Abstract
Background
Diet and obesity influence prostate cancer risk and progression–effects that may be mediated through the gut microbiome.
Objective
Explore relationships between diet, gut microbes and Gleason sum in overweight and obese prostate cancer patients enrolled in a presurgical weight loss trial.
Design
Randomized Controlled Trial (NCT01886677) secondary analysis.
Participants/setting
In 2013–2014, 40 prostate cancer patients in the southeastern United States were randomized and equally allocated to weight loss and wait-list control arms while they awaited prostatectomy; stool samples were collected on a subset of 22.
Intervention
Registered dietitians and exercise physiologists provided semi-weekly in-person and telephone-based guidance on calorie-restricted diets and exercise to promote ~0.91 kg/week weight loss.
Main outcome measures
Baseline and follow-up 24-hour dietary recalls were conducted and analyzed (ASA24) for macronutrients, micronutrients, and food groups. Microbiome analysis targeting the V4 region of the 16S rRNA gene was performed on fecal samples. Biopsy Gleason sum data were accessed from diagnostic pathology reports.
Statistical analyses performed
Associations between dietary factors and operational taxonomic units (OTUs) were determined by beta diversity analysis. Wilcoxon Signed Rank and Mann-Whitney U-testing assessed within- and between- arm differences. Associations between Gleason sum and OTUs, and diet and OTUs, were analyzed using Spearman correlations.
Results
At baseline, Proteobacteria (median: 0.06, interquartile range: 0.01–0.16) were abundant, with four orders positively associated with Gleason sum. Gleason sum was associated with Clostridium (ρ= 0.579, p=0.005) and Blautia (ρ= −0.425, p=0.049). Increased red meat consumption from baseline was associated with Prevotella (ρ= −0.497, p=0.018) and Blautia (ρ= 0.422, p=0.039). Men who increased poultry intake had decreased Clostridiales abundance (p=0.009).
Conclusions
This hypothesis-generating study provides a starting point for investigating the relationships between the fecal microbiome, diet and prostate cancer. Adequately powered studies are required to further explore and validate these findings.
Keywords: Prostatic neoplasms, gastrointestinal microbiome, microbiota, weight loss, diet, obesity, Proteobacteria
Introduction
Obesity is a risk factor for several types of cancer and has been associated with more aggressive disease and poor outcomes in prostate cancer.1, 2 It is hypothesized that weight loss could be an effective preventive measure for obesity-associated cancers.3 Recently there has been conjecture regarding the role of the gut microbiome in potentially mediating the impact of negative energy balance.
The human digestive tract is inhabited by thirty to forty trillion bacteria and other single cell organisms.4 These organisms are integral in fermenting unabsorbed macronutrients into beneficial short chain fatty acids and vitamins that can be absorbed and utilized by the host.5, 6 Because the microbiome is a dynamic ecosystem containing thousands of taxa with unique metabolic processes and behaviors, it is important to characterize the microbiome in host disease states, as has been documented in obesity,7–10 metabolic syndrome,11 diabetes,12, 13 colorectal cancer,14, 15 and breast cancer.16 Longitudinal studies have been proposed to describe a microbiota composition that may precede diseases such as prostate cancer, hypothesizing that the effects of dietary factors and subsequent changes in the microbiome composition could prevent disease onset and/or progression.17
The concept of dysbiosis, or an unstable microbial community, has been hypothesized to contribute to chronic diseases through several mechanisms.18 Though the human gut harbors pathogens and responds rapidly to them as in the case of food poisoning, it is possible that non-pathogenic bacteria may be deleterious to community homeostasis.19 The inflammation associated with dysbiosis can manifest locally as in gastrointestinal diseases, but also results in intestinal permeability which can allow dietary nutrients and bacterial metabolites to enter the circulatory system.20
While diet is known to affect various hormones and metabolic pathways, the relationship between diet and the microbiome has yet to be fully understood. Short-term dietary changes have been shown to cause significant shifts in microbiome composition;21–25 however, many artifacts from mode of birth and breastfeeding in infancy,26 frequency and latency of antibiotic use,27 habitual diet,28 and other factors prevent characterization of a single healthy or ideal microbiome.29 More evidence is needed to establish desirable levels of various bacterial species and further characterize diets that could potentially promote and/or maintain that balance. It is unknown whether a signature of fecal microbiota exists in prostate cancer as has been observed in colorectal cancer.30
The objectives of this study were to describe the microbiota in men with clinically-confirmed prostate cancer and to investigate the longitudinal relationships between weight, dietary factors and microbiota during a presurgical weight loss trial conducted at The University of Alabama at Birmingham from 2013 to 2014.
Subjects and Methods
Participants in this study were men with prostate cancer enrolled in a presurgical weight loss trial (NCT01886677) approved by the Institutional Review Board of the University of Alabama at Birmingham (UAB), and described previously.31 Forty men were recruited through the urology clinics at UAB and the Urology Centers of Alabama from 2013 to 2014. Microbiome analysis became available midway through the trial; the remaining subset of 22 men who had complete diet and microbiome data at baseline and follow-up visits were included in this secondary analysis. Study participants had to be overweight or obese (BMI≥25 kg/m2), have no medical conditions that would affect weight status or the ability to pursue unsupervised exercise, received no other treatment for their prostate cancer, and scheduled for surgery at least 23 days after study enrollment and their baseline appointment. All participants provided written informed consent.
