Abstract
Helicobacter pylori is a human gastric pathogen associated with gastric and duodenal ulcers as well as specific gastric cancers. H. pylori infects approximately 50% of the world's population, and infections can persist throughout the lifetime of the host. Motility and chemotaxis have been shown to be important in the infection process of H. pylori. We sought to address the specific roles of chemotaxis in infection of a mouse model system. We found that mutants lacking cheW, cheA, or cheY are all nonchemotactic and infect FVB/N mice with an attenuated phenotype after 2 weeks of infection. If infections proceeded for 6 months, however, this attenuation disappeared. Histological and culture analysis revealed that nonchemotactic mutants were found only in the corpus of the stomach, while the wild type occupied both the corpus and the antrum. Further analysis showed that nonchemotactic H. pylori isolates had an increased 50% infectious dose and were greatly outcompeted when coinfected with the wild type. If nonchemotactic mutants were allowed to establish an infection, subsequent infection with the wild type partially displaced the nonchemotactic mutants, indicating a role for chemotaxis in maintenance of infection. The data presented here support four roles for chemotaxis in H. pylori mouse infections: (i) establishing infection, (ii) achieving high-level infection, (iii) maintaining an infection when there are competing H. pylori present, and (iv) colonizing all regions of the stomach.
Helicobacter pylori is a motile, chemotactic bacterium that colonizes the stomachs of ∼50% of the world's population (13). Infection with H. pylori can persist throughout the lifetime of the host and can cause symptoms ranging from mild gastritis to gastric and duodenal ulcers to cancers, such as mucosa-associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma (4, 39).
Motility and chemotaxis, directed swimming, are survival factors for many bacterial species. Both motility and chemotaxis aid commensal and pathogenic infections, but little is known of the precise benefits of these processes. For example, the 50% lethal dose (LD50) of Vibrio anguillarum nonchemotactic mutants for trout is increased 400-fold (28) (for reviews see references 22 and 30). In contrast, some nonchemotactic mutants of V. cholerae colonize infant mice better than does the wild type, in part at least because they occupy a larger portion of the gastrointestinal tract (11, 20). In most of these cases, however, it is not known why motility mutants fare differently than the wild type. H. pylori is an excellent organism with which to ascertain motility's role in infection, because this bacterium is thought to lack a significant environmental niche outside of the human host and, thus, likely uses motility and chemotaxis within the host. In fact, disruption of genes involved in either motility or chemotaxis attenuates colonization of mice and piglets (16, 17, 19, 24, 29). Although these findings indicate that motility and chemotaxis play important roles during infection, these experiments did not define their contributions to the establishment and/or maintenance of infection.
Chemotaxis has been extensively studied in the model organism Escherichia coli. E. coli encodes several chemoreceptors that sense environmental conditions and relay this information to a histidine kinase, CheA, through the coupling protein CheW. CheA acts to phosphorylate the response regulator CheY. CheY, in turn, interacts with the flagellar motor in its phosphorylated form to alter both the rotational direction of the flagellum and the swimming path of the bacterium (5, 37). Loss of any of the proteins that act downstream of the receptors results in a nonchemotactic phenotype in E. coli. The two published H. pylori genomic sequences contain cheW (HP0391/JHP990), cheA (HP0392/JHP989), and cheY (HP1067/JHP358) (2, 38). Disruption of cheW, cheA, or cheY renders H. pylori nonchemotactic in vitro (9, 19, 31). Furthermore, cheY mutants do not colonize piglets and HSD-ICR mice, and cheA mutants are unable to colonize the latter (19). These studies suggest that chemotaxis is required at some stage in H. pylori infection.
To further define the role of chemotaxis during H. pylori infection, we constructed strains with deletions in cheA, cheY, and cheW. In contrast to previous reports (19), we show that nonchemotactic mutants are moderately attenuated for mouse infection. Further analysis showed that nonchemotactic mutants do not infect all regions of the stomach and that they have a distinct disadvantage when the wild type is present. Our data support a model in which chemotaxis guides H. pylori to the mucosa efficiently and helps it locate the antrum. In this model, chemotaxis helps H. pylori compete with the wild type for a limiting nutrient or niche.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Helicobacter pylori strains SS1 and G27 are motile human clinical isolates. SS1 infects mice consistently to high levels. All E. coli and H. pylori strains used are presented in Table 1.
TABLE 1.
