pérez. 2003. PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol. Rev. 26:419-432. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Servant, F., C. Bru, S. Carrère, E. Courcelle, J. Gouzy, D. Peyruc, and D. Kahn. 22November2003, posting date. ProDom: automated clustering of homologous domains. Brief. Bioinformatics 3:246-251. [Online.] http://prodes.toulouse.inra.fr/prodom/2002.1/html/home.php. [DOI] [PubMed] [Google Scholar]
- 34.Shelton, A. M., J.-Z. Zhao, and R. T. Roush. 2002. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu. Rev. Entomol. 47:845-881. [DOI] [PubMed] [Google Scholar]
- 35.Smith, R. A., and G. A. Couche. 1991. The phylloplane as a source of Bacillus thuringiensis variants. Appl. Environ. Microbiol. 57:311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song, F., J. Zhang, A. Gu, Y. Wu, L. Han, K. He, Z. Chen, J. Yao, Y. Hu, G. Li, and D. Huang. 2003. Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl. Environ. Microbiol. 69:5207-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uribe, D., W. Martinez, and J. Cerón. 2003. Distribution and diversity of cry genes in native strains of Bacillus thuringiensis obtained from different ecosystems from Colombia. J. Invertebr. Pathol. 82:119-127. [DOI] [PubMed] [Google Scholar]