After completing baseline appointments, men were randomized to either weight loss or wait-list control conditions and completed follow-up visits 1–2 days prior to surgery. Men randomized to the weight loss arm were prescribed a healthful energy-restricted diet and encouraged to exercise 30 minutes/day to promote weight loss of 0.91 kilograms per week. Diet and exercise guidance was provided through semi-weekly in-person and telephone contact with a registered dietitian and an exercise physiologist. Men in the control arm were offered the intervention after prostatectomy. All medications were recorded at baseline and follow-up appointments.32, 33 Two-24 hour dietary recalls (one weekend and one week day) were obtained by a registered dietitian using a multiple pass method at baseline and follow-up and entered into the ASA24 (2011. Bethesda, MD: National Cancer Institute).34,35 Food groups were derived from the United States Department of Agriculture MyPyramid equivalents Database and macro- and micronutrients were derived from United States Department of Agriculture Food and Nutrient Database for Dietary Studies.36 Healthy Eating Index (HEI) 2010 is an index of diet quality that indicates conformance to 2010 Dietary Guidelines for Americans. Scores are derived from adequacy and moderation variables that include calorie-adjusted food groups and specific nutrients;37 scores were calculated using SAS, Version 9.4 (SAS Institute, Inc., Cary, NC, USA). Variables and values for all nutrients were analyzed with and without vitamin and mineral supplements. The average intake variables of the two recalls at each time point were used for reporting and analyses. Anthropometrics were obtained using standard procedures,38 and prostate cancer Gleason sum data on diagnostic biopsies were obtained from the patients’ medical records. Gleason sum is a score that pathologists use to classify both the major and minor histological pattern of prostatic tumor, with “1” assigned to tumors that are least aggressive and “5” to tumors that are most aggressive; a Gleason sum adds scores for major and minor patterns together.39
Fecal samples were collected after a bowel movement on sterile wipes and stored in sealed plastic bags; participants were instructed to store them in their freezer until the time of their appointment, and were subsequently stored at −80°C until batch analyzed. Microbiome analysis methods were previously reported by Demark-Wahnefried et al.31 Microbe DNA was extracted using a ZR Fecal DNA Miniprep, and the V4 region of the 16S rRNA gene was PCR-amplified and sequenced using the Illumina Miseq, as previously described.40, 41 Analyses were performed with the Quantitative Insight into Microbial Ecology (QIIME) suite, version 1.7,42 and a wrapper for QIIME (i.e., QWRAP).40 Operational taxonomic units (OTUs) were selected with uclust (a QIIME algorithm to cluster gene sequences) at 97% similarity; which is indicative of most 16S rRNA genes at the genus level. OTUs were aligned with PyNAST (in QIIME), and OTUs with abundance < 0.0005% were filtered.43 Taxonomic assignments to OTUs were performed with the Ribosomal Database Project classifier,44 informed by the May 2013 version of the Greengenes 16S database.45 The final filtered OTU table was used to generate taxonomy charts and to calculate alpha and beta diversity. Within-sample (alpha) diversity measures were calculated using observed species, Shannon Index, and whole tree Phylogenetic Diversity.29, 46 Beta diversity was calculated using Bray Curtis and weighted and unweighted Unifrac clustering of donors based on their microbial composition.47 Principal coordinates analysis (PCoA) was performed by QIIME to visualize the dissimilarity matrix (beta-diversity) among the samples. Samples in PCoA plots were colored based on the grouping variables used in beta diversity analysis.
Statistics
Dietary intake variables were analyzed longitudinally by calculating magnitude of change from baseline to follow-up and dichotomizing the total sample based on median change values. For baseline analyses, absolute intake amounts were dichotomized. Individual nutrients and food groups with significantly different clustering (as visualized on PCoA plots and statistically with beta diversity metrics, PERMANOVA p<0.05) between halves of intake change were tested for differences in individual taxa frequencies. Operational Taxonomic Units (OTUs) were grouped by phyla, classes, orders, families, genera and species and tested for significant differences on all taxonomic levels individually in frequency between groups (Kruskal Wallis p<0.05 after false discovery rate [FDR] correction). Taxonomic units with significant differences were retained for further analysis of associations between diet and microbiome composition. Median tests were used for post-hoc analyses of dichotomized dietary variables that were found to be significantly associated with changes in OTUs.
Associations between nutritional factors and taxonomic groups were examined at baseline and longitudinally using Spearman correlations on continuous variables. Due to the small sample size and non-normal distribution of microbiome data, medians are reported and tested for between and within arm differences. Baseline anthropometrics, dietary factors, alpha diversity and specific OTUs were compared between study arms using Mann Whitney U-tests. Longitudinal changes in anthropometrics, dietary factors and specific OTUs were analyzed using Wilcoxon Signed Rank tests within study arms. P-values of 0.05 or less were considered statistically significant.
Results
Participant characteristics
Men participating in this substudy were on average 60.9 years old, weighed 98.9 kg, had a BMI of 31.6 kg/m2, and were on the study 53 days. Sixty-eight percent of participants were non-Hispanic white males (n=15), and 32% (n=7) were non-Hispanic black males. The majority (n=13, 59%) of men had biopsy Gleason sums of seven, while three had scores of eight, and six had a score of six. There were no differences in race or Gleason sum between study arms (Fisher’s exact tests p=1.000, p=0.781 for race and Gleason sum, respectively). Five men in each arm were taking medications known to affect the microbiome throughout the study. In the intervention arm, men took metformin (n=1); proton pump inhibitors (PPIs) (n=2); and non-steroidal anti-inflammatory drugs (NSAIDs) (n=2). In the control arm, men took metformin (n=1); metformin and PPI (n=1); NSAID and PPI (n=2); and NSAID (n=1). Antibiotic use was not reported by any participants while on the study.