Plasmids and bacterial strains used in this studya
| Strain or plasmid | Relevant characteristic(s) | Antibiotic resistance | Reference or source |
|---|---|---|---|
| E. coli | |||
| DH10B | Cloning strain | Gibco BRL | |
| H. pylori | |||
| G27 | Wild type, type I | 12 and Nina Salama | |
| SS1 | Wild type, type I | 25 and Janie O'Rourke | |
| G27 cheW mutant | G27 ΔcheW::aphA3 | Kn | This study |
| SS1 cheW mutant | SS1 ΔcheW::aphA3 | Kn | This study |
| SS1 cheA mutant | SS1 ΔcheA::cat forward | Cm | This study |
| SS1 cheA mutant | SS1 ΔcheA::cat reverse | Cm | This study |
| LC143 | SS1 rdxA::cat | Cm, Mt | This study |
| LC144 | SS1 rdxA::aphA3 | Kn, Mt | This study |
| SS1 cheY mutant | SS1 ΔcheY::cat102 | Cm | This study |
| SS1 cheY complemented | SS1 ΔcheY::cat102 rdxA::cheY | Cm, Mt | This study |
| Plasmids | |||
| pBluescript KS+ (pBS) | Cloning vector | Ap | Stratagene |
| pBS-Kan | pBS with aphA3 from Campylobacter coli | Ap, Kn | Nina Salama |
| pBS-cat | pBS with cat gene from C. coli | Ap, Cm | 33 and Nina Salama |
| pKT10 | pBS with 2,653-bp H. pylori cheW and flanking sequence inserted into EcoRV site | Ap | This study |
| pKT11 | pKT10 with 334-bp deletion in cheW replaced by aphA3: ΔcheW::aphA3 | Ap, Kn | This study |
| pKT20 | pBS with 6,879-bp H. pylori cheA and flanking sequence inserted into EcoRV site | Ap | This study |
| pKT22 | pKT20 with 1,638-bp deletion in cheA and cat inserted in the opposite orientation of cheA | Ap, Cm | This study |
| pKT23 | pKT20 with 1,638-bp deletion in cheA and cat inserted in the same orientation as cheA | Ap, Cm | This study |
| pRdxA | pBC-SK− with 5′ and 3′ regions of H. pylori rdxA locus flanking a polylinker | Cm | 35 and Johannes Kusters |
| pLC292 | 0.75-kb KpnI-SacI fragment from | Ap | This study |
| pRdxA cloned into the KpnI-SacI sites of pBS-SK+ | |||
| pLC307 | 0.8-kb HincII fragment of pBS-cat cloned into the SmaI site of pLC292 | Ap, Cm | This study |
| pLC308 | 1.4-kb fragment from pBS-Kan using KANupstrm and KANdowns primers cloned into the SmaI site of pLC292 | Ap, Kn | This study |
| pKO126 | pBS::cheY | Ap | 3 |
| pKO127 | pKO126::cat-mut (cheY102) | Ap, Cm | This study |
| pKO140 | pLC292::cheY | Ap | This study |
| pCat-mut | pBScat mutagenized to place restriction sites (BamHI, SmaI, AscI) between the end of cat and its transcriptional terminators to create cat-mut | Ap, Cm | This study |
The following plasmids were constructed with both H. pylori SS1 and G27 genes cloned into them: pKT10, pKT11, pKT20, pKT22, pKT23. Antibiotic abbreviations are as follows: Ap, ampicillin; Cm, chloramphenicol; Kn, kanamycin; Mt, metronidazole.
E. coli DH10B was grown in Luria-Bertani (LB) broth as described previously (6), without the addition of NaOH at 37°C. H. pylori was cultured on Columbia horse blood agar (CHBA) as previously described (29) or in Brucella broth plus 10% (vol/vol) fetal bovine serum (FBS) (BB10; Gibco). All H. pylori growth was at 37°C under microaerobic conditions accomplished either by Campygen packs (Oxoid) or a gas mixture of 5 to 10% O2, 10% CO2, and 80 to 85% N2. Soft-agar assays for H. pylori were performed in Brucella broth plus 2.5% (vol/vol) FBS, 0.35% (wt/vol) agar. H. pylori retrieved from mice were grown on CHBA plates supplemented with 200 μg of bacitracin/ml and 10 μg of nalidixic acid/ml. Selective antibiotics were added at the following concentrations for E. coli and H. pylori, respectively: kanamycin, 30 and 15 μg/ml; chloramphenicol, 20 and 10 μg/ml; ampicillin, 100 μg/ml (E. coli only); and metronidazole, 9 μg/ml (H. pylori only).
All chemicals were from Sigma or Fisher, media components were from Difco or BBL, and molecular biology reagents were from New England Biolabs or Stratagene. All sequencing was carried out at the University of California—Berkeley.
Cloning and mutagenesis.
All primers for cloning and inverse PCR mutagenesis (iPCR) were designed from the two published H. pylori genome sequences (2, 38) and are listed in Table 2. Plasmids and corresponding strains are listed in Table 1.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| cheW4 | 5′ GCTGTCTTTAGCAAAAGGCAACTC |
| cheW5 | 5′ TCTCTGATGATGGCAAAGGGTTAG |
| cheWdel3up | 5′ GTTTATCCATAGCCCCTATGGG |
| cheWdel3dn | 5′ GACTCGTGTGTAACCCAATGGG |
| KANupstrm | 5′ GGCCGGATCCGATAAACCCAGCGAACC |
| KANdowns | 5′ GGCCAAGCTTTTTAGACATCTAAATC |
| cheAup1 | 5′ TATCGCTCAAAGTCTCTGGC |
| cheAdown1 | 5′ TCACTGAAGCTGTGGATGGG |
| cheAdelup1 | 5′ GGCTTGGCTGATCAAAAAATTGGC |
| cheAdeldn1 | 5′ GGCGACTCTAAAAATGCGATTGAG |
| rdxAstart | 5′ CGCCATTCTTGCAAGATGTTTG |
| rdxAend | 5′ CTCGCTTCTGCCACCCTCTT |
| cheY1 | 5′ GAAGGGATCCTTACAAATAAGAACGCTC |
| LccheY2 | 5′ GCTCTAGATCAATCGTTTGTCCCTAAAACAACC |
| cheY3 | 5′ GGAAGCTGCAGGTTTCTTTATCGTCAAACGC |
| cheY4 | 5′ GCTCATTGAACGCTCCATTTAGC |
| 126del1 | 5′ CCAGTAGTTTCAAAGTGCTTC |
| 126del2 | 5′ CCAACGATTGAGTGTTAAAGCC |
Cloning and mutagenesis of cheW.
cheW and flanking sequences were amplified from H. pylori SS1 and G27 genomic DNA (QIAGEN DNeasy) using primers cheW4 and cheW5. The identity of the resulting 2.65-kb PCR fragment was verified by sequencing, and then it was cloned into EcoRV-cut pBluescript KS+ to generate pKT10 (Table 1). A 334-bp deletion was made in the cheW coding sequence by iPCR using primers cheWdel3up and cheWdel3dn. aphA3 was amplified by PCR from pBS-Kan with primers KANupstrm and KANdowns, and the resulting product was ligated with the cheW iPCR product to generate pKT11. pKT11 was used to transform H. pylori G27 and SS1 by natural transformation as previously described (33), using the cheW gene from that strain. Kanamycin-resistant colonies were colony purified twice, and the ΔcheW::aphA3 genetic architecture was verified by PCR amplification of chromosomal DNA using primers cheW4 and cheW5. Southern blotting was used to verify that there was only a single copy of aphA3 (Amersham Pharmacia).