Microbiome composition and alpha-diversity
At baseline, alpha diversity did not differ between study arms (Mann-Whitney U-test p=0.332, p=0.401, p=0.365) for observed species, Shannon Index, and whole tree Phylogenetic Diversity, respectively). Four phyla present in fecal samples represented 1% or more of the microbiome composition. Median proportions of Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria were 0.61, 0.19, 0.06 and 0.02, respectively (Table 1). Additionally, fifteen genera, which represented greater than 0.01% relative abundance in all samples are presented in Table 1. At baseline, no between-arm differences in beta diversity or OTUs were observed; moreover, the most abundant OTUs and Firmicutes to Bacteroidetes (F:B) ratio did not differ within study arms over time.
Table 1.
Baseline and follow-up fecal microbe compositiona of 22 men with prostate cancer over the duration of a presurgical weight loss trial.
| Weight loss group (n=11) | Control group (n=11) | Between Groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Follow-up | Changeb | Baseline | Follow-up | Changeb | Changec | |||||
| Median | IQR | Median | IQR | p-value | Median | IQR | Median | IQR | p-value | p-value | |
| Actinobacteria | 0.0236 | 0.0115, 0.0880 | 0.0348 | 0.0072, 0.0466 | 0.859 | 0.0179 | 0.0115, 0.0313 | 0.0296 | 0.0194, 0.0457 | 0.062 | 0.270 |
| Corynebacterium | 0.0037 | 0.0003, 0.0116 | 0.0011 | 0.0001, 0.0078 | 0.929 | 0.0030 | 0.0007, 0.0227 | 0.0054 | 0.0008, 0.0684 | 0.182 | 0.898 |
| Bifidobacterium | 0.0026 | 0.0005, 0.0052 | 0.0018 | 0.0001, 0.0161 | 0.374 | 0.0031 | 0.0007, 0.0102 | 0.0092 | 0.0072, 0.0182 | 0.110 | 0.401 |
| Bacteroidetes | 0.1845 | 0.1180, 0.4433 | 0.2578 | 0.1770, 0.3510 | 0.859 | 0.1911 | 0.0754, 0.3706 | 0.1572 | 0.1272, 0.2604 | 0.534 | 0.699 |
| Bacteroides | 0.2078 | 0.0623, 0.3674 | 0.1603 | 0.1112, 0.2630 | 0.213 | 0.1150 | 0.0159, 0.1723 | 0.0636 | 0.0497, 0.2789 | 0.374 | 0.847 |
| Prevotella | 0.0059 | 0.0003, 0.0305 | 0.0038 | 0.0002, 0.0325 | 0.424 | 0.0142 | 0.0041, 0.0269 | 0.0188 | 0.0025, 0.0439 | 0.859 | 0.652 |
| Firmicutes | 0.6508 | 0.5294, 0.7729 | 0.5899 | 0.4636, 0.7398 | 0.424 | 0.5702 | 0.3658, 0.7935 | 0.7164 | 0.5925, 0.7861 | 0.110 | 0.151 |
| Blautia | 0.1124 | 0.0563, 0.2068 | 0.1678 | 0.0257, 0.2425 | 0.534 | 0.0505 | 0.0294, 0.0887 | 0.0586 | 0.0293, 0.1989 | 0.477 | 0.519 |
| Faecalibacterium | 0.0703 | 0.0507, 0.0921 | 0.0566 | 0.0381, 0.1296 | 1.000 | 0.0669 | 0.0246, 0.0972 | 0.0290 | 0.0079, 0.0660 | 0.286 | 0.332 |
| Roseburia | 0.0500 | 0.0178, 0.0879 | 0.0485 | 0.0012, 0.0566 | 0.155 | 0.0280 | 0.0123, 0.0575 | 0.0108 | 0.0031, 0.0604 | 0.790 | 0.847 |
| Ruminococcus | 0.0207 | 0.0056, 0.0439 | 0.0190 | 0.0108, 0.0392 | 0.790 | 0.0192 | 0.0111, 0.0535 | 0.0120 | 0.0065, 0.0298 | 0.328 | 0.478 |
| Clostridium | 0.0058 | 0.0013, 0.0098 | 0.0056 | 0.0034, 0.0077 | 0.929 | 0.0073 | 0.0010, 0.0108 | 0.0054 | 0.0005, 0.0126 | 1.000 | 0.217 |
| SMB53 | 0.0053 | 0.0009, 0.0113 | 0.0060 | 0.0002, 0.0204 | 0.424 | 0.0052 | 0.0004, 0.0185 | 0.0044 | 0.0023, 0.0096 | 0.929 | 0.847 |
| Finegoldia | 0.0031 | 0.0006, 0.0088 | 0.0015 | 0.0007, 0.0074 | 0.534 | 0.0032 | 0.0005, 0.0105 | 0.0100 | 0.0019, 0.0514 | 0.075 | 0.974 |
| Streptococcus | 0.0030 | 0.0016, 0.0309 | 0.0052 | 0.0022, 0.0589 | 0.722 | 0.0061 | 0.0015, 0.0254 | 0.0054 | 0.0027, 0.0178 | 0.790 | 0.519 |
| Allobaculum | 0.0007 | 0.0002, 0.0019 | 0.0013 | 0.0003, 0.0474 | 0.374 | 0.0006 | 0.0001, 0.0148 | 0.0008 | 0.0002, 0.0080 | 1.000 | 0.797 |
| Lactobacillus | 0.0004 | 0.0003, 0.0037 | 0.0009 | 0.0003, 0.0044 | 0.859 | 0.0021 | 0.0006, 0.0113 | 0.0028 | 0.0007, 0.0041 | 0.424 | 0.974 |
| Proteobacteria | 0.0223 | 0.0087, 0.0816 | 0.0328 | 0.0201, 0.1392 | 0.657 | 0.1434 | 0.0239, 0.1910 | 0.0513 | 0.0172, 0.0899 | 0.131 | 0.133 |
| Escherichia | 0.0004 | 0.0002, 0.0049 | 0.0013 | 0.0003, 0.0050 | 0.859 | 0.0043 | 0.0008, 0.0273 | 0.0029 | 0.0004, 0.0078 | 0.477 | 0.300 |
| F:B Ratiod | 1.81 | 1.20, 5.25 | 2.81 | 1.62, 5.29 | 0.790 | 4.44 | 1.88, 17.19 | 3.78 | 1.63, 5.67 | 0.155 | 0.365 |
Four major phyla with species representing >0.01% of average composition of all study samples.