Cloning and mutagenesis of cheA and cheY.
cheA was mutagenized in a manner similar to that used for cheW, but primers cheAup1 and cheAdown1 were used for the cloning and primers cheAdelup1 and cheAdeldn1 were used for the iPCR deletion. cat was cut from pBS-cat using HincII and was ligated into the cheA deletion. The orientation of the chloramphenicol resistance cassette was determined by restriction digestion, and the plasmids were named pKT22 (cat oriented opposite to cheA) and pKT23 (cat oriented the same as cheA). Both plasmids were used to transform H. pylori strain SS1 (33). The genetic architecture of the ΔcheA::cat mutations were verified using primers cheAup1 and cheAdown1, and a Southern blot was performed using a probe for the cat gene.
cheY was mutagenized by the same method as that for cheW, starting from pKO126 and using iPCR with primers 126del1 and 126del2. Because there are several genes downstream of cheY, we used a cat gene without transcriptional terminators. cat-mut was digested from pCat-mut using SmaI and HincII and was ligated with iPCR-cheY to create pKO127, in which the cat gene orientation is the same as that for cheY. H. pylori transformation and mutant verification was carried out as described for cheW. In addition, reverse transcription-PCR (RT-PCR) showed that a gene downstream of cheY (copA) was still transcribed, supporting the suggestion that this is a nonpolar mutation (data not shown).
Complementation of cheY.
cheY and its promoter were amplified from pKO126 using primers cheY1 and LCcheY2. The PCR fragment was digested with BamHI and XbaI and was cloned into BamHI-XbaI-cut pLC292, an ampicillin-resistant version of pRdxA, to create pKO140. pKO140 was used to transform H. pylori SS1 to metronidazole resistance, as described above, using cell-extract-treated plasmid (15). Genomic DNA from SS1 rdxA::cheY was used to transform SS1 ΔcheY::cat to metronidazole resistance to create ΔcheY::cat rdxA::cheY. Verification of the rdxA::cheY insertion and retention of cheY::cat was done by PCR using primers that flank the mutations (rdxAstart/rdxAend and cheY3/cheY4).
Creation of rdxA::cat and rdxA::kan.
A HincII-fragment from pBScat bearing the cat gene was cloned into SmaI-cut pLC292 to create pLC307. A PCR fragment bearing aphA3, as described above in the cheW cloning section, was ligated to SmaI-cut pLC292 to create pLC308. These plasmids were used to transform H. pylori to chloramphenicol or kanamycin resistance as described above, and the chromosomal architecture was verified by PCR with primers rdxAstart and rdxAend.
Mouse inoculations.
All animal protocols were approved by the Institutional Animal Use and Care Committee. H. pylori SS1 (laboratory passaged along with the isogenic mutants) and its isogenic mutants were grown overnight in shaking BB10 liquid culture. The resulting bacterial concentration of the culture was determined by measuring the optical density at 600 nm (OD600) (3 × 108 bacteria/ml/OD). Four- to 6-week-old male FVB/N mice (Charles River) were infected once with approximately 1 × 107 to 5 × 107 CFU in 1 ml of BB10 by oral gavage (20 gauge, 38 mm length; Popper). Coinfections were performed with 1 × 107 to 5 × 107 CFU/ml of each strain. All inocula were plated to obtain the exact concentration of bacteria. Mice were sacrificed usually at 2 weeks postinoculation, and the stomachs were processed as described previously (29). Half of the stomach was homogenized with a sterile pestle in BB10 and plated to determine the CFU/gram of stomach. To obtain coinfection and superinfection data, the stomach homogenates were plated onto two media that differentiate between the two strains.
Superinfections were performed by infection with 1 × 107 to 5 × 107 CFU of the primary infecting strain, followed 1 week later by the same amount of the second strain. Mock superinfection controls were performed by administering 1 ml of BB10 as the second inoculum.
The 50% infectious dose (ID50) measurements were performed by infecting mice as described above with serial dilutions covering a target range of 25 to 2.5 × 106 CFU/ml. C57BL/6 mice (Taconic Labs) were used in addition to FVB/N. Mice were sacrificed after 3 days. Based on the number of infected mice at each inoculation dose, the ID50 was determined by the Reed-Meunch calculation (32).
Histology.
Infected male FVB/N mice were sacrificed 3 weeks after infection. Following gastrectomy, one-half of each stomach was placed in a histology cassette with sponge (Fisher) and was fixed using buffered formalin (Fisher). Stomachs were embedded, sectioned, and stained by Histo-Tec (Hayward, Calif.). For each stomach, one Warthin-Starry- and one hematoxylin-and-eosin-stained section were examined using a 100× oil immersion lens. The examiner was blind to the identity of the infecting stain. Five fields containing stained bacteria were evaluated in the antrum and corpus for each animal.
Statistical analysis.
All statistical analyses were performed using the two-tailed student's t test available at http://graphpad.com/quickcalcs/ttest.cfm. P < 0.05 was considered statistically significant.
RESULTS
ΔcheW, ΔcheY, and ΔcheA mutants are not chemotactic.
ΔcheW, ΔcheY, and ΔcheA mutants were constructed in H. pylori strains G27 and the mouse-infecting strain SS1 by internal deletion and insertion of an antibiotic resistance cassette, resulting in ΔcheW::aphA3, ΔcheY::cat, and ΔcheA::cat strains. These mutants have the appropriate chromosomal architecture by PCR analysis and Southern blotting, and all mutants are motile by microscopic investigation (data not shown). Additionally, none of the mutants had an in vitro growth defect in either of two types of growth analyses. For the first, we grew each strain singly in BB10 broth. For the second, we mixed equal amounts of the mutant and the wild type in BB10 broth and ascertained the ratio of the two strains over several days of growth (data not shown).