Wilcoxon Signed Rank Test was used to determine differences over time within groups.
Mann-Whitney U Test was used to determine difference between groups over time.
Firmicutes : Bacteroidetes Ratio.
Gleason sum and microbiota
Correlation coefficients with corresponding p-values for associations between OTUs and Gleason sum are reported in Table 2. Gleason sum was positively associated with the phylum Deferribacteres (p=0.032) and several Proteobacteria taxa including Caulobacterales (p=0.048), Burkholderiales (p=0.025), and Pasteurellales (p=0.028). Gleason sum also was positively associated with Clostridium (p=0.005) and inversely associated with Blautia (p=0.049), both of the phylum Firmicutes.
Table 2.
Spearman rank correlations between biopsy Gleason sum and baseline operational taxonomic unitsa of fecal microbes in 22 men with prostate cancer.
| Operational Taxonomic Unit | ρ | p-value |
|---|---|---|
| [Thermi] | 0.572 | 0.005 |
| Bacteroidetes;c__Flavobacteriia;o__Flavobacteriales | 0.434 | 0.044 |
| Deferribacteres | 0.458 | 0.032 |
| Firmicutes;c__Clostridia;o__Clostridiales;f__Clostridiaceae;g__Clostridium | 0.579 | 0.005 |
| Firmicutes;c__Clostridia;o__Clostridiales;f__Lachnospiraceae;g__Blautia | −0.425 | 0.049 |
| Proteobacteria;c__Alphaproteobacteria;o__Caulobacterales | 0.425 | 0.048 |
| Proteobacteria;c__Alphaproteobacteria;o__Rhizobiales | 0.423 | 0.05 |
| Proteobacteria;c__Betaproteobacteria;o__Burkholderiales | 0.476 | 0.025 |
| Proteobacteria;c__Gammaproteobacteria;o__Pasteurellales | 0.469 | 0.028 |
Operational Taxonomic Unit levels are abbreviated as follows: c__ class, o__order, f__family, g__genus
Between-arm and baseline correlative analyses
Nutrient and food group intake and body weight status are reported in Table 3. At baseline, there were no between-arm differences in nutrients, food groups or diet quality scores. The weight loss arm reported decreasing total calories, total carbohydrates, and total grains while there were no reported changes in the control arm. Diet quality did not change for either arm, though the weight loss arm did experience significant weight loss. Beta diversity analysis indicated no significant effect of weight loss (Weighted Unifrac FDR p=0.258) or self-reported physical activity (Weighted Unifrac FDR p=0.235) on the composition of fecal microbiota. Beta diversity analyses of nutrients and food groups in the entire sample revealed differences in several taxa, which were further analyzed for associations with dietary factors.
Table 3.
Baseline and follow-up weight status and dieta composition of 22 men with prostate cancer over the duration of a presurgical weightloss trial.
| Weight loss group (n=11) | Control group (n=11) | Between Groups |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Follow-up | Changec | Baseline | Follow-up | Changec | Changed | |||||
| Median | IQR | Median | IQR | p | Median | IQR | Median | IQR | p | p | |
| Body Weight (kg) | 97.2 | 89.3, 99.6 | 88.4 | 84.5, 96 | 0.003 | 100.9 | 90.8, 110.7 | 94.6 | 91.4, 99.7 | 0.230 | 0.040 |
| Body Mass Index (kg/m2) | 30.2 | 28.2, 33.5 | 28.6 | 26.7, 31.8 | 0.003 | 31.3 | 27.1, 33.5 | 29.6 | 27.5, 32.9 | 0.213 | 0.056 |
| Total Calories (kcal) | 1238 | 1171, 2251 | 1151 | 824, 1308 | 0.050 | 1596 | 977, 1726 | 1582 | 1231, 1805 | 0.859 | 0.101 |
| Total Protein (g) | 58.7 | 41.8, 71.3 | 52.2 | 43.3, 62.2 | 0.248 | 72.2 | 52, 82 | 63.4 | 48.5, 90.3 | 0.929 | 0.847 |
| Total Fat (g) | 60.7 | 43.6, 82.4 | 36.4 | 22.6, 61.7 | 0.075 | 85.3 | 43.2, 92.8 | 65.5 | 52.2, 86.6 | 0.477 | 0.438 |
| Total Carbohydrate (g) | 137.4 | 83.0, 263.2 | 88.7 | 84.1, 153.6 | 0.041 | 150.3 | 99.8, 188.7 | 153.5 | 102.1, 220.6 | 0.477 | 0.056 |
| Total Sugar (g) | 46.9 | 33.9, 102.1 | 45.4 | 28.2, 59.3 | 0.477 | 55.5 | 34.9, 66.2 | 54.7 | 36.3, 108.2 | 0.286 | 0.270 |
| Total Fiber (g) | 13.0 | 10.9, 22.9 | 10.0 | 6.6, 11.9 | 0.075 | 11.3 | 8.9, 14.5 | 10.0 | 7.2, 14.0 | 0.374 | 0.365 |
| Total Grains (oz. eq.) | 4.96 | 1.42, 5.63 | 2.84 | 0.87, 4.07 | 0.050 | 3.48 | 2.76, 5.34 | 4.67 | 3.52, 5.47 | 0.248 | 0.028 |
| Total Vegetables (cup eq.) | 1.45 | 1.01, 2.08 | 1.02 | 0.50, 1.53 | 0.328 | 1.74 | 0.52, 2.45 | 1.54 | 0.66, 1.93 | 0.374 | 0.748 |
| Total Fruit (cup eq.) | 0.45 | 0.11, 1.48 | 0.73 | 0.01, 1.16 | 0.477 | 0.51 | 0.05, 1.47 | 0.64 | 0.18, 0.87 | 0.721 | 0.438 |
| Total Dairy (cup eq.) | 0.46 | 0.13, 0.88 | 0.77 | 0.29, 1.47 | 0.285 | 0.90 | 0.36, 1.56 | 0.67 | 0.41, 1.29 | 0.424 | 0.193 |
| Total Meat, protein foods (oz. eq.) | 3.72 | 2.69, 5.92 | 2.43 | 1.86, 4.53 | 0.286 | 4.92 | 3.83, 6.46 | 4.36 | 2.89, 7.02 | 0.790 | 0.478 |
| Healthy Eating Index 2010 b | 64.9 | 60.8, 72.73 | 64.0 | 62.9, 69.6 | 0.657 | 65.0 | 60.22, 68.41 | 64.7 | 55.49, 68.31 | 0.534 | 0.748 |
Two 24 hour dietary recalls were obtained at baseline and at follow-up and entered into National Cancer Institute ASA24 dietary assessment software.
The Healthy Eating Index 2010 is a diet quality assessment tool that measures conformance to 2010 Dietary Guidelines for Americans.
Wilcoxin Signed Rank Test was used to determine differences over time within groups.
Mann-Whitney U Test was used to determine difference between groups over time.
Baseline associations between dietary intake and microbiota indicate that men who consumed more calories did so from all macronutrients and food groups (baseline correlation data not shown). Positive correlations were observed between Bacteroidetes and protein intake. Protein intake was inversely correlated with the F:B ratio and Actinobacteria. Campylobacterales (phylum Proteobacteria) was positively associated with fiber and sugar, dark green vegetables and fruit, and Verrucomicrobia (Akkermansia Muciniphila represented >99% of this phylum) was positively correlated with orange vegetables at baseline. HEI-2010 scores were inversely associated with Lactobacillus (phylum Firmicutes) and positively correlated with Campylobacterales and Verrucomicrobiales (data not shown).
Beta-diversity and PERMANOVA analyses
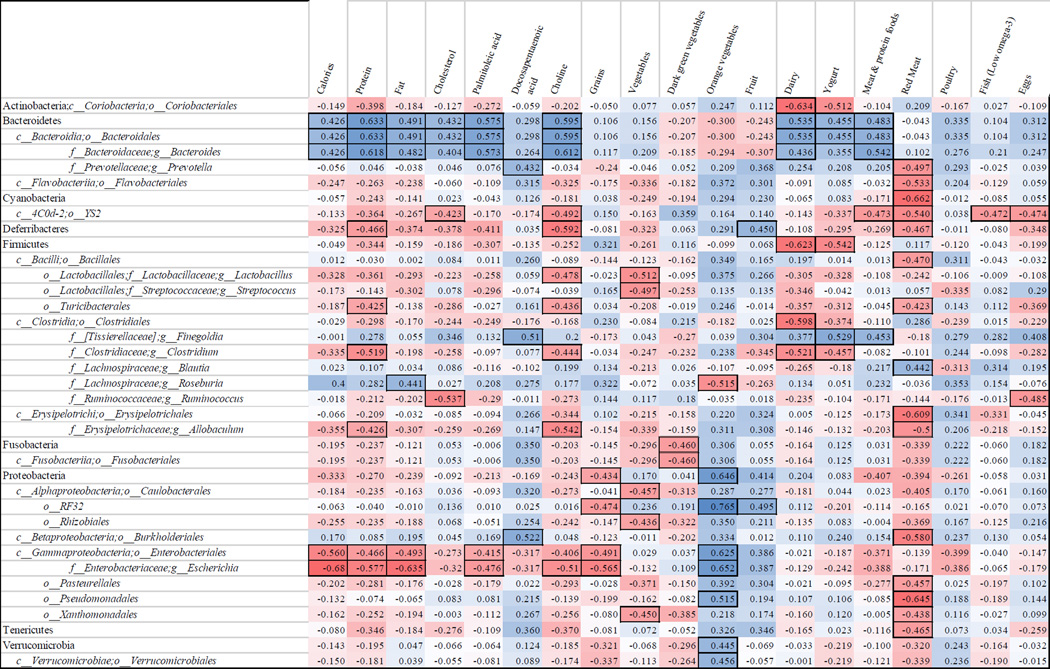
Changes in nutrients and food groups resulting in dissimilar microbiota were evaluated by Bray-Curtis, Unweighted Unifrac and Weighted Unifrac clustering analysis (See Supplementary Figure). Clustering by nutrients and food groups with significant differences for all three methods were poultry, meat, and lutein and zeaxanthin. The food groups orange vegetables, eggs, potatoes, milk, and low omega-3 fish; nutrients lycopene, vitamin D, vitamin B12, choline, cholesterol, alcohol, and caffeine; and the fatty acids eicosapentaenoic acid, paullinic acid, and palmitoleic acid were significantly different for at least one beta diversity metric. Taxa level analyses for each of these variables were conducted and 43 taxa were significantly different, with FDR corrected p < 0.05 (see supplementary data). Comparing high and low intakes of eggs at baseline (essentially non-consumers vs. consumers of whole eggs, median 0.378 ounces/day including mixed dishes), 26 OTUs differed between groups. Seventeen of the OTUs were in the phylum Proteobacteria, and were lower for egg consumers than non-consumers. Post-hoc analyses indicated men who increased poultry intake had lower levels of Clostridiales than men who had no or negative change in intake (Median test, p=0.009). Correlations between changes in dietary variables and changes in OTUs are shown in Figure 1. Bacteroides were positively and Escherichia (phylum Proteobacteria) were negatively associated with change in total calories (p<0.048, p<0.001 for Bacteroides and Escherichia, respectively), protein (p=0.002, p=0.005), and fat (p=0.023, p=0.001). Change in red meat consumption was inversely associated with several small OTUs including Flavobacteriales (phylum Bacteroidetes) (p=0.011), YS2 (phylum Cyanobacteria) (p=0.009), Deferribacteres (p=0.028), Bacillales (p=0.027) and Turicibacterales (p=0.05) (phylum Firmicutes), Burkholderiales (p=0.005), Pasteurellales (p=0.033), Pseudomonadales (p=0.001) and Xanthomonadales (p=0.042) (phylum Proteobacteria), and Tenericutes (p=0.029). Change in choline intake was associated with several OTUs in addition to those related to protein food consumption, notably its inverse correlation to Lactobacillus (p=0.024), Clostridium (p=0.038) and Escherichia (p=0.015).