To test whether the G27 and SS1 ΔcheW::aphA3, ΔcheY::cat, or ΔcheA::cat mutants were chemotactic in soft agar, low-density agar plates were inoculated with each mutant strain of H. pylori as previously described (29). After 5 days of growth, the mutants had not increased in colonial diameter compared to the wild-type parents (data not shown). Because these mutants retained motility but could not form the expanding colony, we concluded they were nonchemotactic, as found by others with different strain backgrounds (9, 19, 31).
Nonchemotactic strains of H. pylori infect FVB/N mice.
To determine whether our nonchemotactic H. pylori mutants could infect mice, male inbred FVB/N mice were infected with wild-type H. pylori strain SS1 or its isogenic ΔcheW, ΔcheY, or ΔcheA mutant for 2 weeks. We chose FVB/N mice because they have been used in other studies with H. pylori (18, 21). In contrast to previous results using defined nonchemotactic mutants of SS1 H. pylori and HSD-ICR mice (19), we found that our nonchemotactic mutants infected all mice (Fig. 1). ΔcheW mutants reisolated from mouse stomachs retained their nonchemotactic phenotype according to the soft-agar assay 1 week, 2 weeks, 2 months, and 6 months postinfection (see experiment below; data not shown), suggesting that the ability of nonchemotactic mutants to infect mice is not due to reversion to the chemotactic phenotype.
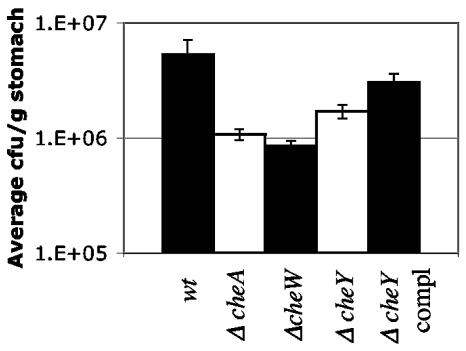
FIG. 1.
Nonchemotactic H. pylori mutants infect FVB/N mice to high but significantly lower levels than wild-type (wt) H. pylori. Wild-type (n = 12), ΔcheW (n = 11), ΔcheA (n = 18), ΔcheY (n = 4), and cheY-complement (n = 4) H. pylori SS1 strains were administered orally to mice. ΔcheW and ΔcheA were tested in at least two separate infections. ΔcheA and ΔcheW mutants colonized all mice significantly less well than the wild type (P < 0.01). Results from both ΔcheA strains (cat inserted in both orientations) were averaged together, as they were not statistically different from each other (P > 0.1). Error bars represent the standard errors of the means (SEM).
Although SS1 ΔcheW, ΔcheY, or Δ cheA each infected mice, the numbers of H. pylori in the stomachs were significantly lower than levels achieved by wild-type bacteria. Mice infected with the wild type had an average of 3.0 × 106 CFU/gram of stomach after 2 weeks, while mice infected with ΔcheW, ΔcheA, or ΔcheY mutants had 8.3 × 105, 1.1 × 106, and 1.7 × 106 CFU/gram of stomach, respectively (Fig. 1). Introduction of a wild-type copy of cheY into the cheY deletion strain increased the level of infection to nearly wild-type levels, indicating that the attenuation in infection of the ΔcheY strain is due to loss of cheY. These complementation data are more striking in the coinfection experiments (presented below), where both the cheY-complemented strain and wild-type H. pylori are recovered at nearly the same levels. These results indicate that nonchemotactic mutant H. pylori strains have a subtle but significant attenuation for mouse infection in single-strain infections. This ability of nonchemotactic mutants to infect allowed us to tease out how chemotaxis aids infection.
Nonchemotactic mutants are outcompeted by wild-type H. pylori when coinfected.
To address whether the nonchemotactic mutant H. pylori infection defect observed in singe-strain infections would be altered by the presence of wild-type bacteria, we carried out coinfection experiments. Mice were coinfected with approximately equal numbers of wild-type and nonchemotactic bacteria, and at 2 weeks postinfection we determined the competitive index. In all experiments, the wild type outcompeted ΔcheW and ΔcheY mutant bacteria (Table 3). In contrast, when two nonchemotactic (ΔcheW ΔcheA or ΔcheW ΔcheY) bacterial strains were coinfected, recovery of both strains was similar, indicating that these different nonchemotactic strains are indistinguishable in infection ability and thus likely share the same defect (Table 3). The inability of the nonchemotactic H. pylori mutants to be recovered from mice when coinoculated with the wild type shows that there is a strong need for chemotaxis when wild-type H. pylori is present.
TABLE 3.
Coinfections of mice with nonchemotactic and wild-type H. pyloria
| Exp (n) | Strain | Inoculum dose (input) (CFU) | Avg CFU/g of stomach (output) | Input ratio | Competitive index (output ratio/input ratio) |
|---|---|---|---|---|---|
| 1 (4) | cheW | 2.76 × 106 | 4.82 × 105 | 1.13 | 0.9 |
| cheA | 2.43 × 106 | 4.80 × 105 | |||
| 2 (5) | cheW | 2.5 × 107 | 1.8 × 105 | 1.39 | 0.3 |
| cheY | 1.8 × 107 | 4.26 × 105 | |||
| 3 (5) | Wild type | 1.8 × 107 | 3.6 × 106 | 0.3 | <0.001 |
| cheW | 5.3 × 106 | <100 | |||
| 4 (6) | Wild type | 1.32 × 107 | 4.07 × 106 | 2.26 | 0.009 |
| cheW | 2.98 × 107 | 8.1 × 104 | |||
| 5A (5) | Wild type | 2.53 × 107 | 2.42 × 106 | 2.39 | <0.001 |
| cheW | 6.05 × 107 | 616 | |||
| 5B (5) | Wild type | 2.53 × 107 | 2.86 × 106 | 0.8 | <0.001 |
| cheW | 2.02 × 107 | 188 | |||
| 5C (5) | Wild type | 2.53 × 107 | 2.16 × 106 | 0.27 | <0.001 |
| cheW | 6.7 × 106 | 1500 | |||
| 6 (4) | Wild type | 8.6 × 107 | 5.74 × 106 | 1 | 0.008 |
| cheY | 8.6 × 107 | 4.59 × 104 | |||
| 7 (4) | Wild type | 8.6 × 107 | 1.49 × 106 | 1.28 | 1.03 |
| cheY/complement | 1.1 × 108 | 1.96 × 106 |
Each strain listed under Strain was mixed and coinoculated into mice for 2-week infections. Competitive index is the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type).