Figure 1.
Spearman’s rank correlations between change in nutrient and food groups with change in fecal microbes in 22 men with prostate cancer over the duration of a presurgical weightloss trial. a,b
a The first row lists food groups and nutrients that associated with microbe changes in beta diversity tests. The first column lists taxa that changed with changes in nutrient intake. The numbers in the grid are Spearman’s rank correlation coefficients. Squares are colored on a red to white to blue scale from negative to positive (−1 to 1). Squares in boxes have p values <0.05. Operational Taxonomic Unit levels are abbreviated as follows: c__ class, o__order, f__family, g__genus.
b Two 24 hour dietary recalls were obtained at baseline and at follow-up and entered into National Cancer Institute ASA24 dietary assessment software.
Discussion
In this first reported study to investigate changes in the microbiota that occur with dietary change in men with prostate cancer, fecal microbes reflected dietary factors and changes expected over the course of the study. Baseline characteristics of the microbiome, namely elevated Proteobacteria levels were fairly unique and were comparable only to previous reports of ileal Crohn’s Disease,48 which was not present in any of our participants. At baseline, Bacteroides were elevated compared to Prevotella. The relative abundance of Prevotella or Bacteroides is driven by the presence of high plant fiber vs refined, fat and protein based diets, respectively.49 Previous work has established three enterotypes based on statistical clustering across multiple geographical regions with differing enrichment of only three genera within two phyla: the Bacteroides and Prevotella (phylum Bacteroidetes) and Ruminoccocus (phylum Firmicutes).50 It could be hypothesized that diet quality would be reflected in the microbial composition given the abundance of fiber digesting Prevotella, among several other species. While increased Bacteroides may not be directly associated with disease risk, it is reflective of diet quality. While these prostate cancer patients may have changed their diet from prediagnosis, their diet quality, as measured by the HEI-2010, was comparable to nationally representative adult cancer survivors of the same age.51
As reflected in the lack of change in diet quality, men in this study decreased calories from all food sources, rather than replacing energy dense foods with nutrient dense choices. Several genera in the phylum Firmicutes were abundant across all participants. The unexpected correlations with several Firmicutes and Proteobacteria genera and biopsy Gleason sum at diagnosis is noteworthy; a case-control study of hospitalized adults prior to C. difficile infection found that individuals who developed the infection had altered levels of several OTUs,52 reiterating that dysbiosis allows opportunistic bacteria to increase in abundance. In this sample of men with prostate cancer, we hypothesize that elevated levels of opportunistic Proteobacteria contribute to systemic inflammation which may potentiate disease onset or progression.
It has been observed that obese individuals have greater F:B ratios compared to lean individuals.8 Similarly, decreases in the F:B ratio have been documented in individuals losing weight,22 and an increase in F:B ratio can be observed with short-term overfeeding, or positive energy balance.24 In our study, the F:B ratio did not differ between study arms or over time within study arms. Though our intervention period was brief and although several men lost 5 kilograms or more, their follow-up BMIs were still in the overweight or obese category.