Introduction of a wild-type copy of cheY into the ΔcheY H. pylori mutant led to elimination of the competition defect (Table 3), indicating that the inability of ΔcheY bacteria to compete with the wild type is due to the loss of cheY and not a polar or other secondary effect. The results of the complemented cheY in these competition assays suggests that the slightly smaller numbers of the complemented cheY mutant compared to that of the wild type in the single-strain infections is likely due to the small number of animals used in that former experiment.
Based on both the in vitro chemotaxis assays and the results of mouse infections, our data suggest that ΔcheA, ΔcheW, and ΔcheY strains behave similarly in vitro and in vivo. The remainder of our experiments were carried out only with the ΔcheW SS1 mutant, because it is representative of all nonchemotactic mutants. Furthermore, we chose to use the ΔcheW mutant because cheW is predicted to be at the end of its operon (2, 38) and is least likely to exhibit any subtle polar effects.
ΔcheW H. pylori strains have an initial colonization defect.
The next infection aspect we examined was whether nonchemotactic H. pylori strains have an initial colonization defect by determining the 50% infectious dose (ID50) for the ΔcheW mutant and comparing it to that of the wild type. Serial dilutions of ΔcheW and wild-type H. pylori SS1 were administered to FVB/N mice, and the percentage of animals infected at each dose was determined. The ID50 of the ΔcheW strain was 4.7 × 104 CFU and was 200 CFU for the wild-type strain (Table 4). This >100-fold increase in the ID50 suggests the ΔcheW mutant is considerably impaired for establishing infection.
TABLE 4.
ID50 of wild-type and nonchemotactic H. pylori in FVB/N micea
| Strain | Doseb (n) | % Infected |
|---|---|---|
| Wild-type SSI | 2.0 × 104 (5) | 100 |
| 2.0 × 103 (4) | 100 | |
| 2.0 × 102 (5) | 40 | |
| SS1 cheW | 8.6 × 105 (5) | 80 |
| 8.6 × 104 (5) | 100 | |
| 8.6 × 103 (5) | 0 | |
| 8.6 × 102 (5) | 0 |
The data is from one experiment but is characteristic of two separate experiments.
Dose in CFU.
To verify that the results obtained were not unique to FVB/N mice, we analyzed the ID50 of wild-type and nonchemotactic H. pylori for C57BL mice, a commonly used H. pylori mouse model strain. The wild type and isogenic ΔcheW mutants were administered at doses of 104 to 107 and 105 to 108, respectively, and bacterial levels were determined 3 days postinoculation. All animals were infected at all doses (data not shown). These results indicate the ID50 of wild-type SS1 in C57BL mice is less than 104, and that for the ΔcheW strain is less than 105. These findings suggest that C57BL mice are similar to FVB/N mice in their susceptibility to infection and their ability to allow nonchemotactic mutants to infect, and this suggestion argues that our findings are applicable to other mouse strains.
ΔcheW mutants are partially displaced by the wild type.
To determine whether bacterial chemotaxis is important for infection maintenance, we asked whether an established infection with nonchemotactic bacteria could be displaced upon superinfection with wild-type H. pylori. We reasoned that if chemotaxis plays an ongoing role during maintenance, superinfection with wild-type bacteria would displace the nonchemotactic mutant. Alternatively, if chemotaxis were no longer required after initiation, the nonchemotactic mutant would remain after superinfection.
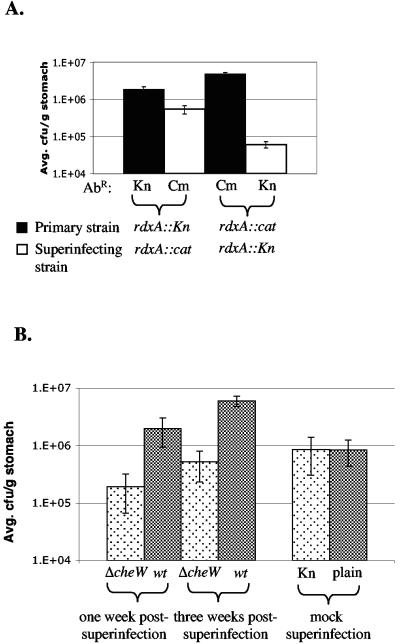
As a control for these experiments, we determined how two H. pylori strains with equal infecting abilities would behave during superinfections. We chose two strains, each marked at the rdxA locus with a different antibiotic resistance. Previous results in our laboratory showed that these rdxA mutants infect mice similarly (S. M. Williams and K. M. Ottemann, unpublished data). Mice were infected first with one marked H. pylori strain, the infection was allowed to establish for 1 week, and the mice were superinfected with the second strain. Regardless of the order in which each strain was inoculated, the initial infecting strain was recovered at significantly higher numbers than the superinfecting strain (Fig. 2A). The superinfecting strain, however, was able to infect. These results suggest that, in the context of two H. pylori strains with equal ability to infect, the initially infecting strain has an advantage over the superinfecting strain.
FIG. 2.
H. pylori recoveries from superinfections reveal a partial displacement of ΔcheW by the wild type (wt). AbR denotes the antibiotic resistance of the indicated strain. Kn, kanamycin; Cm, chloramphenicol. (A) rdxA::aphA3 (rdxA::Kn) infection followed 1 week later with rdxA::cat (n = 5). Colonization levels were significantly different (P < 0.01). In the reciprocal experiment, rdxA::cat was superinfected with rdxA::Kn (n = 4) (P < 0.01). Error bars represent standard errors of the means (SEM). (B) Mice infected initially with ΔcheW and superinfected 1 week later with wild-type H. pylori for 1 week (n = 13) (P < 0.01) or 3 weeks postsuperinfection (n = 5) (P < 0.01). The 1-week experiment was repeated twice with similar results, but only one data set is shown. The ΔcheW output from the 3-week infection was not significantly different from that of the 1-week infection (P > 0.05), but ΔcheW outputs from the superinfection experiment were different from those of the mock superinfection where mice were gavaged with BB10 (n = 6) (P < 0.01). Mock superinfection outputs plated on selective and nonselective media are included to show that the selective media do not confer lower counts from the outputs.
In contrast, superinfection of an established ΔcheW H. pylori infection with wild-type bacteria resulted in significantly higher recovery of the superinfecting wild-type strain (Fig. 2B). Similar results were obtained when the infection was allowed to persist for 3 weeks postsuperinfection, indicating that our observations likely represent a steady state. Additionally, mice superinfected with the wild type had significantly less ΔcheW mutant than mock-superinfected mice that were gavaged with broth subsequent to the initial infection with ΔcheW. Plating of ΔcheW mutants recovered from mock superinfections on both selective and nonselective media showed that the selective medium does not confer lower counts from the outputs. These observations suggest that wild-type bacteria are able to displace some nonchemotactic mutants at early time points postinfection and argue that chemotaxis plays an ongoing role during the maintenance of infection during the first month.
ΔcheW H. pylori persists as well as the wild type in long-term infections.
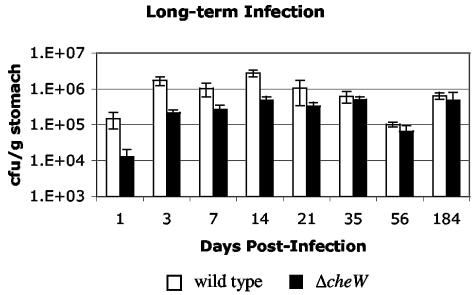
Our superinfection experiments demonstrate that bacterial chemotaxis is not only needed for establishing colonization but also plays a role in maintenance during the first month of infection. To determine whether chemotaxis plays a role in the maintenance of long-term infection, we compared the ability of nonchemotactic and wild-type bacteria to persist in the host over extended periods of time as single infecting strains. Mice were infected with either wild-type or ΔcheW H. pylori, and infection levels were monitored at 1 to 184 days postinfection (Fig. 3). Similar to the results shown in Fig. 1, the ΔcheW mutant was recovered at significantly smaller numbers than wild-type bacteria at early time points (P < 0.05 at 14 days). However, after 2 or 6 months of infection, the outputs of each bacterial strain were not significantly different from each other (P > 0.5 at 6 months). These data suggest that while chemotaxis is important for the initial establishment and early maintenance of infection, it is not required for long-term persistence in the host when the wild type is not present.
FIG. 3.
Six-month infection with wild-type and ΔcheW H. pylori. Shown are averages of all mice at different time points over 6 months for single-strain infections with wild-type H. pylori or ΔcheW H. pylori. For all experiments n = 4, except for day-21 wild type (n = 3) and day 184 ΔcheW (n = 6). Error bars represent the standard errors of the means (SEM).
ΔcheW H. pylori is not found in the antral mucosa.
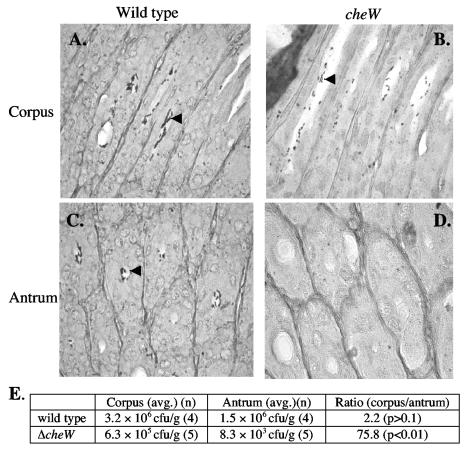
Early in infection, there is less ΔcheW mutant than wild-type H. pylori. To analyze whether nonchemotactic mutants and the wild type occupy the same gastric niches, we analyzed histological sections of mice infected for 3 weeks with either the wild type or the ΔcheW strain. Although bacteria exhibiting typical H. pylori morphology were less prevalent in histological sections from mice singly infected with the ΔcheW mutant than the wild type, we detected both strains in the oxyntic mucosa (corpus). Both strains were seen primarily as foci containing 4 to 20 bacterial cells in the lumen of the glands (Fig. 4A and B). In the antrum, however, we could find only wild-type H. pylori (Fig. 4C and D).
FIG. 4.
Histological samples of oxyntic (corpus) and antral mucosa of FVB/N mice infected with H. pylori. After 3 weeks of infection, both wild-type (A) and cheW (B) H. pylori are visible in the glands of the oxyntic mucosa (corpus). In contrast, the wild type (C) but not the ΔcheW mutants (D) can be located in the antrum. Arrowheads mark bacteria in the glands. (E) Colony counts from the corpus and antrum of stomachs infected with wild-type or ΔcheW H. pylori for 2 weeks. P values refer to the difference in counts between corpus and antrum.
To further support these observations, we infected mice with either the ΔcheW mutant or wild-type H. pylori, sacrificed them at 2 weeks postinfection, physically subdivided the stomach into corpus or antrum as done previously (1), and cultured each portion separately. Our findings support those of the histology: we found the wild type at similar numbers in the corpus and the antrum, while the ΔcheW mutant was barely detectable in the antrum (Fig. 4E). These observations suggest that nonchemotactic mutants occupy only a subset of H. pylori's normal gastric habitats.
DISCUSSION
Nonchemotactic mutants are attenuated in their ability to infect mice, yet they are able to establish infection.