Actinobacteria are gram positive, primarily composed of the beneficial genus Bifidobacterium, 21 and have been recognized as the third most abundant phylum in healthy adults.29 However, Proteobacteria were more abundant in our study participants both before and after the intervention. Increases in Proteobacteria have been characteristic of dysbiosis, often accompanied by a decrease in Bacteroidetes and Actinobacteria. Gram negative bacteria, which comprise the Bacteroidetes and Proteobacteria phyla, contain lipopolysaccharide membranes, but are not constitutively pathogenic.53 These phyla appear to increase and decrease with over- and underfeeding, respectively, suggesting that their concentrations may be a proxy for energy balance.24 Proteobacteria are generally elevated in obesity but do not exponentially outnumber Actinobacteria as in this study. An abundance of potentially pathogenic gram negative bacteria is associated with dysbiosis due to translocation of bacteria,12 their structural proteins,54 lipopolysaccharide55 and their metabolic byproducts into the bloodstream. Gram negative bacteria have also been observed in atherosclerotic and periodontic plaque,56 suggesting that translocation through the gut lining may promote other chronic disease states. Additionally, intestinal permeability through various mechanisms can allow for excessive absorption of nutritive and non-nutritive metabolites, which is often implicated in obesity.12 Taken together, these findings suggest the potential for a microbiome signature of aggressive prostate cancer. While intriguing, given that this study was performed in a relatively circumscribed geographic area, such findings need to be validated with larger studies from diverse populations. Furthermore, such studies also would warrant the inclusion of healthy controls as well as sufficient numbers of men with either benign variants or precursor lesions (e.g., high grade prostatic intraepithelial neoplasia).
The relationship between nutrient and food group changes and microbe composition suggests larger scale studies are warranted. The decrease in Clostridiales by participants who increased poultry consumption, and decreases in several taxa observed with increased red meat consumption may suggest a microbiome altering effect of those food sources, whether directly or through transfer of antibiotic resistance genes in those foods.57
While this is one of the first studies to investigate changes in the microbiota in response to dietary change in men with prostate cancer, and is strengthened by being part of a randomized controlled trial, there are some limitations which should be acknowledged. The first limitation is that it is a post-hoc analysis and therefore findings should be viewed from the standpoint of hypothesis-generation rather than hypothesis testing. Secondly, our intervention period was brief and may not have allowed sufficient time for some populations of microbiota to shift. Thirdly, this study was conducted in the southeastern United States and the dietary and environmental exposures may be unique to this region; therefore, generalizability cannot be assumed. Other exposures that are known to affect the microbiome, such as metformin, PPIs and NSAIDs, were taken by almost half of the study participants throughout the study. Though this may have affected their microbe composition at both time points, it is unknown whether these medications mediate microbe composition change associated with diet change. Additionally, dietary data were obtained from self-report rather than direct observation. Finally, our sample size is modest and while it rivals many other reported longitudinal studies in this arena, larger studies are needed for validation.
Conclusion
In summary, this longitudinal study utilized beta diversity by differences in food and nutrient consumption to characterize the relationship between diet and microbes. Moreover, our data serve as a starting point and document that overweight and obese men with prostate cancer may be dysbiotic and manifest a unique microbiome profile. Further investigation is warranted to determine if a microbiotic signature of prostate cancer exists, whether it is causative, and if it can be manipulated with diet and exercise to serve in efforts aimed toward cancer prevention and control.
Supplementary Material
Acknowledgments
This study was supported by the National Cancer Institute R21 (CA161263), P30 (CA13148), and R25 (CA047888). The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165, UL1TR001417), and Heflin Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
No authors report a conflict of interest.
Contributor Information
Andrew D. Frugé, Cancer Prevention and Control Training Program, Department of Nutrition Sciences, University of Alabama at Birmingham (UAB), 205-996-7367 (fruge@uab.edu) 348 Webb Nutrition Sciences Bldg., 1675 University Blvd, Birmingham, AL, 35294.
Travis Ptacek, Department of Microbiology, UAB, Birmingham, AL, 205-934-3727 (tptacek@uab.edu).
Yuko Tsuruta, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-2418 (tsuru@uab.edu).
Casey D. Morrow, Department of Cell, Developmental & Integrative Biology, UAB, Birmingham, AL 205-934-5705 (caseym@uab.edu).
Maria Azrad, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-9751 (maria123@uab.edu).
Renee A. Desmond, Division of Preventive Medicine, UAB School of Medicine (SOM), Birmingham, AL 205-934-3722 (desmond@uabmc.edu).
Gary R. Hunter, Department of Human Studies, UAB, Birmingham, AL 205-934-8338 (ghunter@uab.edu).
Soroush Rais-Bahrami, Department of Urology, UAB SOM, Birmingham, AL 205-996-8765 (sraisbahrami@uabmc.edu).
Wendy Demark-Wahnefried, Department of Nutrition Sciences, UAB, Birmingham, AL 205-975-4022 (demark@uab.edu).