We have shown that nonchemotactic H. pylori mutants are able to establish and maintain infection in mice, although they are defective compared to wild-type bacteria. This conclusion was reached from the compilation of five different experiments: (i) nonchemotactic mutant bacteria are recovered in smaller numbers than the wild type from single-strain infections, (ii) nonchemotactic mutants have a low competitive index when coinfected with the wild type, (iii) nonchemotactic mutants have a higher ID50 than the wild type, (iv) wild-type bacteria partially displace nonchemotactic mutants, and (v) nonchemotactic mutants do not colonize all regions of the stomach. The higher ID50 of nonchemotactic H. pylori strains indicates that these bacteria are impaired in their ability to establish initial colonization of the gastric mucosa. Once this obstacle has been overcome, however, nonchemotactic mutants survive and multiply within the mouse stomach to levels that are only slightly below those of the wild type as long as no competing bacteria are present.
Given the multiple roles described here for chemotaxis during colonization and infection, there are several possible explanations for the attenuation of the nonchemotactic mutants. The initial defect in colonization may be due to an inability of nonchemotactic bacteria to efficiently find and penetrate the mucous layer. During longer term infection, the nonchemotactic mutants may be less able to survive in the gastric mucosa. For example, without the ability to sense directional cues, they may swim to regions of the gastric mucosa that are sloughed off, be unable to locate limited nutrients, or be more susceptible to the host immune response. Such defects would lead to a decrease in bacterial number by either leading to greater bacterial clearance or a reduced bacterial growth rate. Recent work has shown that pH is important for spatial orientation of H. pylori in the gastric mucosa (34). Perhaps relocalization of nonchemotactic H. pylori throughout the mucous would result in greater sloughing of the microbes, although this hypothesis remains to be tested.
Additionally, one of the reasons for the smaller numbers of cheW mutant bacteria is that they are only in a portion of the stomach, the corpus, and not both the antrum and corpus at 3 weeks postinfection. The reason that the nonchemotactic bacteria do not colonize the antrum at this stage is not yet known, but it may be due to reasons like those stated above, such as that the nonchemotactic mutants cannot locate this niche, they grow more slowly within it, or they are eliminated from it.
During any type of mixed infection, nonchemotactic mutants are always outcompeted by the wild type. This outcome is strongly accentuated when the two strains are coinfected at the outset. In this case, very little to no mutant was detected, suggesting that if the wild type is present at the beginning of an infection, it establishes infection exclusively. Consistent with these results, Kavermann and coworkers found that cheA was necessary for infection of Mongolian gerbils in a signature-tagged mutagenesis coinfection experiment in which this mutant competed with other mutants (23). However, we found that when a nonchemotactic mutant precedes the wild type, the two strains can coexist, albeit with the wild-type strain dominating. The observation that the presence of wild-type bacteria greatly exacerbates the defect of the nonchemotactic mutant may give us clues about the signals that guide H. pylori chemotaxis. One simple explanation is that the wild type is better able to locate and utilize a limiting nutrient; thus, the nonchemotactic mutant has a growth disadvantage. Mathematical modeling studies suggest that two H. pylori strains can coexist if they occupy different niches, and thus it would be interesting to analyze whether these coexisting strains are found in different locations (10).
Superinfection with the wild type partially displaces nonchemotactic H. pylori.
We utilized superinfection experiments to reveal the more subtle defects of the nonchemotactic bacterial mutants during the early stages of an established infection. Other work on superinfections utilized nonisogenic strains. A paper by Ayraud et al. describes superinfections of C57BL/6 mice using different human clinical H. pylori isolates (7). They found that only one bacterial strain emerged, and it was usually the primary strain. These results are supported by Danon et al., where some evidence is provided that an established H. pylori strain can prevent colonization by a challenging strain (14). In contrast, with two different mouse-adapted strains of H. pylori that colonize distinct stomach regions, Akada et al. showed that both strains can simultaneously infect, and superinfection does not alter colonization of either strain (1). In agreement with all of these studies, our superinfection experiments show that dual-bacteria infection can be established, but the initially infecting H. pylori strain dominates these mixed infections if both strains are equally fit. These findings suggest that the initial strain has an as-yet-undefined advantage over subsequent strains. Our observation that nonchemotactic H. pylori mutants do not prevent superinfection by wild-type strains but actually are displaced by them underscores the importance of chemotaxis in maintaining an established infection.
Nonchemotactic mutants are able to persist for a 6-month course of infection.
Although superinfection experiments conducted early during the course of infection suggest that there is an ongoing need for bacterial chemotaxis during the maintenance of infection, data from single-strain infections show that nonchemotactic H. pylori mutants are able to persist as well as the wild type for up to 6 months. In fact, over that length of infection, the wild-type H. pylori levels decreased, resulting in very similar numbers of both strains. The ability of the nonchemotactic H. pylori mutant to survive in the host for a long duration supports the idea that if infection can occur, i.e., if the ID50 is overcome, nonchemotactic bacterial mutants can survive well and may not be substantially impaired in maintaining long-term colonization, provided they are not challenged by wild-type H. pylori. Superinfection experiments carried out at longer time points may help to determine whether chemotaxis is still necessary during persistent infection.
FVB/N and C57BL mice are permissive for H. pylori infection.
In addition to the information about the behavior of nonchemotactic H. pylori mutants, this study supports previous findings that the FVB/N mouse strain is particularly susceptible to infection by H. pylori, as shown by its relatively low ID50 (29). FVB/N mice have been used in several H. pylori studies, including examining host parietal cell response upon exposure to H. pylori (27) and using transgenic FVB/N mice to study H. pylori adherence to the Lewis B antigen (18). Additionally, wild-type FVB/N mice have been used to examine the stability of the cag pathogenicity island in vivo (36). Previous work on nonchemotactic H. pylori mutants used a different mouse strain, HSD-ICR (19). HSD-ICR mice are outbred and require two doses of 107 to 108 CFU of H. pylori SS1 administered on two successive days to obtain consistent infection (19, 26). This mouse strain difference may underlie our different experimental findings in that HSD-ICR mice appear to be more resistant to infection by H. pylori than FVB/N mice. There were, however, other differences between the studies of Foynes et al. and ours, including pretreatment of the bacteria with acid prior to infection in the HSD-ICR experiments, which may have contributed to our divergent findings (19).
Originally, Salama et al. reported that the ID50 for wild-type H. pylori strain SS1 in C57BL/6NTac mice is ∼105 (33). Additional experiments, however, revealed a bacterial strain other than SS1 was inadvertently used (N. Salama, personal communication). We found that the ID50 for H. pylori SS1 in C57BL mice is less than 104. In FVB/N mice, our results and those of others indicate that a single inoculum with ∼500 CFU of H. pylori strain SS1 is sufficient for infection of 100% of animals (29). The H. pylori ID50 has been established for CD1 mice at 1.4 × 104 (8). These data suggest that FVB/N and possibly C57BL mice are particularly susceptible to infection by H. pylori and represent convenient model systems in which to study the more subtle aspects of pathogenesis.
In summary, we have shown that nonchemotactic H. pylori strains establish infection poorly, are outcompeted by wild-type microbes, are slow to achieve wild-type infection levels, and do not localize to the antral mucosa. Nonchemotactic mutants have a strong defect in establishing infection, suggesting that chemotaxis guides the bacteria from the harsh stomach lumen to the desirable mucous layer. If given in high doses, however, nonchemotactic mutants are able to establish and maintain infection at almost wild-type levels for at least 6 months. Nonetheless, without chemotaxis the mutants only weakly populate the antrum. Mixed infections support a model in which chemotaxis is needed to find nutrients or niches that are plentiful enough when the nonchemotactic mutant is the sole H. pylori isolate in the stomach but that become limiting when a more fit strain (the wild type) inhabits the same environment.
Acknowledgments
We thank Catherine Beckwith and Corrine Davies (Stanford University) for assistance with the histology; members of the Ottemann laboratory, Nina Salama, Doug Berg, Lalita Ramakrishnan, and Fitnat Yildiz, for helpful discussions and comments on the manuscript; and Andrew Woodruff for creating pCat-mut.
We also thank the Burroughs Welcome Fund Career Award (3295 to K.M.O.), the Ellison Medical Foundation New Scholar Award in Infectious Disease (ID-NS-0030-01 to K.M.O.), and the National Institutes of Health (AI050000 to K.M.O.) for funding.
Editor: D. L. Burns
REFERENCES
- 1.Akada, J. K., K. Ogura, D. Dailidiene, G. Dailide, J. M. Cheverud, and D. E. Berg. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149:1901-1909. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. Dejonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merber, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trast. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature (London) 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Andermann, T. M., Y.-T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors promote Helicobacter pylori infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 1994. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA 272:65-69. [PubMed] [Google Scholar]
- 5.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 7.Ayraud, S., B. Janvier, L. Salaun, and J. L. Fauchere. 2003. Modification in the ppk gene of Helicobacter pylori during single and multiple experimental murine infections. Infect. Immun. 71:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51:15-32. [DOI] [PubMed] [Google Scholar]
- 9.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriology 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaser, M. J., and D. Kirschner. 1999. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc. Natl. Acad. Sci. USA 96:8359-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, S. M., and A. Camilli. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 101:5018-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 14.Danon, S. J., B. J. Luria, R. E. Mankoski, and K. A. Eaton. 1998. RFLP and RAPD analysis of in vivo genetic interactions between strains of Helicobacter pylori. Helicobacter 3:254-259. [DOI] [PubMed] [Google Scholar]
- 15.Donahue, J. P., D. A. Israel, R. M. J. Peek, M. J. Blaser, and G. G. Miller. 2000. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37:1066-1074. [DOI] [PubMed] [Google Scholar]
- 16.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 17.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falk, P. G., L. Bry, J. Holgersson, and J. I. Gordon. 1995. Expression of a human alpha-1,3/4-fucosyltransferase in the pit cell lineage of FVB/N mouse stomach results in production of Le-b-containing glycoconjugates: a potential transgenic mouse model for studying Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 92:1515-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freter, R., and P. C. O'Brien. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: fitness and virulence of nonchemotactic Vibrio cholerae mutants in infant mice. Infect. Immun. 34:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guruge, J. L., P. G. Falk, R. G. Lorenz, M. Dans, H.-P. Wirth, M. J. Blaser, D. E. Berg, and J. I. Gordon. 1998. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 95:3925-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 23.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 26.McColm, A. 1997. Nonprimate animal models of H. pylori infection, p. 241. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Totowa, N.J.
- 27.Mills, J. C., A. J. Syder, C. V. Hong, J. L. Guruge, F. Raaii, and J. I. Gordon. 2001. A molecular profile of the mouse gastric parietal cell with and without exposure to Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:13687-13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole, R., Milton, D. L., and Wolf-Watz, H. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 29.Ottemann, K. M., and A. Lowenthal. 2002. Helicobacter pylori uses motility for both initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 31.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 32.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 33.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. The vacuolating cytotoxin of Helicobacter pylori plays a role during colonization of a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber, S., M. Konradt, C. Groll, P. Scheid, G. Hanauer, H. O. Werling, C. Josenhans, and S. Suerbaum. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101:5024-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smeets, L. C., J. J. E. Bijlsma, S. Y. Boomkens, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sozzi, M., M. Crosatti, S. K. Kim, J. Romero, and M. J. Blaser. 2001. Heterogeneity of Helicobacter pylori cag genotypes in experimentally infected mice. FEMS Microbiol. Lett. 203:109-114. [DOI] [PubMed] [Google Scholar]
- 37.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. I. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magansanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 38.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Lftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Tizegerald, N. Lee, M. D. Adams, E. K. Kichey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Person, J. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Wathey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 338:539-547. [DOI] [PubMed] [Google Scholar]
- 39.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]