References
- 1.Yuan C, Cao Y, Chavarro J, et al. Prediagnostic Body-mass Index, Smoking and Prostate Cancer Survival: A Cohort Consortium Study of Over 10,000 White Men with Prostate Cancer. Cancer Epidemiol Biomarkers Prev. 2015;24:759–760. [Google Scholar]
- 2.Allott EH, Masko EM, Freedland SJ. Obesity and Prostate Cancer: Weighing the Evidence. Eur Urol. 2013;63:800–809. doi: 10.1016/j.eururo.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolin KY, Colditz GA. Can weight loss prevent cancer? Br J Cancer. 2008;99:995–999. doi: 10.1038/sj.bjc.6604623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. bioRxiv. 2016 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson HL, Campbell BJ. Review article: dietary fibre-microbiota interactions. Aliment Pharmacol Ther. 2015;42:158–179. doi: 10.1111/apt.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015;3:207–215. doi: 10.1016/S2213-8587(14)70134-2. [DOI] [PubMed] [Google Scholar]
- 8.Kasai C, Sugimoto K, Moritani I, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS. ONE. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran CP, Shanahan F. Gut microbiota and obesity: role in aetiology and potential therapeutic target. Best Pract Res Clin Gastroenterol. 2014;28:585–597. doi: 10.1016/j.bpg.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Kovatcheva-Datchary P, Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract Res Clin Gastroenterol. 2013;27:59–72. doi: 10.1016/j.bpg.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects. Med. 2013;34:39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Hartstra AV, Bouter KEC, Bäckhed F, Nieuwdorp M. Insights Into the Role of the Microbiome in Obesity and Type 2 Diabetes. Diabetes Care. 2015;38:159–165. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- 14.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10 doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goedert JJ, Gong Y, Hua X, et al. Fecal Microbiota Characteristics of Patients with Colorectal Adenoma Detected by Screening: A Population-based Study. EBioMedicine. 2015;2:597–603. doi: 10.1016/j.ebiom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedert JJ, Jones G, Hua X, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amirian ES, Petrosino JF, Ajami NJ, Liu Y, Mims MP, Scheurer ME. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infect Agents Cancer. 2013;8:42–42. doi: 10.1186/1750-9378-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract—identification of keystone and foundation taxa. Microbiome. 2015;3:44. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira CM, Vieira AT, Vinolo MAR, Oliveira FA, Curi R, Martins FdS. The Central Role of the Gut Microbiota in Chronic Inflammatory Diseases. J Immunol. Res. 2014;2014:689492. doi: 10.1155/2014/689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouhnik Y, Raskine L, Simoneau G, et al. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: a double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am J Clin Nutr. 2004;80:1658–1664. doi: 10.1093/ajcn/80.6.1658. [DOI] [PubMed] [Google Scholar]
- 22.Simoes CD, Maukonen J, Scott KP, Virtanen KA, Pietilainen KH, Saarela M. Impact of a very low-energy diet on the fecal microbiota of obese individuals. Eur J Nutr. 2014;53:1421–1429. doi: 10.1007/s00394-013-0645-0. [DOI] [PubMed] [Google Scholar]
- 23.Graessler J, Qin Y, Zhong H, et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13:514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- 24.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu GD, Chen J, Hoffmann C, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science (New York, N.y.) 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 31.Demark-Wahnefried W, Nix JW, Hunter GR, et al. Feasibility outcomes of a presurgical randomized controlled trial exploring the impact of caloric restriction and increased physical activity versus a wait-list control on tumor characteristics and circulating biomarkers in men electing prostatectomy for prostate cancer. BMC Cancer. 2016;16:1–12. doi: 10.1186/s12885-016-2075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers MA, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178.e171–179.e171. doi: 10.1016/j.cmi.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015 doi: 10.1038/nature15766. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Keyzer W, Huybrechts I, De Vriendt V, et al. Repeated 24-hour recalls versus dietary records for estimating nutrient intakes in a national food consumption survey. Food Nutr. Res. 2011;55 doi: 10.3402/fnr.v55i0.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subar AF, Kirkpatrick SI, Mittl B, et al. The Automated Self-Administered 24-Hour Dietary Recall (ASA24): A Resource for Researchers, Clinicians and Educators from the National Cancer Institute. J Acad Nutr Diet. 2012;112:1134–1137. doi: 10.1016/j.jand.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodner-Montville J, Ahuja JKC, Ingwersen LA, Haggerty ES, Enns CW, Perloff BP. USDA Food and Nutrient Database for Dietary Studies: Released on the web. J Food Comp Anal. 2006;19(Supplement):S100–S107. [Google Scholar]
- 37.National Cancer Institute Applied Research Program. National Cancer Institute; 2011. Risk Factor Monitoring and Methods. Healthy Eating Index-2010. http://riskfactor.cancer.gov/tools/hei. [Google Scholar]
- 38.Lohman T, Roache A, Martorell R. Anthropometric Standardization Reference Manual. Med Sci Sports Exerc. 1992;24:952. [Google Scholar]
- 39.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 40.Kumar R, Eipers P, Little RB, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet. 2014;82:18.18.11–18.18.29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome. 2015;3:17. doi: 10.1186/s40168-015-0080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 47.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Letters. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang FF, Liu S, John EM, Must A, Demark-Wahnefried W. Diet quality of cancer survivors and noncancer individuals: Results from a national survey. Cancer. 2015;121:4212–4221. doi: 10.1002/cncr.29488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincent C, Stephens DA, Loo VG, et al. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome. 2013;1:1–11. doi: 10.1186/2049-2618-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuijt TJ, van der Poll T, Wiersinga WJ. Gut Microbiome and Host Defense Interactions during Critical Illness. In: Vincent J-L, editor. Annual Update in Intensive Care and Emergency Medicine. Vol. 2012. Springer Berlin Heidelberg; 2012. pp. 29–40. [Google Scholar]
- 54.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-Like Receptor–Gut Microbiota Interactions: Perturb at Your Own Risk! Annu Rev Physiol. 2012;74:177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 55.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-Induced Dysbiosis of the Intestinal Microbiota and the Effects on Immunity and Disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muszer M, Noszczynska M, Kasperkiewicz K, Skurnik M. Human Microbiome: When a Friend Becomes an Enemy. Arch Immunol Ther Exp. 2015;63:287–298. doi: 10.1007/s00005-015-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Devirgiliis C, Barile S, Perozzi G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011;6:275–284. doi: 10.1007/s12263-011-0226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.